Abstract
Moyamoya disease (MMD) is a rare chronic cerebrovascular disease mainly found in individuals of East Asian ethnicity, and its pathogenesis is largely unknown. Transfer RNA-derived fragments (tRFs) are novel biological entities involved in many biological processes; however, whether tRFs contribute towards MMD pathogenesis remains unexplored. In the present study, deep sequencing technology was used to identify alterations in tRF expression profiles between patients with MMD and healthy controls. The sequencing findings were validated using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Subsequently, the putative target genes of tRFs were predicted using miRNA target prediction algorithms. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed to further evaluate potential functions of tRFs. The sequencing results demonstrated that 38 tRFs were differentially expressed between patients and controls, of which 22 were upregulated and 16 were downregulated. RT-qPCR analysis confirmed the validity of the sequencing results. GO and KEGG pathway enrichment analyses indicated that 15 pathways were associated with the selected tRFs. These pathways were mainly involved in angiogenesis and metabolism, both of which are physiopathological fundamentals of MMD. The results provided a novel insight into the mechanisms underlying MMD pathogenesis, and demonstrated that tRFs may serve as potential therapeutic targets for the future treatment of MMD.
Keywords: transfer RNA, tRNA-derived fragment, pathogenesis, moyamoya disease, vascular disorders
Introduction
Moyamoya disease (MMD) is a cerebrovascular disease, which was first described by Takeuchi and Shimizui in 1957 (1). It is characterized by progressive stenosis or occlusion of the distal portions of the internal carotid arteries, with compensatory development of an aberrant vascular network at the base of the brain. The incidence of MMD is higher in East Asian countries compared with the rest of the world (2,3). Its initial clinical manifestations include transient ischemic attack, infarction, hemorrhage, headaches and epilepsy. Genetic factors are likely to be involved in the development of MMD; epidemiological investigations have revealed that 7–12% of patients in Japan have a familial occurrence (4). In addition, studies have revealed that non-coding RNAs, including micro(mi)RNAs and long non-coding (lnc)RNAs exert an important regulatory role in this disease (5,6). However, the etiology and pathogenesis of MMD remain unclear.
With the rapid progress and wide use of next generation sequencing technologies, several independent studies have revealed the existence of transfer RNA (tRNA)-derived fragments (tRFs) (7–9). These derivatives were originally viewed as tRNA degradation products, but accumulating evidence suggests that they serve roles in various biological processes, including regulation of gene expression, cell proliferation and tumor suppression (10–12). For example, a study on mouse sperm indicated that 5′-tiRNAs (tRNA-derived stress-induced RNAs), a subset of tRFs, may alter expression profiles and modify RNA, and act as a paternal epigenetic factor regulating intergenerational inheritance of certain metabolic diseases (13). Accumulation of 5′-tiRNAs due to tRNA hypomethylation could specifically repress translation and activate stress pathways, eventually leading to human neurodevelopmental disorders (14). However, the potential function of tRFs in MMD has yet to be elucidated.
In the present study, a prospective case-control study was performed to investigate and compare the expression profiles of tRFs in patients with MMD and healthy controls. Furthermore, bioinformatics analyses were used to further investigate the molecular basis of tRFs in MMD.
Materials and methods
Patient samples
The present study was conducted in accordance with the Declaration of Helsinki, and was approved by the Research Ethics Board of Beijing Tiantan Hospital, Capital Medical University (Beijing, China). Written informed consent was obtained from all individuals who were included in the study. Between March 2017 and April 2017, three adult patients who were diagnosed with MMD and underwent neurosurgical treatment at the Department of Neurosurgery, Beijing Tiantan Hospital were enrolled for the RNA-sequencing experiment. Cerebral digital subtraction angiography was performed in all patients. MMD was diagnosed according to the criteria put forward by the Japanese Ministry of Health and Welfare in 2012 (15). Patients diagnosed with quasi-MMD, hypertension, hyperlipidemia or diabetes, and patients taking certain drugs (including antibiotics, antiplatelet drugs and antiepileptic drugs, among others) were excluded from the study to prevent potential interference to the experimental results. Overall, three male patients with MMD (age range, 30–51 years) were included in this study, which were compared with three age-matched healthy male volunteers (age range, 25–29 years). The duration from disease onset to hospital admission in patients with MMD was >2 weeks. Due to ethical issues, blood vessels could not be obtained from the patients; therefore, peripheral blood was used as an alternative, which is consistent with other studies on MMD (5,6). In the present study, all the blood samples were collected prior to surgical treatment. To verify the RNA sequencing results, five patients (three females, two males) with MMD (age range, 30–42 years) were included in the study, and were compared with five healthy male volunteers (age range, 26–32 years). Samples were collected between March 2017 and April 2017. A total of 2.5 ml peripheral blood was collected from each participant.
RNA extraction and small RNA-sequencing
A total of 2.5 ml peripheral blood was collected from each participant in K2EDTA-coated Vacutainer® tubes (BD Biosciences, Franklin Lakes, NJ, USA). Total RNA was extracted from the six blood samples using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocol. Subsequently, RNA was quantified using a NanoDrop™ ND-1000 spectrophotometer (NanoDrop; Thermo Fisher Scientific, Inc., Wilmington, DE, USA). RNA integrity was assessed by 1% agarose gel electrophoresis. RNA samples were firstly pretreated using the rtStar™ tRF&tiRNA Pretreatment kit (cat. no. AS-FS-005; Arraystar Inc., Rockville, MD, USA) to remove any RNA modifications, which may interfere with small RNA sequencing library construction. The pretreated total RNA samples were subjected to sequencing library preparation using a commercial kit (NEB Next® Multiplex Small RNA Library Prep Set for Illumina®; cat. no. 7580; New England BioLabs, Inc., Ipswich, MA, USA). The procedure was conducted according to the manufacturer's protocol, which included 3′- and 5′-adapter ligation, cDNA synthesis and library polymerase chain reaction (PCR) amplification. The completed libraries were quantified on an Agilent Bioanalyzer 2100 (Agilent Technologies, Inc., Santa Clara, CA, USA), and then sequencing was performed using an Illumina NextSeq 500 system (Illumina, Inc., San Diego, CA, USA), according to the manufacturer's protocol.
Data analysis
Analysis of the sequence reads was performed using the Solexa pipeline (Off-Line Base Caller software version 1.8, Illumina, Inc.). The sequencing quality was examined using FastQC software version 0.11.5 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The trimmed reads were first compared to the genomic tRNA database (http://gtrnadb.ucsc.edu/) using Novo Align software version 2.07.11 (http://www.novocraft.com/main/index.php) to identify known mature- and pre-tRNAs. The unidentified reads were further aligned to several other small RNA databases (mRNA/ribosomal RNA/small nuclear RNA/small nucleolar RNA/Piwi-interacting RNA/miRNA), including GtRNA database (http://gtrnadb.ucsc.edu), tRFs database (http://genome.bioch.virginia.edu/trfdb/) and Mintbase (https://cm.jefferson.edu/MINTbase/). The tRF expression levels were measured and normalized to the number of transcripts per million of total aligned tRNA reads. Expression profiles between patients and healthy controls were compared by calculating the fold change (i.e. the ratio of the group means) for each tRF. Statistical significance was determined using Student's t-test. In total, 38 tRFs with fold change ≥2.0 and P<0.05 were considered significantly differentially expressed. Volcano plot and heat map were constructed to analyze the differentially expressed tRFs between the two groups.
Validation using reverse transcription-quantitative PCR (RT-qPCR)
RT-qPCR is considered the gold standard for gene expression quantification (16,17). To verify the RNA sequencing results, four tRFs were randomly selected and their expression levels were detected in five patients with MMD and five healthy controls (An independent cohort, n=5). Total RNA was extracted from peripheral blood using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). RNA samples were pretreated using the rtStar™ tRF&tiRNA Pretreatment kit (cat. no. AS-FS-005; Arraystar Inc.) to remove RNA modifications, and were then reverse transcribed to cDNA using the rtStar™ First-Strand cDNA Synthesis kit (cat. no. AS-FS-003; Arraystar Inc.) according to the manufacturer's protocol. qPCR was performed using ViiA 7 Real-time PCR system, using Arraystar SYBR® Green Real-time qPCR Master mix (cat. no. AS-MR-005-25; Applied Biosystems; Thermo Fisher Scientific, Inc.) with specific primers (Table I). The thermocycling conditions were the following: Initial denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 10 sec and 60°C for 60 sec. U6 was used as an internal control to assess the efficiency of the reaction. RT-qPCR experiments were performed in triplicate. Data were analyzed using the 2−ΔΔCq method (17); a standard dilution curve was used to determine amplification efficiency.
Table I.
Primers for reverse transcription-quantitative polymerase chain reaction.
| Gene | Primers |
|---|---|
| U6 | F:5′-GCTTCGGCAGCACATATACTAAAAT-3′ |
| R:5′-CGCTTCACGAATTTGCGTGTCAT-3′ | |
| AS-tDR-000586 | F:5′-TCTCCCACATGGTCTAGCGGT-3′ |
| R:5′-TGCTCTTCCGATCTGAAAACCA-3′ | |
| AS-tDR-000924 | F: 5′-AGAGTTCTACAGTCCGACGATC-3′ |
| R:5′-CCGATCTACTGAGCTATCCG-3′ | |
| AS-tDR-007725 | F:5′-CAGTCCGACGATCTCCCTGT-3′ |
| R:5′-ATCTGAGAGCGCCGAATCC-3′ | |
| AS-tDR-011363 | F:5′-CAGTCCGACGATCCTGTCACG-3’ |
| R: 5′-TTCCGATCTGCCCCGGTCT-3′ |
U6 was used as the internal control. F, forward; R, reverse.
Bioinformatics analysis
As with miRNAs, tRFs can interact with Argonaute (AGO) proteins (18). Previous studies have demonstrated that tRFs are able to target RNAs similarly to miRNAs (19,20). In addition, it has been proposed that tRF seed sequences have similar matching profiles to miRNAs (21–23). Therefore, miRNA target-predicting algorithms including miRanda (http://www.microrna.org/microrna/home.do; August 2010 release) and TargetScan (version 71; http://www.targetscan.org/vert_71/), were used to identify the potential target genes of the selected tRFs, based on the presence of binding sites in the 3′untranslated region. Subsequently, the predicted target genes were inputted into the Database for Annotation, Visualization and Integrated Discovery (http://david.abcc.ncifcrf.gov/) using Gene Ontology (GO; http://www.geneontology.org/) to analyze their molecular functions. Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.ad.jp/kegg/) database was used to identify significant pathways for these target genes. DAVID (version 6.8; http://david.ncifcrf.gov/) software was used to generate the pathway map.
Statistical analysis
Data are presented as the means ± standard deviation. Data analysis was performed using R software version 3.3.1 (R Project for Statistical Computing, Vienna, Austria). Differences between two groups were analyzed using the Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression profiles of tRFs
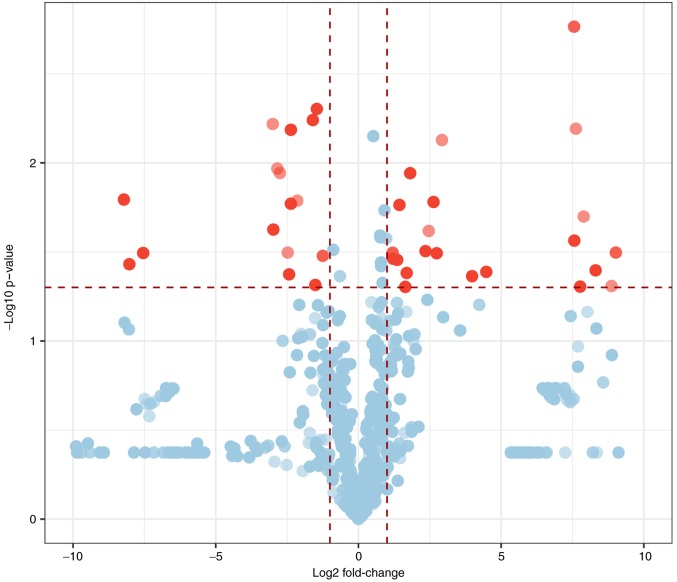
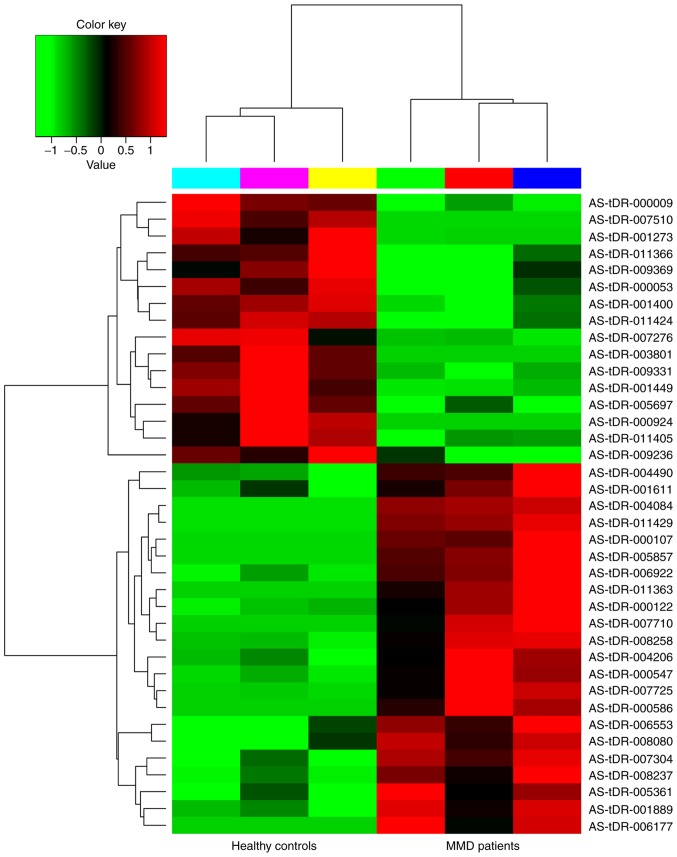
The clinical characteristics of patients with MMD are summarized in Table II, including age, sex, initial presentation, disease duration, Suzuki stage (24) and Modified Rankin Score (25). All individuals were of Han Chinese ethnicity. Differentially expressed tRFs in patients with MMD vs. healthy controls were identified by RNA sequencing. A total of 22 tRFs were upregulated and 16 tRFs were downregulated (fold change ≥2.0; P<0.05). Notably, 26 tRFs were significantly differentially expressed by at least 5-fold between the two groups (fold change ≥5.0; P<0.05); with 15 upregulated and 11 downregulated tRFs. Among the differentially expressed tRFs, 5′-tiRNA derived from tRNAGlu(TTC) was the most upregulated, whereas i-tRF from tRNAGln(CTG) and tRF-1 from pre-tRNAGln(CTG) were the most downregulated. More upregulated tRFs were observed compared with downregulated tRFs. The 38 upregulated and downregulated tRFs ranked by fold change are summarized in Tables III and IV. A volcano plot was constructed to identify differentially expressed tRFs (Fig. 1). The heatmap revealed systematic variations between the MMD and control groups in terms of tRF expression (Fig. 2).
Table II.
Clinical characteristics of patients with moyamoya disease.
| Patient no. | Age (years) | Sex | Clinical presentation | Duration from disease onset to hospital admission (months) | Suzuki stage (I to VI) | mRS |
|---|---|---|---|---|---|---|
| 1 | 51 | Male | Intracerebral hemorrhage | 6 | R, III; L, III | 2 |
| 2 | 38 | Male | Limb weakness | 12 | R, II; L, II | 2 |
| 3 | 30 | Male | Intraventricular hemorrhage | 6 | R, III; L, II | 3 |
L, left; mRS, Modified Rankin Score; R, right.
Table III.
Summary of the 22 tRFs upregulated in patients with moyamoya disease vs. healthy controls.
| tRF ID | Mintbase ID | tRF sequence (5′-3′) | Amino acid-anticodon | Fold change | P-value |
|---|---|---|---|---|---|
| AS-tDR-000586 | tRF-36-86J8WPMN1E8Y7ZD | TCCCACATGGTCTAGCGGTTAGGATTCCTGGTTTTC | Glu-TTC | 518.143 | 0.031859 |
| AS-tDR-007710 | N/A | GCCCGGATGATCCTCAGTGGTCTGGGGTGCAGGCTTC | SeC-TCA | 466.968 | 0.049086 |
| AS-tDR-011363 | N/A | CTGTCACGCGGGAGACCGGGGC | Asp-GTC | 317.306 | 0.040083 |
| AS-tDR-005857 | tRF-37-EH623K7SIR3DR2M | ACGCGGGAGACCGGGGTTCGATTCCCCGACGGGGAGC | Asp-GTC | 237.194 | 0.019999 |
| AS-tDR-006177 | tRF-16-K1JKE1B | CCACCCAGGGACGCCA | Asn-GTT | 217.935 | 0.049457 |
| AS-tDR-011429 | N/A | TAGGGAGGTTATGATTAACT | Arg-TCG | 196.348 | 0.006424 |
| AS-tDR-000107 | tRF-22-WEK879652 | TCGATCCCGGGTTTCGGCACCA | Phe-GAA | 188.997 | 0.027295 |
| AS-tDR-004084 | tRF-18-S8V0JUDR | GTCGGTAGAGCATGAGAC | Lys-CTT | 187.974 | 0.001717 |
| AS-tDR-007725 | N/A | TCCCTGTGGTCTAGTGGTTAGGATTCGGCGCTCTC | Glu-CTC | 22.298 | 0.040897 |
| AS-tDR-000547 | tRF-36-87R8WP9N1EWJQ7D | TCCCTGGTGGTCTAGTGGTTAGGATTCGGCGCTCTC | Glu-CTC | 15.778 | 0.043218 |
| AS-tDR-007304 | N/A | GCATTGGTGGTTCAGTGGTAGAATTCTCGCCTT | Glu-TTC | 7.596 | 0.007439 |
| AS-tDR-005361 | tRF-17-9LON4V3 | TGGTAGAATTCTCGCCT | Gly-CCC | 6.683 | 0.032141 |
| AS-tDR-006553 | tRF-18-P6KP6HD2 | GCCCACCCAGGGACGCCA | Asn-GTT | 6.191 | 0.016570 |
| AS-tDR-008080 | N/A | GCCCGGATGATCCTCAGTGGTCTGGGGTGCAGGCT | SeC-TCA | 5.507 | 0.024088 |
| AS-tDR-004206 | tRF-18-1SS2PMX | AGTCGGTAGAGCATCAGA | Lys-TTT | 5.097 | 0.031258 |
| AS-tDR-006922 | tRF-17-884U1D2 | TCCGGCTCGAAGGACCA | Tyr-GTA | 3.513 | 0.011413 |
| AS-tDR-001611 | tRF-22-8XF6RE98N | TCCTAAGCCAGGGATTGTGGGT | Arg-CCT | 3.227 | 0.041503 |
| AS-tDR-000122 | tRF-17-K5KKOV2 | CCCACCCAGGGACGCCA | Asn-GTT | 3.119 | 0.049625 |
| AS-tDR-001889 | tRF-16-941QKSD | TGGTGAGTATCCCCGC | Asp-GTC | 2.707 | 0.017218 |
| AS-tDR-008258 | N/A | TGGTTCAGTGGTAGAAT | Gly-GCC & Gly-CCC | 2.561 | 0.035012 |
| AS-tDR-008237 | N/A | CTAAGCCAGGGATTGTGGGC | Arg-CCT | 2.316 | 0.034470 |
| AS-tDR-004490 | tRF-21-JOWU1M70E | CATAATCTGAAGGTCGTGAGT | Met-CAT | 2.293 | 0.031923 |
tRFs, transfer RNA-derived fragments.
Table IV.
Summary of the 16 tRFs downregulated in patients with moyamoya disease vs. healthy controls.
| tRF ID | Mintbase ID | tRF sequence (5′-3′) | Amino acid-anticodon | Fold change | P-value |
|---|---|---|---|---|---|
| AS-tDR-007510 | N/A | ACTCTGGACTCTGAATCT | Gln-CTG | 297.887 | 0.016055 |
| AS-tDR-000924 | tRF-16-PS5P4PE | GCCCGGATAGCTCAGT | Lys-TTT | 260.969 | 0.037030 |
| AS-tDR-003801 | tRF-17-MMUKLYK | CGGTCTAAGGCGCTGCG | Leu-CAG | 186.513 | 0.032042 |
| AS-tDR-011424 | N/A | ATATCCAACCTTCGGCTATAGGG | Thr-CGT | 8.018 | 0.006046 |
| AS-tDR-011366 | tRF-19-WB86N7HU | TCGAATCCCACTTCTGACA | Leu-CAG & Leu-CAA | 7.902 | 0.023668 |
| AS-tDR-005697 | tRF-18-BS6PDFD2 | AACCGGGCAGAAGCACCA | Val-CAC | 7.168 | 0.010754 |
| AS-tDR-000053 | tRF-31-79MP9P9NH57SD | GTTTCCGTAGTGTAGTGGTTATCACGTTCGC | Val-AAC | 6.730 | 0.011393 |
| AS-tDR-009236 | N/A | GGCTCGAAGGACTTCGTCTGTAA | Tyr-GTA | 5.578 | 0.031881 |
| AS-tDR-009369 | tRF-21-WD8YQ84VD | TCGACTCCTGGCTGGCTCGCC | Arg-ACG | 5.389 | 0.042221 |
| AS-tDR-001449 | tRF-32-79MP9P9NH57SJ | GTTTCCGTAGTGTAGTGGTTATCACGTTCGCC | Val-AAC & Val-CAC | 5.162 | 0.016935 |
| AS-tDR-001400 | tRF-32-6978WPRLXN4VQ | GGCTCGTTGGTCTAGGGGTATGATTCTCGCTT | Pro-CGG & Pro-TGG & Pro-AGG | 5.159 | 0.006515 |
| AS-tDR-011405 | N/A | TTGAGATACTGAGTAGTCTGGTG | Arg-TCG | 4.422 | 0.016291 |
| AS-tDR-000009 | tRF-31-P4R8YP9LON4VD | GCATGGGTGGTTCAGTGGTAGAATTCTCGCC | Gly-GCC | 3.029 | 0.005745 |
| AS-tDR-007276 | N/A | GTTTCCGTAGTGTAGTGGTTATCACGTTCGCT | Val-CAC & Val-AAC | 2.853 | 0.048408 |
| AS-tDR-009331 | N/A | TCGACTCCTGGCTGGCTCG | Arg-ACG | 2.749 | 0.004973 |
| AS-tDR-001273 | tRF-35-86V8WPMN1E8Y7Z | TCCCATATGGTCTAGCGGTTAGGATTCCTGGTTTT | Glu-TTC | 2.377 | 0.033231 |
tRFs, transfer RNA-derived fragments.
Figure 1.
Volcano plot of the differentially expressed tRFs. Red dots represent tRFs differentially expressed in patients with MMD vs. healthy controls (fold change ≥2.0; P<0.05). MMD, moyamoya disease; tRFs, transfer RNA-derived fragments.
Figure 2.
Heat map of the differentially expressed tRFs in patients with MMD vs. healthy controls. Red and green indicate upregulation and downregulation, respectively. MMD, moyamoya disease; tRFs, transfer RNA-derived fragments.
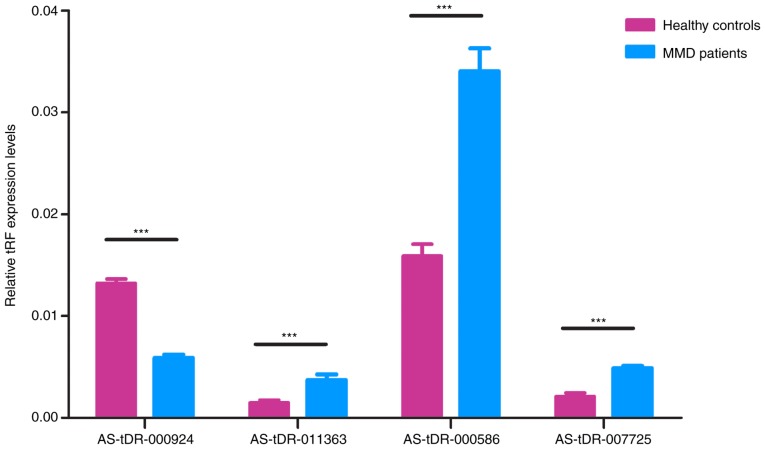
RT-qPCR validation of RNA sequencing results
Four tRFs (AS-tDR-000586, AS-tDR-000924, AS-tDR-007725 and AS-tDR-011363) were randomly selected for validation of RNA sequencing results using RT-qPCR in MMD and control samples (n=5). The expression profiles of these tRFs were in agreement with the results obtained from RNA sequencing. There were significant differences in tRF expression levels in patients with MMD compared with healthy controls (P<0.05; Fig. 3). These findings indicated that the RNA sequencing data were reliable enough to warrant further analysis.
Figure 3.
Reverse transcription-qPCR validation of four randomly selected tRFs in an independent cohort of patients with MMD and healthy controls. The qPCR results were consistent with the RNA sequencing results, demonstrating the reliability of the tRF expression profile data. Data are presented as the means ± standard deviation (n=5). ***P<0.05. MMD, moyamoya disease; qPCR, quantitative polymerase chain reaction; tRFs, transfer RNA-derived fragments.
tRF target gene prediction
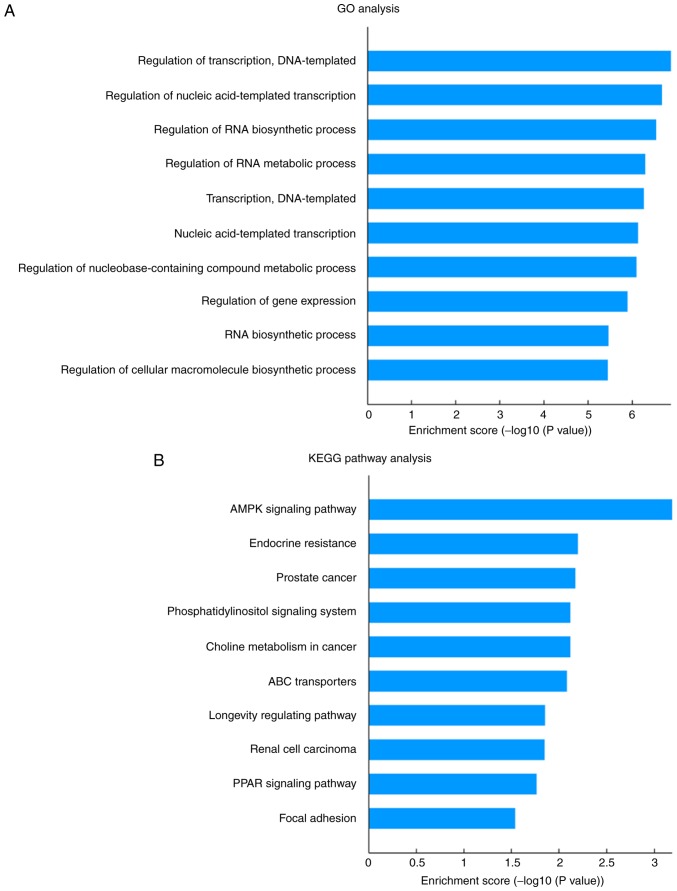
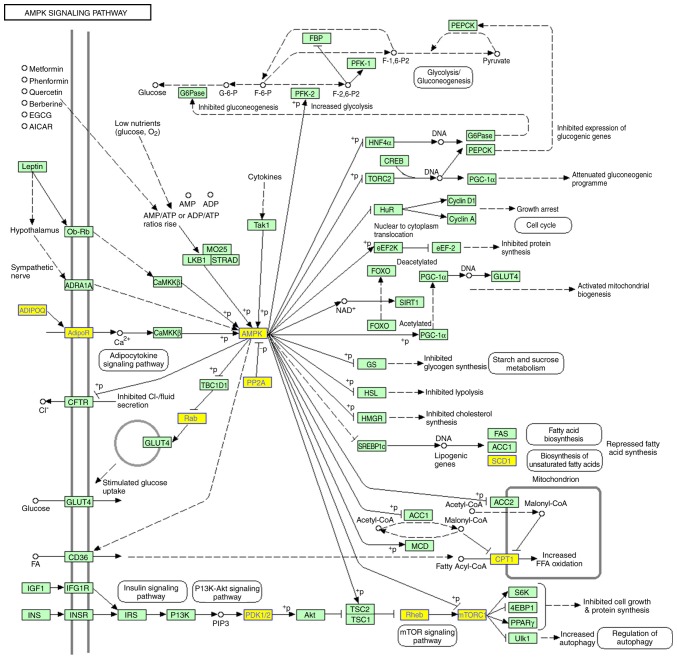
In order to investigate the function of these tRFs, their putative target genes were predicted using miRanda and TargetScan. GO analysis of these predicted tRF targets revealed 220 significantly enriched GO terms for biological process. The top five GO biological process terms were ‘regulation of transcription, DNA-templated’, ‘regulation of nucleic acid-templated transcription’, ‘regulation of RNA biosynthetic process’, ‘regulation of RNA metabolic process’ and ‘transcription, DNA-templated’ (Fig. 4A). KEGG pathway analysis revealed 15 significantly enriched pathways that corresponded to the target genes (P<0.05; Fig. 4B). These pathways were mainly angiogenesis- and metabolism-associated pathways, including ‘AMPK signaling pathway’, ‘phosphatidylinositol signaling system’, ‘ABC transporters’ and ‘endocrine resistance’. As shown in Fig. 5, a number of genes involved in the AMP-activated protein kinase (AMPK) signaling pathway were targets of tRFs.
Figure 4.
Bioinformatics analysis of tRF target genes. Top 10 significantly enriched (A) GO terms and (B) KEGG pathways for the predicted tRF target genes. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; tRFs, transfer RNA-derived fragments.
Figure 5.
Transfer RNA-derived fragment target genes in the Kyoto Encyclopedia of Genes and Genomes AMPK pathway map generated using DAVID software. Target genes are highlighted in yellow. AMPK, AMP-activated protein kinase.
Discussion
MMD is a complex disease with unknown pathophysiology. Several factors, including genetic, acquired and environmental factors, have been implicated in MMD (26). Previous studies have demonstrated that familial MMD is inherited in an autosomal dominant manner with incomplete penetrance; therefore, genetic factors may serve a pivotal role in MMD development and progression (26,27). Through the extensive development of high-throughput sequencing, tRFs have become a highly studied topic in the field of biology and medicine. Numerous studies have been conducted to explore their biogenesis and biological functions (11,13,28,29); however, to the best of our knowledge, there are no reports demonstrating tRF expression profiles in patients with MMD, and the association between tRFs and MMD. In the present study, the expression of tRFs in patients with MMD and healthy controls were explored; 38 tRFs were differentially expressed and maybe linked to the development of the disease.
tRFs are a type of small non-coding RNA, which were first identified in 1977 in the urine and serum of patients with cancer (30,31). Due to their size, tRFs are broadly classified into two groups. tiRNAs are 30–40 nucleotides (nt) long and are generated by specific cleavage at the anticodon loop of full-length tRNAs. Another class of tRFs is 18–22 nt long and consists of four subtypes: tRF-1, tRF-3, tRF-5 and i-tRF. Although the function of tRFs remains largely undefined, increasing evidence suggests that tRFs can act in canonical miRNA pathways (19,22,32–34). For example, Maute et al revealed that CU1276, a tRF-3 derived from tRNAGly(GCC), is abundantly expressed in normal germinal center B cells and manifests the same functional features as an miRNA (22). In addition, it has been demonstrated that this particular tRF can bind AGO proteins and post-transcriptionally repress replication protein A1 in a sequence-specific miRNA-like manner, demonstrating a clear regulatory role for tRFs. In 2014, Park et al compared the expression levels of miRNAs in patients with advanced breast cancer and healthy controls; it was revealed that miRNA-1280 is differentially expressed in patients and could reflect tumor status, highlighting its potential as a clinical biomarker. Subsequently, miRNA-1280 has been confirmed to be a tRF from tRNALeu rather than a classical miRNA (34). In addition, tRFs have been reported to have roles in cerebrovascular diseases. By using deep sequencing to profile small RNA transcripts in rat brain tissues, Li et al revealed that the expression of tRFs is significantly enhanced following ischemia (28). Two specific tRFs derived from tRNAVal and tRNAGly could suppress angiogenesis by inhibiting the function of vascular endothelial cells, thus suggesting that these tRFs may have important roles in cerebral ischemic stroke.
It had previously been demonstrated that miRNAs and lncRNAs are differentially expressed in patients with MMD, and both are considered to be associated with MMD pathogenesis (5,6). In the present study, tRF expression profiles in peripheral blood samples were investigated, and it was revealed that tRF expression levels in patients with MMD were altered compared with in healthy controls. Amongst the differentially expressed tRFs, 5′-tiRNA derived from tRNAGlu(TTC) was the most significantly upregulated tRF, whereas i-tRF from tRNAGln(CTG) and tRF-1 from pre-tRNAGln(CTG) were significantly downregulated. The results suggested that these tRFs may have the potential to serve as novel candidate biomarkers for MMD. In addition, tRFs from tRNAGly and tRNAVal were upregulated in patients with MMD, which was in accordance with a previous study performed in rat ischemic brain (28). This may be due to progressive stenosis or occlusion of the intracranial vessels in patients with MMD leading to reduced blood supply to the brain and cerebral ischemia. By using GO enrichment analysis, it was identified that these tRFs were mainly involved in biological regulation. Notably, it has previously been reported that tRFs can act as signaling molecules and modulators invarious biological processes (9,35,36).
AMPK is a stress-activated protein kinase, which can be activated by nutrient deprivation, including hypoxia/ischemia, vigorous exercise and starvation (37,38). The activation of AMPK appears to be crucial for angiogenesis (38), the main process through which new blood vessels are formed. Adiponectin, an AMPK activator, can promote angiogenesis in response to tissue ischemia by activating AMPK signaling in endothelial cells (39). Vascular endothelial growth factor (VEGF) is an endothelial cell-specific mitogen, which has a critical role in angiogenesis (39). Nagata et al identified that suppression of AMPK signaling could inhibit endothelial cell migration to VEGF and in vitro differentiation into tube-like structures under hypoxic conditions, further suggesting that AMPK signaling is essential for angiogenesis in response to ischemic stress (37). Notably, aberrant angiogenesis of blood vessel endothelial cells is considered to be the pathological basis of the onset of MMD (40). In the present study, pathway enrichment analysis revealed the ‘AMPK signaling pathway’ as the most enriched pathway, which was composed of several tRF target genes, indicating that tRFs may be involved in the pathogenesis of MMD.
The phosphatidylinositol signaling system is another affected pathway identified in the present study. Phosphoinositide 3-kinases (PI3Ks) are a conserved family of enzymes that phosphorylate the 3′-OH of the inositol ring of phosphatidylinositol (41). The PI3K pathway has been reported to be closely correlated with angiogenesis (41). Activation of the PI3K/protein kinase B pathway can increase VEGF production. In addition, this pathway can regulate the expression of other angiogenic factors, including nitric oxide and angiopoietins (41). The PI3K pathway also serves a pivotal role in regulating the proliferation of vascular smooth muscle cells (42).
Other pathways identified in the present study are also closely associated with vascular diseases, including ‘ABC transporters’, ‘ErbB signaling pathway’ and ‘Hippo signaling pathway’. There are many signaling pathways that are associated with MMD; therefore, it is difficult to verify each pathway individually. Nevertheless, the results suggested that tRFs may be involved in the development and progression of MMD, and provided novel therapeutic strategies for clinical treatment of the disease. However, one of the limitations of the present study was the small sample size. Future studies with a larger sample size are required to better understand the potential role of tRFs in the pathophysiology of MMD. Additionally, the functions of tRFs in MMD are based on bioinformatics prediction; therefore, further in vitro and in vivo experiments are required to validate the findings.
In conclusion, the expression profile of tRFs in patients with MMD was revealed using small RNA sequencing, which also identified tRFs that were expressed in an MMD-specific manner. In addition, bioinformatics analyses were used to predict tRFs that may be involved in the pathophysiology of MMD. The results of the present study provided novel insight into the mechanisms underlying MMD pathogenesis, and suggested tRFs that may serve as potential therapeutic targets for the future treatment of MMD.
Acknowledgements
The authors would like to thank Dr Meng Wang (Capital Institute of Pediatrics, Beijing, China) for proofreading the manuscript.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81371292) and the ‘13th Five-Year Plan’ National Science and Technology Supporting Plan (grant no. 2015BAI12B04).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
CW conceived the study, reviewed the literature, and wrote the initial and final drafts of the manuscript. MZ, JW, DZ and SW collected, analyzed and interpreted the data. JZ designed the present study, critically revised the manuscript and gave the final approval for publication. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was conducted in accordance with the Declaration of Helsinki, and was approved by the Research Ethics Board of Beijing Tiantan Hospital, Capital Medical University. Written informed consent was obtained from all individuals who were included in the study.
Patient consent for publication
All patients within this study provided consent for the publication of their data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Takeuchi K, Shimizu K. Hypoplasia of the bilateral internal carotid arteries. Brain Nerve. 1957;9:37–43. [Google Scholar]
- 2.Kim JS. Moyamoya disease: Epidemiology, clinical features, and diagnosis. J Stroke. 2016;18:2–11. doi: 10.5853/jos.2015.01627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Acker G, Fekonja L, Vajkoczy P. Surgical management of moyamoya disease. Stroke. 2018;49:476–482. doi: 10.1161/STROKEAHA.117.018563. [DOI] [PubMed] [Google Scholar]
- 4.Kuriyama S, Kusaka Y, Fujimura M, Wakai K, Tamakoshi A, Hashimoto S, Tsuji I, Inaba Y, Yoshimoto T. Prevalence and clinicoepidemiological features of moyamoya disease in Japan: Findings from a nationwide epidemiological survey. Stroke. 2008;39:42–47. doi: 10.1161/STROKEAHA.107.490714. [DOI] [PubMed] [Google Scholar]
- 5.Dai D, Lu Q, Huang Q, Yang P, Hong B, Xu Y, Zhao W, Liu J, Li Q. Serum miRNA signature in Moyamoya disease. PLoS One. 2014;9:e102382. doi: 10.1371/journal.pone.0102382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao F, Yu L, Zhang D, Zhang Y, Wang R, Zhao J. Long noncoding RNAs and their regulatory network: Potential therapeutic targets for adult moyamoya disease. World Neurosurg. 2016;93:111–119. doi: 10.1016/j.wneu.2016.05.081. [DOI] [PubMed] [Google Scholar]
- 7.Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, Daub CO, Kai C, Kawai J, Yasuda J, et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: TRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martens-Uzunova ES, Olvedy M, Jenster G. Beyond microRNA-novel RNAs derived from small non-coding RNA and their implication in cancer. Cancer Lett. 2013;340:201–211. doi: 10.1016/j.canlet.2012.11.058. [DOI] [PubMed] [Google Scholar]
- 11.Kumar P, Kuscu C, Dutta A. Biogenesis and function of transfer RNA-related fragments (tRFs) Trends Biochem Sci. 2016;41:679–689. doi: 10.1016/j.tibs.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson P, Ivanov P. TRNA fragments in human health and disease. FEBS Lett. 2014;588:4297–4304. doi: 10.1016/j.febslet.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng GH, Peng H, Zhang X, Zhang Y, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351:397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 14.Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33:2020–2039. doi: 10.15252/embj.201489282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Research Committee on the Pathology, Treatment of Spontaneous Occlusion of the Circle of Willis, Health Labour Sciences Research Grant for Research on Measures for Infractable Diseases Guidelines for diagnosis and treatment of moyamoya disease (spontaneous occlusion of the circle of Willis) Neurol Med Chir (Tokyo) 2012;52:245–266. doi: 10.2176/nmc.52.245. [DOI] [PubMed] [Google Scholar]
- 16.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Shigematsu M, Kirino Y. tRNA-derived short non-coding RNA as interacting partners of argonaute proteins. Gene Regul Syst Bio. 2015;9:27–33. doi: 10.4137/GRSB.S29411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karaiskos S, Grigoriev A. Dynamics of tRNA fragments and their targets in aging mammalian brain. F1000Res 5. ISCB Comm J. 2016:2758. doi: 10.12688/f1000research.10116.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar P, Anaya J, Mudunuri SB, Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014;12:78. doi: 10.1186/s12915-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, Dalla-Favera R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci USA. 2013;110:1404–1409. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyoshi K, Miyoshi T, Siomi H. Many ways to generate microRNA-like small RNAs: Non-canonical pathways for microRNA production. Mol Genet Genomics. 2010;284:95–103. doi: 10.1007/s00438-010-0556-1. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki J, Takaku A. Cerebrovascular ‘moyamoya’ disease. Disease showing abnormal net-like vessels in base of brain. Arch Neurol. 1969;20:288–299. doi: 10.1001/archneur.1969.00480090076012. [DOI] [PubMed] [Google Scholar]
- 25.van Swieten JC, Koudstaal PJ, Visser MC, Schouten HJ, van Gijn J. Interobserver agreement for the assessment of handicap in stroke patients. Stroke. 1988;19:604–607. doi: 10.1161/01.STR.19.5.604. [DOI] [PubMed] [Google Scholar]
- 26.Scott RM, Smith ER. Moyamoya disease and moyamoya syndrome. N Engl J Med. 2009;360:1226–1237. doi: 10.1056/NEJMra0804622. [DOI] [PubMed] [Google Scholar]
- 27.Mineharu Y, Liu W, Inoue K, Matsuura N, Inoue S, Takenaka K, Ikeda H, Houkin K, Takagi Y, Kikuta K, et al. Autosomal dominant moyamoya disease maps to chromosome 17q25.3. Neurology. 2008;70:2357–2363. doi: 10.1212/01.wnl.0000291012.49986.f9. [DOI] [PubMed] [Google Scholar]
- 28.Li Q, Hu B, Hu GW, Chen CY, Niu X, Liu J, Zhou SM, Zhang CQ, Wang Y, Deng ZF. tRNA-derived small non-coding RNAs in response to ischemia inhibit angiogenesis. Sci Rep. 2016;6:20850. doi: 10.1038/srep20850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivanov P. Emerging roles of tRNA-derived fragments in viral infections: The case of respiratory syncytial virus. Mol Ther. 2015;23:1557–1558. doi: 10.1038/mt.2015.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borek E, Baliga BS, Gehrke CW, Kuo CW, Belman S, Troll W, Waalkes TP. High turnover rate of transfer RNA in tumor tissue. Cancer Res. 1977;37:3362–3366. [PubMed] [Google Scholar]
- 31.Speer J, Gehrke CW, Kuo KC, Waalkes TP, Borek E. tRNA breakdown products as markers for cancer. Cancer. 1979;44:2120–2123. doi: 10.1002/1097-0142(197912)44:6<2120::AID-CNCR2820440623>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 32.Karaiskos S, Naqvi AS, Swanson KE, Grigoriev A. Age-driven modulation of tRNA-derived fragments in Drosophila and their potential targets. Biol Direct. 2015;10:51. doi: 10.1186/s13062-015-0081-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sobala A, Hutvagner G. Transfer RNA-derived fragments: Origins, processing, and functions. Wiley Interdiscip Rev RNA. 2011;2:853–862. doi: 10.1002/wrna.96. [DOI] [PubMed] [Google Scholar]
- 34.Park IH, Kang JH, Lee KS, Nam S, Ro J, Kim JH. Identification and clinical implications of circulating microRNAs for estrogen receptor-positive breast cancer. Tumour Biol. 2014;35:12173–12180. doi: 10.1007/s13277-014-2525-5. [DOI] [PubMed] [Google Scholar]
- 35.Yeung ML, Bennasser Y, Watashi K, Le SY, Houzet L, Jeang KT. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: Evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 2009;37:6575–6586. doi: 10.1093/nar/gkp707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013;10:1798–1806. doi: 10.4161/rna.27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagata D, Mogi M, Walsh K. AMP-activated protein kinase (AMPK) signaling in endothelial cells is essential for angiogenesis in response to hypoxic stress. J Biol Chem. 2003;278:31000–31006. doi: 10.1074/jbc.M300643200. [DOI] [PubMed] [Google Scholar]
- 38.Fisslthaler B, Fleming I. Activation and signaling by the AMP-activated protein kinase in endothelial cells. Circ Res. 2009;105:114–127. doi: 10.1161/CIRCRESAHA.109.201590. [DOI] [PubMed] [Google Scholar]
- 39.Ouchi N, Shibata R, Walsh K. AMP-activated protein kinase signaling stimulates VEGF expression and angiogenesis in skeletal muscle. Circ Res. 2005;96:838–846. doi: 10.1161/01.RES.0000163633.10240.3b. [DOI] [PubMed] [Google Scholar]
- 40.Chmelova J, Kolar Z, Prochazka V, Curik R, Dvorackova J, Sirucek P, Kraft O, Hrbac T. Moyamoya disease is associated with endothelial activity detected by anti-nestin antibody. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2010;154:159–162. doi: 10.5507/bp.2010.024. [DOI] [PubMed] [Google Scholar]
- 41.Karar J, Maity A. PI3K/AKT/mTOR pathway in angiogenesis. Front Mol Neurosci. 2011;4:51. doi: 10.3389/fnmol.2011.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morello F, Perino A, Hirsch E. Phosphoinositide 3-kinase signalling in the vascular system. Cardiovasc Res. 2009;82:261–271. doi: 10.1093/cvr/cvn325. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.