Abstract
As part of the SAMPL5 blind prediction challenge, we calculate the absolute binding free energies of six guest molecules to an octa-acid (OAH) and to a methylated octa-acid (OAMe). We use the double decoupling method via thermodynamic integration (TI) or Hamiltonian replica exchange in connection with the Bennett acceptance ratio (HREM-BAR). We produce the binding poses either through manual docking or by using GalaxyDock-HG, a docking software developed specifically for this study. The root mean square deviations for our most accurate predictions are 1.4 kcal mol−1 for OAH with TI and 1.9 kcal mol−1 for OAMe with HREM-BAR. Our best results for OAMe were obtained for systems with ionic concentrations corresponding to the ionic strength of the experimental solution. The most problematic system contains a halogenated guest. Our attempt to model the σ-hole of the bromine using a constrained off-site point charge, does not improve results. We use results from molecular dynamics simulations to argue that the distinct binding affinities of this guest to OAH and OAMe are due to a difference in the flexibility of the host. We believe that the results of this extensive analysis of host-guest complexes will help improve the protocol used in predicting binding affinities for larger systems, such as protein-substrate compounds.
Keywords: Binding free energy simulations, Thermodynamic integration, Hamiltonian replica exchange, Bennett acceptance ratio, Double decoupling method, Molecular dynamics simulations, GalaxyDock-HG
Introduction
Improving the methods used for the characterization of solubility [1, 2] and binding affinity [3, 4] is a crucial step in advancing drug design [5]. Free energy simulations (FES) have been used extensively in predicting binding affinities for receptor–ligand complexes. Both alchemical methods, (energy computed via exponential averaging approach [6], the Bennett acceptance ratio [7, 8] or thermodynamic integration [9]) and non-alchemical methods, based on the potential of mean force [10, 11], have been shown to be effective. They provide accurate assessments, especially if the binding pose or the experimental binding energy are known [12, 13]. But precisely computing the binding free energy becomes a challenging endeavor when no prior knowledge is available, and the assessment is made “blindfolded”.
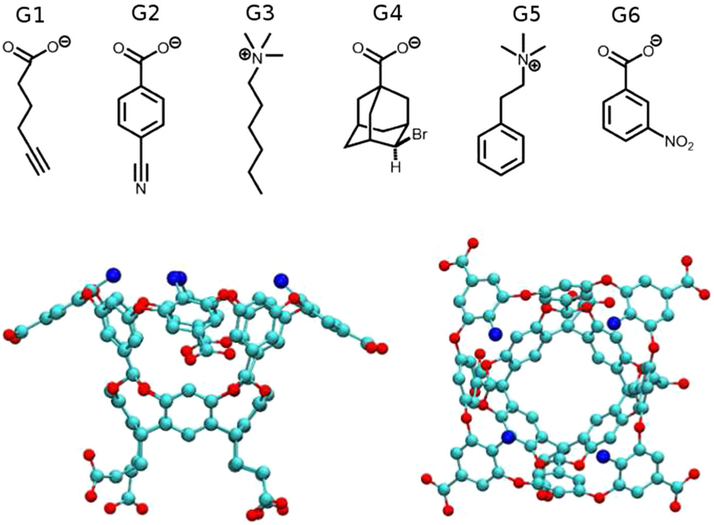
The statistical assessment of the modeling of proteins and ligands (SAMPL) blind challenge is a wonderful venue for testing the accuracy and precision of computational methods and for advancing new concepts and approaches [14–22]. Since its third edition, cucurbituril (CB) and CB-based molecules have been part of the host category (SAMPL3: CB7, CB8, and acyclic CB6, SAMPL4: CB7, SAMPL5: CBClip) [17–19]. Host-guest complexes consisting of such small molecule receptors and their ligands are well-suited for FES due to their fast convergence rates and to the simplicity of identifying the driving forces of the interactions. Octa-acids (OAH), a new type of water-soluble deep-cavity cavitands [23], were introduced as hosts in the SAMPL4 challenge [18, 24]. They exhibit eight carboxylates around the outer surface making it highly hydrophilic, whereas the interior is hydrophobic and consists of three rows of aromatic rings [25, 26]. Their ability to function as hosts was tested in SAMPL4 with a structurally diverse set of nine guests [18]. The strength of association was determined through a combination of 1H NMR and isothermal titration calorimetry (ITC). The same techniques were applied in SAMPL5 to determine the binding affinity of six guests (G1–G6) to OAH and to a methylated OAH (OAMe) [27] (see Fig. 1). Experiments were conducted at 25° C, in 10 mM sodium phosphate buffer for G1–G5 and 50 mM sodium phosphate buffer for G6, at pH 11.5 for ITC and pH 11.3 for NMR. The OAMe-G4 system was investigated at 5°C. In both SAMPL3 and SAMPL4, the lowest root mean square deviation (RMSD) for CB hosts was obtained through the solvated interaction energy (SIE) method with implicit solvent [28]. In SAMPL4, FES with explicit solvent [29] gave the best results for the OAH host. Both approaches used the general amber force field (GAFF) [30] to parametrize the molecules.
Fig. 1.
OAH/OAMe and guests in the charged state. The hosts are represented as ball-and-stick, carbon atoms are colored in cyan and oxygen atoms are colored in red; blue spheres represent the methyl groups in OAMe; hydrogen atoms are not shown
Here, we will discuss our approach to calculating absolute binding free energies of the OAH and OAMe hosts and their guests, and our results submitted to the SAMPL5 challenge. We give details on the implemented protocol, and we introduce GalaxyDock-HG, a modified protein-ligand docking program, used to obtain binding poses for the complexes. Results from a similar approach applied to the CBClip host can be found in Ref. [31]. We will also comment on the outcomes of the two studies, and on the possible sources of discrepancy.
Methods
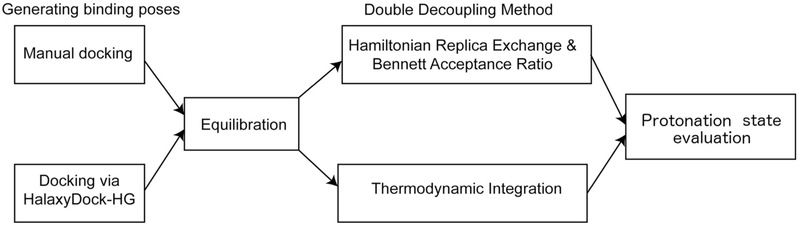
The protocol implemented for calculating the absolute binding free energy values is depicted in Fig. 2.
Fig. 2.
The protocol for calculating the absolute binding free energy values
System preparation
Parameters for the hosts and guests were generated by using the CHARMM General Force Field (CGENFF) for organic molecules [32]. The σ-hole parameters for G4 were generated similarly to work in Ref. [33]. All parameters are included in the Supporting Information. The two hosts had each a net charge of −8, due to presence of carboxylate groups and the high experimental pH. Similarly, four of the guests (G1, G2, G4 and G6) contained carboxylate groups and had a charge of −1. The remaining two guests (G3 and G5) contained a trimethyl ammonium ((N(CH3)4)+) group, and had a charge of +1. The guests were also modeled as neutral, to account for the possibility of changing protonation state upon binding. The free energy of “neutralizing” the various guests at the experimental pH was estimated by a series of quantum mechanical (QM) computations (see details in Supporting Information, QM Calculations subsection). For the anionic guests, we computed the free energy of binding a proton in the bulk aqueous phase by using QM methods, and then corrected this value by using the Henderson-Hasselbalch equation to obtain a binding free energy of the proton at the experimental pH. Because the cationic guests have no labile protons, yet are still acid-like (due to their ability to decrease hydroxide concentration in solution), we instead calculated the free energy of binding a hydroxide ion to the quaternary ammonium. We then applied the Henderson-Hasselbalch equation to account for the high concentration of hydroxide at pH 11.5. This analysis revealed that despite the fact that quaternary ammonium salts are known to readily dissociate, there may be significant populations of G3 and G5 that are associated with hydroxy ions, due to the extremely high pH of the experimental conditions (see Table 10 in Supporting Information).
To account for this effect, we chose to model “neutralized” versions of G3 and G5 by using a hydroxide ion restrained to the (N(CH3)4)+ group, at the QM-optimized distance of rN-O = 2.962 Å. The O—N—C angle (between the oxygen atom in the hydroxide ion, N atom in the tetramethylammonium group and the connecting C atom) was kept at 180° (see Supporting Information Figure 1).While this model fails to account for the proper, dynamic nature of the hydroxide association to the cationic guests, it does depict the statistically likely presence of the hydroxide in proximity of the quaternary ammonium group. We believe that this is a better approach than disregarding the effect entirely.
Guests and host-guest complexes were then solvated in TIP3P explicit solvent [34] in a cubic box with edges of 50 Å. We first modeled the guest in the cavity and then solvated the complex, therefore the cavity remained dry. We added enough Na+ and Cl− ions to either neutralize the system (i.e., bring the total charge of the system to zero; we call this system type “1”) or to reach the ionic strength of the experimental conditions (system type “2”) (see Supporting Information Table 1).
We used the CHARMM [35] version c39b2 [36] to perform energy minimization of the systems (55,000 steps) with constrained heavy atoms in both the host and the guest. We then annealed the systems with harmonically restrained heavy atoms (force constants of 1 kcal mol−1 Å −2) for 142,500 steps to 298 K. This was followed by equilibration for 500 ps, with heavy atoms in the host and guest harmonically restrained with force constants of 0.5 kcal mol−1 Å −2. The temperature was maintained constant by employing the Nosé-Hoover thermostat [37], and pressure was controlled by using the Langevin piston barostat [38]. Water geometry was kept rigid with SHAKE [39] and a 1-fs timestep was used.
Subsequent molecular dynamics (MD) simulations were unrestrained, unless the guest left the cavity immediately after equilibration, in which case we employed short (2–50 ns) restrained MD simulations; all MD simulations started from the bound state. The resulting structures were then used in calculations of the binding free energy. We applied distance restraints (from the nuclear Overhauser effect (NOE) module) between guest and host with force constants of 75 kcal mol−1 Å −2. We continued running unrestrained MD simulations for 400 ns to 2.5 μs per system (see Table 2 in Supporting Information). These last simulations were used in the analysis of the host dynamics.
Binding poses
The initial binding poses were generated in two ways: either by manually placing the guests in the cavity and keeping their charged groups atop the cavity, or through GalaxyDock-HG, a docking program that we developed specifically for the host-guest challenge. When performing the former, we first simulated the unrestrained host for 50 ns in an NPT ensemble, neutralized with 8 Na ions. We then placed the amphiphilic guests with the hydrophobic tail into the pocket of the host, and the hydrophilic head group towards the portal where it remains solvated. This reduces the overall hydrophobicity of the cavity and inhibits dimerization of cavitands and the formation of a capsule [24]. This orientation is especially favorable for G4, as it has been suggested that halogenated guests prefer a position with the halogen in the cavity, which enhances the CH· · ·Br-R interaction with the benzal hydrogen atoms [25, 26].
GalaxyDock-HG finds the guest binding poses through global optimization of the AutoDock4 scoring function [40–42] with the conformational space annealing (CSA) algorithm [43, 44]. GalaxyDock-HG was developed based on the Galaxy-Dock protein-ligand docking program [41]. The main differences between GalaxyDock-HG and GalaxyDock are as follows: (1) energy was evaluated in the continuous space instead of interpolating energy values at the grid points because the host-guest systems were small enough to run global optimization efficiently in the continuous space, and (2) the initial set of conformations for CSA (the initial bank) was generated by randomly perturbing initial structures instead of using the geometry-based predocking method of GalaxyDock, because more intensive pre-docking for small systems was deemed superfluous. Unlike the CBClip [31], the structural flexibility of the octa-acid and of the methylated octa-acid was ignored during docking since they are more rigid than CBClip, and disconsidering their flexibility did not affect the docking performance. A total of 50 conformations were selected as the initial bank after local energy minimization, and the bank was evolved by the CSA algorithm as implemented in GalaxyDock. More details on the method can be found in Supporting Information. Up to 3 final binding poses were selected based on their energy following the clustering, and we simulated the lowest-energy structure.
Free energy calculations
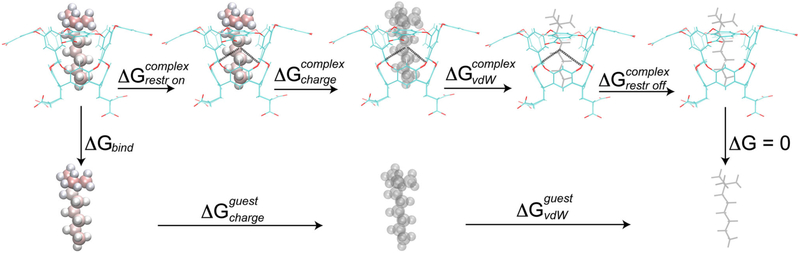
We calculated the values for the absolute binding free energy by subjecting the structures resulting from MD simulations to the double-decoupling method (DDM) [4, 45, 46]. As a first step of the DDM, the solvated guest is transferred from solution into gas phase. In the second step, the bound guest is transferred into gas phase, i.e., it ceases all interaction with the host and with the solvent. Combining these steps provides us with the binding free energy by using the thermodynamic cycle depicted in Fig. 3. DDM is a conformational-dependent method, therefore the guest is restrained with respect to the host, such that it does not drift from the binding site. This was performed by using one distance restraint, two angle restraints and three dihedral restraints. The force constants had the following values: 5 kcal mol−1 Å−2, 20 and 20 kcal mol−1 rad−2 for distance, angle and dihedral geometrical harmonic restraints, respectively. The free energy spent to turn the restraints off () between the guest and the host was calculated analytically, as described in Ref. [46] and detailed in Supporting Information. We calculated the binding free energy for both the charged and the neutral guests. To calculate the absolute binding free energies, we used the c39b2 version of the CHARMM package [35, 36], with CGenFF. We used either Thermodynamic integration (TI) [9] or the Hamiltonian replica exchange [47–49] post-processed with the Bennett acceptance ratio (HREM-BAR) [50–53]. We chose two alchemical methods due to being the most robust methods, albeit the most computationally demanding, to calculate the entities of interest [54]. We intended to compare TI, which is a more established method and is easier to implement, with HREM-BAR, which requires more computational resources. Based on work by Boresch and Bruckner [55], as well as our previous results in SAMPL3 [51], it was expected that BAR would outperform TI. However, as pointed out in Ref. [56], TI can be competitive with the right numerical quadrature scheme. Moreover, the TI approach uses soft cores [57, 58], while the HREM-BAR scheme is based on serial insertion [59], which was used to speed up the simulations.
Fig. 3.
Illustration of the thermodynamic cycle used in DDM
TI generates intermediate states between the initial (0) and final (1) states via combining each state’s potential function by a mixing factor λ. For each » value, it is necessary to conduct an independent simulation. We used 16 » points for estimating each free energy difference associated with turning off electrostatic (ΔGcharge) and van der Waals interactions (ΔGvdW). Simulations were run in an NVT ensemble. At each point we performed an equilibration for 20 ps, followed by 200 ps of production. The HREM-BAR simulations used 12 » points and 20 » points for estimating ΔGvdW and ΔGcharge, respectively. Each HREM-BAR simulation was run for 1 ns, with a total of 32 ns for each system. For both TI and HREM-BAR calculations, the free energy attributed to turning on the restraints () was computed with the PERT module of CHARMM using TI with 19 » values. Each » point was equilibrated for 20 ps followed by production for 100 ps. The results were integrated with the trapezoidal rule. For each host-guest complex, we repeated the simulations for at least three times, and, in some cases, for as many as twelve times. All FES used Particle Mesh Ewald and 14 Å cutoffs. We used a timestep of 1 fs in both approaches.
Calculating the free energy of neutralization
The guests exist in solution as a mixture of species with different charges. The charge of the guests can change depending on environmental conditions (e.g., pH), or upon binding to a host. To evaluate the free energy of modifying the charge of the bound guest ( in Fig. 4), we first computed the free energy of changing the charge while free in the bulk solution () by performing a series of QM calculations at the M06-2X/6-31+G(d) level of theory with SMD implicit solvent. For the anionic guests, this consisted in calculating the free energy of deprotonating a carboxylic acid in the bulk aqueous phase, whereas for the cationic guests, we computed the free energy of binding of a hydroxide ion to the guest. For further methodological details of these calculations, please see Supporing Information. We then estimated the binding free energy for both the ionic ( or ) and the neutralized form (). With knowledge of , (or ) and , and by using the thermodynamic cycle in Figure 4, we evaluated . Finally, we could estimate the correct free energy of binding by including contributions from both charged and neutral species. This analysis revealed that the ionic guests are charged in bulk and binding does not shift their pKa to the neutral state. It also indicates that inclusion of the “neutralized” states of the cationic guest helped improve our results.
Fig. 4.
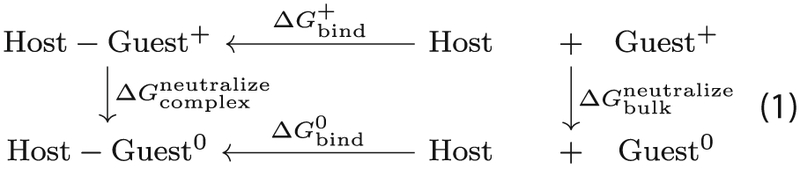
Thermodynamic cycle used for calculating the free energy necessary for the guest to change protonation state while in complex (), illustrated for a positively charged guest. To evaluate we first compute the free energy of binding of the charged guest () and of the neutral guest () to the host through DDM and evaluate the free energy of changing protonation state in bulk () through QM calculations
Results and discussion
FES results
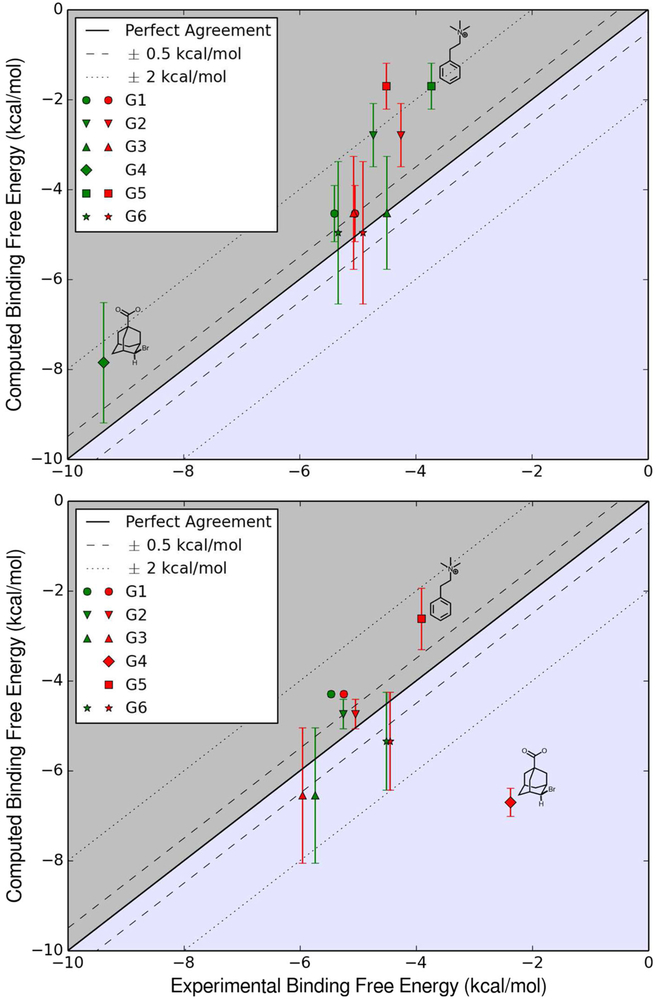
By combining different methods and solution concentrations with corrections for the protonation state we obtained seven sets of binding free energy values for each host and its cohort of guests: TI, TI-ps, HBAR, HBAR-ps, HBAR-ps1, HBAR-ps2 and “All”. Binding free energy values for each set are given in Table 1. These values were selected from the lowest computed binding free energy values for each system type by TI and by HREM-BAR, which are given in Tables 6 and 7 (Supporting Information, respectively. By selecting the lowest binding free energy value obtained for each system irrespective of the protonation state we obtained the TI and the HBAR sets. We then reassessed them by estimating the protonation state of the guest when bound to the host: “protonation state” -corrected, or ps-corrected (TI-ps, HBAR-ps), therefore these sets include values for G1−, G2−, G3°, G4−, G5° and G6−. We did not discern between values for systems with different ionic concentration in the TI and TI-ps sets. But when accounting for the salt concentration effect upon the binding free energy, the HBAR-ps set was split into HBAR-ps1 and HBAR-ps2, which contained values computed for systems of type “1” and type “2”, respectively. The “all” set combined all resulting free energy values, including higher binding free energy values not submitted in previous sets. This combination of data did not improve our errors with respect to experimental values. Computational results were compared to the ITC and the NMR experimental data. The correlation between experimental and computed values is plotted in Fig. 5. The errors (differences between experimental and computed values) pertaining to each experimental set are plotted in Supporting Information Figure 4.
Table 1.
Absolute binding free energy values for OAH-guest and OAMe-guest complexes (in kcal mol−1)
| Complex | † | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OAH–G1 | −7.6 ± 0.7 | −4.5 ± 0.6 | −6.4 ± 0.7 | −4.5 ± 0.2 | −4.2 ± 1.0 | −4.5 ± 0.2 | −3.4 ± 1.1 | −5.40 | −5.05 | −5.22 |
| OAH–G2 | −2.7 ± 0.7 | −2.7 ± 0.7 | −3.2 ± 0.2 | −3.2 ± 0.2 | −2.2 ± 0.2 | −3.2 ± 0.2 | −2.7 ± 0.3 | −4.73 | −4.26 | −4.49 |
| OAH–G3 | −4.8 ± 1.1 | −4.5 ± 1.2 | −3.4 ± 0.0 | −3.4 ± 0.0 | −3.4 ± 0.0 | −3.4 ± 0.0 | −4.0 ± 0.5 | −4.50 | −5.07 | −4.78 |
| OAH–G4 | −8.1 ± 1.5 | −7.8 ± 1.3 | −5.0 ± 0.1 | −5.0 ± 0.1 | −5.0 ± 0.1 | −4.4 ± 0.5 | −5.5 ± 1.1 | −9.38 | – | −9.38 |
| OAH–G5 | −1.7 ± 0.1 | −1.6 ± 0.5 | −1.7 ± 0.1 | −1.6 ± 0.5 | −1.6 ± 0.5 | −1.6 ± 0.5 | −1.6 ± 0.5 | −3.73 | −4.51 | −4.12 |
| OAH–G6 | −4.9 ± 1.5 | −4.9 ± 1.5 | −4.1 ± 0.1 | −4.1 ± 0.1 | −4.1 ± 0.1 | −4.1 ± 0.1 | −4.5 ± 0.2 | −5.34 | −4.91 | −5.12 |
| RMSD* | 1.6 | 1.4 | 2.2 | 2.2 | 2.3 | 2.4 | 2.1 | |||
| r* | 0.7 | 0.9 | 0.4 | 0.7 | 0.7 | 0.5 | 0.8 | |||
| τ* | 1.0 | 0.8 | 0.8 | 1.0 | 1.0 | 0.8 | 0.7 | |||
| OAMe–G1 | −6.1 ± 0.9 | −4.2 ± 0.8 | −6.4 ± 0.7 | −6.4 ± 0.7 | −4.2 ± 0.4 | −4.2 ± 0.0 | −3.7 ± 0.5 | −5.47 | −5.25 | −5.36 |
| OAMe–G2 | −7.1 ± 0.6 | −7.1 ± 0.6 | −5.3 ± 0.3 | −5.3 ± 0.3 | −5.3 ± 0.3 | −4.7 ± 0.3 | −6.1 ± 1.1 | −5.26 | −5.05 | −5.15 |
| OAMe–G3 | −6.5 ± 1.5 | −6.5 ± 1.5 | −6.5 ± 1.5 | −6.5 ± 1.5 | −6.5 ± 1.5 | −6.5 ± 1.5 | −6.5 ± 1.5 | −5.74 | −5.96 | −5.85 |
| OAMe–G4 | −9.9 ± 2.4 | −9.9 ± 2.4 | −10.5 ± 1.0 | −10.5 ± 1.0 | −10.5 ± 1.0 | −6.6 ± 0.3 | −7.8 ± 1.2 | – | −2.38 | −2.38 |
| OAMe–G5 | −2.6 ± 0.6 | −2.6 ± 0.6 | −2.9 ± 0.7 | −2.9 ± 0.7 | −2.6 ± 0.6 | −2.6 ± 0.6 | −2.6 ± 0.6 | − | −3.91 | −3.91 |
| OAMe–G6 | −7.0 ± 0.3 | −7.0 ± 0.3 | −7.0 ± 0.3 | −7.0 ± 0.3 | −7.0 ± 0.3 | −5.3 ± 1.0 | −6.0 ± 0.5 | −4.52 | −4.45 | −4.48 |
| RMSD* | 3.4 | 3.4 | 3.5 | 3.5 | 3.5 | 1.9 | 2.5 | |||
| r* | −0.3 | −0.4 | −0.4 | −0.4 | −0.5 | 0.0 | −0.2 | |||
| τ* | −0.2 | −0.2 | 0.0 | 0.0 | −0.2 | 0.0 | 0.0 | |||
| MSD** | −1.2 | −0.7 | −0.6 | −0.5 | −0.1 | 0.3 | 0.0 | |||
| MAD** | 2.3 | 2.2 | 2.3 | 2.3 | 2.4 | 1.9 | 2.2 | |||
| RMSDtot ** | 3.1 | 3.0 | 3.2 | 3.1 | 3.2 | 2.4 | 2.7 |
Rows 2–7 pertain to OAH-guest complexes and rows 9–14 correspond to OAMe-guest complexes. We included the root mean square deviation for each set (RMSD), the overall mean signed deviation (MSD) and mean absolute deviation (MAD) and the root mean square deviation for each method (RMSDtot)
Average experimental value from ITC and NMR experiments. The NMR data are missing the OAH-G4 binding free energy, and the ITC data are missing OAMe-G4 and OAMe-G5 binding free energies
RMSD, Pearson’s coefficient (r) and Kendall rank coefficient (τ) computed with respect to the average of the two experimental sets
MSD, MAD and RMSDtot computed with respect to the average of the two experimental sets
Fig. 5.
Top Computed for OAH and guests versus ITC and NMR values. Bottom Computed for OAMe and guests versus ITC and NMR values. Values from the ITC data set are represented by green symbols and from the NMR data set are represented in red. Above the ’perfect agreement’ line, the values are underestimated, and below the line values are overestimated
To simplify the analysis, we used the average of the ITC and and the NMR data as our reference values for some metrics and combined the data for OAH and OAMe to calculate an overall mean signed deviation (MSD), mean absolute deviation (MAD) and root mean square deviation (RMSDtot). The resulting metrics are given in the last three rows of Table 1. The overall best result was given by HBAR-ps2 with an RMSD of 2.4 kcal mol−1 (see column 7 of Table 1). The second best result is given by TI-ps with a RMSD of 3.0 kcal mol−1 (see third column of Table 1). This indicates that BAR overall slightly outperforms TI. This finding agrees with previous experiences in SAMPL3, where CGenFF also obtained a similar RMSD for host-guest systems (2.6 kcal mol−1) [51].
A comparison of the protonation state-corrected results (RMSD of 2.4 and 3.0 kcal mol−1 for BAR and TI) with the raw results without corrections (RMSD of 3.2 and 3.1 kcal mol−1 for BAR and TI, see HBAR and TI results in columns two and four of Table 1) highlights the influence of protonation states on the binding. For BAR, the RMSD improves by 0.8 kcal mol−1. The comparison between HBAR-ps2 (RMSD of 2.4 kcal mol−1) and HBAR-ps1 (RMSD of 3.2 kcal mol−1) demonstrates the relative importance of reproducing the ionic strength of the experimental conditions. While HBAR-ps1 only contains enough counter ions to neutralize the system, HBAR-ps2 contains more ions in order to reproduce ionic strength. This indicates that the influence of ions on the host-solvent or guest-solvent is not negligible.
In terms of systematic error (MSD), the results of the different methods range between −1.2 and 0.3 kcal mol−1. TI exhibits the largest absolute MSD with −1.2 kcal mol−1. The MSD of TI decreases upon incorporation of the effect of protonation states to −0.7 kcal mol−1. However, this MSD is still larger than the MSD of BAR without correcting for the protonation state (−0.6 kcal mol−1). This could be an effect of systematic errors in the numerical quadrature schemes, but additional analysis would be required to confirm that. The MSD of BAR (−0.6 kcal mol−1) improves upon inclusion of the protonation state and ionic strength effects to +0.3 kcal mol−1. Thus it is possible to infer that some over-correction took place. However, considering that the average standard deviations are larger than this difference, it is probably not statistically significant.
When considering the convergence of the different methods in terms of the standard deviations, one can see that the TI results are less converged than the BAR results (average standard deviations of 1.0 and 0.5 kcal mol−1). This may be attributed to the very short length of the TI simulations (0.2 ns per » point) compared to the BAR calculations (1.0 ns per » point). Furthermore, the BAR results are based on Hamiltonian replica exchange, therefore it is probable that the sampling of the BAR simulations is significantly improved compared to TI. This difference in sampling can probably explain most of the different behavior.
When analyzing data corresponding to each host, the TI-ps data set exhibited the lowest RMSD values for OAH: 1.4 kcal mol−1 when compared to the averaged experimental values (last column in Table 1). For each experimental set, the RMSD were 1.3 kcal mol−1 when compared to the ITC data, and 1.4 kcal mol−1 compared to the NMR data (which do not include G4). This set also scored the highest value for Pearson’s correlation coefficient (r = 0.9) among our submitted sets, and a reasonable value for the Kendall correlation coefficient (τ = 0.8). All τ values for the OAH-guest complexes are between 0.7 and 1. For OAMe, the HBAR-ps2 data set registered the lowest RMSD value of 1.9 kcal mol−1 (0.8 kcal mol−1 compared to the ITC data set, which did not include values for G4 and G5, and 1.9 kcal mol−1 compared to the NMR data). The HBAR-ps2 set registered negative values for both correlation coefficients, r = −0.09 and τ = −0.07. All submitted sets for the OAMe host scored negative or mostly low or negative r and τ values. This can be attributed to few data points, out of which OAMe-G4 was grossly over-estimated. We continued the HREM-BAR simulations for both the OAH and the OAMe systems following the conclusion of the SAMPL5 challengs (see Table 7 in Supporting Information, italicized values) and the RMSD for OAH decreased from 1.4 to 1.2 kcal mol−1. The lowest-RMSD set changed from TI-ps to HREM. This result also shows that HREM-BAR was more successful than TI in obtaining data that are closer to the experimental values. However, one should keep in mind that the simulation time for each » point for TI was quite short. Additional TI simulations with equilibration and production times increased to 50 and 500 ps, respectively, showed that errors with respect to experimental value were improved (results in Table 8 Supporting Information), indicating that longer TI simulation are required to obtain more accurate results.
The low RMSD values obtained with HBAR-ps2 reflect the need to reproduce the ionic strength of the experimental solution. ΔGbind values for the best-performing set are plotted versus the ITC and NMR experimental data sets for each host in Fig. 5. The TI-ps data set underestimated values of the binding energies of the guests to OAH, whereas the HBAR-ps2 data set was evenly-distributed. Most values were within a 2 kcal mol−1 deviation from perfect agreement. In both sets, the most striking outliers were G4 and G5. For G4 we underestimated its ΔGbind to OAH by at least 1.2 kcal mol−1, and overestimated its ΔGbind to OAMe by at least 4.3 kcal mol−1 in each data set we submitted. G4 registers the largest difference (7 kcal mol−1) in ΔGbind between the two hosts. For all other guests, the difference between ΔGbind in OAH and in OAMe is ≤ 1, 0.1, 0.6, 1.0, 0.2 and 0.6 kcal mol−1. G4 is the strongest binder to OAH and the weakest binder to OAMe, with binding constants of 7.43 × 106 M−1 and 5.57 × 101 M—1. G4 is also the only guest for which there is only one experimental value for each host.
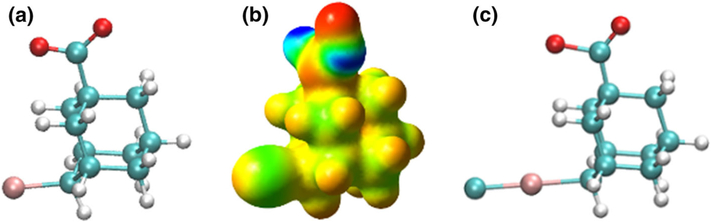
This discrepancy led us to investigate further the manner in which G4 binds to the two hosts. When we first parametrized G4 (Fig. 6a), we did not take into account the σ-hole [60] of the bromine (Fig. 6b). We reparametrized G4 (Fig. 6c) and added a lonepair to the bromine group, similarly to this work [33] (see Supporting Information for the parameter files). The resulting OAH-G4 and OAMe-G4 complexes were solvated and we added 25 mM of NaCl, which reproduces the ionic strength of 10 mM Na3PO4 at pH 11. We employed the same protocol as for the previous systems and recalculated the binding free energies, included in Supporting Information Table 9. ΔGbind became more positive, underestimating the binding affinity for both hosts.
Fig. 6.
a Ball-and-stick representation of the initially-parametrized G4, b electrostatic potential surface for G4, c ball- and-stick representation of G4 with lonepair
The binding free energy for OAH-G4 was initially underestimated by −1.5 kcal mol−1 in the TI-ps set and for OAMe-G4 was overestimated by 4.3 kcal mol−1 in the HBAR-ps2 set. When re-computing ΔGbind for the OAH-G4 complex, the difference increased to −4.7 kcal mol−1 by TI and to −5.5 kcal mol−1 by HREM-BAR. As for OAMe-G4, the energy values become underestimated by −2.3 kcal mol−1 by TI and −0.8 kcal mol−1 by HREM-BAR. We combined these new results with the rest of the TI-ps for OAH complexes and HBAR-ps2 for OAMe, and re-evaluated the RMSD. The RMSD values changed from 1.3 to 2.4 kcal mol−1 for OAH, and from 1.9 to 1.8 kcal mol−1 for OAMe. This showed that the addition of the lonepair improved the RMSD for OAMe only marginally, whereas for OAH it seemed to be quite detrimental.
Guest ranking by binding free energy was not consistent among experimental (ITC, NMR) nor computational methods (TI, HREM-BAR, GalaxyDock-HG). But, as Table 12 (Supporting Information) shows, there was somewhat consensus as to what the strongest and weakest binders are.
MD simulations
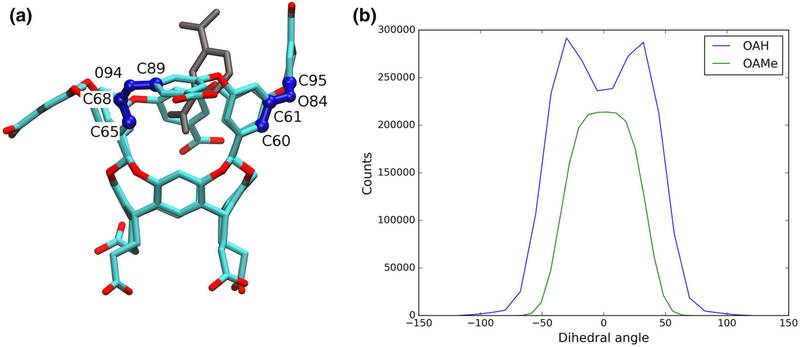
We used long unrestrained all-atom MD simulations to analyze the difference in the dynamics of the hosts. Details on the systems and simulation times for each system are given in Tables 1 and 3 (Supporting Information), respectively. We generated 53.4 μs of all-atom trajectories, 28.7 μs for the OAH systems and 24.7 μs for the OAMe systems. We investigated the flexibility of the two hosts by analyzing the eight dihedral angles that include the ether bond along the portal to the cavity. The third row of aromatic rings can adopt an ‘up’ or a ‘down’ conformation (Fig. 7a), changing the values of the dihedral angles involving the connecting ether bonds. The flexibility of the portal to the cavity was predicted experimentally [26, 61]. We show that the angles adopt more extreme values in the up conformation. In the down conformation, the dihedral angles have values from −50 to +50, whereas in the up conformation, the angles range from −105 to +105. The different angle distribution between OAH and OAMe indicates that values for the angles associated with the ‘up’ conformation are missing in the OAMe systems. Since the only difference between the two hosts are the methyl groups around the portal in OAMe, we inferred that they hinder the flexibility of the ether bonds. We believe that this rigidity could explain the low binding affinity of the voluminous G4 to OAMe.
Fig. 7.
a OAH with two analogous dihedral angles and the corresponding atoms: the angle in the down conformation has a value of 40.8° and the angle in the up conformation has a value of 104.8°. b Dihedral angle distribution of the values of all eight dihedral angles around the portal for OAH and OAMe. As exemplified in a, the difference between “up” and “down” is that the values become more extreme
Conclusions
The binding free energies for two hosts and a cohort of six guests were computed by employing alchemical techniques with the DDM (Fig. 3). Since DDM is very sensitive to the initial orientation of the guest relative to the host, we made efforts to generate sensible binding poses and developed a new docking program for small molecular receptors and ligands, GalaxyDock-HG, based on the protein-ligand analog [44]. To calculate the binding free energies, we developed a specific protocol (Fig. 2) and used two different approaches: TI with softcore potentials and HREM-BAR, which used serial deletion. Both methods performed well and our results submitted to the SAMPL5 challenge were consistently ranked near the top by RMSD with respect to the experimental data [19]. We evaluated the protonation state of the bound guests, which estimated G1, G2, G4 and G6 to be charged in solution, as well as when bound to the hosts. G3 and G5 were found to be “neutral” in solution and “neutral” when complexed to the host, due to the high concentration of hydroxide at the elevated experimental pH.
We consistently obtained large errors with respect to experimental data for G4, a halogen-adamantane carboxylic acid, with either host. There is evidence of carceroisomerism [26, 62, 63] for halogen-adamantane guests preferring a binding position with the halogen oriented towards the interior of the cavity [26]. By placing G4 in this orientation within the host, our results indicated strong binding to both OAH and OAMe, albeit slightly underestimated for OAH-G4, and grossly overestimated for OAMe-G4. Our first hypothesis was that this error was due to the poorly-parametrized bromine: it is a large atom that we represented as a point charge. By modeling the associated σ-hole and attaching a lonepair to the bromine group, ΔGbind became more positive for both OAH and OAMe, and overall results did not improve. We hypothesized further that the four methyl groups bordering the portal of the host might inhibit the binding of G4 into the cavity.
Experiments have shown that halogen-adamantanes (chloro-, bromo-, iodo-) bind to octa-acids more strongly than adamantane, and that neither adamantane, nor halogen-adamantane bind to OAMe [61]. The analysis of the dihedral angles that include the ether bonds between the second and third row of aromatic rings shows that OAMe is more rigid than OAH. The presence of the methyl groups prohibits the dynamics of the dihedral angles and hinders the flexibility of the host, which can be crucial in ligand binding. Even though GalaxyDock-HG provided us with a binding pose of the guest inside the cavity, similar to the manually-placed guest, this starting position might be incorrect.
Under-estimating DGbind for the OAH-G4 system might also point towards either a need for further reparametrization of the guest or for a different approach: guests like G4 might benefit from the use of advanced force-fields with anisotropy and charge penetration terms. In Ref. [64], Jiao et al. used the double decoupling method with a polarizable potential energy function and explicit-water to calculate the binding affinities of benzamidine and diazamidine to trypsin, and obtained very good agreement with experiment. Electrostatics and polarization play important roles in molecular recognition and need to be accounted for in modeling binding events. Also, our study was conducted with a ‘dry’ cavity. Including water molecules when performing the simulations could lead to more accurate results [24]. Lastly, ranking the binding ability of ligands through computational methods has been known to be in need of improvement [65]. Neither method that we used was able to correctly rank the guest binding ability, but, interestingly enough, the experimental methods themselves also ranked the binding affinities differently.
The lowest RMSD values for the host-guest systems were 1.4 kcal mol−1 for the OAH host via TI and 1.9 kcal mol−1 for the OAMe host with the HREM-BAR method (data set HBAR-ps2). The low RMSD of the HBAR-ps2 set reflects the need to use an ionic concentration that reflects the ionic strength of the experimental solution. This finding is also supported by a previous SAMPL3 study [51]. The TI-ps set had the best RMSD for the OAH host since HREM-BAR simulations pertaining to this host were not completed by the end of the SAMPL5 challenge. When obtaining more data from HREM-BAR simulations, the lowest RMSD for the OAH host was obtained for the HREM-BAR set (1.2 kcal mol−1). Therefore, although TI is easy to implement, extensive data collection is necessary and additional data indicate that one should run longer simulations to obtain accurate results. FES on the CBClip host and guest compounds obtained better RMSD by TI (by 0.9 kcal mol−1) rather than by HREM-BAR [31]. The CBClip guests were larger, and contained some complex aromatic molecules. The results were somewhat surprising, but further analysis indicated that the better results might be due to the use of softcore potentials for estimating the van der Waals component of the binding free energy, whereas the HREM-BAR method uses serial deletion. Another explanation is that TI might benefit from fortuitous error cancellation. In future studies, we would employ the HREM-BAR method with softcore potentials for appropriate sampling and obtaining accurate results. We attribute the relatively low RMSD values of our results to the implemented protocol, producing reasonable binding poses and accurately modeling the ionic concentrations of the solutions.
Despite the growth in computational power and resources, binding free energy calculations are still laborious and it is difficult to predict values within the typical experimental uncertainty of 1 kcal mol−1. In order for computational methods to be complementary to experiments, techniques need to be improved to provide more reliable and faster results. These difficulties demonstrate the continued need for the computational community to blindly assess their methods through means such as provided by the SAMPL challenge.
Supplementary Material
Acknowledgments
The authors would like to thank Tim Miller, Richard Venable and John Legato for technical assistance. We would also like to thank Richard Pastor for very helpful discussions concerning the importance of accurately evaluating the ionic concentration. We extend our gratitude to Andrew C. Simmonett and Michael Lerner for helpful comments on the manuscript. This work was partially supported by the intramural research program of the National Heart, Lung and Blood Institute (NHLBI) of the National Institutes of Health and employed the high-performance computational capabilities of the LoBoS and Biowulf Linux clusters at the National Institutes of Health. (http://www.lobos.nih.gov and (http://biowulf.nih.gov). Florentina Tofoleanu, Juyong Lee and Frank Pickard have been supported by the NHLBI Intramural Lenfant Biomedical Fellowship.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10822-016-9965-5) contains supplementary material, which is available to authorized users.
References
- 1.Mobley DL, Dill KA, Chodera JD (2008) Treating entropy and conformational changes in implicit solvent simulations of small molecules. J Phys Chem B 112:938–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mobley DL, Bayly CI, Cooper MD, Shirts MR, Dill KA (2009) Small molecule hydration free energies in explicit solvent: an extensive test of fixed-charge atomistic simulations. J Chem Theory Comput 5(2):350–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oostenbrink C, van Gunsteren WF (2005) Free energies of ligand binding for structurally diverse compounds. Proc Natl Acad Sci USA 102(19):6750–6754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mobley DL, Graves AP, Chodera JD, McReynolds AC, Shoichet BK, Dill KA (2007) Predicting absolute ligand binding free energies to a simple model site. J Mol Biol 371(4):1118–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jorgensen WL (2004) The many roles of computation in drug discovery. Science 303(5665):1813–1818 [DOI] [PubMed] [Google Scholar]
- 6.Severance DL, Essex JW, Jorgensen WL (1995) Generalized alteration of structure and parameters: a new method for free-energy perturbations in systems containing flexible degrees of freedom. J Comput Chem 16:311–327 [Google Scholar]
- 7.Bennett CH (1976) Efficient estimation of free energy differences from Monte Carlo data. J Comput Phys 22:245–268 [Google Scholar]
- 8.Shirts MR, Bair E, Hooker G, Pande VS (2003) Equilibrium free energies from nonequilibrium measurements using maximum-likelihood methods. Phys Rev Lett 91:140601. [DOI] [PubMed] [Google Scholar]
- 9.Kirkwood JG (1935) Statistical mechanics of fluid mixtures. J Chem Phys 3:300–313 [Google Scholar]
- 10.Woo H-J, Roux B (2005) Calculation of absolute protein-ligand binding free energy from computer simulations. Proc Natl Acad Sci USA 102:6825–6830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Velez-Vega C, Gilson MK (2013) Overcoming dissipation in the calculation of standard binding free energies by ligand extraction. J Comput Chem 34(27):2360–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gumbart JC, Roux B, Chipot C (2013) Standard binding free energies from computer simulations: what is the best strategy? J Chem Theory Comput 9:794–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jo S, Jiang W, Lee HS, Roux B, Im W (2013) Charmm-gui ligand binder for absolute binding free energy calculations and its application. J Chem Inf Model 53(1):267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicholls A, Mobley DL, Guthrie JP, Chodera JD, Bayly CI, Cooper MD, Pande VS (2008) Predicting small-molecule solvation free energies: an informal blind test for computational chemistry. J Med Chem 51(4):769–779 [DOI] [PubMed] [Google Scholar]
- 15.Guthrie JP (2009) A blind challenge for computational solvation free energies: introduction and overview. J Phys Chem B 113(14):4501–4507. doi: 10.1021/jp806724u [DOI] [PubMed] [Google Scholar]
- 16.Geballe MT, Skillman AG, Nicholls A, Guthrie JP, Taylor PJ (2010) The SAMPL2 blind prediction challenge: introduction and overview. J Comput Aided Mol Des 24(4):259–279 [DOI] [PubMed] [Google Scholar]
- 17.Muddana HS, Varnado CD, Bielawski CW, Urbach AR, Isaacs L, Geballe MT, Gilson MK (2012) Blind prediction of host-guest binding affinities: a new SAMPL3 challenge. J Comput Aided Mol Des 26(5):475–487. doi: 10.1007/s10822-012-9554-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muddana HS, Fenley AT, Mobley DL, Gilson MK (2014) The sampl4 host-guest blind prediction challenge: an overview. J Comput Aided Mol Des 28(4):305–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin J, Henriksen NM, Slochower DR, Chiu MW, Mobley DL, Gilson MK (2016) Overview of the SAMPL5 host-guest challenge: are we doing better? J Comput Aided Mol Des. doi: 10.1007/s10822-016-9974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellingson BA, Geballe MT, Wlodek S, Bayly CI, Skillman AG, Nicholls A (2014) Efficient calculation of SAMPL4 hydration free energies using OMEGA, SZYBKI, QUACPAC, and Zap TK. J Comput Aided Mol Des 28(3):289–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beckstein O, Fourrier A, Iorga BI (2014) Prediction of hydration free energies for the SAMPL4 diverse set of compounds using molecular dynamics simulations with the OPLS-AA force field. J Comput Aided Mol Des 28(3):265–276 [DOI] [PubMed] [Google Scholar]
- 22.Gallicchio E, Chen H, Chen H, Fitzgerald M, Gao Y, He P, Kalyanikar M, Kao C, Lu B, Niu Y, Pethe M, Zhu J, Levy RM (2015) BEDAM binding free energy predictions for the SAMPL4 octa-acid host challenge. J Comput Aided Mol Des 29(4):315–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gan H, Benjamin CJ, Gibb BC (2011) Nonmonotonic assembly of a deep-cavity cavitand. J Am Chem Soc 133(13):4770–4773 [DOI] [PubMed] [Google Scholar]
- 24.Gibb CLD, Gibb BC (2014) Binding of cyclic carboxylates to octa-acid deep-cavity cavitand. J Comput Aided Mol Des 28(4):319–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laughrey ZR, Upton TG, Gibb BC (2006) A deuterated deep-cavity cavitand confirms the importance of C–H· · · X–R hydrogen bonds in guest binding. Chem Commun 9:970–972 [DOI] [PubMed] [Google Scholar]
- 26.Gibb CLD, Stevens ED, Gibb BC (2001) CH · · · XR (X=Cl, Br, and I) hydrogen bonds drive the complexation properties of a nanoscale molecular basket. J Am Chem Soc 123(24):5849–5850 [DOI] [PubMed] [Google Scholar]
- 27.Gibb BC et al. (2016) SAMPL5 experimental paper. J Comput Aided Mol Des [Google Scholar]
- 28.Hogues H, Sulea T, Purisima EO (2014) Exhaustive docking and solvated interaction energy scoring: lessons learned from the sampl4 challenge. J Chem Inf Model 28(4):417–427 [DOI] [PubMed] [Google Scholar]
- 29.Mikulskis P, Cioloboc D, Andrejić M, Khare S, Brorsson J, Genheden S, Mata RA, Söderhjelm P, Ryde U (2014) Free-energy perturbation and quantum mechanical study of sampl4 octa-acid host-guest binding energies. J Comput Aided Mol Des 28(4):375–400 [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general amber force field. J Comput Chem 25(9):1157–1174 [DOI] [PubMed] [Google Scholar]
- 31.Lee J, Tofoleanu F, König G, Pickard FC IV, Huang J, Damjanović A, Baek M, Seok C, Brooks BR (2016) CBClip and guests. Same Issue [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, MacKerell AD Jr (2010) CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem 31(4):671–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gutirrez IS, Lin FY, Vanommeslaeghe K, Lemkul JA, Armacost KA, Brooks CL III, MacKerell AD Jr. (2016) Parametrization of halogen bonds in the CHARMM general force field: improved treatment of ligandprotein interactions. Bioorg Med Chem. doi: 10.1016/j.bmc.2016.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79(2):926–935 [Google Scholar]
- 35.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M (1983) CHARMM—a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem 4:187 [Google Scholar]
- 36.Brooks BR, Brooks CL III, Mackerell AD Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodošček M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M (2009) CHARMM: the biomolecular simulation program. J Comput Chem 30:1545–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoover WG (1985) Canonical dynamics—equilibrium phase-space distributions. Phys Rev A 31:1695. [DOI] [PubMed] [Google Scholar]
- 38.Feller SE, Yhang YH, Pastor RW, Brooks BR (1995) Constant pressure molecular dynamics simulation: the Langevin piston method. J Chem Phys 103:4613 [Google Scholar]
- 39.van Gunsteren WF, Berendsen HJC (1977) Algorithms for macromolecular dynamics and costraint dynamics. Mol Phys 34:1311–1327 [Google Scholar]
- 40.Huey R, Morris GM, Olson AJ, Goodsell DS (2007) A semiempirical free energy force field with charge-based desolvation. J Comput Chem 28(6):1145–1152 [DOI] [PubMed] [Google Scholar]
- 41.Shin W-H, Kim J-K, Kim D-S, Seok C (2013) GalaxyDock2: protein-ligand docking using beta-complex and global optimization. J Comput Chem 34(30):2647–2656 [DOI] [PubMed] [Google Scholar]
- 42.Shin W-H, Lee GR, Seok C (2015) Evaluation of GalaxyDock based on the community structure activity resource 2013 and 2014 benchmark studies. J Chem Inf Model 56(6):988–995 [DOI] [PubMed] [Google Scholar]
- 43.Lee J, Scheraga HA, Rackovsky S (1997) New optimization method for conformational energy calculations on polypeptides: conformational space annealing. J Comput Chem 18(9):1222–1232 [Google Scholar]
- 44.Shin W-H, Heo L, Lee J, Ko J, Seok C, Lee J (2011) Ligdockcsa: proteinligand docking using conformational space annealing. J Comput Chem 32(15):3226–3232 [DOI] [PubMed] [Google Scholar]
- 45.Gilson MK, Given JA, Bush BL, McCammon JA (1997) The statistical-thermodynamic basis for computation of binding affinities: a critical review. Biophys J 72:1047–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boresch S, Tettinger F, Leitgeb M, Karplus M (2003) Absolute binding free energies: a quantitative approach for their calculation. J Phys Chem B 107:9535–9551 [Google Scholar]
- 47.Fukunishi H, Watanabe O, Takada S (2002) On the Hamiltonian replica exchange method for efficient sampling of biomolecular systems: application to protein structure predictions. J Chem Phys 116(20):9058–9067 [Google Scholar]
- 48.Itoh SG, Okumura H, Okamoto Y (2010) Replica-exchange in van der waals radius space: overcoming steric restrictions for biomolecules. J Chem Phys 132:134105. [DOI] [PubMed] [Google Scholar]
- 49.Itoh SG, Okumura H (2013) Hamiltonian replica-permutation method and its applications to an alanine dipeptide and amyloid-β(29–42) peptides. J Comput Chem 34:2493–2497 [DOI] [PubMed] [Google Scholar]
- 50.König G, Bruckner S, Boresch S (2009) Unorthodox uses of Bennett acceptance ratio method. J Comput Chem 30:1712–1718 [DOI] [PubMed] [Google Scholar]
- 51.König G, Brooks BR (2012) Predicting binding affinities of host-guest systems in the SAMPL3 blind challenge: the performance of relative free energy calculations. J Comput Aided Mol Des 26:543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.König G, Pickard FC IV, Mei Y, Brooks BR (2011a) Predicting hydration free energies with a hybrid QM/MM approach: an evaluation of implicit and explicit solvent models in sampl4. J Comput Aided Mol Des 28:245–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.König G, Hudson PS, Boresch S (2011b) Multiscale free energy simulations: an efficient method for connecting classical md simulations to QM or QM/MM free energies using non-boltzmann bennett reweighting schemes. J Chem Theory Comput 10(4):1406–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mobley DL, Dill KA (2009) Binding of small-molecule ligands to proteins: “What you see” is not always “what you get”. Structure 17(4):489–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bruckner S, Boresch S (2011) Efficiency of alchemical eree energy simulations I: practical comparison of the exponential formula, thermodynamic integration and Bennett’s acceptance ratio method. J Comput Chem 32:1303–1319 [DOI] [PubMed] [Google Scholar]
- 56.Bruckner S, Boresch S (2011) Efficiency of alchemical free energy simulations II: improvements for thermodynamic integration. J Comput Chem 32:1320–1333 [DOI] [PubMed] [Google Scholar]
- 57.Zacharias M, Straatsma TP, Lennard-Jones JA (1994) Separation-shifted scaling, a new scaling method for lennard-jones interactions in thermodynamic integration. J Chem Phys 100:9025 [Google Scholar]
- 58.Beutler TC, Mark AE, van Schaik RC, Gerber PR, van Gunsteren WF (1994) Avoiding singularities and numerical instabilities in free energy calculations based on molecular simulations. Chem Phys Lett 222:529–539 [Google Scholar]
- 59.Boresch S, Bruckner S (2011) Avoiding the van der Waals endpoint problem using serial atomic insertion. J Comput Chem 32(11):2449–2458 [DOI] [PubMed] [Google Scholar]
- 60.Murray JS, Lane P, Politzer P (2009) Expansion of the σ-hole concept. J Mol Model 15(6):723–729 [DOI] [PubMed] [Google Scholar]
- 61.Laughrey ZR, Gibb CLD, Senechal T, Gibb BC (2003) Guest binding and orientation within open nanoscale hosts. Chem Eur J 9(1):130–139 [DOI] [PubMed] [Google Scholar]
- 62.van Wageningen AMA, Timmerman P, van Duynhoven JPM, Verboom W, van Veggel FCJM, and Reinhoudt DN (1997) Calix[4]arene-based (hemi)carcerands and carceplexes: synthesis, functionalization, and molecular modeling study. Chem Eur J 3(4):639–654 [Google Scholar]
- 63.Paek K, Ihm H, Yun S, Lee HC (1999) Carceroisomerism and twistomerism in C 4ν tetraoxatetrathiahemicarceplexes. Tetrahedron Lett. 40(50):8905–8909 [Google Scholar]
- 64.Jiao D, Golubkov PA, Darden TA, Ren P (2008) Calculation of proteinligand binding free energy by using a polarizable potential. Proc Natl Acad Sci USA 105(17):6290–6295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gilson MK, Zhou H-X (2007) Calculation of protein-ligand binding affinities. Annu Rev Biophys Biomol Struct 36:21–42 [DOI] [PubMed] [Google Scholar]
- 66.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA Jr., Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Keith T, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas O, Foresman JB, Ortiz JV, Cioslowski J. and Fox DJ (2010) Gaussian 09, Revision B.01, Gaussian, Inc., Wallingford [Google Scholar]
- 67.Zhao Y, Truhlar DG (2007) The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, non-covalent interactions, excited states, and transition elements: two new functionals and systematic testing of four M06-class functionals and 12 other function. Theor Chem Acc 120:215–241 [Google Scholar]
- 68.Zhao Y, Truhlar DG (2008) Density functionals with broad applicability in chemistry. Acc Chem Res 41:157–167 [DOI] [PubMed] [Google Scholar]
- 69.Marenich AV, Cramer CJ, Truhlar DG (2009) Universal solvation model based on solute electron density and on a continuum model of the solvent defined by the bulk dielectric constant and atomic surface tensions. J Phys Chem B 113(18):6378–6396 [DOI] [PubMed] [Google Scholar]
- 70.Hariharan PC, Pople JA (1974) Accuracy of AH n equilibrium geometries by single determinant molecular orbital theory. Mol Phys 27(1):209–214 [Google Scholar]
- 71.Casasnovas R, Ortega-Castro J, Frau J, Donoso J, Muoz F (2014) Theoretical pKa calculations with continuum model solvents, alternative protocols to thermodynamic cycles. Int J Quantum Chem 114(20):1350–1363 [Google Scholar]
- 72.Liptak MD, Shields GC (2001) Accurate pK(a) calculations for carboxylic acids using complete basis set and Gaussian-n models combined with CPCM continuum solvation methods. J Am Chem Soc 123(30):7314–7319 [DOI] [PubMed] [Google Scholar]
- 73.Perrin DD (1982) Ionization constants of inorganic acids and bases in aqueous solution, 2nd edn. Pergamon, Oxford [Google Scholar]
- 74.Majstorovic V (2011) Changes in high-molecular weight compounds during beech litter decomposition. Masters thesis [Google Scholar]
- 75.Cullen W, Turega S, Hunter CA, Ward MD (2015) pH-dependent binding of guests in the cavity of a polyhedral coordination cage: reversible uptake and release of drug molecules. Chem Sci 6:625–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.