Abstract
Background
Telomeres are nucleoprotein structures present at the terminal region of the chromosomes. Mutations in genes coding for proteins involved in telomere maintenance are causative of a number of disorders known as telomeropathies. The genetic origin of these diseases is heterogeneous and has not been determined for a significant proportion of patients.
Methods
This article describes the genetic characterization of a cohort of patients. Telomere length was determined by Southern blot and quantitative PCR. Nucleotide variants were analyzed either by high-resolution melting analysis and Sanger sequencing of selected exons or by massive sequencing of a panel of genes.
Results
Forty-seven patients with telomere length below the 10% of normal population, affected with three telomeropathies: dyskeratosis congenita (4), aplastic anemia (22) or pulmonary fibrosis (21) were analyzed. Eighteen of these patients presented known pathogenic or novel possibly pathogenic variants in the telomere-related genes TERT, TERC, RTEL1, CTC1 and ACD. In addition, the analyses of a panel of 188 genes related to haematological disorders indicated that a relevant proportion of the patients (up to 35%) presented rare variants in genes related to DNA repair or in genes coding for proteins involved in the resolution of complex DNA structures, that participate in telomere replication. Mutations in some of these genes are causative of several syndromes previously associated to telomere shortening.
Conclusion
Novel variants in telomere, DNA repair and replication genes are described that might indicate the contribution of variants in these genes to the development of telomeropathies. Patients carrying variants in telomere-related genes presented worse evolution after diagnosis than the rest of patients analyzed.
Electronic supplementary material
The online version of this article (10.1186/s13023-019-1046-0) contains supplementary material, which is available to authorized users.
Keywords: Telomere, Dyskeratosis congenita, Pulmonary fibrosis, Aplastic anemia, DNA repair, Telomeropathies
Background
Chromosome ends are protected by nucleoprotein structures, the telomeres. In humans, telomere DNA is composed by multiple repetitions of the TTAGGG hexanucleotide [1]. The 3′ end of telomeres is formed by a G-rich single stranded extension that invades the upstream double-stranded region to form telomeric loops (T-, D-loop). Telomere DNA is bound by a protein complex named shelterin [2] and this nucleo-protein structure protects chromosome ends from degradation and is critical for genome stability (recently reviewed in [3]).
Telomeres cannot be completely synthesized during DNA replication, which is known as the end-replication problem and results in progressive shortening of the telomeres as cells proliferate. In most organisms, telomere DNA is elongated after each round of DNA replication by a specialized complex with reverse transcriptase activity known as telomerase [4]. The enzymatic activity of the complex is provided by the TERT (telomerase reverse transcriptase) protein while the RNA component TR (telomerase RNA, encoded by TERC) is used as template. Essential components of the complex are also DKC (Dyskeratosis congenita), NOP10, NHP2 and GAR1 proteins, which bind and stabilize TR and are required for telomerase complex assembly [5].
Telomere replication, elongation and protection are impaired by the strong secondary and tertiary structure of their DNA [6]. The high content of Guanines of the telomeres makes them prone to form secondary structures such as G-quadruplexes. In addition, the T- and D-loops impair the access of the replicative machinery. These structures are solved by helicases and structure-specific endonucleases.
In the absence of a functional telomerase complex, telomeres are progressively shortened. One of the main functions of telomere structure is to avoid the recognition of telomeres as damaged DNA which is achieved by inhibitory interactions between shelterin components and DNA-repair proteins [7]. When several telomeres reach critically short length the structure of the nucleoprotein complex can no longer be maintained. In that case, telomeres are recognized as damaged DNA and a response is triggered that can result in cell-cycle arrest, cell apoptosis or senescence [8].
The presence of functional mutations in the genes coding for proteins of the shelterin and telomerase complexes and auxiliary proteins causes a number of diseases named telomeropathies or telomere biology disorders that may affect different organs [9, 10]. Telomeropathies are very heterogeneous diseases depending on the gene mutated and the specific mutations, their penetrance and the existence of anticipation effects. Therefore, patients with the same mutation can present different manifestations. Some patients present severe symptoms at an early age such as those of dyskeratosis congenita (DC) or the related Hoyeraal-Hreidarsson, Resvesz and Coats plus syndromes [11]. Other diseases may appear later such as Aplastic Anemia, (AA) (20–30 years) [12] or pulmonary fibrosis (40–60 years) (PF) [13]. Mutations in 13 different genes have been identified as causative of these diseases but a large number of patients remain genetically undiagnosed [14].
Variants in the gene coding for one helicase, RTEL1 (OMIM 608833) are also present in patients with telomeropathies [15]. Others helicases and nucleases, like SLX4, BLM, WRN and RecQL4, have been shown to participate in telomere preservation [16]. Actually, mutations in some of these genes also result in telomere shortening [16, 17]. The term secondary telomeropathies has been recently proposed for diseases caused by mutations in this group of genes [18].
The reduced number of patients of these rare diseases and the number of putative candidate genes makes their molecular diagnosis challenging. In this article, the molecular analysis of 47 patients treated in Spanish hospitals was performed. Twenty-six of them were diagnosed of DC or AA and twenty-one of PF. The results obtained identified possibly pathogenic variants at the telomere-associated genes TERT, TERC, ACD, CTC1 and RTEL1 in heterozygosis. In addition, a significant proportion of patients presented rare variants in genes coding for proteins involved in DNA repair and in the resolution of complex DNA structures.
Methods
Patients
Patients with severe telomere shortening (percentile < 10) who presented dyskeratosis congenita (DC), aplastic anemia (AA) or pulmonary fibrosis (PF) (sporadic and familial forms) were included. These patients were diagnosed, treated and followed in Spanish reference health care provider centres for these rare diseases. DC was diagnosed in patients with the characteristic mucocutaneous symptoms, nail dystrophy, abnormal skin pigmentation, oral leukoplakia and bone marrow failure. The diagnosis of AA was based on bone marrow and blood cells counts. The diagnosis of PF was established in accordance to international guidelines [19]. The more relevant characteristics of the patients are summarized in Additional file 1: Table S1. All samples were collected after obtaining informed consent for genetic analysis, including for research. Clinical and demographic data were collected.
Genetic analyses
The existence of sequence variants in the TERT, TERC and DKC1 genes was determined in six patients by PCR amplification of exons, high resolution melting analyses (HRM) and sequencing of candidate exons as previously described [20]. Later on, genetic analysis of 41 patients was made by massive parallel sequencing using a panel of genes related to haematological disorders, as shown in Additional file 1: Table S2 (MBFSv1.1 panel). Detailed protocols are presented in Additional file 1: Supplemental Material.
Telomere length determination
Telomere length was determined by two different methods, Southern blots of enzymatically digested DNA (Additional file 1: Figure S1) and quantitative PCR. Both methods have been previously used for determination of telomere length from patients with inherited bone marrow failure [21]. Detailed protocols are presented in Additional file 1: Supplemental Material.
Ethical statement
All the participants in this study provided written informed consent for genetic analysis. The investigation was conducted in accordance with the ethical standards of the Declaration of Helsinki and the guidelines of the concerned hospitals.
Results
Single nucleotide variants in genes related to telomere biology
The possible genetic bases of the diseases were studied in patients that presented symptoms related to dyskeratosis congenita (DC), either mucocutaneous symptoms or, more often, aplastic anemia (AA) or sporadic or familial cases of pulmonary fibrosis (PF). Patients with these diseases present significant telomere shortening. According to previous reports [12, 22], patients with telomere length below the 1 % of the age-mated population in the case of DC and < 10% in AA and PF where included in this study. Beside clinical symptoms, DC/AA and PF patients differed at the age of presentation of the disease, 12.2 + 12.2 years for DC/AA and 58.3 + 10.2 years for PF (Additional file 1: Table S1).
First analyses were made in six patients by PCR amplification of exons of the DKC, TERT and TERC genes using the previously described High-resolution melting and DNA sequencing method [20] (Patients indicated by an asterisk in Tables 1, 4 and Additional file 1: Table S1). Subsequently, taking advantage of newer technologies, DNA samples of 41 patients were analyzed by massive sequencing of a panel of genes related to haematological disorders including all the genes previously associated to telomere-related diseases (Additional file 1: Table S2). Out of these patients, 23 were diagnosed of DC/AA and 18 of PF.
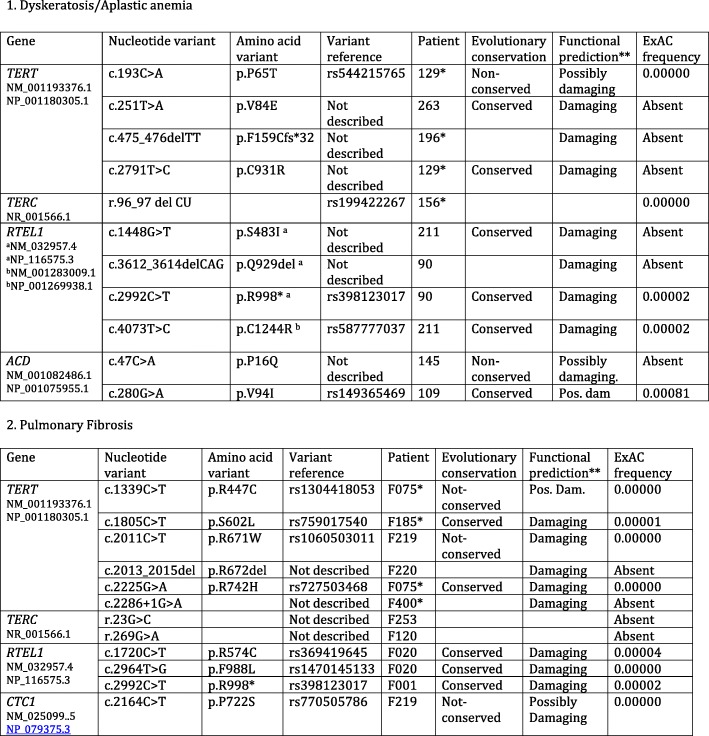
Table 1.
Mutations found in telomere-related genes
*Patients analyzed by exon amplification and Sanger sequencing
**The prediction shown is a summary of the ones made by the programs SIFT ensemble 66, Polyphen-2 v2.2.2, MutationAssessor, release 2, FATHMM v2.3, CADD v1.3 and dbscSNV1.1.
Table 4.
Genes coding for proteins involved in telomere biology and DNA repair in which rare variants were found in the different patients and clinical evolution
| Patient | Telomere biology genes | DNA repair genes | Clinical evolution |
|---|---|---|---|
| Pulmonary Fibrosis | |||
| F001 | RTEL1 | BRCA2 | Deceased |
| F010 | Stable | ||
| F020 | RTEL1 | FANCM, RAD51C | Lung transplant |
| F021 | FANCM | Deceased | |
| F025 | Lung transplant | ||
| F044 | ATM; RECQL4 | Disease progressing | |
| F075* | TERT | N.D. | Deceased |
| F106 | POT1 | BRCA1 | Lung transplant |
| F120 | TERC, NHP2 | SLX4 | Deceased |
| F179 | BRCA2 | Disease progressing | |
| F185* | TERT | N.D. | Deceased |
| F188 | RECQL4 | Disease progressing | |
| F200 | NHP2 | ATR | Deceased |
| F213 | Disease progressing | ||
| F219 | TERT, CTC1, RTEL1 | FANCU | Deceased |
| F220 | TERT | FANCA | Lung transplant |
| F242 | Lung transplant | ||
| F244 | ATR | Disease progressing. | |
| F253 | TERC , CTC1 | RAD51, RAD51C | Deceased |
| F382 | NBN | Disease progressing | |
| F400* | TERT | N.D. | Waiting transplant |
| Dyskeratosis/Aplastic anemia | |||
| 26 | DKC1 Ref 19 | Mre11A | Deceased |
| 36 | DKC1 Ref 19 | ATR, BRCA2 | Disease progressing |
| 44 | FANCF | Stable | |
| 47 | ATR, FANCD2, FANCL, RECQL4, SLX4 | Recovered | |
| 48 | WRN, | N.D. | |
| 52 | FANCM, BLM | HSC transplant | |
| 62 | SLX4 | HSC transplant | |
| 69 | ATM | Stable | |
| 86 | RECQL4 | Stable | |
| 90 | RTEL1 | BRCA2 | HSC transplant |
| 109 | ACD | SLX4, Rad51C | HSC transplant |
| 121 | ATM | HSC transplant | |
| 129* | TERT | N.D. | Deceased |
| 134 | ATR, Mre11A | Stable | |
| 145 | ACD | BRCA2, SLX4 | Deceased |
| 156* | TERC | N.D. | Deceased |
| 169 | FANCM | Deceased | |
| 181 | RECQL4 | Stable | |
| 196* | TERT | N.D. | Deceased |
| 198 | ATM, FANCL, PALB2, RAD50 | Disease progressing | |
| 204 | ATM, RECQL4 | Stable | |
| 205 | Stable | ||
| 211 | RTEL1 | PALB2 | HSC transplant |
| 223 | NHP2 | ATM | Stable |
| 263 | TERT, RTEL1 | BRCA2, SLX4 | HSC transplant |
| 282 | SLX4 | N.D. | |
N.D. Not determined. Boldface: genes that present variants with a frequency between 0.001 and 0.003 in the ExAC database.
Nucleotide variants were further analysed when their frequency in the general population was lower than one in a thousand. The results obtained for genes previously related to telomere shortening are shown in Table 1. These studies allowed the identification of single nucleotide variants (SNVs) and indels at TERT, TERC, RTEL1, ACD and CTC1 genes in heterozygosis. Histograms of the Sanger sequencing reaction confirming these variants are shown in Additional file 1: Figure S2.
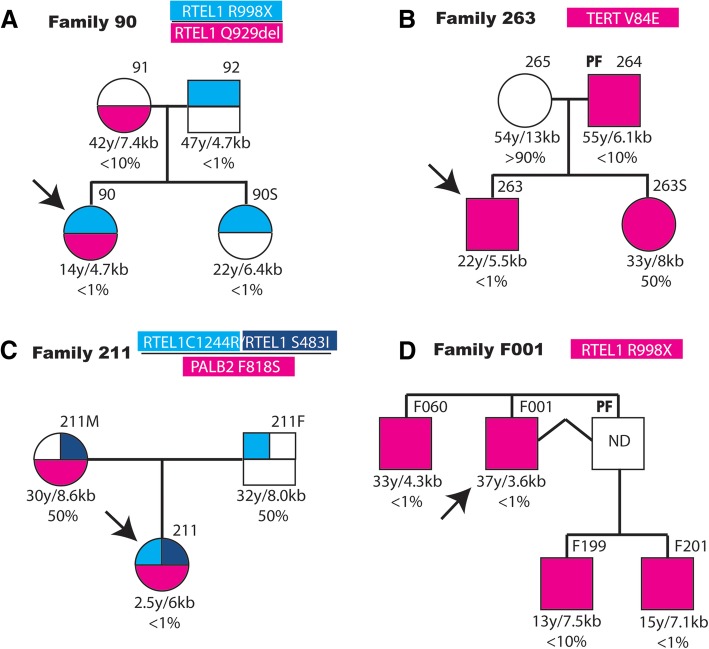
TERT variants were found in eight patients (Table 1). AA patient 129 presented two missense variants, p.P65T and p.C931R. The p.P65T variant was inherited from the mother of the patient, which also presented short telomeres. The p.C931R variant was not present in the parents of the patient. AA patient 196 presented in heterozygosis a deletion of two nucleotides at positions c.475_476 that produced a frame shift mutation after the amino acid residue 158. One stop codon will be found 31 residues downstream, resulting in a 189 amino acids-long protein. This variant was inherited from the father of the patient, which presented PF. A son of the patient that also presented this variant, died before birth. AA patient 263 carried in heterozygosis the TERT p.V84E variant. His father also carries the variant in heterozygosis and presents PF and short telomeres. A sister that carries the variants is presently unaffected at the age of 33 years (Fig. 1).
Fig. 1.
Familiar inheritance of variants in telomere-related genes in patients with aplastic anemia and pulmonary fibrosis. The family of four affected patients is represented in panels (a-d). The probands (indicated by arrows) of families (a-c) are affected of aplastic anemia and that of family (d) of pulmonary fibrosis. The number of the family and the variants analyzed are shown in the upper part of each diagram. In the cases where several variants were studied, they are shown in the heading in the same relative order and colour as in the diagrams. The presence of the variants is represented as colour-filled squares or circles and that of the presence of the reference allele as empty figures. The number assigned to each patient or relative is indicated in the upper, right part of each symbol. The age of each individual, the determined telomere size, in kb, and the corresponding percentage of the age-matched population are shown under each symbol. Relatives affected by pulmonary fibrosis are indicated in the upper left part of their symbol (PF). Patient F001 in panel (d) had a deceased identical twin brother as indicated by joining them with a broken line
PF patient F075 presented two missense variants at TERT, p.R447C and p.R742H. The variant p.R742H has been already reported [23] while the p.R447C variant is described for the first time. The mother and a brother of this patient died of PF. PF patient F185 presented the p.S602 L amino acid variant. PF patient F219 presented the p.R671W variant, reported to be pathogenic [24]. PF patient F220 presented an in-frame deletion at amino acids 671_672 that has not been previously described. A variant in a splicing consensus site was found in patient F400.
Variants in TERC were found in three patients (Table 1). AA patient 156 presented in heterozygosis a deletion of two bases (r.96_97delCU) that has been previously described [25]. The mother of the patient died with PF and liver cirrhosis, a brother carrying this variant also presented liver cirrhosis and short telomeres. However, a sister that carries the variant is presently healthy at the age of 40 years even if her telomeres are short. PF patient F120 presented the r.269G > A variant, that has not been previously reported. PF patient F253 presented the undescribed r.23G > C variant. PF patient F219 presented the p.P722S variant in CTC1 that has not been previously described.
Four patients presented rare variants in RTEL1 in heterozygosis (Table 1). AA patient 90 presented the p.R998* variant recently reported as pathogenic [26]. The father and one sister of the patient carried this variant, and presented short telomeres (Fig. 1). A representative Southern blot of telomeres including relatives of patient 90 is shown in Additional file 1: Figure S1. In addition, patient 90 and mother carried the p.Q929del variant, recently described [26]. AA patient 211 presented the new pS483I variant and the described p.C1244R variant [27]. In addition, patient 211 presented the amino acid variant p.F818S in PALB2 protein, also involved in telomere maintenance, as will be discussed later. The RTEL1 variant p.C1244R was inherited from the father and the RTEL1 variant p.S483I and the PALB2 variant from the mother and both parents presented average telomere length (Fig. 1). PF patient F001 presented the p.R998* pathogenic variant. This RTEL1 variant was present in the three analyzed relatives of patient F001, all with short telomeres (Fig. 1). PF patient F020 presented the described p.F988 L [15] and the non-described p.R574C variants.
Nucleotide variants in ACD, coding for the TPP1 shelterin protein, were observed in two patients. AA patient 145 presented the p.P16Q amino acid variant and AA patient 109 the p.V94I variant, both in heterozygosis. The variant of patient 145 is inherited from the mother that reports several relatives affected of DC. Patients 109 and 145 presented chromosome 7 monosomy.
Single nucleotide variants in genes related to DNA repair and homologous recombination
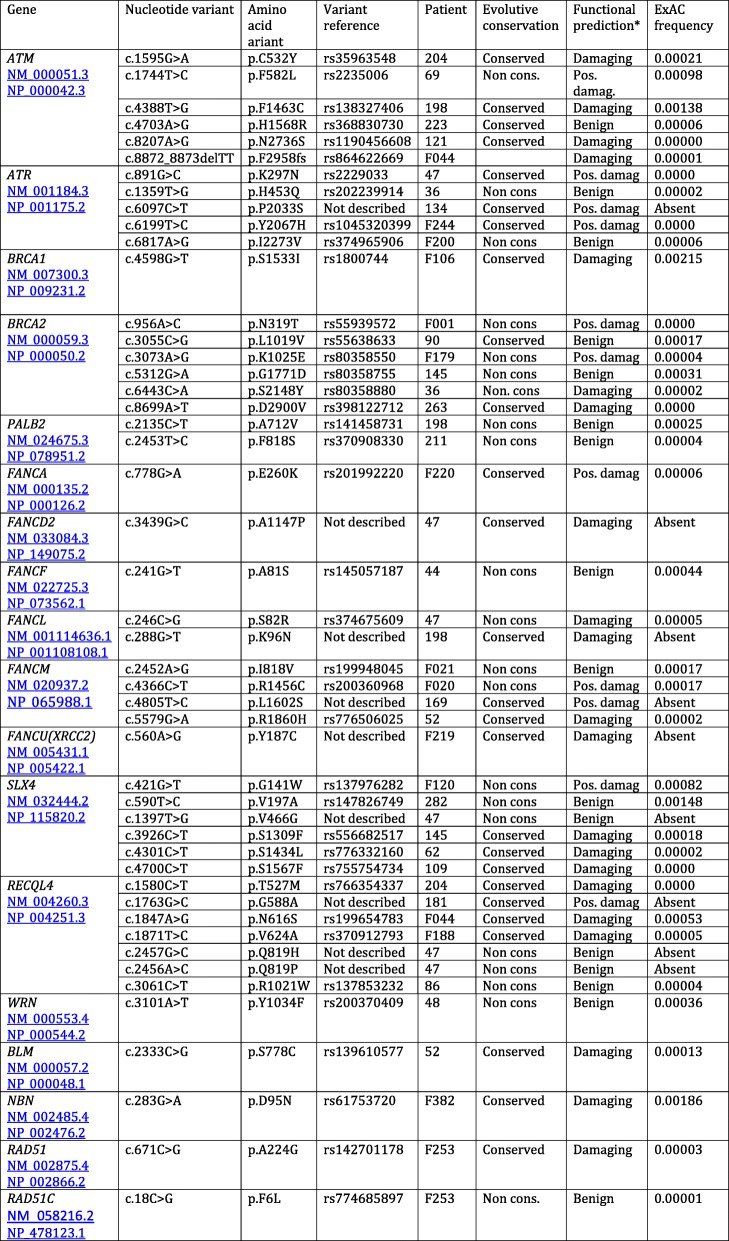
Massive sequencing identified a number of rare SNVs (frequency lower than one in a thousand) in genes coding for proteins involved in DNA repair by homologous recombination, as shown in Table 2. Among these genes are those coding for the protein kinases ATM (Ataxia-telangiectasia mutated) and ATR (Ataxia telangiectasia and Rad3-related), mutated in the genetic-instability syndromes Ataxia Telangiectasia and Seckel syndrome, respectively. Rare variants in these proteins were found in 35% of the DC/AA and 17% of the PF patients (Table 2). Fanconi Anemia is also due to cytogenetic instability, hypersensitivity to DNA crosslinking agents and defective DNA repair. This disease can be caused by mutation in genes that code for components of a protein complex involved in DNA repair. Rare variants in several of these genes (FANCA, FANCD2, FANCF, FANCL, FANCM, FANCU) were found in 22% of the DC/AA and PF patients (Table 2). The DNA- associated proteins BRCA1 and BRCA2 (Breast Cancer 1 and 2) also play an important role in DNA break repair and recombination in association with PALB2 (Partner and Localizer of BRCA2). Rare variants in the BRCA1, BRCA2 or PALB2 genes were found in 26% of the DC/AA and 17% of the PF patients (Table 2). RecQL4 is a DNA helicase (RecQ-like, type 4) involved in DNA damage repair and rare variants were found in 17 and 11% of DC/AA and PF patients, respectively. Variants in the gene coding for the structure-specific-endonuclease subunit SLX4, involved in DNA repair and replication, were found in 22% of the DC/AA and 6% of the PF patients (Table 2). Mutations in SLX4 have been associated with Fanconi Anemia (group P). Finally, single rare variants were found in other genes coding for proteins involved in DNA repair such as WRN, BLM, RAD51, RAD51C and NBN in 9 and 17% of the DC/AA and PF patients, respectively. The variants found in these genes for each patient are shown in Table 3. Some of these variants are described for the first time as indicated (“Not described” in Table 3).
Table 2.
Frequency of rare variants in genes coding for proteins involved in DNA-Repair
| Dyskeratosis/Anemia | Pulmonary Fibrosis | |
|---|---|---|
| ATM/ATR | 8/23 (35%) | 3/18 (17%) |
| BRCA1, BRCA2, PALB2 | 6/23 (26%) | 3/18 (17%) |
| FANC-A, −D2, −F, −L, −M, −U | 5/23 (22%) | 4/18 (22%) |
| RECQL4 | 4/23 (17%) | 2/18 (11%) |
| SLX4 | 5/23 (22%) | 1/18 (6%) |
| WRN, BLM, NBN, RAD51, RAD51C | 2/23 (9%) | 3/18 (17%) |
Table 3.
Variants in DNA repair genes
*The prediction shown is a summary of the ones made by the programs SIFT ensemble 66, Polyphen-2 v2.2.2, Mutation Assessor, release 2, FATHMM v2.3, CADD v1.3 and dbscSNV1.1.
The variants present in each patient and their clinical evolution from diagnosis are summarized in Table 4. Two patients with previously described DKC1 mutations, 26 and 36 [20], are included. This table includes some variants whose frequency is slightly higher than one in a thousand (indicated in bold letters). One of them is a variant in the gene POT1 (pQ301H) present at a frequency of 0.002 in the ExAC database. The change is possibly damaging and is present in patient F106. A second variant in NHP2 (p.R101Q) is present at a frequency of 0.002 at the ExAC database. The change is possibly damaging, and has been detected in three patients (F200, F120 and 223). Among the proteins involved in DNA repair, two patients (F20, 109) present the p.T287A variant at the RAD51C gene (frequency of 0.003 in the ExAC database). Other two patients (26 and 134) present the p.A492D variant at the MRE11A gene (frequency 0.002 in ExAC).
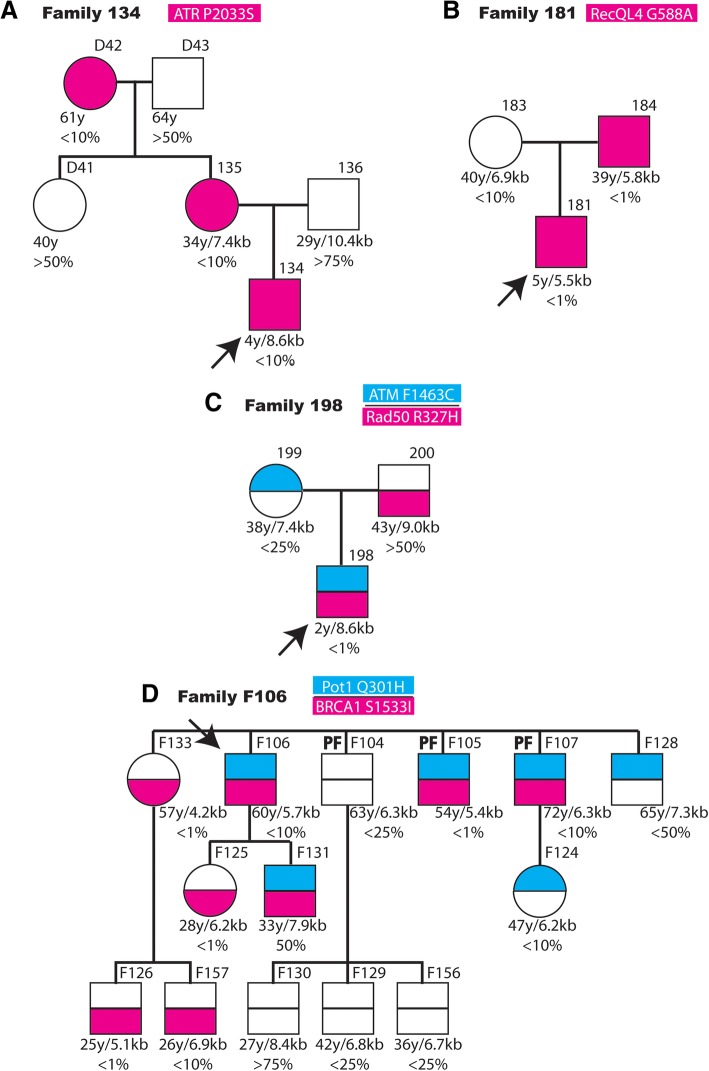
The possible correlation between the presence of rare variants in proteins involved in DNA repair or replication, telomere length and clinical manifestation was investigated in the family of several patients (Fig. 2). AA patient 134 carries one variant in the ATR gene not previously described. The variant is inherited from his mother and grandmother, both relatives have short telomeres. The father and maternal grandfather, that do not carry this variant, present an average telomere size.
Fig. 2.
Familiar inheritance of variants in genes related to DNA repair and replication in patients with aplastic anemia and pulmonary fibrosis. Schematic representation of four families of patients (indicated by arrows) affected by aplastic anemia (a-c) or pulmonary fibrosis (d). The number of each family and the variants studied are shown in the upper part of each panel. The presence of the rare variant is indicated as coloured square or circles and the presence of the reference variant by open figures. In panels (c and d) where several variants were studied, their relative position in the diagrams and colour are indicated in the upper part. The number of each family member and its possible pulmonary fibrosis diagnosis (PF) is indicated in the upper part of each symbol. The age, telomere length (kb) and the corresponding percentage of the age-matched population are indicated in the lower part of each symbol
AA patient 181 present a variant in RECQL4 not described previously. This variant is present in the father with very short telomeres. The mother does not carry this variant and has longer telomeres even that below the average of the population (10%).
AA patient 198 presents a novel variant in ATM and a rare variant in RAD50. The ATM variant is inherited from the mother and the RAD50 p.R850Q variant from the father (Fig. 2). The father present telomeres of average size and the mother of size bellow the average. However, the son presents relatively shorter telomeres than the parents.
The PF patient F106 carries a rare variant in BRCA1 and a relatively rare variant in POT1 (frequency 0.002). The patient has four brothers and three have been also diagnosed of PF. Two of them carry the same variants in POT1 and BRCA1 and short telomeres. A sister carries only the BRCA1 variant and very short telomeres although is presently asymptomatic. Two of the brothers carry exclusively the POT1 mutation and present longer telomeres (25–50% of the population) although one of them has developed pulmonary fibrosis. The patient has a daughter, which inherited the BRCA1 variant and present very short telomeres. Two nephews and a niece carry the BRCA1 variant and have short telomeres. One niece inherited the POT1 change and presents short telomeres.
Discussion
The genetic variants present in patients with symptoms of DC, AA or PF have been studied in a population of patients from Spanish hospitals. Heterozygous variants were found in genes previously related to telomeropathies in 17 patients, 8 diagnosed of DC/AA and 9 of PF (Table 1). Two variants in ACD were found in the only two AA patients that also presented chromosome 7 monosomy, previously associated to the presence of short telomeres in AA patients as a consequence of chromosomal instability [28].
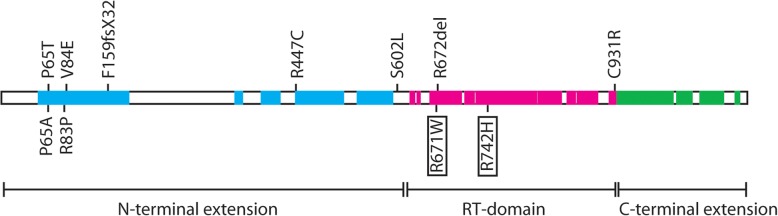
Several of the variants found had been reported previously as pathogenic. The TERT variants p.R671W and p.R742H have been described associated to PF [23, 24]. The TERC variant r.96_97delCU was described in a DC patient transmitted with a dominant pattern of inheritance [25]. The RTEL1 variants p.Q929del [26] pF988 L [29], p.R998* [26] and p.C1244R [27] were described in DC patients. The RTEL1 p.R574C variant has been described as a clinical variant. The other variants found in this study have not been previously reported and according to the actual guidelines should be considered as variants of unknown significance (VUS). However, we speculate that some of them could be pathogenic because they are closely related to described pathogenic variants, as schematically shown in Fig. 3. This is the case of TERT p.P65T variants since the p.P65A variant has been found in patients of acute myeloblastic leukemia (AML) [26]. The TERT p.V84E variant is close to the p.R83P variant, found in patients with AA and myelodisplastic syndrome [30]. In addition, the V84 residue is conserved through evolution and the V > E change is considered as damaging. Other variants produced changes potentially damaging in conserved residues located in functional domains of the proteins like the TERT variants p.F159fsX32 and p.S602 L that affect residues located in the conserved N-terminal domain (Fig. 3). The p.F159fsX32 variant would produce a truncated protein and a situation of functional hemizygosis. The variants p.R672del and pC931R are located in the reverse transcriptase and C-terminal domains of TERT, respectively (Fig. 3). In the case of the RTEL1 gene, the pS483I variant in a conserved residue is close to the p.P484L variant associated to PF [31].
Fig. 3.
Schematic representation of the location of TERT variants on the conserved and functional domains of the protein. Conserved regions of TERT are represented as filled boxes and coloured according to their participation in the protein functional domains (shown in the lower part of the scheme). Regions of the N-terminal extension, involved in RNA binding, are coloured in blue. Regions of the reverse transcriptase domain (RT-domain) are shown in cyan and those of the C-terminal extension in green. The location of the new amino acid variants described in the article is shown in the upper part of the figure. Two previously described variants close to new variants (P65A and R83P) are shown in the lower part of the figure. The location of two variants found in this project and previously described as pathogenic are shown in the lower part inside boxes
The gene panel used for the analyses of the DNA of 41 patients by massive sequencing detected the presence of numerous variants in genes coding for proteins involved in DNA repair such as ATM, ATR, BRCA1, BRCA2 and several genes of the FANC complex, mutated in Fanconi Anemia. Variants in genes coding for DNA helicases and nucleases involved in the resolution of DNA complex structures, such as SLX4, RECQL4, WRN, BLM, RAD51, RAD51C or NBN were also found. Variants in these genes have not been related to telomeropathies previously, with the exception of SLX4 [32].
The proteins ATM and ATR play a signalling role in DNA double-strand break repair pathways and their continued activation can result in cell cycle arrest, replicative senescence or apoptotic cell death. However, specific interactions with shelterin protein, ATM/TRF2, and ATR/TRF1, inhibit their activation to preserve telomere structure. Therefore, it is conceivable that functional variants at ATM and ATR could affect telomere structure and stability. Indeed, cells of Ataxia telangiectasia patients with homozygous mutations in ATM present telomere shortening and increase frequency of chromosome end-to-end fusions (CEFs) [33].
BRCA1 and BRCA2 proteins also participate in DNA damage repair by homologous recombination and non-homologous end joining [34] and play a protective role at telomeres [35]. BRCA1/2 mutations result in shortening of the telomere single-stranded overhang and increase telomere length variability [36]. BRCA2 heterozygous cell lines present frequently CEFs [37]. Germline mutations in BRCA1 and BRCA2 are associated with greatly increased frequency of breast and ovarian cancers and telomere shortening was associated with genetic anticipation in hereditary breast cancer [38].
Proteins mutated in Fanconi Anemia (FA) patients are involved in repair of DNA-inter-strand crosslinks and have been also involved in telomere maintenance (reviewed by Sarkar and Liu [39]). FA patients are reported to present increased CEFs and overall shorter telomeres [40, 41].
Replication of telomeric DNA presents specific challenges due to structure of the DNA with unconventional regions including T loop and G quadruplexes and requires the participation of a large number of proteins (reviewed by Martinez and Blasco [6]). Among them are proteins with DNA helicase activity, like RTEL1, BLM, WRN or RecQL4, endonuclease complexes, like the one formed by SLX4, SLX1, MU881 and XPF and recombinases like Rad51 and Rad51C.
Mutations in these genes cause diseases with some symptoms resembling those of telomeopathies, as mentioned in the Background section. Many of these diseases show telomere fragility [42], telomere loss and/or increased CEFs [43]. These proteins are located to telomeres through interactions with TRF1 of TRF2 shelterin proteins [44–48] . In addition, TERT expression in cells isolated from patients of these diseases rescue some of their alterations [49, 50]. Variants in SLX4 have been found with increased frequency in AA patients [32].
The existence of overlapping symptoms in diseases caused by mutations in genes coding for proteins involved in telomere-maintenance and in DNA-repair and replication proteins have been recently reviewed [18]. In this article we describe the presence of rare variants in many of these genes in patients that present short telomeres and telomeropathies such as DC, AA and PF. We consider the possibility that the presence of one or more variants in these genes in heterozygosis, some times in addition to variants in telomere maintenance genes, could have a cumulative pathogenic effect. This association could be extensive to some more frequent variants in genes involved in primary telomeropathies, as those observed in NHP2 or POT1. This possible pathogenic association need to be confirmed by functional studies and analyzing more extensive cohorts of patients.
The small number of patients analyzed prevents a statistical analysis of the possible genotype-phenotype correlation. There seems to be, however poor outcome in patients that carry variants in genes related to telomere biology, as shown in Table 4. For example, nine AA patients carried variants in telomere genes and five of them died in the course of the study and the other four required hematopoietic stem cell transplantation (HSCT). In contrast, only one patient died and three required HSCT of the sixteen that presented short telomeres but not rare variants in telomere related genes. Similar evolution was observed in PF patients. Nine carried mutations in telomere genes and four died and four required lung transplant. Of the 12 patients without mutations in these genes, two died and three required lung transplant. In particular, the two patients that carried two TERT variants evolved very quickly and both died in a few months after the diagnosis of the disease. One of these patients was diagnosed of AA and the other of PF. Among the patients with worst evolution without mutations in telomerase genes, three presented variants in FANCM (two died and one required HSCT). However, the analysis of a larger cohort of patients will be required to confirm or discard these proposals.
Conclusions
Using massive sequencing of a panel of genes and Sanger sequencing of selected exons we identified novel variants in telomere biology genes in a series of patients of DC, AA and PF form Spanish hospitals. In additions we describe the existence of frequent rare variants in genes involved in DNA damage response and the resolution of complex DNA structures in this population of patients. Achieving a molecular diagnosis from patients of these telomere biology diseases would be important for accurate genetic counselling and effective treatment of the patients.
Additional file
Supplemental methods. Table S1A. Clinical characteristics of the Dyskeratosis congenita, Aplastic anemia patients. Table S1B. Clinical characteristics of Pulmonary fibrosis patients. Table S2. Genes included in the panel used for massive sequencing. Table S3. Description of the control population used for the estimation of normal mean, telomere length and percentile for each age-range. Figure S1. Representative Southern blot used for telomere length determination. Figure S2. Histograms of the sequences obtained by Sanger sequencing of the regions of genes related to telomere biology that presented SNVs or indels. (DOC 3474 kb)
Acknowledgments
We are grateful to AFEFPI (Asociación de Familiares y Enfermos de Fibrosis Pulmonar Idiopática) for patients’ support. We acknowledge support of the publication fee by the CSIC Open Access Publication Support initiative through its Unit of Information Resources for Research (URICI).
Funding
Funded by grants PI14–01495 and PI17–01401 (Fondo de Investigaciones Sanitarias, Instituto de Salud Carlos III, Spain supported by FEDER funds) and by one ACCI project from CIBERER and one grant to the FPI cohort from CIBERES.
Availability of data and materials
Additional data and materials can be provided upon request to the authors.
Authors’ contributions
Conception and design of the work: EGA-S, JS, MM-M, RP, LS. Generation of DNA sequencing data: EGA-S, EG, LP-B, EV, PM, JC, LI, PL, RP, LS. Clinical diagnosis and follow up of the patients: LP-C, AR-L, AC, IB-S, LIG-G, AM-N, MM-G, AMG-M, CR-V, MB-O, GPN, VL-F, M-LU, CD-H, CV, SM, BL-M. Data analysis and interpretation: EGA-S, EG, JC, JS, MM-M, RP, LS. Critical revision of the article: All the authors. All authors approved the final version of the manuscript to be published.
Ethics approval and consent to participate
This study was conducted in accordance with the principles of the Helsinki Declaration and the guidelines of the concerned hospitals.
Consent for publication
All the participants in this study provided written informed consent for genetic analysis, including for research.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Elena G. Arias-Salgado, Email: egarcia@iib.uam.es
Eva Galvez, Email: eva.galvez@salud.madrid.org.
Lurdes Planas-Cerezales, Email: lurdesplanas@hotmail.com.
Laura Pintado-Berninches, Email: lpintado@iib.uam.es.
Elena Vallespin, Email: elena.vallespin@salud.madrid.org.
Pilar Martinez, Email: pmartinez.hulp@salud.madrid.org.
Jaime Carrillo, Email: jcarrillo@genomica.com.
Laura Iarriccio, Email: liarriccio@iib.uam.es.
Anna Ruiz-Llobet, Email: aruizl@sjdhospitalbarcelona.org.
Albert Catalá, Email: acatala@hsjdbcn.org.
Isabel Badell-Serra, Email: Ibadell@santpau.cat.
Luis I. Gonzalez-Granado, Email: luisignacio.gonzalez@salud.madrid.org
Andrea Martín-Nalda, Email: amnalda@yahoo.es.
Mónica Martínez-Gallo, Email: mmartinez@vhebron.net.
Ana Galera-Miñarro, Email: anam.galera@carm.es.
Carmen Rodríguez-Vigil, Email: carmenrvi@hotmail.com.
Mariana Bastos-Oreiro, Email: bastos.mariana@yahoo.com.
Guiomar Perez de Nanclares, Email: GUIOMARPEREZDEMANCLARESLEAL@osakidetza.eus.
Virginia Leiro-Fernández, Email: virginia.leiro.fernandez@sergas.es.
Maria-Luz Uria, Email: muria@vhebron.net.
Cristina Diaz-Heredia, Email: crdiaz@vhebron.net.
Claudia Valenzuela, Email: claudiavale@hotmail.com.
Sara Martín, Email: smartins@bellvitgehospital.cat.
Belén López-Muñiz, Email: belenlmb@yahoo.es.
Pablo Lapunzina, Email: plapunzina.hulp@salud.madrid.org.
Julian Sevilla, Email: julian.sevilla@salud.madrid.org.
María Molina-Molina, Email: mariamolinamolina@hotmail.com.
Rosario Perona, Email: rperona@iib.uam.es.
Leandro Sastre, Phone: +34 915854437, Email: lsastre@iib.uam.es.
References
- 1.McElligott R, Wellinger RJ. The terminal DNA structure of mammalian chromosomes. EMBO J. 1997;16(12):3705–3714. doi: 10.1093/emboj/16.12.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19(18):2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 3.Maciejowski J, de Lange T. Telomeres in cancer: tumour suppression and genome instability. Nat Rev Mol Cell Biol. 2017;18(3):175–186. doi: 10.1038/nrm.2016.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt JC, Cech TR. Human telomerase: biogenesis, trafficking, recruitment, and activation. Genes Dev. 2015;29(11):1095–1105. doi: 10.1101/gad.263863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egan ED, Collins K. Specificity and stoichiometry of subunit interactions in the human telomerase holoenzyme assembled in vivo. Mol Cell Biol. 2010;30(11):2775–2786. doi: 10.1128/MCB.00151-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez P, Blasco MA. Replicating through telomeres: a means to an end. Trends Biochem Sci. 2015;40(9):504–515. doi: 10.1016/j.tibs.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Karlseder J, Hoke K, Mirzoeva OK, Bakkenist C, Kastan MB, Petrini JH, et al. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2004;2(8):E240. doi: 10.1371/journal.pbio.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426(6963):194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 9.Savage SA. Human telomeres and telomere biology disorders. Prog Mol Biol Transl Sci. 2014;125:41–66. doi: 10.1016/B978-0-12-397898-1.00002-5. [DOI] [PubMed] [Google Scholar]
- 10.Bertuch AA. The molecular genetics of the telomere biology disorders. RNA Biol. 2016;13(8):696. doi: 10.1080/15476286.2015.1094596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perona R, Iarriccio L, Pintado-Berninches L, Rodriguez-Centeno J, Manguan-Garcia C, Garcia E, et al. Molecular diagnosis and precission therapeutic approaches for telomere biology disorders. In: Larramendy M, Soloneski S, et al., editors. Telomeres: INTECH. 2016. pp. 77–117. [Google Scholar]
- 12.Townsley DM, Dumitriu B, Young NS. Bone marrow failure and the telomeropathies. Blood. 2014;124(18):2775–2783. doi: 10.1182/blood-2014-05-526285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Armanios M. Telomerase and idiopathic pulmonary fibrosis. Mutat Res. 2012;730(1–2):52–58. doi: 10.1016/j.mrfmmm.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barbaro P, Ziegler DS, Reddel RR. The wide-ranging clinical implications of the short telomere syndromes. Intern Med J. 2016;46(4):393. doi: 10.1111/imj.12868. [DOI] [PubMed] [Google Scholar]
- 15.Walne AJ, Vulliamy T, Kirwan M, Plagnol V, Dokal I. Constitutional mutations in RTEL1 cause severe dyskeratosis congenita. Am J Hum Genet. 2013;92(3):448–453. doi: 10.1016/j.ajhg.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doksani Y, de Lange T. The role of double-strand break repair pathways at functional and dysfunctional telomeres. Cold Spring Harb Perspect Biol. 2014;6(12):a016576. doi: 10.1101/cshperspect.a016576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alter BP, Giri N, Savage SA, Rosenberg PS. Telomere length in inherited bone marrow failure syndromes. Haematologica. 2014;100(1):49–54. doi: 10.3324/haematol.2014.114389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Opresko PL, Shay JW. Telomere-associated aging disorders. Ageing Res Rev. 2016;33:52–66. doi: 10.1016/j.arr.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, et al. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. 2011;183(6):788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carrillo J, Martinez P, Solera J, Moratilla C, Gonzalez A, Manguan-Garcia C, et al. High resolution melting analysis for the identification of novel mutations in DKC1 and TERT genes in patients with dyskeratosis congenita. Blood Cells Mol Dis. 2012;49(3–4):140–146. doi: 10.1016/j.bcmd.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 21.Gadalla SM, Cawthon R, Giri N, Alter BP, Savage SA. Telomere length in blood, buccal cells, and fibroblasts from patients with inherited bone marrow failure syndromes. Aging (Albany NY) 2010;2(11):867–874. doi: 10.18632/aging.100235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alter BP, Baerlocher GM, Savage SA, Chanock SJ, Weksler BB, Willner JP, et al. Very short telomere length by flow fluorescence in situ hybridization identifies patients with dyskeratosis congenita. Blood. 2007;110(5):1439–1447. doi: 10.1182/blood-2007-02-075598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petrovski S, Todd JL, Durheim MT, Wang Q, Chien JW, Kelly FL, et al. An exome sequencing study to assess the role of rare genetic variation in pulmonary fibrosis. Am J Respir Crit Care Med. 2017;196(1):82–93. doi: 10.1164/rccm.201610-2088OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diaz de Leon A, Cronkhite JT, Katzenstein AL, Godwin JD, Raghu G, Glazer CS, et al. Telomere lengths, pulmonary fibrosis and telomerase (TERT) mutations. PLoS One. 2010;5(5):e10680. doi: 10.1371/journal.pone.0010680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vulliamy T, Marrone A, Szydlo R, Walne A, Mason PJ, Dokal I. Disease anticipation is associated with progressive telomere shortening in families with dyskeratosis congenita due to mutations in TERC. Nat Genet. 2004;36(5):447–449. doi: 10.1038/ng1346. [DOI] [PubMed] [Google Scholar]
- 26.Cardoso SR, Ellison ACM, Walne AJ, Cassiman D, Raghavan M, Kishore B, et al. Myelodysplasia and liver disease extend the spectrum of RTEL1 related telomeropathies. Haematologica. 2017;102(8):e293–e2e6. doi: 10.3324/haematol.2017.167056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Guen T, Jullien L, Touzot F, Schertzer M, Gaillard L, Perderiset M, et al. Human RTEL1 deficiency causes Hoyeraal-Hreidarsson syndrome with short telomeres and genome instability. Hum Mol Genet. 2013;22(16):3239–3249. doi: 10.1093/hmg/ddt178. [DOI] [PubMed] [Google Scholar]
- 28.Calado RT, Cooper JN, Padilla-Nash HM, Sloand EM, Wu CO, Scheinberg P, et al. Short telomeres result in chromosomal instability in hematopoietic cells and precede malignant evolution in human aplastic anemia. Leukemia. 2012;26(4):700–707. doi: 10.1038/leu.2011.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kannengiesser C, Borie R, Menard C, Reocreux M, Nitschke P, Gazal S, et al. Heterozygous RTEL1 mutations are associated with familial pulmonary fibrosis. Eur Respir J. 2015;46(2):474–485. doi: 10.1183/09031936.00040115. [DOI] [PubMed] [Google Scholar]
- 30.Vulliamy TJ, Kirwan MJ, Beswick R, Hossain U, Baqai C, Ratcliffe A, et al. Differences in disease severity but similar telomere lengths in genetic subgroups of patients with telomerase and shelterin mutations. PLoS One. 2011;6(9):e24383. doi: 10.1371/journal.pone.0024383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stuart BD, Choi J, Zaidi S, Xing C, Holohan B, Chen R, et al. Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nat Genet. 2015;47(5):512–517. doi: 10.1038/ng.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collopy LC, Walne AJ, Vulliamy TJ, Dokal IS. Targeted resequencing of 52 bone marrow failure genes in patients with aplastic anemia reveals an increased frequency of novel variants of unknown significance only in SLX4. Haematologica. 2014;99(7):e109–e111. doi: 10.3324/haematol.2014.105320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smilenov LB, Morgan SE, Mellado W, Sawant SG, Kastan MB, Pandita TK. Influence of ATM function on telomere metabolism. Oncogene. 1997;15(22):2659–2665. doi: 10.1038/sj.onc.1201449. [DOI] [PubMed] [Google Scholar]
- 34.Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer. 2016;16(2):110–120. doi: 10.1038/nrc.2015.21. [DOI] [PubMed] [Google Scholar]
- 35.Min J, Choi ES, Hwang K, Kim J, Sampath S, Venkitaraman AR, et al. The breast cancer susceptibility gene BRCA2 is required for the maintenance of telomere homeostasis. J Biol Chem. 2012;287(7):5091–5101. doi: 10.1074/jbc.M111.278994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uziel O, Yerushalmi R, Zuriano L, Naser S, Beery E, Nordenberg J, et al. BRCA1/2 mutations perturb telomere biology: characterization of structural and functional abnormalities in vitro and in vivo. Oncotarget. 2016;7(3):2433–2454. doi: 10.18632/oncotarget.5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bodvarsdottir SK, Steinarsdottir M, Bjarnason H, Eyfjord JE. Dysfunctional telomeres in human BRCA2 mutated breast tumors and cell lines. Mutat Res. 2012;729(1–2):90–99. doi: 10.1016/j.mrfmmm.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 38.Martinez-Delgado B, Gallardo M, Tanic M, Yanowsky K, Inglada-Perez L, Barroso A, et al. Short telomeres are frequent in hereditary breast tumors and are associated with high tumor grade. Breast Cancer Res Treat. 2013;141(2):231–242. doi: 10.1007/s10549-013-2696-6. [DOI] [PubMed] [Google Scholar]
- 39.Sarkar J, Liu Y. Fanconi anemia proteins in telomere maintenance. DNA Repair (Amst) 2016;43:107–112. doi: 10.1016/j.dnarep.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Callen E, Samper E, Ramirez MJ, Creus A, Marcos R, Ortega JJ, et al. Breaks at telomeres and TRF2-independent end fusions in Fanconi anemia. Hum Mol Genet. 2002;11(4):439–444. doi: 10.1093/hmg/11.4.439. [DOI] [PubMed] [Google Scholar]
- 41.Joksic I, Vujic D, Guc-Scekic M, Leskovac A, Petrovic S, Ojani M, et al. Dysfunctional telomeres in primary cells from Fanconi anemia FANCD2 patients. Genome Integr. 2012;3(1):6. doi: 10.1186/2041-9414-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferrarelli LK, Popuri V, Ghosh AK, Tadokoro T, Canugovi C, Hsu JK, et al. The RECQL4 protein, deficient in Rothmund-Thomson syndrome is active on telomeric D-loops containing DNA metabolism blocking lesions. DNA Repair (Amst) 2013;12(7):518–528. doi: 10.1016/j.dnarep.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blagoev KB, Goodwin EH, Bailey SM. Telomere sister chromatid exchange and the process of aging. Aging (Albany NY) 2010;2(10):727–730. doi: 10.18632/aging.100206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghosh AK, Rossi ML, Singh DK, Dunn C, Ramamoorthy M, Croteau DL, et al. RECQL4, the protein mutated in Rothmund-Thomson syndrome, functions in telomere maintenance. J Biol Chem. 2012;287(1):196–209. doi: 10.1074/jbc.M111.295063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Opresko PL, von Kobbe C, Laine JP, Harrigan J, Hickson ID, Bohr VA. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J Biol Chem. 2002;277(43):41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 46.Zimmermann M, Kibe T, Kabir S, de Lange T. TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling. Genes Dev. 2014;28(22):2477–2491. doi: 10.1101/gad.251611.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bower BD, Griffith JD. TRF1 and TRF2 differentially modulate Rad51-mediated telomeric and nontelomeric displacement loop formation in vitro. Biochemistry. 2014;53(34):5485–5495. doi: 10.1021/bi5006249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarkar J, Wan B, Yin J, Vallabhaneni H, Horvath K, Kulikowicz T, et al. SLX4 contributes to telomere preservation and regulated processing of telomeric joint molecule intermediates. Nucleic Acids Res. 2015;43(12):5912–5923. doi: 10.1093/nar/gkv522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crabbe L, Jauch A, Naeger CM, Holtgreve-Grez H, Karlseder J. Telomere dysfunction as a cause of genomic instability in Werner syndrome. Proc Natl Acad Sci U S A. 2007;104(7):2205–2210. doi: 10.1073/pnas.0609410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du X, Shen J, Kugan N, Furth EE, Lombard DB, Cheung C, et al. Telomere shortening exposes functions for the mouse Werner and Bloom syndrome genes. Mol Cell Biol. 2004;24(19):8437–8446. doi: 10.1128/MCB.24.19.8437-8446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods. Table S1A. Clinical characteristics of the Dyskeratosis congenita, Aplastic anemia patients. Table S1B. Clinical characteristics of Pulmonary fibrosis patients. Table S2. Genes included in the panel used for massive sequencing. Table S3. Description of the control population used for the estimation of normal mean, telomere length and percentile for each age-range. Figure S1. Representative Southern blot used for telomere length determination. Figure S2. Histograms of the sequences obtained by Sanger sequencing of the regions of genes related to telomere biology that presented SNVs or indels. (DOC 3474 kb)
Data Availability Statement
Additional data and materials can be provided upon request to the authors.