Summary
Background
Wilms tumour is the most common childhood renal cancer and is genetically heterogeneous. While several Wilms tumour predisposition genes have been identified, there is strong evidence that further predisposition genes are likely to exist. Our study aim was to identify new predisposition genes for Wilms tumour.
Methods
In this exome sequencing study, we analysed lymphocyte DNA from 890 individuals with Wilms tumour, including 91 affected individuals from 49 familial Wilms tumour pedigrees. We used the protein-truncating variant prioritisation method to prioritise potential disease-associated genes for further assessment. We evaluated new predisposition genes in exome sequencing data that we generated in 334 individuals with 27 other childhood cancers and in exome data from The Cancer Genome Atlas obtained from 7632 individuals with 28 adult cancers.
Findings
We identified constitutional cancer-predisposing mutations in 33 individuals with childhood cancer. The three identified genes with the strongest signal in the protein-truncating variant prioritisation analyses were TRIM28, FBXW7, and NYNRIN. 21 of 33 individuals had a mutation in TRIM28; there was a strong parent-of-origin effect, with all ten inherited mutations being maternally transmitted (p=0·00098). We also found a strong association with the rare epithelial subtype of Wilms tumour, with 14 of 16 tumours being epithelial or epithelial predominant. There were no TRIM28 mutations in individuals with other childhood or adult cancers. We identified truncating FBXW7 mutations in four individuals with Wilms tumour and a de-novo non-synonymous FBXW7 mutation in a child with a rhabdoid tumour. Biallelic truncating mutations in NYNRIN were identified in three individuals with Wilms tumour, which is highly unlikely to have occurred by chance (p<0·0001). Finally, we identified two de-novo KDM3B mutations, supporting the role of KDM3B as a childhood cancer predisposition gene.
Interpretation
The four new Wilms tumour predisposition genes identified—TRIM28, FBXW7, NYNRIN, and KDM3B—are involved in diverse biological processes and, together with the other 17 known Wilms tumour predisposition genes, account for about 10% of Wilms tumour cases. The overlap between these 21 constitutionally mutated predisposition genes and 20 genes somatically mutated in Wilms tumour is limited, consisting of only four genes. We recommend that all individuals with Wilms tumour should be offered genetic testing and particularly, those with epithelial Wilms tumour should be offered TRIM28 genetic testing. Only a third of the familial Wilms tumour clusters we analysed were attributable to known genes, indicating that further Wilms tumour predisposition factors await discovery.
Funding
Wellcome Trust.
Introduction
Wilms tumour is a childhood kidney tumour that affects one in 10 000 children. Its histology is similar to that of the developing kidney and is typically triphasic, with blastemal, stromal, and epithelial components.1 Biphasic tumours, with two of the three components, and monomorphic tumours consisting of only one component also occur but are rarer.1
Wilms tumour is primarily a non-familial condition, with only about 2% of affected individuals having a relative with the tumour.2 Given its rarity, inherited causes—rather than chance—are assumed to underlie familial clusters, and several have been reported, including constitutional mutations in the genes WT1, CTR9, and REST.2, 3, 4 Wilms tumour is also known to be associated with many genetic conditions, including the WAGR, Denys-Drash, Beckwith-Wiedemann, Simpson-Golabi-Behmel, Perlman, mosaic variegated aneuploidy, hereditary hyperparathyroidism-jaw tumour, Li-Fraumeni, DICER1, and Bohring-Opitz syndromes, Fanconi anaemia, and PIK3CA-related overgrowth spectrum.2, 3, 4, 5, 6, 7, 8, 9, 10, 11 These conditions are diverse in their clinical and histological associations, inheritance patterns, and mutational mechanisms of pathogenicity. The underlying predisposition genes have many different functions and are involved in diverse biological processes. The identification of these genes and investigations into their role in Wilms tumour predisposition have led to fundamental insights into developmental, cellular, and oncological mechanisms and have important clinical implications for individuals with Wilms tumour and their families.2, 3, 4, 5, 6, 7, 8, 9, 10, 11 Furthermore, many causes of familial and syndromic Wilms tumour also contribute to non-familial, non-syndromic Wilms tumour.2, 4, 5
Research in context.
Evidence before this study
Wilms tumour is a rare childhood kidney tumour. We searched PubMed for papers in English with the terms “Wilms” AND “genetic” OR “mutation” OR “familial” OR “syndrome”, yielding 2801 papers that we reviewed to identify those relevant to genetic predisposition to Wilms tumour. This review identified 17 genes previously shown to predispose to Wilms tumour and showed that further Wilms tumour predisposition genes must exist, because many syndromic cases and familial clusters have not been explained.
Added value of this study
To our knowledge, this study is the largest exome sequencing study to date of individuals with Wilms tumour, involving 890 individuals, including 91 individuals from 49 familial Wilms tumour pedigrees. We identified four new Wilms tumour predisposition genes, TRIM28, FBXW7, KDM3B, and NYNRIN. We showed that FBXW7 and KDM3B are pleiotropic cancer predisposition genes, and that KDM3B and NYNRIN might also cause non-malignant phenotypes, particularly intellectual disability. Our study identified TRIM28 as a major Wilms tumour predisposition gene, making a similar contribution to familial and unselected Wilms tumour as those of constitutional WT1 and REST mutations. We also found an association between TRIM28 mutations and epithelial histology and a strong parent-of-origin-effect, because all inherited TRIM28 mutations were maternally transmitted. Functional enrichment analyses revealed remarkable diversity in the biological pathways affected by Wilms tumour predisposition genes. We also found limited overlap between the 21 constitutionally mutated Wilms tumour predisposition genes and 20 genes somatically mutated in Wilms tumour.
Implications of all the available evidence
This study provides new insights into the causes of Wilms tumour and describes the overall landscape of Wilms tumour predisposition. Wilms tumour shows remarkable genetic heterogeneity and aetiological complexity, which have substantial clinical impact. Genetic testing should be made available to individuals with Wilms tumour, but will need to encompass both broad genetic testing, for example by exome sequencing, and testing for 11p15 epigenetic abnormalities. Moreover, our findings suggest that more Wilms tumour predisposition genes are likely to exist, which will have relevance for future research and clinical testing.
Research over the past 25 years has led to tremendous advances in our knowledge of Wilms tumour predisposition. However, available evidence suggests that our knowledge is still incomplete and that further predisposition factors remain to be discovered. In particular, the cause of many familial clusters is still unknown.4 In this study, we aimed to use exome sequencing to identify new Wilms tumour predisposition genes and to characterise and contextualise the genetic landscape of such predisposition.
Methods
Study design and participants
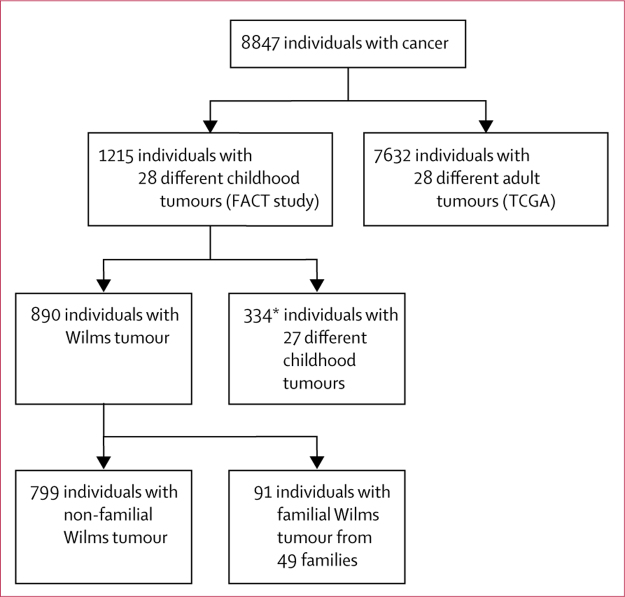
In this exome sequencing study, we included participants recruited to the Factors Associated with Childhood Tumours (FACT) Study. All children with a childhood solid tumour from the UK were eligible for participation in the FACT study; children with familial childhood cancer anywhere in the world were also eligible for participation in the FACT study. We analysed lymphocyte DNA from 1215 individuals with 28 different childhood tumours, of whom 1206 had one childhood tumour and nine individuals had two different childhood tumours (appendix). This cohort included 890 individuals with Wilms tumour: 799 had non-familial disease and 91 were from 49 familial Wilms tumour pedigrees in which two or more individuals had Wilms tumour due to an unknown genetic or epigenetic cause (figure 1; appendix). Most participants with Wilms tumour were from the UK and, therefore, were likely to have been treated with chemotherapy before surgery. We used constitutional (germline) exome data from 7632 individuals with 28 different adult cancers available from The Cancer Genome Atlas (TCGA) on May 13, 2014 (figure 1; see appendix for the types and number of adult cancers interrogated). As reference data, we used the Exome Aggregation Consortium (ExAC) data, version 3, accessed on Nov 13, 2015 (excluding the TCGA samples),12 and the ICR1000 UK exome series.13 We generated and analysed the ICR1000 UK exome series and childhood cancer sample data from the FACT participants by use of consistent sequencing and analytical processes.
Figure 1.
Cancer cohorts investigated
Wilms tumour families are pedigrees in which two or more individuals had Wilms tumour. FACT=Factors Associated with Childhood Tumours. TCGA=The Cancer Genome Atlas. *Nine individuals had two different childhood tumours.
The FACT study was approved by the London Multicentre Ethics Committee (05/MRE02/17), and its collaborators are listed in the appendix. Written informed consent was obtained from all participants, their parents or guardians, or both, as appropriate (age cutoff for consent was 18 years).
Procedures
We did exome sequencing in samples from all childhood cancer probands and 119 individuals from 49 familial Wilms tumour pedigrees who did not have Wilms tumour, by using 50 ng genomic DNA and the Nextera DNA sample preparation kit (Illumina, San Diego, CA, USA) or 1·5 μg genomic DNA and the TruSeq exome enrichment kit (Illumina). The captured libraries were amplified by PCR with the supplied paired-end PCR primers. Sequencing was done with HiSeq 2000 (Illumina) or HiSeq 2500 (Illumina). We used the OpEx v1.0 pipeline to do variant calling in childhood cancer, adult cancer (TGCA), and ICR1000 exome data.14 We also reannotated the variants in the ExAC data with the CAVA tool in OpEx, to ensure variant calling consistency across the different cohorts.14 We used the protein-truncating variant prioritisation method to prioritise potential disease-associated genes for follow-up; this is a proven strategy for identifying tumour suppressor genes in outbred populations, which we have used to identify several other cancer predisposition genes.3, 4, 10 We validated variants in TRIM28, FBXW7, NYNRIN, and KDM3B genes by use of Sanger sequencing in the probands and any available relatives, designing primers with BatchPrimer3. We used the QIAGEN Multiplex PCR kit (QIAGEN, Hilden, Germany) to prepare PCRs, and the resulting amplicons were bidirectionally sequenced with BigDye Terminator cycle sequencing kits (Thermo Fisher Scientific, Waltham, MA, USA) and an ABI 3730 sequencer (Life Technologies, Carlsbad, CA, USA). We analysed sequencing traces with Mutation Surveyor software and by visual inspection with Chromas, version 2.13. We validated the CDC73 mutation with the TruSight Cancer panel (Illumina). We did in-silico analyses of variant pathogenicity with Alamut Visual, version 2.9.0.
Statistical analysis
We used the methods described by Akawi and colleagues15 to obtain the probability of a family in our study having two protein-truncating variants in a given gene. The method uses the frequency of rare protein-truncating variants (allele frequency <0·001) in ExAC and the number of observed protein-truncating variants in a given gene to estimate the probability of an individual having two of these variants in that gene. The baseline prevalence of having two protein-truncating variants per gene is calculated as the proportion of rare protein-truncating variants squared. We observed two individuals with two protein-truncating variants and nine individuals with a single protein-truncating variant in NYNRIN among 844 individuals with Wilms tumour from the 890 included in this study. We used the R function to calculate the probability of observing two individuals with two NYNRIN protein-truncating variants: analyse_inherited_enrichment from the R package recessiveStats with hgnc=”NYNRIN”, chrom=“14”, counts$biallelic_lof=2, counts$lof_func=9, and cohort_n=844.
We used a binomial test—dbinom function in R—to calculate the probability of all ten TRIM28 mutations with known inheritance being maternally inherited, assuming the baseline probability of inheriting the variant from either parent was 0·5.
We did a functional enrichment analysis with use of g:Profiler (version r1665_e85_eg32).16 We used the 21 predisposition genes described for Wilms tumour as our query set. We looked for enrichment among Gene Ontology molecular function terms and pathway gene sets from the Kyoto Encyclopedia of Genes and Genomes, requiring the size of the functional category to have a minimum of five genes and using the Benjamini-Hochberg correction for multiple testing p value as the significance threshold. The false discovery rate q values presented in this study are the Benjamini-Hochberg critical values.
For the somatic cancer driver comparisons, we used 20 genes reported to be somatically mutated in Wilms tumour. These included 17 established genes reported in more than one publication and three newly reported genes (ACTB, BCOR, NONO) with at least three somatic mutations in the TARGET discovery series.17 We used the COSMIC cancer gene census to establish which of the 21 Wilms tumour predisposition genes, and which of the 20 somatically mutated Wilms tumour driver genes, were also somatically mutated in other cancer types.
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. SM, SY, EH, and NR had full access to all the data. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
We used the protein-truncating variant prioritisation method with dominant and recessive inheritance models to identify genes with different protein-truncating variants in 890 individuals with Wilms tumour (figure 1). The three genes with the strongest signal in the prioritisation analyses were TRIM28, FBXW7, and NYNRIN. We did Sanger sequencing to validate protein-truncating variants and rare non-synonymous variants of these genes in probands and any available samples from relatives to further evaluate their status as bona fide Wilms tumour predisposition genes.
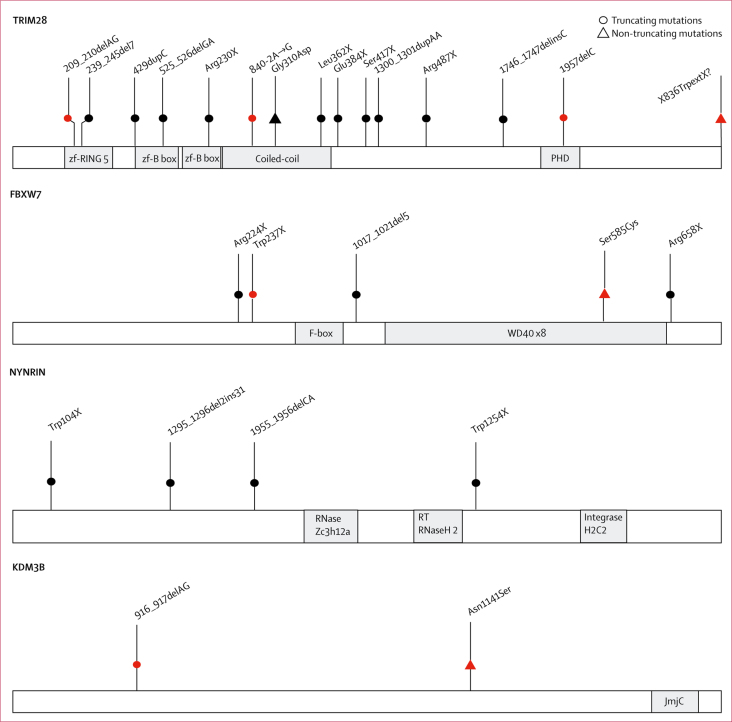
We identified pathogenic truncating mutations of TRIM28 in 17 individuals with Wilms tumour from 13 families (figure 2, table, appendix). At least three of these truncating mutations had arisen de novo. Protein-truncating variants in TRIM28 are extremely rare in the general population, because the gene is highly intolerant to truncating variation, with a pLI score of 1·0 (pLI >0·9 indicates extreme intolerance to protein-truncating variants).12 We found no other cancers in individuals carrying TRIM28 mutations. We also did not find any TRIM28 protein-truncating variants in 334 individuals with 27 other childhood cancers or in 7632 individuals with adult cancers, suggesting that TRIM28 pathogenic mutations primarily predispose to Wilms tumour. Of note, the TRIM28 mutations in two families in our study (ID_0498 and ID_0506) were independently, and coincidentally, reported while we were preparing this manuscript.18 We identified a de-novo stop-loss mutation in another family (ID_7574), which we assumed to be pathogenic. Finally, in family ID_0477, which included six cases of Wilms tumour, we identified TRIM28 929G→A, leading to the protein change Gly310Asp (figure 2, table, appendix). We believe this mutation to be pathogenic because it is absent from public and in-house datasets, it segregates with Wilms tumour in the family, it is predicted to be deleterious by in silico tools, and it is at a crucial residue within the coiled-coil domain of TRIM28 that is reported to interact with AMER1, which is encoded by a gene somatically mutated in Wilms tumour.19
Figure 2.
Schematic representations of TRIM28, FBXW7, NYNRIN, and KDM3B
Schematic representations of encoded proteins are shown, with functional domains in grey. The position of cancer-predisposing mutations is shown above the protein. Red symbols denote de novo mutations.
Table.
Molecular and clinical features of individuals with mutations in TRIM28, FBXW7, NYNRIN, KDM3B, or CDC73
| Sex | Mutations (protein change) | Inheritance | Tumour type, age at diagnosis (months) | Unilateral or bilateral | Histology | Other | Status, age (years) | |
|---|---|---|---|---|---|---|---|---|
| TRIM28 | ||||||||
| ID_0477_01 | F | 929G→A (Gly310Asp) | Maternal | WT, 24 | Unilateral | Epithelial predominant | .. | .. |
| ID_0477_02 | M | 929G→A (Gly310Asp) | Maternal | WT, 84 | Unilateral | Epithelial | .. | .. |
| ID_0477_03 | F | 929G→A (Gly310Asp) | Maternal | WT, 93 | Unilateral | NA | .. | .. |
| ID_0498_01 | M | 1746_1747delinsC | Maternal | WT, 8 | Unilateral | Epithelial | .. | Alive, 30 |
| ID_0498_02 | F | 1746_1747delinsC | Maternal | WT, 5 | Unilateral | Epithelial | .. | Alive, 29 |
| ID_0498_03 | F | 1746_1747delinsC | NA | WT, 6 | Unilateral | Epithelial | .. | .. |
| ID_0487_01 | M | 429dupC | Maternal | WT, 15 | Unilateral | Epithelial predominant | .. | Alive, 19 |
| ID_0487_02 | M | 429dupC | NA | WT, 18 | Unilateral | NA | .. | .. |
| ID_0506_01 | M | 525_526delGA | Maternal | WT, 39 | Unilateral | Epithelial | .. | Alive, 23 |
| ID_0506_02 | F | 525_526delGA | Maternal | WT, 8 | Bilateral | Epithelial | .. | Alive, 20 |
| ID_7487_01 | F | 239_245del7 | Maternal | WT, 118 | Unilateral | Epithelial predominant, diffuse anaplasia | .. | Died, 12 |
| ID_1982 | M | 1957delC | De novo | WT, 11 | Bilateral | Epithelial predominant | .. | Alive, 15 |
| ID_6530 | M | 209_210delAG | De novo | WT, 15 | Unilateral | Epithelial and blastemal | Autism, speech delay | Alive, 6 |
| ID_1969 | M | 840–2A→G | De novo | WT, 118 | Unilateral | Epithelial and blastemal | .. | Alive, 19 |
| ID_7574 | M | 2508A→G (X836TrpextX?)* | De novo | WT, 13 | Unilateral | Epithelial predominant | Autism, intellectual disability | .. |
| ID_0902 | F | 1250C→A (Ser417X) | Maternal | WT, 12 | Unilateral | Epithelial predominant | .. | .. |
| ID_0692 | F | 1459C→T (Arg487X) | NA | WT, 13 | Bilateral | NA | .. | Alive, 36 |
| ID_6671 | F | 688C→T (Arg230X) | NA | WT, 10 | Bilateral | Epithelial predominant | Chronic kidney disease | Alive, 6 |
| ID_0796 | F | 1085T→A (Leu362X) | NA | WT, 61 | Unilateral | NA | .. | Alive, 33 |
| ID_0866 | F | 1300_1301dupAA | NA | WT, 90 | Unilateral | Epithelial predominant | .. | Alive, 29 |
| ID_0936 | M | 1150G→T (Glu384X) | NA | WT, 8 | Unilateral | NA | .. | .. |
| FBXW7 | ||||||||
| ID_0811 | M | 710G→A (Trp237X) | De novo | WT, 76 | Unilateral | NA | Osteosarcoma at 39 years | Died, 39 |
| ID_2084_01 | M | 1972C→T (Arg658X) | NA | WT, 42 | Unilateral | Focal anaplasia | Relapse at 66 months | .. |
| ID_0592 | F | 1017_1021del5 | Paternal | WT, 28 | Unilateral | NA | hypotonia | Alive, 18 |
| ID_1227 | F | 670C→T (Arg224X) | NA | WT, 73 | .. | NA | .. | Died, 7 |
| ID_7520 | M | 1753A→T (Ser585Cys) | De novo | Rhabdoid, 40 | .. | Extra-renal rhabdoid with INI1 loss | Two febrile convulsions | Alive, 5 |
| NYNRIN | ||||||||
| ID_0493_01 | M | 1955_1956delCA | Paternal | WT, 24 | Unilateral | Blastemal predominant | Inguinal hernia | .. |
| ID_0493_01 | .. | 3761G→A (Trp1254X) | Maternal | .. | .. | .. | .. | .. |
| ID_0493_02 | M | 1955_1956delCA | Paternal | WT, 24 | Unilateral | Triphasic | .. | .. |
| ID_0493_02 | .. | 3761G→A (Trp1254X) | Maternal | .. | .. | .. | .. | .. |
| ID_6049 | M | 311G→A (Trp104X) | Maternal | WT, 34 | Unilateral | Triphasic | Epilepsy, hypothyroidism, intellectual disability | Alive, 11 |
| ID_6049 | .. | 1295_1296del2ins31 | Paternal | .. | .. | .. | .. | .. |
| KDM3B | ||||||||
| ID_7225 | F | 3422A→G (Asn1141Ser) | De novo | WT, 49 | Bilateral | NA | Hyperpigmentation | .. |
| ID_2086 | M | 916_917delAG | De novo | Hepatoblastoma, 131 | .. | NA | Autism, abnormal pigmentation, intellectual disability | .. |
| CDC73 | ||||||||
| ID_6491_01 | F | 878dupA | Paternal | WT, 192 | Unilateral | Epithelial predominant | Convergent strabismus | Alive, 21 |
| ID_6491_02 | M | 878dupA | NA | WT, 96 | Unilateral | NA | .. | Alive, 48 |
Pedigrees and chromatograms are shown in the appendix. F=female. WT=Wilms tumour. M=male. NA=not available.
The stop codon (X) at position 836 is changed to Trp, extending the protein by an unknown number of amino acids (?).
We established that ten of the TRIM28 mutations had been inherited, and that in all cases, the mutation had been transmitted from the mother, a significant association (p=0·00098). Pathology information was available for 16 tumours, of which 14 were epithelial or epithelial-predominant (table).
We identified truncating FBXW7 mutations in four individuals with Wilms tumour, of which one was de novo (in ID_0811), one had been inherited from an unaffected father (in ID_0592), and two were of unknown provenance (in ID_2084 and ID_1227; figure 2, table, appendix). FBXW7 is highly intolerant to protein-truncating variants (pLI=1·00) and these data suggest that FBXW7 is a Wilms tumour predisposition gene. Two of the four individuals with truncating FBXW7 mutations have died (table). Additionally, ID_2084 was treated for Wilms tumour at 3·5 years of age, but relapsed when he was 5·5 years old. We did not find truncating FBXW7 mutations in individuals with other childhood or adult cancers. However, we identified a de novo non-synonymous mutation, 1753A→T (protein change Ser585Cys), in a child with an extra-renal rhabdoid tumour (ID_7520), which we assumed to be pathogenic.
We identified biallelic truncating mutations in NYNRIN in three children from two families (ID_0493 and ID_6049; figure 2, table, appendix). Each parent was heterozygous for one of the mutations. These mutations were absent from ExAC and the ICR1000 series. We found no individuals with two NYNRIN truncating mutations in the ICR1000 series and no homozygous protein-truncating variants in ExAC (individual-level data is not available for ExAC, therefore it is not possible to know if anyone had two different protein-truncating variants). Additionally, the probability of finding two different families with the same phenotype and two truncating NYNRIN mutations by chance is 4·0 × 10−9. One of the affected children had an inguinal hernia and another had epilepsy, hypothyroidism, and intellectual disability (table). It is unclear whether any of these additional clinical features are related to the biallelic NYNRIN mutations. We did not identify biallelic NYNRIN protein-truncating variants in individuals with other childhood or adult cancers.
In addition to the agnostic protein-truncating variant prioritisation analyses, we reviewed the exome data in genes proposed as possible childhood cancer predisposition genes, identified through a systematic review of 19 171 genes for links to Mendelian disease. This review led us to the identification of two de-novo KDM3B mutations, a non-synonymous mutation in a child with Wilms tumour and a hyperpigmented lesion on her buttock (ID_7225) and a truncating mutation in a child with hepatoblastoma, hyperpigmentation and hypopigmentation, autism, and intellectual disability (ID_2086; figure 2, table, appendix). In 2018, Diets and colleagues20 reported a KDM3B truncating mutation in a girl with acute myeloid leukaemia, mild intellectual disability, and hip dysplasia and a de novo non-synonymous KDM3B mutation in a boy with Hodgkins lymphoma and moderate intellectual disability. KDM3B is highly intolerant to both protein-truncating variants (pLI=1·00) and non-synonymous variation (Z=4·99; the Z score is the deviation of observation from expectation for non-synonymous variants). Taken together, these data provide strong evidence that KDM3B is a childhood cancer predisposition gene.
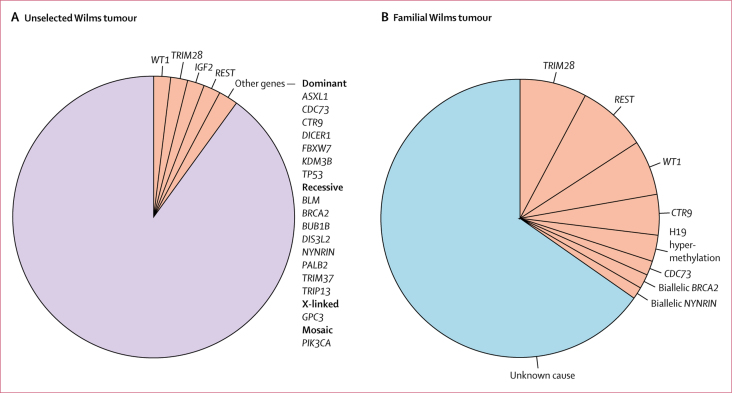
These new discoveries bring the number of constitutionally mutated genes confirmed as Wilms tumour predisposition genes to 21. We estimate that, together, these constitutional events contribute to about 10% of unselected Wilms tumour (figure 3). Four contributors—WT1, TRIM28, REST, and 11p15 epimutations and uniparental disomy that result in biallelic IGF2 expression—each account for about 2%.2, 4, 5 The remaining 17 are very rare and, together, probably account for no more than 2% of unselected Wilms tumours.2, 3, 4, 6, 7, 8, 9, 10, 11 Functional enrichment analysis highlighted nucleic acid metabolism, chromosome organisation, chromatin or histone modification, and negative regulation of cellular processes as important pathways underlying Wilms tumour predisposition (appendix).
Figure 3.
Contribution of constitutional mutations to unselected and familial Wilms tumour
(A) About 10% of unselected Wilms tumours are due to constitutional mutations in one of 21 genes (pink). (B) A third of familial Wilms tumours are explicable by known Wilms tumour predisposition factors (pink) and two thirds are of unknown cause (blue).
We have investigated 65 families with two or more cases of Wilms tumour over the last 20 years, including the 49 familial Wilms tumour pedigrees in this study (appendix). In two families, we found a constitutional predisposing mutation in one individual with Wilms tumour, but not their affected relative. We have identified causative constitutional mutations in 22 (35%) of the remaining 63 families (figure 3). The most common of which were mutations in REST (five [8%] of 63), TRIM28 (five [8%] of 63), and WT1 (four [6%] of 63). CTR9 mutations were present in three families and H19 hypermethylation was found in two families. Biallelic BRCA2 mutations, biallelic NYNRIN mutations, and a CDC73 mutation were found in one family each. We identified the CDC73 mutation through this present study (table, appendix). CDC73 is an established Wilms tumour predisposition gene but, to our knowledge, has not previously been associated with familial Wilms tumour. We did not find any cause in two thirds of the families (41 [65%] of 63).
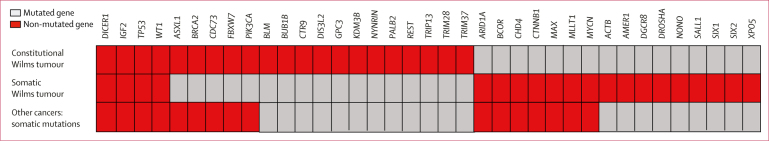
Finally, we assessed the overlap between the 21 Wilms tumour predisposition genes and 20 somatically mutated Wilms tumour driver genes (figure 4). Only four genes—WT1, IGF2, TP53, and DICER1—promoted Wilms tumour oncogenesis in both contexts, and all four were also somatically altered in other cancers. A further five constitutionally mutated genes—PIK3CA, FBXW7, ASXL1, BRCA2, and CDC73—were somatically mutated in other cancer types but have not been proven to be somatic drivers in Wilms tumour. The remaining 12 Wilms tumour predisposition genes are not known to be somatically mutated cancer drivers.
Figure 4.
Overlap of constitutionally and somatically mutated Wilms tumour genes
Discussion
To our knowledge, this is the largest exome sequencing study of individuals with Wilms tumour to date, including 890 affected individuals. Our analyses found three autosomal dominant Wilms tumour predisposition genes—TRIM28, FBXW7, and KDM3B—and one autosomal recessive Wilms tumour predisposition gene, NYNRIN. Constitutional TRIM28 mutations join constitutional WT1 and REST mutations as a relatively common contributor to Wilms tumour predisposition, accounting for about 8% of familial Wilms tumour and about 2% of unselected Wilms tumour.2, 4 We found a strong association between TRIM28 mutations and epithelial Wilms tumour, with most individuals with a TRIM28 mutation having Wilms tumour of predominantly epithelial histology. This suggests that TRIM28 mutations make a sizeable contribution to epithelial Wilms tumour, and we recommend that all children with this rare favourable subtype of Wilms tumour should be offered TRIM28 gene testing.
Familial Wilms tumour pedigrees due to TRIM28 mutations showed incomplete penetrance and a strong parent-of-origin effect, because all inherited mutations were maternally transmitted. TRIM28 is not imprinted, but it is located close to PEG3, which is imprinted and paternally expressed.21 At the organism level, PEG3 promotes growth but at the cellular level, it is a putative tumour suppressor.21 A possible explanation for the maternal bias of TRIM28 mutations is that somatic inactivation of the paternal wild-type TRIM28 allele, through mitotic recombination, also leads to loss of the paternally expressed PEG3, thus promoting tumourigenesis. There is precedence for this model in cancer predisposition: SDHD mutations predispose to phaeochromocytoma almost exclusively when inherited paternally.22 The combination of somatic loss of the maternal wild-type SDHD allele and maternally expressed growth inhibitory genes at the IGF1–H19 imprinting region has been proposed as the explanation for this pattern.22 However, a factor against this model for TRIM28 is that the loss of heterozygosity in the tumour from ID_506_01 did not appear to include PEG3.18 Given the prevalence of TRIM28 mutations in Wilms tumour, confirmation of the parent-of-origin bias we observed should be possible in the near future. If this parent-of-origin effect is supported, DNA methylation analyses at the PEG3 imprinting control region and loss of heterozygosity and PEG3 expression analyses in tumours from individuals with TRIM28 mutations might help to provide a mechanistic explanation. From a clinical perspective, establishing if the penetrance of TRIM28 mutations is influenced by the parent-of-origin of the mutation is important, because this would have considerable impact on cancer risks and genetic counselling.
We provided evidence that constitutional FBXW7 mutations predisposed to Wilms tumour and to other malignancies. Two of the five individuals carrying FBXW7 mutations had a malignancy other than Wilms tumour. ID_0811 developed osteosarcoma as an adult, after having Wilms tumour. ID_7520 had a rhabdoid tumour and did not have Wilms tumour. The assessment of additional individuals with rhabdoid tumour and de-novo FBXW7 mutations would be useful to further support the role of FBXW7 in rhabdoid tumour predisposition. Furthermore, a woman with Hodgkin lymphoma, adult Wilms tumour, early-onset breast cancer, and a constitutional FBXW7 deletion was reported in 2015,23 and a man with renal cell cancer and a constitutional t(3;4)(q21;q31) translocation disrupting FBXW7 was reported in 2009.24 These data suggest that individuals with FBXW7 mutations might be at risk of multiple childhood and adult cancers and will require ongoing close monitoring. Notably, we believe that the in-frame FBXW7 variant reported25 in an individual with Wilms tumour is not pathogenic because it is not rare and the child also had a pathogenic WT1 mutation.
KDM3B also appears to be a pleiotropic cancer predisposition gene. The four KDM3B pathogenic mutations reported to date have been associated with four different cancers: Wilms tumour and hepatoblastoma in our study, and acute myeloid leukaemia and Hodgkin lymphoma in the study by Diets and colleagues.20 Large-scale, broad mutation testing of FBXW7 and KDM3B in individuals with cancer will probably be required to establish the full spectrum of associated cancers, because of the rarity of truncating variants and the challenges in interpreting non-synonymous variation in these genes. There are indications that KDM3B and NYNRIN mutations might cause non-malignant phenotypes, particularly intellectual disability. More data on the contribution of these genes to non-malignant conditions will probably become available over the next decade through extensive exome and genome sequencing being done in children with developmental disorders.
The four genes we reported here have different functions, and it is unclear why or how they predispose to Wilms tumour. TRIM28 encodes a multidomain protein involved in the regulation of many cellular processes, including transcriptional repression, p53 degradation, pluripotency maintenance, autophagosome formation, epithelial-mesenchyme transition, and the DNA damage response.26 TRIM28 is highly expressed in many cancers, and its inactivation has not been previously associated with oncogenesis. This might explain why inactivating TRIM28 mutations seem to predispose to Wilms tumour alone. The mechanisms underlying this Wilms tumour predisposition are not known, but it is notable that TRIM28 is a major binding partner of AMER1, which is encoded by a gene that is frequently somatically mutated in Wilms tumour.19
FBXW7 encodes the substrate recognition component of the E3-ubiquitin ligase SCF complex, which is responsible for recognising and binding phosphorylated substrates and regulating their turnover through proteosome degradation.27 FBXW7 is an established tumour suppressor gene and frequently mutated in many cancers, particularly endometrial and gastrointestinal cancers.27 FBXW7 is not a confirmed somatic driver in Wilms tumour because only one confirmed somatic FBXW7 point mutation has thus far been reported.25 This situation is similar to that of PIK3CA. Constitutional mosaic PIK3CA mutations predispose to Wilms tumour, whereas somatic mutations at the same residue are common in many cancers but have not been reported in Wilms tumour.9
KDM3B encodes a histone H3 demethylase that specifically catalyses the demethylation of H3K9Me1 and H3K9Me2 residues, and is required for normal somatic growth in mice.28 Tumour-suppressive and tumour-promoting KDM3B activities have been proposed in leukaemia, although somatic driver KDM3B mutations have not been reported. Finally, there is very little known about the functions of NYNRIN, though NYN domains are thought to be involved in RNA processing and NYNRIN has been implicated in microRNA–mRNA regulation.29
The diverse functions of these four new Wilms tumour predisposition genes mirror the broad range of biological processes in which known Wilms tumour predisposition genes operate, as shown by our functional enrichment analysis (appendix). Functional exploration of these genes was beyond the scope of our study, but we hope our results might encourage such assessments, which will probably provide novel insights into oncogenesis and kidney development.
Our analyses were designed to identify tumour suppressor genes in which constitutional truncating mutations predisposed to cancer, but were not designed to identify other mechanisms of cancer predisposition. For example, it is very possible that non-truncating coding variation might be contributing to familial and non-familial Wilms tumour, and future analyses to investigate this would be worthwhile. Non-coding, epigenetic, and mosaic abnormalities are all known to be relevant to Wilms tumour predisposition but were not investigated in our study. Notably, none of the known Wilms tumour predisposition genes are within the regions on chromosomes 2p24, 11q14, 5q14, 22q12, and Xp22 identified in a genome-wide association study30 of Wilms tumour, and the causal mechanisms underlying the associations in that study are unknown. Additionally, the mutations at 17q21 responsible for FWT1-linked families have not yet been discovered.2
Genetic predisposition to Wilms tumour exhibits remarkable heterogeneity, and this is particularly noteworthy because childhood cancers are generally assumed to be aetiologically simpler than adult cancers. Furthermore, our study provides strong evidence that further genetic, genomic, or epigenetic Wilms tumour predisposition factors exist, because only a third of the familial Wilms tumour pedigrees we investigated have been explained. Any further familial Wilms tumour genes discovered will be highly likely to contribute also to non-familial Wilms tumour.
Our study reveals new insights into the complexity, mechanisms, and clinical implications of Wilms tumour predisposition. Although our understanding of the genetic landscape of Wilms tumour predisposition is still far from complete, the available knowledge has considerable scientific and clinical use. Given the extensive heterogeneity and the absence of family history or additional clinical features in many individuals with a mutation, we believe routine genetic testing in all individuals with Wilms tumour would be scientifically and clinically valuable.
Acknowledgments
Acknowledgments
We thank the families for their participation and the many doctors, nurses, and counsellors who recruited them to the FACT study. The FACT collaborators are listed in the appendix. This study was funded by the Wellcome Trust (100210/Z/12/Z). We acknowledge support of the National Institute for Health Research Clinical Research Network (NIHR CRN), the Children's Cancer and Leukaemia Group (CCLG), and the Royal Marsden-ICR NIHR Biomedical Research Centre. We thank Jessie Bull for assistance in FACT recruitment and Ann Strydom for assistance in preparing the manuscript.
Contributors
SM, SY, ER, and NR designed the study. SM, EP-P, SS, and SH did the molecular studies. SM, SY, EH, AE, MC, and ER handled data management, data analyses, or both. JA, SB, TC, RFa, RFu, AG, RG, JH, SL, FM, JN, MR, JS, DW, and DY contributed to the sample and data collection coordinated by AZ and MW-P. NR, SM, and SY wrote the manuscript with input from the other authors.
Declaration of interests
NR reports personal fees from AstraZeneca and Genomics, outside the submitted work. ER reports personal fees from Foresite Capital. JA reports personal fees from TC Biopharm and holds founder shares in Autolus Therapeutics. All other authors declare no competing interests.
Supplementary Material
References
- 1.Gadd S, Huff V, Huang CC. Clinically relevant subsets identified by gene expression patterns support a revised ontogenic model of Wilms tumor: a Children's Oncology Group Study. Neoplasia. 2012;14:742–756. doi: 10.1593/neo.12714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scott RH, Stiller CA, Walker L, Rahman N. Syndromes and constitutional chromosomal abnormalities associated with Wilms tumour. J Med Genet. 2006;43:705–715. doi: 10.1136/jmg.2006.041723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanks S, Perdeaux ER, Seal S. Germline mutations in the PAF1 complex gene CTR9 predispose to Wilms tumour. Nat Commun. 2014;5:4398. doi: 10.1038/ncomms5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahamdallie SS, Hanks S, Karlin KL. Mutations in the transcriptional repressor REST predispose to Wilms tumor. Nat Genet. 2015;47:1471–1474. doi: 10.1038/ng.3440. [DOI] [PubMed] [Google Scholar]
- 5.Scott RH, Douglas J, Baskcomb L. Constitutional 11p15 abnormalities, including heritable imprinting center mutations, cause nonsyndromic Wilms tumor. Nat Genet. 2008;40:1329–1334. doi: 10.1038/ng.243. [DOI] [PubMed] [Google Scholar]
- 6.Reid S, Schindler D, Hanenberg H. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 7.Astuti D, Morris MR, Cooper WN. Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat Genet. 2012;44:277–284. doi: 10.1038/ng.1071. [DOI] [PubMed] [Google Scholar]
- 8.Russell B, Johnston JJ, Biesecker LG. Clinical management of patients with ASXL1 mutations and Bohring-Opitz syndrome, emphasizing the need for Wilms tumor surveillance. Am J Med Genet A. 2015;167:2122–2131. doi: 10.1002/ajmg.a.37131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gripp KW, Baker L, Kandula V. Nephroblastomatosis or Wilms tumor in a fourth patient with a somatic PIK3CA mutation. Am J Med Genet A. 2016;170:2559–2569. doi: 10.1002/ajmg.a.37758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yost S, de Wolf B, Hanks S. Biallelic TRIP13 mutations predispose to Wilms tumor and chromosome missegregation. Nat Genet. 2017;49:1148–1151. doi: 10.1038/ng.3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schultz KAP, Williams GM, Kamihara J. DICER1 and associated conditions: identification of at-risk individuals and recommended surveillance strategies. Clin Cancer Res. 2018;24:2251–2261. doi: 10.1158/1078-0432.CCR-17-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lek M, Karczewski KJ, Minikel EV. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruark E, Munz M, Renwick A. The ICR1000 UK exome series: a resource of gene variation in an outbred population. F1000Res. 2015;4:883. doi: 10.12688/f1000research.7049.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruark E, Munz M, Clarke M. OpEx—a validated, automated pipeline optimised for clinical exome sequence analysis. Sci Rep. 2016;6:31029. doi: 10.1038/srep31029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akawi N, McRae J, Ansari M. Discovery of four recessive developmental disorders using probabilistic genotype and phenotype matching among 4,125 families. Nat Genet. 2015;47:1363–1369. doi: 10.1038/ng.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reimand J, Arak T, Adler P. g:Profiler—a web server for functional interpretation of gene lists (2016 update) Nucleic Acids Res. 2016;44:W83–W89. doi: 10.1093/nar/gkw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gadd S, Huff V, Walz AL. A Children's Oncology Group and TARGET initiative exploring the genetic landscape of Wilms tumor. Nat Genet. 2017;49:1487–1494. doi: 10.1038/ng.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halliday BJ, Fukuzawa R, Markie DM. Germline mutations and somatic inactivation of TRIM28 in Wilms tumour. PLoS Genet. 2018;14:e1007399. doi: 10.1371/journal.pgen.1007399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim WJ, Wittner BS, Amzallag A. The WTX tumor suppressor interacts with the transcriptional corepressor TRIM28. J Biol Chem. 2015;290:14381–14390. doi: 10.1074/jbc.M114.631945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diets IJ, Waanders E, Ligtenberg MJ. High yield of pathogenic germline mutations causative or likely causative of the cancer phenotype in selected children with cancer. Clin Cancer Res. 2018;24:1594–1603. doi: 10.1158/1078-0432.CCR-17-1725. [DOI] [PubMed] [Google Scholar]
- 21.He H, Kim J. Regulation and function of the peg3 imprinted domain. Genomics Inform. 2014;12:105–113. doi: 10.5808/GI.2014.12.3.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hensen EF, Jordanova ES, van Minderhout IJ. Somatic loss of maternal chromosome 11 causes parent-of-origin-dependent inheritance in SDHD-linked paraganglioma and phaeochromocytoma families. Oncogene. 2004;23:4076–4083. doi: 10.1038/sj.onc.1207591. [DOI] [PubMed] [Google Scholar]
- 23.Roversi G, Picinelli C, Bestetti I. Constitutional de novo deletion of the FBXW7 gene in a patient with focal segmental glomerulosclerosis and multiple primitive tumors. Sci Rep. 2015;5:15454. doi: 10.1038/srep15454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuiper RP, Vreede L, Venkatachalam R. The tumor suppressor gene FBXW7 is disrupted by a constitutional t(3;4)(q21;q31) in a patient with renal cell cancer. Cancer Genet Cytogenet. 2009;195:105–111. doi: 10.1016/j.cancergencyto.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Williams RD, Al-Saadi R, Chagtai T. Subtype-specific FBXW7 mutation and MYCN copy number gain in Wilms' tumor. Clin Cancer Res. 2010;16:2036–2045. doi: 10.1158/1078-0432.CCR-09-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Czerwinska P, Mazurek S, Wiznerowicz M. The complexity of TRIM28 contribution to cancer. J Biomed Sci. 2017;24:63. doi: 10.1186/s12929-017-0374-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeh CH, Bellon M, Nicot C. FBXW7: a critical tumor suppressor of human cancers. Mol Cancer. 2018;17:115. doi: 10.1186/s12943-018-0857-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Z, Chen X, Zhou S, Liao L, Jiang R, Xu J. The histone H3K9 demethylase Kdm3b is required for somatic growth and female reproductive function. Int J Biol Sci. 2015;11:494–507. doi: 10.7150/ijbs.11849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng L, Luo DY. Identification of key genes and construction of microRNA-mRNA regulatory networks in bladder smooth muscle cell response to mechanical stimuli using microarray expression profiles and bioinformatics analysis. World J Urol. 2018;36:241–247. doi: 10.1007/s00345-017-2132-3. [DOI] [PubMed] [Google Scholar]
- 30.Turnbull C, Perdeaux ER, Pernet D. A genome-wide association study identifies susceptibility loci for Wilms tumor. Nat Genet. 2012;44:681–684. doi: 10.1038/ng.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.