Abstract
background: The ATP-binding cassette (ABC) transporters family is one of the largest families of membrane proteins existing in all living organisms. Pyrethroid resistance has become the largest unique obstacle for mosquito control worldwide. ABC transporters are thought to be associated with pyrethroid resistance in some agricultural pests, but little information is known for mosquitoes. Herein, we investigated the diversity, location, characteristics, phylogenetics, and evolution of ABC transporter family of genes in the Anopheles sinensis genome, and identified the ABC transporter genes associated with pyrethroid resistance through expression profiles using RNA-seq and qPCR. Results: 61 ABC transporter genes are identified and divided into eight subfamilies (ABCA-H), located on 22 different scaffolds. Phylogenetic and evolution analyses with ABC transporters of A. gambiae, Drosophila melanogaster, and Homo sapiens suggest that the ABCD, ABCG, and ABCH subfamilies are monophyly, and that the ABCC and ABCG subfamilies have experienced a gene duplication event. Both RNA-seq and qPCR analyses show that the AsABCG28 gene is uniquely significantly upregulated gene in all three field pyrethroid-resistant populations (Anhui, Chongqing, and Yunnan provinces) in comparison with a laboratory-susceptible strain from Jiangsu province. The AsABCG28 is significantly upregulated at 12-h and 24-h after deltamethrin exposure in three-day-old female adults. Conclusion: This study provides the information frame for ABC transporter subfamily of genes, and lays an important basis for the better understanding and further research of ABC transporter function in insecticide toxification. The AsABCG28 gene is associated with pyrethroid detoxification, and it functions at later period in the detoxification process for xenobiotics transportation.
Keywords: Anopheles sinensis, ATP-Binding Cassette (ABC) transporters, pyrethroid resistance, deltamethrin, gene expression, RNAseq analysis
1. Introduction
ATP-binding cassette (ABC) transporters constitute one of the largest families of membrane proteins, and have been found in all living organisms from bacteria to human being. The majority of these proteins function as the ATP-dependent transport of various substrates (e.g., sugars, amino acids, lipids, peptides, vitamins, sterols, hormones, metal ions, xenobiotics, and chemotherapeutic drugs) across lipid membranes [1,2]. ABC transporters share highly conserved domains with all members containing the highly conserved cytosolic nucleotide-binding domains (NBDs, also called ATP-binding cassettes), and most members also containing less conserved transmembrane domains (TMDs) [3]. The NBDs contain several conserved motif sequences, such as ABC-signature (LSGGQ), Walker A-motif, Walker B-motif, Q-loop, and H-motif, which bind and hydrolyze ATP to energize the transport cycle. The TMDs usually consist of 5–6 membrane-spanning helices that provide substrate specificity and translocation [4]. The ABC signature motif can be uniquely used to identify ABC transporters [5]. In eukaryotes, ABC transporters are either full transporters containing four domains (two NBDs and two TMDs) in one polypeptide, or half-transporters only containing two domains (one TMD and one NBD), and the later need to homo- or heterodimerize to form a functional ABC transporter with at least four domains: two NBDs and two TMDs [6]. Based on the sequence similarity of the NBDs, the ABC transporter family of genes is classified into eight subfamilies termed from ABCA to ABCH [1]. Up to now, the ABC transporter genes have been identified in some insect species at genome-wide level, including Drosophila melanogaster (56 ABC transporter genes) [1], Bombyx mori (51) [7], Apis mellifera (41) [4,7], Anopheles gambiae (52) [7], and Tribolium castaneum (73) [8].
Many mosquito species are main disease vectors, and chemical control with insecticides is still the most critical measure in the mosquito control program. The pyrethroid is the only insecticide recommended by the World Health Organization (WHO) in the program, and is widely used for the treatment of mosquito net and indoor residual spraying [9]. However, the significant increasing of pyrethroid resistance has become the unique, largest obstacle for the mosquito control worldwide [10,11,12]. The molecular mechanisms responsible for pyrethroid resistance are complicated, including the alteration of the target sites in the para-type sodium channel, which leads knockdown resistance, and the over expression of detoxification enzymes, such as P450s, esterases, and GSTs, which leads metabolic resistance [13,14,15]. In recent years, the role of ABC transporters has also been reported to be associated with pyrethroid resistance in some species of agricultural and medical importance [4]. In mosquitoes, microarray experiments revealed that four ABC transporter genes were significantly upregulated in a pyrethroid-resistant strain of the dengue vector Aedes aegypti. Quantitative PCR showed that the ABCB4 gene was upregulated approximately seven times in an A. aegypti-resistant strain compared to the susceptible strain [16]. RNA-seq analysis revealed that the AGAPOO8436 gene (ABCC subfamily) was significantly upregulated in a deltamenthrin-resistant strain of A. gambiae [17]. More recently, the expression profile analysis of ABC transporter genes in permethrin-treated larvae and adults of A. stephensi showed that AnstABCBmember6 and AnstABCG4 were significantly upregulated, which indicates that these ABC transporters might be relevant to insecticide defense [18,19,20]. However, the expression profile of ABC transporter genes involved in pyrethroid resistance has never been investigated in different field populations with pyrethroid resistance in a given species.
The mosquito species Anopheles sinensis is a major malaria vector in China and other Southeast Asian countries [21]. Due to extensive and continued application of pyrethroid insecticides, pyrethroid resistance is now widespread in A. sinensis in China as well as South Korea [22]. We have deeply sequenced and assembled the genome of A. sinensis [15,21,23], and we are conducting comprehensive research on the molecular mechanism of pyrethroid resistance using the species as model. Some families of genes have been identified at whole-genome level and their association with pyrethroid resistance have been investigated or summarized, such as CCEs [14], P450s [15], CSPs [23], CPs [24], OBPs [25], and iGluRs [26]. However, little information has been known about the ABC transporter genes and their association with pyrethroid resistance in A. sinensis. In the present study, we identified and classified the ABC transporter genes at whole-genome level in A. sinensis, and characterized these ABC transporters, including motif pattern, genomic distribution, gene structure, and phylogenetic relationship. More importantly, we compared the expression profiles of these ABC transporter genes from A. sinensis pyrethroid-resistant field populations and pyrethroid-susceptible laboratory strain by RNA-seq and RT-qPCR, analyzed the timescale transcriptional response of representative ABC transporter genes after pyrethroid treatment of pyrethroid-resistant laboratory strain, and identified and discussed the ABC transporter genes associated with pyrethroid resistance. The study confirms that ABC transporters play an important role in pyrethroid resistance, and provides the necessary basis to further elucidate the mechanism of ABC transporters in pyrethroid resistance in A. sinensis.
2. Results and Discussion
2.1. Diversity and Genomic Location of the A. sinensis ABC Transporter Genes
A total of 61 putative ABC transporter genes are identified from A. sinensis genome. Of them, 58 genes have evidences of mRNA expression with EST sequences, and other three genes all have full-length protein-coding sequences extracted from the genome, although they do not have expression sequence support. The nomenclature of all these genes is in reference of that of A. gambiae [27], as well as the motif and phylogenetic analyses in the present study. AsABCA8 is the longest ABC transporter gene—encoding 1993 amino acids (a.a.)—and AsABCG24 is the shortest one, encoding 528 a.a. The information of genome position, exon number, gene length, amino acid size, and EST of the 61 ABC transporter genes identified are summarized in Table 1. The ABC transporter number of A. sinensis is similar to that in the D. melanogaster genome (56 ABC transporter genes)1, but more than that in A. mellifera (41) [4,7], B. Mori (51) [7], and A. gambiae genomes (52) [7], and less than that in T. castaneum (73) [8] (Table 2).
Table 1.
Information summary of 61 ABC transporter genes identified in the A. sinensis genome.
| Gene Name | Gene Annotation ID | Genome Location a | Number of Exon | Gene Length (bp) (Exon + Intron) | Amino Acid Number (AA) | EST Support b |
|---|---|---|---|---|---|---|
| ABCA Subfamily (10 genes) | ||||||
| AsABCA1 | ASI10006665 | scaffold26:2898415-2904542(−) | 10 | 4839+1289 | 1613 | + |
| AsABCA2 | ASI10006666 | scaffold26:2907038-2912368(−) | 7 | 5304+3051 | 1768 | + |
| AsABCA3 | ASI10001787 | scaffold79:3171-8550(+) | 7 | 4956+424 | 1652 | + |
| AsABCA4 | ASI10005446 | scaffold46:1576155-1586398(−) | 7 | 4965+5279 | 1655 | + |
| AsABCA5 | ASI10011585 | scaffold5:2204987-2211927(−) | 8 | 4989+1952 | 1663 | + |
| AsABCA6 | ASI10011586 | scaffold5:2214289-2220237(−) | 6 | 4887+1062 | 1629 | + |
| AsABCA7 | ASI10017076 | scaffold14:35853362-35861993(−) | 7 | 4941+1825 | 1647 | + |
| AsABCA8 | ASI10008148 | scaffold9:4353984-4361438(+) | 9 | 5979+1476 | 1993 | + |
| AsABCA9 | ASI10012001 | scaffold5:6985656-6991716(−) | 14 | 5007+1054 | 1669 | + |
| AsABCA10 | ASI10008529 | scaffold116:3498475-3502806(−) | 5 | 3417+915 | 1139 | + |
| ABCB Subfamily (5 genes) | ||||||
| AsABCB1 | ASI10016633 | scaffold14:31503016-31505870(−) | 4 | 2547+308 | 849 | + |
| AsABCB2 | ASI10010010 | scaffold15:2635069-2641163(+) | 11 | 3882+2213 | 1294 | + |
| AsABCB3 | ASI10009929 | scaffold15:1271785-1274177(−) | 4 | 2181+212 | 727 | + |
| AsABCB4 | ASI10006641 | scaffold26:2544630-2550018(+) | 7 | 2280+3109 | 760 | + |
| AsABCB5 | ASI10016463 | scaffold14:28845118-28848788(−) | 9 | 1965+1706 | 655 | + |
| ABCC Subfamily (16 genes) | ||||||
| AsABCC1 | ASI10014633 | scaffold14:5983931-5989564(+) | 7 | 3534+2100 | 1178 | + |
| AsABCC2 | ASI10014632 | scaffold14:5970014-5980211(−) | 6 | 4140+6058 | 1380 | + |
| AsABCC3 | ASI10014742 | scaffold14:7100932-7109199(+) | 9 | 4347+3921 | 1449 | + |
| AsABCC4 | ASI10004082 | scaffold75:426548-436333(−) | 12 | 4404+5382 | 1468 | + |
| AsABCC6 | ASI10004081 | scaffold75:419144-426092(−) | 11 | 3972+2977 | 1324 | - |
| AsABCC7a | ASI10001723 | scaffold85:324079-329862(+) | 7 | 4236+1548 | 1412 | + |
| AsABCC7b | ASI10002723 | scaffold140:852869-858944(+) | 7 | 4236+1840 | 1412 | + |
| AsABCC9 | ASI10007114 | scaffold1:2805165-2810461(−) | 6 | 4512+785 | 1504 | + |
| AsABCC10 | ASI10007112 | scaffold1:2792623-2797620(−) | 7 | 4521+477 | 1507 | + |
| AsABCC11 | ASI10007115 | scaffold1:2812936-2817655(−) | 4 | 4212+508 | 1404 | + |
| AsABCC12 | ASI10003245 | scaffold6:579715-584317(−) | 3 | 4416+187 | 1472 | + |
| AsABCC13 | ASI10001014 | scaffold123:207484-225544(−) | 18 | 4017+14044 | 1339 | + |
| AsABCC14 | ASI10009681 | scaffold4:5523940-5529838(−) | 11 | 3453+2446 | 1151 | + |
| AsABCC15 | ASI10007113 | scaffold1:2799112-2804048(−) | 7 | 4515+422 | 1505 | + |
| AsABCC16 | ASI10009679 | scaffold4:5516323-5523342(−) | 9 | 2199+4821 | 733 | + |
| AsABCC17 | ASI10004083 | scaffold75:439279-443721(−) | 4 | 4203+240 | 1401 | + |
| ABCD Subfamily (2 genes) | ||||||
| AsABCD1 | ASI10017457 | scaffold14:39675247-39680568(−) | 6 | 2226+3096 | 742 | + |
| AsABCD2 | ASI10013565 | scaffold16:5022027-5030607(−) | 8 | 2085+6496 | 695 | + |
| ABCE Subfamily (1 gene) | ||||||
| AsABCE1 | ASI10014410 | scaffold14:2830364-2832333(+) | 3 | 1827+143 | 609 | + |
| ABCF Subfamily (3 genes) | ||||||
| AsABCF1 | ASI10006319 | scaffold100:2782009-2783929(−) | 2 | 1815+106 | 605 | + |
| AsABCF2 | ASI10011705 | scaffold5:3835256-3838673(−) | 2 | 2805+613 | 935 | + |
| AsABCF3 | ASI10009140 | scaffold7:4751204-4753620(+) | 4 | 2175+242 | 725 | + |
| ABCG Subfamily (21 genes) | ||||||
| AsABCG2 | ASI10004617 | scaffold28:1623903-1626178(−) | 5 | 1899+377 | 633 | + |
| AsABCG3 | ASI10008396 | scaffold116:2141408-2144367(+) | 6 | 2286+674 | 762 | + |
| AsABCG6 | ASI10005115 | scaffold77:2634493-2634706(−) | 6 | 1599+331 | 533 | + |
| AsABCG7 | ASI10009696 | scaffold4:5791835-5801142(−) | 7 | 2091+7217 | 697 | + |
| AsABCG10 | ASI10002998 | scaffold68:338877-350631(−) | 9 | 2073+9682 | 691 | + |
| AsABCG14 | ASI10017482 | scaffold14:39864271-39876401(−) | 8 | 1806+10325 | 602 | + |
| AsABCG13 | ASI10017484 | scaffold14:39904223-39909126(+) | 7 | 2297+2607 | 869 | + |
| AsABCG17a | ASI10002994 | scaffold68:308541-310459(−) | 3 | 1779+140 | 593 | + |
| AsABCG17b | ASI10002995 | scaffold68:315600-317518(−) | 3 | 1779+140 | 593 | + |
| AsABCG18 | ASI10002993 | scaffold68:304413-306415(−) | 4 | 1782+241 | 594 | + |
| AsABCG19 | ASI10002992 | scaffold68:299889-301953(−) | 5 | 1800+265 | 600 | + |
| AsABCG20 | ASI10002991 | scaffold68:295550-298589(−) | 5 | 1710+1330 | 570 | + |
| AsABCG21 | ASI10011562 | scaffold5:2012784-2021328(+) | 7 | 1839+6706 | 613 | + |
| AsABCG22 | ASI10002996 | scaffold68:318211-320277(−) | 5 | 1794+273 | 598 | + |
| AsABCG23 | ASI10002997 | scaffold68:321662-335687(−) | 7 | 2100+11926 | 700 | + |
| AsABCG11 | ASI10003001 | scaffold68:361453-367680(−) | 8 | 1995+4233 | 665 | + |
| AsABCG24 | ASI10005113 | scaffold77:2627164-2629127(−) | 6 | 1584+380 | 528 | - |
| AsABCG25 | ASI10005114 | scaffold77:2629922-2632124(−) | 7 | 1791+412 | 597 | - |
| AsABCG26 | ASI10003266 | scaffold6:809083-811434(−) | 7 | 1839+513 | 613 | + |
| AsABCG27 | ASI10014141 | scaffold16:12792427-12799169(−) | 5 | 1962+4781 | 654 | + |
| AsABCG28 | ASI10015886 | scaffold14:21804555-21808651(+) | 6 | 1833+17432 | 611 | + |
| ABCH Subfamily (3 genes) | ||||||
| AsABCH1 | ASI10006388 | scaffold100:3673129-3675938(−) | 7 | 2328+482 | 776 | + |
| AsABCH2 | ASI10003817 | scaffold91:793497-800786(+) | 11 | 2133+5157 | 711 | + |
| AsABCH3 | ASI10017471 | scaffold14:39764080-39771407(+) | 10 | 2172+5156 | 724 | + |
a “+” indicates the positive strand, and “−” the negative strand. b “+” indicates that the gene is supported by transcription sequence, and “−” not.
Table 2.
Gene number comparison of eight ABC transporter subfamilies among genomes of A. sinensis, five other insect species, and human being.
| ABC Subfamily | A. sinensis | A. gambiae | D. melanogaster | A. mellifera | B. Mori | T. castaneum | H. sapiens |
|---|---|---|---|---|---|---|---|
| ABCA | 10 | 9 | 10 | 3 | 6 | 10 | 13 |
| ABCB | 5 | 5 | 8 | 5 | 8 | 6 | 11 |
| ABCC | 16 | 13 | 14 | 9 | 15 | 35 | 12 |
| ABCD | 2 | 2 | 2 | 2 | 2 | 2 | 4 |
| ABCE | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ABCF | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| ABCG | 21 | 16 | 15 | 15 | 13 | 13 | 5 |
| ABCH | 3 | 3 | 3 | 3 | 3 | 3 | 0 |
| Total | 61 | 52 | 56 | 41 | 51 | 73 | 48 |
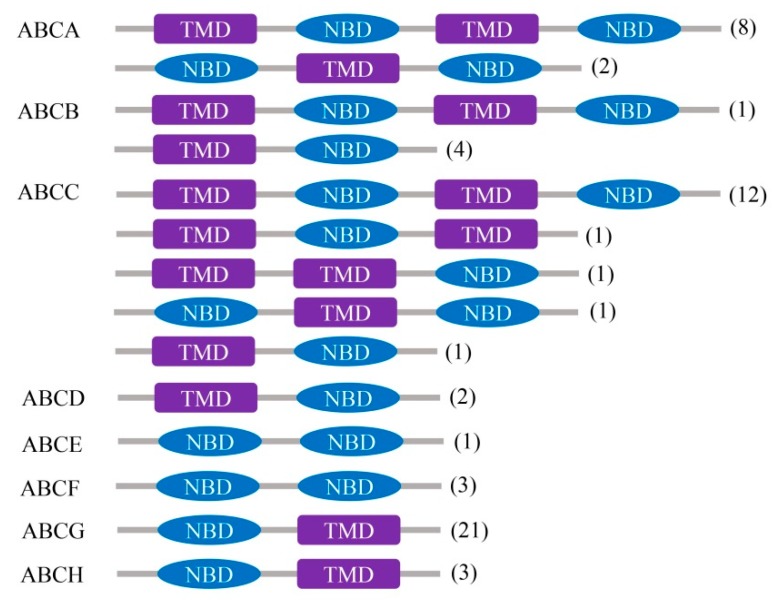
All 61 A. sinensis ABC transporter genes possess one or two conserved and characteristic NBDs (Figure 1). The ABCA, ABCB, and ABCC subfamilies contain eight, one, and 12 full transporter genes (two NBDs and two TMDs), whereas the ABCD, ABCE, ABCF, ABCG, and ABCH subfamilies have no full transporter genes. The ABCA has two additional genes with two NBDs and one TMD, ABCB has four additional half-transporter genes with one NBD and one TMD, and ABCC has two additional genes with one NBD and two TMDs, one additional gene with two NBDs, and one TMD, and one half-transporter gene, respectively. The ABCD, ABCG, and ABCH subfamilies have two, 21, and three genes with all of them being half-transporter; however, the ABCE and ABCF subfamilies have one and three genes, respectively, all of them only contain two NBDs (without half-transporter).
Figure 1.
Conserved domains of A. sinensis ABC transporters. Gray bars represent the amino acid sequences of each structure type of ABC transporter genes. The subfamily names of the ABC transporters are marked on the left side of the structure types, and the gene numbers of each structure type are denoted in brackes on the right side of the gray bars. The purple-filled rectangular boxes denote transmembrane domain (TMD) positions on the amino acid sequences, and the blue-filled oval boxes indicate nucleotide binding domain (NBD) positions. The TMD and NBD domains of each A. sinensis ABC transporter were identified using the program Pfam.
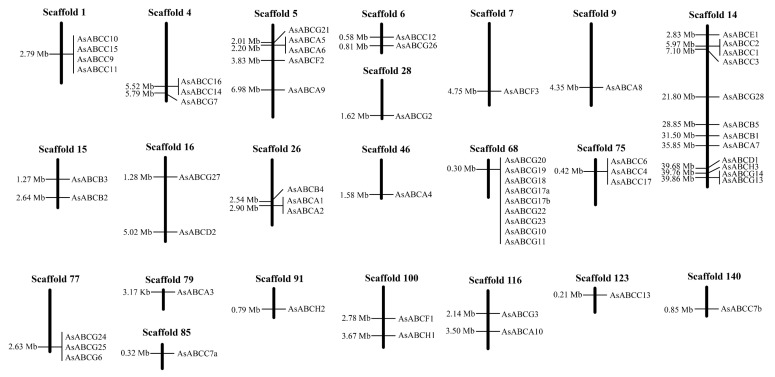
Genome mapping reveals that the 61 genes are located on 22 different scaffolds (Figure 2, Table 1). Among them, 19 genes (31.1% of all ABC transporter genes) are tandemly arranged into four clusters with at least three genes, 10 (16.4%) into five clusters with two genes, and 32 (52.5%) are present as singletons. For ABCC subfamily, four genes (4/16, 25.0%) were clustered on scaffold 1, three (3/16, 18.8%) on scaffold 75, two on scaffold 4, and two on scaffold 14; the other five genes are singly distributed. For ABCG subfamily, nine genes (9/21, 42.9%) are clustered on scaffold 68, three (3/21, 14.3%) on scaffold 77, two (2/21, 9.5%) on scaffold 14, and the remaining seven genes are singletons. For ABCA subfamily, there are four genes in two clusters belonging to scaffold 5 and scaffold 26, respectively, and the remaining six genes are singly distributed. The structure analysis shows that the exon number of the 61 A. sinensis ABC transporter genes is high complication ranging from two to 18 (Table 1). The majority of them have seven exons (27.9%, 17/61), followed by six (13.1%, 8/61), four (9.8%, 6/61), and five (9.8%, 6/61). In addition, nine genes have more than ten exons (14.8%, 10/61) and 12 genes have less than five exons (19.7%, 12/61). A total of 360 introns are predicted in all A. sinensis ABC transporter genes, whose sizes range from 13 to 9820 bp (444 bp in average), but the majority of them (64.2%, 231/360) displays sizes from 60 to 98 bp (Figure S1).
Figure 2.
Distribution of A. sinensis ABC transporter genes on scaffolds. The number of left side on scaffold represents physical locations, and on the right side are gene names.
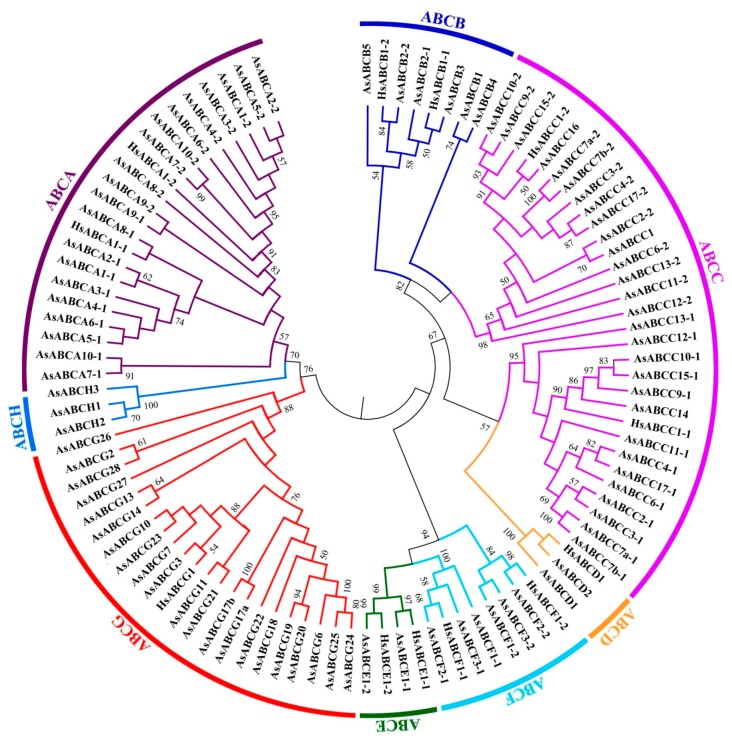
2.2. Phylogenetics and Characteristics of the A. sinensis ABC Transporter Genes
Phylogenetic analysis reveals that the A. sinensis ABC transporter genes can be classified into eight subfamilies (A–H) with high bootstrap values at most of the respective brancges (Figure 3). The ABCD, ABCG, and ABCH subfamilies have two, 21, and three genes, respectively, and their branches have 100%, 88%, and 100% bootstrap values of support, respectively. These suggest that these three subfamilies are monophyly. The ABCA subfamily has only 57% bootstrap value of support, and it appears to be closer in phylogenetics with ABCH, both together having a 70% bootstrap value. The ABCB, ABCC, and ABCF subfamilies appear to be paraphyly with ABCB, having closer relationship with ABCC, ABCC with ABCB and ABCD, and ABCF with ABCE. We separately investigated phylogenetic relationships of ABC transporter genes in each subfamily with inclusion of the ABC transporter sequences of A. gambiae, D. melanogaster, and H. sapiens to further understand the evolution and characteristics.
Figure 3.
Phylogenetic relationships of the A. sinensis ABC transporter genes. The relationships were inferred based on the amino acid sequences of nucleotide binding domains (NBD) with maximum likelihood method using MEGA5.0, and those genes with two NBDs were treated as two separate gene taxa with each taxon using one different NBD amino acid sequence. The bootstrap values calculated from 1000 replicates were marked on each corresponding node. ABCA, ABCB, ABCC, ABCD, ABCE, ABCF, ABCG, and ABCH represent eight different subfamilies in the ABC transporter family.
ABCA subfamily (Figure S2A). Ten ABCA subfamily genes were identified in the A. sinensis genome. Phylogenetic analysis reveals that the A. gambiae ABCA genes each have one ortholog with that of A. sinensis, except for the AGAP001523 that is sister with AsABCA7 + AsABCA10. These three genes (their clade with 100% bootstrap value support) form a sister group with DmCG31731 that is involved in programmed cell death in D. melanogaster metamorphosis [28]. In A. sinensis, the ABCA subfamily contains eight full transporters (2TMDs and 2NBDs) and two incomplete ABC transporters (1TMD and 2NBDs). Interestingly, all nine ABCA transporters in A. gambiae are full transporters [7], the same as the water flea, Daphnia pulex [29]. However, both full and half-transporters present in the ABCA subfamily of other insect species, such as D. melanogaster, T. castaneum, A. mellifera, and B. Mori [7]. All ABCA transporters are full transporters in human [1], but no ABCA transporter has been found in the yeast Saccharomyces cerecisiae [30]. In the plant Arabidopsis thaliana, the ABCA subfamily consists of 11 half-transporters and only one full transporter [31]. Thus, the ABCA subfamily shows great variation among species.
ABCB subfamily (Figure S2B). In A. sinensis, five ABCB subfamily genes were identified, and each has distinct clades according to phylogenetic analysis. The A. sinensis ABCB subfamily consists of one full transporter and four half-transporters, which is consistent with A. gambiae and A. mellifera [4]. ABCB1 (MDR1/P-glycoprotein) was the first characterized human ABC transporter associated with multidrug resistance (MDR) phenotype to cancer, and later studies revealed additional MDRs in the ABCB subfamily [32]. In D. melanogaster, three ABCB full transporters were found and named Mdr49, Mdr50, and Mdr65. Mdr65 is involved in chemical protection of fruit fly brain by creating a blood–brain barrier (BBB) and Mdr49 performs a critical role in germ cell migration, while no further information is available on Mdr50 [33,34]. Furthermore, it was demonsterated that colchicine exposure and heat shock increased the expression of Mdr49 in D. melanogaster larvae, and exposure of D. melanogaster flies to polycyclic aromatic hydrocarbons led to an induced Mdr49 expression, suggesting that Mdr49 is involved in resistance to insecticides and other chemical compounds [35]. As shown in Figure S2B, AsABCB2, a full transporter, is clearly clustered with A. gambiae AGAP005639 and D. melanogaster DmMdr49 in phylogenetic analysis (bootstrap value of 100%). Particularly, D. melanogaster and other arthropod ABCB FT/P-gp have also frequently been linked to insecticide transporter and/or resistance [4,36], whether AsABCB2 is involved in xenobiotic resistance requires further investigation.
ABCC subfamily (Figure S2C). In A. sinensis, there are 16 ABCC subfamily genes including 12 full transporters (two TMDs and two NBDs), 1 half-transporter (one TMD and one NBD), and three short form transporters (NBD-TMD-NBD, TMD-NBD-TMD, and TMD-TMD-NBD). In D. melanogaster, all 14 ABCC transporters are full transporters, whereas other insects such as A. gambiae, Ap. mellifeta, B. Mori and T. castaneum consist of different numbers of full, half, and incomplete ABCC transporters [7], implying that the ABCC subfamily diversely evolves in insects. In our phylogenetic analysis, two ABCCs (AsABCC7a and AsABCC7b) cluster together with high bootstrap support. Both genes show 99.72% identity in their amino acid sequences and have identical exon–intron structure (Figure S1), indicating they might have arisen by recent duplication events. AsABCC12 clusters with A. gambiae AGAP007917, which is a putative ortholog of D. melanogaster CG7806 and HsABCC10/MRP7. HsABCC10/MRP7 has been porved to confer a HsABCC1/MRP1-type multidrug resistance phenotype in cellular models [37]. According to the phylogenetic analysis, AsABCC6 and AsABCC17 group together with a cluster of D. melanogaster ABCC transporters CG10505. CG10505 is regulated by heavy metals via metal-responsive transcription factor 1, and has been reported to be involved in biochemical detoxification of zinc and copper [38]. In addition, AsABCC11 clusters with A. gambiae AGAP008436 (99% of bootstrap value), which has been detected with overexpression in a deltamenthrin-resistant strain of A. gambiae [17]. Thus, the potential roles of mosquito ABCC subfamily in detoxification is an important subject that needs to be developed.
ABCD subfamily (Figure S2D). The ABCD subfamily members are involved in the import of fatty acids and/or fatty acy1-CoAs into peroxisome [39]. Up to now, all identified ABCD transporters are known to be half-transporter and assembled as homodimers to function as transporters at the cellular level. In A. sinesis, we identified two ABCD subfamily genes, similar to the other species including A. gambiae, A. mellifera, B. Mori, D. melanogaster, and T. castaneum [4]. Based on phylogenetic analysis, each As-ABCD gene shows a clear homologous relationship to those of A. gambiae, D. melanogaster, and H. sapiens, indicating that the ABCD subfamily is evolutionarily conserved.
ABCE and ABCF subfamilies (Figure S2E). The ABCE and ABCF subfamilies consist of a pair of linked NBDs, but lack TMDs [40]. Therefore, both subfamilies are not involved in transmembrane functions, but they play a fundamental role in biological processes. In human, ABCE1 was identified as an inhibitor of RNase L and functioned in viral infection, tumor cell proliferation, antiapoptosis, translation initiation, elongation, termination, and ribosome biosynthesis, while human ABCF protein only participate in gene regulation system and ribosome assembly [41,42]. In insects, little has been known about both subfamilies. Recently, the injection of dsRNA specific for TcABCE-3A or TcABCF-2A of T. castaneum resulted in a lethal phenotype with 100% mortality in penultimate larvae, suggesting that they are active in cell viability [8]. In A. sinensis, only one ABCE gene and three ABCF genes were identified, as shown in most eukaryotes [4]. Phylogenetic analysis shows that each gene of both subfamilies grouped a unique cluster together with those of A. gambiae, D. melanogaster, and H. sapiens, indicating that they are highly conserved during evolution and probably have analogous functions as their human orothologus.
ABCG subfamily (Figure S2F). The ABCG subfamily only consists of half-transporters in most metazoans species, while several additional ABCG full transporters (also called pleiotropic drug resistance proteins (PDRs)) have been found in plant and fungi [43,44]. Twenty-one ABCG genes are identified in A. sinensis, suggesting that ABCG is the largest ABC subfamily in this species. All A. sinensis ABCG genes are half-transporters that possess a typical reverse domain architecture (NBD-TMD), and need to dimerize to form a functional transporters. The phylogenetic analysis reveals that AsABCG17a and AsABCG17b are neighbouring genes with 100% of bootstrap support. Both genes exhibit 100% amino acid identity and show a head-to-tail orientation (Table 1), suggesting they are the result of a tandem duplication.
In insects, the D. melanogaster ABCG genes white, scarlet, and brown are well studied and characterized as pigment precursor transporters into pigment cells [45,46]. The role of the T. castaneum and B. Mori ortholog of D. melanogaster white has also been experiemtally validated in eye pigmentation [8,47]. Phylogenetic analysis reveals that D. melanogaster white clusters with A. gambiae AGAP000553 and A. sinensis AsABCG27, D. melanogaster scarlet clusters with A. gambiae AGAP000133 and A. sinensis AsABCG28, and D. melanogaster brown clusters with A. gambiae AGAP007655 and A. sinensis AsABCG26. However, whether all these ABC genes in A. sinensis are involved in the transportation of pigment precursor requires further investigation.
Most evidence has approved that ABCG subfamily was also involved in pesticide transporter and/or resistance [4]. Pedra et al. found an ABCG-type transporter gene that was significantly overtranscribed in both DDT-resistant 91-R and Wisconsin strains compared to the susceptible Canton-S flies [48]. You et al. showed by RNA-seq that ABCG transporters were more highly expressed in a Plute xylostella chlorpyrifos-resistant strain compared to a susceptible strain [49]. Yang et al. revealed by microarray gene expression studies that an ABCG transporter gene was highly overexpressed in adult Bemisia tabaci flies of a thiametaoxam resistant strain [50,51]. Jones et al. determined that one ABCG gene, ABCG4 (AGAP001333), was overexpressed (2-fold) in an A. arabiensis DTT-resistant stain than the susceptible strain [52]. Recently, Epis et al. investigated the expression profile of ABC transporter genes in A. stephensi, the results showed that AnstABCG4 (AGAP001333) gene was upregulated expression in permethrin-trated larvae and adult mosquitoes compared to those of nontreated mosquitoes [19]. In our ABCG subfamily, A. sinensis AsABCG28 clusters with A. gambiae AGAP001333 with a 99% bootstrap value, indicating that AsABCG28 might function in the defense against insectcide.
ABCH subfamily (Figure S2G). The ABCH subfamily was first identified in D. melanogaster and found in arthropods and zebrafish [1,4,53]. The domain architecture of the ABCH and ABCG genes is similar. In A. sinensis, three ABCH genes are found and process a reverse domain organization (NBD-TMD). Phylogenetic analysis shows that the ABCH genes are originated from a commom ancestor among the three insects. However, their physiological functions were poorly understood. Silva et al. deterimented that a ABCH gene was over expressed in adult aphids Myzus persicae upon exposure to pirimicarb [54]. You et al. found that an ortholog of D. melanogaster CG9990 was the most upregulated ABCH transporter in a chlorpyrifos-resistant strain of Plutella xylostella compared to the susceptible stain [35]. Based on the ABCH phylogenetic analysis, AsABCH2 is well clustered with D. melanogaster CG9990, highlighting the potential role of ABCH subfamily member in detoxification.
2.3. Ka/Ks of the Ortholog Genes in A. sinensis and A. gambiae
To better understand whether ABC transporter genes in A. sinensis and A. gambiae were under different evolutionary constraints, the pairwise Ka/Ks for each orthologous group was calculated. The results show that the purify selection is the main selection pressure driving the diversities of ABC transporters (all Ka/Ks<<1).
2.4. RNA-seq Expression Profile and qPCR Verification of ABC Transporters Associated with Pyrethorid Resistance
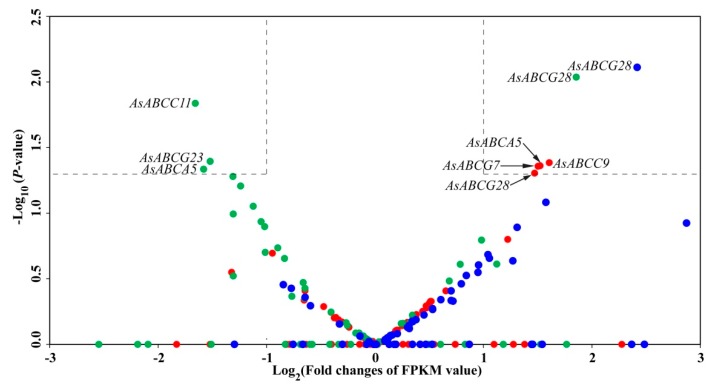
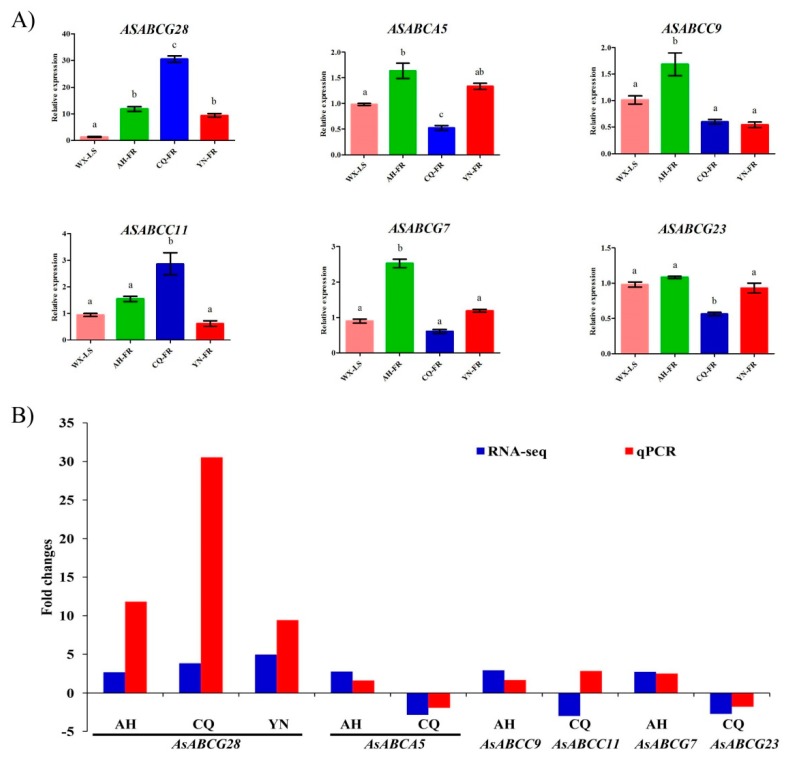
The expression profiles analysis of all ABC transporters from A. sinensis shows that six genes are significantly differentially expressed in at least one pyrethroid-resistant population (Figure 4, Table S1). AsABCG28 is the uniquely significantly upregulated gene in the three resistant populations in comparison with susceptible WX-LS, with 2.69-fold upregulation (in AH-FR), 3.84-fold upregulation (CQ-FR), and 4.96-fold upregulation (YN-FR). The AsABCC9, AsABCG7, and AsABCA5 are significantly upregulated in AH-FR by 2.96-fold, 2.75-fold, and 2.79-fold, respectively. Although the AsABCA5 is also significantly upregulated in AH-FR, it is downregulated in CQ-FR. In contrast, AsABCC11 and AsABCG23 are significantly downregulated by 2.97-fold and 2.70-fold, respectively.
Figure 4.
Expression profile of ABC transporter genes in three pyrethroid-resistent field populations (AH-FR, CQ-FR, and YN-FR) compared with a laboratory-susceptible strain (WX-LS), detected by RNA-seq. Genes with FPKM value ≥ 2 (i.e., ≥ 1 in Log2, showing on X-axis) and p-value < 0.05 (i.e., > 1.30 in –Log10, showing on Y-axis) are considered to be significantly upregulated, whereas those with FPKM value ≤ 0.5 (≤ −1 in Log2) and p-value < 0.05 are regarded as downregulated. Vertical dotted lines mark the 1 and −1 of Log2 (fold changes of FPKM value) values on the X-axis, and horizontal dotted lines denote the p-value = 0.05 of –Log10 (p-value) on the Y-axis. There are six genes that are significantly differently expressed in different populations, in which four genes are upregulated (AsABCG28 commonly in three populations) and three genes are downregulated (AsABCA5 is upregulated in AH-FR but downregulated in CQ-FR). The red-, green-, and blue-filled circles indicate the AH-FR, CQ-FR, and YN-FR, respectively.
In order to validate the results of the RNA-seq, these six genes with significant expression difference are selected for verification using qPCR analysis (Figure 5A) with gene-specific primers designed in the present study (Table 3). The results show that expression fold changes are basically consistent with those in RNA-seq analysis for five of these six genes tested (Figure 5B). The AsABCG28 is also significantly upregulated in all three resistant populations in comparison to the susceptible strain WX-LS. The AsABCG28 shares 76% a.a. identity with AGAP001333 of A. gambiae, which has been reported to be significantly overexpressed in DDT resistance strain in A. arabiensis (as ABCG4) [52], and in thiamethoxam resistant strain in sweetpotato whitefly, Bemisia tabaci (as ABCG4) [55]. These findings imply that AsABCG28 is an essential member involving defense against insecticide. AsABCC9 and AsABCG7 are significantly upregulated in AH-FR compared with WX-LS, which is the first report that these two genes are associated with pyrethroid resistance. AsABCC11, the homolog of A. gambiae AGAP008436, is significantly upexpressed in CQ-FR based on the qPCR analysis, consistent with earlier study that showes that AGAP008436 is upregulated by 2.88-fold in detamethrin resistant strain in A. gambiae [17]. However, it is downexpressed in CQ-FR based on RNA-seq analysis. These results might countribute to the different batches of samples and/or the different splicing of the target gene. The AsABCG23 is uniquely significantly downregulated in CQ-RF according to the qPCR analyses, consistent with the RNA-seq detection. The AsABCA5 is significantly upexpressed in AH-FR compared with WX-LS, which agrees with an earlier study that shows that one ABCA gene is significantly upregulated in a chlorpyrifos-resistant strain of Lepidoptera Pl. xylostella [49]. However, the gene is downexpressed in CQ-FR, but no significant difference is observed in YN-FR. These findings indicated that the expression variation of AsABCA5 might be due to the difference of geographical distribution in the mosquitoes investigated. Whether or not AsABCA5 is related to pyrethroid resistance need to further investigate.
Figure 5.
qPCR verification of six genes significantly differentially expressed in RNA-seq analysis. (A) The relative expression levels of these genes are shown as the mean ± SD of three biological replicates in qPCR analysis. The S7 (RPS7) and L49 (RPL49) genes were used as internal reference for expression normalization. The population/strain pairs marked with same letters are not significantly different in expression (p-value ≥ 0.05), and the remaining pairs are significantly different (p-value < 0.05), determined by one-way ANOVA. (B) Comparison of expression fold-changes between RNA-seq and qPCR analyses for the six genes in three field populations comparative with the laboratory-susceptible strain WX-LS. AH: Anhui population; CQ: Chongqing; YN: Yunnan.
Table 3.
Primer sequences used for qPCR.
| Gene Name | Forward Primer (5′–3′) | Reverse Primer (5′–3′) | Size (Base Pairs) |
|---|---|---|---|
| AsABCG28 | CAACCTGTACTCCACCACCC | TAACGACCAGACCGAGCAAC | 140 |
| AsABCA5 | GGGAGTTCAAGTGTCTCGGG | TGTACTGTTTGACGTCCGCT | 129 |
| AsABCC9 | TGCGTAGCCGGTTGACTATC | AATGAGACAGCTCCAGTGCC | 126 |
| AsABCG7 | ATCTGAACCTCGGCGGAAAC | CCCAGAACTGGTGGAACTCG | 198 |
| AsABCB1 | GTAAAACCGGCGAAGTGCTG | GCGTCATGGTGAGAAACACG | 177 |
| AsABCB2 | GTGAGATCCCGATGCAGGAG | TTCTGACCACCGGACAGTTG | 130 |
| AsABCB3 | TTCTCGGTTCGGTCATTCGG | ACACAAACTCTACGCCCTCG | 150 |
| AsABCB4 | GTTTCACGACACCATTCGGC | AACTTAAGGCCACGCTCTCC | 156 |
| AsABCB5 | ACGGGTTGAACATGGGTGAG | GATAGCACGGGCGATAGCAA | 176 |
| AsABCC2 | GGAGAGGTGCTGATCGATGG | CTGGGTAGTCCTCGAAGGGA | 142 |
| RPS7 | CGGAGAAGATGGCATGGGAGAT | ATAGTGAGCATAGGCCCGGTTA | 148 |
| RPL49 | GGAGCCGGTCGGTGATATGT | TTCCTTCTCGGTCGGCTTCG | 121 |
2.5. ABC Transporters Expression Subject to Deltamethrin Treatment
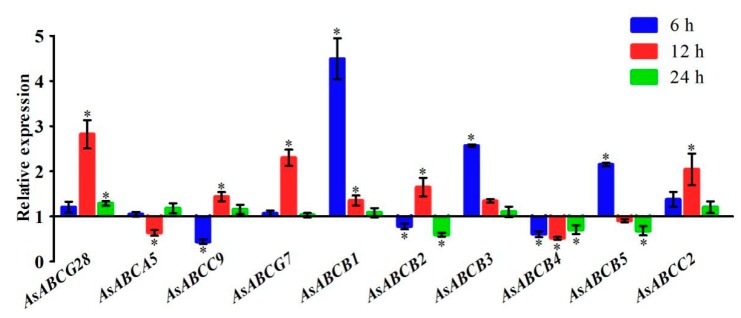
A total of ten ABC transporter genes in A. sinensis (AsABCG28, AsABCA5, AsABCC9, AsABCG7, AsABCB1, AsABCB2, AsABCB3, AsABCB4, AsABCB5, and AsABCC2) were chosen to investigate the expression changes on timescale subject to deltamethrin treatment using three-day-old female adults of the laboratory deltamethrin-resistant strain WX-LR (Figure 6). The ten genes included four genes that showed significantly upexpression in RNA-seq analysis, and six genes that have been reported to be overexpressed in response to insecticide stress. The qPCR results show that three ABCB genes (AsABCB1, AsABCB5, and AsABCB3) are significantly overexpressed at 6-h after deltamethrin exposure. AsABCB1 has the highest of overexpression (4.5-fold), and is also significantly upregulated in both larvae and adults of the mosquito A. stephensi after permethrin treatment (as AnstABCBmember6) [19,20]. The AsABCB5 had 2.2-folds overexpression, and is also overexpressed in ivermectin-resistant Rhipicephalus microplus upon exposure to ivermectin (as RmABCB10) [56]. The AsABCB3 had 2.6-fold overexpression, but no overexpression is observed in A. stephensi exposed to permethrin (as AnstABCB3) [19,20]. The detoxification process can be subdivided into four different phases (0–III), and involves a diversity of enzymatic complexes. ABC transporters are thought to act in phases 0 and III of the detoxification process [4]. In phase 0, ABC transporter genes belonging to the ABCB subfamily can directly extrude a wide range of xenobiotics out of cells, therefore avoiding the accumulation of these xenobiotics in the cells or organism [19]. The findings herein support the hypothesis that ABCB genes are involoved in early detoxification process after insecticide treatment.
Figure 6.
Expression changes of ten ABC transporter genes after pyrethroid treatment for A. sinensis female adults. The ten genes include four genes (AsABCG28, AsABCA5, AsABCC9, and AsABCG7) that are significantly upregulated in RNA-seq and qPCR analyses in different resistant population, and six genes (AsABCB1, AsABCB2, AsABCB3, AsABCB4, AsABCB5, and AsABCC2) that have been reported to be associated with pyrethroid resistance. The expression levels (means ± SD) were determined at 6-h, 12-h, and 24-h postpyrethroid treatment using qPCR with three biological replicates. The samples marked with “*” were significantly differently expressed (p-value < 0.05) compared with the untreated samples at the same time, and the samples without “*” were not significantly differently expressed.
Seven genes, inculding two ABCG genes (AsABCG28 and AsABCG7), two ABCC genes (AsABCC9 and AsABCC2), and three ABCB genes (AsABCB1, AsABCB2, and AsABCB3), show significantly upregulated expression 12-h after deltamethrin exposure (Figure 6). Most importantly, the AsABCG28 has the highest overexpression (2.8 folds), and it is also significantly upregulated at 24-h. The AnstABCG4, the orthology of AsABCG28 in A. stephensi showed similar expression pattern in permethrin-exposed larvae and adults [18,19,20]. Earlier reports showed that ABCG subfamily of genes participated in the transportion of xenobiotics modified by detoxifying enzymes in phase III [4,36]. Our research reveals that the AsABCG28 is associated with pyrethroid detoxification, and it functions at later period in the detoxification process for xenobiotics transportion. In addition, the AsABCC2 had 2.0-fold overexpression, and it is also significantly upregulated in pyrethroid-resistant strain of A. aegypti (as ABCC2) [16]. The AsABCB2 had 1.6-fold overexpression at 12 h, but was downexpressed at 6-h and 24-h. This gene is significantly upregulated in A. stephensi at 1-h and 24-h after permethrin exposure (as AnstABCB2) [20], and in a susceptible strain of A. aegypti at 48-h post-temephos treatment (as AaegP-gp) [57]. A growing body of studies have indicated that the ABCB full transporter genes (P-gp) are associated with insecticide transport and/or resistance, including organophosphates [57], organochlorines [52], carbamates [58], and pyrethroids [59,60]. These results suggest that the AsABCC2 and AsABCB2 might participate in deltanethrin defense at the later phase of the detoxification.
The AsABCB4 is significantly downregulated at all three time points investigated. However, microarray analysis showed that the ABCB4 was significantly upregulated in different populations of DDT-resistant A. gambiae mosquitoes (as AGP006364) [61], and it was not differentially expressed or downregulated in both adults and larvae of A. stephensi (as AnstABCB4) [18,19,20]. The AsABCB4 has a higher copy number in pyrethroid (permethrin and deltamethrin)-resistant A. aegypti populations compared to a laboratory-susceptible strain (as ABCB4) [16]. In addition, AsABCB2 at 6 h and 24 h, AsABCC9 at 6 h, AsABCA5 at 12 h, and AsABCB5 at 24 h herein also show significant downregulation. These findings could not reveal the regulation and function of the downregulation of these genes.
3. Materials and Methods
3.1. Mosquito Samples and Sequence Data
Two laboratory A. sinensis strains, WX-LS and WX-LR, susceptible and resistant to pyrethroid, respectively, were established in the Institute of Entomology and Molecular Biology, Chongqing Normal University, China. Field pyrethroid-resistant mosquitoes were collected from rice (Oryza sativa) fields in different locations of three geographical populations: Anhui (AH), Chongqing (CQ) and Yunnan (YN) provinces, China, between July and September, 2013.
A. sinensis genome data, including genome sequence and annotation, predicted gene set of sequences, and deduced amino acid sequences of the gene set, were obtained from the Institute of Entomology and Molecular Biology, Chongqing Normal University, China (publication in preparation). Two sets of transcriptome data were downloaded from the National Center for Biotechnology Information (NCBI) as an EST database (Accession numbers: SRA073189 and GAFE01000001-GAFE01028133). The ABC transporter amino acid sequences of A. gambiae, D. melanogaster and Homo sapiens were retrieved from NCBI as well (Table S2).
3.2. Identification and Characteristcs Analyses of A. sinensis ABC Transporter Genes
To identify putative ABC transporter genes, we firstly used the highly conserved NBD motif sequences of all ABC transporter genes from D. melanogaster and A. gambiae [7] as queries to conduct BLASTP searching against A. sinensis amino acid database with an e-value cut-off at 1e-6. Secondly, the Hidden Markov Model (HMM) for ABC transporter conservative motif ABC_tran (Pfam 00005) was downloaded from Pfam (http://pfam.xfam.org) and used as query to search the A. sinensis amino acid database using the program hmmsearch within the HMMER3 software Suite30, with an e-value threshold of 1e-10. Thirdly, all putative ABC transporter protein sequences obtained above were used as queries to perform TBLASTN searching against the A. sinensis genome with an e-value cut-off at 1e-5. Finally, the FGENSH+ (http://www.softberry.com) software was used to predict the genes with best matched individual ABC protein sequences in TBLASTN search results.
To further confirm the reliability of our results, all predicted ABC transporter genes were used as queries to perform BLASTN searches against the A. sinensis EST database [21,62]. A 95% or greater identity and minimum cut-off E-value (≤e-20) were employed to discriminate between duplicate genes. In addition, all genes were further analyzed by the program Pfam (http://pfam.xfam.org) to idientify its NBD and TMD domains. A BLASTN search using all obtained ABC sequences as quries was further conducted against the A. sinensis genome database to determine the location of ABC transporter genes on A. sinensis scaffolds. The structure of the ABC transporter genes were generated with GSDS (http://gsds.cbi.pku.edu.cn/) using intron/exon sites obtained from the A. sinensis genome database.
3.3. Phylogenetic Analysis of A. sinensis ABC Transporter Genes
The NBD sequences of preliminary gene models were extracted using the Prosite facility (http://prosite.expasy.org/prosite.html) with predicted protein sequences and the prosite profile PS50893. The NBD sequences were then aligned using ClustalW [63] and constructed phylogenetic relationships using maximum-likelihood method with Jones–Taylor–Thornton substitution model. The bootstrap values were calculated with 1000 replicates using the program package MEGA 5.0 [64]. Full-length sequences from A. sinensis, A. gambiae, D. melanogaster, and human ABC transporter protein sequences were used to carry out the phylogenetic analysis of each subfamily of ABC transporter genes using the same methodology as mentioned above.
3.4. Estimation of Synonymous Substitutions and Nonsynonymous Substitutions
The ABC transpoter sequences of each orthologs pair from A. sinensis and A. gambiae were first aligned using ClustalW software [63]. The files of the sequence alignments were then convented to PHYLIP alignment using MEGA 6. Finally, the convented files were uploaded to the YN00 program of PAML to estimate synonymous substitutions (Ka) and nonsynonymous substitution (Ks) [65]. Ka/Ks ratios of >1, <1, and =1 represent positive selection, stabilizing selection, and neutral selection, respectively.
3.5. RNA-seq Analysis and qPCR Verification of ABC Transporters Associated with Pyrethroid Resistance
The field-collected mosquito larvae and pupae were reared to adulthood at 27 ± 1 °C with 70 ± 10% relative humidity in local laboratories. The female adults of three days post emergence were identified to species morphologically, and then the A. sinensis mosquitoes were used to bioassay with 0.05% deltamethrin (Sigma-Aldrich, St. Louis, MO, USA) test papers according to the standard WHO tube bioassay [66]. More than 1000 mosquitoes per population collected from multiple sites in field were tested with 20 mosquitoes per tube, and paraffin oil-treated papers without deltamethrin were used as the control. After 60 min exposure to 0.05% deltamethrin, mosquitoes were transferred to recovery cups and maintained on a 6% sucrose solution for 24 h [15]. The mosquitoes alive 24 h after the recovery period were defined as resistant mosquitoes [66]. At least 100 resistant adult mosquitoes three days old from each population were preserved in RNAlater (Qiagen Shanghai, China) for RNA extraction and RNA-seq library construction, respectively. Three pyrethroid-resistant populations from Anhui (AH-FR), Chongqing (CQ-FR), and Yunnan (YN-FR) were used in the RNA-seq analysis and quantitative real-time Polymerase Chain Reaction (qPCR) verification, and the laboratory pyrethroid-susceptible strain (WX-LS) originally collected from Wuxi, Jiangsu province was handled with the same procedure.
Prior to RNA extraction, two or three legs of each individual mosquito were used to extract genomic DNA for molecular identification using the ribosomal DNA internal transcribed spacer rDNA-ITS2-based method [67]. Samples of 15 mosquitoes were pooled and prepared total RNA for each RNA-seq sample using Trizol Reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer protocol. Three biological replicates were set for three field-resistant populations and WX-LS strain. The quantity of total RNA was assessed using a NanoDrop spectrophotometer (NanoDrop Technologies, Thermo Fisher Scientific Inc., Waltham, MA, USA). RNA quality was assessed using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). The cDNA for each library was synthesized and amplified using the Mint-2 cDNA synthesis kit (Evrogen, Moscow, Russia). Illumina HiSeq™ 2000 was used for cDNA library sequencing at Beijing Genomics Institute (BGI), following the standard Illumina procedures.
TopHat was used to map reads obtained from Illumina sequencing to the A. sinensis genome [68]. Cufflinks was used to determined expressions in terms of fragments per kb per million reads (FPKM) [69]. Differential accumulation of transcripts between deltamethrin-resistant and -susceptible mosquitoes was assessed by the Cuffdiff program within Cufflinks. To minimize the impact of sequencing length and nucleotide composition, the FPKM for each gene of each sample was calculated to determine the expression level [70]. The normalized gene expression level of A. sinensis ABC transporter genes each in the three resistant populations was compared with the WX-LS strain. The genes that had the FPKM fold changes of ≥ 2 [log2 (fold change) ≥ 1] and the p-value ≤ 0.05 [−Log10 (p-value) ≥ 1.301] were considered as significantly upregulated between the resistant and susceptible strains. In contrast, the genes with the FPKM fold change ≤ 2 and the p-value ≤ 0.05 [−Log10 (p-value) ≥ 1.301] were regarded as significantly downregulated.
The ABC transporter genes to be significantly differentially expressed detected by RNA-seq analyses were chosen to perform qPCR. The expression confirmation of selected genes were investigated in the three deltamethrin resistant populations (AH-FR, CQ-FR, and YN-FR) using the laboratory-susceptible strain (WX-LS) as reference, and the qPCRs were carried out with three biological replicates (three mosquitoes per sample) and three technical replicates. Total RNA was extracted from four individual mosquitoes as the same conditions as RNA-seq with TRIzol Reagent (Invitrogen) following the manufacturer’s instructions. Complementary DNA was synthesized from 1.0 μg RNA using PrimScript TM RT Reagent Kit with gDNA Eraser (TaKaRa, Dalian, China) and stored at −20 °C. Real-time PCR was performed using CFX Connect Real-Time PCR System (Bio-Rad, USA). Each reaction with a final volumn of 15 μL was mixed with 7.5 μL of 2 x qPCR mix (Bio-Rad, Hercules, CA, USA), 0.5 μL each of gene-specific primers and 1 μL the cDNAs templates, and 5.5 μL of double distilled water. Running conditions were as follows: 94 °C for 3 min, 40 cycles of 95 °C for 5 s, 60 °C for 15 s, and 72 °C for 15 s. Gene-specific primes were designed using Primer Premier 5.0 [71]. The ribosomal protein S7 (RPS7) and the ribosomal protein L49 (RPL49) were used as internal controls. The relative expression levels of each gene in four populations/strain were normalized in comparison with RPS7 and RPL49 CT values using the 2−ΔΔCt method [72]. All data were shown as mean ± SD (standard deviation). The statistical significance of the gene expressions was calculated using one-way Analysis of Variance (ANOVA) and a value of p < 0.05 was considered statistically significant.
3.6. Expression Changes of Pyrethroid Resistance-Relative ABC Transporter Genes Subject to Deltamethrin Treatment
To analyse the expression profiles of ABC transporter genes after deltamenthrin treatment, the bioassays were conducted on three days old female mosquitoes following the standard WHO tube test [66]. Batches of 20–25 mosquitoes from A. sinensis strain WX-LR were exposed to test papers impregnated with 0.05% deltamethrin (Sigma-Aldrich, St. Louis, MO, USA) for one hour. After exposure, the mosquitoes were then transferred to recovery cups and supplied with 6% glucose solution. Tests with non-impregnated control papers were used as controls. Survivors and nonexposed mosquitoes were collected at different times (6-h, 12-h, and 24-h) postrecovery. The mosquitoes were stored in RNAlater (Qiagen Shanghai, China) and kept at −80 °C for molecular analysis.
Based on the RNA-seq and qPCR analyses in the present study and previously published literature [4,20], the expression of selective ABC transporter genes from A. sinensis were investigated at three time-points (6-h, 12-h, and 24-h) after recovery. The relative gene expressions of all selective genes were determined by qPCR as mentioned above. Three biological replicates were run for each sample on a plate. The expression levels of target genes were calculated relative to the internal reference genes. For each target gene, the expression level of the control group was considered as the basal level (set equal to 1). The relative expression level in the treated group was showed as means ± SD. The expression level between treated and control groups were calculated using a Student’s t-test and a value of p < 0.05 was considered statistically significant.
4. Conclusions
The study provides useful insights into the diversity, location, characteristics and phylogenetics of ABC transporter family of genes in A. sinensis genome, and ABC transporter genes associated with pyrethroid resistance. The ABCD, ABCG, and ABCH subfamilies are monophyly, and the ABCC and ABCG subfamilies experience gene duplication event. The AsABCG28 gene is associated with pyrethroid detoxification, and it functions at later period in the detoxification process for xenobiotics transportion. This study provides the information frame for ABC transporter subfamily of genes, and lays an important basis for the better understanding and further research of ABC transporter function in insecticide toxification.
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/20/6/1409/s1.
Author Contributions
Q.H. preformed the experiments and wrote the original draft; F.S. analyzed the data; Z.Y. provided materials; Y.Z. and W.F. contributed reagents; and B.C. supervisered the research.
Funding
This research was supported by the following, the National Natural Science Foundation of China (31672363, 31872262), the National Key Program of Science and Technology Foundation Work of China (2015FY210300), and the Scientific and Technological Research Program of Chongqing Municipal Education Commission (KJ1600338).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Dean M., Rzhetsky A., Allikmets R. The human ATP-Binding cassette (ABC) transporter superfamily. Genome Res. 2001;11:1156–1166. doi: 10.1101/gr.GR-1649R. [DOI] [PubMed] [Google Scholar]
- 2.Higgins C.F. ABC transporters: From microorganisms to man. Annu. Rev. Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 3.Rees D.C., Johnson E., Lewinson O. ABC transporters: The power to change. Nat. Rev. Mol. Cell Biol. 2009;10:218–227. doi: 10.1038/nrm2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dermauw W., Leeuwen T.V. The ABC gene family in arthropods: Comparative genomics and role in insecticide transport and resistance. Insect Biochem. Mol. Biol. 2014;45:89–110. doi: 10.1016/j.ibmb.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Strauss A.S., Wang D., Stock M., René R.G., Groth M., Boland W., Burse A. Tissue-specific transcript profiling for ABC transporters in the sequestering larvae of the phytophagous leaf beetle Chrysomela populi. PLoS ONE. 2014;9:e98637. doi: 10.1371/journal.pone.0098637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hollenstein K., Dawson R.J., Locher K.P. Structure and mechanism of ABC transporter proteins. Curr. Opin. Struct. Biol. 2007;17:412–418. doi: 10.1016/j.sbi.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Liu S., Zhou S., Tian L., Guo E., Luan Y., Zhang J., Li S. Genome-wide identification and characterization of ATP-binding cassette transporters in the silkworm, Bombyx mori. BMC Genom. 2001;12:476–491. doi: 10.1186/1471-2164-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broehan G., Kroeger T., Lorenzen M., Merzendorfer H. Functional analysis of the ATP-binding cassette (ABC) transporter gene family of Tribolium castaneum. BMC Genom. 2013;14:6–25. doi: 10.1186/1471-2164-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.WHO . Pesticides and Their Aapplication for the Control of Vectors and Pests of Public Health Importance. World Health Organization; Geneva, Switzerland: 2006. [Google Scholar]
- 10.Phillips R.S. Current status of malaria and potential for control. Clin. Microbiol. Rev. 2001;14:208–226. doi: 10.1128/CMR.14.1.208-226.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemingway J., Field L., Vontas J. An overview of insecticide resistance. Science. 2002;298:96–97. doi: 10.1126/science.1078052. [DOI] [PubMed] [Google Scholar]
- 12.Ranson H., N’guessan R., Lines J., Moiroux N., Nkuni Z., Corbel V. Pyrethroid resistance in african anopheline mosquitoes: What are the implications for malaria control? Trends Parasitol. 2011;27:91–98. doi: 10.1016/j.pt.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Hemingway J., Hawkes N.J., Mccarroll L., Ranson H. The molecular basis of insecticide resistance in mosquitoes. Insect Biochem. Mol. 2004;34:653–665. doi: 10.1016/j.ibmb.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 14.Wu X.M., Xu B.Y., Si F.L., Li J.J., Yan Z.T., Yan Z.W., He X., Chen B. Identification of carboxylesterase genes associated with pyrethroid resistance in the malaria vector Anopheles sinensis (Diptera: Culicidae): Carboxylesterase genes in Anopheles sinensis. Pest Manag. Sci. 2018;74:159–169. doi: 10.1002/ps.4672. [DOI] [PubMed] [Google Scholar]
- 15.Yan Z.W., He Z.B., Yan Z.T., Si F.L., Zhou Y., Chen B. Genome-wide and expression-profiling analyses suggest the main cytochrome P450 genes related to pyrethroid resistance in the malaria vector, Anopheles sinensis (Diptera Culicidae) Pest Manag. Sci. 2018;74:1810–1820. doi: 10.1002/ps.4879. [DOI] [PubMed] [Google Scholar]
- 16.Bariami V., Jones C.M., Poupardin R., Ranson H. Gene amplification, ABC transporters and cytochrome P450s: Unraveling the molecular basis of pyrethroid resistance in the dengue vector, Aedes aegypti. PLoS Negl. Trop. Dis. 2012;6:e1692. doi: 10.1371/journal.pntd.0001692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonizzoni M., Afrane Y., Dunn W.A., Atieli F.K., Zhou G., Zhong D., Li J., Githeko A., Yan G. Comparative transcriptome analyses of deltamethrin resistant and -susceptible Anopheles gambiae mosquitoes from Kenya by RNASeq. PLoS ONE. 2012;7:e44607. doi: 10.1371/journal.pone.0044607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Epis S., Porretta D., Mastrantonio V., Comandatore F., Sassera D., Rossi P., Cafarchia C., Otranto D., Favia G., Genchi C., et al. ABC transporters are involved in defense against permethrin insecticide in the malaria vector Anopheles stephensi. Parasite Vector. 2014;7:349–355. doi: 10.1186/1756-3305-7-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Epis S., Porretta D., Mastrantonio V., Urbanelli S., Sassera D., Marco L.D., Mereghetti V., Montagna M., Ricci I., Favia G., et al. Temporal dynamics of the ABC transporter response to insecticide treatment: Insights from the malaria vector Anopheles stephensi. Sci. Rep. 2014;4:7435–7439. doi: 10.1038/srep07435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mastrantonio V., Ferrari M., Epis S., Negri A., Scuccimarra G., Favia G., Porretta D., Urbanelli S., Bandi C. Gene expression modulation of ABC transporter genes in response to permethrin in adults of the mosquito malaria vector Anopheles stephensi. Acta Trop. 2017;171:37–43. doi: 10.1016/j.actatropica.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 21.Chen B., Zhang Y.J., He Z.B., Li W.S., Si F.L., Tang Y., He Q.Y., Qiao L., Yan Z.T., Fu W.B., et al. De novo transcriptome sequencing and sequence analysis of the malaria vector Anopheles sinensis (Diptera: Culicidae) Parasite Vector. 2014;7:314–325. doi: 10.1186/1756-3305-7-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin Q., Li Y., Zhong D., Zhou N., Chang X., Cui C., Yan L., Chen X.G. Insecticide resistance of Anopheles sinensis and An. vagus in Hainan Island, a malaria-endemic area of China. Parasites Vectors. 2014;7:92–100. doi: 10.1186/1756-3305-7-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mei T., Fu W.B., Li B., He Z.B., Chen B. Comparative genomics of chemosensory protein genes (CSPs) in twenty-two mosquito species (Diptera: Culicidae): Identification, characterization, and evolution. PLoS ONE. 2018;13:e0190412. doi: 10.1371/journal.pone.0190412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu B.Q., Qiao L., He Q.Y., Zhou Y., Ren S., Chen B. Genome-wide identification, characterization and evolution of cuticular protein genes in the malaria vector Anopheles sinensis (Diptera: Culicidae) Curr. Opin. Insect Sci. 2018;25:739–750. doi: 10.1111/1744-7917.12483. [DOI] [PubMed] [Google Scholar]
- 25.He X., He Z.B., Zhang Y.J., Zhou Y., Xian P.J., Qiao L., Chen B. Genome-wide identification and characterization of odorant-binding proteins of genes in the malaria vector Anopheles sinensis (Diptera: Culicidae) Curr. Opin. Insect Sci. 2016;23:366–376. doi: 10.1111/1744-7917.12333. [DOI] [PubMed] [Google Scholar]
- 26.Wang T.T., Si F.L., He Z.B., Chen B. Genome-wide identification, characterization and classification of ionotropic glutamate receptor genes (iGluRs) in the malaria vector Anopheles sinensis (Diptera: Culicidae) Parasite Vector. 2018;11:34–45. doi: 10.1186/s13071-017-2610-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roth C.W., Holm I., Graille M., Dehoux P., Rzhetsky A., Wincker P., Weissenbach J., Brey P.T. Identification of the Anopheles gambiae ATP-binding cassette transporter superfamily genes. Mol. Cells. 2003;15:150–158. [PubMed] [Google Scholar]
- 28.Dutta S. Ph.D. Thesis. University of Maryland, Department Mol. Cell Bio.; College Park, MD, USA: 2008. Genetic Regulation of Autophagic Cell Death in Drosophila melanogaster; p. 130. [Google Scholar]
- 29.Sturm A., Cunningham P., Dean M. The ABC transporter gene family of Daphnia pulex. BMC Genom. 2009;10:170–187. doi: 10.1186/1471-2164-10-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Decottignies A., Goffeau A. Complete inventory of the yeast ABC proteins. Nat. Genet. 1997;15:137–145. doi: 10.1038/ng0297-137. [DOI] [PubMed] [Google Scholar]
- 31.Sanchez-Fernandez R., Davies T.G.E., Coleman J.O.D., Rea P.A. The Arabidopsis thaliana ABC Protein Superfamily, a Complete Inventory. J. Biol. Chem. 2001;276:30231–30244. doi: 10.1074/jbc.M103104200. [DOI] [PubMed] [Google Scholar]
- 32.Gottesman M.M., Fojo T., Bates S.E. Multidrug resistance in cancer: Role of ATP-dependent transporters. Nat. Rev. Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 33.Mayer F., Mayer N., Chinn L., Pinsonneault R.L., Kroetz D., Bainton R.J. Evolutionary conservation of vertebrate blood-brain barrier chemoprotective mechanisms in Drosophila. J. Neurosci. 2009;29:3538–3550. doi: 10.1523/JNEUROSCI.5564-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ricardo S., Lehmann R. An ABC transporter controls export of a Drosophila germ cell attractant. Science. 2009;323:943–946. doi: 10.1126/science.1166239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vache C., Camares O., Cardoso-Ferreira M.C., Dastugue B., Creveaux I., Vaury C., Bamdad M. A potential genomic biomarker for the detection of polycyclic aromatic hydrocarbon pollutants: Multidrug resistance gene 49 in Drosophila melanogaster. Environ. Toxicol. Chem. 2007;26:1418–1424. doi: 10.1897/06-552R.1. [DOI] [PubMed] [Google Scholar]
- 36.Buss D.S., Callaghan A. Interaction of pesticides with p-glycoprotein and other ABC proteins: A survey of the possible importance to insecticide, herbicide and fungicide resistance. Pestic. Biochem. Physiol. 2008;90:141–153. doi: 10.1016/j.pestbp.2007.12.001. [DOI] [Google Scholar]
- 37.Hopper-Borge E., Xu X., Shen T., Shi Z., Chen Z.S., Kruh G.D. Human multidrug resistance protein 7 (ABCC10) is a resistance factor for nucleoside analogues and epothilone B. Cancer Res. 2009;69:178–184. doi: 10.1158/0008-5472.CAN-08-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yepiskoposyan H., Egli D., Fergestad T., Selvaraj A., Treiber C., Multhaup G., Georgiev O., Schaffner W. Transcriptome response to heavy metal stress in Drosophila reveals a new zinc transporter that confers resistance to zinc. Nucleic Acids Res. 2006;34:4866–4877. doi: 10.1093/nar/gkl606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker A. Peroxisomal ABC transporters. FEBS Lett. 2006;580:1139–1155. doi: 10.1016/j.febslet.2005.12.095. [DOI] [PubMed] [Google Scholar]
- 40.Kerr I.D. Sequence analysis of twin ATP binding cassette proteins involved in translational control, antibiotic resistance, and ribonuclease L inhibition. Biochem. Biophys. Res. Commun. 2004;315:166–173. doi: 10.1016/j.bbrc.2004.01.044. [DOI] [PubMed] [Google Scholar]
- 41.Tian Y., Han X., Tian D.L. The biological regulation of ABCE1. Iubmb Life. 2012;64:795–800. doi: 10.1002/iub.1071. [DOI] [PubMed] [Google Scholar]
- 42.Paytubi S., Wang X., Lam Y.W., Izquierdo L., Hunter M.J., Jan E., Hundal H.S., Proud C.G. ABC50 promotes translation initiation in mammalian cells. J. Biol. Chem. 2009;284:24061–24073. doi: 10.1074/jbc.M109.031625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kovalchuk A., Driessen A.J. Phylogenetic analysis of fungal ABC transporters. BMC Genom. 2010;11:177–197. doi: 10.1186/1471-2164-11-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verrier P.J., Bird D., Buria B., Dassa E., Forestier C., Geisler M., Klein M., Kolukisaoglu U., Lee Y., Martinoia E. Plant ABC proteins a unified nomenclature and updated inventory. Trends Plant Sci. 2008;13:151–158. doi: 10.1016/j.tplants.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Sullivan D.T., Grillo S.L., Kitos R.J. Subcellular localization of the first three enzymes of the ommochrome synthetic pathway in Drosophila melanogaster. J. Exp. Zool. 1974;188:225–233. doi: 10.1002/jez.1401880210. [DOI] [PubMed] [Google Scholar]
- 46.Mackenzie S.M., Brooker M., Gill T.R., Cox G.B., Howells A.J., Ewart G.D. Mutations in the white gene of Drosophila melanogaster affecting ABC transporters that determine eye colouration. Biochim. Biophys. Acta. 1999;1419:173–185. doi: 10.1016/S0005-2736(99)00064-4. [DOI] [PubMed] [Google Scholar]
- 47.Komoto N., Quan G., Sezutsu H., Tamura T. A single-base deletion in an ABC transporter gene causes white eyes, white eggs, and translucent larval skin in the silkworm w-3(oe) mutant. Insect Biochem. Mol. Biol. 2009;39:152–156. doi: 10.1016/j.ibmb.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 48.Pedra J.H.F., Mcintyre L.M., Scharf M.E., Pittendrigh B.R. Genome-wide transcription profile of field- and laboratory-selected dichlorodiphenyltrichloroethane (DDT)-resistant Drosophila. Proc. Natl. Acad. Sci. USA. 2004;101:7034–7039. doi: 10.1073/pnas.0400580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.You M., Yue Z., He W., Yang X., Yang G., Xie M., Zhan D., Baxter S.W., Vasseur L., Gurr G.M., et al. A heterozygous moth genome provides insights into herbivory and detoxification. Nat. Genet. 2013;45:220–225. doi: 10.1038/ng.2524. [DOI] [PubMed] [Google Scholar]
- 50.Yang N., Xie W., Jones C.M., Bass C., Jiao X., Yang X., Liu B., Li R., Zhang Y. Transcriptome profiling of the whitefly Bemisia tabaci reveals stage-specific gene expression signatures for thiamethoxam resistance. Insect Mol. Biol. 2013;22:485–496. doi: 10.1111/imb.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang N., Xie W., Yang X., Wang S., Wu Q., Li R., Pan H., Liu B., Shi X., Fang Y., et al. Transcriptomic and proteomic responses of sweetpotato whitefly, Bemisia tabaci, to thiamethoxam. PLoS ONE. 2013;8:e61820. doi: 10.1371/journal.pone.0061820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones C.M., Toé H.K., Sanou A., Namountougou M., Hughes A., Diabaté A., Dabiré R., Simard F., Ranson H. Additional selection for insecticide resistance in urban malaria vectors: DDT resistance in Anopheles arabiensis from Bobo-Dioulasso, Burkina Faso. PLoS ONE. 2012;79:e45995. doi: 10.1371/journal.pone.0045995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Annilo T., Chen Z., Shulenin S., Costantino J., Thomas L., Lou H., Stefanov S., Dean M., Rzhetsky A., Allikmets R. Evolution of the vertebrate ABC gene family: Analysis of gene birth and death. Genomics. 2006;88:1–11. doi: 10.1016/j.ygeno.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 54.Silva A.X., Jander G., Samaniego H., Ramsey J.S., Figueroa C.C. Insecticide resistance mechanisms in the green peach aphid Myzus persicae (Hemiptera: Aphididae) I: A transcriptomic survey. PLoS ONE. 2012;7:e36366. doi: 10.1371/journal.pone.0036366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tian L., Song T., He R., Zeng Y., Xie W., Wu Q. Genome-wide analysis of atp-binding cassette (ABC) transporters in the sweetpotato whitefly, Bemisia tabaci. BMC Genom. 2017;18:330–345. doi: 10.1186/s12864-017-3706-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pohl P.C., Klafke G.M., Carvalho D.D., Martins J.R., Daffre S., da Silva Vaz I., Jr., Masuda A. ABC transporter efflux pumps: A defense mechanism against ivermectin in Rhipicephalus (Boophilus) microplus. Int. J. Parasitol. 2011;41:1323–1333. doi: 10.1016/j.ijpara.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 57.Figueira-Mansur J., Ferreira-Pereira A., Mansur J.F., Franco T.A., Alvarenga E.S., Sorgine M.H., Neves B.C., Melo A.C., Leal W.S., Masuda H., et al. Silencing of P-glycoprotein increases mortality in temephos-treated Aedes aegyptilarvae. Insect Mol. Biol. 2013;22:648–658. doi: 10.1111/imb.12052. [DOI] [PubMed] [Google Scholar]
- 58.Luo L., Sun Y.J., Yang L., Huang S., Wu Y.J. Avermectin induces P-glycoprotein expression in S2 cells via the calcium/calmodulin/NF-kB pathway. Chem. Biol. Interact. 2013;203:430–439. doi: 10.1016/j.cbi.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 59.Srinivas R., Shamsundar G.S., Jayalakshmi S.K., Sreermalu K. Effect of insecticides and inhibitors on P-glycoprotein ATPase (M-type) activity of resistant pest Helicoverpa armigera. Curr. Sci. 2005;88:1449–1452. [Google Scholar]
- 60.Simmons J., D’Souza O., Rheault M., Donly C. Multidrug resistance protein gene expression in Trichoplusia ni caterpillars. Insect Mol. Biol. 2013;22:62–71. doi: 10.1111/imb.12003. [DOI] [PubMed] [Google Scholar]
- 61.Fossog T.B., Poupardin R., Costantini C., Awono-Ambene P., Wondji C.S., Ranson H., Antonio-Nkondjio C. Resistance to DDT in an urban setting: Common mechanisms implicated in both M and S forms of Anopheles gambiae in the city of Yaoundé Cameroon. PLoS ONE. 2013;8:e61408. doi: 10.1371/journal.pone.0061408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu G., Zhong D., Cao J., Zhou H., Li J., Liu Y., Bai L., Xu S., Wang M.H., Zhou G., et al. Transcriptome profiling of pyrethroid resistant and susceptible mosquitoes in the malaria vector Anopheles sinensis. BMC Genom. 2014;15:448–461. doi: 10.1186/1471-2164-15-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 64.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 66.WHO . Test Procedures for Insecticide Resistance Monitoring in Malaria Vectors, Bio-Efficacy and Persistence of Insecticides on Treated Surfaces. World Health Organization; Geneva, Switzerland: 1998. [Google Scholar]
- 67.Gao Q., Beebe N.W., Cooper R.D. Molecular identification of the malaria vectors Anopheles anthropophagus and Anopheles sinensis (Diptera: Culicidae) in central China using polymerase chain reaction and appraisal of their position within the Hyrcanus group. J. Med. Entomol. 2004;41:5–11. doi: 10.1603/0022-2585-41.1.5. [DOI] [PubMed] [Google Scholar]
- 68.Trapnell C., Pachter L., Salzberg S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 71.Lalitha S. Primer premier 5. Biotech. Softw. Internet. Rep. 2000;1:270–272. doi: 10.1089/152791600459894. [DOI] [Google Scholar]
- 72.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Anal. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.