Abstract
Isotype switching involves a region-specific, nonhomologous recombinational deletion that has been suggested to occur by nonhomologous joining of broken DNA ends. Here, we find increased donor/acceptor homology at switch junctions from PMS2-deficient mice and propose that class switching can occur by microhomology-mediated end-joining. Interestingly, although isotype switching and somatic hypermutation show many parallels, we confirm that PMS2 deficiency has no major effect on the pattern of nucleotide substitutions generated during somatic hypermutation. This finding is in contrast to MSH2 deficiency. With MSH2, the altered pattern of switch recombination and hypermutation suggests parallels in the mechanics of the two processes, whereas the fact that PMS2 deficiency affects only switch recombination may reflect differences in the pathways of break resolution.
The adaptive immune response utilizes two processes that enable an antibody, after initial encounter with the antigen, to bind that antigen more effectively as well as to recruit powerful effector pathways leading to antigen neutralization. One of these processes, somatic hypermutation, introduces nucleotide substitutions at high frequency into the Ig V gene, allowing the production and subsequent selection of lymphocytes that make antibodies with increased affinity for the antigen (1). The other process, class-switch recombination, allows a B lymphocyte that initially expresses an IgM antibody to develop into an IgG-, IgA-, or IgE-expressing cell, changing the antibody's effector activity (2, 3). Switch recombination is exceptional among programmed gene rearrangements in that it is neither evidently a homologous nor a site-specific recombination event; rather, it is region-specific.
Although hypermutation and switching are distinct processes and can occur independently in the activated B cell, they share a number of features—a circumstance that previously led us to propose that aspects of their mechanism are in common (4). Thus, both processes are (i) targeted to the Ig gene loci, (ii) linked to transcription, (iii) associated with DNA breaks, (iv) associated with related consensus sequences (GAGCT), and (v) found with diminished efficiency and increased consensus targeting in mice deficient in MSH2. This suggestion of a shared component to the mechanisms of hypermutation and switch recombination was greatly strengthened by the demonstration that deficiency in a single protein (activation-induced deaminase; see ref. 5) is sufficient to abolish both processes while leaving most other features unaffected.
However, although hypermutation and class-switch recombination show many parallels, a clear distinction is in their sensitivity to deficiency in components of the Ku/DNA-PKcs complex: switching is substantially inhibited by deficiency in these proteins, whereas hypermutation is essentially unaffected (6–9). Presumably, this distinction reflects the fact that the parallels between hypermutation and switching are likely to relate to the initiation of the two events (and possibly the generation of DNA breaks) rather than to the pathway of resolution of the initiating DNA lesion.
We and others have noted that class-switch recombination is diminished in MSH2-deficient mice, and we noted that μ-switch breaks were more focused on consensus motifs (4, 10). An effect of MSH2 deficiency on the nature of the switch junctions could simply reflect that MSH2 plays a role in correcting the nucleotide mismatches that are generated as a consequence of the switch recombination itself; indeed, nucleotide substitutions are frequently observed adjacent to the break junctions (11). This hypothesis ascribes a function to MSH2 that falls within its conventional role in mismatch repair. However, an alternative hypothesis, which we favor, is that MSH2 is actively involved in assisting the switch recombination process and is not just contributing by way of repairing the errors left in its wake. As an approach toward discriminating these two hypotheses, we here investigate the effect of PMS2 deficiency on class-switch recombination. The reasoning is that if the effect of MSH2 deficiency on the nature of switch recombination junctions is solely a consequence of the lack of resultant mismatch repair, then the switch junctions in PMS2-deficient mice should be similarly affected because mismatch repair depends on MSH2 (possibly in conjunction with MSH3 or MSH6) working through PMS2. We confirm the observation of Schrader et al. (10) that PMS2 deficiency (like MSH2 deficiency; refs. 4 and 10) leads to diminished isotype switching. This in itself does not necessarily imply a role for PMS2 or mismatch repair in the mechanics of the switch recombination process itself (see, for example, ref. 12). However, we find that PMS2-deficiency does have an effect on the actual switch recombination events that is quite distinct from that of MSH2-deficiency. The results point to the ability of a microhomology-mediated process to participate in the resolution of switch breaks.
Materials and Methods
Mice.
PMS2-deficient mice (13) were kindly provided by S. Baker (Univ. of California, Berkeley). Pms2+/− animals were intercrossed in our barrier unit to generate homozygous mice. The mice were genotyped with a PCR-based assay on tail DNA.
Analysis of Mutation.
Mutation analysis was performed as described (14). Briefly, germinal center [CD45R(B220)+ PNAhi] B cells were sorted from Peyer's patches, and the JH4 3′-flanking sequence of endogenous rearrangements of VHJ558 family members were amplified by PCR. The resulting products were cloned into M13mp19 and sequenced by using a ABI377 sequencer (Applied Biosystems). Sequences were aligned and analyzed by using PREGAP and GAP4 software (15). A 403-bp segment was analyzed for mutations (excluding the VDJ junction and the 5′-terminal 16 residues of the JH4 segment). All analysis of hotspots and mutation bias were performed on restricted data after elimination of clonally related sequences identified on the basis of shared VDJ rearrangements. The maximum PCR error for the described conditions was estimated (by sequencing clones from fractionated PNAlow B cells) to be less than 0.8 × 10−3.
In Vitro Cultures.
Single cell suspensions of spleen cells (after red blood cell lysis) were cultured at 1 × 106 cells/ml in RPMI medium 1640 supplemented with 10% FCS and 50 μg/ml Escherichia coli lipopolysaccharide (LPS; Sigma) with or without 50 ng/ml IL4 (R&D Systems). Cultures were performed for 4 days before flow cytometric analysis and DNA extraction. Flow cytometric analysis of surface IgG1, IgG2b, or IgG3 expression was performed by using biotinylated anti-Ig antibodies (PharMingen), FITC-conjugated streptavidin (Dako), and phycoerythrin-conjugated rat RA3–6B2 monoclonal antimouse CD45R(B220) (GIBCO). Activated lymphocytes were gated on the basis of forward and side scatter.
Analysis of Switch Recombination Junctions.
Genomic DNA was prepared from day-4 LPS/IL-4 spleen cultures after proteinase K digestion. PCR conditions have been described by using the following primers for the first round of the nested PCR: Sμ1 (5′-TAGTAAGCGAGGCTCTAAAAAGCAT, nucleotides 5031–5055 of MUSIGCD07 or 5′-AACTCTCCAGCCACAGTAATGACC, nucleotides 4942–4965) and either Sγ3.1 (5′-CTACTGAGTTCCTGTGCTTG, nucleotides 1050–1069 of MUSIGCD18) Sγ1.1 (CTGTAACCTACCCAGGAGACC, 8918–8938 of MUSIGHANB) or Sγ2b CATCTCTGGAAGCTTTCCATG, 1151–1171 of MMU85373). The second round of the nested PCR was performed by using Sμ2 (5′-ATCGAATTCGCTTGAGCCAAAATGAAGTAGACT, nucleotides 5140–5163 of MUSIGCD07, or 5′-ACGCTCGAGAAGGCCAGCCTCATAAAGCT, nucleotides 4972–4994) and either Sγ3.2 (5′-CCGGAATTCTTGACCTGGTACCCTAGC; nucleotides 1035–1052 of MUSIGCD18), Sγ1.2 (5′-GTCGAATTCCCCCATCCTGTCACCTATA nucleotides 8868–8886 of MUSIGHANB), or Sγ2b.2 (GCTGAATTCACATGCCGCTCTCCCCAGGTATC, nucleotides 1092–1114 of MMU85373) before purification of the DNA, and cloning into pBluescript after digestion with EcoRI and sequencing. For Sγ3 an alternative pair of primers (termed primers β) about 290 bases upstream were also used [first round: GGTTCCTCTGCTCAGGAATTAC, nucleotides 749–770 of MUSIGCD18 (or nucleotides 2632–2653 of MUSIGHANA) and CCGGAATTCCCTGACCCAGGAGCTGCATAAC, nucleotides 726–742 of MUSIGCD18 (or nucleotides 2603–2625 of MUSIGHANA)]. Switch junctions were identified by using BLAST with the low-complexity filter disabled. Donor/acceptor homology was monitored by measuring the number of nucleotides of perfect identity between donor and acceptor sequences at the switch junction.
Results
Switch Junctions Exhibit Microhomology in PMS2-Deficient Mice.
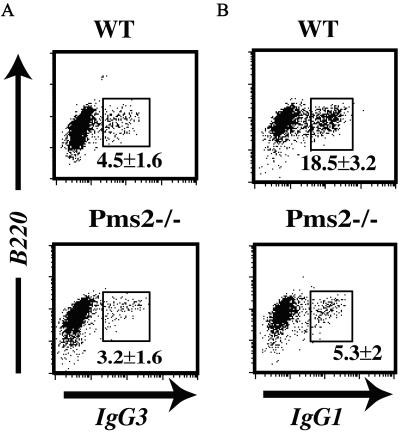
Spleen cells from PMS2-deficient (or Pms2+/+ control) mice were cultured in vitro in the presence of the B cell mitogen LPS or with LPS + IL4 to analyze Ig class switch recombination. There was no difference between the cultures from PMS2-deficient mice and control mice with regard to the proportion of cells induced to blast as judged by scatter analysis. However, in keeping with the results of Schrader et al. (10), the B cells from PMS2-deficient exhibited a reduced level of switching—especially with respect to IgG1 (Fig. 1).
Figure 1.
Diminished in vitro class switching on LPS ± IL-4 stimulation of B cells from PMS2-deficient mice. Expression of surface IgG3 (A) and IgG1 (B) from Pms2−/− mice and litter-matched wild-type controls. The average percentage of B220+ cells that are sIgG+ is shown in each panel based on analysis of groups of 4–6 mice.
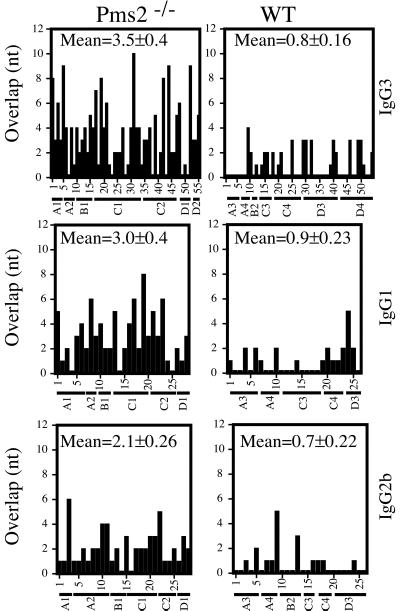
To ascertain whether the molecular process of switch recombination was affected in PMS2-deficient mice, IgG3 switch junctions in mitogen-stimulated splenic B cells were cloned after PCR amplification. The sequences obtained were compared with the germline Sμ and Sγ3 sequences by using BLAST with the low-complexity filter disabled. In an initial analysis, it was striking that the precise location of the switch junction in the sequences obtained from a PMS2-deficient mouse (but not from a control) could not be pinpointed because the germline Sμ and Sγ3 sequences were identical in the region of the junction (compare mouse A1 to mouse A3, Fig. 2). We therefore screened a much larger number of switch junctions from multiple PMS2-deficient as well as litter-matched wild-type mice and, after automated BLAST alignment, computed the overlap by the number of nucleotides of uninterrupted identity between Sμ donor and Sγ acceptor at the junction. The average length of overlap in the IgG3 switch junctions from PMS2-deficient mice was 3.5 nt, as opposed to 0.8 nt in wild-type littermates. The skewing to increased donor/acceptor homology was also evident in switches to IgG1 and, although to a somewhat lesser extent, in IgG2b switch junctions (Fig. 2). Furthermore, of 55 IgG3 switch junctions from PMS2-deficient mice, 7 exhibited a donor/acceptor overlap at the junction of eight nucleotides or more; in contrast, none of the 54 IgG3 switch junctions from wild-type mice showed more than even a four-nucleotide overlap (Fig. 2).
Figure 2.
Increased microhomology at switch junctions from PMS2-deficient mice. The histograms depict the number of nucleotides of donor/acceptor identity at the junction for each switch junction cloned. The provenance of each clone is indicated on the x axis with the mouse from which each clone derives designated by a letter (A, B, and C, specifying the litter) and a digit. Identity was determined by the number of bases of continuous overlap at the switch junction. The mean donor/acceptor overlap at the switch junctions (±SEM) is shown in each panel.
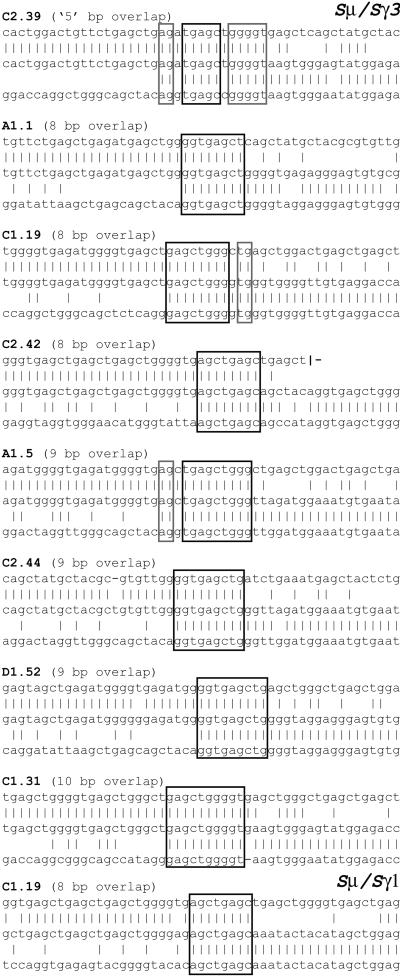
A listing of the switch junctions exhibiting longer donor/acceptor homology is provided in Fig. 3. It is particularly striking to note the presence of these very long overlaps given that, in the regions analyzed in this work, the longest stretch of identity between germline Sμ and germline Sγ3 is provided by a single example of a 12-nucleotide sequence. Although we have used stringent criteria to measure the extent of donor/acceptor overlap at switch junctions (only counting uninterrupted stretches of perfect donor/acceptor identity), several junctions from Pms2−/−, but not control mice, reveal more extensive overlaps, in which the homology is interrupted by a single mismatch (see, for example, clone C2.39 in Fig. 3). The increased proportion of switch junctions from PMS2-deficient mice that exhibit microhomology is even more striking if such relaxed criteria of homology are used (data not shown).
Figure 3.
Switch junctions with overlaps of eight nucleotides or more (all from PMS2-deficient mice). Overlap was determined by identifying the longest region at the switch junction of perfect uninterrupted donor/acceptor identity. Clone C2.39 (which is therefore determined as exhibiting an overlap of only five nucleotides) is nevertheless included as an example of a junction from a PMS2-deficient mouse (of a type not found in control mice), in which the donor/acceptor homology (highlighted in gray) extends beyond the longest region of perfect donor/acceptor identity. The germline Sμ sequence is not precisely known in the region further 3′ of that depicted for clone C2.42.
Thus, the effects of deficiency in PMS2 and in MSH2 on switch recombination are quite different. In MSH2-deficient mice, we did not observe any tendency toward increased donor/acceptor homology at IgG switch junctions, but instead noted a markedly increased preference for the Sμ switch junction breaks to be located at consensus motifs (4). In PMS2-deficient animals, we see a marked skewing toward increased donor/acceptor homology (Figs. 2 and 3) but do not find any increased tendency for Sμ breakpoints to occur at a GAGCT consensus sequence (Table 1). The difference between the effects of PMS2- and MSH2-deficiency on class-switch recombination supports the suggestion that at least one of these proteins is acting outside its conventional role in mismatch repair.
Table 1.
Location of the Sμ breakpoints in IgG3 switch junctions
| PMS2−/− | WT | |
|---|---|---|
| Within GAGCT consensus | 22 | 22 |
| Outside GAGCT consensus | 33 | 32 |
| Total no. of breakpoints | 55 | 54 |
WT, wild type.
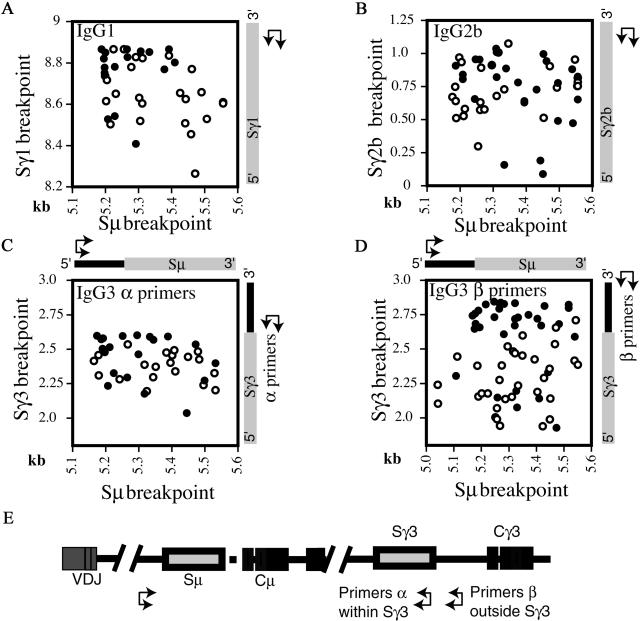
The increased reduction in switching to IgG1 as compared with IgG3 in PMS2-deficient mice could reflect the fact that Sγ3 has greater overall homology to Sμ than does Sγ1 (16). In the IgG1 switches investigated, the distribution of Sγ1 breakpoints in PMS2-deficient mice is shifted toward the more 3′ part of Sμ, and is more shifted toward the 5′ part of Sγ1; this reflects a shift toward increased donor/homology while retaining a similar size of switch junction PCR product (Fig. 4A). The extent to which such a shift is detected will presumably depend on the local distribution of homology between Sμ and Sγ over the respective switch regions as well as on the priming positions of the oligonucleotides used for the PCR amplification. Indeed, the effect of PMS2 deficiency on the overall distribution of IgG3 breakpoints is even more apparent when the PCR amplification is performed by using an Sγ3 nesting pair of oligonucleotides that primes back from downstream of Sγ3 rather than from within Sγ3. This allows detection of both switch junctions in which the γ3 breakpoint lies downstream of the repetitive (more Sμ-homologous) Sγ3 region as well as junctions in which the γ3 breakpoint lies within Sγ3 (contrast Fig. 4 C and D).
Figure 4.
Scatter analysis of the μ/γ breakpoints derived from in vitro stimulated spleen cells for IgG1 (A), IgG2b (B), and IgG3 (C and D). The x axis indicates the position of the Sμ breakpoint and the y axis of the Sγ breakpoint. Open circles denotes breakpoints from Pms2−/− mice, filled circles from their litter-matched controls. The number on the axis reflects the position of the breakpoints and are derived from MUSIGCD07 (Sμ), a composite number based on MUSIGCD18 and MUSIGHANA (Sγ3), MUSIGHANB (Sγ1), and MMU85373 (Sγ2b). (E) The scheme for PCR amplification. Switching to IgG3 was analyzed by sequencing switch junctions cloned after PCR amplification by using one set of nested PCR oligonucleotides priming forward from upstream of Sμ and another of two sets of nested oligonucleotides priming back from either within (Primers α) or downstream (Primers β) of Sγ3.
Hypermutation Profile Is Unaffected in PMS2-Deficient Mice.
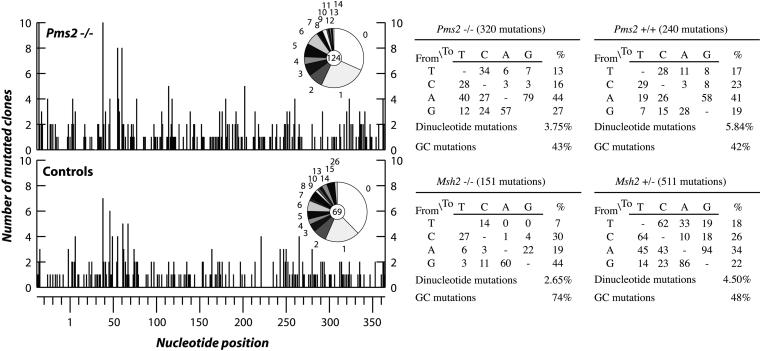
As discussed in the introduction, there are several parallels between class switch recombination and somatic hypermutation. In particular, the idea of an active role of MSH2 in switch recombination follows our earlier suggestion that the altered spectrum of nucleotide substitutions observed during Ig hypermutation in MSH2-deficient mice reflects an active role for MSH2 in the hypermutation process itself (17). Thus, in Msh2−/− mice, the nucleotide substitutions introduced by somatic hypermutation are more focused on the intrinsic hotspots (17, 18), which we interpreted as implicating MSH2 in assisting mutation creation at sites distinct from the intrinsic hotspots. If MSH2 is indeed acting to assist the generation of nucleotide substitutions rather than simply fulfilling its conventional mismatch repair role, then—as in class switch recombination—one would expect different effects to result from deficiency in either MSH2 or PMS2. The data of Frey et al. (18) suggest that this is indeed the case, because these authors did not observe any effect of PMS2 deficiency on the spectrum of nucleotide substitutions introduced during somatic hypermutation. However, several other groups have also analyzed the mutation spectrum in PMS2-deficient mice, and the results have not been concordant with either no effect reported or effects noted variously on dinucleotide mutations, preferential targeting of G/C or a decreased ratio of mutation in A as opposed to T (19–22). Because it was important to ascertain whether deficiency in PMS2 and MSH2 have similar or differing effects on the mutation spectrum in hypermutation, we analyzed the mutation spectrum in PMS2-deficient mice by using the same strategy that we previously exploited for MSH2-deficient mice. Our results (Fig. 5) confirm those of Frey et al. (18). Thus, the increase in mutations in G/C as opposed to A/T and the concomitant hotspot-focusing of mutation that was observed in MSH2-deficient mice (17, 18) is not also observed in PMS2-deficient mice. Furthermore, we did not detect any increase in dinucleotide mutations in either the Pms2−/− or Msh2−/− mismatch-repair-deficient mice.
Figure 5.
Analysis of hypermutation in Pms2−/− mice and age-matched controls. For each nucleotide position in the 403-bp interval of the JH4–CH intron analyzed, the histogram depicts the number of PCR-amplified clones that carry a mutation at that position. Clonally related sequences were excluded by comparison of the CDR3 region. Nucleotide position number 1 corresponds to the first base of the JH4–CH intron. Pie graphs show the proportion of sequences with nos. of mutations per clone. The central circle shows the total number of clones analyzed. The nucleotide substitution preferences deduced from these data for Pms2−/− and Pms2+/− mice are shown in tabular form, with the data in the “%” column corrected for base composition. The results for Msh2−/− and Msh2+/− mice are included for comparison and are taken from Rada et al. (17).
Discussion
The donor/acceptor homology seen at many of the switch junctions from PMS2-deficient mice indicate that if switch recombination does indeed proceed by a DNA end-joining mechanism (23), then joining of the broken DNA ends can be mediated by a microhomology-mediated pathway. Such a joining mechanism could well involve an MRE11/RAD50/p95 complex (24, 25). It is notable that, although the prevalence of donor/acceptor homology is striking in the switch junctions from PMS2-deficient mice, such homology is also detectable—although to a much lesser extent—in switch junctions from wild-type animals. (The mean donor/acceptor overlap length of 0.8 in IgG3 junctions from wild-type mice and the ratio of the number of junctions with four-base overlaps to those with one-base overlaps in this data set is significantly above what would be expected for randomly generated joints, calculated on the basis of the germline Sμ and Sγ3 sequences.) Thus, the microhomology-mediated end-joining that appears to make a major contribution to switching in the absence of PMS2 could also make a variable contribution under other conditions. This is consistent with the findings of Pan and Hammarström (personal communication) who found increased homology at switch junctions in B cells from patients suffering from ataxia–telangiectasia.
PMS2 and MSH2 deficiency have differential effects on switch recombination, suggesting that at least one of these proteins is acting outside its conventional role in mismatch repair. Mismatch repair as judged by in vitro assays and microsatellite stability is defective in cells from mice deficient in either MSH2 or PMS2 (13, 26, 27). Thus, were MSH2 (possibly in conjunction with MSH3 or MSH6) simply acting to recognize base mismatches at the recombination junctions and thereby recruit PMS2/MLH1 to effect repair, then we would have anticipated similar skewing effects resulting from deficiency in either MSH2 or PMS2 because, in both situations, mismatch repair is inoperative. However, what we observe is a markedly increased propensity for switch junctions to show donor/acceptor homology in PMS2-deficient mice (this work), as opposed to a distinct bias in MSH2-deficient mice where we saw no such increased donor/acceptor homology but rather a tendency for Sμ breaks to be focused on consensus motifs (4).
That MSH2 and PMS2 deficiency should have different effects on switch recombination is readily understandable if, in addition to acting in mismatch repair, MSH2 has—as previously proposed (4)—an active role in facilitating switch recombination, such as assisting the removal of nonhomologous 3′ ends during break repair (28). The altered pattern of switch recombination in Pms2−/− mice could be a consequence of PMS2 deficiency leading to an altered abundance of other DNA repair proteins—see, for example, ref. 29. A variant on this theme (suggested to us by Claude-Agnes Reynaud) would be that, in the absence of PMS2, MSH2 could be diverted to partake in a complex such as the yeast RAD1/RAD10/MSH2 complex that is implicated in the removal of nonhomologous 3′ ends (28); this could lead to increased microhomology at switch junctions. However, most straightforwardly, the microhomology seen at switch junctions from Pms2−/− mice could simply indicate that mismatch repair is normally needed to resolve switch junctions which lack microhomology. In the absence of mismatch repair, a bias toward junctions displaying microhomology would thereby ensue. A similar bias in mice rendered mismatch repair deficient through ablation of MSH2 might not be observed because of the additional participation of MSH2 in class switching at an earlier stage in the process, consistent with the known interactions of MSH2 with early intermediates at double-strand breaks (30).
Mindful of the parallels between hypermutation and switch recombination, we were motivated to ask whether the contrasting effects of PMS2 and MSH2 deficiencies on class switch recombination also applied to hypermutation. The data of Frey et al. (17) clearly indicate that they do apply to hypermutation, with the mutation spectrum in PMS2-deficient mice being unaffected, whereas the spectrum is biased toward mutation in G + C and is strongly hotspot-focused in MSH2-deficient mice (17, 18). However, others have seen effects of PMS2 deficiency on mutation spectrum (20–22). Here we confirm the findings of Frey et al. (18) by using a strategy to analyze hypermutation that provides large databases of mutations uncomtaminated by skewing the effects of clonal amplification or antigen selection. The results therefore support the proposals (4, 17) that MSH2 (probably together with MSH6 but not MSH3; ref. 31) is actively involved in hypermutation and in class-switch recombination, in both cases acting outside its conventional role in mismatch repair. There is, however, a notable difference between the effects of PMS2-deficiency on switch recombination and on hypermutation: the pattern of switch junctions is affected, but the mutation spectrum in hypermutation is not. This might reflect the fact that although there are parallels in the initiation of hypermutation and class-switching, resolution of the initiating DNA lesion will likely proceed by quite distinct pathways in the two processes.
Acknowledgments
We thank Sean Baker for provision of PMS2-deficient mice, John Jarvis for help with cell culture, and Janet Stavnezer, and Qiang Pan and Lennart Hammarström for communicating results before publication. This work was supported in part by the Leukaemia Research Fund. C.M. and C.R. were supported by the Association for International Cancer Research.
Abbreviation
- LPS
lipopolysaccharide
References
- 1.Parham P. Immunol Rev. 1998;162:1–293. [Google Scholar]
- 2.Stavnezer J. Adv Immunol. 1996;61:79–146. doi: 10.1016/s0065-2776(08)60866-4. [DOI] [PubMed] [Google Scholar]
- 3.Lorenz M, Radbruch A. Curr Top Microbiol Immunol. 1996;217:151–169. doi: 10.1007/978-3-642-50140-1_11. [DOI] [PubMed] [Google Scholar]
- 4.Ehrenstein M R, Neuberger M S. EMBO J. 1999;18:3484–3890. doi: 10.1093/emboj/18.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 6.Rolink A, Melchers F, Andersson J. Immunity. 1995;5:319–330. doi: 10.1016/s1074-7613(00)80258-7. [DOI] [PubMed] [Google Scholar]
- 7.Casellas R, Nussenzweig A, Wuerffel R, Pelanda R, Reichlin A, Suh H, Qin X F, Besmer E, Kenter A, Rajewsky K, Nussenzweig M C. EMBO J. 1998;17:2404–2411. doi: 10.1093/emboj/17.8.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manis J P, Gu Y, Lansford R, Sonoda E, Ferrini R, Davidson L, Rajewsky K, Alt F W. J Exp Med. 1998;187:2081–2089. doi: 10.1084/jem.187.12.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bemark M, Sale J E, Kim H J, Berek C, Cosgrove R A, Neuberger M S. J Exp Med. 2000;192:1509–1514. doi: 10.1084/jem.192.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schrader C E, Edelmann W, Kucherlapati R, Stavnezer J. J Exp Med. 1999;190:323–330. doi: 10.1084/jem.190.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunnick W, Hertz G Z, Scappino L, Gritzmacher C. Nucleic Acids Res. 1993;21:365–372. doi: 10.1093/nar/21.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vora K A, Tumas-Brundage K M, Lentz V M, Cranston A, Fishel R, Manser T. J Exp Med. 1999;189:471–482. doi: 10.1084/jem.189.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker S M, Bronner C E, Zhang L, Plug A W, Robatzek M, Warren G, Elliott E A, Yu J, Ashley T, Arnheim N. Cell. 1995;82:309–319. doi: 10.1016/0092-8674(95)90318-6. [DOI] [PubMed] [Google Scholar]
- 14.Jolly C J, Klix N, Neuberger M S. Nucleic Acids Res. 1997;25:1913–1919. doi: 10.1093/nar/25.10.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonfield J K, Smith K F, Staden R. Nucleic Acids Res. 1995;23:4992–4999. doi: 10.1093/nar/23.24.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanton L W, Marcu K B. Nucleic Acids Res. 1982;10:5993–6006. doi: 10.1093/nar/10.19.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rada C, Ehrenstein M R, Neuberger M S, Milstein C. Immunity. 1998;9:135–141. doi: 10.1016/s1074-7613(00)80595-6. [DOI] [PubMed] [Google Scholar]
- 18.Frey S, Bertocci B, Delbos F, Quint L, Weill J-C, Reynaud C-A. Immunity. 1998;9:127–134. doi: 10.1016/s1074-7613(00)80594-4. [DOI] [PubMed] [Google Scholar]
- 19.Cascalho M, Wong J, Steinberg C, Wabl M. Science. 1998;279:1207–1210. doi: 10.1126/science.279.5354.1207. [DOI] [PubMed] [Google Scholar]
- 20.Winter D B, Phung Q H, Umar A, Baker S M, Tarone R E, Tanaka K, Liskay R M, Kunkel T A, Bohr V A, Gearhart P J. Proc Natl Acad Sci USA. 1998;95:6953–6958. doi: 10.1073/pnas.95.12.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim N, Bozek G, Lo J C, Storb U. J Exp Med. 1999;190:21–30. doi: 10.1084/jem.190.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong Q, Maizels N. Mol Immunol. 1999;36:83–91. doi: 10.1016/s0161-5890(99)00027-9. [DOI] [PubMed] [Google Scholar]
- 23.Kenter A L. J Exp Med. 1999;190:307–310. doi: 10.1084/jem.190.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haber J E. Cell. 1998;95:583–586. doi: 10.1016/s0092-8674(00)81626-8. [DOI] [PubMed] [Google Scholar]
- 25.Paull T T, Gellert M. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 26.de Wind N, Dekker M, Berns A, Radman M, te Riele H. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 27.Narayanan L, Fritzell J A, Baker S M, Liskay R M, Glazer P M. Proc Natl Acad Sci USA. 1997;94:3122–3127. doi: 10.1073/pnas.94.7.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugawara N, Paques F, Colaiacovo M, Haber J E. Proc Natl Acad Sci USA. 1997;94:9214–9219. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang D K, Ricciardiello L, Goel A, Chang C L, Boland C R. J Biol Chem. 2000;275:18424–18431. doi: 10.1074/jbc.M001140200. [DOI] [PubMed] [Google Scholar]
- 30.Evans E, Sugawara N, Haber J E, Alani E. Mol Cell. 2000;5:789–799. doi: 10.1016/s1097-2765(00)80319-6. [DOI] [PubMed] [Google Scholar]
- 31.Wiesendanger M, Kneitz B, Edelmann W, Scharff M D. J Exp Med. 2000;191:579–584. doi: 10.1084/jem.191.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]