Abstract
Neural stem cells (NSCs) are a class of self-renewing and undifferentiated progenitor cells that retain the ability to differentiate to neurons, astrocytes and oligodendrocytes. MicroRNAs (miRNAs) are small noncoding RNAs that serve crucial roles in regulating a number of cellular processes, including cell proliferation, differentiation and apoptosis. Our previous GeneChip data indicated that the expression of miR-329-3p was increased in neurons compared with NSCs. However, whether miRNA-329-3p participates in regulating NSC function remains to be elucidated. In the present study, it was identified that the expression of miR-329-3p was upregulated in NSCs during neuronal differentiation, whereas expression of transcription factor E2F1 (E2F1), a putative target gene of miR-329-3p, was downregulated. Using luciferase reporter assays, it was confirmed that miR-329-3p regulated E2F1 expression. As differentiation has been demonstrated to limit the proliferative capacity of NSCs, the effects of miR-329-3p and E2F1 modulation on NSC proliferation were examined. Forced overexpression of miR-329-3p or RNA-mediated silencing of E2F1 inhibited NSC proliferation, and overexpression of miR-329-3p also inhibited E2F1 expression. Notably, ectopic expression of E2F1 reversed the inhibition of NSC proliferation induced by miR-329-3p overexpression. These results indicated that miR-329-3p may serve crucial roles in regulating the proliferation of NSCs, at least in part via inhibition of E2F1 expression. These data improve the understanding of the microRNA-mRNA regulatory network that controls NSC proliferation.
Keywords: neural stem cell, microRNA, microRNA-329-3p, transcription factor E2F1, proliferation
Introduction
Neural stem cells (NSCs) are undifferentiated self-renewing progenitor cells that are able to differentiate into neurons, astrocytes or oligodendrocytes (1). Previous studies have identified NSC transplantation as a promising regenerative therapy for a number of central nervous system disorders, including spinal cord injuries, neurodegenerative diseases, traumatic brain injury and ischemic brain injury (2,3). However, this potentially powerful technology is underdeveloped for clinical application at present, and additional studies are required to explore the mechanisms that regulate NSC proliferation and differentiation.
MicroRNAs (miRNAs) are small noncoding RNAs, measuring 21–22 nucleotides in length, that regulate gene expression by binding to partially complementary sequences in the 3′untranslated region (UTR) of one or more target mRNAs (4). miRNAs have been demonstrated to serve crucial roles in numerous cell processes, including proliferation, differentiation and apoptosis (5–7). Consistent with this, miRNAs are abundant in the central nervous system, where they have critical roles in neurogenesis and neuronal physiology (8,9). In addition, previous data have indicated that miRNAs are involved in regulating stem cell proliferation and differentiation by controlling the expression of a number of stem cell regulators (10–12).
Based on a pilot miRNA microarray study, it was hypothesized that miR-329-3p may be expressed at decreased levels in NSCs compared with terminally differentiated neurons. However, the expression levels and role of this miRNA in NSC biology remain unclear. The present study identified that miR-329-3p expression was upregulated during NSC differentiation, while that of transcription factor E2F1 (E2F1), which was confirmed to be a miR-329-3p target gene, was decreased. Downregulation of E2F1 inhibited the proliferation of NSCs. Transfection of NSCs with miR-329-3p mimic inhibited E2F1 expression and cell proliferation, which was reversed by the restoration of E2F1 expression. These results indicated that miR-329-3p may serve an important role in regulating NSC proliferation by targeting E2F1 mRNA. The results of the present study may contribute to the development of regenerative therapies based on NSC transplantation.
Materials and methods
Cell culture and transfection
The NSCs used in the present study were derived from the hippocampus and the subventricular zone of the brain of Sprague-Dawley rats at 14.5 days of gestation, and were purchased from Cyagen Biosciences, Inc. (Guangzhou, China; RASNF-01001). The cells were cultured in Dulbecco's modified Eagle's medium/F12 medium (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) with 2% B27 supplement without vitamin A (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 20 ng/ml basic fibroblast growth factor (PeproTech, Inc., Rocky Hill, NJ, USA), 20 ng/ml epidermal growth factor (PeproTech, Inc.), and 2 mM L-glutamine (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Cells were maintained at 37°C in a 5% CO2 humidified atmosphere and the culture medium was changed every 2–3 days. The cells were allowed to proliferate to form neurospheres.
NSCs were transfected with miR-329-3p-specific or negative control (NC) mimics, E2F1-targeting or NC small interfering RNAs (siRNAs) (50 nmol/l) (GenePharma Co., Ltd., Suzhou, China), and E2F1 overexpression or control vectors (Generay Biotech Co., Ltd., Shanghai, China) using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.), in accordance with the manufacturer's protocol. Subsequent experiments took place 24 h after transfection. The mimic and siRNA sequences are presented in Table I. The transfection efficiency was evaluated by reverse transcription quantitative polymerase chain reaction (RT-qPCR) as described subsequently.
Table I.
Sequences of mimics, siRNAs and primers used in the present study.
| A, Mimic and siRNA sequences | ||
|---|---|---|
| Name | Direction | Sequences |
| miR-329-3p mimic | Sense | 5′-AACACACCCAGCUAACCUUUUU-3′ |
| Antisense | 5′-AAAGGUUAGCUGGGUGUGUUUU-3′ | |
| E2F1 siRNA | Sense | 5′-GCAGAAACGACGCAUCUAUTT-3′ |
| Antisense | 5′-AUAGAUGCGUCGUUUCUGCTT-3′ | |
| B, Reverse transcription-quantitative polymerase chain reaction primers | ||
| Name | Direction | Sequences |
| miR-329-3p | Forward | 5′-GCGCGAACACACCCAGCTAACCTTTTT-3′ |
| U6 | Forward | 5′-CTCGCTTCGGCAGCACA-3′ |
| Reverse | 5′-AACGCTTCACGAATTTGCGT-3′ | |
| E2F1 | Forward | 5′-GAAGAAGACCGGTTGTCACC-3′ |
| Reverse | 5′-GAAATCCAGAGGGGTCAAGTC −3′ | |
| GAPDH | Forward | 5′-ATTCCATGGCACCGTCAAGGCTGA-3′ |
| Reverse | 5′-TTCTCCATGGTGGTGAAGACGCCA-3′ | |
miR, microRNA; si, small interfering; E2F1, transcription factor E2F1.
Cell differentiation
NSCs were cultured for 24 h in culture dishes which had been coated with extracellular matrix proteins (Matrigel; Corning Incorporated, Corning, NY, USA) at 37°C overnight, and neuronal differentiation was then induced by changing the medium to neurobasal medium supplemented with 2% B27 (both from Gibco; Thermo Fisher Scientific, Inc.) and 2 mM l-glutamine (Sigma-Aldrich; Merck KGaA).
GeneChip microarray assay
miRNAs were isolated from NSCs and differentiated neurons with the miRNeasy kit (Qiagen GmbH, Hilden, Germany), as described by Zou et al (13). GeneChip microarray assays were performed by CapitalBio Technology (Beijing, China), also as previously described (13). The expression of key miRNAs was further verified by RT-qPCR.
RT-qPCR
Total RNA was isolated from NSCs using TRIzol® reagent (Takara Biotechnology Co., Ltd., Dalian, China), in accordance with the manufacturer's protocol. For miR-329-3p detection, miRNAs were reverse transcribed using the miRcute Plus miRNA First-Strand cDNA Synthesis kit (Tiangen Biotech Co., Ltd., Beijing, China) and amplified by qPCR using a miRcute Plus miRNA SYBR Green qPCR Detection kit (Tiangen Biotech Co., Ltd.). The reverse transcription of the miRNAs consisted of 60 min at 42°C and 3 min at 95°C. The PCR cycle conditions included an initial denaturation step at 95°C for 15 min, followed by 40 cycles of 94°C for 20 sec and 60°C for 34 sec. For mRNAs, RNA was reverse transcribed using a PrimeScript™ RT Reagent kit (Takara Biotechnology Co., Ltd.) and amplified using SYBR Premix Ex Taq™ II (Takara Biotechnology Co., Ltd.), in accordance with the manufacturer's protocol. The reverse transcription process consisted of 15 min at 37°C and 5 sec at 85°C, followed by storage at 4°C. The qPCR cycle conditions included an initial denaturation step at 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 34 sec. RT-qPCR was performed using an ABI 7500 system (Thermo Fisher Scientific, Inc.). GAPDH and U6 snRNA were used as internal controls for the detection of mRNA and miRNA. The 2−ΔΔCq method was used to calculate relative gene expression (14). Primer sequences were synthesized by Sangon Biotech Co., Ltd., (Shanghai, China), with the exception of a universal reverse primer for miR-329-3p (Tiangen Biotech Co., Ltd.). The primer sequences are listed in Table I.
Western blot analysis
Western blot analysis was performed in accordance with standard methods. Cells were lysed on ice for 30 min in lysis buffer containing phenylmethylsulfonyl fluoride (Beyotime Institute of Biotechnology, Haimen, China) and centrifuged at 12,000 × g for 20 min at 4°C. The supernatants were collected and protein was quantified using a BCA Protein Assay kit (Wanleibio, Co., Ltd., Shanghai, China). A total of 30 µg/lane total protein were separated by 12% SDS-PAGE and electrotransferred to polyvinylidene fluoride membranes. The membranes were blocked by incubation in 2.5% (wt/vol) skimmed milk powder in TBS containing 0.1% Tween-20 (TBST) for 2 h at room temperature, washed with TBST 3 times, and then incubated at 4°C overnight with rabbit anti-rat polyclonal E2F1 antibody (1:500 dilution; cat. no. WL02394; Wanleibio Co., Ltd) or rabbit anti-rat polyclonal β-actin antibody (1:1,000 dilution; cat. no. WL0002c; Wanleibio Co., Ltd) diluted in TBST. The membranes were washed again in TBST and incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:5,000 dilution; cat. no. AS014; ABclonal Biotech Co., Ltd., Woburn, MA, USA) for 2 h at room temperature. Finally, the membranes were washed 3 times for 10 min each and the protein bands were visualized using an enhanced chemiluminescence detection system. Densitometric analysis was performed using Image-Pro Plus 6.0 (Media Cybernetics, Inc., Rockville, MD, USA).
Cell proliferation assay
A Cell Counting Kit-8 assay (CCK-8; Beyotime Institute of Biotechnology) and a 5-ethynyl-2′-deoxyuridine (EdU) incorporation assay (Guangzhou RiboBio Co., Ltd., Guangzhou, China) were used to measure NSCs proliferation, in accordance with the manufacturers' protocols. For the CCK-8 assay, NSCs were plated at 5×103/100 µl/well (n=5/condition) in 96-well plates for 24 h and then transfected with the mimics or siRNAs using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.). Following incubation for 0, 24, 48 or 72 h at 37°C, 10 µl CCK-8 reagent was added to each well and the plates were incubated at 37°C for an additional 2 h. The absorbance at 450 nm was measured using a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA). For the EdU incorporation assay, the cells were plated at 1×104 cells/well in 96-well plates, transfected with the mimics or siRNAs for 48 h using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.), and incubated in culture medium containing 50 µM EdU for 4 h. The cells were washed with PBS for 5 min, fixed in 4% paraformaldehyde for 30 min, incubated in 50 µl 2 mg/ml glycine solution for 5 min, washed with PBS for 5 min, permeabilized with 100 µl 0.5% Triton X-100/PBS for 10 min, washed again with PBS, and then incubated in 100 µl of 1X Apollo® staining solution (Guangzhou RiboBio Co., Ltd.) for 30 min. The cells were washed 3 times for 10 min each with 0.5% Triton X-100/PBS, incubated with 100 µl 1X Hoechst 33342 for 30 min, and then washed twice with PBS. All of these procedures were performed at room temperature. Finally, the cells were observed under a fluorescence microscope (Olympus Corporation, Tokyo, Japan; magnification, ×400).
Prediction of miRNA target genes
Potential targets for miR-329-3p were predicted using the bioinformatics databases TargetScan (http://www.targetscan.org/) and miRDB (http://mirdb.org/index.html) (15,16).
Dual-luciferase reporter assay
The wild-type (wt) or mutated (mt) E2F1 3′-UTR sequence was cloned into the pmirGLO Luciferase reporter vector (Generay Biotech Co., Ltd.) and transfected into 293 cells (American Type Culture Collection, Manassas, VA, USA). Cells were seeded at 1×105 cells per well in 24-well plates and co-transfected at 37°C with 60 nM miR-329-3p or control mimic together with 1 µg wt or mt E2F1 3′-UTR vectors using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 24 h from transfection, the cells were given fresh medium and cultured for an extra 24 h. Subsequently, luciferase activity was measured using the Dual-Luciferase Reporter reagent (Promega Corporation, Madison, WI, USA), in accordance with the manufacturer's protocol. Relative luciferase activity was calculated as follows: (Firefly luciferase activity/Renilla luciferase activity) ×100%.
Statistical analysis
GraphPad Prism 7.0 software (GraphPad Software, Inc., La Jolla, CA, USA) was used for all analyses. Data are presented as the mean ± standard deviation. Differences between groups were analyzed using unpaired Student's t-test and one- or two-way analysis of variance, followed by a Dunnett's or Sidak's multiple comparisons test, as appropriate. P<0.05 was considered to indicate a statistically significant difference.
Results
Differential expression of miRNAs in NSCs and neurons
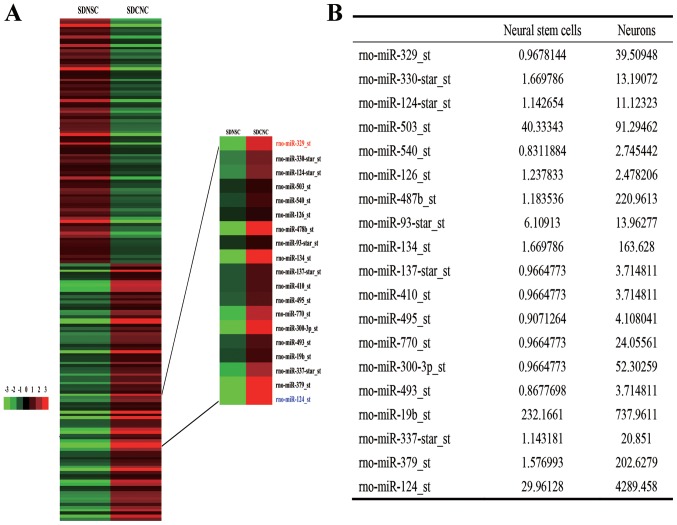
Analysis of the GeneChip data indicated that miR-329-3p was expressed at increased levels in differentiated rat cortical neurons compared with in the starting population of NSCs (Fig. 1A and B).
Figure 1.
Expression of miR-329-3p in rat NSCs and differentiated neurons. (A) Heat maps demonstrating differentially expressed miRNAs in SDNSC and SDCNC detected by microRNA array. Green and red represent low and high expression, respectively. (B) Results of a GeneChip microarray assay indicating that miR-329-3p was expressed at increased levels in neurons compared with in NSCs. miR/miRNA, microRNA; NSC, neural stem cells; SDNSC, Sprague-Dawley rat neural stem cells; SDCNC, Sprague-Dawley rat cortical neurons; rno, Rattus norvegicus.
miR-329-3p is upregulated and E2F1 is downregulated during NSC differentiation
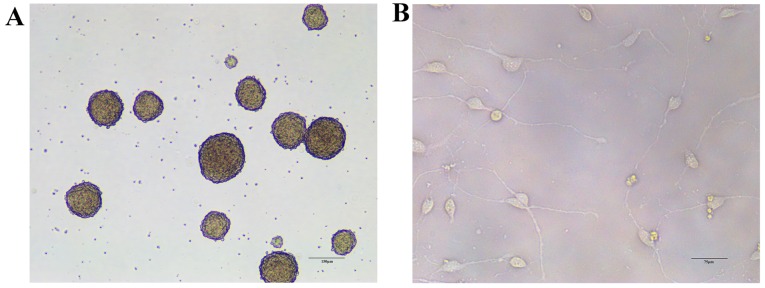
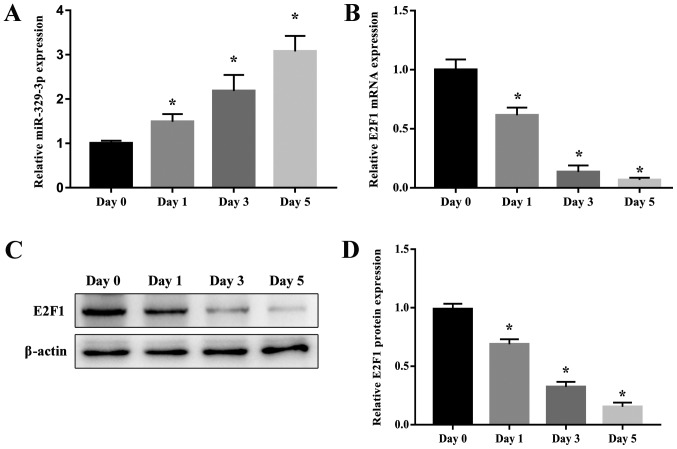
To validate the results of the GeneChip microarray assay, RT-qPCR analysis of miR-329-3p levels in NSCs on day 0 and after 1, 3, and 5 days of culture in medium that promotes neuronal differentiation was performed. The morphology of NSCs and cells differentiated from NSCs is presented in Fig. 2. The results demonstrated that miR-329-3p expression was gradually upregulated during the differentiation process (Fig. 3A). The E2F1 expression was also measured in differentiating NSCs at the mRNA and protein levels using RT-qPCR and western blot analysis, respectively. Expression of E2F1 mRNA (Fig. 3B) and protein (Fig. 3C and D) was identified to decrease over the course of neuronal differentiation, which contrasted with the increase in miR-329-3p expression observed.
Figure 2.
Morphology of NSCs and cells differentiated from NSCs. (A) A photomicrograph of neurospheres in culture. (B) A photomicrograph of NSCs induced to differentiate for 3 days. NSCs, neural stem cells.
Figure 3.
Expression of miR-329-3p and E2F1 during NSC differentiation. (A) RT-qPCR analysis of miR-329-3p on days 0, 1, 3, and 5 after induction of neuronal differentiation. (B) RT-qPCR analysis of E2F1 mRNA expression and (C) western blot analysis of E2F1 protein expression on days 0, 1, 3, and 5 after induction of neuronal differentiation. GAPDH and β-actin were probed as internal controls for (B) and (C) respectively. (D) Quantification of protein expression. U6 was measured as an internal control for miR-329-3p. Data are presented as the mean ± standard deviation. *P<0.05 vs. day 0. miR, microRNA; E2F1, transcription factor E2F1; NSC, neural stem cell; RT-qPCR, reverse transcription quantitative polymerase chain reaction.
Upregulation of miR-329-3p suppresses NSC proliferation
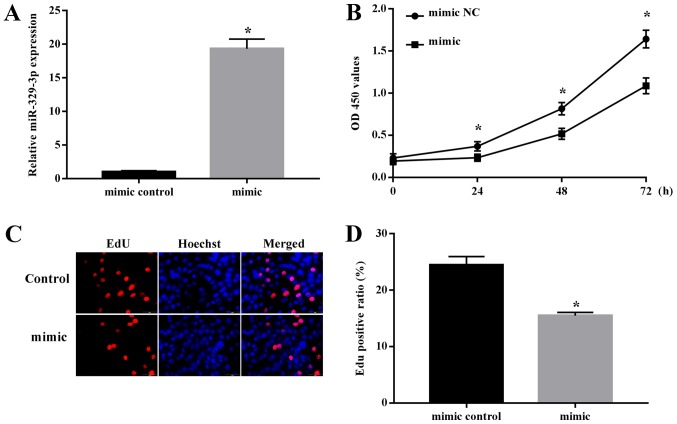
Previous data have suggested that the differentiation of NSCs is accompanied by a concomitant decrease in their ability to proliferate (17). As miR-329-3p levels gradually increased during NSC differentiation, we hypothesized that this miRNA may be involved in suppressing NSC proliferation. To examine this, NSCs were transfected with miR-329-3p mimics or NC mimic and cell proliferation was examined. RT-qPCR analysis confirmed that miR-329-3p expression was significantly increased in NSCs transfected with the miR-329-3p mimic compared with in those with the NC mimic (Fig. 4A). Cell proliferation was evaluated by the colorimetric CCK-8 assay and the fluorescent EdU incorporation assay. Each of these assays indicated that proliferation was decreased in cells transfected with the miR-329-3p mimic compared with the level with the control mimic (Fig. 4B-D), demonstrating that miR-329-3p overexpression represses NSC proliferation.
Figure 4.
Regulation of NSC proliferation by miR-329-3p. NSCs were transfected with a control mimic or miR-329-3p mimics for 48 h and then analyzed. (A) Reverse transcription quantitative polymerase chain reaction analysis of miR-329-3p expression level. (B) Cell Counting Kit-8 assay of NSC proliferation. (C) EdU assay of NSC proliferation. Red and blue staining indicates EdU and Hoechst staining, respectively. Scale bar, 50 µm. (D) Quantification of EdU-positive cells. U6 was used as an internal control for miR-329-3p. Data are presented as the mean ± standard deviation. *P<0.05 vs. negative control mimic. NSC, neural stem cell; miR, microRNA; NC, negative control; OD, optical density; EdU, 5-ethynyl-2′-deoxyuridine.
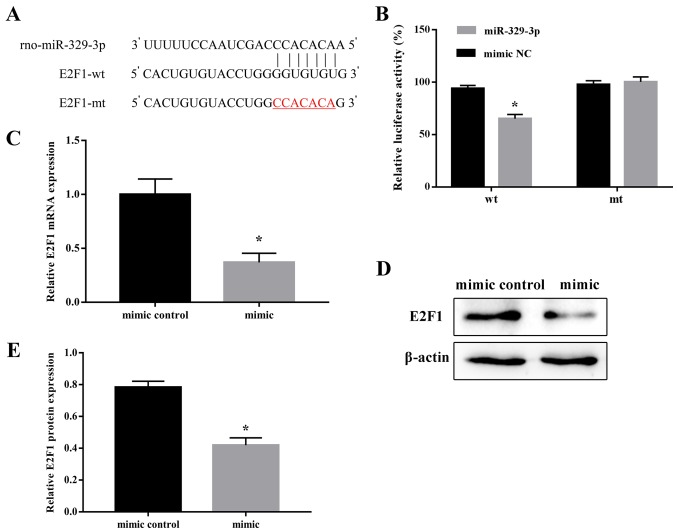
E2F1 is an miR-329-3p target gene
Screening of the bioinformatic databases TargetScan (http://www.targetscan.org/) and miRDB (http://mirdb.org/index.html) identified a potential miR-329-3p-binding sequence in the 3′-UTR of E2F1 (Fig. 5A). To determine whether E2F1 was a direct target of miR-329-3p, a dual-reporter assay was performed, in which luciferase expression was driven by the wt or mt form of the E2F1 3′-UTR sequence. For this, 293 cells were co-transfected with the wt or mt E2F1 3′-UTR reporter vector together with an miR-329-3p or NC mimic, and luciferase activity was measured after 48 h. It was identified that the miR-329-3p mimic significantly inhibited luciferase expression driven by the wt, but not the mt, 3′-UTR sequence (Fig. 5B). These results confirmed that miR-329-3p directly regulated E2F1 expression. To additionally define the targeting association between miR-329-3p and E2F1, the expression of E2F1 was examined following transfection of the miR-329-3p mimic in NSCs. As expected, transfection of the miR-329-3p mimic decreased the expression of E2F1 mRNA and protein in NSCs (Fig. 5C-E).
Figure 5.
Direct targeting of E2F1 by miR-329-3p. (A) Putative miR-329-3p-binding sequence in the wt E2F1 3′-UTR. (B) Luciferase reporter assay of miR-329-3p targeting of E2F1 in 293 cells co-transfected with wt or mt E2F1 3′-UTR constructs and control or miR-329-3p mimics. (C) Reverse transcription quantitative polymerase chain reaction analysis and (D) western blot analysis of E2F1 mRNA and protein expression, respectively, in neural stem cells transfected with control or miR-329-3p mimics. GAPDH and β-actin were used as internal controls for (C) and (D) respectively. (E) Quantitative analysis of protein expression. Data are presented as the mean ± standard deviation. *P<0.05 vs. negative control mimic. E2F1, transcription factor E2F1; miR, microRNA; wt, wild-type; mt, mutated; UTR, untranslated region; rno, Rattus norvegicus.
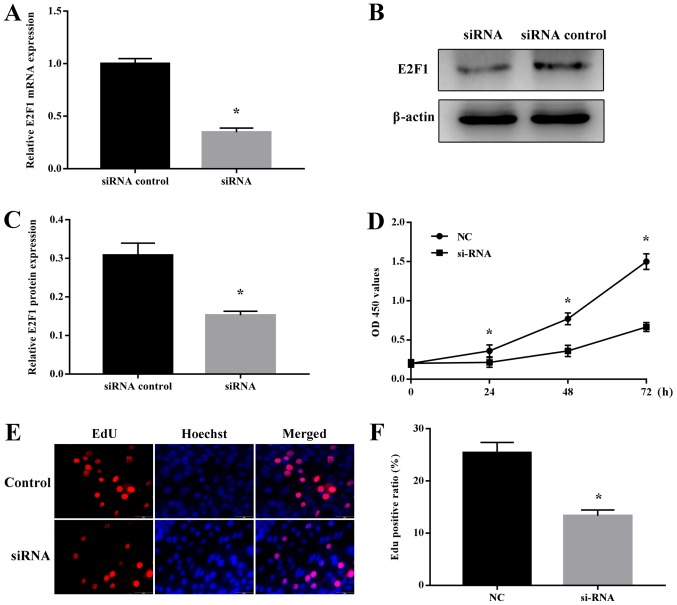
E2F1 regulates NSC proliferation
Several studies have demonstrated that E2F1 is involved in the maintenance of stem cells (17–19). To investigate the potential role of E2F1 in NSC proliferation, cells were first transfected with E2F1-specific or NC siRNA, and it was confirmed that E2F1 mRNA and protein expression was specifically decreased by the E2F1-targeting siRNA (Fig. 6A-C). Notably, analysis of cell proliferation using the CCK-8 and EdU assays revealed that E2F1 siRNA specifically inhibited the proliferation of NSCs (Fig. 6D-F). Therefore, E2F1 appeared to serve a vital role in regulating NSC proliferation.
Figure 6.
Regulation of NSC proliferation by E2F1. NSCs were transfected with a control siRNA or E2F1-targeting siRNA for 48 h and then analyzed. (A) Reverse transcription quantitative polymerase chain reaction analysis of E2F1 mRNA expression and (B) western blot analysis of E2F1 protein expression. GAPDH and β-actin were probed as internal controls for (A) and (B) respectively. (C) Quantification of protein expression. (D) Cell Counting Kit-8 assay of NSC proliferation. (E) EdU assay of NSC proliferation. Red and blue staining indicates EdU and Hoechst nuclear staining, respectively. Scale bar, 50 µm. (F) Quantification of the EdU-positive cells. Data are presented as the mean ± standard deviation. *P<0.05 vs. siRNA control. NSCs, neural stem cells; E2F1, transcription factor E2F1; siRNA, small interfering RNA; NC, negative control; OD, optical density; EdU, 5-ethynyl-2′-deoxyuridine.
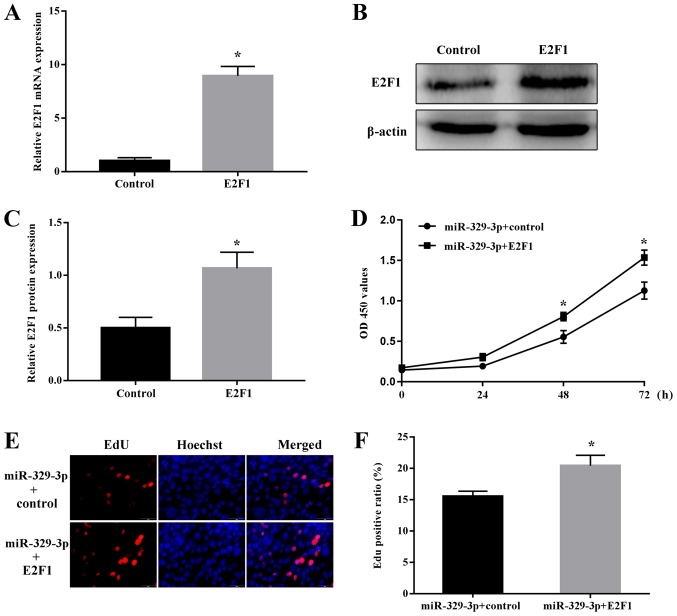
miR-329-3p regulates NSC proliferation by targeting E2F1
Finally, the functional effects of miR-329-3p-mediated regulation of E2F1 in NSCs were examined. Cells were co-transfected with miR-329-3p together with the E2F1 overexpression vector (omitting the 3′-UTR sequence targeted by miR-329-3p) or E2F1 vector control. Overexpression of E2F1 was confirmed at the mRNA (Fig. 7A) and protein levels (Fig. 7B and C). Notably, ectopic expression of E2F1 reversed the decrease in NSC proliferation induced by transfection with the miR-329-3p mimic, as measured using the CCK-8 (Fig. 7D) and EdU incorporation assays (Fig. 7E and F).
Figure 7.
miR-329-3p-mediated regulation of NSC proliferation via E2F1. (A) Reverse transcription quantitative polymerase chain reaction analysis of E2F1 mRNA expression and (B) western blot analysis of E2F1 protein expression. GAPDH and β-actin were probed as internal controls for (A) and (B) respectively. (C) Quantitative analysis of protein expression. (D) Cell Counting Kit-8 assay of NSC proliferation. (E) EdU assay of NSC proliferation. Red and blue indicate EdU and Hoechst nuclear staining, respectively. Scale bar, 50 µm. (F) Quantification of EdU-positive cells. Data are presented as the mean ± standard deviation. *P<0.05 vs. control. miR, microRNA; NSC, neural stem cell; E2F1, transcription factor E2F1; OD, optical density; EdU, 5-ethynyl-2′-deoxyuridine.
Discussion
NSCs have the capacity to proliferate, self-renew and differentiate into neurons, astrocytes or oligodendrocytes (20–22). The proliferation and differentiation of NSCs are modulated by multiple regulators, including miRNAs (17,23). For example, NSC proliferation and differentiation are regulated by miR-125b, miR-378, miR-506-3p and miR-1297 via modulation of Nestin, nuclear receptor subfamily 2 group E member 1, transcription factor E2-alpha and transcription factor HES-1 expression, respectively (11,24–26). It has also been demonstrated that multiple miRNAs interact cooperatively to regulate cellular processes. For example, miR-124, miR-128 and miR-137 were demonstrated to orchestrate biological roles in NSCs by controlling the expression of the common target gene Sp1 transcription factor (27). In addition, the abnormal expression of miRNAs has been demonstrated to be associated with neurological diseases including neuroblastoma and glioma (28,29).
In the GeneChip analysis in the present study, it was identified that miR-329-3p was upregulated in differentiated neurons compared with the level in NSCs. Previous studies have suggested that miR-329-3p serves a key role in the proliferation of various types of cancer (28–30). For example, Yang et al (28) demonstrated that miR-329 inhibited the growth and invasion of neuroblastoma cells by regulating lysine-specific histone demethylase 1A expression. Xiao et al (29) also demonstrated that miR-329 expression was downregulated in glioma cells and that its overexpression inhibited cell proliferation. In addition, Kang et al (30) indicated that downregulation of miR-329 promoted the development of human breast cancer. However, the role of miR-329-3p in NSC biology remains unclear. Therefore, miR-329-3p was selected for examination in the present study. As the proliferative capacity of NSCs has been demonstrated to decrease during differentiation (17), the present study initially examined the expression of miR-329-3p during the differentiation of NSCs, and it was identified that its expression gradually increased during this process, which was consistent with the data from the GeneChip analysis. Then, miR-329-3p was overexpressed in NSCs, and it was demonstrated that this overexpression suppressed NSC proliferation.
E2F1 is a member of the E2F family of transcription factors and serves crucial roles in cell proliferation (17). E2F1 has been identified to be closely associated with the growth of a variety of tumors (31–33). For example, Lv et al (31) demonstrated that E2F1 was involved in the growth and invasion of hepatoma cells caused by aflatoxin B1. In addition, Sun et al (32) revealed that the expression of E2F1 was increased in lung cancer tissues and asserted that E2F1 may be considered as a novel biomarker for the prognosis of lung cancer. Zhang et al (33) also indicated that E2F1 promoted the expression of lncRNA RGMB antisense RNA 1 (RGMB-AS1) by binding to its promoter region, and that upregulated lncRNA RGMB-AS1 promoted the proliferation, migration and invasion of papillary thyroid carcinoma. It has also been suggested that E2F1 increased cell proliferation by promoting transition through the G1/S checkpoint of the cell cycle (34,35). Exiting the cell cycle is a crucial step in cell differentiation (36), and E2F1 serves a key role in this process (37,38). In concordance with this, NSC differentiation is known to be accompanied by a decrease in proliferation (17). In the present study, it was identified that E2F1 expression was decreased during NSC differentiation. The data demonstrating that the downregulation of E2F1 inhibited the proliferation of NSCs was also consistent with the upregulation of miR-329-3p. As the overexpression of E2F1 restored the proliferative capacity of miR-329-3p-overexpressing NSCs, the results from the present study confirmed that miR-329-3p modulated NSC proliferation by repressing E2F1 expression.
In conclusion, the present study revealed a novel role for miR-329-3p in regulating NSC proliferation, in part through the inhibition of E2F1 expression. These data provide insight into the miRNA-mRNA regulatory network that controls NSC proliferation, and suggest a new avenue of study into the functions of miR-329-3p and E2F1 in the nervous system. In addition, the results may contribute to the development of novel therapeutic strategies based on NSC transplantation.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- CCK-8
Cell Counting Kit-8
- E2F1
transcription factor E2F1
- EdU
5-ethynyl-2′-deoxyuridine
- miRNA/miR
microRNA
- mt
mutated
- NSC
neural stem cell
- RT-qPCR
reverse-transcription quantitative polymerase chain reaction
- UTR
untranslated region
- wt
wild-type
Funding
The present study was supported by the Science and Technology Plan Project of Liaoning Province (grant no. 201602857).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
DL and GT designed the experiments. DL, YS, XD and YH performed the experiments, and analyzed and collected the data. DL wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Liu S, Yin F, Zhang J, Wicha MS, Chang AE, Fan W, Chen L, Fan M, Li Q. Regulatory roles of miRNA in the human neural stem cell transformation to glioma stem cells. J Cell Biochem. 2014;115:1368–1380. doi: 10.1002/jcb.24786. [DOI] [PubMed] [Google Scholar]
- 2.Makri G, Lavdas AA, Katsimpardi L, Charneau P, Thomaidou D, Matsas R. Transplantation of embryonic neural stem/precursor cells overexpressing BM88/Cend1 enhances the generation of neuronal cells in the injured mouse cortex. Stem Cells. 2010;28:127–139. doi: 10.1002/stem.258. [DOI] [PubMed] [Google Scholar]
- 3.Mosher KI, Andres RH, Fukuhara T, Bieri G, Hasegawa-Moriyama M, He Y, Guzman R, Wyss-Coray T. Neural progenitor cells regulate microglia functions and activity. Nat Neurosci. 2012;15:1485–1487. doi: 10.1038/nn.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le MT, Xie H, Zhou B, Chia PH, Rizk P, Um M, Udolph G, Yang H, Lim B, Lodish HF. MicroRNA-125b promotes neuronal differentiation in human cells by repressing multiple targets. Mol Cell Biol. 2009;29:5290–5305. doi: 10.1128/MCB.01694-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu X, Li Z, Shen J, Wu WK, Liang J, Weng X, Qiu G. MicroRNA-10b promotes nucleus pulposus cell proliferation through RhoC-Akt pathway by targeting HOXD10 in intervetebral disc degeneration. PLoS One. 2013;8:e83080. doi: 10.1371/journal.pone.0083080. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Yu X, Li Z, Yu J, Chan MT, Wu WK. MicroRNAs predict and modulate responses to chemotherapy in colorectal cancer. Cell Prolif. 2015;48:503–510. doi: 10.1111/cpr.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santra M, Chopp M, Santra S, Nallani A, Vyas S, Zhang ZG, Morris DC. Thymosin beta 4 up-regulates miR-200a expression and induces differentiation and survival of rat brain progenitor cells. J Neurochem. 2016;136:118–132. doi: 10.1111/jnc.13394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan Z, Lu M, Qiao C, Zhou Y, Ding JH, Hu G. MicroRNA-7 enhances subventricular zone neurogenesis by inhibiting NLRP3/Caspase-1 axis in adult neural stem cells. Mol Neurobiol. 2016;53:7057–7069. doi: 10.1007/s12035-015-9620-5. [DOI] [PubMed] [Google Scholar]
- 9.Zhang JF, Shi LL, Zhang L, Zhao ZH, Liang F, Xu X, Zhao LY, Yang PB, Zhang JS, Tian YF. MicroRNA-25 negatively regulates cerebral ischemia/reperfusion injury-induced cell apoptosis through Fas/FasL pathway. J Mol Neurosci. 2016;58:507–516. doi: 10.1007/s12031-016-0712-0. [DOI] [PubMed] [Google Scholar]
- 10.Bian S, Hong J, Li Q, Schebelle L, Pollock A, Knauss JL, Garg V, Sun T. MicroRNA cluster miR-17-92 regulates neural stem cell expansion and transition to intermediate progenitors in the developing mouse neocortex. Cell Rep. 2013;3:1398–1406. doi: 10.1016/j.celrep.2013.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui Y, Xiao Z, Han J, Sun J, Ding W, Zhao Y, Chen B, Li X, Dai J. MiR-125b orchestrates cell proliferation, differentiation and migration in neural stem/progenitor cells by targeting nestin. BMC Neurosci. 2012;13:116. doi: 10.1186/1471-2202-13-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garg N, Po A, Miele E, Campese AF, Begalli F, Silvano M, Infante P, Capalbo C, De Smaele E, Canettieri G, et al. microRNA-17-92 cluster is a direct Nanog target and controls neural stem cell through Trp53inp1. EMBO J. 2013;32:2819–2832. doi: 10.1038/emboj.2013.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou D, Chen Y, Han Y, Lv C, Tu G. Overexpression of microRNA-124 promotes the neuronal differentiation of bone marrow-derived mesenchymal stem cells. Neural Regen Res. 2014;9:1241–1248. doi: 10.4103/1673-5374.135333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Wong N, Wang X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. ELife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palm T, Hemmer K, Winter J, Fricke IB, Tarbashevich K, Sadeghi Shakib F, Rudolph IM, Hillje AL, De Luca P, Bahnassawy L, et al. A systemic transcriptome analysis reveals the regulation of neural stem cell maintenance by an E2F1-miRNA feedback loop. Nucleic Acids Res. 2013;41:3699–3712. doi: 10.1093/nar/gkt070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pulikkan JA, Dengler V, Peramangalam PS, Peer Zada AA, Müller-Tidow C, Bohlander SK, Tenen DG, Behre G. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood. 2010;115:1768–1778. doi: 10.1182/blood-2009-08-240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han BW, Feng DD, Li ZG, Luo XQ, Zhang H, Li XJ, Zhang XJ, Zheng LL, Zeng CW, Lin KY, et al. A set of miRNAs that involve in the pathways of drug resistance and leukemic stem-cell differentiation is associated with the risk of relapse and glucocorticoid response in childhood ALL. Hum Mol Genet. 2011;20:4903–4915. doi: 10.1093/hmg/ddr428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 21.Brett JO, Renault VM, Rafalski VA, Webb AE, Brunet A. The microRNA cluster miR-106b~25 regulates adult neural stem/progenitor cell proliferation and neuronal differentiation. Aging (Albany NY) 2011;3:108–124. doi: 10.18632/aging.100285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi Y, Sun G, Zhao C, Stewart R. Neural stem cell self-renewal. Crit Rev Oncol Hematol. 2008;65:43–53. doi: 10.1016/j.critrevonc.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 24.Huang Y, Liu X, Wang Y. MicroRNA-378 regulates neural stem cell proliferation and differentiation in vitro by modulating Tailless expression. Biochem Biophys Res Commun. 2015;466:214–220. doi: 10.1016/j.bbrc.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Jiaqi C, Zhaoying C, Huimin C. MicroRNA-506-3p regulates neural stem cell proliferation and differentiation through targeting TCF3. Gene. 2016;593:193–200. doi: 10.1016/j.gene.2016.08.026. [DOI] [PubMed] [Google Scholar]
- 26.Zheng J, Yi D, Shi X, Shi H. miR-1297 regulates neural stem cell differentiation and viability through controlling Hes1 expression. Cell Prolif. 2017;50 doi: 10.1111/cpr.12347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santos MC, Tegge AN, Correa BR, Mahesula S, Kohnke LQ, Qiao M, Ferreira MA, Kokovay E, Penalva LO. miR-124, −128, and −137 orchestrate neural differentiation by acting on overlapping gene sets containing a highly connected transcription factor network. Stem Cells. 2016;34:220–232. doi: 10.1002/stem.2204. [DOI] [PubMed] [Google Scholar]
- 28.Yang H, Li Q, Zhao W, Yuan D, Zhao H, Zhou Y. miR-329 suppresses the growth and motility of neuroblastoma by targeting KDM1A. FEBS Lett. 2014;588:192–197. doi: 10.1016/j.febslet.2013.11.036. [DOI] [PubMed] [Google Scholar]
- 29.Xiao B, Tan L, He B, Liu Z, Xu R. MiRNA-329 targeting E2F1 inhibits cell proliferation in glioma cells. J Transl Med. 2013;11:172. doi: 10.1186/1479-5876-11-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang H, Kim C, Lee H, Rho JG, Seo JW, Nam JW, Song WK, Nam SW, Kim W, Lee EK. Downregulation of microRNA-362-3p and microRNA-329 promotes tumor progression in human breast cancer. Cell Death Differ. 2016;23:484–495. doi: 10.1038/cdd.2015.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lv J, Yu YQ, Li SQ, Luo L, Wang Q. Aflatoxin B1 promotes cell growth and invasion in hepatocellular carcinoma HepG2 cells through H19 and E2F1. Asian Pac J Cancer Prev. 2014;15:2565–2570. doi: 10.7314/APJCP.2014.15.6.2565. [DOI] [PubMed] [Google Scholar]
- 32.Sun CC, Zhou Q, Hu W, Li SJ, Zhang F, Chen ZL, Li G, Bi ZY, Bi YY, Gong FY, et al. Transcriptional E2F1/2/5/8 as potential targets and transcriptional E2F3/6/7 as new biomarkers for the prognosis of human lung carcinoma. Aging (Albany NY) 2018;10:973–987. doi: 10.18632/aging.101441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z, Li SY, Zhang LB. LncRNA RGMB-AS1 is activated by E2F1 and promotes cell proliferation and invasion in papillary thyroid carcinoma. Eur Rev Med Pharmacol Sci. 2018;22:1979–1986. doi: 10.26355/eurrev_201804_14725. [DOI] [PubMed] [Google Scholar]
- 34.DeGregori J, Kowalik T, Nevins JR. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/MCB.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inoshita S, Terada Y, Nakashima O, Kuwahara M, Sasaki S, Marumo F. Regulation of the G1/S transition phase in mesangial cells by E2F1. Kidney Int. 1999;56:1238–1241. doi: 10.1046/j.1523-1755.1999.00705.x. [DOI] [PubMed] [Google Scholar]
- 36.Ma Q, Hu QS, Xu RJ, Zhen XC, Wang GH. Protease Omi facilitates neurite outgrowth in mouse neuroblastoma N2a cells by cleaving transcription factor E2F1. Acta Pharmacol Sin. 2015;36:966–975. doi: 10.1038/aps.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McClellan KA, Slack RS. Novel functions for cell cycle genes in nervous system development. Cell Cycle. 2006;5:1506–1513. doi: 10.4161/cc.5.14.2980. [DOI] [PubMed] [Google Scholar]
- 38.Tsume M, Kimura-Yoshida C, Mochida K, Shibukawa Y, Amazaki S, Wada Y, Hiramatsu R, Shimokawa K, Matsuo I. Brd2 is required for cell cycle exit and neuronal differentiation through the E2F1 pathway in mouse neuroepithelial cells. Biochem Biophys Res Commun. 2012;425:762–768. doi: 10.1016/j.bbrc.2012.07.149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.