Abstract
Quantitative proteome analysis allows comparisons of protein or phosphoprotein levels across multiple cell types or conditions. A number of experimental approaches have been described toward quantitative proteomics. In this chapter, we focus on Tandem Mass Tag (TMT) isobaric labeling of peptides for global, relative quantitation of proteins and phosphopeptides. To date, there has been no published protocol describing chemical labeling of small amounts of peptides specifically extracted from small tumor samples, for which rigorous sample preparation is necessary to ensure reproducible TMT labeling.
Keywords: Proteomics, Phosphoproteomics, Relative quantitation of proteins, Tandem mass tags, TMT, Peptide labeling, MaxQuant
1. Introduction
Relative quantitation of protein expression is a sought-after application. Proteomic analysis can reveal aspects of the proteome, such as posttranslational modifications, which are not captured by transcriptomic approaches and which may be important in disease states. A sensitive measure of the proteome can be achieved by using Stable Isotope Labeling of Amino acids in Cell cultures (SILAC) [1]. In this approach, cells are grown in culture in the presence and absence of isotopically labeled amino acids, such as arginine and lysine. After about five passages to ensure complete incorporation of labeled amino acids into the proteome, the cell populations from the experimental (labeled with the heavy amino acids) and control (unlabeled) groups are harvested. Normalized protein extracts from each sample are mixed and processed for bottom-up mass spectrometry (MS), which is used to quantify peptides using full scan MS. The MS intensities of the peptides containing heavy and light amino acids are measured and their ratio is used to quantify the relative amount of a specific protein in the experimental and control groups. This is an extremely reproducible method which eliminates intraexperimental variability caused by differential sample preparations. It is, however, limited to cell culture labeling and comparison of two samples typically.
Peptide labeling by Tandem Mass Tags (TMT) emerged in 2003 as a method to simultaneously compare and accurately measure relative abundances of peptides in up to ten samples by using tandem mass spectrometry (MS/MS) [2]. This approach has the added advantage of not being limited to labeling in cell culture and can label peptides extracted from small amounts of tissue, such as samples isolated from glioblastoma (GBM) specimens. The utility of TMT labeling in the oncology space is exemplified by its ability to successfully compare and quantify key proteins and phosphopeptides in molecular subtypes of breast cancer [3]. In this chapter, we will provide a comprehensive, step-by-step protocol for labeling and analyzing limited amounts of GBM samples using the 10-plex TMT approach.
In most cases, proteins are isolated from cell culture or tissues by one of two methods: (a) solubilization with detergent and/or chaotropic agents, fractionation with denaturing SDS-PAGE, and in-gel digestion [4, 5]; or (b) digestion of unfractionated proteins in solution using optimized protocols [6], followed by peptide fractionation with high-performance liquid chromatography (HPLC). Both methods result in peptides that can be processed using similar workflows. After preliminary quantification, TMT labeling, and mixing of peptides, nano-liquid chromatography coupled to tandem MS (nano-LC-MS/MS) analysis is performed using a high-resolution mass spectrometer. In the experiments described in this chapter, a Thermo Scientific Q Exactive High Field (HF) mass spectrometer is used for all experiments involving TMT quantitation of peptides. The resulting mass spectra are submitted for analysis using MaxQuant Proteomics software [7], freely available at www.maxquant.org. The Andromeda search engine [8] (free to download at: www.andromeda-search.org) embedded in Maxquant is used to identify peptide spectra and roll them into protein identifications. Generated “.txt” files contain precise information about reporter ion intensities of TMT-labeled peptides. Labeling efficiencies, mixing of the TMT-labeled peptide pools (close to TMT channel medians of 1:1:1:1 etc. to ensure equal mixing of all samples), as well as the final analysis of TMT reporter ion intensities can be easily achieved by importing MaxQuant analyzed data into a second, freely available, and complementary program called Perseus [9, 10] (available at www.perseus-framework.org).
2. Materials
All solutions are made with Milli-Q water except for buffers for the HPLC system coupled to the mass spectrometer which uses HPLC grade water.
2.1. Standard, Quantified Yeast Peptides
MS compatible yeast protein extract digest: 100 μg (Promega, Madison, WI 53711, USA). For the current study, we generated our own in-house yeast tryptic peptides which were taken from stocks of 5 μg aliquots stored at −80 °C prior to use.
2.2. TMT Labeling Reagents
TMT10-plex Mass Tag Labeling Kit (Thermo Scientific, Rockford, IL, USA).
Acetonitrile Optima Grade: Water content of 0.001% (see Note 1).
HEPES buffer (0.2 M, pH 8.5).
Hydroxylamine: 5% (w/w).
2.3. De-salting (Peptide Cleanup)
SepPak Vac 1 cc (100 mg), tC18 Cartridges (Waters Corporation, Milford, MA, USA).
Methanol conditioning solution: 90% (v/v).
Equilibration and washing solution: water–trifluoroacetic acid (TFA), 100:0.1 (v/v).
Elution solution: acetonitrile–water–trifluoroacetic acid (TFA), 70:30:0.1 (v/v).
Self-packed Stage-Tips [11] using Empore C18 High Performance Extraction Disks (3M, St. Paul, MN, USA). Empore disks have chromatographic materials such as C18 immobilized in a Teflon meshwork. Self-packed Stage tips are made by placing a small plug of the C18 material into a 200 μL pipette tip.
Stage Tip Equilibration and wash buffer: Water with 0.1% formic acid (FA) (v/v).
Stage Tip Elution buffer: acetonitrile–water–formic acid (FA), 70:30:0.1 (v/v).
2.4. Hydrophilic Interaction Liquid Chromatography (HILIC) Fractionation of Peptides
A narrow-bore, HPLC system equipped with Agilent 1100 series dual pump and detector coupled to a Gilson fraction collector, model 203B.
TSK gel amide-80 column: 4.6 mm ID × 25 cm and 5 μm beads (TOSOH Bioscience, LLC, King of Prussia, PA, USA).
Mobile phase A: Acetonitrile with 0.1% TFA.
Mobile phase B: Water with 0.1% TFA.
2.5. High Resolution Mass Spectrometer Coupled to Nanoflow LC (LC-MS)
Q Exactive High Field (HF) mass spectrometer (see Note 2).
EASY-nLC nanoflow HPLC system (Thermo Scientific).
Mobile phase A: Water with 0.1% formic acid (FA) (v/v).
Mobile phase B: Acetonitrile with 0.1% FA (v/v).
Reprosil C18, 3 μm beads (Dr. Maisch GmbH, 72119 Ammerbuch-Entringen, Germany).
Nanospray emitter: PicoFrit SELF/P 360 μm OD × 75 μm ID, 10 μm tip ID (New Objective, Woburn, MA, USA).
2.6. Software for Analysis
MaxQuant (Version 1.5.5.1).
Perseus (Version 1.5.6.0) Proteomic Analysis Software.
3. Methods
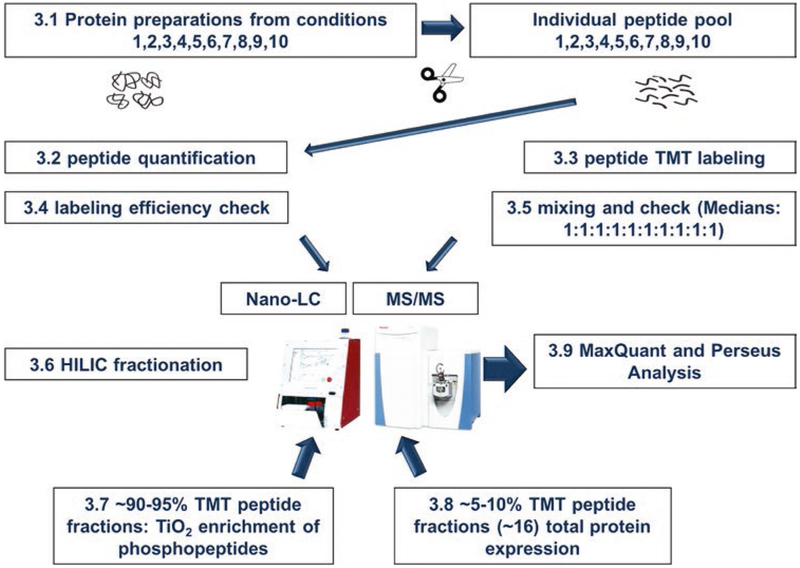
The schematic in Fig. 1 illustrates the steps of TMT labeling. All steps are carried out at room temperature.
Fig. 1.
Schematic workflow of TMT labeling procedures
3.1. Generation of Peptides
Prepare peptides (see Note 3) by in-solution digestion of proteins, as previously described [6]. If a cell pellet is not obtained, protein can be precipitated from lysates free of detergents using five volumes of cold acetone [12] and made ready for proteolysis.
Alternative protocols for preparing peptides by in-gel digestion are also available [5].
3.2. Preliminary Quantitation of Peptides
For this step, small amounts of peptides (in the range of 2 pg to 200 ng) are used (see Note 4).
Analyze standard yeast peptide amounts of 2, 20, 200, 2000, 20,000, 200,000 pg by LC-MS/MS.
Subject each raw file to MaxQuant analysis.
Use protein.txt file to obtain total intensity of all the identified proteins in each of the experiment.
Plot log10 total intensity of protein (x axis) vs. log10 of yeast peptide used (y axis) in each experiment (Fig. 2).
Use the constructed standard curve to calculate peptide concentration in experimental samples.
Fig. 2.
Quantitation of small amounts (ng to pg) of peptides using a MS-based assay. Standard amounts of a known mixture of peptides are analyzed by LC-MS/MS, and total matched peptide spectral intensities obtained for identified proteins are plotted against the amount analyzed as functions of log10. The resulting standard curve is then used for determining concentrations of unknown amounts of peptide pools generated from samples. AUC area under the curve
3.3. TMT Labeling
If using the TMT Mass Tag Labeling Kit (Thermo Scientific), follow the manufacturer’s instructions. During the initial solubilization of each label, it is important to use anhydrous acetonitrile, as water inactivates the stored label (see also Note 1). We have effectively reconstituted each TMT label in solvent volumes four times higher than recommended by the manufacturer’s instructions. This dilution has enabled accurate samplings of small volumes of TMT label and at this more dilute reconstitution, label has been stable for about 8 months, stored at −80 °C.
After each label is solubilized, test the labeling kit for labeling efficiency of each channel using quantitated, in-house yeast peptides as outlined above, before using it for important experimental samples. Typically, for labeling low amounts of sample peptide, test 100 ng amounts of yeast tryptic peptides with TMT 10-channel labeling reagents (TMT-126, 127N, 127C, 128N 128C, 129N, 129C, 130N, 130C, 131) for testing.
Dry peptides by vacuum centrifugation.
Add 10 μL of acetonitrile to 100 ng of peptide.
Sonicate and vortex to ensure solubilization (see Note 5).
Add 17 μL of 0.2 M HEPES buffer (pH 8.5) to the peptide solution (see Notes 6 and 7).
Add TMT label from each channel to the peptide pools. For TMT labeling of small amounts of peptides (e.g., 100 ng), we found that we need to use TMT label-peptide 150:1 (w/w) to achieve labeling efficiencies of 95% and above. For labeling peptide amounts >1 μg, the ratio of TMT label-peptide (w/w) is 10:1. The volume of labeling reagent is 3 μL, for a total reaction volume of 30 μL. Allow labeling to proceed for 1 h.
After 1 h, add 2 μL 5% hydroxylamine solution for 15 min to quench the labeling reaction.
Centrifuge the TMT-labeled peptides under vacuum to near dryness.
TMT-labeled peptide pools are cleaned up with Sep-Pak or Stage Tips, depending on the starting amounts (see Note 8).
Sample cleanup with Sep-Pak solid phase columns: Wash and condition column with conditioning solution. Equilibrate with 0.1%TFA. Apply sample, taking care not to introduce air into the sample bed. Wash away nonspecifically bound salts, additives, with equilibration buffer. Elute peptides with 1 mL 70% Acetonitrile/0.1% TFA.
Sample clean-up with Stage-Tip Disks: Wet the disks with 50 μL 100% methanol solution using centrifugal force (2–5 min spin at 300–500 × g depending on the size of the disks packed in the pipette tip). Centrifugation is required for all the ensuing steps. Wash the disk with 40 μL of 0.1% formic acid. Load peptide sample dissolved in 20 μL 0.1% formic acid. Elute peptides with 20 μL 70% acetonitrile in 0.1% formic acid. Do not dry the Stage-Tip during the steps described above.
From the pool of purified TMT-labeled peptides, about 1% is analyzed by LC-MS/MS with MS1 resolution set at 120,000 and MS/MS resolution set at 60,000.
Raw data are analyzed by MaxQuant with “Variable Modifications” set for TMT 10-plex 126, 127N, 127C, 128N 128C, 129N, 129C, 130N, 130C, 131 to be at N-termini, as well as lysine for database searching and peptide identification. At the beginning of the MaxQuant analysis, these modifications are introduced as “Standard” modifications under the “Configuration” tab of MaxQuant. To achieve this, duplicate all of the 10-plex modifications and one by one, rename and save the newly created modifications as Standard modification under the “Modifications Type.” Finally, choose “Modify Table” and “Save Changes.” Restart MaxQuant and you will now see and can choose the TMT 10-plex candidates under the “Variable modifications” option of Group-Specific parameters.
3.4. Labeling Efficiency Calculations
After MaxQuant analysis, open the “evidence.txt” file as an Excel table. For each of the TMT experiments, count the rows of peptides modified by TMT at the N-termini of the peptides.
Count the rows of peptides that are modified by TMT at lysines (see Note 9). Percentages of the tryptic peptides that are labeled at lysines and/or N-termini are listed.
A very small percent of peptides that are N-terminally blocked (acetylated) are found to be TMT-derivatized at the C-terminal lysines whenever the peptide ends with lysine. These are grouped under Lys TMT (with N-Acetyl Lys) Peptide # (see Note 10).
Finally, calculate the percentage labeling by accounting for all the N-terminal TMT, lysine TMT and finally, Lys and N-terminal TMT-derivatized peptides (see Note 11), compared to number of total peptides identified for the specific LC-MS/MS Experiment.
A typical result of labeling efficiency that includes a check for 10-plex TMT-labeled standard yeast peptide samples can be seen in detail in Table 1. In general, when TMT labeling samples contain ~100 ng of peptides, labeling efficiency around 95% is acceptable; for samples ranging from 20 to 100 μg peptide amounts, we typically achieve over 95% labeling efficiencies.
After the labeling efficiency of the TMT Mass Tag Labeling Kit is established, peptide samples from experimental materials are TMT-labeled and the labeling efficiency for each sample is determined as described above.
Table 1.
Labeling check for small amounts of peptides
| TMT label | 126 | 127N | 127C | 128N | 128C | 129N | 129C | 130N | 130C | 131 |
|---|---|---|---|---|---|---|---|---|---|---|
| Amount analyzed | 100 ng | 100 ng | 100 ng | 100 ng | 100 ng | 100 ng | 100 ng | 100 ng | 100 ng | 100 ng |
| Total peptide # | 4216 | 4045 | 4547 | 4332 | 4858 | 4121 | 4519 | 4724 | 5172 | 5204 |
| Unmodified peptide # | 227 | 230 | 201 | 189 | 238 | 243 | 239 | 217 | 255 | 252 |
| N-Acetyl TMT # | 75 | 50 | 61 | 58 | 80 | 69 | 84 | 74 | 84 | 79 |
| N-Acetyl TMT % | 1.8% | 1.2% | 1.3% | 1.3% | 1.6% | 1.7% | 1.9% | 1.6% | 1.6% | 1.5% |
| N term TMT # | 3516 | 3458 | 3937 | 3736 | 4151 | 3456 | 3861 | 4015 | 4717 | 4444 |
| N term TMT % | 84% | 86% | 87% | 86% | 85% | 84% | 85% | 85% | 91% | 85% |
| Lys TMT # | 2680 | 2635 | 2949 | 2739 | 3097 | 2561 | 2898 | 3017 | 3360 | 3411 |
| Lys TMT % | 64% | 65% | 65% | 63% | 64% | 62% | 64% | 64% | 65% | 66% |
| Either N term or Lys TMT # | 3989 | 3815 | 4346 | 4143 | 4620 | 3878 | 4280 | 4507 | 4917 | 4952 |
| All TMT % peptides (%) | 95% | 94% | 96% | 96% | 95% | 94% | 95% | 95% | 95% | 95% |
TMT label incorporation results are shown for small amounts of peptides. The labeled peptides were reconstituted in 20 μL 0.1% formic acid, desalted using Stage Tips and eluted in 20 μL of 40% acetonitrile/0.1% formic acid, dried, and reconstituted in 10 μL 0.1% formic acid. Eight microliter were analyzed by LC-MS/MS. This optimized experiment for 100 ng of quantified yeast peptide pools is used for determining sample amounts for scaled up labeling
3.5. Mixing of the Ten Channels and Evaluation of the Mixing
A pilot experiment is performed to calibrate the mixing of the ten TMT samples.
1–5% of peptide pool from each TMT channel is mixed equally by peptide mass.
Dry the mixed sample using a vacuum centrifuge until a small drop is seen at the bottom of the tube (see Note 12).
Following this, the peptide mixture is analyzed by LC-MS/MS, with MS1 resolution set at 120,000 and MS/MS resolution set at 60,000. The resulting RAW file is analyzed by MaxQuant with Reporter Ion MS2 activation in “Group Specific Parameters “Type” (see Note 13); under “Isobaric labels” click on the correct TMT plex used (e.g., 10-plex TMT).
After MaxQuant analysis, under “combined, txt folder,” upload the “peptides.txt” into the Perseus analysis software using “Generic matrix upload.”
Select “Reporter Ion Intensity” obtained from ten channels and move these values into the “Main” expression field.
From the uploaded peptides, remove the ones derived from “Reverse” and “Potential contaminant databases” by using the “Filter rows based on categorical column” pull-down arrow.
Next, “Filter rows based on valid values” so that number of valids in each row, in total, is “Greater than 0.” Minimum number should be Min. number of Reporter Ion Intensities listed; e.g., ten for ten channels.
Under the “Basic” pull-down menu and “Summary statistics (rows)” obtain the mean for all the peptides found in all channels.
Under “Normalization,” “Divide” Rows by the Mean.
Under “Basic” and “Summary statistics (columns),” obtain the Median for all the Reporter Intensities.
From these values obtained, make a correction to the mixing to achieve Median 1 for all the channels, and reanalyze the mix.
A typical result of the pilot mixing of ten channels is shown in Table 2.
Table 2.
Medians of TMT ratios for mixing equal amounts of TMT labeled peptides
| Median ratios for each label | Channel IDs |
|---|---|
| 1.0 | Reporter intensity 126 |
| 1.0 | Reporter intensity 127N |
| 1.1 | Reporter intensity 127C |
| 1.0 | Reporter intensity 128N |
| 1.0 | Reporter intensity 128C |
| 0.9 | Reporter intensity 129N |
| 1.0 | Reporter intensity 129C |
| 1.0 | Reporter intensity 130N |
| 0.9 | Reporter intensity 130C |
| 0.9 | Reporter intensity 131 |
The mixing is optimized until the medians for each TMT channel are close to 1:1:1 etc.
3.6. Hydrophilic Interaction Liquid Chromatography (HILIC) or Basic pH Reversed-Phase (BPRP) Fractionation of Peptides
Once the TMT-labeled peptide test portions are successfully mixed in approximately equal amounts, prepare a large-scale mix and dry the mix to near dryness (see Note 12).
The sample is now ready for fractionation using liquid chromatography. Hydrophilic interaction liquid chromatography (HILIC) or basic pH reverse phase (BPRP) chromatographic fractionation of TMT-labeled peptides is strongly recommended when global analysis is planned, which is best suited for peptide pools of more than 30 μg. Sample fractionation increases depth of proteome coverage, and reduces ratio compression due to cofragmentation of coeluting, nearly isobaric peptides in MS/MS (see Note 13).
Dissolve TMT peptides in 90% acetonitrile with 0.1% TFA (see Note 14).
Centrifuge briefly and inject 30–200 μg of TMT peptide mix on the HILIC column equilibrated with 90% acetonitrile with 0.1% TFA.
A gradient of 90% acetonitrile with 0.1% TFA to water with 0.1% TFA is introduced over 65 min.
Collect fractions every 4 min; over 65 min of HPLC separation, approximately 16 fractions are generated.
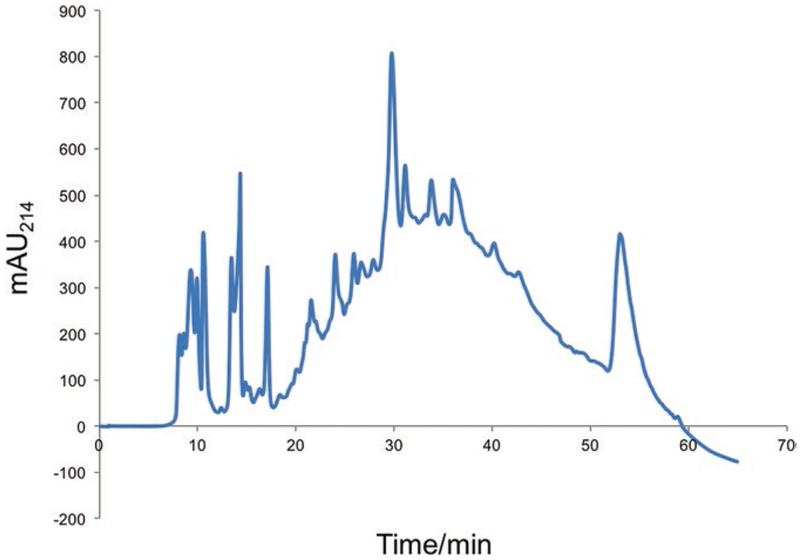
A typical separation of TMT labeled peptide using HILIC separation is shown in Fig. 3.
Fig. 3.
HILIC separation of 50 μg TMT-labeled peptide mixture. UV absorption at 214 nm (in mAU) is the “x” axis and elution time is the “y” axis. During the course of the chromatographic separation, a fraction is collected every 4 min, resulting in nearly 15 fractions of TMT peptide pools for subsequent total protein and phospho-peptide TiO2 enrichment analyses
3.7. Phosphopeptide Enrichment
3.8. Total Proteome Quantitation
3.9. Final Data Analysis
All data are analyzed with MaxQuant Proteomics software platform integrated with the Andromeda search engine.
Downstream data analysis is done by Perseus Proteomics software.
3.10. Example Using this Protocol
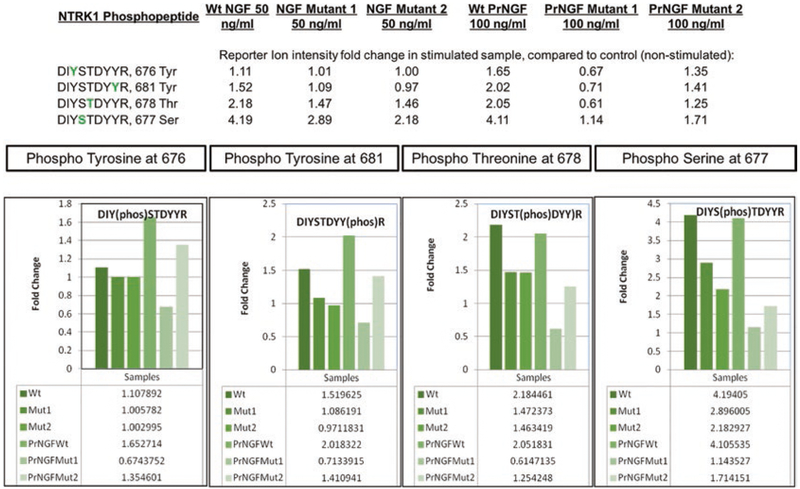
We have successfully used the method described in this chapter to investigate stimulation of PC12 cells by Nerve Growth Factor (NGF). Twenty microgram of peptide material was collected from six cell cultures treated either with wild-type (wt) or one of two mutant versions of mature NGF or pro-NGF (PrNGF). In Fig. 4, we can follow the effect of NGF stimulation on the high affinity nerve growth factor receptor (NTRK1) phosphopeptides containing amino acids 674–682 by comparing reporter ion intensities in stimulated and unstimulated control samples.
Fig. 4.
Phosphopeptide quantitation. Relative quantitation of four phosphopeptides in the high-affinity nerve growth factor receptor (NTRK1) that are stimulated by two different doses (50 and 100 ng/mL) of wild type and mutant NGF or proNGF (PrNGF) ligand, compared to control (unstimulated) sample. After TMT labeling, fractionation and enrichment steps, differentially phosphorylated peptides are identified and quantitated by comparing the reporter ion intensities in each of the ten TMT channels. Fold change of the phosphopeptides in each treated sample, compared to the untreated sample is then determined
4. Notes
Water content in acetonitrile should be minimized since the shelf-life of TMT reagent is affected adversely by the presence of water. Store reconstituted TMT labels in sealed pouches containing desiccant packs, at −80 °C. Just before usage, the labels are brought to room temperature.
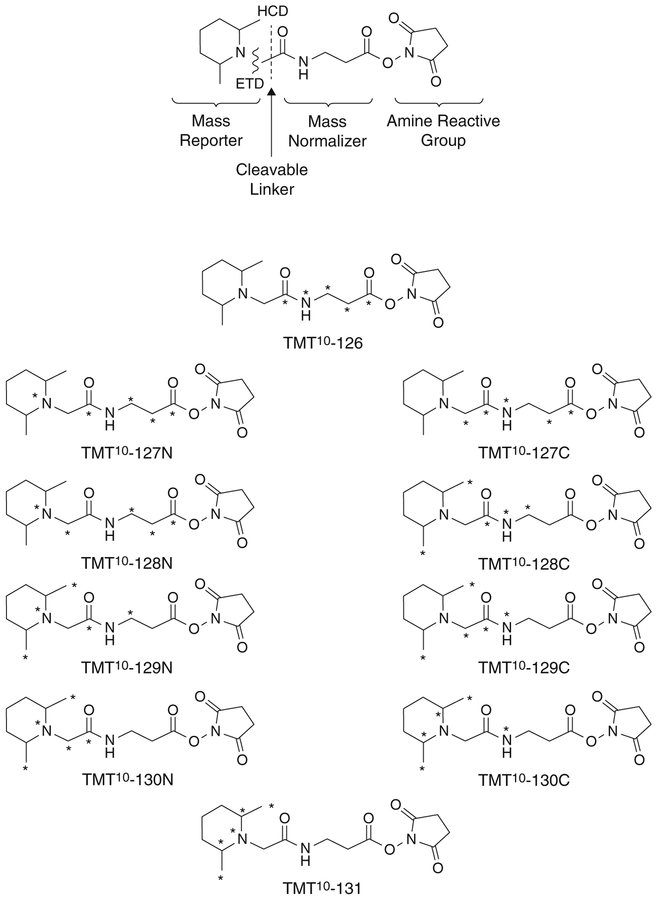
The high MS/MS resolution (60,000 at m/z 200) at fast scan speeds of the Q Exactive HF mass spectrometer makes it well-suited for use with 10-plex TMT, which relies on small (several mDa) mass differences between four pairs of the reporter ions (127N, 127C, 128N, 128C, 129N, 129C, 130N, 130C) (Fig. 5). Much lower resolution will suffice for 6-plex TMT which does not rely on very small mass differences between reporter ions.
Up to ten different peptide samples can be prepared at the same time and then combined for a 10-plex TMT labeling experiment.
Commercially available kits consume much more of the peptide sample; for example, the Pierce Quantitative Colorimetric Peptide Assay (Thermo Scientific, Rockford, IL, USA) uses ~300 ng to 20 μg of peptide for constructing the standard curve.
It is good practice to use acetonitrile during the solubilization of sample peptides, especially when labeling <1 μg of peptides. The organic solvent assists in keeping peptides from sticking to polypropylene surfaces [4].
HEPES buffer (0.2 M, pH 8.5) is essential to ensure correct pH during the reaction between the label and the peptide.
We have experienced incomplete labeling of relatively small amounts of peptides when using TEAB (triethyl ammonium bicarbonate) buffer. HEPES buffer is more effective for use in TMT labeling of peptides.
“Sep-Pak” columns are used when the recovered peptide amount is above 10 μg. Otherwise, for TMT-labeled peptide purifications, we routinely use self-packed Stage-Tip disks. One layer or plug has the capacity to clean up 4–5 μg of peptide material, while two layers of stacked plugs have the capacity to clean up 10 μg of peptide material.
Lysine residue can be modified by TMT as an internal (missed cleavage) amino acid or as a C-terminal tryptic peptide amino acid.
For tryptic peptides that have N-terminal acetyl group and C-terminal arginine with no uncleaved internal lysines, there is no TMT derivatization.
It is good practice to break down the number and percentage labeling of TMT peptides as labeled at the N-terminus only, lysines only, or both (Table 1). By inspecting the percentage labeling of each label position, we can detect any deterioration of the TMT label kit. The percentage of peptides labeled at the N-terminus is usually higher than the percentage labeled at the lysines. When this ratio is reversed, reduced effectiveness of the TMT reagent is suspected.
For sample handling steps involving drying of peptide pools by vacuum centrifugation, never dry the sample completely. When samples are dried to completion, it is difficult to recover peptides and solubilize them [4]. A small drop (1 μL) should still be visible after reducing the sample volume by evaporation.
If MS3 is going to be used to measure the reporter ion intensities, activate MS3 under Group Specific Parameter “Type” in MaxQuant settings. MS3 can greatly reduce TMT ratio compression due to cofragmentation of contaminating peptides that coelute with and are nearly isobaric to the main peptides of interest, and consequent contribution of TMT reporter ion signal from the contaminants [14].
During HILIC chromatography, the peptide pools are dissolved in starting HPLC solvent of around 90% acetonitrile/0.1% TFA. It is necessary to ensure the presence of water (10%) in the buffer, since pure organic buffer will not solubilize all of the peptides.
Fig. 5.
Tandem mass tag reagents detailing mass reporter, normalizer, and amine reactive groups, as well as the positions of stable isotope labels for each reagent
Acknowledgments
This work was supported by Shared Instrumentation Grant S10 RR027990 and NINDS P30 NS050276 to Thomas A. Neubert. We thank Giovanna Testa, Thorsten Kranz, Moses Chao, Antonino Cattaneo, and David Chiu for providing experiments which made this chapter possible.
References
- 1.Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1:376–386 [DOI] [PubMed] [Google Scholar]
- 2.Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Hamon C (2003) Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem 75:1895–1904 [DOI] [PubMed] [Google Scholar]
- 3.Huang FK, Zhang G, Lawlor K, Nazarian A, Philip J, Tempst P, Dephoure N, Neubert TA (2017) Deep coverage of global protein expression and phosphorylation in breast tumor cell lines using TMT 10-plex isobaric labeling. J Proteome Res 16:1121–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erdjument-Bromage H, Lui M, Lacomis L, Grewal A, Annan RS, McNulty DE Carr SA, Tempst P (1998) Examination of micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J Chromatogr A 826:167–181 [DOI] [PubMed] [Google Scholar]
- 5.Sebastiaan Winkler G, Lacomis L, Philip J, Erdjument-Bromage H, Svejstrup JQ, Tempst P (2002) Isolation and mass spectrometry of transcription factor complexes. Methods 26:260–269 [DOI] [PubMed] [Google Scholar]
- 6.Villen J, Gygi S (2008) The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc 3:1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotech 26:1367–1372 [DOI] [PubMed] [Google Scholar]
- 8.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10:1794–1805 [DOI] [PubMed] [Google Scholar]
- 9.Cox J, Mann M (2012) 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics 13(suppl 16):S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein M, Geiger T, Mann M, Cox J (2016) The Perseus computational platform for comprehensive analysis of (prote) omics data. Nat Methods 13:731–740 [DOI] [PubMed] [Google Scholar]
- 11.Rappsilber J, Mann M, Ishihama Y (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2:1896–1906 [DOI] [PubMed] [Google Scholar]
- 12.Simpson DM, Beynon RJ (2010) Acetone precipitation of proteins and the modification of peptides. J Proteome Res 9:444–450 [DOI] [PubMed] [Google Scholar]
- 13.Zhang G, Neubert TA (2011) Comparison of three quantitative phosphoproteomic strategies to study receptor tyrosine kinase signaling. J Proteome Res 10:5454–5462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ting L, Rad R, Gygi S, Haas W (2011) MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Methods 8:937–940 [DOI] [PMC free article] [PubMed] [Google Scholar]