Highlights
-
•
Natural factors had major effects on community dynamics of microbial target groups.
-
•
Conversely, “Foxy-2” exposed no major effect on rhizosphere microbial communities.
-
•
Archaeal community had greater rhizosphere competence than “Foxy-2” in clayey soil.
-
•
Compatibility of indigenous soil nitrifying prokaryotes with “Foxy-2” was verified.
Keywords: Maize rhizosphere, Fusarium oxysprorum f.sp. strigae inoculation, Bacterial and archaeal amoA gene abundance and community composition, Rhizosphere competence
Abstract
Fusarium oxysporum f.sp. strigae (Fos) is an effective biocontrol agent (BCA) against the parasitic weed Striga hermonthica. It acts in the rhizosphere of several tropical cereals, where it may interfere with indigenous microbial populations. To test this impact, we assessed in a 2-season field experiment at two contrasting tropical agro-ecological sites the response of nitrifying and total indigenous prokaryotic communities in the rhizosphere of maize to the exposure of the Fos-BCA “Foxy-2”. At early leaf development (EC30), flowering (EC60) and senescence (EC90) stage of maize, rhizosphere samples were obtained and subjected to community analysis of bacterial and archaeal amoA (ammonia monooxigenase) (AOB, AOA) and 16S rRNA genes. Abundance and community composition of all studied genes were predominantly influenced by soil type, crop growth stage and seasonality. No major effect of “Foxy-2” was found. Notably, total archaeal community relative to bacteria dominated in the clayey soil which was linked to its strong soil organic carbon (SOC) background. Compared to bacterial nitrifiers, domination of nitrifying archaea increased towards senescence stage which was explained by biochemical differences in organic resource availability between the crop growth stages. During the short rain season, the higher archaeal abundance was mainly driven by increased availability of organic substrates, i.e., extractable organic carbon. Our findings suggested that archaea had greater rhizosphere competence than “Foxy-2” in soils with higher clay and SOC contents. We verified that “Foxy-2” in maize rhizospheres is compatible with nitrifying prokaryotes under the given environments, in particular in clayey soils dominated by archaea.
1. Introduction
The fungal strain Fusarium oxysporum f.sp strigae (“Foxy-2”) has been acknowledged as a potent biological control agent (BCA) against Striga hermonthica which is parasitic to several cereals cultivated in Sub-Saharan Africa (Schaub et al., 2006, Venne et al., 2009, Elzein et al., 2010). “Foxy-2” proliferates in the crop rhizosphere and has been mainly studied regarding its virulence and mode of action (Schaub et al., 2006, Ndambi et al., 2011, Avedi et al., 2014).
The rhizosphere is a hot spot of microbial activities interacting with plants (Grayston et al., 1998), but no understanding is currently available regarding the interactions of “Foxy-2” with indigenous microorganisms colonizing the roots of cereals and environmental factors including site-specific soil and climatic conditions. This is of particular relevance since “Foxy-2” is often delivered via seed coating and proliferates subsequently along the roots where it directly interacts with other rhizosphere organisms. Thus, prior to broad-scale application of the BCA “Foxy-2” in the field, its compatibility with non-target rhizosphere microorganisms needs to be thoroughly assessed under contrasting environmental conditions (Miedaner et al., 2001, Edel-Hermann et al., 2009, Martin-Laurent et al., 2013). In the rhizosphere, “Foxy-2” must co-exist with indigenous microbial populations as well as maintain its efficacy under a range of environmental factors including seasonal alternations considering rainfall and temperature patterns, varying soil types and also crop growth stages (Girvan et al., 2003, Beed et al., 2007, Bell et al., 2009, Raaijmakers et al., 2009, Watson, 2013).
Abundance and composition of rhizosphere microbial communities are mainly shaped by rhizodeposition which is the transfer of plant-derived carbon (C) and nitrogen (N) compounds below ground (Høgh-Jensen and Schjoerring, 2001, Kandeler et al., 2002, Rasche et al., 2006a, Wichern et al., 2008, Jones et al., 2009, Fustec et al., 2011). Rhizodeposition is influenced by external factors such as soil type (properties), plant species and their growth stages, as well as climatic conditions (Rasche et al., 2006a, Hai et al., 2009, Hayden et al., 2010). Climatic conditions are of particular importance as rainfall and temperature control crop physiology, photosynthesis activity and consequently rhizodeposition shaping the root-associated microbial community (Waring and Running, 1998, Bell et al., 2009, Rasche et al., 2011).
It remains speculative, if an inoculation and proliferation of “Foxy-2” in the crop rhizosphere results in competition with indigenous rhizosphere for root exudates as critical energy sources and if such interactions are influenced by environmental factors. This assumption is corroborated by earlier studies showing bacterial populations affected by fungi leading to a distinct selection of competitive community members in the rhizosphere (Marschner et al., 2001, Strange, 2005, Cavagnaro et al., 2006). Musyoki et al. (2015) showed that “Foxy-2” inoculated into soils was compatible with nitrifying prokaryotes.
Many BCAs exhibit beneficial effects under laboratory set-ups, while such effects become inconsistent once they are evaluated under greenhouse or even field conditions (Lugtenberg and Kamilova, 2009, Avedi et al., 2014). Advanced understanding of the influence of “Foxy-2” on non-target, functionally relevant rhizosphere prokaryotes under contrasting field conditions is therefore not only essential to validate previous studies under controlled conditions (Musyoki et al., 2015). It is also important to evaluate the extent of a “Foxy-2” impact against acknowledged factors (e.g., seasonality, soil type, crop growth stage) that determine the dynamics of rhizosphere communities (Doohan et al., 2003, Rasche et al., 2006a, Rasche et al., 2011).
Bacteria and archaea are ubiquitous in soils and responsible for the decomposition and mineralization (i.e., N cycle) of organic matter (Widmer et al., 2006, Raybould and Vlachos, 2011). A critical component of the microbial driven N cycle is the nitrification step which is catalyzed by key enzymes such as the amoA gene encoding the α-subunit of ammonia monooxygenase (Nicol et al., 2008, Zhang et al., 2013). It has been extensively reported that abundance and community composition of ammonia-oxidizing prokaryotes (i.e., bacteria (AOB), archaea (AOA)) respond sensitively to environmental change including the exposure to fungi (Rasche et al., 2011, Raybould and Vlachos, 2011, Wessén and Hallin, 2011, Martin-Laurent et al., 2013, Pereira e Silva et al., 2013). Although these studies acknowledge the use of AOB and AOA abundance as bioindicators for soil ecosystem disturbance surveys, the effects of BCAs (e.g., “Foxy-2”) on dynamics of these functionally relevant groups in soils are yet to be understood.
The objective of this study was therefore to assess the response of ammonia-oxidizing prokaryotes to “Foxy-2” exposure and to assay these presumed effects at different growth stages of maize cultivated in contrasting environments during two cropping seasons. In addition, we have evaluated these effects against those caused by an N-rich organic input which was supposed to compensate any resource competition between “Foxy-2” and non-target rhizosphere microbial communities (Ayongwa et al., 2011, Musyoki et al., 2015). The major hypothesis was that under field conditions, natural factors such as crop growth stage, soil type (properties) and climatic conditions (i.e., rainfall and temperature patterns) expose a greater influence on the abundance and community composition of resident prokaryotic populations than the BCA “Foxy-2” in a maize rhizosphere (Buée et al., 2009, Nihorimbere et al., 2011).
2. Material and methods
2.1. Fungal biocontrol agent
The fungal strain Fusarium oxysporum f.sp. strigae (“Foxy-2”) used in this study was isolated from diseased S. hermonthica collected in North Ghana (Abbasher et al., 1995). Identification of the isolate was done by the Julius-Kühn-Institute, Berlin, Germany (accession number: BBA-67547-Ghana). Since then, the isolate is being preserved at −80 °C at the Institute of Plant Production and Agroecology in the Tropics and Subtropics, University of Hohenheim, Stuttgart, Germany.
2.2. Study site description
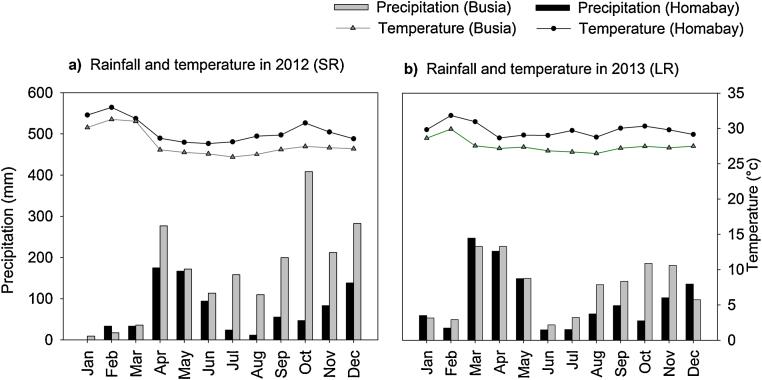
The field experiments were carried out in post-entry quarantine facilities (PEQ) at Agricultural Training Centre field stations in Western Kenya (Avedi et al., 2014). Two study sites (Busia, 0°26′S–34°15′E; 1200 m above sea level (a.s.l.); Homabay, 0°40′–0°S and 0°34°50′E; 1305 m a.s.l.) were chosen because of the reported high S. hermonthica infestation in these areas (De Groote et al., 2005). The sites were fallow for a year before the experiment was established. The fallow in Busia consisted of short grasses (e.g., Digitaria scalarum), while the fallow at Homabay consisted of grasses (Digitaria scalarum), and weeds such as black nightshade (Solanum nigrum) and thorn apples (Datura stramonium). The study areas have bimodal rainfall patterns with two growing seasons, the first rainy season with long rains (LR) from April to August and the second rainy season with short rains (SR) from September to January. Busia received 121 and 231 mm precipitation per month during the LR and SR season, respectively, and had a mean temperature of 27 °C during both seasons (Fig. 1). Homabay received 216 and 77 mm of rainfall per month during the LR and SR season, respectively, while the mean annual temperature was 29 °C in both seasons (Fig. 1). Initial soil characterization revealed that the soil at Homabay has a clayey texture (49% clay, 19% silt, 32% sand) and contained 0.22 and 2.87% total nitrogen and carbon, respectively, while Busia soil has a clay loam texture (33% clay, 22% silt, 45% sand) and contained 0.19 and 1.57% total nitrogen and carbon, respectively.
Fig. 1.
Monthly rainfall and temperature distribution during the short rain (SR) season (September 2012–January 2013) and the long rain (LR) season (March–August 2013).
2.3. Field experiment and rhizosphere sampling
The study covered two seasons (first season: SR; September 2012 to January 2013; second season: LR; April 2013 to August 2013). Maize variety WH507, commonly preferred by farmers in the study area due to its less susceptibility to S. hermonthica and also recommended for an integrated S. hermonthica control, was planted in 3 × 2.7 m2 plots with a spacing of 30 by 70 cm (Avedi et al., 2014). The experiment was laid out in a randomized complete block design (RCBD) with three replicates and comprised of three treatments: (i) uncoated maize and S. hermonthica (C, control), (ii) coated maize with “Foxy-2” and S. hermonthica (F + S), and (iii) coated maize with “Foxy-2”, S. hermonthica and Tithonia diversofolia residues as additional N source (F + S + T). Maize seeds were coated with “Foxy-2” (1.15 × 105 colony forming units per seed) as described by Musyoki et al. (2015).
Land was prepared by hand digging and two maize seeds per hill were planted at a depth of approximately 3 cm. One Table spoonful of a S. hermonthica seed-sand mixture (1:4 ratio with approximately 1000 S. hermonthica seeds) was placed in every planting hole (Avedi et al., 2014). At sowing, all plots received a blanket application of 60 kg P2O5 ha−1 season−1 as diammonium phosphate (DAP) (NH4)2HPO4) to avoid any phosphorus limitation. In addition, mineral N fertilizer was split applied to treatments C + S and F + S as calcium ammonium nitrate (CaNH4NO3) at a rate of 120 kg N ha−1 growing season−1 with 1/3 and 2/3 added 3 and 8 weeks after sowing, respectively (Chivenge et al., 2011, Muema et al., 2015). For treatment F + S + T, N was applied as fresh T. diversifolia leaf and stem material (5 t dry weight ha−1 to supply similar levels to 120 kg of inorganic N) which was hand-incorporated to a soil depth of 0–15 cm at the onset of each rainy season. Two weeks after germination, seedlings were thinned to 1 per hole. Hand weeding was done after every 2 weeks for all weeds except S. hermonthica.
Rhizosphere samples (approximately 50 g) were collected according to standard procedures (Milling et al., 2004) at EC30 (early leaf development stage), EC60 (flowering stage), and EC90 (senescence stage) by shaking the roots of three plants per plot to remove non-rhizosphere soil. Rhizophere soil samples were scraped off the roots of sampled plants. The rhizosphere soil samples of the three representative plants per plot were then mixed to form one composite sample. Soils were freeze-dried and stored in a dark and dry place until further analysis.
2.4. Microbial abundance
Four hundred milligrams of freeze-dried rhizosphere soil was used for DNA extraction from each of the three replicates per treatment. Soil DNA was extracted using the FastDNA® Spin for Soil Kit (MP Biomedicals, Solon, Ohio, USA) following the manufacturer’s instructions. Quality of extracted DNA was checked using 1.5% (w/v) agarose gel. DNA extracts were quantified (Nanodrop ND-1000, NanoDrop Technologies, Wilmington, USA) and stored at −20 °C for further analysis.
Abundance of total and ammonia oxidizing prokaryotic communities was estimated by DNA-based quantitative PCR (qPCR) using bacterial and archaeal 16S rRNA genes (total community) as well as bacterial and archaeal genes encoding ammonia-monooxygenase (amoA genes) as molecular markers (Table 1). For standard preparation, amplicons from each target gene were purified (Invisorb Fragment CleanUp kit, Stratec Molecular GmbH, Berlin, Germany), ligated into the StrataClone™ PCR cloning vector pSC-A (Stratagene, La Jolla, CA, USA) and ligation products were transformed with StrataClone Solopack competent cells (Startagene) (Rasche et al., 2011). The qPCR assays were carried out in a 25 μl reaction containing 12.5 μl of Power SYBR® green master mix (Applied Biosystems, Foster City, CA, USA), 1 μl primer (each 0.4 μM), 0.25 μl T4 gene 32 protein (500 ng μl−1, MP Biomedicals) and 10 ng template DNA. The qPCRs were performed on a StepOnePlus™ Real-Time PCR System (Applied Biosystems) and were started with 10 min at 95 °C, followed by amplification cycles specific for each target gene (Table 1). Melting curve analysis of amplicons was conducted to confirm that fluorescence signals originated from specific amplicons and not from primer-dimers or other artifacts. Each DNA sample was processed in triplicate reactions, whereas the standard curves were generated using duplicate serial dilutions of isolated plasmid DNA containing the genes studied (Rasche et al., 2011). Gene copy numbers and reaction efficiencies (total bacteria 94% ± 8, total archaea 97% ± 3, AOB 96% ± 7, AOA 96% ± 9) were calculated using the Stepone software version 2.2.2 (Applied Biosystems) and presented per gram of dry soil.
Table 1.
Description of primer sets, PCR ingredients and amplification details used for quantitative PCR and T-RFLP analysis.
| Target group | Primer (reference) | qPCR | T-RFLP |
|---|---|---|---|
| All bacteria (16 S rRNA) | Eub338 (Lane, 1991) | 95 °C 5 min | |
| Eub518 (Muyzer et al., 1993) | 40 cycles: 95 °C 30 s, 55 °C 35 s, 72 °C 45 s | ||
| 8f (Weisburg et al., 1991) | 95 °C 5 min | ||
| 1520r (Edwards et al., 1989) | 40 cycles: 95 °C 1 min, 58 °C 30 s, 72 °C 1 min; 72 °C 10 min | ||
| All archaea (16 S rRNA) | Ar109f (Lueders and Friedrich, 2000) | 95 °C 5 min | 95 °C 5 min |
| Ar912r (Lueders and Friedrich, 2000) | 40 cycles: 95 °C 30 s, 52 °C 35 s, 72 °C 45 s, 78 °C 20 s | 35 cycles: 95 °C 1 min, 52 °C 30 s, 72 °C 1 min, 72 °C 10 min | |
| Ammonia oxidizing bacteria (AOB) | AmoA-1f (Rotthauwe et al., 1997) | 95 °C 5 min | 95 °C 5 min |
| AmoA-2r (Rotthauwe et al., 1997) | 45 cycles: 95 °C 30 s, 57 °C 45 s, 72 °C 45 s, 78 °C 20 s | 40 cycles: 94 °C 30 s, 53 °C 30 s, 72 °C 1 min, 72 °C 10 min | |
| Ammonia oxidizing archaea (AOA) | Arch-amoAf (Francis et al., 2005) | 95 °C 5 min | 94 °C 5 min |
| Arch-amoAr (Francis et al., 2005) | 45 cycles: 95 °C 30 s, 53 °C 45 s, 72 °C 45 s, 78 °C 20 s | 35 cycles: 94 °C 30 s, 53 °C 45 s, 72 °C 10 min | |
2.5. Microbial community composition
Prokaryotic 16S rRNA and amoA genes were PCR-amplified as described in Rasche et al. (2011) (Table 1). Following the results of qPCR analysis (no treatment effects (“Foxy-2” and “organic inputs”) over the two seasons), we conducted T-RFLP analysis only in the second season based on the hypothesis that microbial community composition is less sensitive in responding to environmental changes than community abundance (Ritz et al., 2009, Wessén and Hallin, 2011). All forward primers were labeled with 6-carboxyflourescein at their 5′ ends. Replicate amplicons of the two genes were pooled, purified (Sephadex G-50, GE Healthcare Biosciences, Waukesha, WI, USA) according to Rasche et al. (2006b), and 200 ng of each purified amplicon were digested with a 5 U combination of enzymes AluI and RsaI (New England Biolabs, Ipswich, USA). Reactions were incubated at 37 °C for 4 h, and purified (Sephadex G-50). An aliquot of 2 μl was mixed with 17.75 μl HiDi formamide (Applied Biosystems) and 0.25 μl internal 500 ROX™ size standard (Applied Biosystems). Labeled terminal-restriction fragments (T-RFs) were denatured at 95 °C for 3 min, chilled on ice and detected on an ABI 3130 automatic DNA sequencer (Applied Biosystems). Peak Scanner™ software package (version 1.0, Applied Biosystems) was used to compare relative lengths of T-RFs with the internal size standard and to compile electropherograms into numeric data set, in which fragment length and peak height > 50 fluorescence units were used for profile comparison. T-RFLP profiles used for statistical analyses were normalized according to Dunbar et al. (2000).
2.6. Soil chemical analysis
Soil chemical analyses were performed on all soil samples taken at EC30 and EC90. Total carbon (TC), total nitrogen (Nt), extractable organic C (EOC), extractable N (EON) and pH of soils were recorded on bulk soils. The pH analysis was conducted in a soil water ratio of 1:2.5 using a pH meter (inoLab® Labor-pH-Meter, WTW GmbH, Weilheim, Germany). TC and Nt was quantified by dry combustion (vario MAX CN analyzer, Elementar Analysensysteme GmbH, Hanau, Germany). For EOC measurement, 5 g of soil were extracted with 20 ml 0.5 M K2SO4, shaken horizontally (250 rpm) for 30 min and filtered (Rotilabo-Rundfilter AP55.1 (retention of 2–3 μm), Carl Roth GmbH, Karlsruhe, Germany). EOC concentration in filtered extract was measured on an Analytik Jena Multi N/C 2100 analyzer (Analytik Jena AG, Jena, Germany). Ammonium (NH4+) and nitrate (NO3−) were extracted with 1 M KCl (soil to extractant ratio (w/v) of 1:4), shaken on a horizontal shaker for 30 min at 250 rpm and filtered (Rotilabo-Rundfilter AP55.1, Carl Roth GmbH). Concentrations of NH4+ and NO3− were measured on an auto-analyzer (Bran & Luebbe, Norderstedt, Germany) (Mulvaney, 1996), while EON was determined as the difference between Nt and mineral N (NH4+ and NO3−) according to Rousk and Jones (2010).
2.7. Statistical analysis
Statistical analyses were performed using Statistical Analysis Software program (SAS Institute, 2015). The analysis was done using a generalized linear mixed model assuming a negative binomial error distribution and a log link function. The fixed effects were; “Treatment” (“Foxy-2”, organic N addition (T. diversifolia), S. hermonthica, control), “Site” (soil properties), “Season” (SR 2012/2013, LR 2013), and “Growth stage” of maize (EC30, EC60, EC90) and all their two and three way interactions. The random effect included the blocking factor and replicates nested within blocks. The full model was fitted using restricted log pseudo-likelihood in the SAS GLIMMIX procedure (SAS Institute, 2015). We included a variance-covariance matrix to account for temporal auto-correlation in the residuals for all observations made within a block. The model was fitted separately for all variables across sites (soil properties) (Table 2). Due to the observed significant site effect the model was further fitted separately per site to account for site differences. Adjusted means and their associated standard errors at 95% confidence limits were estimated on the original data scale. Wherever, the two- or three-way interactions were significant, we decomposed them in terms of their simple effects slices. This enabled us to test adjusted means for significance between pairs of treatment, season or crop growth stages at fixed values of the other terms in the interaction effect. The means were compared using the PDIFF option of the LSMEANS as well as the letters (letter display) from the SAS generalized linear mixed models procedure. For soil chemical properties, NH4+ and EON were log base 10 transformed, while NO3− was square root transformed. Therefore, no standard errors are shown for these values as back transformed data was used for comparison purposes with the non-transformed chemical data (Table 4) (Piepho, 2009).
Table 2.
Analysis of variance to determine significant effects of factors ‘Site’ (ST), “Season” (SS), “Treatment” (T) and “Growth stage” (EC) and their interactions on soil chemical properties and abundance of the two assayed genes.
| Factor | Total bacteria | Total archaea | Bacterial amoA gene | Archaeal amoA gene | EOC (mg kg−1) | EON (mg kg−1) | NH4+ (mg kg−1) | NO3+ (mg kg−1) | pH (H2O) |
|---|---|---|---|---|---|---|---|---|---|
| Season (SS) | n.s. | *** | *** | *** | *** | *** | *** | *** | *** |
| Treatment (T) | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | * | *** |
| Site (ST) | n.s. | *** | n.s. | n.s. | * | ** | *** | *** | *** |
| Growth stage (EC) | *** | *** | *** | *** | *** | *** | *** | *** | *** |
| SS × T | n.s. | * | n.s. | n.s. | n.s. | * | n.s. | n.s. | *** |
| SS × ST | *** | n.s. | *** | n.s. | n.s. | *** | * | *** | *** |
| ST × T | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | ** | n.s. | *** |
| EC × T | n.s. | n.s. | *** | n.s. | ** | n.s. | ** | * | *** |
| EC × ST | *** | *** | *** | ** | *** | * | *** | ** | *** |
| SS × T × ST | * | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | * | *** |
| EC × T × ST | n.s. | n.s. | *** | n.s. | n.s. | n.s. | n.s. | ** | *** |
Significance levels: n.s.: P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001.
Table 4.
Dynamics of soil chemical properties as driven by factors “Site”, “Season”, “Treatment” and “Growth stage” and their interactions. Values are given as average (n = 3) along with standard error (SE). Different letters within a column show significant differences within sites and growth stages EC30 and EC90 (P < 0.05). Values without SE have been back transformed.
| Site | Growth stage | Treatment | EOC (mg kg−1) |
EON (mg kg−1) |
NH4+ (mg kg−1) |
NO3− (mg kg−1) |
pH (H2O) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SR | LR | SR | LR | SR | LR | SR | LR | SR | LR | |||
| Busia | EC30 | C + S | 242a | 273a | 22ab | 118a | 71a | 35b | 27a | 40b | 5.48a | 5.33c |
| F + S | 264a | 247a | 14b | 203a | 97a | 81a | 29a | 101a | 5.49a | 5.41b | ||
| F + S + T | 294a | 264a | 31a | 132a | 67a | 52ab | 36a | 55b | 5.35b | 5.43a | ||
| SE | 12.54 | 12.43 | 0.02 | 0.02 | ||||||||
| EC90 | C + S | 281a | 197b | 37a | 48.7a | 13a | 13a | 0.27a | 36a | 5.64c | 4.91b | |
| F + S | 283a | 256a | 34a | 92.3a | 13a | 21a | 1.07a | 15b | 5.70b | 4.86c | ||
| F + S + T | 301a | 224ab | 26a | 69.9a | 14a | 16a | 0.61a | 20b | 5.81a | 4.99a | ||
| SE | 9.73 | 13.46 | 0.03 | 0.02 | ||||||||
| Homa Bay | EC30 | C + S | 294a | 295a | 204a | 148a | 123a | 76a | 34a | 18a | 7.04b | 6.68a |
| F + S | 263a | 226b | 142ab | 150a | 143a | 89a | 38a | 14a | 6.91c | 6.17b | ||
| F + S + T | 266a | 281ab | 92b | 198a | 99a | 82a | 16b | 14a | 7.26a | 6.11c | ||
| SE | 10.04 | 14.13 | 0.05 | 0.09 | ||||||||
| EC90 | C + S | 273a | 156a | 48a | 25a | 4b | 3ab | 9a | 5a | 7.35b | 7.59a | |
| F + S | 221ab | 172a | 33a | 22a | 4b | 2b | 5a | 5a | 7.93a | 7.22c | ||
| F + S + T | 214b | 178a | 40a | 29a | 13a | 4a | 2a | 5a | 7.27c | 7.41b | ||
| SE | 15.45 | 4.29 | 0.10 | 0.05 | ||||||||
Treatments: C + S = uncoated maize + S. hermonthica, F + S = coated maize (with “Foxy-2”) + S. hermonthica, F + S + T = coated maize + S. hermonthica + Tithonia diversofolia; Season: SR = short rains, LR = Long rains; Growth stage: EC30 = Early leaf development stage, EC90 = Senescence stage; SE: Standard error.
Pearson’s linear correlation coefficients were calculated for assessing the relations between abundance of nitrifying and total prokaryotes with soil chemical properties.
“Treatment”, “Site”, and “Growth stage” effects as well as their interaction effects on T-RFLP data sets for each gene were tested using permutation multivariate analysis of variance (PERMANOVA) (Anderson and Walsh, 2013). Factor effects were further assayed based on Bray-Curtis similarity coefficients (Rees et al., 2005, Rasche et al., 2011, Rasche et al., 2014). A similarity matrix was generated for all possible pairs of samples for each target gene. The similarity matrix was used for analysis of similarity (ANOSIM) to test the hypothesis that composition of studied microbial communities was altered by factors “Treatment”, “Site”, and “Growth stage”. ANOSIM is based on rank similarities between the sample matrix and produces a test statistic ‘R’ (Rees et al., 2005). A ‘global’ R was first calculated in ANOSIM, which evaluated the overall effect of a factor in the data set. This step was followed by a pairwise comparison, whereby the magnitude of R indicated the degree of separation between two tested communities. An R score of 1 indicated a complete separation, while 0 indicated no separation (Rees et al., 2005). For graphical visualization of the distinct effect of factor “Growth stage” and “Site” on the composition of analyzed prokaryotic communities, canonical analysis of principal coordinates (CAP) was performed on resemblance matrix data generated based on Bray-Curtis similarity coefficients (Clarke, 1993, Anderson et al., 2008, Muema et al., 2015). Moreover, to test the influence of soil chemical properties on the community composition shifts of assayed genes, distance-based linear models (DISTLM) were used (Permanova+ software package in Primer v6) (Anderson et al., 2008). This procedure calculates a linear regression between the community composition and log transformed soil chemical data using the Shannon diversity index (H’) to evaluate how much of the variation in the microbial community composition is explained by variation in the soil chemical data (Legender and Andersson, 1999, Boj et al., 2011). PERMANOVA, ANOSIM, CAP and DISTLM analyses were conducted with Primer6 for windows (version 6.1.13) (Primer-E Ltd., Ply-mouth, UK) with PERMANOVA+ version 1.0.6 as add-on for Primer6 software (Anderson et al., 2008).
3. Results
3.1. Microbial abundance
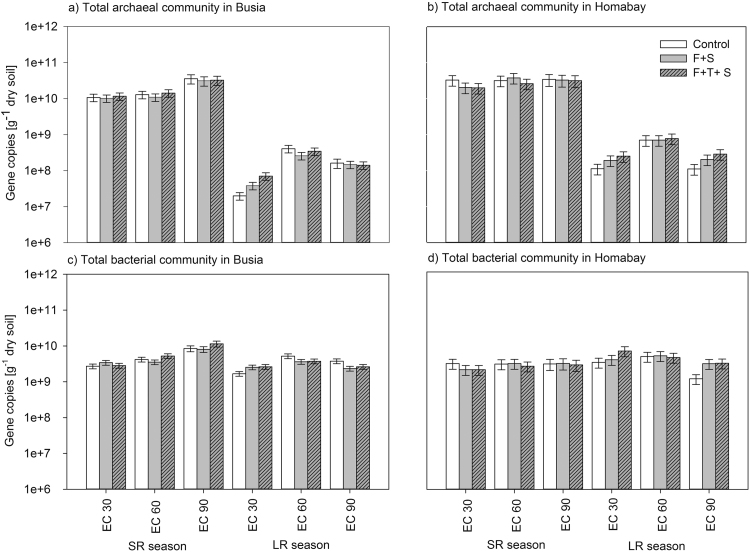
Abundance of archaeal 16S RNA genes was influenced by factors “Site”, “Season” and “Growth stage” (Table 2 and Fig. 2a and b). Overall, Homabay site had higher 16S RNA archaeal gene copies than Busia site (P < 0.001, Fig. 2a and b). A clear distinction of archaeal abundance was revealed in response to the three crop growth stages, where EC90 showed generally highest 16S RNA archaeal gene copies during the SR season, while archaeal abundance was dominating during EC60 during the LR season (P > 0.001) (Fig. 2a and b). Furthermore, a higher archaeal 16S RNA gene abundance was found in Homabay at EC30 and EC60 in both seasons compared to Busia (P < 0.001) (Fig. 2a and b).
Fig. 2.
Abundance of the total archaeal and bacterial community at Busia (a, c) and Homabay (b, d) sites as determined during the two cropping seasons. Values are given as average (n = 3) along with standard error (SE). Treatments are: C + S, uncoated maize and S. hermonthica, F + S, coated maize with “Foxy-2” and S. hermonthica, F + S + T, coated “Foxy-2”, S. hermonthica and T. diversofolia.
Total bacterial abundance was most significantly influenced by factor ‘Growth stage’ resulting in lowest bacterial 16S RNA gene copies at EC30 (Fig. 2c and d). Additionally, an interaction was found between factors “Season” and “Site”, where higher bacterial 16S rRNA gene copies were measured at Busia during SR season (P < 0.001) (Table 2 and Fig. 2c). The interaction of “Growth stage” and “Site” revealed a higher significant increase in bacterial 16S RNA gene copies from EC30 to EC60 and EC90 at Busia during SR season compared to LR and Homabay (P < 0.001) (Table 2 and Fig. 2c and d).
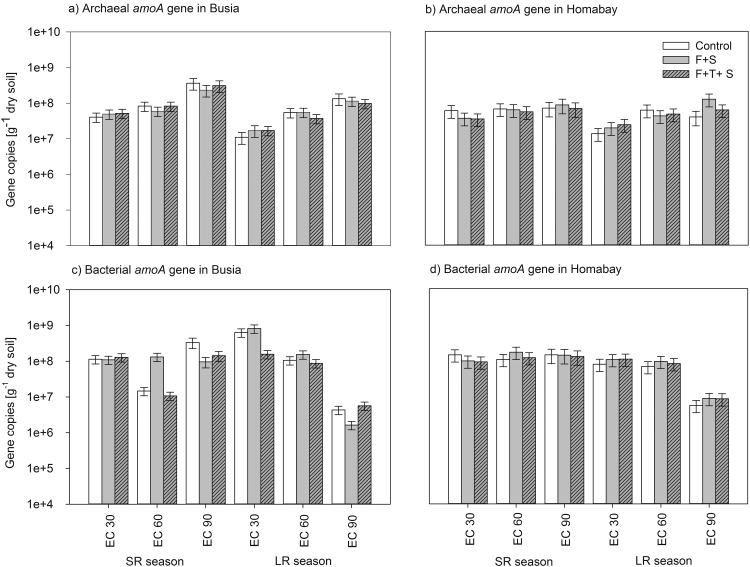
Abundance of archaeal amoA gene abundance (AOA) was influenced by factors “Season” and “Growth stage” and significant interactions were found between “Growth stage” and “Site” (P < 0.05) (Table 2). SR season showed higher AOA abundance in comparison to LR season particularly at Busia, where also highest gene copies were found at EC90 in both seasons (P < 0.01) (Fig. 3a).
Fig. 3.
Abundance of the archaeal and bacterial amoA genes at Busia (a, c) and Homabay (b, d) sites as determined during during the two cropping seasons. Values are given as average (n = 3) along with standard error (SE). Treatments are: C + S, uncoated maize and S. hermonthica, F + S, coated maize with “Foxy-2” and S. hermonthica, F + S + T, coated “Foxy-2”, S. hermonthica and T. diversofolia.
Abundance of bacterial amoA gene abundance (AOB) was influenced by factors “Season” and “Growth stage” revealing a significant interactions of “Growth stage” “Treament” and “Site” (P < 0.001) (Table 2). A major trend was that during the LR season, AOB revealed a constant abundance decrease from EC30 to EC90 and more strongly so at Busia (Fig. 3c). Generally, all genes tended to show a higher abundance in the treatments F + S (“Foxy-2”) and its combination with Tithonia diversifolia (F + S + T) than the control (C) at EC30 during the LR season in both sites (P > 0.05) (Figs. 2 a,b,c,d and 3 a,b,d).
3.2. Microbial community composition
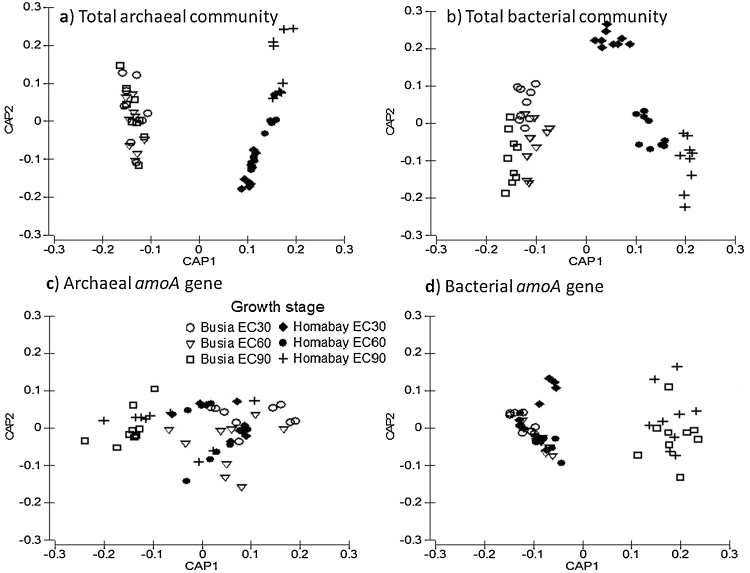
Analysis of similarity (ANOSIM) of T-RFLP data revealed significant effects of factors “Site” and “Growth stage” but not “Treatment” on the community composition of all studied genes (P < 0.05) (Table 3). A subsequent PERMANOVA analysis revealed significant interactions (P < 0.001) as evidenced by the effect of “Growth stage” on total archaeal communities at Homabay but not at Busia site which was further proved by pairwise ANOSIM (EC30 versus EC60 (R = 0.234), EC60 versus EC90 (R = 0.44), EC30 versus EC90 (R = 0.694) (P < 0.01)) (Fig. 4a). For total bacterial community, effect of “Growth stage” was more pronounced at Homabay (Fig. 4b) as was evidenced by pairwise ANOSIM (EC30 versus EC60 (R = 0.735), EC30 versus EC90 (R = 0.941) and EC60 versus EC90 R = 0.268). Likewise, the AOB (amoA gene) community composition revealed similar trends (Fig. 4d). Conversely, AOA (amoA gene) community composition differences by “Growth stage” were observed to a greater extent at Busia mainly between EC30 and EC90 (R = 0.9) (P < 0.001) (Fig. 4c).
Table 3.
Global R values for the main factors “Site”, “Growth stage” and “Treatment” as obtained from the analysis of similarity of T-RFLP fingerprints generated from the four studied genes.
| Prokaryotic group | Factor |
||
|---|---|---|---|
| Site | Growth stage | Treatment | |
| Total bacteria | 0.791a*** | 0.780***b | −0.038n.s. |
| Total archaea | 0.950*** | 0.708*** | −0.046n.s. |
| Bacterial amoA gene | 0.172** | 0.400*** | −0.009n.s. |
| Archaeal amoA gene | 0.648*** | 0.132** | −0.015n.s. |
R indicates the degree of separation between two populations, with a score of 1 indicating complete separation and 0 indicating no separation.
Significance levels: n.s.: P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001.
Fig. 4.
Canonical analysis of principal coordinates (CAP) for visual presentation of prokaryotic community composition (TRLFP) as shaped by “Site” and “Growth stage”. Prokayotic communities are (a) total archaea, (b) total bacteria, (c) ammonia-oxidizing archaea and (d) ammonia-oxidizing bacteria.
3.3. Soil chemical properties
EOC and EON were influenced by factors “Season” “Growth stage” and “Site” with significant interactions of factor ‘Growth stage’ and “Site” (P < 0.05) (Table 2). EOC was highest at EC90 at Busia during SR season (P < 0.05) (Table 4), while EON was largest at Homabay site at EC30 of SR season (P < 0.05) (Table 4). EON values were highest at Busia at EC30 during LR season (Table 4). For mineral N values, seasonal effects were evidenced by higher NH4+ concentrations at EC30 during SR season (P < 0.001) (Table 4). No consistent effect of factors ‘Treatment’, “Site” and “growth stage” on NO3− contents was observed due to several interactions (Table 2). Soil pH was influenced by all four factors with significant interactions (P < 0.001) (Table 2). For example, soil at Homabay had a higher pH compared to Busia, and SR season induced mostly higher soil pH at both sites (Table 4).
3.4. Correlations between microbial abundance and soil chemical properties
Extractable organic carbon (EOC) was positively correlated with total bacterial abundance and AOB abundance (P < 0.05), whereas extractable organic nitrogen (EON) negatively correlated with AOA (P < 0.001) (Table 5). Mineral N (ammonium (NH4+) and nitrate (NO3−)) revealed a negative correlation with AOA abundance (P < 0.01). In addition, NO3− correlated negatively with total bacterial abundance (P < 0.05). Soil pH positively correlated with total archaeal abundance (P < 0.001).
Table 5.
Pearson’s linear correlation coefficients between abundance of the two genes and soil chemical data obtained in Homabay and Busia sites at EC30 and EC90.
| Prokaryotic group | EOC (mg kg−1) | EON (mg kg−1) | NH4+ (mg kg−1) | NO3+ (mg kg−1) | pH (H2O) |
|---|---|---|---|---|---|
| Total bacteria | 0.253* | n.s. | n.s. | −0.325* | n.s. |
| Total archaea | n.s. | n.s. | n.s. | n.s. | 0.365** |
| Bacterial amoA gene | 0.263* | n.s. | n.s. | 0.320* | n.s. |
| Archaeal amoA gene | n.s. | −0.387** | −0.418** | −0.374* | n.s. |
Significance levels: n.s.: P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001.
3.5. Regressions between microbial community composition and soil chemical properties
Variance in total bacterial community composition was explained by alterations of extractable organic carbon (EOC), extractable organic nitrogen (EON) and ammonium (NH4+) concentrations (Table 6). Changes in total archaeal community composition were explained by alterations of all measured soil chemical properties, with a greater variation explained by nitrate (NO3−) (Table 6). AOB community composition changes were explained all measured soil chemical properties, except pH (Table 6). However, a greater percentage of AOB was explained by NH4+ (46%) (Table 6). Community composition changes of AOA were mainly explained by soil pH (Table 6).
Table 6.
Regression analyses between community composition of the two studied genes and soil chemical data taken at EC30 and EC90 across both study sites.
| Prokaryotic group | EOC (mg kg−1) | EON (mg kg−1) | NH4+ (mg kg−1) | NO3+ (mg kg−1) | pH (H2O) |
|---|---|---|---|---|---|
| Total bacteria | 0.280a***b | 0.186a**b | 0.383*** | 0.026n.s. | 0.000n.s. |
| Total archaea | 0.107* | 0.107* | 0.231*** | 0.316*** | 0.168** |
| Bacterial amoA gene | 0.344*** | 0.245** | 0.459*** | 0.273** | 0.001n.s. |
| Archaeal amoA gene | 0.000n.s | 0.009n.s. | 0.010n.s. | 0.0715n.s. | 0.295** |
R2 indicates the proportion of variation in Shannon diversity explained by log values of the chemical data.
Significance levels: n.s.: P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001.
4. Discussion
4.1. Dominance of environmental factors over presence of BCA
Previous studies on non-target effects of beneficial Fusarium spp. strains as biocontrol agents (BCAs) on soil microbial communities have been restricted to controlled conditions with short investigation periods rather than field conditions (Edel-Hermann et al., 2009, Karpouzas et al., 2011, Musyoki et al., 2015). Generally, controlled conditions do not represent the natural conditions that prevail in the rhizopshere of crops grown in the field. In this respect, the tripartite interaction between Fusarium spp. BCAs, indigenous microbial communities and host crop (rhizosphere) have been suggested to vary with environmental conditions (Goh et al., 2009, Mendes et al., 2013). To account for this, we carried out field experiments during two cropping seasons to study the effects of the BCA “Foxy-2” relative to the acknowledged effects of natural factors including soil properties (site), crop growth stage and seasonality (rainfall patterns) on the abundance (qPCR) and community composition (T-RFLP) of total and nitrifying prokaryotes in rhizospheres of maize grown in contrasting soils. Our results revealed that site, crop growth stage and seasonal variations controlled the abundance and community composition of prokaryotic nitrifiers to a greater extent than “Foxy-2” inoculation. This central outcome further reinforced the need for extended field studies under different conditions for better understanding of rhizospheric interaction effects between indigenous communities and potential biological control agents (Musyoki et al., 2015).
In the current study, Busia and Homabay soils had clay contents ranging from 33 to 49%, respectively, which were higher than that of the sandy soil (22%) used for a previous study by Musyoki et al. (2015), where a promoting effect nitrifying archaea was reported in a sandy but not in a clay soil. These soil texture differences may have partially masked “Foxy-2” related shifts in the abundance of total and also nitrifying prokaryotes. Notably, total archaeal abundance was higher in the Homabay soil than in that of Busia, while total bacteria, nitrifying archaea (AOA) and bacteria (AOB) were not influenced by soil type (site). This higher abundance and also a clear distinction of the composition of the total archaeal community in the clayey Homabay soil was mainly attributed to the higher soil pH and higher clay contents promoting a larger soil organic carbon (SOC) background at Homabay, an explanation which is in agreement with earlier studies (Gerzabek et al., 2002, Shen et al., 2008, Morimoto et al., 2011). Likewise, we have recently observed that clayey soils hampered the proliferation of “Foxy-2” (Zimmermann et al., 2015). It is likely that the archaeal community colonized similar resource niches like “Foxy-2” which has been reported to proliferate better in low nutrient conditions of e.g. sandy soils (Larkin and Fravel, 2002, Erguder et al., 2009, Zimmermann et al., 2015). We therefore speculated that indigenous rhizosphere archaea were involved in the suppression of “Foxy-2” as was not only corroborated by negative correlations between the BCA and indigenous total archaea (J. Zimmermann, personal communication), but also by earlier studies stating that clayey soils evolve a high natural suppression potential against microorganisms (Toyota et al., 1996, Larkin and Fravel, 2002). This hypothesized suppressive effect by archaea became obvious during the later stages of the vegetation period in both seasons (i.e., SR (short rains), LR (long rains)), where abundance of total archaea but also that of AOA increased significantly. We assumed that this archaeal dominance at the later crop growth stages increased their competitive and hence suppressive abilities over “Foxy-2”. This niche-based resource competition may have been particularly evident at EC90, when “Foxy-2” was in its saprophytic stage and, similar to indigenous rhizosphere archaea, further increasing the capitalization on available indigenous SOC as central resource (Kandeler et al., 2002, Garbeva et al., 2004, Elzein et al., 2010, Musyoki et al., 2015). Moreover, advanced decomposition of organic matter derived from the fallow period prior to the field experiment setup in the SR season may have contributed to this abundance boost of AOA over AOB since AOA are generally more adapted to acquisition of organic derived nutrients over their bacterial counterparts (Wessén et al., 2010).
4.2. Foxy-2 did not induce a resource limitation for bacteria and archaea
Clear indications of resource limitation for bacterial and archaeal abundances were found under the tested field conditions. This was most obvious at EC30 during LR, where abundance of total and nitrifying bacteria and archaea increased with application of Tithonia diversifolia (TD) residues over the control treatment (C). However, this apparent resource limitation was not induced by presence of “Foxy-2” (F) as there was firstly a positive effect of the BCA on nitrifying (amoA) and total bacterial (16S) as well as their archaeal abundances, and the positive effect was even greater when combined with TD. This stimulating effect was not observed during the initial SR which was traced back to the decomposition of organic matter derived from the fallow period prior to the field experiment setup in the SR season. In addition, the high rainfall amount during the SR rain season (149 mm month−1) in comparison to the lower rainfall amount during the LR season (109 mm month−1) may have contributed to indirect changes through rhizodeposition, consequently affecting the organic resource availability in the rhizosphere (Bell et al., 2009, Rasche et al., 2011). This resource excess was reflected by overall higher total bacterial and archaeal abundances during the SR season.
Furthermore, AOB abundance dominated at the earlier growth stages, particularly at EC30 which was linked to the provision of easily degradable rhizodeposits including EOC, as well as nitrogenous metabolites such as extractable organic nitrogen (EON) and nitrates (NO3+) correlating positively with AOB, but negatively with AOA abundance (Hai et al., 2009, Aira et al., 2010, Neal et al., 2012). Similarly, a strong community differentiation was found between AOB and AOA at EC30 and EC90 corroborating the colonization of distinct ecological niches. In this respect, we found that ammonium (NH4+) contents explained more than 40% of the AOB community shift, while soil pH was the only parameter explaining the AOA community shift. Our findings agreed with earlier studies stating that niche differentiation between AOA and AOB is mainly driven by alterations of soil N and pH conditions (Valentine, 2007, Shen et al., 2008, Erguder et al., 2009, Wessén et al., 2010), which were not consistently significantly influenced by presence of “Foxy-2”.
5. Conclusion and outlook
Our field study revealed that “Foxy-2” application did not impose a negative effect on the abundance and community composition of total and nitrifying prokaryotes. This is an important prerequisite concerning the registration of “Foxy-2” as a potential S. hermonthica BCA. The major observation that crop growth stage, seasonal variations and soil properties controlled total and nitrifying prokaryote abundance and community composition to a greater extent than “Foxy-2” inoculation demonstrates the need to include field studies over several seasons to obtain a more detailed, site-and seasonality-specific understanding on non-target effects of potential BCAs on indigenous microbial communities colonizing the rhizosphere of S. hermonthica affected crops.
Notably, we found clear indications that particularly archaea and “Foxy-2” colonized similar ecological niches for organic resource acquisition in the rhizospheres of maize grown in clayey soils. In that respect, we postulate that archaeal domination in clayey soils is a considerable regulator of “Foxy-2” proliferation due to their greater rhizosphere competence over that of “Foxy-2”. It needs, however, to be tested if this potential out competition of archaea holds true under other environmental conditions than those tested in this study. Therefore, progressive studies should emphasize surveys in distinct agro-ecological zones in which not only the dynamics of indigenous rhizosphere communities under the presence of “Foxy-2” will be assayed. These studies should also consider local adaptation mechanisms of “Foxy-2” which might induce a higher resource competition potential against natural prokaryotic and also fungal communities such as sandy nutrient limited soils. Under such conditions, the use of N-rich organic residue inputs may be considered to compensate any resource competition.
Acknowledgments
The study was financed by the Bill & Melinda Gates Foundation (BMGF, Project “Integrated Striga Management in Africa (ISMA)) and Fiat Panis Foundation. Ms. Musyoki is grateful to the Food Security Centre (FSC) of the University of Hohenheim funded by the German Academic Exchange Program (DAAD) for providing the scholarship. Special thanks go to Dr. Joseph Ogutu and Dr. Juan Carlos Laso Bayas (Department of Bioinformatics, University of Hohenheim) for their support in statistical analysis. The authors are further grateful to Professor Dr. Ludwig Hölzle (Institute of Environmental and Animal Hygiene and Veterinary Medicine, University of Hohenheim) for access to his ABI 3130 Genetic Analyzer.
References
- Abbasher A.A., Kroschel J., Sauerborn J. Microorganisms of Striga hermonthica in northern Ghana with potential as biocontrol agents. Biocontrol Sci. Technol. 1995;5:157–162. [Google Scholar]
- Aira M., Brandón M.G., Lazcano C., Bååth E., Domínguez J. Plant genotype strongly modifies the structure and growth of maize rhizosphere microbial communities. Soil Biol. Biochem. 2010;42:2276–2281. [Google Scholar]
- Anderson M.J., Walsh D.C. PERMANOVA, ANOSIM, and the Mantel test in the face of heterogeneous dispersions: what null hypothesis are you testing? Ecol. Monogr. 2013;83:557–574. [Google Scholar]
- Anderson M.J., Gorley R.N., Clarke K.R. PRIMER-E; Plymouth, UK: 2008. Permanova+ for Primer: Guide to Softaware and Statistical Methods. [Google Scholar]
- Avedi E.K., Ochieno D.M.W., Ajanga S., Wanyama C., Wainwright H., Elzein A., Beed F. Fusarium oxysporum f.sp. strigae strain Foxy 2 did not achieve biological control of Striga hermonthica parasitizing maize in Western Kenya. Biol. Control. 2014;77:7–14. [Google Scholar]
- Ayongwa G.C., Stomph T.J., Belder P., Leffelaar P.A., Kuyper T.W. Organic matter and seed survival of Striga hermonthica—mechanisms for seed depletion in the soil. Crop Prot. 2011;30:1594–1600. [Google Scholar]
- Beed F.D., Hallett S.G., Venne J., Watson A.K. Biocontrol using Fusarium oxysporum: a critical component of integrated Striga management. In: Ejeta G., Gressel J., editors. Integrating New Technologies for Striga Control: Towards Ending the Witch-hunt. World Scientific Publishing Co. Pte. Ltd.; Singapore: 2007. pp. 283–300. [Google Scholar]
- Bell C.W., Acosta-Martinez V., McIntyre N.E., Cox S., Tissue D.T., Zak J.C. Linking microbial community structure and function to seasonal differences in soil moisture and temperature in a Chihuahuan Desert grassland. Microb. Ecol. 2009;58:827–842. doi: 10.1007/s00248-009-9529-5. [DOI] [PubMed] [Google Scholar]
- Boj E., Fortiana J., Esteve A., Claramunt M.M., Costa T. Actuarial applications of distance-based generalized linear models. ASTIN. 2011:1–12. [Google Scholar]
- Buée M., De Boer W., Martin F., van Overbeek L., Jurkevitch E. The rhizosphere zoo: an overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil. 2009;321:189–212. [Google Scholar]
- Cavagnaro T.R., Jackson L.E., Six J., Ferris H., Goyal S., Asami D., Scow K.M. Arbuscular mycorrhizas, microbial communities, nutrient availability, and soil aggregates in organic tomato production. Plant Soil. 2006;282:209–225. [Google Scholar]
- Chivenge P., Vanlauwe B., Gentile R., Six J. Organic resource quality influences short-term aggregate dynamics and soil organic carbon and nitrogen accumulation. Soil Biol. Biochem. 2011;43:657–666. [Google Scholar]
- Clarke K.R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993;18:117–143. [Google Scholar]
- De Groote H., Wangare L., Kanampiu F., Odendo M., Friesen D. Potential markets for herbicide resistant maize seed for Striga control in Africa. Background Paper for a Poster Presented at the European Association of Agricultural Economists Congress; Copenhagen, Denmark; 2005. pp. 23–27. [Google Scholar]
- Doohan F.M., Brennan J., Cooke B.M. Influence of climatic factors on Fusarium species pathogenic to cereals. Eur. J. Plant Pathol. 2003;109:755–768. [Google Scholar]
- Dunbar J., Ticknor L.O., Kuske C.R. Assessment of microbial diversity in four Southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl. Environ. Microbiol. 2000;66:2943–2950. doi: 10.1128/aem.66.7.2943-2950.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edel-Hermann V., Brenot S., Gautheron N., Aimé S., Alabouvette C., Steinberg C. Ecological fitness of the biocontrol agent Fusarium oxysporum Fo47 in soil and its impact on the soil microbial communities. FEMS Microbiol. Ecol. 2009;68:37–45. doi: 10.1111/j.1574-6941.2009.00656.x. [DOI] [PubMed] [Google Scholar]
- Edwards U., Rogall T., Blöcker H., Emde M., Böttger E.C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzein A., Heller A., Ndambi B., De Mol M., Kroschel J., Cadisch G. Cytological investigations on colonization of sorghum roots by the mycoherbicide Fusarium oxysporum f.sp. strigae and its implications for Striga control using a seed treatment delivery system. Biol. Control. 2010;53:249–257. [Google Scholar]
- Erguder T.H., Boon N., Wittebolle L., Marzorati M., Verstraete W. Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol. Rev. 2009;33:855–869. doi: 10.1111/j.1574-6976.2009.00179.x. [DOI] [PubMed] [Google Scholar]
- Francis C.A., Roberts K.J., Beman J.M., Santoro A.E., Oakley B.B. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U. S. A. 2005;102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fustec J., Lesuffleur F., Mahieu S., Cliquet J.B. vol. 2. Springer; Netherlands: 2011. Nitrogen rhizodeposition of legumes; pp. 869–881. (Sustainable Agriculture). [Google Scholar]
- Garbeva P., van Veen J.A., van Elsas J.D. Microbial diversity in soil: selection of microbial populations by plant and soil type and implications for disease suppressiveness. Annu. Rev. Phytopathol. 2004;42:243–270. doi: 10.1146/annurev.phyto.42.012604.135455. [DOI] [PubMed] [Google Scholar]
- Gerzabek M.H., Haberhauer G., Kandeler E., Sessitsch A., Kirchmann H. Response of organic matter pools and enzyme activities in particle size fractions to organic amendments in a long-term field trial. In: Violante A., Huang P.M., Bollag J.M., Gianfreda L., editors. Developments in Soil Science. Elsevier; Amsterdam, The Netherlands: 2002. pp. 329–344. [Google Scholar]
- Girvan M.S., Bullimore J., Pretty J.N., Osborn A.M., Ball A.S. Soil type is the primary determinant of the composition of the total and active bacterial communities in arable soils. Appl. Environ. Microbiol. 2003;69:1800–1809. doi: 10.1128/AEM.69.3.1800-1809.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh K.Y., Daida P., Vujanovic V. Effects of abiotic factors and biocontrol agents on Chlamydospore formatiom in Fusarium graminearum and Fusarium sporotrichioides. Biocontrol Sci. Technol. 2009;19:151–167. [Google Scholar]
- Grayston S.J., Wang S., Campbell C.D., Edwards A.C. Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol. Biochem. 1998;30:369–378. [Google Scholar]
- Høgh-Jensen H., Schjoerring J.K. Rhizodeposition of nitrogen by red clover, white clover and ryegrass leys. Soil Biol. Biochem. 2001;33:439–448. [Google Scholar]
- Hai B., Diallo N.H., Sall S., Haesler F., Schauss K., Bonzi M., Assigbetse K., Chotte J.C., Jean Charles Munch J.C., Michael Schloter M. Quantification of key genes steering the microbial nitrogen cycle in the rhizosphere of sorghum cultivars in tropical agroecosystems. Appl. Environ. Microbiol. 2009;75:4993–5000. doi: 10.1128/AEM.02917-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden H.L., Drake J., Imhof M., Oxley A.P.A., Norng S., Mele P.M. The abundance of nitrogen cycle genes amoA and nifH depends on land uses and soil types in South-Eastern Australia. Soil Biol. Biochem. 2010;42:1774–1783. [Google Scholar]
- Jones D.L., Nguyen C., Finlay R.D. Carbon flow in the rhizosphere: carbon trading at the soil–root interface. Plant Soil. 2009;321:5–33. [Google Scholar]
- Kandeler E., Marschner P., Tscherko1 D., Gahoonia T.S., Nielsen N.E. Microbial community composition and functional diversity in the rhizosphere of maize. Plant Soil. 2002;238:301–312. [Google Scholar]
- Karpouzas D.G., Karatasas A., Spiridaki E., Rousidou C., Bekris F., Omirou M., Ehaliotis C., Papadopoulou K.K. Impact of a beneficial and of a pathogenic Fusarium strain on the finger printing based structure of microbial communities in tomato (Lycopersicon esculentum Mill.) rhizosphere. Eur. J. Soil Biol. 2011;47:400–408. [Google Scholar]
- Lane D. 16S/23S rRNA sequencing. In: Stackebrandt A., Goodfello M., editors. Nucleic Acid Techniques Systematics. 1st ed. Wiley; West Sussex, Chichester, New York, UK: 1991. pp. 115–257. [Google Scholar]
- Larkin R.P., Fravel D.R. Effects of varying environmental conditions on biological control of Fusarium wilt of tomato by nonpathogenic Fusarium spp. Phytopathology. 2002;92:1160–1166. doi: 10.1094/PHYTO.2002.92.11.1160. [DOI] [PubMed] [Google Scholar]
- Legender P., Andersson M.J. Distance-based redundancy analysis: testing multispecies responses in multifactorial elcological experiments. Ecol. Monogr. 1999;69:1–24. [Google Scholar]
- Lueders T., Friedrich M. Archaeal population dynamics during sequential reduction processes in rice field soil. Appl. Environ. Microbiol. 2000;66:2732–2742. doi: 10.1128/aem.66.7.2732-2742.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg B., Kamilova F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009;63:541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- Marschner P., Crowley D.E., Lieberei R. Arbuscular mycorrhizal infection changes the bacterial 16S rDNA community composition in the rhizosphere of maize. Mycorrhiza. 2001;11:297–302. doi: 10.1007/s00572-001-0136-7. [DOI] [PubMed] [Google Scholar]
- Martin-Laurent F., Kandeler E., Petric I., Djuric S., Dimitrios G., Karpouzas D.G. ECOFUN-MICROBIODIV: an FP7 European project for developing and evaluating innovative tools for assessing the impact of pesticides on soil functional microbial diversity—towards new pesticide registration regulation? Environ. Sci. Pollut. Res. 2013;20:1203–1205. doi: 10.1007/s11356-012-1368-0. [DOI] [PubMed] [Google Scholar]
- Mendes R., Garbeva P., Raaijmakers J.M. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microb. Rev. 2013;37:634–663. doi: 10.1111/1574-6976.12028. [DOI] [PubMed] [Google Scholar]
- Miedaner T., Reinbrecht C., Lauber U., Schollenberger M., Geiger H.H. Effects of genotype and genotype-environment interaction on deoxynivalenol accumulation and resistance to Fusarium head blight in rye, tricale and wheat. Plant Breed. 2001;120:97–105. [Google Scholar]
- Milling A., Smalla K., Maidl F., Schloter M., Munch J. Effects of transgenic potatoes with an altered starch composition on the diversity of soil and rhizosphere bacteria and fungi. Plant Soil. 2004;266:23–39. [Google Scholar]
- Morimoto S., Hayatsu M., Hoshino Y.T., Nagaoka K., Yamazaki M., Karasawa T., Takenaka M., Akiyama H. Quantitative analyses of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) in fields with different soil types. Microbes Environ. 2011;26:248–253. doi: 10.1264/jsme2.me11127. [DOI] [PubMed] [Google Scholar]
- Muema E.K., Cadisch G., Röhl C., Vanlauwe B., Rasche F. Response of ammonia-oxidizing bacteria and archaea to biochemical quality of organic inputs combined with mineral nitrogen fertilizer in an arable soil. Appl. Soil Ecol. 2015;95:128–139. [Google Scholar]
- Mulvaney R.L. Nitrogen inorganic forms. In: Sparks D.L., Page A.L., Leoppert R.H., Soltanpour P.N., Tabatabai M.A., Johnston C.T., Sumner M.E., editors. Methods of Soil Analysis. Part 3. SSSA Book Series 5. SSSA and ASA; Madison, WI, USA: 1996. pp. 1123–1184. [Google Scholar]
- Musyoki M.K., Cadisch G., Enowashu E., Zimmermann J., Muema E., Beed F., Rasche F. Promoting effect of Fusarium oxysporum [f.sp. strigae] on abundance of nitrifying prokaryotes in a maize rhizosphere across soil types. Biol. Control. 2015;83:37–45. [Google Scholar]
- Muyzer G., de Waal E.C., Uitterlinden A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndambi B., Cadisch G., Elzein A., Heller A. Colonization and control of Striga hermonthica by Fusarium oxysporum f.sp. strigae, a mycoherbicide component: an anatomical study. Biol. Control. 2011;58:149–159. [Google Scholar]
- Neal A.L., Ahmad S., Gordon-weeks R., Ton J. Benzoxazinoids in root exudates of maize attract Pseudomonas putida to the rhizosphere. PLoS One. 2012;7(4):e35498. doi: 10.1371/journal.pone.0035498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol G.W., Leininger S., Schleper C., Prosser J.I. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 2008;10:2966–2978. doi: 10.1111/j.1462-2920.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- Nihorimbere V., Ongena M., Smargiassi M., Thonart P. Beneficial effect of the rhizosphere microbial community for plant growth and health. Biotechnol. Agron. Soc. Environ. 2011;15:327–337. [Google Scholar]
- Pereira e Silva M.C., Semenov A.V., Schmitt H., van Elsas J.D., Salles J.F. Microbe-mediated processes as indicators to establish the normal operating range of soil functioning. Soil Biol. Biochem. 2013;57:995–1002. [Google Scholar]
- Piepho H.P. Data transformation in statistical analysis of field trials with changing treatment variance. Agron. J. 2009;101:865–869. [Google Scholar]
- Raaijmakers J.M., Paulitz T.C., Steinberg C., Alabouvette C., Möenne-Loccoz Y. The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil. 2009;321:341–361. [Google Scholar]
- Rasche F., Hödl V., Poll C., Kandeler E., Gerzabek H.M., Elsas van J., Sessitch A. Rhizosphere bacteria as affected by transgenic potatoes with antibacterial activities compared with the effects of soil, wild-type potatoes, vegetation stage and pathogen exposure. FEMS Microbiol. Ecol. 2006;72:219–235. doi: 10.1111/j.1574-6941.2005.00027.x. [DOI] [PubMed] [Google Scholar]
- Rasche F., Trondl R., Naglreiter C., Reichenauer T., Sessitsch A. Chilling and cultivar type affect the diversity of bacterial endophytes colonizing sweet pepper (Capsicum annuum L.) Can. J. Microbiol. 2006;52:1036–1045. doi: 10.1139/w06-059. [DOI] [PubMed] [Google Scholar]
- Rasche F., Knapp D., Kaiser C., Koranda M., Kitzler B., Zechmeister-Boltenstern S., Richter A., Sessitsch A. Seasonality and resource availability control bacterial and archaeal communities in soils of a temperate beech forest. ISME J. 2011;5:389–402. doi: 10.1038/ismej.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasche F., Musyoki M.K., Röhl C., Muema E.K., Vanlauwe B., Cadisch G. Lasting influence of biochemically contrasting organic inputs on abundance and community structure of total and proteolytic bacteria in tropical soils. Soil Biol. Biochem. 2014;74:204–2013. [Google Scholar]
- Raybould A., Vlachos D. Non-target organism effects test on Vip3A and their application to the ecological risk assessment for cultivation of MIR162 maize. Transgenic Res. 2011;20:599–611. doi: 10.1007/s11248-010-9442-1. [DOI] [PubMed] [Google Scholar]
- Rees G., Baldwin D., Watson G., Perryman S., Nielsen D. Ordination and significance testing of microbial community composition derived from terminal restriction fragment length polymorphisms: application of multivariate statistics. Antonie Van Leeuwenhoek. 2005;86:339–347. doi: 10.1007/s10482-004-0498-x. [DOI] [PubMed] [Google Scholar]
- Ritz K., Black H.I., Campbell C.D., Harris J.A., Wood C. Selecting biological indicators for monitoring soils: a framework for balancing scientific and technical opinion to assist policy development. Ecol. Indic. 2009;9:1212–1221. [Google Scholar]
- Rotthauwe J.H., Witzel K.P., Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousk J., Jones D.L. Loss of low molecular weight dissolved organic carbon (DOC) and nitrogen (DON) in H2O and 0.5 M K2SO4 soil extracts. Soil Biol. Biochem. 2010;42:2331–2335. [Google Scholar]
- SAS Institute . SAS Institute Inc.; Carey, NC, USA: 2015. SAS System for Windows. Version 9.4. [Google Scholar]
- Schaub B., Marley P., Elzein A., Kroschel J. Field evaluation of an integrated Striga hermonthica management in Sub-Saharan Africa: synergy between Striga-mycoherbicides (biocontrol) and sorghum and maize resistant varieties. J. Plant Dis. Prot. 2006:691–699. [Google Scholar]
- Shen J., Zhang L., Zhu Y., Zhang J., He J. Abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea communities of an alkaline sandy loam. Environ. Microbiol. 2008;10:1601–1611. doi: 10.1111/j.1462-2920.2008.01578.x. [DOI] [PubMed] [Google Scholar]
- Strange R.N. Plant disease: a threat to global food security. Annu. Rev. Phytopathol. 2005;43:83–116. doi: 10.1146/annurev.phyto.43.113004.133839. [DOI] [PubMed] [Google Scholar]
- Toyota K., Young I.M., Ritz K. Effects of soil matric potential and bulk density on the growth of Fusarium oxysporum f.sp. raphani. Soil Biol. Biochem. 1996;28:1139–1145. [Google Scholar]
- Valentine D.L. Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat. Rev. Microbiol. 2007;5:316–323. doi: 10.1038/nrmicro1619. [DOI] [PubMed] [Google Scholar]
- Venne J., Beed F., Avocanh A., Watson A. Integrating Fusarium oxysporum f.sp. strigae into cereal cropping systems in Africa. Pest Manag. Sci. 2009;65:572–580. doi: 10.1002/ps.1741. [DOI] [PubMed] [Google Scholar]
- Waring R.H., Running S.W. 2nd edn. Academic Press; San Diego, CA, USA: 1998. Forest Ecosystems: Analysis at Multiple Scales; p. p.370. [Google Scholar]
- Watson A.K. Biocontrol. In: Joel D.M., Gressel J., Musselman L.J., editors. Parasitic Orobancheceae. Springer Verlag Berlin; Heidelberg: 2013. pp. 469–497. [Google Scholar]
- Weisburg W.G., Barns S.M., Pelletier D.A., Lane D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessén E., Hallin S. Abundance of archaeal and bacterial ammonia oxidizers—possible bioindicator for soil monitoring. Ecol. Indic. 2011;11:1696–1698. [Google Scholar]
- Wessén E., Nyberg K., Jansson J.K., Hallin S. Responses of bacterial and archaeal ammonia oxidizers to soil organic and fertilizer amendments under long-term management. Appl. Soil Ecol. 2010;45:193–200. [Google Scholar]
- Wichern F., Eberhardt E., Mayer J., Joergensen R.G., Müller T. Nitrogen rhizodeposition in agricultural crops: methods, estimates and future prospects. Soil Biol. Biochem. 2008;40:30–48. [Google Scholar]
- Widmer F., Rasche F., Hartmann M., Fliesbach A. Community structures and substrate utilization of bacteria in soils from organic and conventional farming systems of the DOK long-term field experiment. Appl. Soil Ecol. 2006;33:294–307. [Google Scholar]
- Zhang H., Ding W., Yu H., He X. Carbon uptake by a microbial community during 30-day treatment with 13C-glucose of a sandy loam soil fertilized for 20 years with NPK or compost as determined by a GC-C-IRMS analysis of phospholipid fatty acids. Soil Biol. Biochem. 2013;57:228–236. [Google Scholar]
- Zimmermann J., de Klerk M., Musyoki M.K., Viljoen A., Watson A.K., Beed F., Markus Gorfer M., Cadisch G., Rasche F. An explicit AFLP-based marker for monitoring Fusarium oxysporum f.sp. strigae in tropical soils. Biol. Control. 2015;89:42–52. [Google Scholar]