Abstract
There is an increasing emergence of antibiotic-resistant Vibrio alginolyticus, a zoonotic pathogen that causes mass mortality in aquatic animals and infects humans; therefore, there is a demand for alternatives to antibiotics for the treatment and prevention of infections caused by this pathogen. One possibility is through the exploitation of bacteriophages. In the present study, the novel bacteriophage pVa-21 was classified as Myoviridae and characterised as a candidate biocontrol agent against V. alginolyticus. Its morphology, host range and infectivity, growth characteristics, planktonic or biofilm lytic activity, stability under various conditions, and genome were investigated. Its latent period and burst size were estimated to be approximately 70 min and 58 plaque-forming units/cell, respectively. In addition, phage pVa-21 can inhibit bacterial growth in both the planktonic and biofilm states. Furthermore, phylogenetic and genome analysis revealed that the phage is closely related to the giant phiKZ-like phages and can be classified as a new member of the phiKZ-like bacteriophages that infect bacteria belonging to the family Vibrionaceae.
Subject terms: Antimicrobials, Applied microbiology, Bacteriophages
Introduction
Vibrio alginolyticus, a representative of Harveyi clade bacteria, is frequently found in marine environments. This organism can infect a variety of aquatic animals and infection has been linked to several mass mortality cases in major aquaculture species from fish to molluscs and crustaceans1–4. Global warming has caused an increase in sea surface temperature that has undoubtedly led to the unseasonal outbreaks of Vibrio as well as their increased abundance and virulence in marine environments and aquaculture5–8. In aquatic environments, biofilms have been reported as a causative agent of disease recurrence9,10. Indeed, the ability of Vibrio spp. to form biofilms is often correlated with their pathogenicity11,12. Bacterial cells in biofilms are highly tolerant to antibiotics compared with those in the planktonic state13,14, as the biofilm matrix provides bacteria with competitive advantages for survival. Moreover, the misuse or overuse of antimicrobials has led to the emergence of multidrug-resistant bacterial strains15. Unsurprisingly, the isolation of V. alginolyticus strains with multiple antibiotic resistance have been reported from several recent outbreaks16–18. Therefore, there is a growing need for effective alternatives to antibiotics for managing bacterial infections and biofilms.
As viruses of bacteria, bacteriophages (phages) specifically infect and lyse targeted bacteria. Due to their specific antibacterial activities, lytic phages have been demonstrated as alternatives to antibiotic therapy in humans19, veterinary science20, and aquaculture21. Recently, bacteriophage VP01 showed therapeutic potential by significantly reducing the growth of V. alginolyticus and dispersing the biofilms in a concentration-dependent manner22. In addition, V. alginolyticus phages exhibited potential as biocontrol agents in aquatic animals such as the sea cucumber (Apostichopus japonicus)23 and brine shrimp (Artemia salina)24. In the case of biofilm-related outbreaks, phage-infected bacteria existing at the outermost region of the matrix play a pivotal role in spreading the phages through the biofilm complex. Therefore, phages have been considered as alternatives to antibiotics, especially in biofilm eradication.
In theory, bacteria in planktonic or biofilm states can be lysed by a single phage particle as its progeny can infect and lyse adjacent cells. However, phages can be affected and inactivated by environmental factors25,26, which may allow bacterial regrowth to occur. Therefore, it is necessary to investigate and determine the biological characteristics of phages for practical applications and to expand our understanding of phages as alternatives to antibiotics.
With the goal of improving treatment and prevention of V. alginolyticus infection, we isolated and characterised a new lytic phage infecting Vibrio strains. This study focuses on the anti-planktonic and anti-biofilm activities of this phage and its genomic properties.
Results
Isolation and biological properties of phage pVa-21
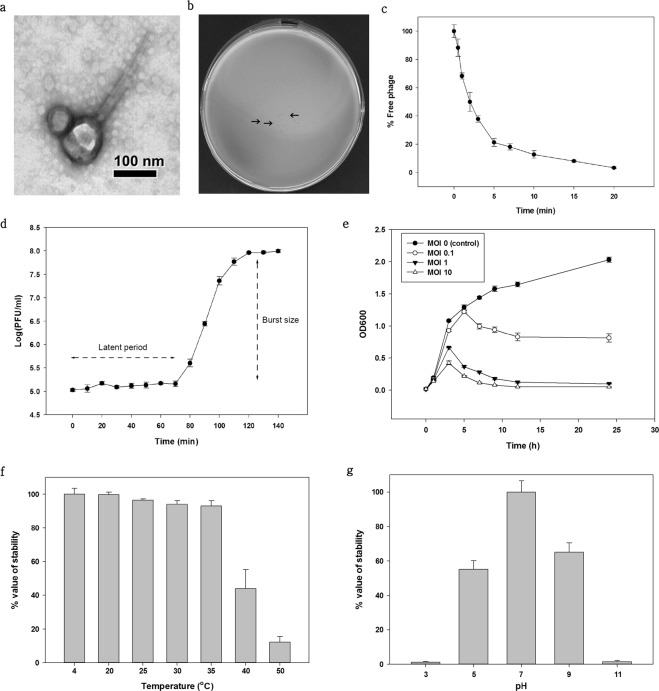
V. alginolyticus phages were isolated from seawater samples after enrichment. Purified phages were examined by transmission electron microscopy (TEM) and classified based on the criteria proposed by Ackermann27. As shown in Fig. 1a, phages were designated as pVa-21 and assigned to the family Myoviridae. It possesses an icosahedral head 87 ± 3 nm in diameter (n = 5) and a contractile tail 240 ± 9 nm in length (n = 5). The host range test was determined against bacteria of the Harveyi clade, which includes major pathogens of aquatic organisms, including V. alginolyticus (n = 5), V. harveyi (n = 5), V. parahaemolyticus (n = 1), V. anguillarum (n = 1), V. campbellii (n = 1), and V. vulnificus (n = 1). Phage pVa-21 was able to infect V. alginolyticus (n = 3) and V. harveyi (n = 1; Table 1). However, the phage did not show infectivity against the ten other bacterial strains tested and phage pVa-21 plaques were very small in size (less than 1 mm; Fig. 1b). The efficiency of plating (EOP) value varied among the Vibrio species, and no strain showed a higher value than the indicator host strain, V. alginolyticus rm-8402. Therefore, in vitro phage infection kinetics on host V. alginolyticus rm-8402 were assessed for adsorption rate, one-step growth curves, and cell lysis at two different multiplicities of infection (MOIs). The percentage of adsorption on the tested strain was 95% after 15 min (Fig. 1c). To identify the growth pattern and burst size of pVa-21, a one-step growth curve was generated. The latent period was found to be approximately 70 min and the burst size, i.e., the number of progeny released after lysis of a single bacterial cell, was approximately 58 virions per cell (Fig. 1d). Kinetics of the planktonic cell lysis test was conducted as shown in Fig. 1e. Strains that were not inoculated with phage pVa-21 (MOI = 0) showed a continuous increase in optical density (OD)600 values during the 24 h of incubation. When the lowest concentration (MOI = 0.1) of phage was applied, the bacterial strain showed partial lysis and growth reached OD600 = 1.0 (7.5 × 108 colony forming units [CFU]/mL) at 24 h; meanwhile, the control group showed OD600 = 2.0 and 1.4 × 109 CFU/mL. In contrast, bacterial growth increased until 3 h and was then completely lysed by the phage at an MOI of 1 and 10. Furthermore, pVa-21 stability was tested at different pH values and temperatures and was estimated by determining the changes in growth based on the number of plaque-forming units (PFU). Short-term thermal stability tests showed that pVa-21 was stable at 4 °C (control), 20 °C, 25 °C, 30 °C, and 35 °C for 1 h, but phage numbers were significantly lower at 40 °C (p < 0.001) and 50 °C (p < 0.001; Fig. 1f). Additionally, pVa-21 showed a significant decrease (p < 0.001) in PFUs under all pH treatment groups except at pH 7 (control group; Fig. 1g).
Figure 1.
Morphology and biological properties of phage pVa-21. (a) Transmission electron micrograph of pVa-21. Scale bar = 100 nm. (b) Plaque morphology of phage pVa-21. The arrow represent plaques. (c) Adsorption assays of pVa-21 with V. alginolyticus strain rm-8402. (d) One-step growth curve of pVa-21 in culture broth of V. alginolyticus strain rm-8402. (e) Planktonic cell lysis kinetics of pVa-21 at an MOI of 0.1, 1, and 10 on V. alginolyticus strain rm-8402. (f) Thermal stability of pVa-21. Phages were incubated for 1 h under different temperatures. (g) pH stability of pVa-21. Phages were incubated for 1 h under different pH values. (f,g) Phage ability to form plaques on host lawns was determined. Relative to control, changes in PFUs were calculated. The results shown in (c–g) represent the mean ± standard deviation of triplicate experiments.
Table 1.
Host range of phage pVa-21 against all bacterial strains used in this study.
| Bacterial species | Strain | EOPa | Source |
|---|---|---|---|
| V. alginolyticus | rm-8402 | 1.00 | Takaoka et al.2 |
| V374 | 0 | Nishibuchi et al.58 | |
| V447 | 0.76 ± 0.09 | Sawabe et al.59 | |
| am-10 | 0.15 ± 0.12 | Seoul National University Aquatic Biomedicine Laboratory Culture collection | |
| SNUFPC 080402 | 0 | ″ | |
| V. harveyi | SFC-BS | 0.70 ± 0.03 | ″ |
| PG-9302 | 0 | ″ | |
| PG-9303 | 0 | ″ | |
| O-3 | 0 | Ishimaru and Muroga60 | |
| O-6 | 0 | Ishimaru and Muroga60 | |
| V. parahaemolyticus | CRS 09–17 | 0 | Jun et al.61 |
| V. anguillarum | HT7601 | 0 | Nishibuchi et al.58 |
| V. campbellii | HUFP 9109 | 0 | Hiroshima University Fish Pathology Laboratory Culture Collection |
| V. vulnificus | HM1-1 | 0 | Nishibuchi et al.58 |
aEOP value is represented as the means ± standard deviation of triplicate replicates.
EOP; efficiency of plating.
Biofilm treatment with phage pVa-21
V. alginolyticus strain rm-8402 was left to form a biofilm for 48 h and then treated with phage pVa-21 (1.6 × 108 PFU/mL). Changes in CFUs, PFUs, and total biofilm biomass by crystal violet staining were measured over a 48-h period following infection. Total biofilm biomass showed a significant reduction (p < 0.001) after 5 h of phage treatment and the viable bacterial cell count inside the biofilm was significantly decreased (p < 0.001) after treatment for 5 h or more (Fig. 2a). During the first 24 h, crystal violet staining intensity and viable cell counts declined rapidly and did not increase over the next 24 h, indicating no biofilm regrowth. As bacterial cell counts decreased after phage treatment, phage concentration increased by more than 10-fold after 24 h compared with its initial concentration. After 48 h, phage concentrations decreased slightly but maintained a concentration of 1.19 × 108 PFU/mL (Fig. 2a). Scanning electron microscopy (SEM) verified that V. alginolyticus rm-8402 formed a biofilm (Fig. 2b) and revealed that the biofilm could only exist within 10 h after phage treatment. At first, bacterial cells located at the edge of the biofilm were disrupted by the phage and washed away after 5 h. Next, the film structure began to disappear at 10 h and then small bacterial aggregates, rather than film structure, were observed at 24 and 48 h after phage treatment (Fig. 2b).
Figure 2.
Infection dynamics of phage pVa-21 on biofilms of V. alginolyticus strain rm-8402. (a) Biofilms (48 h old) formed in 96-well plates were treated with pVa-21 at concentrations of 107 PFU/mL. Total biofilm biomass was measured at OD595 and stained with 1% crystal violet. Viable bacterial cell and phage counts were measured by direct plating on agar. The results represent the mean ± standard deviation of triplicate experiments. (b) Scanning electron micrograph of strain rm-8402 biofilm formation on glass coverslips. Scale bar = 10 μm and 2 μm at 2000× and 5000× magnification, respectively.
Genomic characterisation of phage pVa-21
The whole genome of phage pVa-21 was sequenced and analysed. Generally, genomes of phages belonging to the family Myoviridae consist of double-stranded DNA (dsDNA)28. In line with this, pVa-21 genomic DNA was digested by DNase I but not RNase A, indicating that it is a DNA phage (data not shown). The complete genome sequence of pVa-21 was 231,998 bp long with a GC content of 44.58%, encoding 241 putative open reading frames (ORFs) and no tRNA genes. As shown in Fig. 3, the ORFs of pVa-21 were broadly scattered across the genome and were not clustered according to function, such as those encoding structural, metabolism-related, or lysis proteins. Functional examination of the predicted ORFs indicated that they could be classified into three main categories, nucleotide regulation (e.g., helicase, ribonuclease H, DNA-directed RNA polymerase beta subunit), structure and packaging (e.g., major capsid protein, terminase large subunit, tail fibre protein), and lysis (e.g. lytic transglycosylase). Most genes (211; 87.5%) were located on the positive strand with only 30 genes (12.5%) located on the negative strand. Table S1 lists the general features of the putative ORFs identified in pVa-21.
Figure 3.
Genome map of phage pVa-21. The innermost circles coloured in cyan and purple indicate the positive and negative GC skew, respectively. Black circle indicates GC content. The functional categories of ORFs are indicated by specific colours; grey ORFs represent hypothetical proteins, yellow ORFs represent nucleotide regulation proteins, blue ORFs represent structure and packaging proteins, and red ORFs represent lysis proteins. Scale units are base pairs.
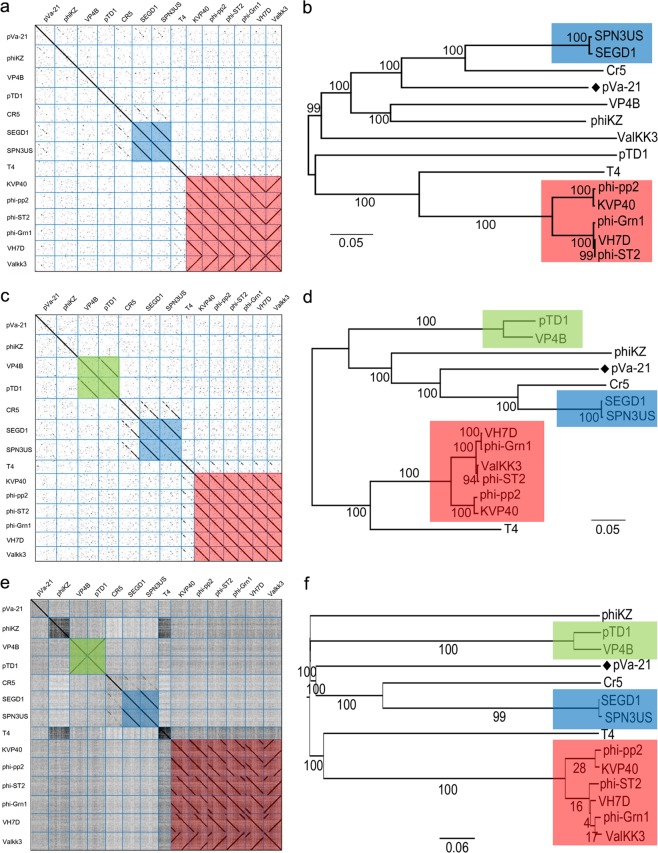
A BLASTn search revealed that phage pVa-21 is related to the phiKZ-like phage group (>65% similarity), which includes the Salmonella phage SPN3US, Cronobacter phage CR5, and enterobacteria phage SEGD1. Comparative analysis was then conducted using phylogeny and the dot plot method (Fig. 4). Terminase large subunit, major capsid protein, and whole genome sequences were used to determine phage relatedness. Dot plot results showed a strong relationship among schizo T4-like phages, while phiKZ-like phages showed clustering; cluster 1 included VP4B and pTD1 and cluster 2 included SPN3US and SEGD1 (Fig. 4a,c,e). Phage pVa-21, newly isolated in this study, showed a weak diagonal pattern with SPN3US, CR5, and SEGD1 for both major capsid protein and terminase large subunit sequences; however, no patterns with T4 and schizo T4-like phages were observed (Fig. 4a,c). The whole genome plot showed clustering as shown on the plot using major capsid protein and terminase large subunit sequences; however, no patterns were observed between pVa-21 and other related phages (Fig. 4e). Phylogenetic analysis revealed that pVa-21 was closely related to the phiKZ-like phages SPN3US, CR5, and SEGD1 (Fig. 4b,d,f) and distantly related to the Pseudomonas bacteriophage phiKZ. As revealed through dot plot analysis, three clusters were generated (SPN3US and SEGD1; pTD1 and VP4B; and schizo T4-like phages). Interestingly, the schizo T4-like Vibrio phage ValKK3 was found clustered with phiKZ-like phages after analysis using terminase large subunit sequences (Fig. 4b).
Figure 4.
Comparative analysis of the phage pVa-21. Analysis was conducted using (a,b) terminase large subunit sequences, (c,d) major capsid protein sequences, (e,f) and whole genome sequences. The dot plot was generated in Gepard52 at a word size of 10. The phylogenetic tree was generated in MEGA 7.0 software54 using (b,d) the neighbour-joining method and (e) VICTOR57 with settings recommended for prokaryotic viruses. (a–e) Clustered phages are indicated with different coloured boxes. Blue box, phiKZ-like Enterobacteria phage SEGD1 and SPN3US; green box, phiKZ-like Vibrio phage VP4B and pTD1; red box, schizo T4-like phages.
By using the artemis comparison tool (ACT), we were able to visualise whole genome comparisons of pVa-21 with VP4B (phiKZ-like Vibrio phage) and KVP40 (schizo T4-like Vibrio phage) as representatives of each group (Fig. S1); homology intensity was found slightly higher with VP4B. Nucleotide homology was also analysed using terminase large subunit, major capsid protein, and whole genome sequences. As shown in Table S2, nucleotide homology was higher with phiKZ-like phages than with schizo T4-like phages based on the terminase large subunit and major capsid protein sequences. Moreover, the gap value between pVa-21 and other phages was shorter with the phiKZ-like phages. Whole genome-wide comparisons, however, revealed no differences in percent identity and gap value. Furthermore, we compared the proteome of phage pVa-21 with two PhiKZ-like Vibrio phages and six schizo T4-like Vibrio phages using CoreGene. A total of 61 proteins were shared with phiKZ-like Vibrio phages while 12 proteins were shared with schizo T4-like Vibrio phages (Table S3).
Overall, the results of these comparative genomic analyses indicate that phage pVa-21 is more homologous with the phiKZ-like phage group rather than the schizo T4-like phage group and can be considered a distant member of phiKZ-like phages. Moreover, several paralogous genes similar to the beta/beta′ subunit of RNA polymerase were found in the pVa-21 genome, which is a distinctive feature of phiKZ-like phages29,30. In addition, three lysis-related proteins were identified in the pVa-21 genome, locus tag pVa21_119 (AQT28060.1), pVa21_134 (AQT28075.1), and pVa21_165 (AQT28106.1). Locus tag pVa21_119, pVa21_134, and pVa21_165 possessed lysis catalytic domains similar to the glycosyl hydrolase 108 domain, lytic transglycosylase domain, and goose egg white lysozyme domain, respectively. All lysis-related proteins were well conserved in phiKZ-like phages while pVa21_165 (AQT28106.1) was found in all phiKZ- and schizoT4-like vibrio phages analysed in this study. Additionally, genomic DNA of phage pVa-21 was examined for the presence of antimicrobial resistance genes, but no such gene was identified.
Discussion
In aquatic environments and fisheries, unseasonal outbreaks of Vibrio spp. are increasing due to global warming. In some cases, bacteria in biofilms showed increased resistance against antibiotics, which is a major obstacle to the current treatment method as it relies on antibiotics. Several studies have also reported a relationship between biofilms and disease recurrence. Notably, V. alginolyticus can form biofilms on polyvinyl-chloride, polyethylene, polystyrene, and glass surfaces that are generally used in aquaculture installations. Consequently, the efficacy of antibiotic therapy is decreasing and new methods are needed in place of antibiotics to control biofilms and multidrug-resistant bacteria effectively.
There have been several reports on controlling Vibrio spp. using bacteriophages21–24,31. However, limited reports are available regarding phages that can infect V. alginolyticus and control biofilm formation, even though this bacterial species can infect a variety of aquatic animals and even humans. In this study, we isolated the novel phage pVa-21, which infects V. alginolyticus, and characterised its biological properties. Phage pVa-21 can infect several fish pathogenic Harveyi clade bacteria. Moreover, pVa-21 was found to be stable up to 35 °C and at a pH range of 7–9, making it suitable for use under normal seawater conditions (i.e., 0–35 °C and pH 7.5–8.4).
As a biocontrol agent, phage therapy should also consider the emergence of resistance as observed for antibiotics. One of the most critical aspects is controlling microbial regrowth. Phage pVa-21 showed considerable anti-planktonic and anti-biofilm effects where planktonic bacteria were lysed after treatment with pVa-21 at an MOI of 1 or 10 (Fig. 1e). Moreover, biofilms were considerably disrupted and did not show regrowth; instead, viable bacterial cells remained in small aggregates (Fig. 2). At an MOI = 0.1, bacterial regrowth was observed. Similarly, a previous study also observed bacterial regrowth after phage treatment, yet additional inoculation with the phage solution could control these regrown bacteria as not all bacteria were resistant to the phages32. Another study suggested that regrown bacterial cells recover susceptibility when subsequently cultured in a phage-free medium, illustrating the transiency of phage resistance33. Indeed, phage pVa-21 formed inhibition zones on lawns of regrown bacteria when spot assays were conducted (data not shown), confirming that susceptiblity of the regrown bacteria and effectiveness of the giant vibriophage pVa-21 were maintained. However, follow up studies should address how pVa-21 prevents V. alginolyticus regrowth and controls mature biofilms for future application in environmental or industrial settings.
The anti-biofilm effect of phages and their enzymes has been previously demonstrated34. A recent study demonstrating the anti-biofilm effect of a phiKZ-like phage infecting V. alginolyticus supports the possibility of implementing phage pVa-21 as a biocontrol agent35. However, owing to specificity, phages cannot control a pathogen if it is not susceptible to the phage. To offset this disadvantage, isolating various types of phages with different host ranges and implementing cocktail therapy to expand antibiotic activity against target pathogens is imperative.
In our study, genetic characterisation revealed that pVa-21 was similar to phiKZ-like phages. Several beta/beta′ RNA polymerase subunits are characteristic of phiKZ-like phages29,30, and phage pVa-21 possessed several virion-associated beta/beta′ RNA polymerase subunits homologous to those of phiKZ. Phylogenetic analysis led to phage pVa-21 clustering with phiKZ-like phages rather than with schizo T4-like phages (Fig. 4). Dot plot and nucleotide homology results between pVa-21 and phiKZ-like Vibrio bacteriophages or schizo T4-like Vibrio bacteriophages also supported that pVa-21 is closely related to the phiKZ-like group rather than the schizo T4-like phage group. Furthermore, pVa-21 was found to exhibit small plaque sizes, which is also a characteristic of phiKZ-like phages36. Based on the currently available databases, no antibiotic resistance, virulence, or temperate phage-related genes were detected in the genome, suggesting that the V. alginolyticus phage pVa-21 can be safely used as a biocontrol agent. However, the majority of ORFs in pVa-21 did not match with predicted functions in GenBank. Thus, to ensure the safe and reliable application of phages in therapeutic settings, further investigation of the phage genome is warranted to gain a deeper understanding of the roles of encoded gene products, as they may produce novel virulence factors or interact undesirably with the host genome37.
In conclusion, the present study characterised the novel phiKZ-like phage pVa-21. Planktonic bacterial cell lysis and biofilm disruption effects of this Vibrio phage help promote the application of bacteriophages in biofilm control. Moreover, the pVa-21 genome is expected to broaden the phiKZ-like phage library. Although the precise biofilm eradication mechanism of pVa-21 remains to be elucidated, our findings support the potential of pVa-21 in controlling the fish pathogen V. alginolyticus. Further studies are needed to isolate other phages for effective pathogen control.
Materials and Methods
Bacterial strains and growth conditions
All bacterial strains used in this study are listed in Table 1. Bacteria were cultured in tryptic soy broth (TSB; Becton Dickinson, Franklin Lakes, NJ) supplemented with 1.5% (w/v) sodium chloride (Daejung Chemicals, Gheonggi-do, South Korea) with shaking at 150 rpm or sub-cultured on tryptic soy agar (Becton Dickinson) at 27 °C.
Phage isolation, purification, and propagation
To isolate phages infecting V. alginolyticus, 65 water samples from the West Sea of South Korea were collected over the span of five months and filtered through 0.2-µm membrane filters (Merck Millipore, Burlington, MA). V. alginolyticus strain rm-8402, which was previously reported as a fish pathogen2, was used as an indicator host strain. To isolate phages, 1% (v/v) of overnight grown (early stationary phase) host strain rm-8402 was used to inoculate a mixture of collected seawater samples and TSB (1:1) and cultured for 24 h at 27 °C. After enrichment, the presence of the phage was verified by spotting the serially diluted culture broth on the bottom agar layered with bacteria. Culture samples that showed inhibition zones were centrifuged at 10,000 × g for 20 min and the resulting supernatant was filtered through a 0.2-μm membrane filter. To confirm the presence of the lytic phage in the filtrate, a double-layer agar method was performed using the filtrate23. After overnight incubation at 27 °C, the plaque was purified five times through single-plaque isolation with a sterile straw to ensure that the isolated phages were descendants from a single virion. The Vibrio phages formed clear and small plaques on V. alginolyticus rm-8402 lawns (Fig. 1a) and were thus selected for further study and designated pVa-21.
Electron microscopy
For phage TEM, the obtained phages were concentrated using polyethylene glycol 8000-NaCl precipitation in sodium chloride-magnesium sulfate (SM) buffer (100 mM NaCl, 50 mM Tris pH 7.5, and 10 mM MgSO4) and then 10 μL of the suspension was spotted on a copper grid. After 2 min, the suspension was removed by absorption onto filter paper and the phages were negatively stained with 2% uranyl acetate for 1 min, followed by three successive washes with water. The grid was air-dried for 10 min and then imaged with a JEM-1010 (Jeol, Tokyo, Japan) operated at 80 kV. Phage dimensions were calculated by measuring the dimensions of five independent phages. The biofilm degradation effects of phage pVa-21 were observed using a ZEISS Sigma field-emission scanning electron microscope (FE-SEM; Carl Zeiss, Oberkochen, Germany) operated at 15 kV. Each biofilm was washed with phosphate-buffered saline (PBS), fixed with 2.5% glutaraldehyde for 1 h, and dehydrated in a graded series of ethanol (50%, 70%, 90%, and 95%; 1 h per step) and three times in 99% ethanol for 1 h. The biofilm was then dried with vacuum desiccator overnight and coated with platinum.
Host range analysis
The host range of the obtained phage was determined using a spot assay and confirmed by the double-layer agar method. Ten microliters of the phage lysate (>107 PFU/mL) was dropped onto the overlaid top agar and mixed with each bacterial strain. The plates were then incubated overnight at 27 °C and checked for the presence of a lysis zone. An EOP assay was conducted to quantify the lytic activity of phage pVa-21. The phage suspension (103 PFU/mL) was then assayed by the double-layer agar method. The total number of plaques was determined after 24 h of incubation and EOP values were calculated by comparing the ratios of PFUs of a susceptible strain to the indicator strain rm-8402 in triplicate.
Adsorption assay and one-step growth curve
The adsorption assay was carried out as described by Lu et al.38. The exponentially growing host strain (1.5 × 108 CFU/mL) was infected with a phage suspension at an MOI (the ratio of virus to bacterial cells) of 0.001 and incubated at 27 °C. Aliquots (100 μL) were taken at 0, 0.5, 1, 2, 3, 5, 7, 10, 15, and 20 min after infection and immediately diluted in 900 μL PBS, followed by centrifugation at 12,000 × g for 5 min. The supernatants were titrated for un-adsorbed free phages using the double-layer agar method. To construct a growth curve, the phage lysate was used to inoculate 10 mL of exponentially growing host strain culture (1.5 × 108 CFU/mL) at an MOI of 0.001. The phage was absorbed for 15 min and then centrifuged at 12,000 × g for 5 min. After the supernatant was discarded, the phage-infected bacterial pellet was re-suspended in 10 mL of preheated TSB and incubated at 27 °C with shaking at 250 rpm. At 10 min intervals, 100-μL aliquots were taken until 140 min and then titres were immediately determined by the double-layer agar method. Titre measurements were carried out in triplicate.
pH and thermal stability assays
For pH stability tests, 10 μL phage suspension (1.3 × 109 PFU/mL) was used to inoculate 1 mL PBS adjusted to pH 3.0, 5.0, 7.0, 9.0, and 11.0 with 1 M NaOH or 1 M HCl. The tubes were then incubated at 27 °C and aliquots were taken after 60 min. For thermal stability tests, 1 mL phage suspension (1.3 × 107 PFU/mL) was incubated at 4 °C, 20 °C, 25 °C, 30 °C, 35 °C, 40 °C, and 50 °C and then 100-μL aliquots were collected after 60 min. Next, aliquot titres were calculated using a 10-fold serial dilution. All tests were performed in triplicate.
Bacterial cell lysis assay
To evaluate the bacteriolytic efficacy of phage pVa-21, bacterial strains showing a turbid or clear lysis pattern in the spot assay were selected. One percent of overnight culture was inoculated into 10 mL of fresh broth to obtain 108 CFU/mL and then the phage was used to inoculate the broth at an MOI of 0, 0.1, 1, and 10. The broth was cultured with vigorous shaking and then OD600 was measured at 0, 1, 3, 5, 7, 9, 12, and 24 h. All tests were performed in triplicate.
Biofilm treatment with pVa-21
To verify the anti-biofilm efficacy of pVa-21, a biofilm was formed according to a previously described method with minor modifications39. Briefly, the biofilm assay was performed in 96-well polystyrene tissue culture microplates (Nunc, Roskilde, Denmark). One percent of overnight culture was inoculated into fresh TSB supplemented with 1% d-glucose (Sigma-Aldrich, St. Louis, MO) and then aliquots (200 μL) were distributed to each microplate well. Two sets of microplates—one for staining and the other for enumeration of bacteria or bacteriophages—were then incubated at 27 °C for 48 h with no shaking. The supernatant of each well was removed and washed twice with PBS to remove all planktonic cells, followed by treatment with 200 μL phage suspension (1.6 × 108 PFU/mL) for 3, 5, 7, 10, 24 and 48 h; changes in PFU were enumerated using the supernatant of each time point. The microplates were then washed twice with PBS and dried. To enumerate viable bacterial cells, biofilm cells were re-suspended in PBS via scraping with a sterile tip and then the suspension was diluted and plated. To quantify the total biomass, formed biofilms were stained with 1% crystal violet for 15 min, after which the wells were washed once more. Crystal violet was dissolved in an ethanol-acetone solution (80:20 v/v) and OD was measured at 595 nm. For SEM, biofilms were formed on glass coverslips (22 × 22 mm) submerged in media in 6-well plates and incubated for 48 h at 27 °C. Then, planktonic cells were removed from the wells, washed with PBS, and inoculated with phage suspension (108 PFU/mL) for 3, 5, 7, 10, 24, and 48 h. At each time point, the glass coverslips were processed as mentioned above and then SEM was performed.
Phage sequencing and genome analysis
Phage genomic DNA was extracted as described previously with minor modifications40. Briefly, the bacteriophage lysate was treated with 10 U DNase I and RNase A (Takara Bio, Kyoto, Japan) to degrade genomic DNA and RNA of the V. alginolyticus host cells according to manufacturer’s instructions. Then, ethylenediaminetetraacetic acid (EDTA) was added to inhibit nucleases. Protease K was also added and incubated at 37 °C for 30 min and then inactivated at 95 °C for 15 min. DNA purification followed conventional phenol-chloroform extraction methods41.
Purified genomic DNA of the phage was then sequenced using an Illumina HiSeq2500 platform (Illumina, San Diego, CA) at Genotech (Daejeon, South Korea). Reads were trimmed and assembled using the CLC Genomic Workbench v6.5.1. Putative ORFs were predicted and annotated using Glimmer v3.0242, Prodigal v1.2043, and protein BLAST. The Rapid Annotation using Subsystem Technology (RAST) server was used for confirmation44. Detection of tRNAs was carried out using tRNAscan-SE v2.045 and the genome map of pVa-21 was drawn using DNA plotter46. The web tool RESFINDER v2.1 was used to search for known antimicrobial resistance coding genes47. Percent nucleotide homology was calculated using EMBOSS Stretcher48 and protein sequence similarities of the phages were analysed using CoreGenes3.5 software49 with the default setting. A dot plot was generated in Gepard50 at a word size of 10.
For phylogenetic analysis, amino acid sequences of the major capsid protein and terminase large subunit were obtained from the Genbank database and aligned using Clustal W51. A phylogenetic tree was constructed using the neighbour-joining method implemented in MEGA v7.052 with 1000 bootstrap replications. The whole genome phylogenetic tree was generated in the Virus Classification and Tree Building Online Resource (VICTOR)53 using the Genome-BLAST Distance Phylogeny (GBDP) method54 under settings recommended for prokaryotic viruses53. The resulting intergenomic distances (including 100 replicates each) were used to infer a balanced minimum evolution tree with branch support via FASTME including Subtree Pruning and Regrafting (SPR) postprocessing55 for the formula D0. The tree was rooted at the midpoint55 and visualised with FigTree56.
Statistical analysis
All analyses were performed with SigmaPlot 12.0 software (Systat Software, Inc. Chicago, IL) using ANOVA with Dunnett’s post-hoc test. P values < 0.05 were considered statistically significant.
Nucleotide sequence accession numbers
The genome sequence of the isolated phage pVa-21 was deposited in GenBank under the accession number KY499642.
Supplementary information
Acknowledgements
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2017R1C1B2004616) and the Cooperative Research Program of Center for Companion Animal Research (PJ013985032018) of the Rural Development Administration, Republic of Korea.
Author Contributions
S.G.K., J.W.J. and D.J. mainly contributed to experimental design. S.G.K., S.Y., H.J.K., S.W.K., J.W.K. and S.J.H. mainly performed the experiments. S.G.K., S.S.G. and S.C.P. analysed the data and wrote the paper.
Data Availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-42681-1.
References
- 1.Sharma K, et al. Vibrio alginolyticus infection in Asian seabass (Lates calcarifer, Bloch) reared in open sea floating cages in India. Aquac. Res. 2012;44:86–92. doi: 10.1111/j.1365-2109.2011.03013.x. [DOI] [Google Scholar]
- 2.Takaoka O, et al. Effect of rotifer enrichment with herbal extracts on growth and resistance of red sea bream, Pagrus major (Temminck & Schlegel) larvae against Vibrio anguillarum. Aquac. Res. 2011;42:1824–1829. doi: 10.1111/j.1365-2109.2010.02783.x. [DOI] [Google Scholar]
- 3.Liu C-H, Cheng W, Hsu J-P, Chen J-C. Vibrio alginolyticus infection in the white shrimp Litopenaeus vannamei confirmed by polymerase chain reaction and 16S rDNA sequencing. Dis. Aquat. Organ. 2004;61:169–174. doi: 10.3354/dao061169. [DOI] [PubMed] [Google Scholar]
- 4.Liu PC, Chen YC, Lee KK. Pathogenicity of Vibrio alginolyticus isolated from diseased small abalone Haliotis diversicolor supertexta. Microbios. 2001;104:71–77. [PubMed] [Google Scholar]
- 5.Cochrane K, De Youg C, Soto D, Bahri T. Climate change implications for fisheries and aquaculture. FAO Fisheries and aquaculture technical paper. 2009;530:212. [Google Scholar]
- 6.Baker-Austin C, et al. Emerging Vibrio risk at high latitudes in response to ocean warming. Nat. Clim. Change. 2013;3:73. doi: 10.1038/nclimate1628. [DOI] [Google Scholar]
- 7.Vezzulli L, et al. Climate influence on Vibrio and associated human diseases during the past half-century in the coastal North. Atlantic. P. Natl. Acad. Sci. USA. 2016;113:E5062–5071. doi: 10.1073/pnas.1609157113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimes NE, et al. Temperature regulation of virulence factors in the pathogen Vibrio coralliilyticus. ISME J. 2012;6:835. doi: 10.1038/ismej.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karunasagar I, Otta SK, Karunasagar I. Biofilm formation by Vibrio harveyi on surfaces. Aquaculture. 1996;140:241–245. doi: 10.1016/0044-8486(95)01180-3. [DOI] [Google Scholar]
- 10.Coquet L, et al. Occurrence and phenotypic characterization of Yersinia ruckeri strains with biofilm-forming capacity in a rainbow trout farm. Appl. Environ. Microb. 2002;68:470–475. doi: 10.1128/AEM.68.2.470-475.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamayo R, Patimalla B, Camilli A. Growth in a biofilm induces a hyperinfectious phenotype in Vibrio cholerae. Infect. Immun. 2010;78:3560–3569. doi: 10.1128/IAI.00048-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faruque SM, et al. Transmissibility of cholera: in vivo-formed biofilms and their relationship to infectivity and persistence in the environment. P. Natl. Acad. Sci. USA. 2006;103:6350–6355. doi: 10.1073/pnas.0601277103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brooun A, Liu S, Lewis K. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 2000;44:640–646. doi: 10.1128/AAC.44.3.640-646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ceri H, Olson ME, Stremick C, Read RR, Morck D. & Buret, A. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 1999;37:1771–1776. doi: 10.1128/jcm.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levy SB. Antibiotic resistance: consequences of inaction. Clin. Infect. Dis. 2001;33:S124–129. doi: 10.1086/321837. [DOI] [PubMed] [Google Scholar]
- 16.Mechri B, Salem IB, Medhioub A, Medhioub MN, Aouni M. Isolation and genotyping of potentially pathogenic Vibrio alginolyticus associated with Ruditapes decussatus larva and juvenile mass mortalities. Aquacul. Int. 2015;23:1033–1047. doi: 10.1007/s10499-014-9862-7. [DOI] [Google Scholar]
- 17.Rameshkumar P, et al. Isolation and characterization of pathogenic Vibrio alginolyticus from sea cage cultured cobia (Rachycentro canadum (linnaeus 1766)) in India. Lett. Appl. Microbiol. 2017;65:423–430. doi: 10.1111/lam.12800. [DOI] [PubMed] [Google Scholar]
- 18.Mohamad Nurliyana, Mohd Roseli Fauzul Aidil, Azmai Mohammad Noor Amal, Saad Mohd Zamri, Md Yasin Ina Salwany, Zulkiply Nor Amalina, Nasruddin Nurrul Shaqinah. Natural Concurrent Infection of Vibrio harveyi and V. alginolyticus in Cultured Hybrid Groupers in Malaysia. Journal of Aquatic Animal Health. 2019;31(1):88–96. doi: 10.1002/aah.10055. [DOI] [PubMed] [Google Scholar]
- 19.Kutter E, et al. Phage therapy in clinical practice: treatment of human infections. Curr. Pharm. Biotechnol. 2010;11:69–86. doi: 10.2174/138920110790725401. [DOI] [PubMed] [Google Scholar]
- 20.Hawkins C, Harper D, Burch D, Änggård E, Soothill J. Topical treatment of Pseudomonas aeruginosa otitis of dogs with a bacteriophage mixture: a before/after clinical trial. Vet. Microbiol. 2010;146:309–313. doi: 10.1016/j.vetmic.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 21.Higuera G, Bastías R, Tsertsvadze G, Romero J, Espejo RT. Recently discovered Vibrio anguillarum phages can protect against experimentally induced vibriosis in Atlantic salmon, Salmo salar. Aquaculture. 2013;392:128–133. doi: 10.1016/j.aquaculture.2013.02.013. [DOI] [Google Scholar]
- 22.Sasikala D, Srinivasan P. Characterization of potential lytic bacteriophage against Vibrio alginolyticus and its therapeutic implications on biofilm dispersal. Microb. pathogenesis. 2016;101:24–35. doi: 10.1016/j.micpath.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, et al. Effect of bacteriophages on Vibrio alginolyticus infection in the sea cucumber, Apostichopus japonicus (Selenka) J. World Aquacult. Soc. 2015;46:149–158. doi: 10.1111/jwas.12177. [DOI] [Google Scholar]
- 24.Kalatzis PG, Bastías R, Kokkari C, Katharios P. Isolation and characterization of two lytic bacteriophages, ϕSt2 and ϕGrn1; phage therapy application for biological control of Vibrio alginolyticus in aquaculture live feeds. PloS one. 2016;11:e0151101. doi: 10.1371/journal.pone.0151101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams, M. H. Bacteriophages. (Interscience Publishers, 1959).
- 26.Weinbauer, M. G., Agis, M., Bonilla-Findji, O., Malits, A. & Winter, C. Bacteriophage in the environment. Bacteriophage: Genetics and Molecular Biology (ed. Grath, S. M. and Sinderen, D. V.) 61–92 (Caister academic press, 2007).
- 27.Ackermann HW. 5500 Phages examined in the electron microscope. Arch. Virol. 2007;152:227–243. doi: 10.1007/s00705-006-0849-1. [DOI] [PubMed] [Google Scholar]
- 28.ICTV Master Species List 2018a v1, https://talk.ictvonline.org/files/master-species-lists/m/msl/7992/ (2018).
- 29.Ceyssens PJ, et al. Development of giant bacteriophage ФKZ is independent of the host transcription apparatus. J. Virol. 2014;88:10501–10510. doi: 10.1128/JVI.01347-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhunchoth A, et al. Two Asian jumbo phages, φRSL2 and φRSF1, infect Ralstonia solanacearum and show common features of φKZ-related phages. Virology. 2016;494:56–66. doi: 10.1016/j.virol.2016.03.028. [DOI] [PubMed] [Google Scholar]
- 31.Letchumanan V, et al. Insights into bacteriophage application in controlling Vibrio species. Front. Microbiol. 2016;7:1114. doi: 10.3389/fmicb.2016.01114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu W, et al. Bacteriophage cocktail for the prevention of biofilm formation by Pseudomonas aeruginosa on catheters in an in vitro model system. Antimicrob. Agents Chemother. 2010;54:397–404. doi: 10.1128/AAC.00669-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan D, Dahl A, Middelboe M. Vibriophages differentially influence biofilm formation by Vibrio anguillarum strains. Appl. Environ. Microbiol. 2015;81:4489–4497. doi: 10.1128/AEM.00518-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.K Chan B, T Abedon S. Bacteriophages and their enzymes in biofilm control. Curr. Pharm. Design. 2015;21:85-–99. doi: 10.2174/1381612820666140905112311. [DOI] [PubMed] [Google Scholar]
- 35.Zhuhua L., Dezan Y. & Yanping Y. Vibrio Harveyi Giant VP4B and Application Thereof. Patent CN103555671A. Washington, DC: Patent Trademark Office (2014).
- 36.ICTV 9th Report, https://talk.ictvonline.org/ictv-reports/ictv_9th_report/ (2011).
- 37.Lima-Mendez G, Toussaint A, Leplae R. A modular view of the bacteriophage genomic space: identification of host and lifestyle marker modules. Res. Microbiol. 2011;162:737–746. doi: 10.1016/j.resmic.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 38.Lu Z, Breidt F, Jr., Fleming H, Altermann E, Klaenhammer T. Isolation and characterization of a Lactobacillus plantarum bacteriophage, ΦJL-1, from a cucumber fermentation. Int. J. Food Microbiol. 2003;84:225–235. doi: 10.1016/S0168-1605(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 39.Snoussi M, et al. Adhesive properties of environmental Vibrio alginolyticus strains to biotic and abiotic surfaces. New Microbiol. 2008;31:489–500. [PubMed] [Google Scholar]
- 40.Kim JH, et al. Complete genome sequence and characterization of a broad-host range T4-like bacteriophage phiAS5 infecting Aeromonas salmonicida subsp. salmonicida. Vet. Microbiol. 2012;157:164–171. doi: 10.1016/j.vetmic.2011.12.016. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., Fritsch, E. F., & Maniatis, T. Molecular cloning: a laboratory manual, 2nd edn. (Cold Spring Harbor Laboratory Press, 1989).
- 42.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hyatt D, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aziz RK, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zankari E, et al. Identification of acquired antimicrobial resistance genes. J. Aantimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rice P, Longden I, Bleasby A. EMBOSS: the european molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 49.Turner D, Reynolds D, Seto D, Mahadevan P. CoreGenes3.5: a webserver for the determination of core genes from sets of viral and small bacterial genomes. BMC Res. Notes. 2013;6:140. doi: 10.1186/1756-0500-6-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krumsiek J, Arnold R, Rattei T. Gepard: A rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics. 2007;23:1026–1028. doi: 10.1093/bioinformatics/btm039. [DOI] [PubMed] [Google Scholar]
- 51.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 52.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meier-Kolthoff JP, Göker M. Bioinformatics. 2017. VICTOR: Genome-based Phylogeny and Classification of Prokaryotic Viruses; pp. 3396–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Farris JS. Estimating phylogenetic trees from distance matrices. Am. Nat. 1972;106:645–667. doi: 10.1086/282802. [DOI] [Google Scholar]
- 56.Rambaut, A. FigTree 1.4.3 - a graphical viewer of phylogenetic trees and a program for producing publication-ready figures, http://tree.bio.ed.ac.uk/software/figtree/ (2006).
- 57.Lefort V, Desper R, Gascuel O. FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 2015;32:2798–2800. doi: 10.1093/molbev/msv150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nishibuchi M, Muroga K, Jo Y. Pathogenic Vibrio isolated from cultured eels-VI. Fish Pathol. 1980;14:125–131. doi: 10.3147/jsfp.14.125. [DOI] [Google Scholar]
- 59.Sawabe T, et al. Vibrio halioticoli sp. nov., a non-motile alginolytic marine bacterium isolated from the gut of the abalone Haliotis discus hannai. Int. J. Syst. Bacteriol. 1998;48:573–580. doi: 10.1099/00207713-48-2-573. [DOI] [PubMed] [Google Scholar]
- 60.Ishimaru K, Muroga K. Taxonomical re-examination of two pathogenic Vibrio species isolated from milkfish and uj jj swimming crab. Fish Pathol. 1997;32:59–64. doi: 10.3147/jsfp.32.59. [DOI] [Google Scholar]
- 61.Jun JW, et al. Bacteriophage therapy of a Vibrio parahaemolyticus infection caused by a multiple-antibiotic–resistant O3: K6 pandemic clinical strain. J. Infect. Dis. 2014;210:72–78. doi: 10.1093/infdis/jiu059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.