Abstract
Background
Oral microbiome has significant impact on both oral and general health. Polyols have been promoted as sugar substitutes in prevention of oral diseases. We aimed to reveal the effect of candies containing erythritol, xylitol or control (sorbitol) on salivary microbiome.
Methods
Ninety children (11.3 ± 0.6 years) consumed candies during 3 years. Microbial communities were profiled using Illumina HiSeq 2000 sequencing and real-time PCR.
Results
The dominant phyla in saliva were Firmicutes (39.1%), Proteobacteria (26.1%), Bacteroidetes (14.7%), Actinobacteria (12%) and Fusobacteria (6%). The microbiome of erythritol group significantly differed from that of the other groups. Both erythritol and xylitol reduced the number of observed bacterial phylotypes in comparison to the control group. The relative abundance of the genera Veillonella, Streptococcus and Fusobacterium were higher while that of Bergeyella lower after erythritol intervention when comparing with control. The lowest prevalence of caries-related mutans streptococci corresponded with the lowest clinical caries markers in the erythritol group.
Conclusions
Daily consumption of erythritol, xylitol or control candies has a specific influence on the salivary microbiome composition in schoolchildren. Erythritol is associated with the lowest prevalence of caries-related mutans streptococci and the lowest levels of clinical caries experience.
Trial registration
ClinicalTrials.gov Identifier NCT01062633.
Electronic supplementary material
The online version of this article (10.1186/s12903-019-0747-z) contains supplementary material, which is available to authorized users.
Keywords: Polyol, Erythritol, Oral microbiome, Saliva, qPCR, Next generation sequencing
Background
The oral microbiome is comprised of around 700 to 1000 microbial species according to the studies using high-throughput technology and has significant impact on both the oral and general health [1–3]. High diversity between individuals has been revealed, although a significant proportion of bacterial sequences of healthy individuals are identical [4–7].
The study of Ling et al. [8] has shown that the salivary microbiome in healthy children is more diverse compared to adults. Keijser et al. [2] have found that, in comparison to adults, children’s saliva appears to include a higher proportion of Firmicutes and Actinobacteria and a lower proportion of Bacteroidetes and Fusobacteria. Geographical differences have been noted as well: in comparison to healthy Americans, a higher relative abundance of Bacteroides (26.64%) and Proteobacteria (19.85%) and lower relative abundance of Firmicutes (38.53%) were found in the Netherlands [2].
Dental caries is associated with an increase in the proportion of acidogenic and aciduric bacteria, especially mutans streptococci. Although Streptococcus sobrinus is less frequently detected than Streptococcus mutans, several studies have reported its association with caries activity. In children, the caries increment with both S. mutans and S. sobrinus is higher than in those with S. mutans alone [9, 10]. At the same time the imbalance between the oral bacteria that involves suboptimal low levels of streptococci and high levels of anaerobic and Gram negative bacteria such as Aggregatibacter actinomycetemcomitans may lead to periodontitis [11, 12].
The microbiome in the oral cavity may be influenced by different factors such as oral hygiene, diet, antimicrobial and physicochemical factors [13]. Application of sugar alcohols (polyols) may be used for prevention of oral diseases to promote sugar substitutes in caries control. The most widely used sugar alcohols are sorbitol (a hexitol) and xylitol (a pentitol). Xylitol has been found to reduce the amount of plaque, the viability and survival of virulent S. mutans [14]. Erythritol (a tetritol) is a newer and therefore less used polyol, while studies suggest its caries-preventive effect [15–21]. Polyols resist fermentation and acidogenesis by the bacteria of dental plaque [22, 23] and are not absorbed via the stomach [24]. It has been recognized that regular use of polyol-containing chewing gums could play a role in preventing caries by increasing salivary flow through mastication, reversing decreases in plaque pH and enhancing remineralization of subsurface enamel lesions [25–28].
Previous studies of the polyols’ effect on oral microbiome have been focused on certain groups of bacteria and routinely performed by using of culture-based techniques. The influence of polyols on total salivary microbiome has not been described.
The aim of this study was to reveal the effect of erythritol, xylitol and sorbitol (control) candies on salivary microbiome after 3 years of a double-blind randomized controlled prospective clinical trial in Estonian schoolchildren. Two molecular approaches were combined, high-throughput sequencing that allows a global systemic view of the human oral microbiome, and real-time PCR with specific primers that provide an accurate and sensitive method for quantification of individual bacteria in total bacterial count [9, 29–31]. Further, associations were made between the changes in the salivary microbiome and clinical caries experience as previously described by Honkala et al. [15] and Falony et al. [17].
Methods
Study population
This study was part of the larger double-blind randomized controlled prospective clinical trial “Effect of erythritol and xylitol on dental caries prevention in children” (ClinicalTrials.gov Identifier NCT01062633) that was carried out at the University of Tartu from 2008 to 2011 and included 485 first- and second-grade schoolchildren from 10 randomly selected schools of south-eastern Estonia (10% of the all schools in this area) [15–17]. All school classes were randomly allocated into 3 intervention groups according to their consumption of erythritol, xylitol or sorbitol (control) candies. The subgroup for microbiological study comprised randomly selected 90 children (n = 30 from each group; 53 girls and 37 boys) with mean ± SD age 11.3 ± 0.6 years (range 10.2–12.5; quartiles 10.8–11.8) (Table 1). Subject recruitment, inclusion and exclusion criteria are more thoroughly described in our previous papers [15–17].
Table 1.
Clinical parameters of the study subjects (n = 90) at the end of the intervention trial (mean ± SD)
| Clinical data | Erythritol | Xylitol | Sorbitol (control) | P values |
|---|---|---|---|---|
| Age (y) | 11.6 ± 0.62 | 11.3 ± 0.6 | 11.1 ± 0.63 | NS |
| DMFT | 2.06 ± 1.71 1 | 3.06 ± 2.18 | 3.62 ± 2.55 1 | 1 0.013 |
| DMFS | 3.2 ± 3.63 2 | 4.37 ± 3.42 | 5.58 ± 4.50 2 | 2 0.025 |
DMFT, number of decayed, missing and filled teeth; DMFS, number of decayed, missing and filled tooth surfaces
1P = 0.013 versus sorbitol
2P = 0.025 versus sorbitol
The study was performed according to Declaration of Helsinki and approved by the Ethics Review Committee on Human Research of the University of Tartu (no. 166/7 17.12.2007). The written agreement forms were signed by parents or caretakers of the participants.
Clinical examination and sample collection
The clinical examination and saliva collection was made by four calibrated examiners and by four dental assistants in the clinic of the Department of Stomatology, Tartu University as described previously [14, 16, 17]. The children were informed not to brush their teeth in the morning of the examination day. Caries status was registered by ICDAS II coding system [32] for analyzing and converted into DMF indices where DMFT indicates number of decayed, missing and/or filled teeth (maximum 28), and DMFS number of decayed, missing and/or filled tooth surfaces (maximum 128) [33]. Salivary samples were collected during the last examination after 3 year consumption of polyols, during 2-min chewing of a piece of paraffin and stored at − 80 °C until analyzed by molecular methods.
Intervention
All children were instructed on oral hygiene during the examination each year. The new toothbrush and fluoridated toothpaste (0.24% sodium fluoride) were given to children twice a year. Throughout the intervention trial (2008–2011), pupils consumed erythritol-, xylitol-, and sorbitol-containing candies. The candies’ consumption was supervised by their teachers and took place during 200 school days per year for 3 years in total. Each participant consumed four small candies three times per school day. Total daily intake of polyol was about 7.5 g. Candies were distributed before the start of the classes (8 a.m.), after school lunch (10:30 a.m.), and at the end of the school day (1:30 or 2:15 p.m.). Candies were not used during the weekends or holidays.
Molecular methods
Salivary DNA was isolated applying the QiaAmp Blood Kit (Qiagen, Hilden, Germany). Real-time PCR was applied to quantify the indicator species Streptococcus mutans, Streprococcus sobrinus, and Aggregatibacter actinomycetemcomitans. Microbial communities were profiled using Illumina® HiSeq 2000 [36]. The details of molecular methods are presented in Additional file 1: Table S6.
Statistical analyses
The statistical analysis of clinical and qPCR data was performed using SIGMASTAT 2.0 (Systat Software, Chicago, USA) statistic software package. According to the data descriptive statistics, Fisher exact test, Bonferroni correction and Mann-Whitney rank sum test were applied to compare the differences in microbiological indices. Spearman rank order correlation test was used to test the associations between microbiological and clinical data. All differences were considered statistically significant if P < 0.05.
In case of principal component (PCA) analysis, only 63 OTUs were retained in the data set. The criterion was that OTU had to be present at least in one sample with relative abundance bigger than 1%. Relative abundance values of phylotypes (OTUs) were Hellinger transformed prior applying principal component analysis. The effect of intervention on bacterial community structure was evaluated applying one-way permutational multivariate analysis (PERMANOVA) followed by between group analysis [45]. Before PERMANOVA, the distance-based test for homogeneity of multivariate dispersions was done. One-way ANOVA was applied to assess differences in diversity indices between study groups. Zero-inflated Gaussian mixture model was applied to detect differentially abundant OTUs and genera between intervention groups using metagenomeSeq software [46].
Results
Clinical indices
The effect of polyols on dental caries indices has been evaluated after 3 years consumption of erythritol, xylitol or sorbitol (control). At the end of the trial, the values of DMFT and DMFS in the subgroup of 90 schoolchildren were significantly lower in the erythritol group versus the sorbitol group (Table 1). Additional clinical data of the full cohort are presented elsewhere [15–17]. The data of the subgroup for the microbiological study may differ somewhat from that of the whole study group since they are based on the random sample of the whole data.
Analysis of the mock community for Illumina sequencing
In order to check sequencing quality, the mock (artificial) community was analyzed along with all clinical samples. Mock community consisted of seven bacterial strains commonly found in the human oral cavity. Results of mock community analysis are presented in Additional file 1: Table S2. All strains were recovered and identified from the obtained data set.
Salivary microbiome composition
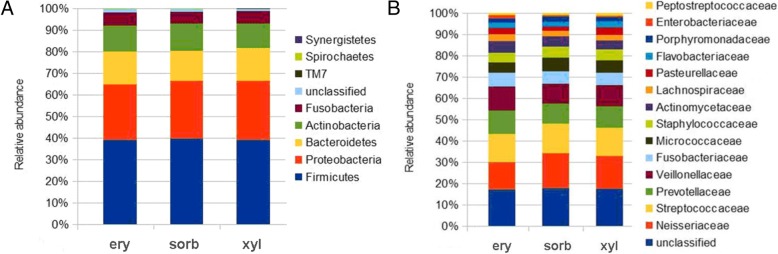
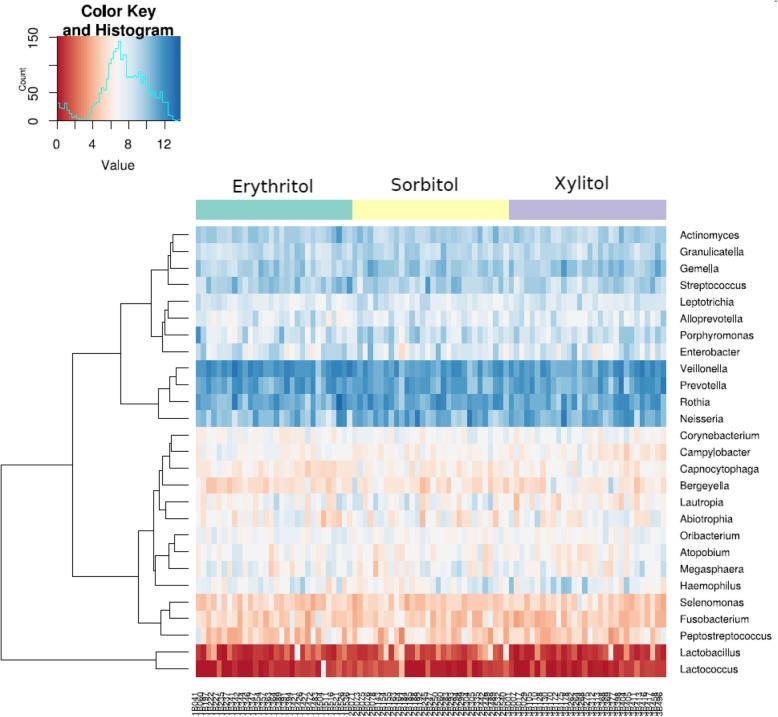
The five dominant phyla (among all obtained sequences) were Firmicutes (39%), Proteobacteria (26%), Bacteroidetes (15%), Actinobacteria (12%) and Fusobacteria (6%) according to RDP classifier. Among the identified 16 different bacterial families the most prevalent were Neisseriaceae, Streptococcaeceae, Prevotellaceae and Veillonellaceae across all intervention groups. The proportions of unclassified phylum and family level sequences were 2 and 18%, respectively. Relative abundance of bacterial phyla and families in the groups are shown on Fig. 1 (a, b). The most abundant phylotypes belonged to Gram negative cocci Veillonella sp. and Neisseria sp., Gram negative anaerobic rod Prevotella sp., and Gram positive cocci Rothia sp. and Gemella sp. (Table 2). Similar results was found when analysing the data on genus level (Fig. 2).
Fig. 1.
Relative abundance of bacterial phyla (a) and families (b) within studied groups. Groups: ery – erythritol, sorb – sorbitol, xyl – xylitol
Table 2.
Proportions of ten most abundant bacterial phylotypes (more than 1%) in saliva microbiomes (mean ± SD)
| OTU | Identification Greengenes | Identification HOMD 16S RefSeq | Erythritol | Xylitol | Sorbitol |
|---|---|---|---|---|---|
| Otu037 | Unclassified Veillonellaceae | Veillonella parvula | 12.4 ± 3.4 | 9.4 ± 3.1 | 7.8 ± 2.8 |
| Otu005 | Veillonella sp. | Veillonella sp. | 10.8 ± 3.4 | 7.9 ± 2.8 | 8.4 ± 3.5 |
| Otu068 | Rothia mucilaginosa | Rothia mucilaginosa | 7.4 ± 4.1 | 7.4 ± 2.7 | 7.9 ± 3.7 |
| Otu070 | Prevotella sp. | Prevotella sp. | 6.5 ± 3.1 | 5.9 ± 3.2 | 6.3 ± 3.2 |
| Otu038 | Rothia mucilaginosa | Rothia mucilaginosa | 5.7 ± 2.7 | 5.5 ± 2 | 7.4 ± 2.8 |
| Otu009 | Prevotella sp. | Prevotella sp. | 6.4 ± 3.5 | 6.7 ± 3.6 | 5.2 ± 2.2 |
| Otu015 | Neisseria sp. | Neisseria sp. | 3.6 ± 2.2 | 5.5 ± 2.6 | 6.6 ± 4.9 |
| Otu018 | Unclassified Neisseriaceae | Neisseria sp. | 2.1 ± 2.2 | 4.2 ± 3.5 | 4 ± 3.7 |
| Otu096 | Gemella sanguinis | Gemella morbillorum | 2.3 ± 1.2 | 3.7 ± 1.9 | 3 ± 1.3 |
| Otu209 | Unclassified Neisseriaceae | Neisseria sp. | 2.2 ± 1.6 | 3.3 ± 2.6 | 2.4 ± 2.2 |
Fig. 2.
Heat map showing the relative abundance of the most predominant genera in saliva. Row represents the relative percentage of each bacterial genus, and column stands for different samples
Influence of polyols on the diversity and structure of saliva microbiome
The intervention groups were different from each other (PERMANOVA, P < 0.001) and the erythritol group was different from the xylitol and sorbitol groups (pair-wise a posteriori comparisons, P < 0.001). Both erythritol and xylitol reduced the number of observed bacterial phylotypes in saliva in comparison to the sorbitol group (ANOVA, P < 0.001, Table 3) The inverted Simpson index, indicator of the community diversity was not significantly different between the test and sorbitol groups (ANOVA, P = 0.075). These results indicated that, compared to the sorbitol, in saliva samples there are less OTUs with high relative abundance after intervention with erythritol and xylitol.
Table 3.
Average species richness and inverted Simpson index values (mean ± SD)
| Treatment groups | Species richness (number of OTUs) 1 | Inverted Simpson Index 2 |
|---|---|---|
| Erythritol | 310 ± 18 | 17.4 ± 4.9 |
| Xylitol | 301 ± 21 | 19.0 ± 3.8 |
| Sorbitol | 327 ± 20 | 20.3 ± 6.1 |
1P < 0.001 (one-way ANOVA)
2P = 0.075 (one-way ANOVA)
Principal component analysis (PCA) was applied to visualize the differences in the microbial communities of saliva samples. The centroid of the erythritol group samples is more distant from the group centroids of other two treatments on PCA plot which is in concordance of the PERMANOVA analysis results (Fig. 3).
Fig. 3.
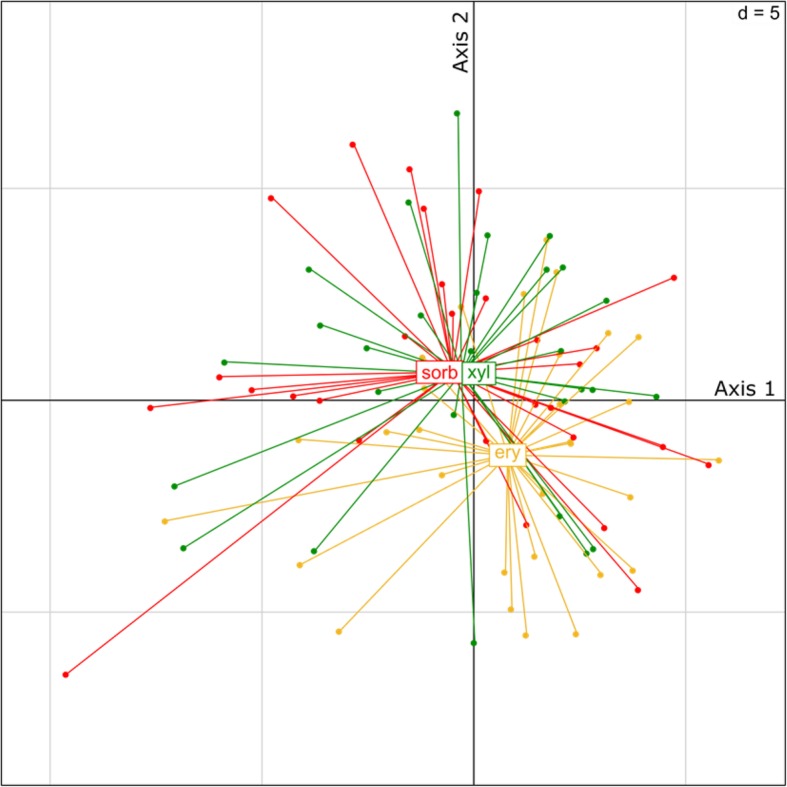
Plot of principal component analysis based on Hellinger transformed OTU relative abundance values. Individual saliva microbiome samples are connected to treatment centroids. First and second principal axes describe 16.7 and 13.3% of overall variation, respectively. Abbreviations: ery - erythritol, sorb - sorbitol, xyl - xylitol
Analysis of differentially abundant OTUs showed that erythritol intervention increased the relative abundance of genus Veillonella and Streptococcus members in saliva compared to both sorbitol and xylitol groups (Additional file 1: Tables S3 and S4). Relative abundance of genus Prevotella and Oribacterium increased in the erythritol group compared to sorbitol, too, while no clear difference appeared in comparison with xylitol group. At the same time the relative abundance of Gemella and Neisseria decreased in case of erythritol in comparison to sorbitol. The relative abundance of Rothia, Actinomyces, Enterobacter and Solobacterium was higher while that of Leptotrichia and Alloprevotella lower for erythritol in comparison to xylitol.
Similar analysis was performed also on the genus level (Table 4 and Additional file 1: Table S5). This analysis confirmed a significantly higher relative abundance of genus Veillonella and Streptococcus members in saliva in erythritol group compared to the xylitol and sorbitol group. In addition, higher abundance of Fusobacterium while a lower abundance of Bergeyella was noted in erythritol group in comparison with xylitol group.
Table 4.
Differentially abundant genera between erythritol and sorbitol groups, and erythritol and xylitol groups based zero-inflated Gaussian mixture model
| Genus | Erythritol compared to sorbitol logFC | Erythritol compared to xylitol logFC |
|---|---|---|
| Bergeyella | 0,47 | 0,83 (P < 0.05) |
| Fusobacterium | −0,34 | −0,93 (P < 0.01) |
| Streptococcus | −0,36 | −0,69 (P < 0.0001) |
| Veillonella | −0,60 (P < 0.001) | −0,50 |
A negative value for fold change (logFC) indicates an increase of in the relative abundance of a particular genus in the erythritol group compared to the sorbitol or xylitol group. Statistically significant fold changes are shown in bold and followed by P value in parenthesis
Prevalence and proportions of indicator bacteria in saliva samples according to real-time PCR
The prevalence and counts of total and three indicator bacteria in saliva samples according to real-time PCR are presented in Table 5. S. sobrinus displayed the lowest prevalence in the erythritol group but also in the xylitol group its prevalence was significantly lower compared to the sorbitol (P = 0.015 and P = 0.011, respectively, versus sorbitol). The counts of S. mutans and S. sobrinus were significantly positively correlated (rs = 0.265, P = 0.0035). Neither the prevalence nor counts of A. actinomycetemcomitans between the groups was statistically significant.
Table 5.
Real-time PCR results: counts (log10 plasmid copies/ml saliva; mean ± SD) and prevalence (%) of total bacteria and three indicator bacteria
| Bacteria spp. | Erythritol | Xylitol | Sorbitol | |||
|---|---|---|---|---|---|---|
| plasmid copies/ml saliva | % | plasmid copies/ml saliva | % | plasmid copies/ml saliva | % | |
| Total bacteria | 9.8 ± 0.4 | 100 | 9.94 ± 0.3 | 100 | 9.8 ± 0.3 | 100 |
| S. mutans | 3.3 ± 1.6 | 83 | 3.6 ± 1.5 | 87 | 3.6 ± 0.96 | 97 |
| S. sobrinus | 0.59 ± 1.4 | 17 1 | 0.72 ± 1.5 | 20 2 | 1.17 ± 1.6 | 37 1, 2 |
| A. actinomycetem-comitans | 1.17 ± 1.8 | 33 | 1.10 ± 2.0 | 27 | 1.35 ± 2.1 | 30 |
1P = 0.015 versus sorbitol; 2 P = 0.011 versus sorbitol
Associations between microbiological and clinical data
We revealed some correlations between bacteria and caries indices. The relative abundance of OTU022 and OTU257 (Leptotrichia) was lower in the erythritol group and positively associated with DMFT (R = 0.22, p = 0.04 and R = 0.21, p = 0.04, respectively). The relative abundance of OTU034 (Enterobacter) was higher in the erythritol group and negatively associated with DMFT (R = − 0.28, p = 0.008) (data not shown). The correlation analysis on genus level confirmed negative association between Enterobacter and caries indicators and revealed positive association between caries indicators and bacterial genera Lactobacillus, Bergeyella and Capnocytophaga (Table 6).
Table 6.
Spearman’s rank-order correlation between clinical and microbiological data. Results presented as correlation coefficient rs; p value
| Clinical data | Bacteria spp. | Presence of bacteria (rs; p) | Counts of bacteria (rs; p) |
|---|---|---|---|
| DMFT | Total bacteria | NS | NS |
| S. mutans | 0.271; 0.004 | 0.308; 0.001 | |
| S. sobrinus | NS | NS | |
| A. actinomycetemcomitans | NS | NS | |
| Enterobacter sp. | −0.28; 0.010 | ||
| Lactobacillus sp. | 0.27; 0.012 | ||
| DMFS | Total bacteria | NS | NS |
| S. mutans | 0.248; 0.009 | 0.306; 0.001 | |
| S. sobrinus | NS | NS | |
| A. actinomycetemcomitans | NS | NS | |
| Bergeyella sp. | 0.21; 0.049 | ||
| Capnocytophaga sp. | 0.23; 0.039 | ||
| Enterobacter sp. | −0.30; 0.005 | ||
| Lactobacillus sp | 0.34; 0.001 |
NS – not significant
Total bacteria, S. mutans, S. sobrinus and A. actinomycetemcomitans were detected using qPCR method while other bacteria using high-throughput sequencing
The correlation analysis between clinical caries experience indices and three indicator bacteria (S. mutans, S. sobrinus and A. actinomycetemcomitans) revealed positive associations between caries indices and S. mutans (Table 6).
Discussion
Our study revealed that the four dominant phyla in the salivary microbiome of schoolchildren, independently of the type of polyol consumed, were Firmicutes, Proteobacteria, Bacteroidetes and Actinobacteria while the most prevalent families were Neisseriaceae, Streptococcaceae, Staphylococcaceae and Veillonellaceae. However, after 3-year consumption of erythritol, xylitol or sorbitol clear differences were shown in the salivary microbiome. Both erythritol and xylitol showed lower numbers of observed bacterial phylotypes in comparison to the sorbitol group. According to PERMANOVA analysis the microbiome of the erythritol group differed significantly from that of the xylitol and sorbitol group. The erythritol intervention showed a higher relative abundance of Veillonella, Streptococcus and Fusobacterium but a lower relative abundance of Bergeyella. The prevalence of caries-related mutans streptococci was also the lowest in the erythritol group that corresponded with the lowest clinical caries markers in this group. To our knowledge this is the first study revealing the effect of different polyols on the salivary microbiome and their association with oral health applying high-throughput sequencing.
Sugar alcohols are noncyclic hydrogenated carbohydrates which not only offer a wide range of sweetness and cooling effect, but also non-cariogenic and less calorigenic properties. Due to their health-promoting benefits they are emerging food ingredients [47]. Previous studies have shown that xylitol, a pentitol type sugar alcohol, can be used as a safe and effective caries-limiting sweetener. It reduces the growth of dental plaque, interferes with the growth of caries-associated bacteria, decreases the incidence of dental caries, and promotes remineralization of caries lesions [19, 48, 49]. This was supported by our study where the children consumed 7.5 g of xylitol per day and the prevalence of mutans streptococci, especially S. sobrinus, was lower in their saliva than in the children consuming control candies. Erythritol, a tetritol-type alditol, is another though less investigated caries-reducing polyol. Unlike sorbitol and xylitol, erythritol is well tolerated, rapidly and almost completely absorbed from the small intestine, not metabolized and excreted unchanged in the urine thereby being almost non-caloric [50, 51]. Erythritol has been shown to significantly reduce the dental plaque weight [16, 18] as well the development of enamel/dental caries [15, 17]. Erythritol reduces growth of the plaque-related biofilm, and streptococci do not produce neither lactic nor other acids from erythritol [16, 18, 19, 52–54].
Recent advances in molecular microbiological techniques have allowed comprehensive surveys of complex bacterial communities, including the microbiota in the oral cavity that is comprised of nearly 700 to 1000 microbial species. There are two types of surfaces in the oral cavity that bacteria can colonize: the hard surfaces of teeth and the soft tissue of the oral mucosa. The microbial population in the saliva originates from both these surfaces and is therefore a mixture of the different microbial consortia [55–57]. The dominant phyla identified in the children’s salivary microbiota upon 3-year polyol consumption (Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria, Fusobacteria) were similar to the dominant phyla identified in observational non-intervention studies [2, 8, 58] that is in alignment with our findings showing that the polyols induced changes on genus level but not phylum level. Illumina technology also revealed Streptococcus and Neisseria [59] as most prevalent genera in the oral microbiome which corresponds partly with our data identifying Neisseria, Veillonella, Prevotella and Rothia, followed by Streptococcus and Gemella as most prevalent genera.
PCA analysis revealed that impact of xylitol on bacterial community is rather similar to the sorbitol, while in the case of erythritol the changes in community structure were clearly visible. The abundance of certain genera (Veillonella, Streptococcus, Fusobacterium) was higher while that of the others was lower (Bergeyella) after erythritol intervention compared to the sorbitol group. The higher abundance of Veillonella in the erythritol group is an intriguing finding since the available data concerning this genus are quite conflicting. Recent metagenomic studies have revealed this microorganism among the most predominant microorganisms in the saliva of healthy individuals [60]. In some studies Veillonella sp. has been related to caries [61, 62] but in the other studies it has been less frequently found in patients with caries [63] which corresponds to our data. Interestingly, co-cultures of Veillonella with S. mutans have been shown to produce more acid than any one of these species separately [64], suggesting that synergistic effects take place. A lack of this synergy because of the low prevalence of mutans streptococci in the erythritol group could potentially explain the lower acid concentrations found in the dental plaque of these children [16]. In another recent polyol study where the oral bacteria were tested using microarray method, no changes in the salivary microbiota took place in the xylitol group while Veillonella atypica showed a significant decrease in the sorbitol group [65], the latter being in accordance with our data.
The genus Streptococcus was more abundant in erythritol group, too. Oral streptococci are highly heterogenic and they are divided into five different groups: Mutans group (prominent members are Streptococcus mutans and Streptococcus sobrinus), Salivarius group (Streptococcus salivarius), Anginosus group (Streptococcus anginosus and Streptococcus intermedius), Sanguinis group (Streptococcus sanguinis and Streptococcus gordonii), and Mitis group (Streptococcus mitis and Streptococcus oralis) [66]. According to the HOMD database the OTU275 was close to oral taxon 423 that is non-cariogenic Streptococcus mitis, thus, the increase of this taxon can be considered as a favorable shift.
Relative abundance of Fusobacteria was quite low (less than 3%) in our study population after polyol consumption. These bacteria co-aggregate with most other oral bacteria that are important bridging organisms between early and late colonizers during plaque formation. Some members of this phylum, such as Leptotrichia can co-aggregate with potential cariogenic bacteria and may significantly associate with dental caries [67]. In accordance, our study showed a positive association between the relative abundance of Leptotrichia and DMFT which decreased after erythritol intervention. Little is known about difficult-to-culture oral bacterium Bergeyella. It has been found from intact enamel surfaces of the children with caries [68] but it has also been associated with extra-oral infections and pregnancy complications [69].
In our study, quantitative real-time polymerase chain reaction was additionally used for rapid and accurate quantification of common oral pathogens like caries-associated S. mutans and S. sobrinus, and periodontitis-associated A. actinomycetemcomitans [9, 70]. qPCR assay has very large dynamic range of target molecule determination because real-time PCR products allows to quantify the amplified products in the log phase of reaction and the overall structure of bacterial communities formed within various oral sites has been revealed [9]. Our data revealed clear association of erythritol intervention with reduced prevalence of S. sobrinus while reduction of S. mutans was slightly above significance level. Both species have been significantly associated with dental caries, general prevalence of S. mutans being higher than that of S. sobrinus while the latter has been associated with more aggressive caries in children [71]. In our substudy also the clinical caries experience markers (DMFT, DMFS) displayed the lowest values in the erythritol group. It has been suggested that an ideal polyol would reduce the counts of the aciduric bacteria without simultaneous increase in counts of periodontal pathogens. In our study neither prevalence nor counts of periodontitis-associated A. actinomycetemcomitans were different between the groups.
As a limitation, periodontal status was not investigated in these children since the periodontal diseases are very infrequent in this age. At the same time, the counts of periodontitis-associated A. actinomycetemcomitans were detected in our subjects but they did not display any deviations from the generally low proportion.
Conclusions
Daily erythritol consumption showed differentiating effects on the salivary microbiome composition in schoolchildren when compared to xylitol or sorbitol (control), while the impact of xylitol and sorbitol on the bacterial community was similar. Erythritol was associated with the lowest prevalence of caries-related mutans streptococci that corresponded to the lowest levels of clinical caries experience markers. This is the first study revealing the effect of polyols on the salivary microbiome and their association with oral health applying Illumina sequencing.
Additional files
Table S1. Specific primers and probes used for real-time PCR and Illumina HiSeq sequencing (V6 hypervariable region of the 16S rRNA gene). Table S2. Mock community analysis results. The initial composition of mock community (strains), phylotype identifications according to Greengenes (GG) and HOMD reference databases, and relative abundance of each phylotype are presented. Table S3. Differentially abundant OTUs between erythritol and control groups based zero-inflated Gaussian mixture model. A negative value for fold change (logFC) indicates an increase in the relative abundance of a particular OTU in the erythritol group compared to the control group. Table S4. Differentially abundant OTUs between erythritol and xylitol groups based zero-inflated Gaussian mixture model. A negative value for fold change (logFC) indicates an increase in the relative abundance of a particular OTU in the erythritol group compared to the xylitol group. Table S5. Differentially abundant genera between erythritol and sorbitol groups, and erythritol and xylitol groups based zero-inflated Gaussian mixture model. A negative value for fold change (logFC) indicates an increase of the relative abundance of a particular genus in the erythritol group compared to the sorbitol or xylitol group. Statistically significant changes are asterisked. Table S6. Details of molecular methods [34, 35, 37–44] (DOC 114 kb)
Acknowledgements
Not applicable.
Funding
This study was supported by Cargill R&D Center Europe (Vilvoorde, Belgium; ClinicalTrials.gov, identifier: NCT01062633), Estonian Research Council (grant No. IUT34–19, PRG548), Estonian Ministry of Education and Research (grant No. KOGU-HUMB) and Enterprise Estonia (grant No. EU48695). The funding bodies had no role in the design of the study and collection, analysis, and interpretation of data, and writing the manuscript, however, three authors were employed by Cargill during the study and preparation of this manuscript.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- DMFS
Decayed, missing and/or filled tooth surfaces
- DMFT
Decayed, missing and/or filled teeth
- DNA
Deoxyribonucleic acid
- ICDAS
International caries detection and assessment system
- OTU
Operational taxonomic unit
- PBS
Phosphate buffered saline
- PCA
Principal component analysis
- qPCR
Quantitative polymerase chain reaction
Authors’ contributions
JŠ study design, data analysis, writing the paper. JT study design, data analysis, writing the paper. RR - acquisition of data, data analysis, writing the paper. RN acquisition of data, data analysis, writing the paper. MS study design, acquisition of data, supervision. JO acquisition of data, data analysis, writing the paper. HN statistical analysis, writing the paper. JKP statistical analysis, writing the paper. KO statistical analysis, writing the paper. KK statistical analysis, writing the paper. EH study design, writing the paper, supervision. SH study design, writing the paper, supervision. KM study design, writing the paper, supervision. PLM study design, writing the paper, supervision. TV statistical analysis, writing the paper. JV study design, data analysis, writing the paper. DB study design, data analysis, writing the paper. PC study design, data analysis, writing the paper, supervision. RM study design, data analysis, writing paper, supervision. All authors have read and approved the final version of this manuscript.
Ethics approval and consent to participate
The study was performed according to Declaration of Helsinki and approved by the Ethics Review Committee on Human Research of the University of Tartu (no. 166/7 17.12.2007). The written agreement forms were signed by parents or caretakers of the participants.
Consent for publication
Not applicable.
Competing interests
Joan Vermeiren, Douwina Bosscher and Peter de Cock were employed by Cargill during the preparation of this manuscript, and Cargill produces erythritol. No potential conflict of interest was reported by other authors.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jelena Štšepetova, Email: jelena.stsepetova@ut.ee.
Jaak Truu, Email: jaak.truu@ut.ee.
Riina Runnel, Email: riina.runnel@ut.ee.
Rita Nõmmela, Email: rita.nommela@ut.ee.
Mare Saag, Email: mare.saag@kliinikum.ee.
Jana Olak, Email: jana.olak@kliinikum.ee.
Hiie Nõlvak, Email: hiie.nolvak@ut.ee.
Jens-Konrad Preem, Email: jens-konrad.preem@ut.ee.
Kristjan Oopkaup, Email: kristjan.oopkaup@ut.ee.
Kaarel Krjutškov, Email: kaarel.krjutshkov@gmail.com.
Eino Honkala, Email: eino.honkala@gmail.com.
Sisko Honkala, Email: sisko.l.honkala@uit.no.
Kauko Mäkinen, Email: kaukokmakinen@gmail.com.
Pirkko-Liisa Mäkinen, Email: makinenpirkkoliisa@gmail.com.
Tero Vahlberg, Email: tero.vahlberg@utu.fi.
Joan Vermeiren, Email: Joan.Vermeiren@gmail.com.
Douwina Bosscher, Email: Douwina_Bosscher@cargill.com.
Peter de Cock, Email: peter_decock@telenet.be.
Reet Mändar, Email: reet.mandar@ut.ee.
References
- 1.Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pockets and other oral sites. Periodontol. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- 2.Keijser BJ, Zaura E, Huse SM, et al. Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res. 2008;87:1016–1020. doi: 10.1177/154405910808701104. [DOI] [PubMed] [Google Scholar]
- 3.Wade WG. The oral microbiome in health and disease. Pharmacol Res. 2013;69:137–143. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Zaura E, Keijser BJ, Huse SM, Crielaard W. Defining the healthy ‘core microbiome’ of the oral microbial communities. BMC Microbiol. 2009;9:259–270. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dewhirst FE, Chen T, Izard J, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–5007. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nasidze I, Li J, Quinque D, Tang K, Stoneking M. Global diversity in the human saliva microbiome. Genome Res. 2009;19:636–643. doi: 10.1101/gr.084616.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–2013. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ling Z, Liu X, Luo Y, et al. Pyrosequencing analysis of the human microbiota of healthy Chinese undergraduates. BMC Genomics. 2013;14:390. doi: 10.1186/1471-2164-14-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi EJ, Lee SH, Kim YJ. Quantitative real-time polymerase chain reaction for Streptococcus mutans and Streptococcus sobrinus in dental plaque samples and its association with early childhood caries. Int J Paediatr Dent. 2009;19:141–147. doi: 10.1111/j.1365-263X.2008.00942.x. [DOI] [PubMed] [Google Scholar]
- 10.Doran KS, Nizet V. Molecular pathogenesis of neonatal group B streptococcal infection: no longer in its infancy. Mol Microbiol. 2004;54:23–31. doi: 10.1111/j.1365-2958.2004.04266.x. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Li L, Yang M, et al. Prevalence and distribution of Aggregatibacter actinomycetemcomitans and its cdtB gene in subgingival plaque of Chinese periodontitis patients. BMC Oral Health. 2014;14:37. doi: 10.1186/1472-6831-14-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kõll-Klais P, Mändar R, Leibur E, Mikelsaar M. Oral microbial ecology in chronic periodontitis and periodontal health. Microb Ecol Health Dis. 2005;17:146–155. doi: 10.1080/08910600500442891. [DOI] [PubMed] [Google Scholar]
- 13.Macrotte H, Lavoie M. Oral microbial ecology and the role of salivary immunoglobulin a. Microb Mol Biol Rev. 1998;62:71–109. doi: 10.1128/mmbr.62.1.71-109.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mäkinen KK. Sugar alcohols, caries incidence and remineralization of caries lesions: a literature review. Int J Dent. 2010;2010:981072. doi: 10.1155/2010/981072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honkala S, Runnel R, Saag M, et al. Effect of erythritol and xylitol on dental caries prevention in children. Caries Res. 2014;48:482–490. doi: 10.1159/000358399. [DOI] [PubMed] [Google Scholar]
- 16.Runnel R, Mäkinen KK, Honkala S, et al. Effect of three-year consumption of erythritol, xylitol and sorbitol candies on various plaque and salivary caries-related variables. J Dent. 2013;41:1236–1244. doi: 10.1016/j.jdent.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Falony G, Honkala S, Runnel R, Olak J, Nõmmela R, Russak S, et al. Long-term effect of erythritol on dental caries development during childhood: a post-treatment survival analysis. Caries Res. 2016;50:579–588. doi: 10.1159/000450762. [DOI] [PubMed] [Google Scholar]
- 18.Mäkinen KK, Saag M, Isotupa KP, Olak J, Nõmmela R, Söderling E, Mäkinen PL. Similarity of the effects of erythritol and xylitol on some risk factors of dental caries. Caries Res. 2005;39:207–215. doi: 10.1159/000084800. [DOI] [PubMed] [Google Scholar]
- 19.Mäkinen K. Sugar alcohol sweeteners as alternatives to sugar with special consideration of xylitol. Med Princ Pract. 2011;20:303–320. doi: 10.1159/000324534. [DOI] [PubMed] [Google Scholar]
- 20.Bonnema A, DeCock P, Eapen A, Bosscher D. The tolerance of erythritol and xylitol based on effective dose methodologies. Am J Physiol Endocrinol Metab. 2016;311:E761. doi: 10.1152/ajpendo.00300.2016. [DOI] [PubMed] [Google Scholar]
- 21.de Cock P, Mäkinen K, Honkala E, Saag M, Kennepohl E, Eapen A. Erythritol is more effective than xylitol and sorbitol in managing Oral health endpoints. Int J Dent. 2016:9868421. [DOI] [PMC free article] [PubMed]
- 22.Willibald-Ettle I, Schiweck H. Advances in sweeteners pp. 134–149. Grenby TH. London: Blackie Academic & Professional; 1996. Properties and applications of isomalt and other bulk sweeteners. [Google Scholar]
- 23.Kandelman D. Sugar, alternative sweeteners and meal frequency in relation to caries prevention: new perspectives. British J Nutr. 1997;77:S121–S128. doi: 10.1079/BJN19970109. [DOI] [PubMed] [Google Scholar]
- 24.Livesey G. Health potential of polyols as sugar replacers, with emphasis on low glycaemic properties. Nutr Res Reviews. 2003;16:163–191. doi: 10.1079/NRR200371. [DOI] [PubMed] [Google Scholar]
- 25.Dye BA, Tan S, Smith V (2007) Trends in oral health status: United States, 1988–1994 and 1999–2004. Vital Health Stat 248:1–92. [PubMed]
- 26.Stookey GK. The effect of saliva on dental caries. JADA. 2008;139(Suppl 2):11S–17S. doi: 10.14219/jada.archive.2008.0347. [DOI] [PubMed] [Google Scholar]
- 27.Ly KA, Milgrom P, Rothen M. Xylitol, sweeteners, and dental caries. Pediatr Dent. 2006;28:154–163. [PubMed] [Google Scholar]
- 28.Deshpande A, Alejandro RJ. The impact of polyol-containing chewing gums on dental caries. J Am Dent Assoc. 2008;139:1602–1614. doi: 10.14219/jada.archive.2008.0102. [DOI] [PubMed] [Google Scholar]
- 29.Belda-Ferre P, Alcaraz LD, Cabrera-Rubio R, Romero H, Simón-Soro A, Pignatelli M, Mira A. The oral metagenome in health and disease. ISME J. 2012;6:46–56. doi: 10.1038/ismej.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu B, Faller LL, Klitgord N, et al. Deep sequencing of the Oral microbiome reveals signatures of periodontal disease. PlosOne. 2012;7:e37919. doi: 10.1371/journal.pone.0037919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hata S, Hata H, Miyasawa-Hori H, Kudo A, Mayanagi H. Quantitative detection of Streptococcus mutans in the dental plaque of Jappanese preschool children by real-time PCR. Lett Appl Microbiol. 2006;42:127–131. doi: 10.1111/j.1472-765X.2005.01821.x. [DOI] [PubMed] [Google Scholar]
- 32.Ismail AI, Sohn W, Tellez M, Amaya A, Sen A, Hasson H, Pitts NB. The international caries detection and assessment system (ICDAS): an intergated system for measuring dental caries. Community Dent Oral Epidmiol. 2007;35:170–178. doi: 10.1111/j.1600-0528.2007.00347.x. [DOI] [PubMed] [Google Scholar]
- 33.Honkala E, Runnel R, Honkala S, Olak J, Vahlberg T, Saag M, Mäkinen KK. Measuring dental caries in the mixed dentition by ICDAS. Int J Dent. 2011;2011:15024. doi: 10.1155/2011/150424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehli EA, Lengyel-Nelson T, Hudziak JJ, Davies GE. Using a commercially available DNA extraction kit to obtain high quality human genomic DNA suitable for PCR and genotyping from 11-year-old saliva saturated cotton spit wads. BMC Res Notes. 2008;1:133. doi: 10.1186/1756-0500-1-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartosch S, Fite A, Macfarlane GT, et al. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecalmicrobiota. Appl Environ Microbiol. 2004;70:3575–3581. doi: 10.1128/AEM.70.6.3575-3581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gloor GB, Hummelen R, Macklaim JM, et al. Microbiome profiling by Illumina sequencing of combinatorial sequence-tagged PCR products. PLoS One. 2010;5:e15406. doi: 10.1371/journal.pone.0015406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hummelen R, Fernandes AD, Macklaim JM, et al. Deep sequencing of the vaginal microbiota of women with hiv. PLoS One. 2010;5:e12078. doi: 10.1371/journal.pone.0012078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foxman B, Luo T, Srinivasan U, et al. The effects of family, dentition, and dental caries on the salivary microbiome. Ann Epidemiol. 2016;26:348–354. doi: 10.1016/j.annepidem.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frank DN. Barcrawl and bartab: software tools for the design and implementation of barcoded primers for highly multiplexed dna sequencing. BMC Bioinformatics. 2009;10:362. doi: 10.1186/1471-2105-10-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schloss PD, Westcott SL, Ryabin T, et al. Introducing mother: open-source, platform-independent, communitysupported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hao X, Jiang R, Chen T. Clustering 16S rRNA for OTU prediction: a method of unsupervised Bayesian clustering. Bioinformatics. 2011;27:611–618. doi: 10.1093/bioinformatics/btq725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina paired-end reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cole JR, Wang Q, Cardenas E, et al. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecology. 2001;26:32–46. [Google Scholar]
- 46.Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods. 2013;10:1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park YC, Oh EJ, Jo JH, Jin YS, Seo JH. Recent advances in biological production of sugar alcohols. Curr Opin Biotechnol. 2016;37:105–113. doi: 10.1016/j.copbio.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 48.Mäkinen KK, Bennett CA, Hujoel PP, et al. Xylitol chewing gums and caries rates: a 40-month cohort study. J Dent Res. 1995;74:1903–1913. doi: 10.1177/00220345950740121501. [DOI] [PubMed] [Google Scholar]
- 49.Alanen P, Isokangas P, Gutman K. Xylitol candies in caries prevention: results of a field study in Estonian children. Community Dent Oral Epidemiol. 2000;28:218–224. doi: 10.1034/j.1600-0528.2000.280308.x. [DOI] [PubMed] [Google Scholar]
- 50.Bornet FR, Blayo A, Dauchy F, Slama G. Plasma and urine kinetics of erythritol after oral ingestion by healthy humans. Regul Toxicol Pharmacol. 1996;24:S280–S285. doi: 10.1006/rtph.1996.0109. [DOI] [PubMed] [Google Scholar]
- 51.de Cock P. Erythritol: a novel noncaloric sweetener ingredient. World Rev Nutr Diet. 1999;85:110–116. doi: 10.1159/000059714. [DOI] [PubMed] [Google Scholar]
- 52.Söderling E. Xylitol, Mutans streptococci, and dental plaque. Adv Dent Res. 2009;21:74–78. doi: 10.1177/0895937409335642. [DOI] [PubMed] [Google Scholar]
- 53.Ghezelbash GR, Nahvi I, Mohammad Rabbani M. Comparative inhibitory effect of xylitol and erythritol on the growth and biofilm formation of oral streptococci. Afr J Microbiol Res. 2012;6:4404–4408. [Google Scholar]
- 54.Hashino E, Kuboniwa M, Alghamdi SA, et al. Erythritol alters microstructure and metabolomic profiles of biofilm composed of Streptococcus gordonii and Porphyromonas gingivalis. Mol Oral Microbiol. 2013;28:435–451. doi: 10.1111/omi.12037. [DOI] [PubMed] [Google Scholar]
- 55.Sakamoto M, Takeuchi Y, Umeda M, Ishikawa I, Benno Y. Application of terminal RFLP analysis to characterize oral bacterial flora in saliva of healthy subjects and patients with periodontitis. J Med Microbiol. 2003;52:79–89. doi: 10.1099/jmm.0.04991-0. [DOI] [PubMed] [Google Scholar]
- 56.Marsh PD, Martin MV. Oral Microbiology, 5th edition. Edinburgh: Churchill Livingstone, Elsevier Limited; 2009.
- 57.Mager DL, Haffajee AD, Socransky S. Effects of periodontitis and smoking on the microbiota of oral mucous membranes and saliva in systemically healthy subjects. J Clinical Periodont. 2003;12:1031–1037. doi: 10.1046/j.0303-6979.2003.00418.x. [DOI] [PubMed] [Google Scholar]
- 58.Crielaard W, Zaura E, Schuller AA, Huse SM, Montijn RC, Keijser BJF. Exploring the oral microbiota of children at various developmental stages of their dentition in the relation to their oral health. BMC Med Genom. 2011;4:22–34. doi: 10.1186/1755-8794-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lazarevic V, Whiteson K, Huse K, Hernandez D, Farinelli L, Østerås M, Schrenzel J, Francois P. Metagenomic study of the oral microbiota by Illumina high-throughput sequencing. J Micobiol Methods. 2009;79:266–271. doi: 10.1016/j.mimet.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barroso E, Martín V, Martínez-Cuesta MC, Peláez C, Requena T. Stability of saliva microbiota during moderate consumption of red wine. Arch Oral Biol. 2015;60:1763–1768. doi: 10.1016/j.archoralbio.2015.09.015. [DOI] [PubMed] [Google Scholar]
- 61.Aas JA, Griffen AL, Dardis SR, et al. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol. 2008;46:1407–1417. doi: 10.1128/JCM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ling Z, Kong J, Jia P, et al. Analysis of oral microbiota in children with dental caries by PCR-DGGE and barcoded pyrosequencing. Microb Ecol. 2010;60:677–690. doi: 10.1007/s00248-010-9712-8. [DOI] [PubMed] [Google Scholar]
- 63.Belstrøm D, Fiehn NE, Nielsen CH, et al. Altered bacterial profiles in saliva from adults with caries lesions: a case-cohort study. Caries Res. 2014;48:368–375. doi: 10.1159/000357502. [DOI] [PubMed] [Google Scholar]
- 64.Noorda WD, Purdell-Lewis DJ, van Montfort AM, Weerkamp AH. Monobacterial and mixed bacterial plaques of Streptococcus mutans and Veillonella alcalescens in an artificial mouth: development, metabolism, and effect on human dental enamel. Caries Res. 1988;22:342–347. doi: 10.1159/000261134. [DOI] [PubMed] [Google Scholar]
- 65.Söderling E, ElSalhy M, Honkala E, et al. Effects of short-term xylitol gum chewing on the oral microbiome. Clin Oral Investig. 2015;19:237–244. doi: 10.1007/s00784-014-1229-y. [DOI] [PubMed] [Google Scholar]
- 66.Kreth J, Merritt J, Qi F. Bacterial and host interactions of Oral streptococci. DNA Cell Biol. 2009;28:397–403. doi: 10.1089/dna.2009.0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aas JA, Barbuto SM, Alpagot T, et al. Subgingival plaque microbiota in HIV positive patients. J Clin Periodontal. 2007;34:189–195. doi: 10.1111/j.1600-051X.2006.01034.x. [DOI] [PubMed] [Google Scholar]
- 68.Jiang W, Ling Z, Lin X, et al. Pyrosequencing analysis of oral microbiota shifting in various caries states in childhood. Microb Ecol. 2014;67:962–969. doi: 10.1007/s00248-014-0372-y. [DOI] [PubMed] [Google Scholar]
- 69.Wang X, Buhimschi CS, Temoin S, Bhandari V, Han YW, Buhimschi IA. Comparative microbial analysis of paired amniotic fluid and cord blood from pregnancies complicated by preterm birth and early-onset neonatal sepsis. PLoS One. 2013;8(2):e56131. doi: 10.1371/journal.pone.0056131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nurelhuda NM, Al-Haroni M, Trovik TA, Bakken V. Caries experience and quantification of Streptococcus mutans and Streptococcus sobrinus in saliva of Sudanese schoolchildren. Caries Res. 2010;44:402–407. doi: 10.1159/000316664. [DOI] [PubMed] [Google Scholar]
- 71.Saraithong P, Pattanaporn K, Chen Z, et al. Streptococcus mutans and Streptococcus sobrinus colonization and caries experience in 3- and 5-year-old Thai children. Clin Oral Investig. 2015;19:1955–1964. doi: 10.1007/s00784-015-1437-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Specific primers and probes used for real-time PCR and Illumina HiSeq sequencing (V6 hypervariable region of the 16S rRNA gene). Table S2. Mock community analysis results. The initial composition of mock community (strains), phylotype identifications according to Greengenes (GG) and HOMD reference databases, and relative abundance of each phylotype are presented. Table S3. Differentially abundant OTUs between erythritol and control groups based zero-inflated Gaussian mixture model. A negative value for fold change (logFC) indicates an increase in the relative abundance of a particular OTU in the erythritol group compared to the control group. Table S4. Differentially abundant OTUs between erythritol and xylitol groups based zero-inflated Gaussian mixture model. A negative value for fold change (logFC) indicates an increase in the relative abundance of a particular OTU in the erythritol group compared to the xylitol group. Table S5. Differentially abundant genera between erythritol and sorbitol groups, and erythritol and xylitol groups based zero-inflated Gaussian mixture model. A negative value for fold change (logFC) indicates an increase of the relative abundance of a particular genus in the erythritol group compared to the sorbitol or xylitol group. Statistically significant changes are asterisked. Table S6. Details of molecular methods [34, 35, 37–44] (DOC 114 kb)
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.