Abstract
Long noncoding RNAs (lncRNAs) serve important roles in the biology of autoimmune diseases and immune-associated disorders. To identify lncRNAs specifically associated with psoriasis, the expression of lncRNAs from biopsies obtained from patients with psoriasis were compared with samples obtained from healthy volunteers using a microarray. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was performed to detect the expression of 10 identified dysregulated lncRNAs. Cis- and trans-regulated target genes of lncRNAs were predicted. The results of microarray analysis indicated that 2,194 lncRNAs and 1,725 mRNAs were significantly dysregulated. Gene Ontology and pathway analyses among the dysregulated genes were performed. Co-expression network analysis was also performed to study molecular interactions. Several identified pathways were associated with psoriasis. Among the 2,194 dysregulated lncRNAs, 1,549 of these had cis- or trans-regulated predicted target genes. Among the 1,725 dysregulated mRNAs, 289 of the cis-regulated target genes and 262 of the trans-regulated target genes may be regulated by the differentially expressed lncRNAs; 10 differentially expressed lncRNAs were randomly selected and then validated. Of these lncRNAs, 7 exhibited the same expression profile as determined via microarray analysis, of which 3 lncRNAs were upregulated and 4 lncRNAs were downregulated. To the best of our knowledge, the present study is the first in which a microarray has been used to investigate the expression profile of lncRNAs associated with psoriasis. Additionally, the expression levels of the 10 aforementioned lncRNAs associated with psoriasis were validated in the present study for the first time using RT-qPCR. The findings demonstrated that lncRNAs may contribute to the pathogenesis of psoriasis and suggested their potential diagnostic and therapeutic value. Furthermore, the findings of the present study suggest that the combined actions of several lncRNAs may contribute to the pathogenesis of psoriasis.
Keywords: long noncoding RNA microarray, psoriasis, expression, function analysis
Introduction
Psoriasis is a lifelong disease that severely reduces the quality of life of patients (1,2). Recently, psoriasis has been suggested to result from the abnormal communication between keratinocytes and immunocytes (3–5). It is also widely accepted that psoriasis has a strong genetic background and numerous psoriasis susceptibility long noncoding (lnc) RNAs has been identified (3,6). Several susceptible genes of psoriasis as detected via genome-wide association analysis, RNA-sequencing (RNA-seq) or microarray comprise noncoding DNA and RNA (7–9); however, little is known regarding the association of specific lncRNAs in the prevalence of this disease.
LncRNAs are >200 nucleotides and can be expressed as alternatively spliced variants; however, they cannot encode proteins (10). LncRNAs include antisense, intronic, intergenic, pseudogenes and retrotransposon transcripts (11). In addition, lncRNAs serve important functional roles in epigenetic, transcriptional or post-transcriptional regulation by acting on cis and trans-regulated target genes (12–14), and form networks of ribonucleoprotein complexes (12). A total of ~1% of the mammalian genome is expressed as mRNA; however, 70–90% is transcribed into lncRNA during development (11,15,16). LncRNAs serve important roles in regulating immune-mediated inflammatory disorders and autoimmunity (17–19). Dysregulation of lncRNAs has been studied in numerous immune diseases, including systemic lupus erythematosus (20), rheumatoid arthritis (21), Crohn's disease, ulcerative colitis (22), multiple sclerosis (23) and psoriasis (7,24,25). This suggests that lncRNAs have crucial roles in the regulation of immune-mediated disease and contribute to the pathogenesis of psoriasis by regulating the expression of protein-coding genes, or by altering chromatin structure (15,26).
LncRNAs are emerging as key molecules in the genesis and progression of psoriasis. Several studies have been conducted on the expression and function of lncRNAs in psoriasis; however, these studies have used RNA sequencing (7,12,27). The understanding of the exact molecular mechanism is limited; no studies have performed an lncRNA microarray to investigate their expression and potential functions.
In the present study, an lncRNA microarray was performed to determine the expression profiles between psoriasis tissue and paired control tissue. In addition, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was conducted to detect 10 dysregulated lncRNAs based on the microarray results. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were conducted to study the potential function of the differentially expressed genes. A co-expression network analysis was also performed to study molecular interactions; cis- and trans-regulated targets of lncRNAs were predicted. The results of the present study revealed that dysregulation of lncRNAs and mRNAs may contribute to the pathogenesis of psoriasis. Furthermore, lncRNAs may be used as potential biomarkers and therapeutic targets for the treatment of psoriasis.
Materials and methods
Tissues
A total of 15 psoriasis specimens were collected from patients with psoriasis (7 males and 8 females; aged 23–39-years-old) from Qilu Hospital of Shandong University (September 2016 to August 2017). Patients did not receive systemic drugs, phototherapy or externally applied drugs during the last 3 months prior to sample collection. All tissues were obtained from the trunks; half of each tissue sample was frozen immediately at −80°C. The remaining tissue was embedded in paraffin for pathological examination. In addition, 15 normal tissues were collected from healthy volunteers (8 males and 7 females; aged 25–40-years-old) and tissue specimens were stored at −80°C; three samples of the psoriasis and normal tissues were used for microarray analysis, and the remaining 12 pairs of tissue were used for RT-qPCR. The present study was approved by the Ethics Committee of Shandong University, Qilu Hospital (Jinan, China). All patients enrolled in the present study provided written informed consent.
LncRNA and mRNA microarray
The microarray [SBC Human (4×180K) competing endogenous (ce)RNA microarray; Shanghai Biotechnology Corporation, Shanghai, China] used in the present study could detect 68,432 lncRNAs and 18,853 mRNAs sourced from the most authoritative databases, including University of California Santa Cruz (https://genome.ucsc.edu/), GENCODE v21/Ensembl (https://www.gencodegenes.org/human/release_21.html), LNCipedia v3.1 (28), Noncode v4 (29) and Lncrnadb v2.0 (30,31). The lncRNA and mRNA probes were designed according to the latest genome version [human/GRCh38 (hg38)].
RNA extraction
Total RNAs from skin samples were extracted using an RNeasy mini kit (cat. no. 74106; Qiagen GmBH, Hilden, Germany) according to the manufacturer's protocols. Following this, RNA integration was investigated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA).
RNA labelling
Total RNAs from skin samples were amplified and labeled using a Low Input Quick Amp WT Labeling kit (cat. no. 5190-2943; Agilent Technologies, Inc.) according to the manufacturer's protocols. Furthermore, a RNeasy mini kit (cat. no. 74106; Qiagen GmBH) was used to purify the labelled cRNAs according to the manufacturer's protocols.
Cy3-labelled hybridization
The RNA of each skin sample in the present study was hybridized with 1.65 µg Cy3-labelled cRNA using a commercialized Gene Expression Hybridization kit (cat. no. 5188-5242; Agilent Technologies, Inc.) according to the manufacturer's protocols. Following this, the slides were washed with a Gene Expression Wash Buffer kit (cat. no. 5188-5327; Agilent Technologies, Inc.) 17 h later.
Data analysis
Each slide was scanned using an Agilent Microarray Scanner (Dye channel, Green; Scan resolution=3 µm; photoelectric multi-plication tube 100%, 20 bit; Agilent Technologies, Inc). The experimental data were collected using Feature Extraction software 10.7 (Agilent Technologies, Inc.), which were then normalized using the limma packages in R (version 3.8) (32).
Function analysis of dysregulated genes
GO analysis was performed to detect the functions of the dysregulated genes. The KEGG database (https://www.genome.jp/kegg/) was used to investigate the functions of these dysregulated genes in the pathways.
Target prediction of dysregulated lncRNAs
As previously described (33), two methods were used to identify target mRNAs of dysregulated lncRNAs. Firstly, cis-acting target mRNAs were searched. Cis-target genes were considered as transcribed within a 10 kbp window upstream or downstream of lncRNAs. Furthermore, target genes were identified according to the sequence of mRNA and RNA duplex energy prediction. BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used in the present study for screening and parameters were set (e-value, 1e-20; identity, ≥95%). RNAplex software (https://omictools.com/rnaplex-tool) and starBase v3.0 (http://starbase.sysu.edu.cn/) were also used in the present study to screen trans-regulated predicted target mRNAs and microRNAs, respectively (e≤-30) (34).
Weighted gene co-expression network analysis
Weighted gene co-expression network analysis (WGCNA) was performed as described in previous studies (35,36).
RT-qPCR
Total RNA extracted from skin samples was reverse transcribed using a ReverTra Ace qPCR kit (cat. no. FSQ-101; Toyobo Life Science, Osaka, Japan) according to the manufacturer's protocols. The expression levels of 10 dysregulated lncRNAs in 12 psoriasis and normal control tissue samples were detected by RT-qPCR using Power SYBR-Green PCR Master Mix (cat. no. 4368708) on the 7900 HT Sequence Detection System (both Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocols. The thermocycling conditions were as follows: Stage 1, 50°C for 2 min; stage 2, 95°C for 10 min; stage 3 (40 cycle) 95°C for 15 sec, 60°C for 1 min, and stage 4, 95°C for 15 sec, 60°C for 15 sec and 95°C for 15 sec. GAPDH was used as an internal control. The primers employed in the present study are listed in Table I. All the experiments were repeated three times (37).
Table I.
Primers used for reverse transcription-quantitative polymerase chain reaction.
| LncRNA, mRNA or gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| ENST00000547006 | CATTGTTGCTTTTCGGGTCT | GGCCACGTAGCAATGACTGT |
| lnc-HSFY2-10:1 | ACCCACGCAGATTTATTCCA | CCTTGGTGGTGGCATTAGTT |
| lnc-PERP-2:7 | CCATTTTTCTGTGGTCAGGTC | GCTGTTGAAGAGCACTGTGAG |
| ENST00000557691 | CATGCTGAGTCTGCAAGGAC | CAAAAACCCCGATGATAGGA |
| lnc-THRSP-6:1 | GTTCCATTGATCCAGCCACT | ACAAGAGGCCACTGACTGCT |
| NONHSAT066260 | CACTGCACTTGGCTGTGATT | GGCCGAGAAGCCTAGAAGAA |
| lnc-MGMT-2:1 | GGAAAAACACAGCAGCCAGT | ACGCCAACACCCTGTAGACT |
| lnc-AP000769.1-1:2 | CACCAGCTTCTCCAATCTTCCT | GGAGTAAGCAAGTCACAATGTTGAG |
| lnc-POLR3E-3:3 | TACTGAGAATGGGGAAGGAAAC | CTCTTGCTTACTGCCTGCTGA |
| lnc-PXDNL-4:1 | CAGGTCAGGCTGTTGGATTCT | AGGATGTGCTTCTTGGGAGTC |
| GAPDH | TGACTTCAACAGCGACACCCA | CACCCTGTTGCTGTAGCCAAA |
LncRNA, long noncoding RNA.
Statistical analyses
Data in the present study were presented as the mean ± standard deviation from three independent experiments. Differences between groups were analyzed using Student's t-test and Benjamini-Hochberg correction with SPSS 17.0 analysis software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Dysregulated lncRNAs
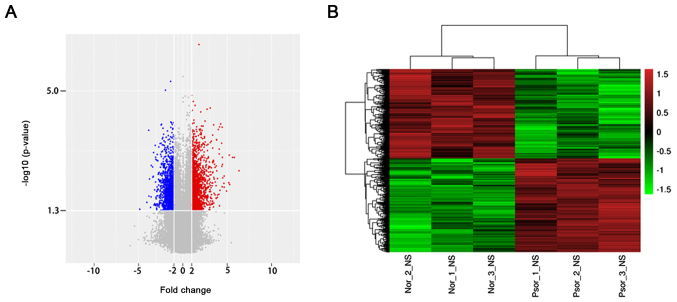
To investigate the dysregulated lncRNAs in psoriasis, an lncRNA microarray analysis was performed using psoriasis lesions and normal tissue. According to the lncRNA microarray results, 2,194 lncRNAs were dysregulated (≥2.0 fold, P<0.05). Among these lncRNAs, 1,123 lncRNAs were upregulated and 1,071 lncRNAs were downregulated (Fig. 1A). Hierarchical clustering analysis was used to arrange specimens in the present study according to the level of expression (Fig. 1B). The top 10 upregulated and downregulated lncRNAs of the microarray results were presented in Table II. Among these, lnc-SLC6A14-1:1 (79.87306773-fold) was the most overexpressed lncRNA and NONHSAT044111 (29.0815707-fold) was the most downregulated lncRNA. The results of microarray analysis revealed that the number of upregulated lncRNAs was greater than that of downregulated lncRNAs.
Figure 1.
Expression profile of lncRNA in psoriasis. (A) Volcano plot of differentially expressed lncRNAs between psoriasis and normal control tissue. (B) Hierarchical clustering analysis of differentially expressed lncRNAs. Red and green colors indicated high and low expression, respectively. LncRNAs, long noncoding RNAs; GO, Gene Ontology.
Table II.
Top 10 differentially expressed lncRNAs in psoriasis tissues compared with normal control.
| A, Upregulated lncRNAs | |||
|---|---|---|---|
| LncRNA | P-value | Fold change | Source |
| lnc-SLC6A14-1:1 | 0.00292 | 79.87 | LNCipedia |
| NR_003062 | 0.00114 | 55.64 | RefSeq |
| lnc-SERPINB3-4:1 | 0.00113 | 48.26 | LNCipedia |
| NONHSAT006518 | 0.00775 | 38.64 | NONCODE |
| lnc-IGFL3-6:1 | 0.00096 | 37.3 | LNCipedia |
| ENST00000472053 | 0.00687 | 36.05 | ENSEMBL |
| NONHSAT006509 | 0.01240 | 30.2 | NONCODE |
| NR_030617 | 0.00942 | 25.21 | RefSeq |
| lnc-RAPGEF2-3:1 | 0.00427 | 25.13 | LNCipedia |
| lnc-RPP40-3:3 | 0.00719 | 24.61 | LNCipedia |
| B, Downregulated lncRNAs | |||
| LncRNA | P-value | Fold change | Source |
| NONHSAT044111 | 0.0344 | 29.08 | NONCODE |
| lnc-PPM1N-1:1 | 0.0600 | 16.66 | LNCipedia |
| lnc-GRHL2-11:1 | 0.0692 | 14.45 | LNCipedia |
| lnc-JAKMIP2-1:1 | 0.0704 | 14.21 | LNCipedia |
| lnc-GGTLC1-2:1 | 0.0734 | 13.62 | LNCipedia |
| NONHSAT025181 | 0.0799 | 12.52 | NONCODE |
| ENST00000447257 | 0.0832 | 12.01 | ENSEMBL |
| ENST00000623414 | 0.0843 | 11.863 | ENSEMBL |
| ENST00000415656 | 0.0943 | 10.61 | ENSEMBL |
| lnc-LINC00273-22:1 | 0.0959 | 10.43 | LNCipedia |
LncRNA, long noncoding RNA.
Dysregulated mRNAs
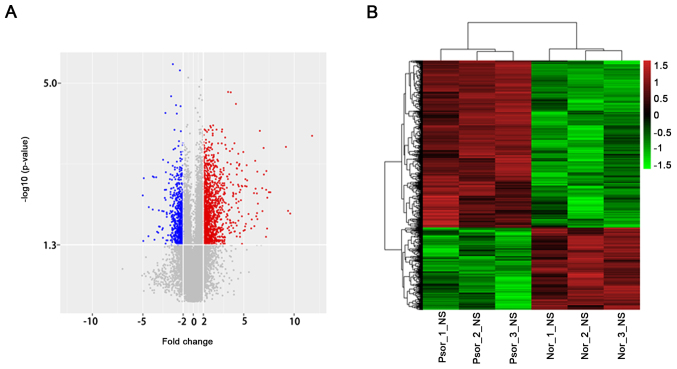
Based on the microarray results, 1,725 dysregulated mRNAs were identified. Among these, 1,157 mRNAs were overexpressed and 568 mRNAs were downregulated (Fig. 2). The top 10 upregulated and downregulated mRNAs were indicated (Table III). DEFB4A (3,505.251346 fold) was the most overexpressed lncRNA and WIF1 (32.27749933 fold) was the most downregulated lncRNA. The microarray results suggested that that there was a greater number of upregulated mRNAs.
Figure 2.
Expression profile of mRNA in psoriasis. (A) Volcano plot of differentially expressed mRNAs between psoriasis and normal control tissue. (B) Hierarchical clustering analysis of differentially expressed mRNAs. Red and green colors indicated high and low expression, respectively.
Table III.
Top 10 differentially expressed mRNAs in psoriasis tissues compared with normal control.
| A, Upregulated mRNAs | |||
|---|---|---|---|
| mRNA | P-value | Fold change | Source |
| DEFB4A | 0.00016 | 3505.25 | RefSeq |
| SERPINB4 | 0.00967 | 780.42 | RefSeq |
| S100A7A | 0.00840 | 672.76 | RefSeq |
| S100A12 | 0.00029 | 582.68 | RefSeq |
| PI3 | 0.00315 | 199.59 | RefSeq |
| SERPINB3 | 0.00314 | 180.61 | RefSeq |
| SPRR2G | 0.00420 | 162.24 | RefSeq |
| S100A9 | 0.02130 | 159.63 | RefSeq |
| LCE3A | 0.00102 | 152.00 | RefSeq |
| IL36A | 0.00276 | 149.66 | RefSeq |
| B, Downregulated mRNAs | |||
| mRNA | P-value | Fold change | Source |
| WIF1 | 0.0310 | 32.28 | RefSeq |
| MT4 | 0.0322 | 31.05 | RefSeq |
| BTC | 0.0333 | 30.05 | RefSeq |
| CACNA1H | 0.0457 | 21.87 | RefSeq |
| CYP2W1 | 0.0470 | 21.29 | RefSeq |
| KRT77 | 0.0615 | 16.27 | RefSeq |
| FAM166B | 0.0617 | 16.20 | RefSeq |
| NEUROD2 | 0.0671 | 14.90 | RefSeq |
| SPINK1 | 0.0693 | 32.28 | RefSeq |
| ZBTB16 | 0.0780 | 31.05 | RefSeq |
Function analysis of dysregulated mRNAs
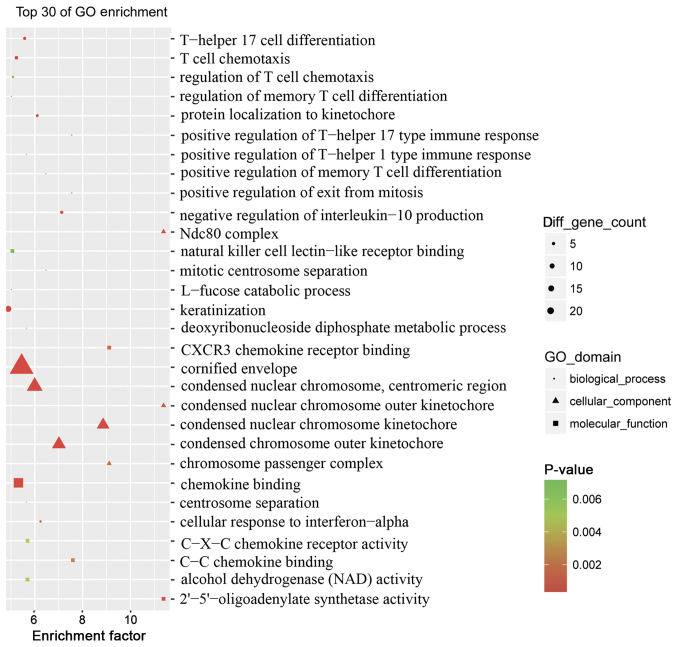
To assess the pathogenesis of psoriasis, the top 30 GO terms between the psoriasis and control groups were obtained via enrichment analysis (Fig. 3). The association of dysregulated mRNAs with ‘molecular function’, ‘biological processes’ and ‘cellular component’ in the GO database was assessed. Numerous dysregulated epigenetic and genetic genes associated with ‘T-helper 17 cell differentiation’, ‘T-cell chemotaxis’, ‘regulation of T-cell chemotaxis’, ‘keratinization’, ‘positive regulation of T-helper 17’ and ‘T-helper 1 type immune response’ were identified. This suggested that a variety of specific genes were involved in the pathogenesis of psoriasis, particularly genes associated with pro-inflammatory cells and molecules.
Figure 3.
Analysis of top 30 Gene Ontology terms between the psoriasis and control groups. The rich factor was calculated using the number of enriched genes divided by the number of all background genes in the corresponding pathway. P-values were calculated using Benjamini-Hochberg correction. P<0.05 was considered to indicate statistically significant over-representation.
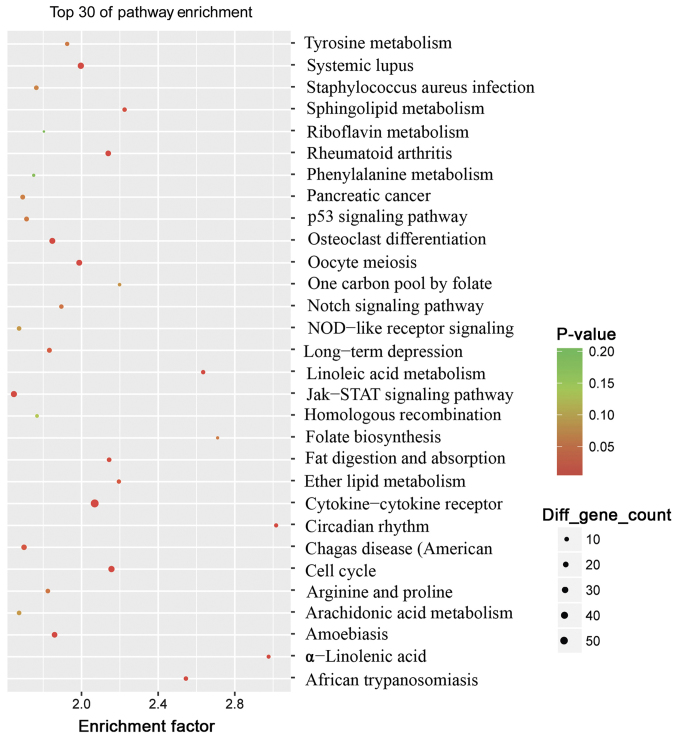
To further investigate the pathogenesis of psoriasis, analysis of the top 30 KEGG pathways between the psoriasis and control groups was performed (Fig. 4). The results revealed that 23 pathways were identified to be significantly enriched (P<0.05). The majority of these pathways were associated with inflammation and lipid metabolism, including the ‘Janus kinase (Jak)-signal transducers and activators of transcription (STAT)’ signaling pathway (associated with 26 genes), the ‘cytokine-cytokine receptor’ interaction signaling pathway (associated with 56 genes), the ‘cell cycle’ signaling pathway (associated with 28 genes), the ‘Toll-like receptor’ signaling pathway (associated with 17 genes), the ‘Wnt’ signaling pathway (associated with 24 genes) and the ‘chemokine’ signaling pathway (associated with 30 genes) (data not shown). These findings also reinforced the fact that psoriasis is a chronic, immune-mediated disorder.
Figure 4.
Analysis of top 30 Kyoto Encyclopedia of Genes and Genomes pathways between the psoriasis and control groups. The rich factor was calculated using the number of enriched genes divided by the number of all background genes in the corresponding pathway. P-values were calculated using Benjamini-Hochberg correction. P<0.05 was considered to indicate statistically significantly over-representation.
Potential targets and function of the dysregulated lncRNAs
To investigate whether the differentially expressed lncRNAs regulate genes and determine the signaling pathways associated with psoriasis, target prediction programs were used to predict the potential targets of the dysregulated lncRNAs (data not shown). It was revealed that 1,549 dysregulated lncRNAs had predicted target genes in the present study. Among these, 1,447 dysregulated lncRNAs had cis-regulated target genes, 397 dysregulated lncRNAs had trans-regulated target genes and 295 dysregulated lncRNAs had trans-and cis-regulated target genes.
Notably, 3,740 mRNAs may be regulated by the dysregulated lncRNAs. Among these, 1,460 mRNAs may be cis-regulated, 2,588 mRNAs may be trans-regulated, and 308 mRNAs may be cis-and trans-regulated. Upon further analysis and integration with the profile of differentially expressed lncRNAs, a total of 489 mRNAs were dysregulated. Furthermore, the results suggested that these mRNAs may be regulated by dysregulated cis- or trans-acting lncRNAs. Among these, 289 dysregulated mRNAs could be regulated by dysregulated cis-acting lncRNAs and 262 dysregulated mRNAs could be regulated by dysregulated trans-acting lncRNAs.
These results indicated that the dysregulated lncRNAs were involved in the onset of psoriasis by regulating their potential target mRNAs and its associated signaling pathways.
Co-expression network of lncRNAs and mRNAs
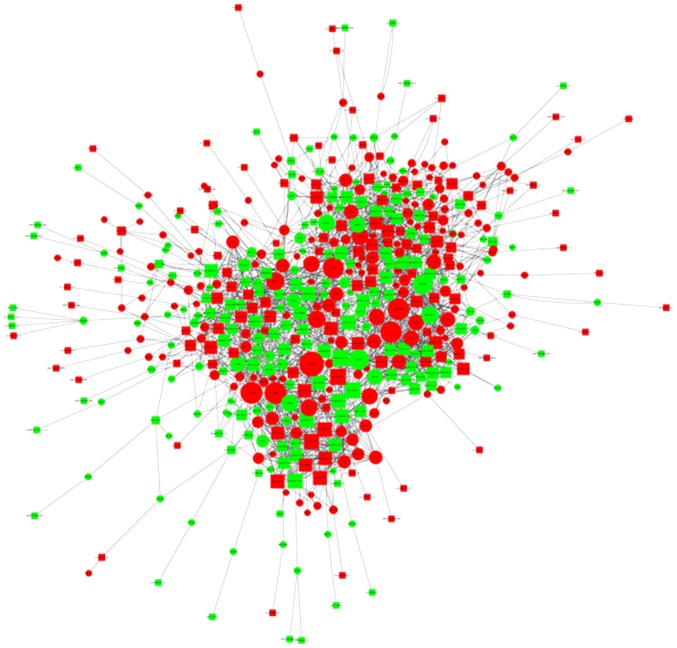
Aided by the microarray data and a weighted gene co-expression network analysis approach, numerous networks of coordinately expressed genes were identified in psoriasis tissues compared with the normal control tissues (Fig. 5). The co-expression results in the present study strongly support a network model in which lncRNAs and mRNAs function together in psoriasis. Furthermore, the findings suggested that lncRNAs do not act alone but rather work in concert with other lncRNAs and mRNAs.
Figure 5.
Analysis of lncRNA-mRNA co-expression in psoriasis. Red represented upregulated genes; green represented downregulated genes; circles represented target mRNA; squares represented lncRNAs; the solid line represented trans regulation; and the dotted line represented cis regulation. The sizes of the circles/squares indicate the number of associated genes correlated. LncRNAs, long noncoding RNAs.
RT-qPCR analysis
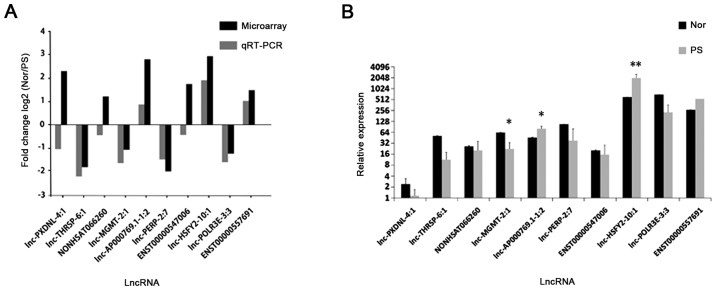
In order to detect the lncRNA and mRNA microarray results, 10 dysregulated lncRNAs were randomly selected for RT-qPCR analysis. Based on the results of the microarray data, 6 upregulated lncRNAs (lnc-AP000769.1–1:2, NONHSAT066260, lnc-PXDNL-4:1, ENST00000557691, lnc-HSFY2-10:1 and ENST00000547006) and 4 downregulated lncRNAs (lnc-MGMT-2:1, lnc-POLR3E-3:3, lnc-THRSP-6:1 and lnc-PERP-2:7) were selected.
The expression levels of dysregulated lncRNAs were validated by RT-qPCR (n=12). The expression levels of 7 lncRNAs were consistent with the microarray results, which exhibited the same dysregulation profiles (up-/down-regulated, ≥2.0 fold) (Fig. 6A). Of note, 3 lncRNAs (lnc-AP000769.1-1:2, ENST00000557691 and lnc-HSFY2-10:1) were upregulated and 4 lncRNAs (lnc-MGMT-2:1, lnc-POLR3E-3:3, lnc-THRSP-6:1 and lnc-PERP-2:7) were downregulated (≥2.0 fold) (Fig. 6A). The expression levels of lnc-MGMT-2:1 (P<0.05), lnc-AP000769.1-1:2 (P<0.05) and lnc-HSFY2-10:1 (P<0.01) were significantly different between the psoriasis and normal tissues (Fig. 6B); however, lnc-PXDNL-4:1 was observed to be downregulated, and the expression levels of NONHSAT066260 and ENST00000547006 in psoriasis and normal tissues exhibited 0.5<fold change<2 (Fig. 6B).
Figure 6.
Results of RT-qPCR. (A) Comparison of fold change [log2 (Nor/Ps)] of lncRNAs between microarray and RT-qPCR results. (B) Relative expression levels of lncRNAs in 12 pairs of Nor and Ps tissue. The expression levels of lncRNA were calculated using the 2−ΔΔCq method, and the results are presented as relative fold changes. Data are expressed as the mean ± standard deviation. Comparisons between two groups were performed using a Student's t test. *P<0.05 and **P<0.01 vs. Nor. Nor, normal control; Ps, psoriasis; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; lncRNAs, long noncoding RNAs.
Discussion
It is well known that psoriasis has a strong genetic predisposition (38–40); however, the underlying genetic mechanisms remain unknown. Previous transcriptomic studies in psoriasis have analyzed the expression levels of mRNA to investigate the functions of their translated proteins, including protein-coding genes associated with the interleukin-17 signaling pathway (41), the nuclear factor (NF)-κB signaling pathway (42,43), the Jak-STAT signaling pathway (44) and the mitogen-activated protein kinase (MAPK) signaling pathway (45,46).
LncRNAs were once considered as transcriptional noise or junk but have recently begun to attract attention in research (15,47). LncRNAs serve important roles in regulating various functions of the immune system (18,48,49). Increasing evidence suggests lncRNA dysregulation may have a vital role in the pathogenesis of psoriasis (8,25,50). At present, few lncRNAs associated with psoriasis have been investigated. Psoriasis associated non-protein coding RNA induced by stress (PRINS), an lncRNA gene, exhibited the highest expression levels in non-psoriatic skin lesions, indicating that it may serve an important role in the susceptibility of psoriasis (51). Széll et al (25) reported that overexpression of PRINS in non-lesional psoriatic skin alters the stress response and contributes to disease pathogenesis. Furthermore, Szegedi et al (52) reported that the dysregulation of PRINS contributed to the onset of psoriasis, resulting in decreased keratinocyte apoptosis. Another lncRNA gene associated with psoriasis, psoriasis susceptibility 1 candidate 3, has been considered to be strongly associated with psoriasis (53). Using RNA-seq, Kretz et al (54) reported terminal differentiation-induced lncRNA, which controls human epidermal differentiation via posttranscriptional regulation. Therefore, similar to mRNAs, lncRNAs may function as proinflammatory or inducers of proliferation in psoriasis by regulating transcriptional regulation or altering the expression levels of their target mRNAs.
To the best of our knowledge, the present study is the first to determine lncRNA and mRNA expression profiles in psoriasis compared with normal healthy volunteers using a microarray. GO and KEGG pathway analyses were conducted to determine the potential functions and co-expression networks of lncRNAs to study the molecular interactions.
To validate the microarray results and predict the possible role of lncRNAs in psoriasis, 10 dysregulated lncRNAs were randomly selected for RT-qPCR analysis. The results of RT-qPCR were consistent with the data of microarray results: 3 lncRNAs (lnc-AP000769.1-1:2, ENST00000557691 and lnc-HSFY2-10:1) were upregulated and 4 lncRNAs (lnc-MGMT-2:1, lnc-POLR3E-3:3, lnc-THRSP-6:1 and lnc-PERP-2:7) were downregulated. By predicting cis- and trans-acting lncRNAs, it was revealed that lnc-AP000769.1-1:2 (upregulated) and lnc-MGMT-2:1 (downregulated) have a common predicted target gene, jak3, which suggested they may be involved in the pathogenesis of psoriasis by regulating jak3. ENST00000557691 (upregulated) and lnc-PERP-2:7 (downregulated) may regulate the MAPK signaling pathway by regulating their predicted target gene, mapk kinase kinase 9. Lnc-HSFY2-10:1 (upregulated) was predicted to regulate microRNA-145 (starBase v3.0). Of note, it has been reported that miR-145 negatively regulates cell proliferation and proinflammatory cytokine release, and inhibits the downstream genes of NF-κB and mechanistic target of rapamycin (mTOR) (55,56). Thus, lnc-HSFY2-10:1 may be involved in the pathogenesis of psoriasis via ceRNA regulation. Lnc-POLR3E-3:3 (downregulated), which possibly regulates interferon regulatory factor 1 (irf1) in trans, may be involved in regulating T regulatory (Treg) cell differentiation, as irf1 has been considered as a regulator of Treg cells by inhibiting Forkhead box P3 in mice (57). Lnc-THRSP-6:1, which has been predicted to potentially regulate C-C motif chemokine ligand 5 (ccl5) (data not shown) via RNAplex software, was downregulated in the present study. This suggested that the predicted target gene proinflammation chemokine, ccl5, may be overexpressed and that lnc-THRSP-6:1 contributes to the pathogenesis of psoriasis. However, the expression levels of lnc-PXDNL-4:1, as determined by RT-qPCR, were not in agreement with the results of microarray analysis in the present study. The lncRNA and mRNA probes were designed according to the latest genome version [human/GRCh38 (hg38)] and the primers were carefully selected; thus, this discrepancy may be due to the small sample size (n=12). This suggests that further investigation using a larger sample is required. In addition, the expression levels of NONHSAT066260 and ENST00000547006 were not significantly different, which indicated that they may not be involved in the onset of psoriasis.
The results microarray and RT-qPCR analyses suggested that lncRNAs were involved in the pathogenesis of psoriasis, possibly by regulating the differentiation and function of Treg cells, the NF-κΒ signaling pathway, the mTOR signaling pathway and the MAPK signaling pathway, the release of cytokines and chemokines, and the JAK-STAT signaling pathway.
GO and KEGG analyses were performed to investigate the biological functions that were enriched among the dysregulated mRNAs. GO analysis identified ‘T-helper 17 cell differentiation’, ‘T-cell chemotaxis’ and the ‘T-helper 17 type immune response’ to be involved in the development of psoriasis. In addition, pathway analysis identified several pathways associated with psoriasis, including the ‘JAK-STAT’ signaling pathway, the ‘cytokine-cytokine’ receptor interaction signaling pathway, the ‘cell cycle’ signaling pathway, the ‘Wnt’ signaling pathway and the ‘MAPK’ signaling pathway. These results further supported the data obtained from the microarray analysis.
Co-expression analyses emphasized the importance of integrating the expression profiles of lncRNA and mRNA to obtain more generalized information. Genes with strong associations are more likely to be co-regulated with their co-expressed genes (58). The results of the co-expression analyses indicated that the dysregulated lncRNA may collaboratively function the pathogenesis of psoriasis.
In addition, it is well reported that the detection of lncRNAs on the array platform is more stable than RNA-seq (59,60). Therefore, the study of lncRNA and mRNA microarray expression in psoriasis is valuable and may improve the understanding of the pathogenesis of psoriasis.
In conclusion, to the best of our knowledge, the present study is the first to conduct a microarray to investigate lncRNA expression in psoriasis. The findings also revealed that numerous lncRNAs were dysregulated in psoriasis; several networks of mRNA and lncRNA were associated with the pathogenesis of psoriasis, including pathways that have not been previously identified. RT-qPCR, target prediction and co-expression analyses revealed that the lncRNAs did not act alone, but rather functioned in concert with other lncRNAs, which may facilitate the development of psoriasis. Despite the limitations of the present study, including the small sample size, and that no functional or mechanical investigations using the biopsies were conducted, the results may provide insight for further into the development and treatment of psoriasis.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81573046), and the Science and Technology Research Project of Shandong (grant no. 2014GSF118125).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JY and JS made substantial contributions to the design and experiments of the present study and should be regarded as co-first authors. MQ, RL and XZ made substantial contributions in analysing and interpreting patient data. JJ and QS made substantial contributions to the clinical diagnosis and histological examination during the experiment. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Shandong University, Qilu Hospital. All patients enrolled in the present study provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Yan S, Xu Z, Lou F, Zhang L, Ke F, Bai J, Liu Z, Liu J, Wang H, Zhu H, et al. NF-κB-induced microRNA-31 promotes epidermal hyperplasia by repressing protein phosphatase 6 in psoriasis. Nat Commun. 2015;6:7652. doi: 10.1038/ncomms8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nestle FO, Kaplan DH, Barker J. Psoriasis. N Engl J Med. 2009;361:496–509. doi: 10.1056/NEJMra0804595. [DOI] [PubMed] [Google Scholar]
- 3.Srivastava A, Nikamo P, Lohcharoenkal W, Li D, Meisgen F, Xu Landén N, Ståhle M, Pivarcsi A, Sonkoly E. MicroRNA-146a suppresses IL-17-mediated skin inflammation and is genetically associated with psoriasis. J Allergy Clin Immun. 2017;139:550–561. doi: 10.1016/j.jaci.2016.07.025. [DOI] [PubMed] [Google Scholar]
- 4.Lowes MA, Suárez-Fariñas M, Krueger JG. Immunology of psoriasis. Annu Rev Immunol. 2014;32:227–255. doi: 10.1146/annurev-immunol-032713-120225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nestle FO, Di Meglio P, Qin JZ, Nickoloff BJ. Skin immune sentinels in health and disease. Nat Rev Immunol. 2009;9:679–691. doi: 10.1038/nri2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harden JL, Krueger JG, Bowcock AM. The immunogenetics of Psoriasis: A comprehensive review. J Autoimmun. 2015;64:66–73. doi: 10.1016/j.jaut.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta R, Ahn R, Lai K, Mullins E, Debbaneh M, Dimon M, Arron S, Liao W. Landscape of long noncoding RNAs in psoriatic and healthy skin. J Invest Dermatol. 2016;136:603–609. doi: 10.1016/j.jid.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsoi LC, Iyer MK, Stuart PE, Swindell WR, Gudjonsson JE, Tejasvi T, Sarkar MK, Li B, Ding J, Voorhees JJ, et al. Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol. 2015;16:24. doi: 10.1186/s13059-014-0570-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan JJ, Qiao M, Li RH, Zhao XT, Wang XY, Sun Q. Downregulation of miR-145-5p contributes to hyperproliferation of keratinocytes and skin inflammation in psoriasis. Br J Dermatol. 2019;180:365–372. doi: 10.1111/bjd.17256. [DOI] [PubMed] [Google Scholar]
- 10.Hrdlickova B, Kumar V, Kanduri K, Zhernakova DV, Tripathi S, Karjalainen J, Lund RJ, Li Y, Ullah U, Modderman R, et al. Expression profiles of long non-coding RNAs located in autoimmune disease-associated regions reveal immune cell-type specificity. Genome Med. 2014;6:88. doi: 10.1186/s13073-014-0088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 12.Tsoi LC, Iyer MK, Stuart PE, Swindell WR, Gudjonsson JE, Tejasvi T, Sarkar MK, Li BS, Ding J, Voorhees JJ, et al. Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol. 2015;16:24. doi: 10.1186/s13059-014-0570-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pacifici M, Kadri F, Jeansonne D, Peruzzi F. Tat-mediated changes of malat1 long non-coding Rna affects the structure and function of Sc35 nuclear speckles domains in neurons. J Neuroimmune Pharmacol. 2013;8:427–428. [Google Scholar]
- 14.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Lan X, Zhang H, Wang Z, Dong W, Sun W, Shao L, Zhang T, Zhang D. Genome-wide analysis of long noncoding RNA expression profile in papillary thyroid carcinoma. Gene. 2015;569:109–117. doi: 10.1016/j.gene.2015.05.046. [DOI] [PubMed] [Google Scholar]
- 16.Eddy SR. Non-coding RNA genes and the modern RNA world. Nat Rev Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- 17.Wu GC, Pan HF, Leng RX, Wang DG, Li XP, Li XM, Ye DQ. Emerging role of long noncoding RNAs in autoimmune diseases. Autoimmun Rev. 2015;14:798–805. doi: 10.1016/j.autrev.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014;35:408–419. doi: 10.1016/j.it.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sigdel KR, Cheng A, Wang Y, Duan L, Zhang Y. The emerging functions of long noncoding RNA in immune cells: Autoimmune diseases. J Immunol Res. 2015;2015:848790. doi: 10.1155/2015/848790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y, Zhang F, Ma J, Zhang X, Wu L, Qu B, Xia S, Chen S, Tang Y, Shen N. Association of large intergenic noncoding RNA expression with disease activity and organ damage in systemic lupus erythematosus. Arthritis Res Ther. 2015;17:131. doi: 10.1186/s13075-015-0632-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Messemaker TC, Frank-Bertoncelj M, Marques RB, Adriaans A, Bakker AM, Daha N, Gay S, Huizinga TW, Toes RE, Mikkers HM, Kurreeman F. A novel long non-coding RNA in the rheumatoid arthritis risk locus TRAF1-C5 influences C5 mRNA levels. Genes Immun. 2016;17:85–92. doi: 10.1038/gene.2015.54. [DOI] [PubMed] [Google Scholar]
- 22.Mirza AH, Berthelsen CH, Seemann SE, Pan X, Frederiksen KS, Vilien M, Gorodkin J, Pociot F. Transcriptomic landscape of lncRNAs in inflammatory bowel disease. Genome Med. 2015;7:39. doi: 10.1186/s13073-015-0162-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santoro M, Nociti V, Lucchini M, De Fino C, Losavio FA, Mirabella M. Expression profile of long non-coding RNAs in serum of patients with multiple sclerosis. J Mol Neurosci. 2016;59:18–23. doi: 10.1007/s12031-016-0741-8. [DOI] [PubMed] [Google Scholar]
- 24.Ahn R, Gupta R, Lai K, Chopra N, Arron ST, Liao W. Network analysis of psoriasis reveals biological pathways and roles for coding and long non-coding RNAs. BMC Genomics. 2016;17:841. doi: 10.1186/s12864-016-3188-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Széll M, Danis J, Bata-Csörgő Z, Kemény L. PRINS, a primate-specific long non-coding RNA, plays a role in the keratinocyte stress response and psoriasis pathogenesis. Pflugers Arch. 2016;468:935–943. doi: 10.1007/s00424-016-1803-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, Costinean S, Negrini M, Miotto E, Croce CM, Patel T. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30:4750–4756. doi: 10.1038/onc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li B, Tsoi LC, Swindell WR, Gudjonsson JE, Tejasvi T, Johnston A, Ding J, Stuart PE, Xing X, Kochkodan JJ, et al. Transcriptome analysis of psoriasis in a large case-control sample: RNA-Seq provides insights into disease mechanisms. J Invest Dermatol. 2014;134:1828–1838. doi: 10.1038/jid.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Volders PJ, Verheggen K, Menschaert G, Vandepoele K, Martens L, Vandesompele J, Mestdagh P. An update on LNCipedia: A database for annotated human lncRNA sequences. Nucleic Acids Res. 2015;43:D174–D180. doi: 10.1093/nar/gkv295. Database Issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie C, Yuan J, Li H, Li M, Zhao G, Bu D, Zhu W, Wu W, Chen R, Zhao Y. NONCODEv4: Exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2014;42:D98–D103. doi: 10.1093/nar/gkt1222. Database Issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quek XC, Thomson DW, Maag JL, Bartonicek N, Signal B, Clark MB, Gloss BS, Dinger ME. lncRNAdb v2.0: Expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015;43:D168–D173. doi: 10.1093/nar/gku988. Database Issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. lncRNAdb: A reference database for long noncoding RNAs. Nucleic Acids Res. 2011;39:D146–D151. doi: 10.1093/nar/gkq1138. Database Issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han L, Zhang K, Shi Z, Zhang J, Zhu J, Zhu S, Zhang A, Jia Z, Wang G, Yu S, et al. LncRNA profile of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis. Int J Oncol. 2012;40:2004–2012. doi: 10.3892/ijo.2012.1413. [DOI] [PubMed] [Google Scholar]
- 34.Tafer H, Hofacker IL. RNAplex: A fast tool for RNA-RNA interaction search. Bioinformatics. 2008;24:2657–2663. doi: 10.1093/bioinformatics/btn193. [DOI] [PubMed] [Google Scholar]
- 35.Sundarrajan S, Arumugam M. Weighted gene co-expression based biomarker discovery for psoriasis detection. Gene. 2016;593:225–234. doi: 10.1016/j.gene.2016.08.021. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, Gu L, Zhang X, Liu M, Jiang H, Cai R, Zhao Y, Cheng B. Global transcriptome and weighted gene co-expression network analyses reveal hybrid-specific modules and candidate genes related to plant height development in maize. Plant Mol Biol. 2018;98:187–203. doi: 10.1007/s11103-018-0763-4. [DOI] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Stawczyk-Macieja M, Szczerkowska-Dobosz A, Rębała K, Purzycka-Bohdan D. Genetic background of skin barrier dysfunction in the pathogenesis of psoriasis vulgaris. Postepy Dermatol Alergol. 2015;32:123–126. doi: 10.5114/pdia.2014.44003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ekman AK, Verma D, Fredrikson M, Bivik C, Enerbäck C. Genetic variations of NLRP1: Susceptibility in psoriasis. Br J Dermatol. 2014;171:1517–1520. doi: 10.1111/bjd.13178. [DOI] [PubMed] [Google Scholar]
- 40.Nikamo P, Lysell J, Ståhle M. Association with genetic variants in the IL-23 and NF-κB pathways discriminates between mild and severe psoriasis skin disease. J Invest Dermatol. 2015;135:1969–1976. doi: 10.1038/jid.2015.103. [DOI] [PubMed] [Google Scholar]
- 41.Lynde CW, Poulin Y, Vender R, Bourcier M, Khalil S. Interleukin 17A: Toward a new understanding of psoriasis pathogenesis. J Am Acad Dermatol. 2014;71:141–150. doi: 10.1016/j.jaad.2013.12.036. [DOI] [PubMed] [Google Scholar]
- 42.Nair RP, Duffin KC, Helms C, Ding J, Stuart PE, Goldgar D, Gudjonsson JE, Li Y, Tejasvi T, Feng BJ, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goldminz AM, Au SC, Kim N, Gottlieb AB, Lizzul PF. NF-κB: An essential transcription factor in psoriasis. J Dermatol Sci. 2013;69:89–94. doi: 10.1016/j.jdermsci.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 44.Johansen C, Rittig AH, Mose M, Bertelsen T, Weimar I, Nielsen J, Andersen T, Rasmussen TK, Deleuran B, Iversen L. STAT2 is involved in the pathogenesis of psoriasis by promoting CXCL11 and CCL5 production by keratinocytes. PLoS One. 2017;12:e0176994. doi: 10.1371/journal.pone.0176994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haase I, Hobbs RM, Romero MR, Broad S, Watt FM. A role for mitogen-activated protein kinase activation by integrins in the pathogenesis of psoriasis. J Clin Invest. 2001;108:527–536. doi: 10.1172/JCI12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mavropoulos A, Rigopoulou EI, Liaskos C, Bogdanos DP, Sakkas LI. The role of p38 MAPK in the aetiopathogenesis of psoriasis and psoriatic arthritis. Clin Dev Immunol. 2013;2013:569751. doi: 10.1155/2013/569751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eades G, Zhang YS, Li QL, Xia JX, Yao Y, Zhou Q. Long non-coding RNAs in stem cells and cancer. World J Clin Oncol. 2014;5:134–141. doi: 10.5306/wjco.v5.i2.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fitzgerald KA, Caffrey DR. Long noncoding RNAs in innate and adaptive immunity. Curr Opin Immunol. 2014;26:140–146. doi: 10.1016/j.coi.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu GC, Pan HF, Leng RX, Wang DG, Li XP, Li XM, Ye DQ. Emerging role of long noncoding RNAs in autoimmune diseases. Autoimmun Rev. 2015;14:798–805. doi: 10.1016/j.autrev.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 51.Sonkoly E, Bata-Csorgo Z, Pivarcsi A, Polyanka H, Kenderessy-Szabo A, Molnar G, Szentpali K, Bari L, Megyeri K, Mandi Y, et al. Identification and characterization of a novel, psoriasis susceptibility-related noncoding RNA gene, PRINS. J Biol Chem. 2005;280:24159–24167. doi: 10.1074/jbc.M501704200. [DOI] [PubMed] [Google Scholar]
- 52.Szegedi K, Sonkoly E, Nagy N, Németh IB, Bata-Csörgo Z, Kemény L, Dobozy A, Széll M. The anti-apoptotic protein G1P3 is overexpressed in psoriasis and regulated by the non-coding RNA, PRINS. Exp Dermatol. 2010;19:269–278. doi: 10.1111/j.1600-0625.2010.01066.x. [DOI] [PubMed] [Google Scholar]
- 53.Holm SJ, Sánchez F, Carlén LM, Mallbris L, Ståhle M, O'Brien KP. HLA-Cw*0602 associates more strongly to psoriasis in the Swedish population than variants of the novel 6p21.3 gene PSORS1C3. Acta Derm Venereol. 2005;85:2–8. doi: 10.1080/00015550410023527. [DOI] [PubMed] [Google Scholar]
- 54.Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi J, Jiang K, Li Z. MiR-145 ameliorates neuropathic pain via inhibiting inflammatory responses and mTOR signaling pathway by targeting Akt3 in a rat model. Neurosci Res. 2018;134:10–17. doi: 10.1016/j.neures.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 56.O'Leary L, Sevinç K, Papazoglou IM, Tildy B, Detillieux K, Halayko AJ, Chung KF, Perry MM. Airway smooth muscle inflammation is regulated by microRNA-145 in COPD. FEBS Lett. 2016;590:1324–1334. doi: 10.1002/1873-3468.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perazzio AS, Oliveira JS, Figueiredo VL, Chauffaille ML. Increase of IRF-1 gene expression and impairment of T regulatory cells suppression activity on patients with myelodysplastic syndrome: A longitudinal one-year study. Leuk Res. 2017;55:6–17. doi: 10.1016/j.leukres.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 58.Azuaje FJ. Selecting biologically informative genes in co-expression networks with a centrality score. Biol Direct. 2014;9:12. doi: 10.1186/1745-6150-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nazarov PV, Muller A, Kaoma T, Nicot N, Maximo C, Birembaut P, Tran NL, Dittmar G, Vallar L. RNA sequencing and transcriptome arrays analyses show opposing results for alternative splicing in patient derived samples. BMC Genomics. 2017;18:443. doi: 10.1186/s12864-017-3819-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Antonini D, Mollo MR, Missero C. Research techniques made simple: Identification and characterization of long noncoding RNA in dermatological research. J Invest Dermatol. 2017;137:e21–e26. doi: 10.1016/j.jid.2017.01.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.