Abstract
Background
In terms of protein production, the internal environment of the host influences the activity of expression elements, thus affecting the expression level of the target protein. Native expression elements from a specific strain always function well in the original host. In the present study, to enhance the endoxylanase (XynA) production level in Corynebacterium glutamicum CGMCC1.15647 with its native expression elements, approaches to reduce host expression obstacles and to promote expression were evaluated.
Results
We identified the signal peptide of CspB2 in C. glutamicum CGMCC1.15647 by MALDI-TOF and applied it along with its promoter for the production of endoxylanase (XynA) in this strain. The native cspB2 promoter and cspB2 signal peptide are superior to the well-used cspB1 promoter and cspA signal peptide for XynA expression in C. glutamicum CGMCC1.15647, and expression in this strain is superior to the expression in C. glutamicum ATCC13032. The highest XynA secretion efficiency level in deep 24-well plates level (2492.88 U/mL) was achieved by disruption of the cell wall protein CspB2 and the protease ClpS, chromosomal integration of xynA and coexisting plasmid expression, which increased expression 11.43- and 1.35-fold compared to that of chromosomal expression and pXMJ19-xynA-mediated expression in the original strain, respectively. In fed-batch cultivation, the highest XynA accumulation (1.77 g/L) was achieved in the culture supernatant after 44 h of cultivation.
Conclusion
Adaptation between the expression elements and the host is crucial for XynA production in C. glutamicum CGMCC1.15647. Strategies including host optimization, chromosomal integration, and coexistence of plasmids were useful for efficient protein production in C. glutamicum.
Electronic supplementary material
The online version of this article (10.1186/s12934-019-1116-y) contains supplementary material, which is available to authorized users.
Keywords: Corynebacterium glutamicum, Endoxylanase, ClpS, Chromosomal expression, Coexisting plasmids
Background
In heterologous protein production, the successful production of recombinant proteins is mainly dependent on the interaction between the expression elements and the host. The ideal bacterial host provides a simplified and stable host environment for the function of synthetic biological circuits [1], such as high expression level of target protein, no inclusion body produced, no degradation of the target protein and easy for secretion. Efforts have been made to increase the protein production levels by optimizing the adaptation between the expression elements and the host in Escherichia coli [2], Bacillus subtilis [3], Saccharomyces cerevisiae [4] and Corynebacterium glutamicum [5]. Due to the following several characteristics such as (1) no endotoxin production and generally regarded as safe status [6], (2) the ability to secrete properly folded protein into the culture, (3) lack of detectable extracellular protease, C. glutamicum is a potential host cell for the secretory production of heterologous proteins, important enzymes and pharmaceutical proteins [7–9]. However, the state-of-the-art of C. glutamicum protein production is in its infancy due to its limited genetic tools compared to those for the most-widely used host, E. coli [10]. To improve the secretory production level of target proteins in C. glutamicum, endogenous expression elements had been explored for protein production in C. glutamicum [11, 12]. In addition to the manipulations based on expression elements, the development of a suitable host that is suitable for the expression elements is also important, including decreasing expression barriers and enhancing the factors that facilitate secretion. Deletion of the cell wall component protein CspB and the penicillin-binding protein resulted in high secretion efficiency of an antibody Fab [13]. Protease deletion is one of the way to improve the stability of heterologous protein in host, which can improve the accumulation level of the target protein. For example, disruption of the five protease genes tppA, pepE, nptB, dppIV and dppV showed 34% higher bovine chymosin production than that in double-disruption of tppA and pepE [14]; the GFP fluorescence intensity was increased by 40.6% in a ClpC disrupted C. glutamicum compared to the intensity in wild type [15]. In addition, recombinant protein can also be expressed from a single chromosomal site. Chromosomal expression of GFP and AprE could be 2.9-fold and 1.5-fold-increased compared to that of expression from the common integration site amyE by random knock-in in B. subtilis [16]. These cases indicated that host optimization would be a useful method to improve protein production, and we believe these approaches could be attempted to improve recombinant protein expression in C. glutamicum.
Endoxylanase (XynA) cleaves the β-1,4-glycosidic bonds present in the main chain of xylan, which is the major constituent of hemicellulose. A better digestibility could be achieved when poultry were fed with the XynA-treaded cereal [17]; addition of XynA could improve the bread volume, reduce stickiness and increase the shelf life [18]; the XynA treated pulp showed superior properties [19, 20]. Reports investigating the secretory production of XynA and its utilization in S. cerevisiae and C. glutamicum were mainly focused on expression elements [21, 22], and the production level could be increased by improving the complementarity between the host and expression elements.
In the present study, we present an efficient system based on native expression elements for the secretory production of XynA in C. glutamicum CGMCCl.15647. In addition to studies on expression elements, the effect of host optimization on the production level of XynA was performed, including cell surface layer protein and protease disruption, integration of the xynA gene into the chromosome, and coexisting plasmid expression. Large-scale production of XynA was also performed using the optimized expression system in fed-batch cultivation.
Results
Identification of the CspB2 protein of C. glutamicum CGMCC1.15647
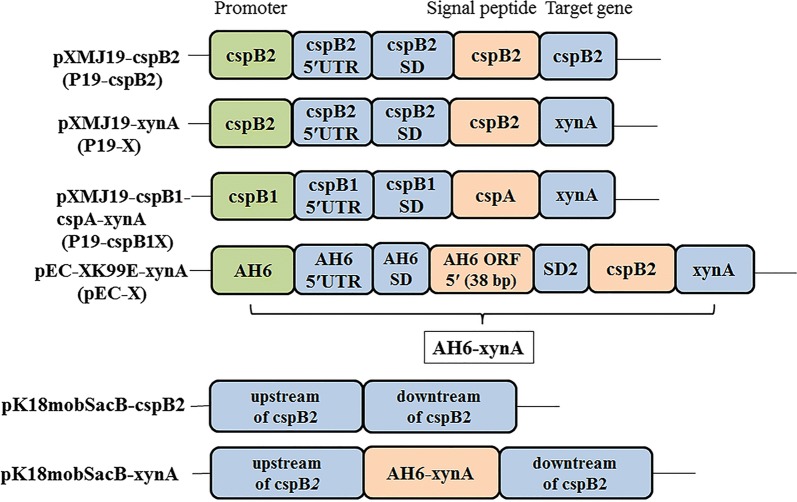
Expression systems based on different backbones, promoters and signal peptides for heterologous protein production in C. glutamicum have been summarized [23, 24], and recombination of these expression elements resulted in different expression levels of the same target protein. Considering the heterologous expression elements may not function well in this strain, we wanted to develop an expression system that is suitable for C. glutamicum CGMCC1.15647 by using its endogenous expression elements. C. glutamicum CGMCC1.15647 was used for EGFP, amylase and ScFv expression in our previous study [25–27]. We believed that the endogenous expression elements from C. glutamicum CGMCC1.15647 would be suitable for protein production in that strain. Then, we analyzed the proteins in the culture supernatant of C. glutamicum CGMCC1.15647 by SDS-PAGE, and the major protein band at approximately 50 kDa (Fig. 1a) was analyzed by MALDI-TOF. The result showed that this major protein was PS2 (Gi|42560027, PS2, Corynebacterium glutamicum) encoded by the cspB gene. To distinguish the cspB gene from C. glutamicum CGMCC1.15647 from other sources, we named it cspB2. Primary homology analysis of the region between the cspB2 promoter, SD sequence and cspB1 with its SD sequence from C. glutamicum ATCC 13869 was performed. The sequences have 91.6% consensus through 501 bp upstream of the start codon, and the − 35 region, − 10 region and SD sequence between the two are the same (Additional file 1: Figure S1A). There were three hydrophobic amino acids that were different between CspB2 and the CspB1 signal peptide: an ‘LVV’ in CspB2 and a ‘VAI’ in CspB1 (Additional file 1: Figure S1B).
Fig. 1.
Identification of CspB2 by SDS-PAGE analysis. a SDS-PAGE analysis of the proteins in the culture supernatant of C. glutamicum CGMCCl.15647. Lane M: protein molecular mass marker; lane Cg47: sample from C. glutamicum CGMCCl.15647. b Identification of CspB2 by SDS-PAGE analysis. Lane M: protein molecular mass marker (Cg47: C. glutamicum CGMCC1.15647, P19-0: pXMJ19, P19-cspB2, pXMJ19-cspB2). The arrow indicates CspB2
To confirm that CspB2 could be expressed and secreted with its promoter and signal peptide at a higher level by plasmid expression than by chromosomal expression, the cspB2 gene with its signal peptide and promoter was cloned into the pXMJ19 vector and transformed into ΔcspB2, which is a CspB2-disrupted strain derived from C. glutamicum CGMCC1.15647. The proteins in the culture medium of the transformant were analyzed by SDS-PAGE after cultivation for 48 h. One major protein that matched the molecular weight of CspB2 appeared in C. glutamicum CGMCC1.15647 and ΔcspB2 harboring pXMJ19-cspB2 (Fig. 1b), while the CspB2-disrupted strain ΔcspB2 lacked this band in the SDS-PAGE gel; we confirmed that this major protein was CspB2 by MALDI-TOF analysis. Figure 1b also shows that the plasmid expression had a higher expression level of CspB2 than did the chromosomal expression in wild-type C. glutamicum CGMCC1.15647.
Secretory production of XynA in C. glutamicum
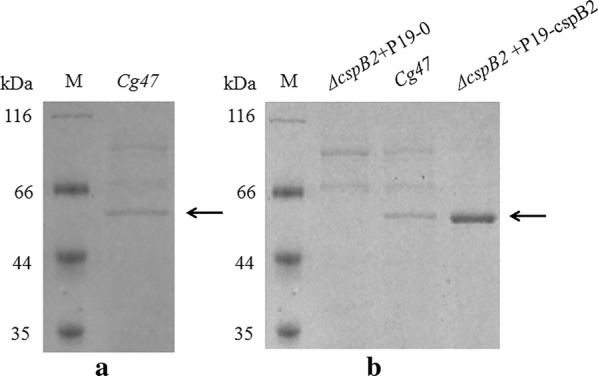
XynA plays a key role in the degradation of xylan, one of the most renewable biomass energies in nature. In the present study, we used the endogenous cspB2 signal peptide with its promoter for XynA production. The wild-type C. glutamicum CGMCCl.15647 was transformed with the pXMJ19 and pXMJ19-xynA plasmid to obtain Cg47+P19-0 (C. glutamicum CGMCCl.15647 carrying pXMJ19) and Cg47+P19-X (C. glutamicum CGMCCl.15647 carrying pXMJ19-xynA), respectively. The strength of expression from the cspB2 promoter and cspB2 signal peptide in C. glutamicum CGMCCl.15647 was compared with that from the previously reported cspB1 promoter and cspA signal peptide, which had been used for Fab expression [13], to choose the optimal construct for XynA production. After cultivation for 48 h, the XynA activity in the culture supernatant of Cg47+P19-X reached 1849.03 U/mL by the XylX6 kit assay (Fig. 2), which was higher than the production (1504.27 U/mL) of Cg47+P19-cspB1X (C. glutamicum CGMCCl.15647 carrying pXMJ19-cspB1-cspA-xynA), and there was a higher XynA activity per OD600 than that under the cspB1 promoter and cspA signal peptide (Fig. 2). This result indicated that XynA could be successfully expressed and secreted by the endogenous cspB2 promoter, 5′ UTR and signal peptide of C. glutamicum CGMCCl.15647, and the endogenous cspB2 promoter and cspB2 signal peptide is superior to the cspB1 promoter and cspA signal peptide for XynA production in C. glutamicum CGMCCl.15647 under the same culture conditions.
Fig. 2.
Comparison of the production of XynA using different promoters, signal peptides and hosts. Secretory production level of XynA by Cg47+P19-0, Cg47+P19-X, Cg47+P19-cspB1X and Cg32+P19-X. (Cg32: C. glutamicum ATCC13032, P19-X: pXMJ19-xynA, P19-cspB1X: pXMJ19-cspB1-cspA-xynA). Error bars represent standard deviations. XynA activity (red), OD600 (green), and XynA activity per OD600 (blue)
To verify the strength of this cspB2 promoter and signal peptide combination in other C. glutamicum subspecies, the pXMJ19-xynA plasmid was transformed into the C. glutamicum ATCC 13032 and the resulting strain Cg32+P19-X (C. glutamicum ATCC 13032 carrying pXMJ19-xynA) was used for XynA production. Under the same culture conditions, the XynA secretion level in strain Cg32+P19-X was 528.93 U/mL (Fig. 2), which is only 28.61% of the activity in Cg47+P19-X, and the XynA activity per OD600 was 35.99% of the activity in Cg47+P19-X (Fig. 2).
Effect of CspB2 and ClpS disruption on XynA secretory expression
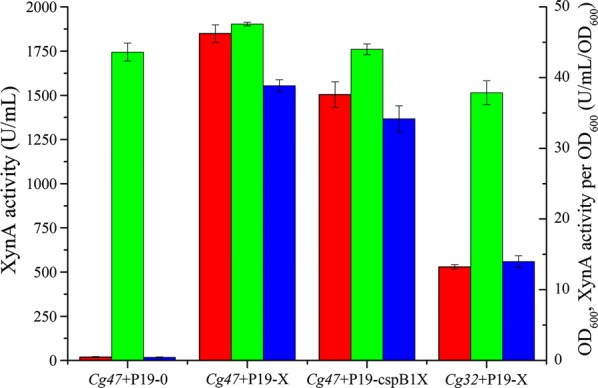
XynA has been expressed in C. glutamicum with different promoters and signal peptides at high levels. Previous authors have mainly focused on plasmid construction, and we believe that the yield of XynA could be further increased by host optimization. The cspB gene encodes the S-layer protein CspB, which acts as a cell wall barrier for Fab secretion in ATCC 13869 [13]. The protease ClpS participates in the degradation of unwanted proteins. To improve the productivity of XynA by the C. glutamicum CGMCCl.15647 strain, we first investigated the effect of cspB2 and clpS deletion mutations on XynA secretory expression. For this purpose, plasmid pXMJ19-xynA was transformed into ΔcspB2 and ΔclpS to obtain strains ΔcspB2+P19-X and ΔclpS+P19-X, respectively. In our study, XynA activity reached 1956.59 U/mL in the cspB2-disrupted strain ΔcspB2+P19-X (Fig. 3), which was 107.56 U/mL greater than the activity in the wild-type strain. It seems that the CspB2 protein forms a physical barrier inhibiting XynA secretion. The ΔclpS mutant ΔclpS+P19-X increased XynA secreted activity by 195.96 U/mL compared to the activity in wild type (Fig. 3). This is probably due to the ClpS participating in or assisting in the degradation of XynA before secretion.
Fig. 3.
Effect of ΔcspB2 and ΔclpS mutations on XynA production. Secretory production level of XynA by ΔcspB2+P19-X, ΔclpS+P19-X and ΔcspB2ΔclpS+P19-X. Error bars represent standard deviations. XynA activity (red), OD600 (green), and XynA activity per OD600 (blue)
Both single mutants of ΔcspB2+P19-X and ΔclpS+P19-X enhanced XynA secretion. To assess the effect of double deletion of cspB2 and clpS on the productivity of XynA, we evaluated the effect of ΔcspB2 and ΔclpS on C. glutamicum CGMCCl.15647. However, the secretion level of XynA was 2036.15 U/mL in ΔcspB2ΔclpS+P19-X (ΔcspB2ΔclpS carrying pXMJ19-xynA) (Fig. 3), indicating that the cspB2 and clpS double mutation had no effect on XynA secretion compared to that of the clpS disruption alone (Fig. 3), but the XynA activity per OD600 increased 10.5% and 29.3% compared to that of Cg47+P19-X and ΔclpS+P19-X, respectively (Fig. 3).
Effect of xynA chromosomal integration and coexistence of plasmids on XynA secretion
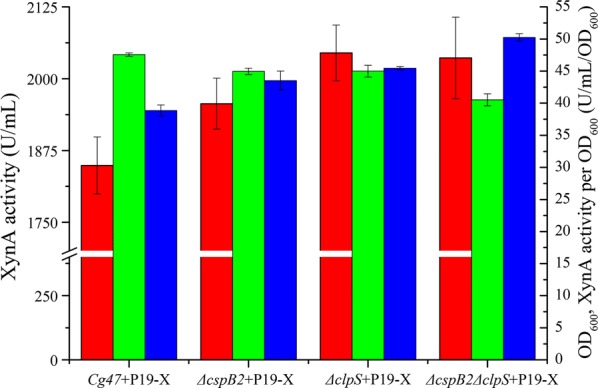
To assess the effect of chromosomal integration of the xynA gene on the production of XynA, the xynA gene with the AH6 promoter and cspB2 signal peptide was integrated into the chromosome of ΔcspB2 and ΔcspB2ΔclpS, yielding ΔcspB2InX and ΔcspB2ΔclpSInX, respectively. The chromosomal expression of XynA activity in ΔcspB2InX and ΔcspB2ΔclpSInX was 218.05 and 256.36 U/mL, respectively, which were much lower than that of plasmid expression as expected (Fig. 4). So it is necessary to combine the plasmid expression for further improving the XynA production. When the pXMJ19-xynA plasmid was transformed into ΔcspB2InX and ΔcspB2ΔclpSInX, the XynA secretion level in the resulting strains ΔcspB2InX+P19-X and ΔcspB2ΔclpSInX+P19-X achieved 2062.67 and 2327.87 U/mL (Fig. 4), respectively, which were higher than that in the relevant unintegrated mutants ΔcspB2+P19-X and ΔcspB2ΔclpS+P19-X, and XynA activity per OD600 in ΔcspB2ΔclpSInX+P19-X was 47.4% higher than that in wild-type Cg47+P19-X (Fig. 4). These results indicated that chromosomal integration contributes to XynA expression and further demonstrated that disruption of ClpS had a positive effect on XynA production. Compared to the original C. glutamicum CGMCCl.15647 strain, the XynA secretion level in ΔcspB2ΔclpSInX+P19-X was increased by 478.84 U/mL.
Fig. 4.
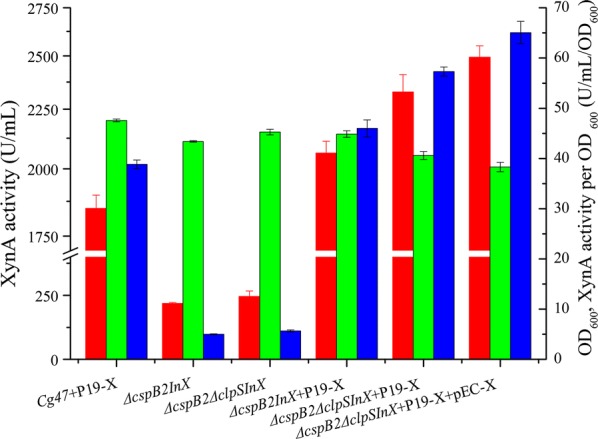
Effect of chromosomal integration and coexistence of plasmids on XynA production. Chromosomal expression of XynA by ΔcspB2InX and ΔcspB2ΔclpSInX; secretory production of XynA by ΔcspB2InX+P19-X, ΔcspB2ΔclpSInX+P19-X, and ΔcspB2ΔclpSInX+P19-X+pEC-X. (pEC-X: pEC-XK99E-xynA). Error bars represent standard deviations. XynA activity (red), OD600 (green), and XynA activity per OD600 (blue)
Generally, multiple plasmids with different replication origins can exist in one host. pXMJ19-xynA and pEC-XK99E-xynA have different antibiotic resistance genes and replicons, so they can coexist in a single C. glutamicum cell. To investigate the production of XynA by compatible plasmids in C. glutamicum, the plasmids pXMJ19-xynA and pEC-XK99E-xynA were cotransformed into ΔcspB2ΔclpSInX to obtain strain ΔcspB2ΔclpSInX+P19-X+pEC-X. This coexisting plasmid construct resulted in lower biomass due to the extra burden of the plasmids (Fig. 4); however, highest activity of XynA reached 2492.88 U/mL were present in ΔcspB2ΔclpSInX+P19-X+pEC-X (Fig. 4), and the XynA activity per OD600 was 67.4% greater than in Cg47+P19-X. Finally, the highest yield of XynA was 11.43- and 1.35-fold greater than that for the chromosomal integration expression strain ΔcspB2InX and the wild-type C. glutamicum CGMCCl.15647 harboring pXMJ19-xynA, respectively, demonstrating that the strategies performed above can be useful for the production of XynA in C. glutamicum CGMCCl.15647.
Enhanced production of XynA by fed-batch cultivation
To achieve large-scale production of XynA, fed-batch cultivation with ΔcspB2ΔclpSInX+P19-X+pEC-X was carried out in a 5-L bioreactor system. The cells continuously grew to an OD600 of 131.52 at 32 h (Fig. 5a) and then decreased gradually. In SDS-PAGE analysis, the XynA band first appeared 8 h after inoculation and increased in density (Fig. 5b). XynA activity could be detected by the XylX6 assay in the culture supernatant at 2 h after inoculation, and it increased quickly in the exponential and stationary phases during cultivation (Fig. 5a). The maximum activity of XynA in the culture supernatant was 3537.24 U/mL at 44 h, which was 12 h after the highest biomass was achieved at 32 h, indicating that XynA accumulated over time. The specific activity of the XynA is 2 U/μg. The highest concentration of XynA that was achieved was 1768.62 mg/L according to the calculation, which was 1.91-fold greater in production yield compared with that of the wild-type C. glutamicum CGMCC1.15647 harboring pXMJ19-xynA (924.52 mg/L), and higher than the previously reported 1.54 g/L [28] obtained with a single plasmid in wild-type C. glutamicum ATCC 13032. The biomass of our system was not the highest achieved, but we obtained a higher XynA concentration than have previous studies, and the proportion of XynA concentration to OD600 was also higher than in previous reports. We believe that the construction of our system for XynA production could be further improved by biomass optimization.
Fig. 5.
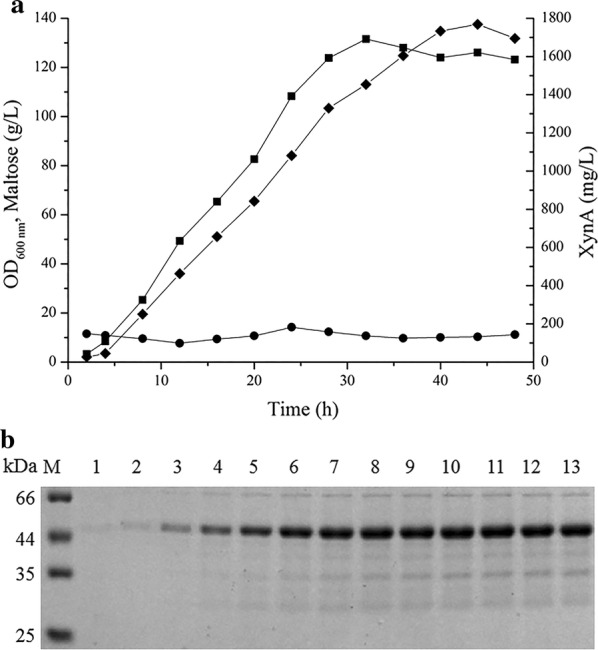
Fed-batch cultivation of ΔcspB2ΔclpSInX+P19-X+pEC-X for XynA production. a Time profiles of cell growth (square), maltose concentration (circle), and XynA concentration (diamond) in the culture supernatant. b SDS-PAGE analysis of the culture supernatant. Lane M: molecular weight markers (kDa); lanes 1–14: samples taken at 2, 4, 8, 12, 16, 20, 24, 28, 32, 36, 40, 44 and 48 h, respectively. Approximately 5 μL culture supernatant was loaded onto each lane
Discussion
The adaptation between the expression elements and the internal environment of the host is crucial for protein production in synthetic biology. Native expression elements would be preferable for function in the internal environment of a specific strain. Here, we identified the native cspB2 promoter and cspB2 signal peptide from C. glutamicum CGMCCl.15647 and applied it for XynA expression, and they were superior to the reported cspB1 promoter and cspA signal peptide elements [29]. It seems that the expression level of the target protein is highly related to the suitability of the expression elements for host because there was a much lower XynA expression level in C. glutamicum ATCC 13032 than in C. glutamicum CGMCC1.15647 with the same expression elements under the same culture conditions.
In addition to plasmid manipulation, another important factor that may affect protein secretion is host optimization, including decreasing barriers that may affect expression and secretion and promoting factors that could facilitate expression and secretion. The XynA expression level could be further increased in C. glutamicum CGMCCl.15647 by disrupting the S-layer protein CspB2 and protease ClpS. In E. coli, ClpS participates in the recognition of the N-terminus of specific proteins during degradation; ClpS is not essential for degradation, but it enhances degradation [30, 31]. XynA from Streptomyces coelicolor A3(2) is a heterologous protein for C. glutamicum, and the regulatory system, including the protease, may manipulate this foreign protein.
Moreover, chromosomal integration and coexisting plasmid transformation were performed to facilitate the expression and secretion of XynA. Considering the suitability between expression elements and the host, we chose the native AH6 promoter and cspB2 signal peptide of C. glutamicum CGMCCl.15647 for xynA integration. Coexistence of pXMJ19-xynA and pEC-XK99E-xynA in the optimized ΔcspB2ΔclpSInX strain (ΔcspB2ΔclpSInX+P19-X+pEC-X) resulted in the highest XynA production, and this coexisting plasmid expression system functioned well because the XynA production level increased with time during Fed-batch cultivation, indicating that coexistence of plasmids would be a useful approach for protein production. Plasmid replication and antibiotic resistance gene expression in coexisting plasmid expression increased the metabolic burden on the host and led to a lower growth rate, and homologous recombination of the xynA genes is another increased danger due to the two used plasmids. However, higher expression levels of the target protein could be achieved despite the extra burden or danger. The approaches performed here for XynA production could be useful for improving heterologous protein production in C. glutamicum.
Conclusion
In summary, we successfully performed optimization of the secretory production of XynA in C. glutamicum by using its native expression elements and further enhanced the production level by chromosomal integration with coexisting plasmids in a cell wall protein (CspB2)- and protease (ClpS)-disrupted mutant (Fig. 6). The highest XynA secretion level reached was 1.77 g/L, which was 14.9% higher than the the recently reported 1.54 g/L [28]. We believe that improvement of the compatibility between the expression elements and the host combined with chromosomal manipulation, not plasmid manipulation alone, will enhance C. glutamicum as a potential host for efficient protein production.
Fig. 6.
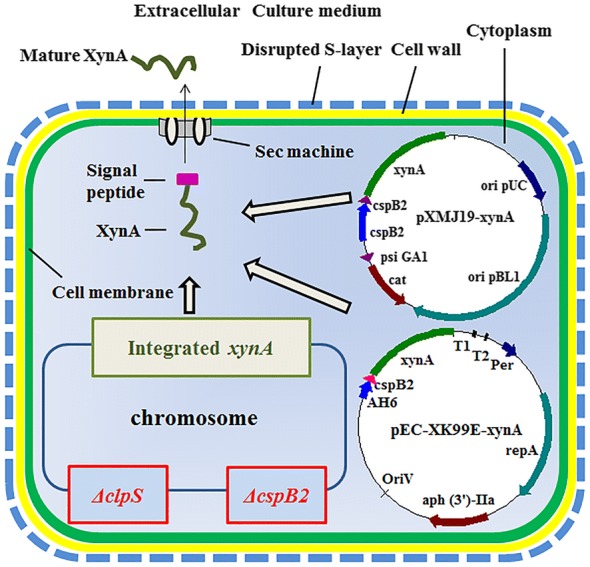
Schematic diagram of optimized C. glutamicum CGMCC 1.15647 for the secretory production of XynA. The chromosomal integrated xynA, plasmids pXMJ19-xynA and pEC-XK99E-xynA contributed to the XynA production by using the native AH6 and cspB2 promoters, and cspB2 signal peptide. In the ClpS and CspB disrupted mutant, protease could make little influence on the recombinant XynA and XynA secretion could encounter less S-layer barrier after the XynA transporting across the membrane by the Sec machine
Materials and methods
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1 and Fig. 7. E. coli was cultivated in LB medium (tryptone, 1 g; yeast extract, 0.5 g; NaCl, 1 g; H2O, 100 mL) at 37 °C. C. glutamicum ATCC 13032 and C. glutamicum CGMCCl.15647 with mutations were cultivated in deep 24-well plates containing 2 mL BHI (brain heart infusion, 37.5 g; H2O 1 L) as seed medium. After cultivation at 30 °C for 12 h with shaking at 230 rpm, the seed culture (200 μL) was inoculated into 2 mL MT medium (maltose, 17.61 g; brain heart infusion, 44.87 g; tryptone, 9 g; MgSO4, 0.6 g; FeSO4·7H2O, 1.39 g; biotin, 1 mg; ascorbic acid, 1 mg; riboflavin, 1 mg; ethyl alcohol, 5 mL; H2O, 1 L) at 30 °C. The concentrations of chloramphenicol and kanamycin used in E. coli and C. glutamicum cultures were 30 and 10 μg/mL and 50 and 30 μg/mL, respectively.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Sources |
|---|---|---|
| Strains | ||
| E. coli DH5α | Lab stock | |
| C. glutamicum ATCC13032 | Wild type | Lab stock |
| C. glutamicum CGMCCl.15647 | Wild type | Lab stock |
| ΔcspB2 | C. glutamicum CGMCCl.15647, cspB2 disruption | This work |
| ΔclpS | C. glutamicum CGMCCl.15647, clpS disruption | Lab stock |
| ΔcspB2ΔclpS | C. glutamicum CGMCCl.15647, cspB2 and clpS disruption | This work |
| ΔcspB2InX | C. glutamicum CGMCCl.15647, cspB2 disruption, xynA integration | This work |
| ΔcspB2ΔclpSInX | C. glutamicum CGMCCl.15647, cspB2 disruption, clpS disruption, xynA integration | This work |
| Cg47+P19-0 | C. glutamicum CGMCCl.15647 harboring pXMJ19 | This work |
| Cg47+P19-X | C. glutamicum CGMCCl.15647 harboring pXMJ19-xynA | This work |
| Cg47+P19-cspB1X | C. glutamicum CGMCCl.15647 harboring pXMJ19-cspB1-cspA-xynA | This work |
| Cg32+P19-X | C. glutamicum ATCC13032 harboring pXMJ19-xynA | This work |
| ΔcspB2+P19-X | ΔcspB2 harboring pXMJ19-xynA | This work |
| ΔclpS+P19-X | ΔclpS harboring pXMJ19-xynA | This work |
| ΔcspB2ΔclpS+P19-X | ΔcspB2ΔclpS harboring pXMJ19-xynA | This work |
| ΔcspB2InX+P19-X | ΔcspB2InX harboring pXMJ19-xynA | This work |
| ΔcspB2ΔclpSInX+P19-X | ΔcspB2ΔclpSInX harboring pXMJ19-xynA | This work |
| ΔcspB2ΔclpSInX+P19-X+pEC-X | ΔcspB2ΔclpSInX harboring pXMJ19-xynA and pEC-XK99E-xynA | This work |
| Plasmids | ||
| pXMJ19 (P19-0) | E. coli–C. glutamicum shuttle vector, Chlr | Lab stock |
| pXMJ19-cspB2 (P19-cspB2) | pXMJ19 derivative, PcspB2, cspB2 signal peptide, cspB2 gene | |
| pXMJ19-cspB1-cspA-xynA (P19-cspB1X) | PcspB1, cspA signal peptide, xynA | This work |
| pXMJ19-xynA (P19-X) | pXMJ19 derivative, PcspB2, cspB2 signal peptide, xynA | This work |
| pEC-XK99E | E. coli–C. glutamicum shuttle vector, Kmr | Lab stock |
| pEC-XK99E-xynA (pEC-X) | pEC-XK99E derivative, PAH6, cspB2 signal peptide, xynA | This work |
| pK18mobSacB | SacB, suicide vector, Kmr | Lab stock |
| pK18mobSacB-cspB2 | pK18mobSacB derivative, for cspB2 disruption | This work |
| pK18mobSacB-xynA | pK18mobSacB derivative, for xynA integration | This work |
Fig. 7.
Brief architecture of vectors for protein production and host mutation
Plasmid manipulation
The primers used are listed in Table 2. Q5 High-Fidelity DNA Polymerase, Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs (Ipswich, MA, USA). Kits for plasmid and genomic isolation were purchased from Sangon (Shanghai, China). For CspB2 expression, the cspB2 gene with its promoter was amplified from the C. glutamicum CGMCCl.14647 chromosome with the primers cspB2F and cspB2R and ligated to the EcoRV and EcoRI sites of the pXMJ19 vector by the CloneExpress II One Step Cloning Kit from Vazyme (Nanjing, China), yielding pXMJ19-cspB2. For XynA expression, two plasmids with different backbones, pEC-XK99E-xynA and pXMJ19-xynA, were constructed. The expression vector pEC-XK99E-xynA consists of the AH6 promoter (Gene ID: NCgl1316), the cspB2 signal peptide, and the xynA gene in pEC-XK99E for XynA expression and was synthesized by Synbio Technologies (Suzhou, China). The pXMJ19-xynA was constructed as follows: the cspB2 signal peptide with its cspB2 promoter was PCR-amplified from the genome of C. glutamicum CGMCCl.15647 by the primers cspB2F and cspR, the xynA gene was PCR-amplified from the pEC-XK99E-xynA clone vector by using the primers xynAF and xynAR, and they were ligated to the EcoRV and EcoRI sites of the pXMJ19 vector, yielding pXMJ19-xynA. To compare the strength of the cspB2 promoter and cspB2 signal peptide of C. glutamicum CGMCCl.15647 with that of the cspB1 promoter and cspA signal peptide that had been used for Fab expression [13], the plasmid pXMJ19-cspB1-cspA-xynA was synthesized by Synbio Technologies using xynA as a reporter gene. pXMJ19-xynA was also transformed into C. glutamicum ATCC 13032 to explore the strength of the cspB2 promoter and the cspB2 signal peptide in another C. glutamicum subspecies. The deletion vector pK18mobSacB-cspB2 for CspB2 knock-out was constructed as follows: the upstream and downstream regions of the cspB2 gene were PCR-amplified from the C. glutamicum CGMCCl.15647 chromosomal DNA template by the primer pairs PS1 + PS2 and PS3 + PS4 and ligated to the EcoRI and HindIII sites of the pK18mobSacB vector, yielding pK18mobSacB-cspB2. The knock-in vector pK18mobSacB-xynA for chromosomal expression of XynA was constructed as follows: the xynA gene with the AH6 promoter and cspB2 signal peptide was amplified from pEC-XK99E-xynA by using the primers A6XF and A6XR and ligated to the XbaI site of pK18mobSacB-cspB2, yielding the xynA knock-in vector pK18mobSacB-xynA.
Table 2.
List of primer oligonucleotide sequences used in this study
| Primer | Sequence |
|---|---|
| cspB2F | cccactaccgagatatccttgaataataattgcaccgcacaggtgatacatg |
| cspB2R | acagccaagctgaattcttagaacttaacgataccggagaggaatgg |
| cspR | agtggtttcctgagcgaatgctg |
| xynAF | tcgctcaggaaaccactgccgagagcacgctcggcgc |
| xynAR | acagccaagctgaattctcagtggtggtggtggtggtgggtgcgggtccagcgttggttg |
| PS1 | ctatgacatgattacgaattctttatacgtttggttatttgccgactg |
| PS2 | gactctagaggatccccggtggaaccgtcagcgtcgt |
| PS3 | ggggatcctctagagtccgctcagaaggcaatcgctgagg |
| PS4 | acgacggccagtgccaagcttattcggccacgaaggcgccg |
| A6XF | tccaccggggatcctctagaagatcttcgagctcaagaaggaac |
| A6XR | gattgccttctgagcggactcagtggtggtggtggtggtgggtgcgggtccagcgttggttg |
Construction of CspB2 and ClpS deletion mutants and XynA integration mutants of C. glutamicum
To analyze the effect of host mutation on XynA production, mutations of the cell surface layer protein CspB2 and the protease ClpS were constructed. The protease ClpS (encoded by clpS, GenBank: CP025533.1)-disrupted mutant ΔclpS was constructed based on homologous recombination in the lab strain. The other mutations were constructed by homologous recombination as described previously [13]. The pK18mobSacB-cspB2 vector was transformed into C. glutamicum CGMCCl.15647, the resulting mutant was designated ΔcspB2 after single-crossover and double-crossover selection. Then, the cspB2 and clpS double mutation was obtained using the same method and designated as ΔcspB2ΔclpS. Then, pK18mobSacB-xynA for integration of xynA into the chromosome was transformed into ΔcspB2 and ΔcspB2ΔclpS, resulting in the desired xynA-integrated mutants ΔcspB2InX and ΔcspB2ΔclpSInX, respectively. The pXMJ19-xynA and pEC-XK99E-xynA plasmids were then transformed into the C. glutamicum mutants alone or together for XynA expression.
Fed-batch cultivation
ΔcspB2ΔclpSInX harboring pXMJ19-xynA and pEC-XK99E-xynA was inoculated into 50 mL BHI as seed medium in a 500-mL flask. After cultivation at 30 °C for 12 h with shaking at 230 rpm, the seed culture (100 mL) was inoculated into 1 L fermentation medium (17.61 g maltose, 44.87 g brain heart infusion, 9 g tryptone, 0.6 g MgSO4, 1.39 g FeSO4·7H2O, 1 mg biotin, 1 mg riboflavin and 1 mg ascorbic acid per liter distilled H2O; pH 7.0) in a 5-L jar bioreactor (Applikon EZ-control, Netherlands), and 10 mL ethyl alcohol was added into the jar upon inoculation. The temperature, dissolved oxygen (DO) concentration and the pH were maintained at 30 °C, 35% (v/v) and 7.0, respectively. To prevent glucose starvation, a maltose solution (400 g/L) was added to the culture medium 12 h after inoculation, and 20 mL was added immediately after sampling. Cell growth was monitored by measuring the optical density at 600 nm (OD600) with a spectrophotometer.
Protein preparation and analysis
After cell cultivation in deep plates for 48 h, the culture supernatant was collected by centrifugation at 12,000g and 4 °C for 5 min for enzyme activity analysis and SDS-PAGE. To identify the secreted CspB2 protein, the protein band at approximately 50 kDa was excised from the SDS-PAGE gel for the culture supernatant of C. glutamicum CGMCCl.15647 and analyzed by MALDI-TOF at Sangon. The activity of XynA was assayed by using the XylX6 kit method (Megazyme, Ireland); the XylX6 assay is highly sensitive and reproducible [32]. One unit of XynA activity is defined as the amount of enzyme required to release 20 nmol of 4-nitrophenol from the XylX6 substrate in 1 min under the defined assay conditions according to the instructions of the XylX6 kit. The protein samples were also analyzed by performing electrophoresis in a 12% (w/v) SDS-PAGE gel. The XynA concentration was measured by using the bicinchoninic acid (BCA) assay.
Additional file
Additional file 1: Figure S1. Comparison of the difference between the cspB1 and cspB2 promoters and signal peptides. (A) Nucleic acid base alignment of the cspB2 and cspB1 signal peptides using ClustalX software. Identical and similar bases are labeled with asterisks (*). The − 35 region, − 10 region and SD sequence are in the black box. (B) Amino acid sequence alignment of the CspB2 and CspB1 signal peptides using ClustalX software. The different residues are shown in the black box.
Authors’ contributions
ZW and LXX designed and performed most experiments. ZW, LXX, YYK, LCL and BZH analyzed data. ZW and LXX mainly wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (21808082, 21878124), the 111 Project (111-2-06), National first-class discipline program of Light Industry Technology and Engineering (LITE2018-24) and the Opening Project of the Key Laboratory of Industrial Biotechnology, Ministry of Education (KLIB-KF201802).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Wei Zhang, Email: zhw.387@163.com.
Yankun Yang, Email: yangyankun@jiangnan.edu.cn.
Xiuxia Liu, Phone: +86 051085329306, Email: liuxiuxia@jiangnan.edu.cn.
Chunli Liu, Email: 823021647@qq.com.
Zhonghu Bai, Phone: +86 051085329306, Email: baizhonghu@jiangnan.edu.cn.
References
- 1.Umenhoffer K, Draskovits G, Nyerges A, Karcagi I, Bogos B, Timar E, Csoergo B, Herczeg R, Nagy I, Feher T, et al. Genome-wide abolishment of mobile genetic elements using genome shuffling and CRISPR/Cas-assisted MAGE allows the efficient stabilization of a bacterial chassis. ACS Synth Biol. 2017;6:1471–1483. doi: 10.1021/acssynbio.6b00378. [DOI] [PubMed] [Google Scholar]
- 2.Hayat SMG, Farahani N, Golichenari B, Sahebkar A. Recombinant protein expression in Escherichia coli: what we need to know. Curr Pharm Des. 2018;24:718–725. doi: 10.2174/1381612824666180131121940. [DOI] [PubMed] [Google Scholar]
- 3.Manabe K, Kageyama Y, Morimoto T, Ozawa T, Sawada K, Endo K, Tohata M, Ara K, Ozaki K, Ogasawara N. Combined effect of improved cell yield and increased specific productivity enhances recombinant enzyme production in genome-reduced Bacillus subtilis Strain MGB874. Appl Environ Microbiol. 2011;77:8370–8381. doi: 10.1128/AEM.06136-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ilmen M, den Haan R, Brevnova E, McBride J, Wiswall E, Froehlich A, Koivula A, Voutilainen SP, Siika-Aho M, la Grange DC, et al. High level secretion of cellobiohydrolases by Saccharomyces cerevisiae. Biotechnol Biofuels. 2011;4:30. doi: 10.1186/1754-6834-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peng F, Wang X, Sun Y, Dong G, Yang Y, Liu X, Bai Z. Efficient gene editing in Corynebacterium glutamicum using the CRISPR/Cas9 system. Microb Cell Fact. 2017;16:201. doi: 10.1186/s12934-017-0814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song Y, Matsumoto K, Yamada M, Gohda A, Brigham CJ, Sinskey AJ, Taguchi S. Engineered Corynebacterium glutamicum as an endotoxin-free platform strain for lactate-based polyester production. Appl Microbiol Biotechnol. 2012;93:1917–1925. doi: 10.1007/s00253-011-3718-0. [DOI] [PubMed] [Google Scholar]
- 7.Freudl R. Beyond amino acids: use of the Corynebacterium glutamicum cell factory for the secretion of heterologous proteins. J Biotechnol. 2017;258:101–109. doi: 10.1016/j.jbiotec.2017.02.023. [DOI] [PubMed] [Google Scholar]
- 8.Ravasi P, Braia M, Eberhardt F, Elena C, Cerminati S, Peiru S, Eugenia Castelli M, Menzella HG. High-level production of Bacillus cereus phospholipase C in Corynebacterium glutamicum. J Biotechnol. 2015;216:142–148. doi: 10.1016/j.jbiotec.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 9.Lee MJ, Kim P. Recombinant protein expression system in Corynebacterium glutamicum and its application. Front Microbiol. 2018;9:2523. doi: 10.3389/fmicb.2018.02523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol. 2014;5:172. doi: 10.3389/fmicb.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki N, Watanabe K, Okibe N, Tsuchida Y, Inui M, Yukawa H. Identification of new secreted proteins and secretion of heterologous amylase by Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2009;82:491–500. doi: 10.1007/s00253-008-1786-6. [DOI] [PubMed] [Google Scholar]
- 12.Teramoto H, Watanabe K, Suzuki N, Inui M, Yukawa H. High yield secretion of heterologous proteins in Corynebacterium glutamicum using its own Tat-type signal sequence. Appl Microbiol Biotechnol. 2011;91:677–687. doi: 10.1007/s00253-011-3281-8. [DOI] [PubMed] [Google Scholar]
- 13.Matsuda Y, Itaya H, Kitahara Y, Theresia NM, Kutukova EA, Yomantas YAV, Date M, Kikuchi Y, Wachi M. Double mutation of cell wall proteins CspB and PBP1a increases secretion of the antibody Fab fragment from Corynebacterium glutamicum. Microb Cell Fact. 2014;13:56. doi: 10.1186/1475-2859-13-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon J, Kimura S, Maruyama J, Kitamoto K. Construction of quintuple protease gene disruptant for heterologous protein production in Aspergillus oryzae. Appl Microbiol Biotechnol. 2009;82:691–701. doi: 10.1007/s00253-008-1815-5. [DOI] [PubMed] [Google Scholar]
- 15.Peng F, Liu X, Wang X, Chen J, Liu M, Yang Y, Bai Z. Triple deletion of clpC, porB, and mepA enhances production of small ubiquitin-like modifier-N-terminal pro-brain natriuretic peptide in Corynebacterium glutamicum. J Ind Microbiol Biotechnol. 2019;46:67–79. doi: 10.1007/s10295-018-2091-8. [DOI] [PubMed] [Google Scholar]
- 16.Jeong DE, So Y, Park SY, Park SH, Choi SK. Random knock-in expression system for high yield production of heterologous protein in Bacillus subtilis. J Biotechnol. 2018;266:50–58. doi: 10.1016/j.jbiotec.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Juturu V, Wu JC. Microbial xylanases: engineering, production and industrial applications. Biotechnol Adv. 2012;30:1219–1227. doi: 10.1016/j.biotechadv.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Butt MS, Tahir-Nadeem M, Ahmad Z, Sultan MT. Xylanases and their applications in baking industry. Food Technol Biotechnol. 2008;46:22–31. [Google Scholar]
- 19.Roselli A, Hummel M, Monshizadeh A, Maloney T, Sixta H. Ionic liquid extraction method for upgrading eucalyptus kraft pulp to high purity dissolving pulp. Cellulose. 2014;21:3655–3666. doi: 10.1007/s10570-014-0344-x. [DOI] [Google Scholar]
- 20.Savitha S, Sadhasivam S, Swaminathan K. Modification of paper properties by the pretreatment of wastepaper pulp with Graphium putredinis, Trichoderma harzianum and fusant xylanases. Bioresour Technol. 2009;100:883–889. doi: 10.1016/j.biortech.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 21.Tian BY, Xu Y, Cai WL, Huang QG, Gao YY, Li X, Huang JZ. Molecular cloning and overexpression of an endo-beta-1,4-xylanase gene from Aspergillus niger in industrial Saccharomyces cerevisiae YS2 Strain. Appl Biochem Biotechnol. 2013;170:320–328. doi: 10.1007/s12010-013-0173-7. [DOI] [PubMed] [Google Scholar]
- 22.Yim SS, Choi JW, Lee RJ, Lee YJ, Lee SH, Kim SY, Jeong KJ. Development of a new platform for secretory production of recombinant proteins in Corynebacterium glutamicum. Biotechnol Bioeng. 2016;113:163–172. doi: 10.1002/bit.25692. [DOI] [PubMed] [Google Scholar]
- 23.Liu X, Yang Y, Zhang W, Sun Y, Peng F, Jeffrey L, Harvey L, McNeil B, Bai Z. Expression of recombinant protein using Corynebacterium glutamicum: progress, challenges and applications. Crit Rev Biotechnol. 2016;36:652–664. doi: 10.3109/07388551.2015.1004519. [DOI] [PubMed] [Google Scholar]
- 24.Liu X, Zhang W, Zhao Z, Dai X, Yang Y, Bai Z. Protein secretion in Corynebacterium glutamicum. Crit Rev Biotechnol. 2017;37:541–551. doi: 10.1080/07388551.2016.1206059. [DOI] [PubMed] [Google Scholar]
- 25.Zhang W, Zhao Z, Yang Y, Liu X, Bai Z. Construction of an expression vector that uses the aph promoter for protein expression in Corynebacterium glutamicum. Plasmid. 2017;94:1–6. doi: 10.1016/j.plasmid.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 26.Liu X, Zhao Z, Zhang W, Sun Y, Yang Y, Bai Z. Bicistronic expression strategy for high-level expression of recombinant proteins in Corynebacterium glutamicum. Eng Life Sci. 2017;17:1118–1125. doi: 10.1002/elsc.201700087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao Z, Liu X, Zhang W, Yang Y, Dai X, Bai Z. Construction of genetic parts from the Corynebacterium glutamicum genome with high expression activities. Biotechnol Lett. 2016;38:2119–2126. doi: 10.1007/s10529-016-2196-y. [DOI] [PubMed] [Google Scholar]
- 28.Choi JW, Yim SS, Jeong KJ. Development of a high-copy-number plasmid via adaptive laboratory evolution of Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2018;102:873–883. doi: 10.1007/s00253-017-8653-2. [DOI] [PubMed] [Google Scholar]
- 29.Itaya H, Kikuchi Y. Secretion of Streptomyces mobaraensis pro-transglutaminase by coryneform bacteria. Appl Microbiol Biotechnol. 2008;78:621–625. doi: 10.1007/s00253-007-1340-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt R, Zahn R, Bukau B, Mogk A. ClpS is the recognition component for Escherichia coli substrates of the N-end rule degradation pathway. Mol Microbiol. 2009;72:506–517. doi: 10.1111/j.1365-2958.2009.06666.x. [DOI] [PubMed] [Google Scholar]
- 31.Wang KH, Sauer RT, Baker TA. ClpS modulates but is not essential for bacterial N-end rule degradation. Gene Dev. 2007;21:403–408. doi: 10.1101/gad.1511907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mangan D, Cornaggia C, Liadova A, McCormack N, Ivory R, McKie VA, Ormerod A, McCleary BV. Novel substrates for the automated and manual assay of endo-1,4-beta-xylanase. Carbohyr Res. 2017;445:14–22. doi: 10.1016/j.carres.2017.02.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Comparison of the difference between the cspB1 and cspB2 promoters and signal peptides. (A) Nucleic acid base alignment of the cspB2 and cspB1 signal peptides using ClustalX software. Identical and similar bases are labeled with asterisks (*). The − 35 region, − 10 region and SD sequence are in the black box. (B) Amino acid sequence alignment of the CspB2 and CspB1 signal peptides using ClustalX software. The different residues are shown in the black box.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.