Abstract
Acetyl-coenzyme A carboxylase 1 (ACC1) serves a major role in fatty acid synthesis. Previous reports have indicated that ACC1 is a promising drug target for treating human diseases, particularly cancers and metabolic diseases; however, the role of ACC1 in liver cancer and normal liver function remains unknown. In the present study, bioinformatics analysis indicated that ACC1 is overexpressed in liver cancer. Kaplan-Meier survival analysis revealed that the expression levels of ACC1 are highly associated with the prognosis of patients with liver cancer. To determine the role of ACC1 in cancer and normal liver cells, ACC1 expression was downregulated in human hepatoma Hep G2 cells and the rat liver cell line BRL 3A using RNA interference technology, which demonstrated that silencing of ACC1 significantly suppressed the cell viability in the two cell lines. Additionally, ACC1 knockdown decreased the mRNA and protein expression levels of the cell proliferation-associated genes MYCN, JUN, cyclin D1 (CCND1) and cyclin A2 (CCNA2) in BRL 3A. Furthermore, the number of cells in division phase (G2/M) was significantly reduced in the interference group, as detected by flow cytometry. Thus, ACC1 may bind and activate CCNA2, CCND1, MYCN and JUN to promote BRL 3A proliferation. In summary, the results of present study indicated that overexpression of ACC1 is significantly associated with the survival time of patients with liver cancer, and may provide insight into the association between ACC1 and cell proliferation in BRL 3A cells.
Keywords: ACC1, liver cancer, Oncomine, cell proliferation, prognosis
Introduction
Acetyl-coenzyme A (CoA) carboxylases (ACCs) are the most highly regulated enzymes in the fatty acid (FA) synthesis pathway; they catalyse the carboxylation of acetyl-CoA into malonyl-CoA, which represents the rate limiting step in de novo FA synthesis (1–4). Additionally, two isoforms of ACC encoded by two different genes in mammalian cells have been described, ACC1 and ACC2; ACC1 is highly enriched in lipogenic tissues (liver and adipose), while ACC2 is mainly expressed in oxidative tissues (heart, skeletal muscle and liver) (5,6). As they are located in a variety specialised tissues, ACC1 and ACC2 serve different metabolic roles. ACC1 generates malonyl-CoA for de novo synthesis of long-chain FAs in the cytosol, while ACC2 generates malonyl-CoA; thus carnitine palmitoyl transferase I is inhibited, preventing FA degradation in the mitochondria (3,5).
A previous study reported that ACC1 is overexpressed in different human cancer cells, and is likely involved in lipogenesis and the development and progression of tumours (7). Knockdown or chemical inhibition of ACC1 in prostate cancer cells has been successful in inducing cell apoptosis (8). Inhibition of ACC1 downregulates epidermal growth factor receptor variant III (EGFRvIII) during human glioblastoma cell proliferation and de novo lipogenesis (9). The interaction between ACC1 and breast cancer 1 indicates the possible role of ACC1 in the susceptibility to breast and ovarian cancers (10). A previous study reported that the molecule is essential for breast cancer cell survival (11). Furthermore, ACC1 regulates endothelial cell migration, and is associated with FA metabolism and the migration of endothelial cells (7). ACCs have been used as targets for treating metabolic diseases, including obesity and diabetes, and its inhibitors have been developed in clinical trials (12–15).
In the present study, the mRNA expression profile of ACC1 in certain types of cancer was investigated using the Oncomine database, and the association between alterations in ACC1 expression and clinical outcomes in numerous types of cancers, including liver, brain and kidney cancer, was analysed. Furthermore, the effects of small interfering RNA (siRNA)-mediated knockdown of ACC1 on the rat liver cell line BRL 3A and human hepatoma Hep G2 cells were determined.
Materials and methods
Oncomine database analysis
The mRNA expression levels of ACC1 in various types of cancers were analysed using the Oncomine database (https://www.oncomine.org/resource/login.html) (16). Cancer tissues were compared with normal tissues using t-tests, and the threshold was set to a P<0.0001, fold change >2 and gene ranking in the top 10%. Roessler liver normal and cancer tissue samples were used in the present study (datasets GSE1898 and GSE4024) (17).
Kaplan-Meier survival analysis
The association between ACC1 expression and survival time of patients was determined using SurvExpress (http://bioinformatica.mty.itesm.mx/SurvExpress) (18). The risk groups were produced using an optimization algorithm from the ordered prognostic index (PI), which is commonly used to generate risk groups: A log-rank test was employed among all values of arranged PI for two groups and the minimum P-value was selected as the cut-off point.
Cell culture
The liver cell lines BRL 3A and Hep G2 were obtained from the American Type Culture Collection (CRL-1442™ and HB-8065™, Manassas, VA, USA). Cells were cultured in Dulbecco's Modified Eagle's medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) at 37°C in an atmosphere containing 5% CO2.
Knockdown of ACC1 with siRNA treatment
BRL 3A and Hep G2 cells were plated with 10% FBS medium at a density of 200,000 cells/well in six-well plates, and incubated overnight at 37°C in a humidified incubator with 5% CO2. The following day, the cells were treated with 50 nmol ACC1-targeting siRNA or an identical concentration of negative control (NC) siRNA (siControl) formulated into lipid complexes using Lipofectamine® RNAiMax (Thermo Fisher Scientific, Inc.) transfection reagent. SiRNA-transfected cells were collected 24, 48 and 72 h post-transfection for quantitative polymerase chain reaction (qPCR) and western blot analysis. human (h)ACC1 siRNAs [sihACC1-(1–3) and sihACC1-2] and siControl were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The same NC siRNA was used for the transfection of BRL 3A and Hep G2 cells; the sequence was random and unrelated to the human or rat genome. The sequences of siControl and siACC1 are presented in Table I.
Table I.
Sequences of ACC1 siRNAs.
| siRNA | Sense | Antisense |
|---|---|---|
| hACC1-1 | 5′-GCUUCUACUUUCUGGAAUUTT-3′ | 5′-AAUUCCAGAAAGUAGAAGCTT-3′ |
| hACC1-2 | 5′-GCUCAUACACUUCUGAAUATT-3′ | 5′-UAUUCAGAAGUGUAUGAGCTT-3′ |
| hACC1-3 | 5′-GCAGCUAUGUUCAGAGAAUTT-3′ | 5′-AUUCUCUGAACAUAGCUGCTT-3′ |
| rACC1 | 5′-GCUGGAGACAGAAAGCUUUTT-3′ | 5′-AAGCUGGAGACAGAAAGCUTT-3′ |
| Control | 5′-UUCUCCGAACGUGUCACGUTT-3′ | 5′-ACGUGACACGUUCGGAGAATT-3′ |
ACC1, acetyl-coenzyme A carboxylase 1; h, human; r, rat; siRNA, small interfering RNA.
RNA isolation and reverse transcription-qPCR (RT-qPCR) analysis
Total RNA (from BRL 3A or HepG2 cells) was extracted using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. RNA (2 µg) was used to synthesise the first strand of cDNA using the GoScript™ Reverse Transcription system (cat. no. A5001; Promega Corporation, Madison, WI, USA). qPCR was performed on a Bio-Rad CFX96 PCR system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) with the SYBR Green Master mix (Qiagen GmbH, Hilden, Germany) and the thermocycling conditions used for all amplifications were: One cycle of 95°C for 2 min and 40 cycles of 95°C for 15 sec, 60°C for 15 sec, and 68°C for 20 sec. mRNA values were normalised to the β-actin housekeeping gene according to the 2−ΔΔCq method (19). Three replicates were performed for each sample. The primers were synthesised by Shanghai Generay Biotech Co., Ltd. (Shanghai, China) and are presented in Table II.
Table II.
Primer sequences used in reverse transcription-quantitative polymerase chain reaction.
| Genes | Forward primer | Reverse primer |
|---|---|---|
| hACC1 | 5′-GCTCCTTGTCACCTGCTTCT-3′ | 5′-CAAGGCCAAGCCATCCTGTA-3′ |
| rACC1 | 5′-TTCTTCTACTGGCGACTGAG-3′ | 5′-TCCCTGCTGATGTATTTGAT-3′ |
| CCND1 | 5′-AAAATGCCAGAGGCGGATGA-3′ | 5′-GAAAGTGCGTTGTGCGGTAG-3′ |
| JUN | 5′-GGCTGTTCATCTGTTTGTCTTCAT-3′ | 5′-CCCTTTTCTTTACGGTCTCGGT-3′ |
| MYC | 5′-ACCCAACATCAGCGGTCG-3′ | 5′-CGTGACTGTCGGGTTTTCCA-3′ |
| CCNA2 | 5′-CTTTTAGTGCCGCTGTCTCTTT-3′ | 5′-GCCCGCATACTGTTAGTGATGT-3′ |
| hβ-actin | 5′-AAATCTGGCACCACACCTTC-3′ | 5′-GGGGTGTTGAAGGTCTCAAA-3′ |
| rβ-actin | 5′-ACATCCGTAAAGACCTCTATGCCAACA-3′ | 5′-GTGCTAGGAGCCAGGGCAGTAATCT-3′ |
ACC1, acetyl-coenzyme A carboxylase 1; CCNA2, cyclin A2; CCND1, cyclin D1; h, human; r, rat.
Cell viability assay
Cells were plated at a density of 10,000 cells/well in 96-well plates. On the second day, cells were transfected with 50 nmol siACC1 or siControl. An MTT assay (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was used to measure the viability of BRL 3A and Hep G2 cells 24, 48 and 72 h post-treatment with siRNA. Briefly, 10% v/v of 5 mg/ml MTT was added to each well, after which the cells were incubated at 37°C for 4 h. The supernatant was discarded, and 0.2 ml of dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA) was added to each of the wells. The wells were gently agitated for 10 min at room temperature, and the absorbance of each well was measured at 570 nm by an ELx808 absorbance reader (BioTek Instruments, Inc., Winooski, VT, USA) (20).
Cell migration assay
Transfected cells (100,000 cells/well) were seeded in 24-well plates. The cell layer was scratched with the tip of a 10-µl pipette. The healing process was observed for 48 and 96 h. The width of the wound was measured 48 and 96 h after scratching in order to evaluate the wound healing ability of the cells. Images of the migrated cells were taken using a microscope (magnification, ×10; Nikon Eclipse 80i; Nikon Corporation, Tokyo, Japan) and the number of cells permeating the septum were counted in five random fields.
Cell cycle analysis
Nuclear DNA content can be quantitatively measured at high speed by flow cytometry. Cells were seeded into six-well plates and transfected with siACC1 or siControl at a final concentration of 50 nmol. After 48 h, the cells were harvested, washed with cold PBS, and then fixed with 70% alcohol at ~20°C overnight. The fixed cells were washed with cold PBS 2–3 times; PBS was discarded and the cells were incubated in 1 ml of PBS containing 50 µg propidium iodide (Sigma-Aldrich; Merck KGaA) and 100 µg RNase A (Sigma-Aldrich; Merck KGaA) for 30 min at 37°C. Samples were then analysed for DNA content by flow cytometry with a FACScan instrument with BD FACStation software version 5.2 (BD Biosciences, Franklin Lakes, NJ, USA).
Western blot analysis
Cells were washed with ice-cold PBS and lysed with radioimmunoprecipitation assay lysis buffer (50 mmol Tris, 150 mmol NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) containing protease inhibitors (Roche Diagnostics, Basel, Switzerland). Protein concentrations were determined by a bicinchoninic acid protein assay kit (Tiangen Biotech Co., Ltd., Beijing, China). SDS loading sample buffer was applied to the proteins, which was subsequently heated at 95°C for 5 min. Proteins (50 µg/lane) were separated by 12% SDS-PAGE and transferred to nitrocellulose membranes (GE Healthcare Life Sciences, Little Chalfont, UK). The membranes were first blocked with 5% non-fat milk in Tris-buffered saline containing 0.1% Tween-20 for 2 h at room temperature and subsequently incubated overnight at 4°C with the following primary antibodies: Rabbit anti-ACC1 (cat. no. BM4414; 1:1,000), rabbit anti-cyclin A2 (CCNA2; cat. no. PB0402; 1:1,000), rabbit anti-n-MYCN (cat. no. PB0769; 1:1,000), rabbit anti-Cyclin D1 (CCND1) (BM0771; 1:1,000) and rabbit anti-JUN (BA0208-2; 1:1,000; all from Boster Biological Technology, Pleasanton, CA, USA). The membrane was further incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (cat. no. 12-348; 1:5,000; Sigma-Aldrich; Merck KGaA) as a secondary antibody for 1 h at room temperature. Bands were visualized with BeyoECL plus reagent (Beyotime Institute of Biotechnology, Haimen, China) and the band density was measured using ImageQuant TL version 1.1 (GE Healthcare Life Sciences) with β-actin (A1978; 1:1,000; Sigma-Aldrich; Merck KGaA) as the internal reference.
Statistical analysis
All data are presented as the mean ± standard deviation of three independent experiments. SPSS 19.0 (IBM Corp., Armonk, NY, USA) was used for statistical analyses. Student's t-tests were used to compare the means of two groups. One-way analyses of variance with Bonferroni's correction was used to compare the means of three or more groups. Survival curves were generated using a Kaplan-Meier analysis and the log-rank test was used to determine P-values. P<0.05 was considered to indicate a statistically significant difference.
Results
Oncomine database analysis
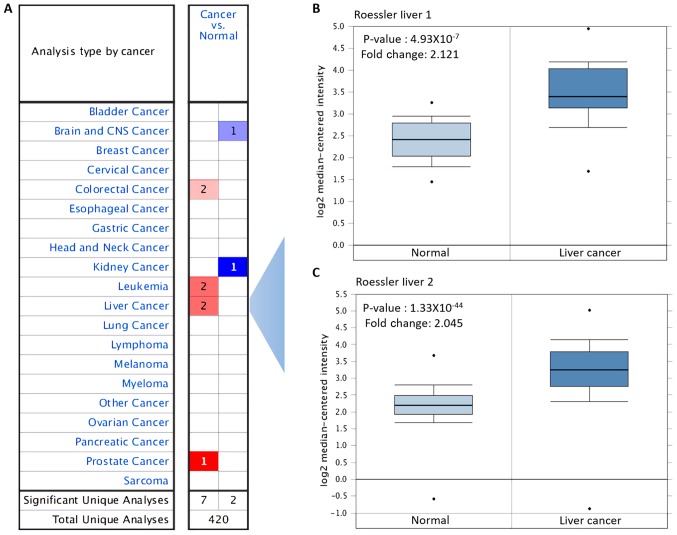
The expression levels of ACC1 mRNA in various types of cancer and normal tissue were investigated using the Oncomine database. Compared with the levels in the corresponding normal tissues, ACC1 was overexpressed in colorectal, leukaemia, liver and prostate cancer, but was downregulated in brain, central nervous system and kidney cancer (Fig. 1A). Overexpression of ACC1 in liver cancer was analysed, with P=4.93×10−7 and fold change, 2.121 reported in Roessler liver; P=1.33×10−44 and fold change, 2.045 in Roessler liver 2 (Table III; Fig. 1B and C).
Figure 1.
Expression of ACC1 in various types of cancers using on the Oncomine database. (A) mRNA expression of ACC1 in different types of cancers compared with in the corresponding normal tissues (red, overexpression; blue, downregulation). (B and C) Expression of ACC1 in liver cancer and normal tissue. ACC1, acetyl-coenzyme A carboxylase 1.
Table III.
Overexpression of ACC1 in various types of cancers.
| Cancer type | P-value | Fold change |
|---|---|---|
| Roessler liver 1 | 4.93×10−07 | 2.121 |
| Roessler liver 2 | 1.33×10−44 | 2.045 |
| Colorectal carcinoma | 1.95×10−09 | 2.297 |
| Sabates-Bellver colon | 2.24×10−05 | 2.487 |
| Coustan-Smith leukemia | 1.36×10−05 | 4.909 |
| Andersson leukemia | 5.08×10−10 | 3.418 |
| Vanaja prostate | 1.86×10−06 | 2.389 |
ACC1, acetyl-coenzyme A carboxylase 1.
Kaplan-Meier survival analysis
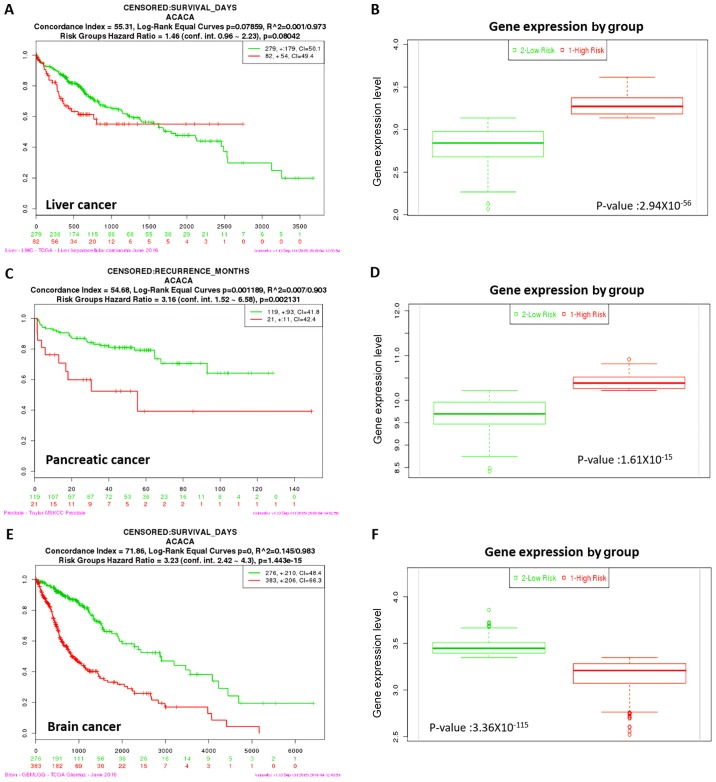
Kaplan-Meier survival analysis was performed to evaluate the prognostic value of ACC1 expression in liver, pancreatic and brain cancer. Overexpression of ACC1 was associated with increased risk, poorer prognosis and shorter overall survival times for liver and pancreatic cancer (Fig. 2). Conversely, downregulation of ACC1 was associated with lower risk, better prognosis and longer overall survival times in brain cancer. Thus, ACC1 may be considered to act as a tumour suppressor gene in brain cancer, and as an oncogene in liver and pancreatic cancer.
Figure 2.
Kaplan-Meier curves of ACC1 in liver, pancreatic and brain cancer. Censored samples are presented as +. The horizontal axis represents the time (day) to event. Outcome events, time scales, concordance indices and P-values of the log-rank test are presented. Red and green curves represent high and low-risk groups, respectively, and the number below the horizontal axis represents the number of individuals not presenting the event of the corresponding risk groups as a function of time. (A and B) High expression levels of ACC1 in liver cancer is associated with high risk, poor prognosis and shorter overall survival times. (C and D) High expression levels of ACC1 in pancreatic cancer indicates high risk, poor prognosis and shorter overall survival times. (E and F) Low expression levels of ACC1 in brain cancer indicates high risk, poor prognosis and shorter overall survival times. ACC1, acetyl-coenzyme A carboxylase 1.
Effects of siRNA-targeting ACC1 on mRNA and protein expression in Hep G2 and BRL 3A cells
To specifically silence ACC1 gene expression in Hep G2 and BRL 3A cells, cells were transfected with siRNA targeting ACC1 mRNA. RT-qPCR and western blotting were performed, and the expression of ACC1 in transfected and control cells after 48 h was detected. As presented in Fig. 3A, human (h)ACC1 mRNA levels in Hep G2 cells were reduced to 0.50±0.04 (sihACC1-1), 0.20±0.03 (sihACC1-2) and 0.40±0.05 (sihACC1-3) relative to those the expression of control cells. The protein levels of hACC1 were determined (Fig. 3B); sihACC1-2 was selected for use in later experiments. Furthermore, rat (r)ACC1 mRNA levels from the rat cell line BRL 3A were significantly reduced to 0.19±0.01 compared with in control cells (Fig. 3C); the protein expression levels of ACC1 were notably suppressed (Fig. 3D).
Figure 3.
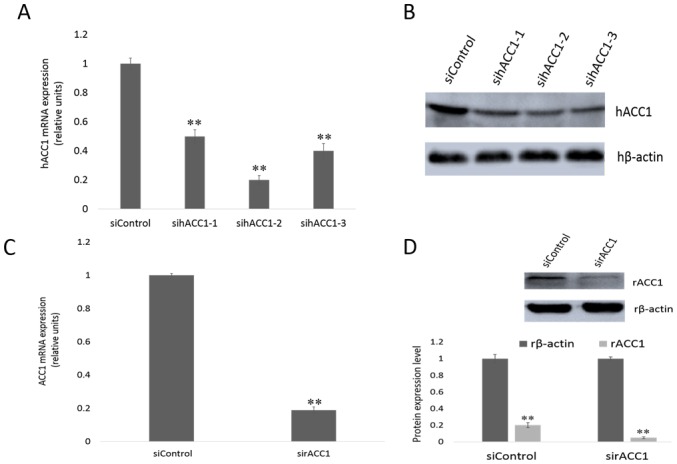
Effects of siRNA-targeting ACC1 on mRNA and protein expression in Hep G2 and BRL 3A cells. (A) mRNA expression of ACC1 in Hep G2 cells following treatment with siRNA-targeting ACC1. The data are presented as the mean ± standard deviation of three independent experiments, **P<0.01 vs. siControl. (B) Protein expression levels were assessed following treatment with siRNA-targeting ACC1 in Hep G2 cells. (C) mRNA expression of ACC1 in BRL 3A cells following treatment with siRNA-targeting ACC1. The data are presented as the mean ± standard deviation of three independent experiments, **P<0.01 vs. siControl. (D) Protein expression levels were assessed following treatment with siRNA-targeting ACC1 in BRL 3A cells. ACC1, acetyl-coenzyme A carboxylase 1; h, human; r, rat; si, small interfering.
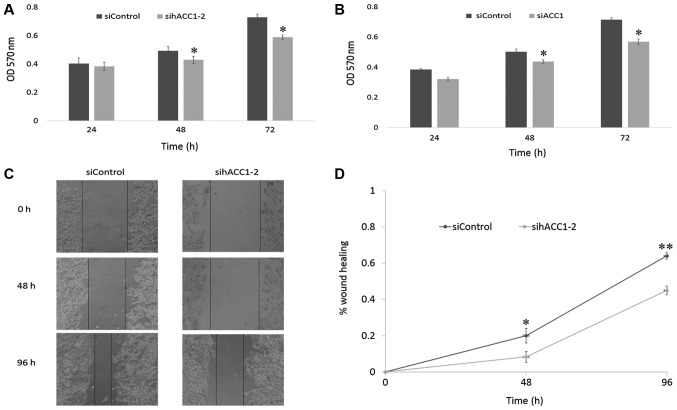
Effects of siRNA-targeting ACC1 on cell viability and migration
To investigate the effects of ACC1 RNAi on cell viability, an MTT assay was used to measure the viability of Hep G2 and BRL 3A cells 24, 48 and 72 h post-transfection with siRNA targeting ACC1. As presented in Fig. 4A and B, cell viability began to decline after 24 h, and significantly decreased at 48 and 72 h in Hep G2 and BRL 3A cells, compared with the siControl. To further investigate the effects of siACC1 on cell migration, Hep G2 cells were treated with sihACC1-2, which significantly reduced cell migration at 48 and 96 h compared with the siControl, respectively (Fig. 4C and D).
Figure 4.
Effects of siRNA-targeting ACC1 on cell viability and migration. (A) Viability of siRNA-transfected and control Hep G2 cells 24, 48 and 72 h post-transfection as assessed with an MTT assay. (B) Viability of siRNA-transfected and control BRL 3A cells 24, 48 and 72 h post-transfection as assessed with an MTT assay. (C and D) Wound healing assays of Hep G2 cells following treatment with sihACC1-2. The data are presented as the mean ± standard deviation of three independent experiments, *P<0.05, **P<0.01 vs. siControl. ACC1, acetyl-coenzyme A carboxylase 1; h, human; OD, optical density; si, small interfering.
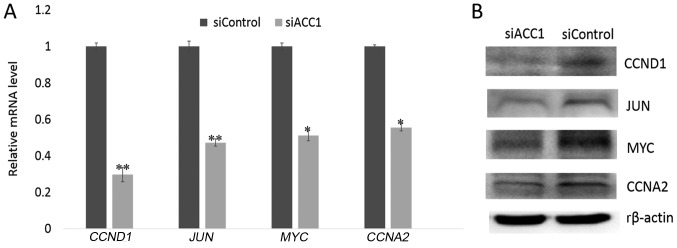
Effects of siRNA-targeting ACC1 on cell proliferation- associated genes
To further investigate the effects of ACC1 RNA interference (RNAi) on cell proliferation, RT-qPCR and western blot analyses were performed to determine the expression of proliferation-associated genes 48 h following interference. The results revealed that the expression of cell proliferation-associated genes, including CCND1, JUN, MYCN and CCNA2, were significantly decreased in siACC1-transfected cells compared with the control group (Fig. 5A); a similar trend in protein was observed (Fig. 5B).
Figure 5.
Effects of siRNA-targeting ACC1 on cell proliferation-associated genes. (A) Expression of cell proliferation-associated genes at the mRNA level as determined by reverse transcription-quantitative polymerase chain reaction analysis. (B) Expression of cell proliferation-related genes at the protein level as assessed by western blot analysis. The data are presented as the mean ± standard deviation of three independent experiments, *P<0.05, **P<0.01 vs. siControl. ACC1, acetyl-coenzyme A carboxylase 1; CCNA2, cyclin A2; CCND1, cyclin D1; r, rat; si, small interfering.
Effects of siRNA-targeting ACC1 on the cell cycle of BRL 3A cells
To investigate how ACC1 affects cell proliferation, flow cytometry was used to analyse the cell cycle distribution of BRL 3A cells. Knockdown of ACC1 expression significantly decreased the percentage of G2/M phase cells (16.73±0.32%) compared with that of control cells (24.10±0.30%; Fig. 6). Furthermore, the percentage of G0/G1 phase cells significantly increased; however, the percentage of S phase remained unchanged. Therefore, ACC1 may modulate BRL 3A cell proliferation by controlling the cell cycle.
Figure 6.
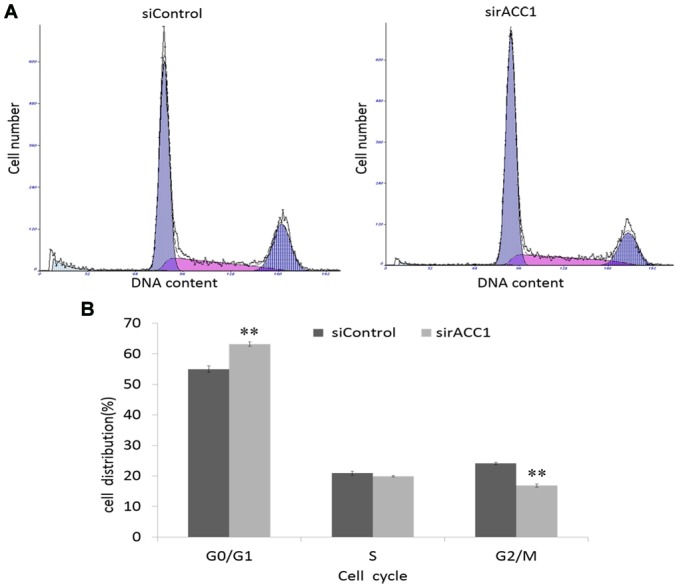
Effects of siRNA-targeting ACC1 on the cell cycle of BRL 3A cells. (A) Cell cycle distribution of siRNA-transfected and control cells 48 h post-transfection as assessed via flow cytometry. (B) Percentage of cells in each phase as measured by flow cytometry. The data are presented as the mean ± standard deviation of three independent experiments, **P<0.01 vs. siControl. ACC1, acetyl-coenzyme A carboxylase 1; si, small interfering.
Discussion
In the present study, the Oncomine database was employed to analyse the association between the overexpression of ACC1 and liver cancer, and determine whether ACC1 affects the survival time of patients with liver cancer. The extent of ACC1 mRNA overexpression in liver cancer was closely associated with the survival time of patients. To determine the role of ACC1 in cancer and normal liver cells, ACC1 expression in Hep G2 and BRL 3A cells was knocked down using RNAi. Downregulation of ACC1 significantly suppressed Hep G2 and BRL 3A cell proliferation and decreased BRL 3A cells in G2/M phase (from 24.10±0.30 to 16.73±0.32%).
To further understand the mechanisms underlying ACC1-mediated BRL 3A cell proliferation, the expression of cell proliferation-associated genes, including MYCN, JUN, CCND1 and CCNA2, was analysed. MYCN is a key regulator of mammalian cell proliferation and is required for oncogenesis (21). Numerous studies have demonstrated that MYCN can induce the transition of liver cells from G0 phase to G1 phase (22,23). CCND1 is another oncogene that drives cell cycle progression, and its presence signifies that liver cells are entering the G1 phase (24). Previous studies have reported that CCND1 is significantly upregulated in liver cells following partial hepatectomy; the protein regulates progression via the G1-phase checkpoint and promotes cell proliferation (25,26). MYCN can induce CCND1-mediated cell cycle progression (27), whilst treatment of MCF7 cells with antisense MYCN can inhibit cell proliferation by decreasing CCND1 expression (28–30). Thus, MYCN may induce CCND1 and enhance the activity of CCND1-cyclin-dependent kinase complexes to drive cell cycle progression and transformation (31). JUN is necessary for cells to progress to G1 phase of the cell cycle and serves as an important regulator for initiating liver regeneration (23,32). Additionally, JUN regulates the transcriptional levels of CCND1, which is required for the efficient proliferation of mouse fibroblasts (33,34). Cyclin A2 (CCNA2), a core component of the cell cycle, is involved in the G2/M transition; thus, it may also affect cell proliferation (35). As a key member of the cyclin family, CCNA2 significantly promotes hepatocyte cell cycle progression (36). The present study revealed that knockdown of ACC1 induced the downregulation of CCNA2 and CCND1, and decreased BRL 3A cells in G2/M phase. This suggested that ACC1 may bind and activate CCNA2, CCND1, MYCN and JUN to promote BRL 3A proliferation. Thus, downregulation of CCNA2 and CCND1 may affect cells decreased in G2/M phase (from 24.10±0.30 to 16.73±0.32%); however, further investigation is required to determine the exact mechanism in detail.
As a master regulator of FA metabolism, ACC1 converts acetyl-CoA to malonyl-CoA, which is a critical substrate for FA synthesis. Compared with normal cells, cancer cells synthesise FAs at higher rates (9). The factors involved in lipid synthesis are also observed in the proliferation, cell growth and viability of certain cancers, including lung (37), colon (38), prostate (8) and breast cancer (11). In LNCaP cells, ACC1 RNAi-mediated silencing induced cell growth inhibition and cytotoxicity (39). In non-small-cell lung cancer cells, ACC inhibition reduces de novo lipid synthesis, and decreases cell growth and viability (40). In human U87 EGFRvIII cells, ACC1/2 knockdown not only suppresses de novo lipogenesis, but notably reduces U87 EGFRvIII cellular proliferation and viability (9). In yeast, inactivation of ACC1 completely inhibits vegetative growth and causes cell death following treatment with FAs; further investigation has revealed that siACC1 induces severe abnormalities in spindle formation, arrests cells in the G2/M phase of the cell cycle, and suppressed cell viability (41,42). However, the importance of increased lipogenesis in tumour cells, and the mechanisms by which interference with this increased lipogenesis promotes tumour cell proliferation, growth and viability require further investigation.
To the best of our knowledge, the present study is the first to determine the association between ACC1 and cell proliferation in the rat liver cell line BRL 3A. ACC1 is not only important for the proliferation of Hep G2 cancer cells, but may also be necessary for the proliferation of BRL 3A cells. ACC1 is highly expressed in liver tissue and hepatocytes, and is the sole regulator of FA synthesis (43). Mutant mice lacking ACC1 exhibit embryonic lethality, which suggests that de novo FA synthesis is essential for embryonic development (44). The present study suggests that ACC1 is a valuable drug target for treating various metabolic pathologies, including hepatic steatosis, non-alcoholic fatty liver disease, metabolic syndrome, obesity and hepatic insulin resistance (7). Thus, ACC1 may be a potential target for the development of novel approaches to liver cancer prevention and therapy. ACC1 can upregulate the cell proliferation-related genes MYCN, JUN, CCND1 and CCNA2 in the rat liver cell line BRL 3A. In future studies, the association between FA synthesis and cell proliferation-related genes should be investigated to improve understanding of the function and targets of ACC1. Additionally, whether ACC1 performs the same functions in vivo merits further investigation by utilising a model of rat partial hepatectomy and knockout of ACC1 via CRISPR-Cas9 technology.
Acknowledgements
The authors' thank members of our laboratory for critical discussions.
Funding
The present study was supported by the Research Fund for the Postdoctoral Program of He'nan, China, Natural Science Foundation of China (grant no. 31572270), the National Fostering Science Foundation Project of Henan Normal University (grant no. 2016PL21) and the Key Scientific Research Projects of Henan Higher Education (grant no. 15A180007).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
CX made substantial contributions to the conception of the present study. BY and LY performed the experiments and wrote the manuscript; QW made substantial contributions to the design of the present study, and interpreted the data. All authors read and approved the final version of the manuscript for publication.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Harwood HJ., Jr Treating the metabolic syndrome: Acetyl-CoA carboxylase inhibition. Expert Opin Ther Targets. 2005;9:267–281. doi: 10.1517/14728222.9.2.267. [DOI] [PubMed] [Google Scholar]
- 2.Kim KH. Regulation of mammalian acetyl-coenzyme A carboxylase. Annu Rev Nutr. 1997;17:77–99. doi: 10.1146/annurev.nutr.17.1.77. [DOI] [PubMed] [Google Scholar]
- 3.McGarry JD, Mannaerts GP, Foster DW. A possible role for malonyl-CoA in the regulation of hepatic fatty acid oxidation and ketogenesis. J Clin Invest. 1977;60:265–270. doi: 10.1172/JCI108764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nugteren DH. The enzymic chain elongation of fatty acids by rat-liver microsomes. Biochim Biophys Acta. 1965;106:280–290. doi: 10.1016/0005-2760(65)90036-6. [DOI] [PubMed] [Google Scholar]
- 5.Munday MR. Regulation of mammalian acetyl-CoA carboxylase. Biochem Soc Trans. 2002;30:1059–1064. doi: 10.1042/bst030a101c. [DOI] [PubMed] [Google Scholar]
- 6.Saggerson D. Malonyl-CoA, a key signaling molecule in mammalian cells. Annu Rev Nutr. 2008;28:253–272. doi: 10.1146/annurev.nutr.28.061807.155434. [DOI] [PubMed] [Google Scholar]
- 7.Glatzel DK, Koeberle A, Pein H, Löser K, Stark A, Keksel N, Werz O, Muller R, Bischoff I, Fürst R. Acetyl-CoA carboxylase 1 regulates endothelial cell migration by shifting the phospholipid composition. J Lipid Res. 2018;59:298–311. doi: 10.1194/jlr.M080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brusselmans K, De Schrijver E, Verhoeven G, Swinnen JV. RNA interference-mediated silencing of the acetyl-CoA-carboxylase-alpha gene induces growth inhibition and apoptosis of prostate cancer cells. Cancer Res. 2005;65:6719–6725. doi: 10.1158/0008-5472.CAN-05-0571. [DOI] [PubMed] [Google Scholar]
- 9.Jones JE, Esler WP, Patel R, Lanba A, Vera NB, Pfefferkorn JA, Vernochet C. Inhibition of Acetyl-CoA Carboxylase 1 (ACC1) and 2 (ACC2) reduces proliferation and de novo lipogenesis of EGFRvIII human glioblastoma cells. PLoS One. 2017;12:e0169566. doi: 10.1371/journal.pone.0169566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magnard C, Bachelier R, Vincent A, Jaquinod M, Kieffer S, Lenoir GM, Venezia ND. BRCA1 interacts with acetyl-CoA carboxylase through its tandem of BRCT domains. Oncogene. 2002;21:6729–6739. doi: 10.1038/sj.onc.1205915. [DOI] [PubMed] [Google Scholar]
- 11.Chajes V, Cambot M, Moreau K, Lenoir GM, Joulin V. Acetyl-CoA carboxylase alpha is essential to breast cancer cell survival. Cancer Res. 2006;66:5287–5294. doi: 10.1158/0008-5472.CAN-05-1489. [DOI] [PubMed] [Google Scholar]
- 12.Abu-Elheiga L, Matzuk MM, Abo-Hashema KA, Wakil SJ. Continuous fatty acid oxidation and reduced fat storage in mice lacking acetyl-CoA carboxylase 2. Science. 2001;291:2613–2616. doi: 10.1126/science.1056843. [DOI] [PubMed] [Google Scholar]
- 13.Harriman G, Greenwood J, Bhat S, Huang X, Wang R, Paul D, Tong L, Saha AK, Westlin WF, Kapeller R, Harwood HJ., Jr Acetyl-CoA carboxylase inhibition by ND-630 reduces hepatic steatosis, improves insulin sensitivity, and modulates dyslipidemia in rats. Proc Natl Acad Sci USA. 2016;113:E1796–E1805. doi: 10.1073/pnas.1520686113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harwood HJ., Jr Acetyl-CoA carboxylase inhibition for the treatment of metabolic syndrome. Curr Opin Investig Drugs. 2004;5:283–289. [PubMed] [Google Scholar]
- 15.Schreurs M, van Dijk TH, Gerding A, Havinga R, Reijngoud DJ, Kuipers F. Soraphen, an inhibitor of the acetyl-CoA carboxylase system, improves peripheral insulin sensitivity in mice fed a high-fat diet. Diabetes Obes Metab. 2009;11:987–991. doi: 10.1111/j.1463-1326.2009.01078.x. [DOI] [PubMed] [Google Scholar]
- 16.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX, Wang XW. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aguirre-Gamboa R, Gomez-Rueda H, Martínez-Ledesma E, Martínez-Torteya A, Chacolla-Huaringa R, Rodriguez- Barrientos A, Tamez-Peña JG, Treviño V. SurvExpress: An online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One. 2013;8:e74250. doi: 10.1371/journal.pone.0074250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Boncler M, Różalski M, Krajewska U, Podsędek A, Watala C. Comparison of PrestoBlue and MTT assays of cellular viability in the assessment of anti-proliferative effects of plant extracts on human endothelial cells. J Pharmacol Toxicol Methods. 2014;69:9–16. doi: 10.1016/j.vascn.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Bouchard C, Staller P, Eilers M. Control of cell proliferation by Myc. Trends Cell Biol. 1998;8:202–206. doi: 10.1016/S0962-8924(98)01251-3. [DOI] [PubMed] [Google Scholar]
- 22.Miquet JG, Freund T, Martinez CS, Gonzalez L, Diaz ME, Micucci GP, Zotta E, Boparai RK, Bartke A, Turyn D, Sotelo AI. Hepatocellular alterations and dysregulation of oncogenic pathways in the liver of transgenic mice overexpressing growth hormone. Cell Cycle. 2013;12:1042–1057. doi: 10.4161/cc.24026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morello D, Lavenu A, Babinet C. Differential regulation and expression of jun, c-fos and c-myc proto-oncogenes during mouse liver regeneration and after inhibition of protein synthesis. Oncogene. 1990;5:1511–1519. [PubMed] [Google Scholar]
- 24.Pauklin S, Vallier L. The cell-cycle state of stem cells determines cell fate propensity. Cell. 2013;155:135–147. doi: 10.1016/j.cell.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelsen CJ, Rickheim DG, Timchenko NA, Stanley MW, Albrecht JH. Transient expression of cyclin D1 is sufficient to promote hepatocyte replication and liver growth in vivo. Cancer Res. 2001;61:8564–8568. [PubMed] [Google Scholar]
- 26.Rickheim DG, Nelsen CJ, Fassett JT, Timchenko NA, Hansen LK, Albrecht JH. Differential regulation of cyclins D1 and D3 in hepatocyte proliferation. Hepatology. 2002;36:30–38. doi: 10.1053/jhep.2002.33996. [DOI] [PubMed] [Google Scholar]
- 27.Mateyak MK, Obaya AJ, Sedivy JM. c-Myc regulates cyclin D-Cdk4 and -Cdk6 activity but affects cell cycle progression at multiple independent points. Mol Cell Biol. 1999;19:4672–4683. doi: 10.1128/MCB.19.7.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carroll JS, Swarbrick A, Musgrove EA, Sutherland RL. Mechanisms of growth arrest by c-myc antisense oligonucleotides in MCF-7 breast cancer cells: Implications for the antiproliferative effects of antiestrogens. Cancer Res. 2002;62:3126–3131. [PubMed] [Google Scholar]
- 29.Daksis JI, Lu RY, Facchini LM, Marhin WW, Penn LJ. Myc induces cyclin D1 expression in the absence of de novo protein synthesis and links mitogen-stimulated signal transduction to the cell cycle. Oncogene. 1994;9:3635–3645. [PubMed] [Google Scholar]
- 30.Perez-Roger I, Kim SH, Griffiths B, Sewing A, Land H. Cyclins D1 and D2 mediate myc-induced proliferation via sequestration of p27(Kip1) and p21(Cip1) EMBO J. 1999;18:5310–5320. doi: 10.1093/emboj/18.19.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oswald F, Lovec H, Möröy T, Lipp M. E2F-dependent regulation of human MYC: Trans-activation by cyclins D1 and A overrides tumour suppressor protein functions. Oncogene. 1994;9:2029–2036. [PubMed] [Google Scholar]
- 32.Riehle KJ, Campbell JS, McMahan RS, Johnson MM, Beyer RP, Bammler TK, Fausto N. Regulation of liver regeneration and hepatocarcinogenesis by suppressor of cytokine signaling 3. J Exp Med. 2008;205:91–103. doi: 10.1084/jem.20070820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herber B, Truss M, Beato M, Müller R. Inducible regulatory elements in the human cyclin D1 promoter. Oncogene. 1994;9:1295–1304. [PubMed] [Google Scholar]
- 34.Musgrove EA, Lee CS, Buckley MF, Sutherland RL. Cyclin D1 induction in breast cancer cells shortens G1 and is sufficient for cells arrested in G1 to complete the cell cycle. Proc Natl Acad Sci USA. 1994;91:8022–8026. doi: 10.1073/pnas.91.17.8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gong D, Pomerening JR, Myers JW, Gustavsson C, Jones JT, Hahn AT, Meyer T, Meyer T, Ferrell JE., Jr Cyclin A2 regulates nuclear-envelope breakdown and the nuclear accumulation of cyclin B1. Curr Biol. 2007;17:85–91. doi: 10.1016/j.cub.2006.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garnier D, Loyer P, Ribault C, Guguen-Guillouzo C, Corlu A. Cyclin-dependent kinase 1 plays a critical role in DNA replication control during rat liver regeneration. Hepatology. 2009;50:1946–1956. doi: 10.1002/hep.23225. [DOI] [PubMed] [Google Scholar]
- 37.Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, Hingorani SR, Tuveson DA, Thompson CB. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 38.Zhan Y, Ginanni N, Tota MR, Wu M, Bays NW, Richon VM, Kohl NE, Bachman ES, Strack PR, Krauss S. Control of cell growth and survival by enzymes of the fatty acid synthesis pathway in HCT-116 colon cancer cells. Clin Cancer Res. 2008;14:5735–5742. doi: 10.1158/1078-0432.CCR-07-5074. [DOI] [PubMed] [Google Scholar]
- 39.Rysman E, Brusselmans K, Scheys K, Timmermans L, Derua R, Munck S, Van Veldhoven PP, Waltregny D, Daniëls VW, Machiels J, et al. De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Res. 2010;70:8117–8126. doi: 10.1158/0008-5472.CAN-09-3871. [DOI] [PubMed] [Google Scholar]
- 40.Svensson RU, Parker SJ, Eichner LJ, Kolar MJ, Wallace M, Brun SN, Lombardo PS, Van Nostrand JL, Hutchins A, Vera L, et al. Inhibition of acetyl-CoA carboxylase suppresses fatty acid synthesis and tumor growth of non-small-cell lung cancer in preclinical models. Nat Med. 2016;22:1108–1119. doi: 10.1038/nm.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hasslacher M, Ivessa AS, Paltauf F, Kohlwein SD. Acetyl-CoA carboxylase from yeast is an essential enzyme and is regulated by factors that control phospholipid metabolism. J Biol Chem. 1993;268:10946–10952. [PubMed] [Google Scholar]
- 42.Al-Feel W, DeMar JC, Wakil SJ. A Saccharomyces cerevisiae mutant strain defective in acetyl-CoA carboxylase arrests at the G2/M phase of the cell cycle. Proc Natl Acad Sci USA. 2003;100:3095–3100. doi: 10.1073/pnas.0538069100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Savage DB, Choi CS, Samuel VT, Liu ZX, Zhang D, Wang A, Zhang XM, Cline GW, Yu XX, Geisler JG, et al. Reversal of diet-induced hepatic steatosis and hepatic insulin resistance by antisense oligonucleotide inhibitors of acetyl-CoA carboxylases 1 and 2. J Clin Invest. 2006;116:817–824. doi: 10.1172/JCI27300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abu-Elheiga L, Matzuk MM, Kordari P, Oh W, Shaikenov T, Gu Z, Wakil SJ. Mutant mice lacking acetyl-CoA carboxylase 1 are embryonically lethal. Proc Natl Acad Sci USA. 2005;102:12011–12016. doi: 10.1073/pnas.0505714102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.