Abstract
The environmental considerations attributing to the escalation of carbon dioxide emissions have raised alarmingly. Consequently, the concept of sequestration and biological conversion of CO2 by photosynthetic microorganisms is gaining enormous recognition. In this study, in an attempt to discern the synergistic CO2 tolerance mechanisms, metabolic responses to increasing CO2 concentrations were determined for Synechococcus elongatus PCC 11801, a fast-growing, novel freshwater strain, using quantitative proteomics. The protein expression data revealed that the organism responded to elevated CO2 by not only regulating the cellular transporters involved in carbon-nitrogen uptake and assimilation but also by inducing photosynthesis, carbon fixation and glycolysis. Several components of photosynthetic machinery like photosystem reaction centers, phycobilisomes, cytochromes, etc. showed a marked up-regulation with a concomitant downshift in proteins involved in photoprotection and redox maintenance. Additionally, enzymes belonging to the TCA cycle and oxidative pentose phosphate pathway exhibited a decline in their expression, further highlighting that the demand for reduced cofactors was fulfilled primarily through photosynthesis. The present study brings the first-ever comprehensive assessment of intricate molecular changes in this novel strain while shifting from carbon-limited to carbon-sufficient conditions and may pave the path for future host and pathway engineering for production of sustainable fuels through efficient CO2 capture.
Subject terms: Proteome, Metabolic engineering
Introduction
Cyanobacteria are gram-negative, oxygenic photoautotrophs that harness sunlight to fix inorganic carbon into biomass. With a rise in carbon dioxide levels and its predicted atmospheric concentration reaching approximately 1072 ppmv (parts per million by volume) by the year 21001, the concept of CO2 uptake and mitigation by photosynthetic microorganisms has gained immense impetus2. Furthermore, to circumvent environmental concerns like diminishing fossil fuel reserves, their increasing costs and mounting CO2 emissions, the focus has recently shifted towards the development of alternative sustainable fuels with abated greenhouse release2,3. Cyanobacteria that typically capture solar energy to synthesize organic molecules are currently also being explored to produce diverse non-natural products using synthetic biology and metabolic engineering tools due to their pliable metabolism4–8. An ideal host cyanobacterium needs to be fast-growing, high light and CO2 tolerant, and genetically amenable9. Although pathway engineering approaches have been demonstrated to produce biochemicals/biofuels from the model cyanobacteria strains, their titers are not comparable to the heterotrophic model organisms like E. coli. In addition, the use of these model cyanobacteria strains in industrial settings poses certain limitations. For instance, the model organisms Synechococcus elongatus PCC 7942 and Synechocystis sp. PCC 6803 (hereafter referred to as Synechococcus 7942 and Synechocystis 6803, respectively) though naturally transformable, are slow-growing and cannot tolerate high light intensity. Other fast-growing strains like Synechococcus sp. PCC 7002 and Synechococcus elongatus UTEX 297310 (hereafter referred to as Synechococcus 7002 and Synechococcus 2973, respectively) either require vitamin B12 as an additional supplement or are not naturally transformable. We have chosen here a recently reported strain from a local freshwater source, Synechococcus elongatus PCC 11801 (hereafter referred to as Synechococcus 11801), a close homolog of the model organism Synechococcus 7942 that possesses all the above-mentioned attributes which qualify it to be a future host strain11. The physiological characterization revealed that this local isolate is tolerant to high levels of CO2, light, temperature, salts and butanol. Additionally, since the strain grew without any supplementation and was found to be naturally transformable, it seems like a promising candidate for further characterization. Despite the high CO2 fixation potential of cyanobacteria, the underlying molecular modulations involved in the sequestration of CO2 are poorly understood. Since proteins are directly in control of various cellular functions, ascertaining protein abundances is likely to provide salient cues to the intracellular rearrangements occurring during environmental perturbations like high CO2 stress. Few studies have reported the proteomics alterations in cyanobacteria and microalgae grown under CO2 replete conditions, but their research was mainly focused either on the effects of high CO2 on CCM (CO2 concentrating mechanisms) or on extracellular proteins12,13, which highlights the paucity of detailed proteomics investigations at high CO2 conditions. Therefore, the present study was undertaken to understand the strategy adopted by Synechococcus 11801 in biological sequestration of CO2 using a high-throughput iTRAQ (isobaric tags for relative and absolute quantitation) based quantitative proteomics approach (Fig. 1A). Furthermore, some of the top hit proteins were validated using MRM (multiple reaction monitoring) assays (Fig. 1B). These proteins displayed a substantially altered expression and represented the major altered physiological pathways. A comprehensive proteomics study on Synechococcus 11801 has not been performed yet and the findings from this study may open up new avenues to develop cyanobacteria cell factories with augmented metabolic fitness.
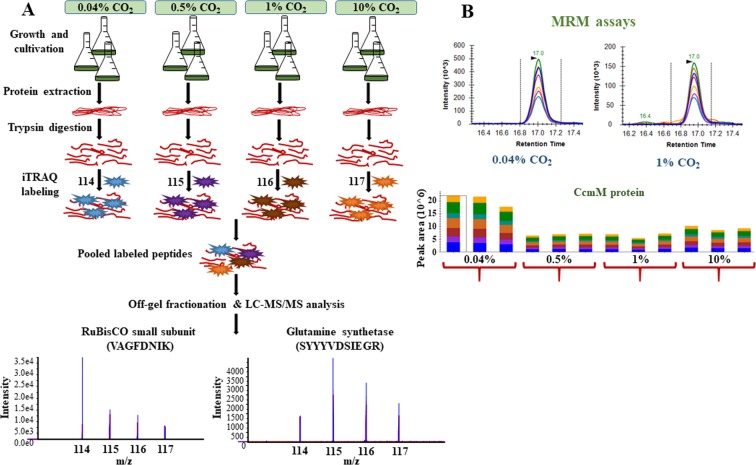
Figure 1.
Quantitative proteomics study of Synechococcus 11801 grown at increasing CO2 levels. (A) Schematic illustration of the overall experimental strategy employed in the discovery-phase global quantitative proteomics study. (B) MRM assays to verify the iTRAQ results. A representative image of the MRM assay of CcmM protein has also been shown.
Results
The strain Synechococcus 11801 can tolerate up to 15% CO2 in the chamber. Its physiological characterization at 0.04% (ambient levels), 0.5%, 1% and 10% CO2 resulted in growth rates of 0.07 h−1, 0.18 h−1, 0.16 h−1 and 0.08 h−1, respectively11 (Fig. 2A). The biochemical characterization under these conditions revealed significant differences in carbon allocation. To understand and validate the molecular and functional mechanisms underlying the process of carbon assimilation, the proteomics study was performed under the respective CO2 conditions. It was centered on the rationale that the strain presented a faster growth rate at 0.5% and 1% CO2 in comparison to the ambient CO2 level of 0.04%. Moreover, after 1% CO2, with an escalation in CO2 levels, a decline in growth rate was observed. The present study investigates the proteomics changes in Synechococcus 11801 under the above-mentioned CO2 conditions using the iTRAQ LC-MS/MS assay followed by validation of a few proteins using targeted proteomics-based MRM assays (Fig. 1).
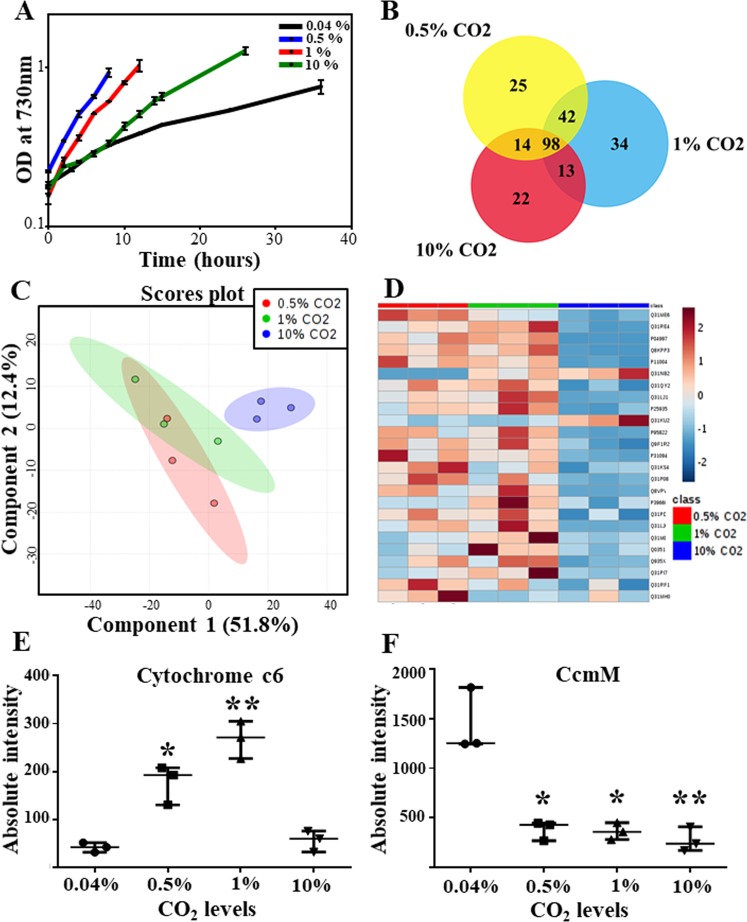
Figure 2.
Bioinformatics analysis of differentially expressed proteins. (A) Growth curve of Synechococcus 11801 at different CO2 concentrations at 38 °C and 400 µE in shake flasks. (B) Venn diagram showing the distribution of differentially expressed proteins among the three different CO2 conditions (0.04%, 0.5% and 10% CO2). (C) PLS-DA plot is showing segregation among the three high CO2 groups based on the protein fold ratio values in each group. The three points in each data group represent the three independent biological replicates. (D) Heatmap demonstrating the fold ratio of 25 differentially altered proteins with respect to the expression at ambient CO2 levels. (E,F) Scatter plots with n = 3 (expressed as median with range) showed an overall change in the expression pattern of cytochrome c6 and CcmM protein, respectively, with an increase in CO2 levels. Unpaired t-test with Welch’s correction was carried out for the comparison of the three high CO2 conditions with the ambient condition, with p-value < 0.05 considered significant. The following p-values were obtained; Cytochrome c6- 0.012 (0.5% CO2 vs. 0.04% CO2), 0.0034 (1% CO2 vs. 0.04% CO2), 0.1993 (10% CO2 vs. 0.04% CO2) and CcmM- 0.0115 (0.5% CO2 vs. 0.04% CO2), 0.0117 (1% CO2 vs. 0.04% CO2), 0.0079 (10% CO2 vs. 0.04% CO2).
High CO2 induced intracellular modulations elucidated by iTRAQ-based quantitative proteomics study
Three independent biological replicate cultures were cultivated for the iTRAQ experiment and the following labeling strategy was employed uniformly throughout the replicates. The iTRAQ reagents 114, 115, 116 and 117 were used to label protein digests from samples grown at 0.04%, 0.5%, 1% and 10% CO2, respectively. The proteomics analysis revealed a total of 861 proteins with at least two peptide signatures, of which 669 proteins were identified in at least two biological replicates. Nonetheless, only those proteins, which were found to be present in all the three replicates, i.e., the 492 proteins (Supplementary Fig. S2) were taken for further investigations. Further, the selection was performed based on the fold ratio values (≥1.5 fold up-regulation or ≤0.66 fold down-regulation) with respect to the ambient condition and for most proteins, the observed differential regulation was seen to be consistent in all three biological replicates. The resulting 248 proteins that showed the same trend (≥1.5 fold up-regulation or ≤0.66 fold down-regulation) across the three replicates were considered for the subsequent statistical and pathway analysis (Supplementary Table S1). These proteins displayed a differential expression pattern in at least one of the three CO2 conditions and may serve as vital cues in the understanding of intracellular proteome adjustments in response to elevated CO2 levels. It was observed that the protein expression levels at 0.5% and 1% CO2 were similar for most of the proteins, as compared to a more drastic protein expression pattern at 10% CO2.
Bioinformatics analysis of differentially expressed proteins during high CO2 stress
The protein list provided in Table 1, which includes some of the key differentially expressed proteins was further subjected to PLS-DA (partial least squares-discriminant analysis) using an online tool MetaboAnalyst 4.014 that segregated the three groups (high CO2 conditions, viz. 0.5%, 1% and 10% CO2) based on the differences among the protein fold ratio values on a 2D-score plot (Fig. 2C). Further, a heatmap was generated displaying the differential expression (fold ratio pattern) of a few of these proteins and is presented in Fig. 2D. Scatter plots of two such proteins showing a significantly altered expression at different CO2 levels have been provided in Fig. 2E,F.
Table 1.
A representative list of proteins from the iTRAQ data (mean ± standard error) showing a differential expression pattern with respect to ambient condition.
| Acc. ID | Locus tag | Protein name | 0.5%/0.04% CO2 | 1%/0.04% CO2 | 10%/0.04% CO2 |
|---|---|---|---|---|---|
| Inorganic carbon uptake and assimilation | |||||
| Q03513 | DOP62_10195 | Carbon dioxide concentrating mechanism protein CcmM | 0.09 ± 0.02 | 0.2 ± 0.05 | 0.07 ± 0.01 |
| Q03511 | DOP62_00505 | Carbon dioxide-concentrating mechanism protein CcmK | 0.11 ± 0.02 | 0.1 ± 0.04 | 0.13 ± 0.1 |
| Q31NB2 | DOP62_10175 | Ribulose 1,5-bisphosphate carboxylase small subunit | 0.03 ± 0 | 0.07 ± 0.01 | 0.09 ± 0.01 |
| Q9F1R2 | NA | HTH-type transcriptional activator CmpR | 0.48 ± 0.06 | NS | 0.12 ± 0.05 |
| Q55107 | DOP62_10270 | Bicarbonate transport ATP-binding protein CmpC | 0.08 ± 0.04 | 0.08 ± 0.03 | 0.04 ± 0.01 |
| P39660 | DOP62_10280 | Bicarbonate-binding protein CmpA | 0.02 ± 0 | 0.04 ± 0.01 | 0.02 ± 0 |
| Q8VPV7 | DOP62_05970 | CO2 hydration protein | 0.27 ± 0.06 | 0.32 ± 0.08 | 0.03 ± 0 |
| Q31ME6 | DOP62_10705 | NAD(P)H-quinone oxidoreductase subunit H | 0.29 ± 0.02 | 0.15 ± 0.01 | 0.06 ± 0.01 |
| Q31NJ5 | DOP62_07505 | NAD(P)H-quinone oxidoreductase subunit I | 0.11 ± 0 | 0.13 ± 0.04 | 0.05 ± 0.02 |
| Q31P07 | DOP62_12075 | NAD(P)H-quinone oxidoreductase subunit J | 0.18 ± 0.06 | 0.15 ± 0.05 | 0.08 ± 0.03 |
| Q31P08 | DOP62_12070 | NAD(P)H-quinone oxidoreductase subunit K | 0.43 ± 0.09 | 0.28 ± 0.1 | 0.05 ± 0.01 |
| Q31LQ7 | DOP62_07410 | NAD(P)H-quinone oxidoreductase subunit M | 0.15 ± 0.06 | 0.26 ± 0.04 | 0.14 ± 0.02 |
| Q31L05 | DOP62_04405 | NAD(P)H-quinone oxidoreductase subunit N | 0.29 ± 0.09 | 0.22 ± 0.03 | 0.07 ± 0.05 |
| Nitrogen absorption and transport | |||||
| P38043 | NA | Nitrate transport protein NrtA | 13.9 ± 5.15 | 11.41 ± 3.7 | 8.68 ± 3.08 |
| Q31PU9 | DOP62_11810 | Glutamate synthase (Ferredoxin) | 4.56 ± 1.54 | 3.8 ± 1.42 | NS |
| Q31L83 | NA | Glutamine synthetase | 11.8 ± 2.14 | 7.91 ± 1.61 | 6.17 ± 2 |
| P39661 | DOP62_10965 | Ferredoxin–nitrite reductase | 6.88 ± 2.72 | 7.11 ± 2.44 | 7.67 ± 2.63 |
| Photosynthesis and carbon fixation | |||||
| Q31LJ0 | DOP62_06080 | Photosystem I P700 chlorophyll a apoprotein A1 | 3.57 ± 0.72 | 4.52 ± 1.44 | 0.38 ± 0.05 |
| Q31LJ1 | DOP62_06085 | Photosystem I P700 chlorophyll a apoprotein A2 | 5.2 ± 0.56 | 8.3 ± 1.92 | NS |
| Q31PI7 | DOP62_06380 | Photosystem I reaction center subunit II | NS | 6.36 ± 2.36 | NS |
| Q31NT9 | DOP62_10920 | Photosystem I reaction center subunit III | 4.47 ± 1.37 | 5.91 ± 2.28 | NS |
| Q31NL7 | DOP62_08820 | Photosystem I reaction center subunit IV | 4.55 ± 2.48 | 8.9 ± 4.09 | 0.12 ± 0.03 |
| P95822 | DOP62_05055 | Photosystem I reaction center subunit XI | 5.9 ± 1.39 | 7.21 ± 1.56 | NS |
| Q31M07 | DOP62_09985 | Photosystem II 12 kDa extrinsic protein | NS | 3.75 ± 1.28 | NS |
| P11004 | DOP62_13040 | Photosystem II CP43 reaction center protein | 2.71 ± 0.51 | 2.28 ± 0.2 | 0.31 ± 0.06 |
| P31094 | DOP62_12825 | Photosystem II CP47 reaction center protein | NS | 1.81 ± 0.18 | 0.25 ± 0.08 |
| Q31RE4 | DOP62_00965 | Photosystem II lipoprotein Psb27 | 2.25 ± 0.19 | 3.73 ± 0.53 | 0.33 ± 0.11 |
| P11472 | DOP62_00570 | Photosystem II manganese-stabilizing polypeptide | NS | NS | 0.38 ± 0.09 |
| P04997 | DOP62_11795, DOP62_01605 | Photosystem II protein D1 2 | NS | 2.21 ± 0.15 | 0.07 ± 0.01 |
| Q31RG2 | DOP62_01060 | Phycobilisome 7.8 kDa linker polypeptide, allophycocyanin-associated, core | 14.39 ± 4.17 | 14.09 ± 4.39 | 5.42 ± 1.99 |
| Q31PD9 | DOP62_11780 | Phycobilisome rod linker polypeptide | 2.67 ± 0.44 | 2.93 ± 0.54 | NS |
| P55020 | DOP62_06590 | Plastocyanin | 10.27 ± 3.38 | 13.63 ± 1.63 | 9.09 ± 2.69 |
| Q8KPP3 | DOP62_12050 | Cytochrome b559 subunit alpha | 2.9 ± 0.35 | 3.13 ± 0.61 | 0.14 ± 0.04 |
| Q54711 | DOP62_05005 | Cytochrome b6 | 6.23 ± 2.53 | NS | NS |
| P25935 | DOP62_11225 | Cytochrome c6 | 19.64 ± 3.81 | 30.36 ± 5.91 | NS |
| P0A2Z8 | DOP62_04935 | ATP synthase epsilon chain | 11.66 ± 0.93 | 13.46 ± 1.27 | 12.98 ± 6.2 |
| Q31RF0 | DOP62_01000 | ATP synthase gamma chain | 4.61 ± 0.97 | 2.57 ± 0.22 | NS |
| Q31RF1 | DOP62_01005 | ATP synthase subunit alpha | 11.82 ± 1.87 | 8.03 ± 1.78 | 4.35 ± 1.91 |
| Q31RF3 | DOP62_01015 | ATP synthase subunit b | 5.53 ± 2.49 | 5.54 ± 1.59 | 3.75 ± 0.31 |
| Q31RF4 | DOP62_01020 | ATP synthase subunit b’ | 14.38 ± 3.66 | 12.09 ± 2.31 | 8.96 ± 2.19 |
| Q31KS4 | DOP62_04930 | ATP synthase subunit beta | 9.82 ± 1.5 | 6.4 ± 1.2 | 3.73 ± 0.76 |
| Q31RF5 | DOP62_01025 | ATP synthase subunit c | NS | NS | 6.5 ± 4 |
| Q31QY2 | DOP62_00835 | Sedoheptulose 1,7-bisphosphatase | 2.51 ± 0.4 | 2.87 ± 0.37 | NS |
| Q9R6W2 | DOP62_10710 | Glyceraldehyde-3-phosphate dehydrogenase | 5.79 ± 1.17 | 4.53 ± 0.68 | NS |
| Q31QU9 | DOP62_00660 | Transketolase | 5.31 ± 1.48 | 3.31 ± 0.85 | 2.58 ± 0.83 |
| Q935X4 | DOP62_03705 | Light-dependent NADPH-protochlorophyllide oxidoreductase | 7.73 ± 2.53 | 10.59 ± 2.72 | NS |
| Q31PM2 | DOP62_07035 | Porphobilinogen deaminase | 16.32 ± 5.01 | 20.42 ± 3.96 | 16.21 ± 2.13 |
| P16891 | DOP62_06580 | Uroporphyrinogen decarboxylase | 11.55 ± 1.97 | 8.03 ± 2.75 | 6.53 ± 1.38 |
| Q31QQ4 | DOP62_00020 | Mg-protoporphyrin IX chelatase | 9.03 ± 1.92 | 6.69 ± 1.66 | 5.4 ± 0.35 |
| Q9Z3G6 | DOP62_06830 | Heme oxygenase (Decyclizing) | 3.59 ± 0.88 | 3.53 ± 0.83 | 3.08 ± 0.65 |
| Q31LA2 | DOP62_05845 | Hydrogenobyrinic acid a,c-diamide cobaltochelatase | 7.13 ± 1.58 | 10.26 ± 1.48 | 5.26 ± 1.89 |
| Q31QJ2 | DOP62_13090 | Glutamate-1-semialdehyde 2,1-aminomutase | 10.02 ± 3.29 | 9.95 ± 2.3 | 6.56 ± 2.61 |
| Photoprotection and ROS response | |||||
| Q31M79 | DOP62_09835 | Flavoprotein | 0.25 ± 0.05 | 0.16 ± 0.04 | 0.15 ± 0.03 |
| Q31NJ4 | DOP62_07510 | NADH dehydrogenase subunit 6 | NS | 0.34 ± 0.16 | 0.18 ± 0.08 |
| Q31KG4 | DOP62_03465 | Chaperon-like protein for quinone binding in photosystem II | NS | 0.49 ± 0.02 | 0.33 ± 0.08 |
| Q7X4K8 | DOP62_04890 | Thioredoxin peroxidase | 0.27 ± 0.02 | 0.25 ± 0.11 | 0.24 ± 0.11 |
| Q31MK3 | DOP62_11435 | Thiosulphate-binding protein | 0.27 ± 0.03 | 0.18 ± 0.07 | 0.13 ± 0.04 |
| Q79PF2 | DOP62_09040 | Glutathione peroxidase | 0.26 ± 0.1 | NS | NS |
| Q31Q49 | DOP62_12620 | Glutathione S-transferase | 0.17 ± 0.06 | 0.2 ± 0.08 | 0.19 ± 0.08 |
| Q31QJ5 | DOP62_13105 | Bacterioferritin comigratory protein | 0.14 ± 0.07 | 0.35 ± 0.17 | 0.4 ± 0.14 |
| Q31L59 | DOP62_05365 | Bacterioferritin comigratory protein | 0.35 ± 0.15 | 0.37 ± 0.01 | NS |
| Glycolysis, TCA cycle and pentose phosphate pathway | |||||
| Q31R18 | DOP62_01165 | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase | 3.87 ± 0.67 | 5.65 ± 1.7 | 5.66 ± 0.96 |
| Q31NZ1 | DOP62_09015 | Dihydrolipoyl dehydrogenase | 3.45 ± 1.26 | 2.26 ± 0.33 | 4.06 ± 1.41 |
| Q31MH0 | DOP62_07935 | Isocitrate dehydrogenase [NADP] | NS | 0.25 ± 0.04 | 0.34 ± 0.1 |
| P21577 | DOP62_01965 | 6-phosphogluconate dehydrogenase, decarboxylating | 0.12 ± 0.03 | 0.08 ± 0.01 | 0.16 ± 0.02 |
| Q31KU2 | DOP62_04740 | Transaldolase | 0.25 ± 0.03 | 0.22 ± 0.03 | NS |
| Response to high CO 2 induced cellular stress | |||||
| P22880 | DOP62_04920 | 10 kDa chaperonin | 7.5 ± 3.65 | 10.31 ± 2.92 | 10.53 ± 1.3 |
| P22879 | DOP62_04915 | 60 kDa chaperonin | 4.93 ± 0.87 | 4.32 ± 0.75 | 5.25 ± 0.42 |
The accession IDs belong to the Synechococcus 7942 UniProt FASTA file. Additionally, the locus tags of Synechococcus 11801 proteins have been mentioned in the table. *NA = Not found, NS = Not significant.
Validation of a few key proteins identified from iTRAQ data using MRM
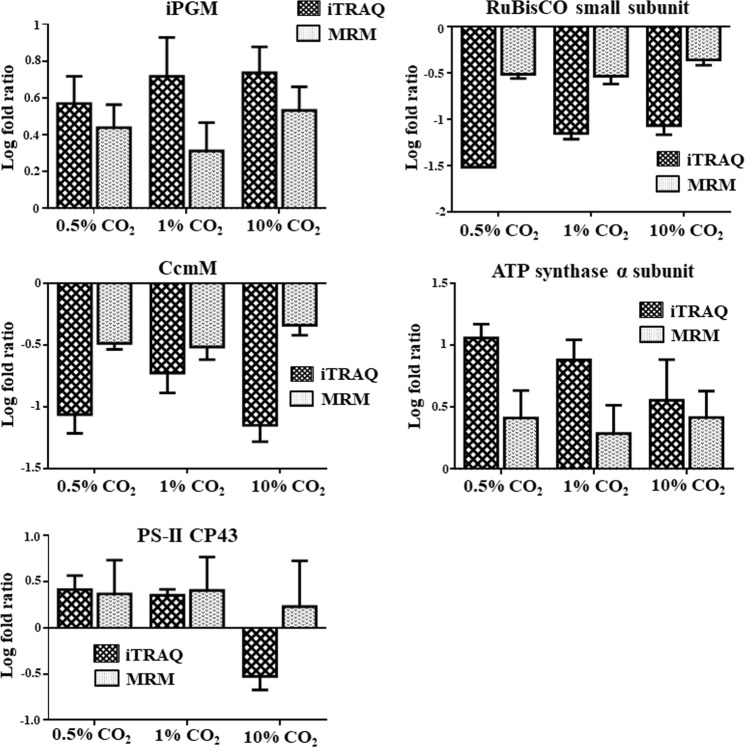
Ten proteins, namely RuBisCO (ribulose-1,5-bisphosphate carboxylase small subunit), carbon dioxide concentrating mechanism protein CcmM, bacterioferritin comigratory protein, SBPase (sedoheptulose 1,7-bisphosphatase), geranylgeranyl reductase, iPGM (2,3-bisphosphoglycerate-independent phosphoglycerate mutase), 60 kDa chaperonin, photosystem II CP43 reaction center protein, ATP synthase α subunit and NrtA (nitrate transport protein) were selected and monitored in all the four CO2 condition samples for MRM analysis. The relative expression levels of these proteins were compared with the iTRAQ quantification values. Both the techniques (iTRAQ and MRM) showed a similar trend in expression levels for most of the proteins, which added more confidence to our data. Proteins like ATP synthase α subunit, iPGM, 60 kDa chaperonin showed an up-regulated expression in both iTRAQ and MRM data (Fig. 3 & Supplementary Fig. S3). Similarly, the down-regulated proteins like CcmM, RuBisCO showed a similar expression trend in both the techniques (Fig. 3). However, for a few proteins like PS-II CP43, the two techniques showed a different trend at 10% CO2. Previous proteomics studies have also reported discrepancies in the expression levels obtained from iTRAQ and MRM analysis15,16. The possible reasons include low abundance of peptides, small sample size and complicated procedure of proteomics analysis without suitable internal standards.
Figure 3.
Validation of a few key proteins showing differential expression in iTRAQ data. (A) MRM-based relative quantification of iPGM, RuBisCO small subunit, CcmM, ATP synthase α subunit and PS-II CP43 have been shown. The log fold ratio values expressed as mean ± standard deviation (n = 3) were compared with the iTRAQ data and a similar trend was observed for most proteins.
Pathway mapping of differentially expressed proteins
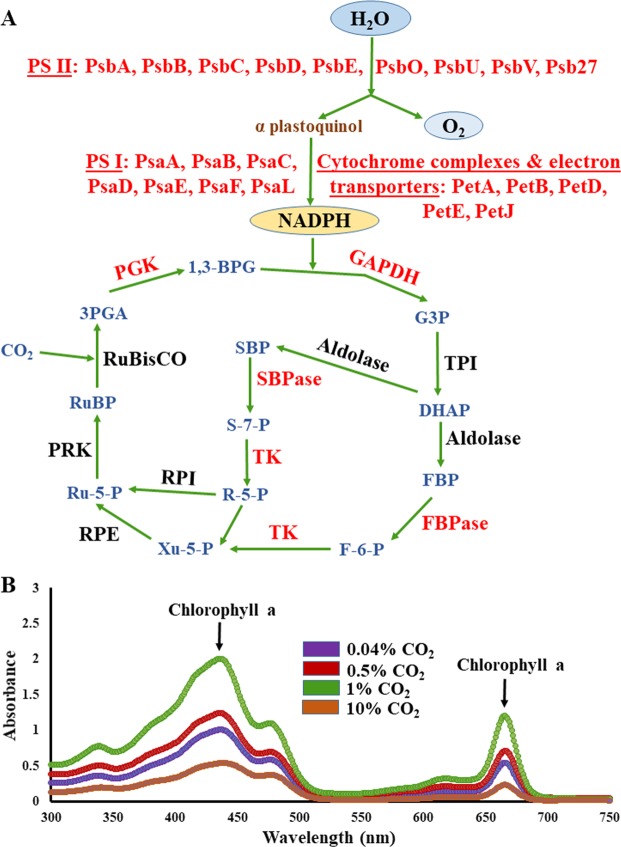
KEGG (Kyoto Encyclopedia of Genes and Genomes) was used to map the 248 differentially expressed proteins to several metabolic pathways and regulatory mechanisms. Major perturbations were observed in cellular transport, central metabolic pathways including glycolysis, PPP (pentose phosphate pathway), TCA cycle (tricarboxylic acid cycle), photosynthesis and carbon fixation (Table 1). Additionally, the expression of several photosynthetic complexes (Fig. 4A) and enzymes belonging to the chlorophyll biosynthesis pathway was perturbed at high CO2. To confirm if the chlorophyll amount was increased under high CO2 stress, we determined its levels using a UV-visible spectrophotometer. There was a progressive increase in chlorophyll levels with the rise in CO2 levels until 1% CO2. At 10% CO2, a decline was observed with respect to the ambient condition (Fig. 4B), consistent with the proteomics results which also revealed a decline in photosynthetic machinery at 10% CO2.
Figure 4.
Effect on photosynthetic machinery with an increase in CO2. (A) Regulation of photosynthetic complexes and Calvin cycle enzymes upon an increase in CO2. The proteins highlighted in red showed a differentially altered expression in iTRAQ data. PsbA: Photosystem II reaction center D1 protein, PsbB: Photosystem II CP47 reaction center protein, PsbC: Photosystem II CP43 reaction center protein, PsbD: Photosystem II reaction center D2 protein, PsbE: Photosystem II cytochrome b559 subunit alpha, PsbO: Photosystem II manganese-stabilizing polypeptide, PsbU: Photosystem II 12 kDa extrinsic protein, PsbV: Photosystem II cytochrome c550, Psb27: Photosystem II lipoprotein, PsaA: Photosystem I chlorophyll a apoprotein A1, PsaB: Photosystem I chlorophyll a apoprotein A2, PsaC: Photosystem I iron-sulfur center, PsaD: Photosystem I reaction center subunit II, PsaE: Photosystem I reaction center subunit IV, PsaF: Photosystem I reaction center subunit III, PsaL: Photosystem I reaction center subunit XI, PetA: Cytochrome f, PetB: Cytochrome b6, PetD: Cytochrome b6-f complex subunit 4, PetE: Plastocyanin, PetJ: Cytochrome c6, GAPDH: Glyceraldehyde 3-phosphate dehydrogenase, G3P: Glyceraldehyde 3-phosphate, TPI: Triosephosphate isomerase, DHAP: Dihydroxyacetone phosphate, FBP: Fructose-1,6-bisphosphate, FBPase: Fructose-1,6-bisphosphatase, F-6-P: Fructose-6-phosphate, TK: Transketolase, Xu-5-P: Xylulose 5-phosphate, RPE: Ribulose-phosphate 3-epimerase, Ru-5-P: Ribulose 5-phosphate, PRK: Phosphoribulokinase, RuBP: Ribulose-1,5-bisphosphate, RuBisCO: Ribulose-1,5-bisphosphate carboxylase/oxygenase, 3PGA: 3-phosphoglycerate, PGK: phosphoglycerate kinase, 1,3-BPG: 1,3-Bisphosphoglycerate, SBP: Sedoheptulose 1,7-bisphosphate, SBPase: Sedoheptulose 1,7-bisphosphatase, S-7-P: Sedoheptulose 7-phosphate, R-5-P: Ribose 5-phosphate, RPI: Ribose phosphate isomerase. (B) The UV-visible spectra recorded with chlorophyll extracts of the Synechococcus 11801 cells grown under 0.04% (ambient), 0.5%, 1% and 10% CO2. The peaks corresponding to the absorption maxima of chlorophyll a (435 and 665 nm) are highlighted.
Discussion
Synechococcus 11801 exhibits a high level of tolerance to different environmental stresses (CO2, light, temperature, and salts) and seems like an encouraging prospect for the production of fuels and other industrially relevant chemicals. Its ability to thrive at exceedingly high CO2 levels was the foundation of our study, which aims at elucidating the cellular mechanism adopted by cyanobacteria in sequestering high levels of CO2. UniProt protein database of the closest neighbour model strain, Synechococcus 7942 that shows 90% conservation with Synechococcus 11801 at the proteome level11, was used for the proteomics analysis. The global proteomics analysis revealed modulations among diverse protein groups and for a better understanding of their roles, they have been categorized into small sub-groups, which are discussed in detail hereafter.
Inorganic carbon uptake and assimilation
One of the most drastic responses towards high CO2 levels comprised of a diminished synthesis of regulatable CCM components involved in the uptake of Ci (inorganic carbon) and enhanced production of nitrogen assimilatory proteins as a feedback mechanism. This decrease in CCM components in cyanobacteria and microalgae at high CO2 is a well-established phenomenon17,18. CCM has evolved to expand the photosynthetic performance of cyanobacteria and factors like CO2 availability strongly influence the biosynthesis of many of its components19 including the five distinct transporters for active Ci uptake20; (i) BCT1, an ABC (ATP-binding cassette) bicarbonate transporter encoded by the cmpABCD operon21, (ii) SbtA, a high-affinity sodium-dependent bicarbonate transporter22 (iii) BicA, a low-affinity sodium-dependent bicarbonate transporter23 (iv) NDH-I4, a low-affinity CO2 uptake system constitutively expressed and (v) NDH-I3, a high-affinity CO2 uptake system inducible under limiting CO2 conditions24,25. CCM provides a growth advantage, predominantly under CO2-limiting conditions wherein the expression levels of inorganic carbon transporters have been reported to be augmented in several omics reports26–29. On the contrary, another proteomics study that was performed to understand the effect of high CO2 on cyanobacteria has revealed that the expression of CCM related proteins is suppressed at high CO2 levels12. This is in accordance with our study wherein we have found a substantial down-regulation of CmpA and CmpC protein at all three high CO2 concentrations, i.e., 0.5%, 1% and 10% (Table 1). The cmpABCD operon is activated under CO2-limited conditions by CmpR, a LysR family transcription regulator30, which too showed a down-regulation in our study. ChpY, a CO2 hydration protein which is a part of the high-affinity CO2 uptake NDH-I3 complex25 also showed a uniform decreased expression with respect to the ambient condition.
Correspondingly, CcmK and CcmM the structural components of carboxysomes which are essential for its assembly and functioning31,32 were seen to be down-regulated in all three CO2 concentrations. Also, the expression of carboxysomal proteins like carbonic anhydrase and RuBisCO small subunit was considerably diminished with the increase in CO2 concentration. Since carboxysomes house most of the cellular RuBisCO, the decline in shell components (CcmK, CcmM) is in agreement with the down-regulation of the enzyme. Interestingly, carbonic anhydrase in plants is also known to exhibit antioxidant activity and plays an additional role in the defense response33. Expression of several subunits of the NDH-1 complex (which is known to be involved in cellular functions like respiration, carbon dioxide uptake, etc.) was also found to be down-regulated at all three high CO2 levels. A recent study by Hirokawa and co-workers had shown improved productivity of 1,3-propanediol (1,3-PDO) in Synechococcus 7942 by disrupting the gene encoding NDH-1 complex subunits34. By eliminating the complex, they observed an increased carbon flux into the Calvin cycle demonstrating it as an attractive route towards an enhanced production of 1,3-PDO and glycerol, further suggesting a suitable target for metabolic engineering.
Nitrogen transport and absorption
It is imperative for cyanobacteria to maintain a harmony between carbon and nitrogen levels for the optimal cell metabolism, therefore, amplified levels of nitrogen assimilatory proteins could be perceived as an attempt to balance the carbon-nitrogen ratio (Table 1). At high CO2, elevated expression of NrtA, NrtC (nitrate transport ATP-binding protein), glutamate synthase (ferredoxin), glutamine synthetase III, glutamine synthetase and ferredoxin–nitrite reductase was observed in a possible effort to maintain the homeostasis of C:N ratio.
The major nitrogen sources for cyanobacteria include ammonium, nitrate, nitrite and urea of which nitrate is predominantly present in the BG-11 medium35. Nitrate assimilation in cyanobacteria encompasses three sequential steps; (i) nitrate uptake into the cell through an ABC-type transporter (that constitutes the products of the nrtA, nrtB, nrtC and nrtD genes)36–38, (ii) a two-step conversion to ammonium in a sequential manner by enzymes NarB (ferredoxin-nitrate reductase) and NirA (ferredoxin nitrite reductase)39,40 and (iii) the incorporation of resulting ammonium generated from nitrate reduction into carbon skeletons via the GS-GOGAT (glutamine synthetase-glutamate synthase) pathway38,41. Moreover, the transporter-encoding genes in Synechococcus 7942 are clustered together and co-transcribed with nirA and narB genes constituting the operon nirA-nrtABCD-narB42,43. Previous studies have delineated stimulatory effects of inorganic carbon on the synthesis of nitrate reductase emphasizing how nitrate utilization is strongly predisposed to the availability of CO244. Moreover, a CO2 enriched environment in Hizikia fusiforme culture boosted nitrate uptake and the activity of nitrate reductase45, further corroborating our findings. In our study, though the expression of NrtA remained exceedingly high in all the conditions, a trend was observed with the increase in CO2 levels as it showed an up-regulation of 13.9 folds in 0.5%, 11.4 folds in 1% and 8.7 folds in 10% CO2 condition. On the other hand, NrtC exhibited a significant increase of 3.7 folds only in 10% CO2 condition. The enzymes involved in GS-GOGAT pathway which is reported to be favored in ammonium-limiting conditions or under high energy supply46,47, viz. glutamine synthetase and ferredoxin-dependent glutamate synthase, were also found to be elevated in our study. Despite that the contents of growth medium were the same for ambient and high CO2 samples, it is understandable that high CO2 environment favors biosynthesis of nitrogen assimilatory proteins as a part of the acclimatization mechanism.
In addition to carbon and nitrogen, expression levels of few other ABC transporters involved in transmembrane ion transport were found to be regulated, i.e., an up-regulation of phosphate binding protein along with a decline in the expression of iron and sulfate-binding protein was observed.
Photosynthesis and carbon fixation
The growth of Synechococcus 11801 at high CO2 levels resulted in a steep rise in its cellular photosynthetic activity wherein an overall surge in expression was observed for photosynthetic proteins, which could be due to the abundant inorganic substrate available for carbon-fixing reactions. Key proteins identified in our study include cytochromes, plastocyanin, reaction centers of PS-I and PS-II (photosystem-I and II), etc. but interestingly few of these proteins showed a significant downshift at 10% CO2. Light-harvesting phycobilisome components like the alpha subunit of allophycocyanin was 3.6 folds up-regulated in 1% CO2 condition whereas the beta subunit showed a 0.17 folds down-regulation at 10% CO2. Previously a proteomics study on Synechocystis 6803 had reported slight down-regulation of subunits of PS-I, PS-II and phycobilisomes upon CO2 downshift26. Similarly, the transcriptomics response of Synechocystis 6803 to carbon limitation studied by Wang and co-workers too revealed a reduced expression of PS-I and PS-II genes28. They further suggested that this reduced requirement for the products of light reactions is due to the limited inorganic substrate available for carbon-fixing reactions.
Interestingly, carbon dioxide has been hypothesized to directly regulate the photosynthetic apparatus in a number of ways, namely by bicarbonate’s involvement in electron donation for the donor side of PS-II48 or by bicarbonate’s role as a proton shuttle stabilizing the binding niche of quinone, stimulating plastoquinol formation49 or by the activation of RuBisCO that requires CO2 (distinct from the CO2 that is fixed) before catalysis50–52. Previous studies also suggest that a short-term CO2 enrichment stimulates photosynthetic rate with a concomitant increase in biomass whereas prolonged exposure to high CO2 may suppress photosynthesis53. Though in our study we subjected the cells to high CO2 stress instead of prolonged exposure to CO2, few photosynthetic proteins showed a strong down-regulation at 10% CO2. Rather than being a direct response to high CO2, this suppression may be attributed to secondary responses linked to carbohydrate accumulation. With the photosynthetic rate exceeding the capacity to utilize the photosynthate for cellular growth, carbohydrate accrual may result into the repression of photosynthetic genes due to a probable feedback inhibition mechanism53. Additionally, reduced nitrogen and RuBisCO content may also contribute to the repression of photosynthesis.
Correspondingly, the proteins associated with carbon fixation like SBPase, glyceraldehyde-3-phosphate dehydrogenase and transketolase were up-regulated at elevated CO2 to facilitate fixation of the copious amount of inorganic carbon available. On numerous occasions, it has been suggested that under an array of growth conditions SBPase exerts a profound control over the Calvin cycle54 and also by increasing the SBPase activity, the photosynthetic rates were markedly increased in plants like tomato and tobacco55,56 suggesting that a higher SBPase activity may be advantageous for photosynthesis and growth. Even slight reductions in SBPase activity results in a substantial decrease in photosynthetic rates57,58, rendering further evidence that SBPase is tightly linked to carbon flux in the Calvin cycle. The enzyme functions at a branch point where assimilated carbon may either be used to regenerate the CO2 acceptor molecule ribulose-1,5-bisphosphate in the Calvin cycle or be assimilated for starch or sucrose and this precise position signifies the prominence of SBPase in the regulation of carbon flow. On the contrary, initial attempts focused at ameliorating carbon fixation were centered on modifying the catalytic properties of RuBisCO, as it was believed to be the key rate-limiting step in the overall photosynthetic incorporation of carbon59,60. However, later it was observed that the levels of RuBisCO had less influence on the control of carbon fixation under a wide range of environmental settings, especially under saturating CO2 conditions, so much so that even halving the activity of RuBisCO did not lead to any substantial diminution in the photosynthetic capacity54. Another protein found in our study, glyceraldehyde-3-phosphate dehydrogenase (encoded by gap2 gene), is essential for the Calvin cycle and is distinct from the other glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase involved in catabolic glucose degradation61,62. Likewise, transketolase’s expression levels were seen to be augmented in our findings because the enzyme apart from taking part in the pentose phosphate pathway is also involved in carbon fixation. An interesting study by Liang et al. has reported an increase in the growth rate and biomass of Synechocystis 6803 upon over-expression of proteins like SBPase and transketolase, highlighting them as potential candidates for an improved growth63.
Akin to proteins of photosynthetic machinery, enzymes involved in the biosynthesis of cofactors such as chlorophyll and porphyrin were markedly up-regulated at high CO2, namely light-dependent NADPH-protochlorophyllide oxidoreductase, uroporphyrinogen decarboxylase, Mg-protoporphyrin IX chelatase, porphobilinogen deaminase, heme oxygenase and hydrogenobyrinic acid a,c-diamide cobaltochelatase (Table 1). These results were found to be consistent with the chlorophyll levels estimated under different CO2 conditions (Fig. 4B). This increase in the synthesis of pigments and cofactors that play a pivotal role in the uptake of light mirrors the undergoing acclimation activity associated with magnified photosynthetic rates. The protein geranylgeranyl reductase, which is involved in chlorophyll and tocopherol biosynthesis64,65 demonstrated a marked 2.33 folds up-regulation at 0.5% CO2 and 0.21 folds down-regulation at 10% CO2. Apart from photosynthesis, proteins belonging to oxidative phosphorylation like ATP synthase also showed pronounced up-regulation at all three high CO2 conditions. All the different subunits of ATP synthase that belong to the same functional complex showed an enhanced expression adding high confidence to the acquired data.
Photoprotection and ROS (reactive oxygen species) response
Flavoprotein (Ortholog of Flv1 found in Synechocystis 6803) that plays a critical role in maintaining cellular redox homeostasis was found to be significantly down-regulated in all three high CO2 conditions (Table 1). It is known to catalyze photoreduction of O2 to H2O without producing ROS at the acceptor side of PS-I, ultimately protecting the oxygen-evolving PS-I complex against photooxidative damage66. Other antioxidant enzymes that protect cells against ROS, for example, glutathione peroxidase, thioredoxin peroxidase, superoxide dismutase, glutathione S-transferase, also demonstrated a decline in their expression. An important group of cyanobacteria proteins that catalyze the reduction of hydroperoxides and confer resistance to oxidative stress is peroxiredoxin67. 1-Cys peroxiredoxin, which is hypothesized to be involved in protecting nucleic acids against oxidative damage68 showed a 0.33 and 0.23 folds down-regulation at 0.5% and 10% CO2, respectively. Bacterioferritin comigratory protein, classified as atypical 2-Cys peroxiredoxin67, which in a previous study along with glutathione peroxidase-reductase system has been shown to be responsible for the detoxification of bentazone-derived peroxides also displayed a strong downshift in our findings69. Another peroxidase found in our study, viz. thioredoxin peroxidase was identified by Yamamoto and co-workers in Synechocystis 6803 as an antioxidant protein involved in reducing H2O2 with an electron transport system-coupled cellular activity70. An excess of excitation energy damages components of the photosynthetic machinery and produces harmful ROS, particularly when anabolism is slowed down due to nutrient deprivation68 and thus photoprotection mechanism is generally enhanced to protect cells from any possible photodamage. This resonates with a comparative proteomics study carried out by Gao et al. wherein they observed an extreme ROS formation by photosynthetic pigments and redox components when Synechocystis 6803 cells were treated with prolonged UV-B (Ultraviolet-B) radiation leading to a reduced light-harvesting ability and an increased expression of antioxidant proteins71. Also, there are numerous studies on plants in which the shared defense mechanisms against environmental pressures like metal toxicity, drought, salinity, chilling, UV-B radiations, etc. lead to increased activity/levels of these antioxidant proteins in response to the enhanced ROS production72,73. On the contrary, in our study, since at high CO2 levels, most of the captured light is utilized in carbon fixation, maintaining a balance between light capture and its utilization, an underproduction of ROS stress-responsive enzymes was observed.
Glycolysis, TCA cycle and PPP
The Synechococcus 11801 cells at high CO2 exhibited a heightened glycolysis pathway with a downshift in the TCA cycle and PPP. Expression levels of glycolytic enzymes belonging to the pay-off phase including phosphoglycerate kinase, 2,3-bisphosphoglycerate-independent phosphoglycerate mutase and pyruvate kinase evinced a strong up-regulation. Another group of proteins that constitute the pyruvate dehydrogenase complex showed an up-regulation at high CO2, but enzymes belonging to the TCA cycle, namely isocitrate dehydrogenase and aconitate hydratase displayed a decline in their expression. Since the flux to acetate does not produce any NADH in comparison to the flux from acetyl-CoA through the TCA cycle which generates NAD(P)H and FADH2, this diversion of carbon flow to acetate, especially in heterotrophs like E. coli. is often regarded as an apt measure to avert additional NAD(P)H accretion74,75. Analogous to TCA cycle, proteins like 6-phosphogluconate dehydrogenase (decarboxylating), transaldolase, glucose-6-phosphate 1-dehydrogenase showed a marked decline in their expression. The oxidative pentose phosphate pathway is predominantly involved in the production of reducing equivalents like NADPH that maintain cellular redox, especially under any oxidative stress. Since the photoprotection mechanism is already repressed, the cellular state of Synechococcus 11801 did not favour the pentose phosphate pathway.
Other proteins and pathways
Several proteins like farnesyl-diphosphate synthase, geranylgeranyl reductase, acetolactate synthase, ketol-acid reductoisomerase that participate in the biosynthesis of terpenoids and branched-chain amino acids showed an up-regulated expression. Various ribosomal proteins, elongation factors and uncharacterized proteins also showed an altered protein expression at high CO2 conditions. Additionally, an up-regulated 3-oxoacyl-[acyl-carrier-protein] synthase 2 protein was seen in our study. This protein catalyzes the condensation reaction of fatty acid synthesis indicating a possible enhanced lipid production, especially since acetyl-coenzyme A synthetase’s expression levels were also found to be increased in our study. High CO2 concentrations in microalgae are known to increase the levels of acetyl CoA, which is a precursor of fatty acid biosynthesis, eventually resulting in lipid accumulation76. Sun and co-workers had shown that genes involved in triacylglycerol biosynthesis were considerably up-regulated at elevated CO2 in Chlorella sorokiniana77. They further suggested that in microalgae an alternative strategy of providing excess CO2 for lipid accumulation may be a preferable approach over the more traditional strategy of nitrogen starvation since nitrogen deprivation may limit cellular growth. In contrast, by growing microalgae under high CO2 condition, both higher growth and lipid content can be achieved and since microalgae can assimilate CO2 from the industrial flue gases, it may be a more advantageous approach for lipid production at a wider scale. They also discovered that under high doses of CO2, expression of genes participating in central carbohydrate metabolic pathways including carbon fixation, glycolysis, and components of the pyruvate dehydrogenase complex were up-regulated, which was consistent with our findings.
At high CO2 levels, the Synechococcus 11801 cells presented paramount changes in proteins participating in carbon-nitrogen uptake and assimilation, photosynthetic complexes, CO2 fixation, glycolysis, TCA cycle, oxidative stress response and translation machinery, as a sequestration prototype towards high CO2. Additionally, in our study, cellular stress-responsive proteins like 10 and 60 kDa chaperonins were seen to be up-regulated in high CO2 conditions. A large number of uncharacterized proteins were also discovered in the study whose expression levels were notably altered under high CO2 conditions. The exact roles played by these unknown proteins remain largely unidentified indicating that our understanding of cyanobacteria’s cellular response towards high CO2 levels is still limited. This opens up new dimensions for exploration to fully comprehend the internal homeostatic response to CO2 stress in cyanobacteria. Moreover, there is a growing body of evidence showing that rising CO2 concentrations can induce rapid adaptive alterations in the genotype composition of photosynthetic species and may even lead to evolutionary changes78. It is very well demonstrated that many cyanobacteria including the model strain Synechococcus 7942 and its fast-growing neighbour strains like Synechococcus 2973 and Synechococcus 11801 produce more biomass in the presence of CO210,11,79. In a host cyanobacteria, the ability to grow at high CO2 conditions is desired as a heterologous expression of any pathway leads to significant redirecting of carbon towards the desired product, generating more demand of carbon that can be fulfilled by cultivating engineered strains in the presence of high CO24,80,81. Therefore, the understanding of intracellular alterations using proteomics may shed light on the strategy adopted by cyanobacteria in sequestering excess CO2. Our results offer novel insights into the global cellular response towards elevated CO2 in the indigenous strain Synechococcus 11801, which may be directly extrapolated to future studies addressing CO2 sequestration mechanisms in close neighbour strains like Synechococcus 7942, Synechococcus 2973 and other closely related photosynthetic organisms whose central metabolism is nearly conserved. In conclusion, the present study provides a global view of the differential cellular responses that occur in Synechococcus 11801 at the proteome level while going from carbon-limiting to carbon-sufficient conditions. Many proteins from our findings may be taken up as prospective targets for host and pathway engineering in Synechococcus 11801 and its close relatives.
Methods
Cultivation conditions of Synechococcus 11801
The strain Synechococcus 11801 was cultured in non-buffered BG-11 medium (initial pH = 7.5) and shake flasks. The culture was grown at 38 °C, 120 rpm and light intensity of 400 µmole photon.m−2.s−1 in a CO2 incubator shaker (Adolf Kuhner AG, LT-X, Birsfelden, Switzerland). The CO2 concentrations in the chamber were varied according to the experimental requirements. Three biological replicates of the cells (OD730 = 0.8) grown under 0.04%, 0.5%, 1% and 10% CO2 were harvested by centrifugation at 8,000 × g for 10 minutes at 4 °C. The cells were washed thrice with ice-cold phosphate buffer, pH = 7.5 and the pellets were stored at −80 °C till use.
Protein extraction, quantification and quality check
The protocol reported by Guerreiro et al.82 was used for protein extraction from the bacterial pellet with minor modifications. In brief, the pellet was resuspended in a lysis buffer having 50 mM ammonium bicarbonate, 8 M urea and a protease inhibitor cocktail. The cell lysis was performed by subjecting the samples to five freeze-thaw cycles using liquid nitrogen followed by five cycles of sonication on ice (30% amplitude for 1 min, 2-sec pulse and 2-sec gap). The supernatant was collected, after the centrifugation was carried out at 12,000 × g for 30 minutes and was further used for proteomics experiments. The protein concentration in each sample was estimated using QuickStart Bradford reagent (Bio-Rad, USA) and for quality check, the samples were run on a 12% SDS-PAGE gel. Further, the protein sample was used for iTRAQ-based quantitative proteomics analysis.
Sample processing for LC-MS/MS analysis (In-solution digestion, iTRAQ labeling and peptide OFFGEL fractionation)
The protein samples extracted from the three biological replicates were used for iTRAQ-based quantitative proteomics analysis. Prior to iTRAQ labeling, 50 µg of the protein sample was taken from all the four conditions, i.e., 0.04% CO2, 0.5% CO2, 1% CO2 and 10% CO2 and in-solution digestion was performed using the enzyme trypsin (Trypsin Gold, Mass spectrometry grade, Promega, Madison, WI, USA). The protein samples were diluted with dissolution buffer provided with iTRAQ kit to decrease the urea concentration followed by reduction with TCEP [tris (2-carboxyethyl) phosphine] at 60 °C for 1 hour. Subsequently, the samples were alkylated using MMTS (methyl methanethiosulfonate) for 30 minutes at room temperature to prevent the reformation of disulphide bonds. Trypsin was added at a 1:30 trypsin: protein ratio and the samples were kept at 37 °C for 16 hours for complete protein digestion.
After in-solution digestion, labeling of the digested peptides was performed using a 4plex-iTRAQ reagent multi-plex kit (AB Sciex, USA) as per the manufacturer’s instructions in a manner that iTRAQ reagents 114, 115, 116 and 117 were used to label trypsin-digested peptides from the samples grown at 0.04%, 0.5%, 1% and 10% CO2, respectively. The labeling reaction was quenched using 100 μL of water followed by incubation for 30 minutes at room temperature. All the labeled samples of the respective replicate set were pooled together and fractionated using Agilent 3100 OFFGEL fractionator (Agilent Technologies, USA) with IPG strip (pH range: 4–7, length: 24 cm) following manufacturer’s instructions. Each fraction was collected separately, and the peptide mixture was further cleaned up using zip-tips having C-18 columns (Millipore, USA) for salt removal before the MS/MS analysis. The labeled, fractionated, zip-tipped fractions were analyzed using Triple-TOF 5600 instrument and the raw.wiff files obtained were investigated using the ProteinPilot software (5.0.1, AB Sciex).
LC-MS/MS analysis for the database-aided protein identification and quantitation
The desalted samples were reconstituted in 0.1% formic acid and analyzed on a Triple-TOF instrument (SCIEX, Framingham, MA) coupled to eksigent ekspertTM nanoLC 425. The data acquisition was performed in a positive ion mode wherein the MS and MS/MS spectra were acquired in the range of 350–1250 and 100–1800 m/z, respectively. The instrument was operated in DDA (data-dependent acquisition) mode wherein maximum 20 precursors per cycle were chosen for fragmentation from each MS spectrum. The tandem MS/MS spectra were obtained in a high sensitivity mode with iTRAQ reagent collision energy adjustment. The raw files obtained in the.wiff format were analyzed using the ProteinPilot software and searched against the Synechococcus 7942 UniProt database. ParagonTM search algorithm was used for protein identification and quantification83. The following parameters were selected for the analysis: Sample type- iTRAQ 4plex (peptide labeled); Cys alkylation- MMTS; Digestion- Trypsin; Instrument- Triple TOF 5600; Special factors- Urea denaturation; ID focus- Biological modifications. Further, the database search was carried out with a Thorough effort while the detected protein threshold confidence was set at 99%, which corresponded to an unused score of 2. The option for FDR (false discovery rate) was also selected (Supplementary Fig. S1). The acquired mass spectrometric data files from the shotgun proteomics experiments have been deposited to the ProteomeXchange Consortium via the PRIDE84 partner repository with the dataset identifier PXD011485.
MRM to validate key findings from the iTRAQ shotgun MS/MS data
Ten differentially expressed proteins from the iTRAQ data, namely RuBisCO, CcmM, bacterioferritin comigratory protein, SBPase, geranylgeranyl reductase, iPGM, 60 kDa chaperonin, PS-II CP43 reaction center protein, ATP synthase α subunit and NrtA were chosen for MRM-based validation. The FASTA file of Synechococcus 7942 was fed into the Skyline software85 to generate in silico trypsin-digested peptides, from which unique peptides to these proteins were filtered. Further refinement was done by choosing precursor and product ions having only +2 and +1 charge, respectively. The peptide length ranged from 8–16 amino acids with no missed cleavages and at least 3 peptides were analyzed from each protein with a minimum of 3 transitions monitored per peptide. A total of 217 transitions were monitored for these 10 proteins in a single method run. Information like peptide sequences, precursor and product m/z, retention time have been provided in Supplementary Table S2. The MRM analysis was carried out on TSQ Altis™ triple quadrupole mass spectrometer (Thermo, USA) coupled to EASY-nLC™ 1200 in a scheduled manner with a retention time window of ± 1.5 minutes. Both the quadrupoles were operated at 0.7 resolution units (FWHM) and a cycle time of 1 second was kept. All the three biological replicates were analyzed and the result files were imported into the Skyline software followed by peak area computation.
Bioinformatics functional analysis to identify perturbed pathways
Pathway identification of the differentially altered proteins was done using KEGG wherein the UniProt Accession IDs were uploaded and matched against the reference species (Synechococcus 7942). Further, the online tool MetaboAnalyst was employed in generating PLS-DA plot and heatmap, which showed the segregation among different CO2 conditions based on the protein fold ratio values. Scatter plots were generated using the GraphPad Prism software.
Measurement of chlorophyll using spectrophotometer
The chlorophyll was extracted from 2 mL of cells at OD730 nm = 0.8 under all the conditions (0.04%, 0.5%, 1% and 10% CO2) using a previously described protocol86. The spectra were recorded from 300–750 nm using UV-visible spectrophotometer (Shimadzu, UV-2600, Singapore).
Supplementary information
Acknowledgements
This work was supported by the Department of Biotechnology grant awarded to S.S. and P.P.W. towards the DBT-Pan IIT Center for Bioenergy (Grant Number BT/EB/PAN IIT/2012). We would also like to acknowledge the MASSFIITB facility at IIT Bombay supported by the Department of Biotechnology (BT/PR13114/INF/22/206/2015) where the targeted proteomics experiments were carried out. Additionally, we would thank Mr. Saicharan Ghantasala and Mr. Sandip Kumar Patel for their help in MRM optimization.
Author Contributions
S.S., P.P.W. and K.M. conceived and designed the study. K.M. performed the proteomics experiments and analyzed the data. D.J. grew the strain and performed physiological studies. M.N. assisted in collecting the iTRAQ mass spectrometry data. K.M. wrote the manuscript. D.J., M.N., C.B.P., P.P.W. and S.S. have critically reviewed and provided expert advice for the improvement of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-42576-1.
References
- 1.Arce GLAF, Carvalho JA, Jr, Nascimento LFC. A time series sequestration and storage model of atmospheric carbon dioxide. Ecol. Model. 2014;272:59–67. doi: 10.1016/j.ecolmodel.2013.09.006. [DOI] [Google Scholar]
- 2.Seth JR, Wangikar PP. Challenges and opportunities for microalgae-mediated CO2 capture and biorefinery. Biotechnol. Bioeng. 2015;112:1281–1296. doi: 10.1002/bit.25619. [DOI] [PubMed] [Google Scholar]
- 3.Machado IM, Atsumi S. Cyanobacterial biofuel production. J. Biotechnol. 2012;162:50–56. doi: 10.1016/j.jbiotec.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Oliver JW, Machado IM, Yoneda H, Atsumi S. Cyanobacterial conversion of carbon dioxide to 2,3-butanediol. Proc. Natl. Acad. Sci. USA. 2013;110:1249–1254. doi: 10.1073/pnas.1213024110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hendry JI, Prasannan CB, Joshi A, Dasgupta S, Wangikar PP. Metabolic model of Synechococcus sp. PCC 7002: Prediction of flux distribution and network modification for enhanced biofuel production. Bioresour. Technol. 2016;213:190–197. doi: 10.1016/j.biortech.2016.02.128. [DOI] [PubMed] [Google Scholar]
- 6.Hendry JI, et al. Rerouting of carbon flux in a glycogen mutant of cyanobacteria assessed via isotopically non-stationary 13C metabolic flux analysis. Biotechnol. Bioeng. 2017;114:2298–2308. doi: 10.1002/bit.26350. [DOI] [PubMed] [Google Scholar]
- 7.Knoot CJ, Ungerer J, Wangikar PP, Pakrasi HB. Cyanobacteria: Promising biocatalysts for sustainable chemical production. J. Biol. Chem. 2018;293:5044–5052. doi: 10.1074/jbc.R117.815886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luan G, Lu X. Tailoring cyanobacterial cell factory for improved industrial properties. Biotechnol. Adv. 2018;36:430–442. doi: 10.1016/j.biotechadv.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Sengupta A, Pakrasi HB, Wangikar PP. Recent advances in synthetic biology of cyanobacteria. Appl. Microbiol. Biotechnol. 2018;102:5457–5471. doi: 10.1007/s00253-018-9046-x. [DOI] [PubMed] [Google Scholar]
- 10.Yu J, et al. Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO2. Sci. Rep. 2015;5:8132. doi: 10.1038/srep08132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaiswal D, et al. Genome features and biochemical characteristics of a robust, fast growing and naturally transformable cyanobacterium Synechococcus elongatus PCC 11801 isolated from India. Sci. Rep. 2018;8:16632. doi: 10.1038/s41598-018-34872-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramanan R, Vinayagamoorthy N, Sivanesan SD, Kannan K, Chakrabarti T. Influence of CO2 concentration on carbon concentrating mechanisms in cyanobacteria and green algae: a proteomic approach. Algae. 2012;27:295–301. doi: 10.4490/algae.2012.27.4.295. [DOI] [Google Scholar]
- 13.Baba M, Suzuki I, Shiraiwa Y. Proteomic analysis of high-CO2-inducible extracellular proteins in the unicellular green alga, Chlamydomonas reinhardtii. Plant Cell Physiol. 2011;52:1302–1314. doi: 10.1093/pcp/pcr078. [DOI] [PubMed] [Google Scholar]
- 14.Chong J, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Narumi R, et al. A strategy for large-scale phosphoproteomics and SRM-based validation of human breast cancer tissue samples. J. Proteome Res. 2012;11:5311–5322. doi: 10.1021/pr3005474. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, et al. iTRAQ-Based Quantitative Proteomic Comparison of Early- and Late-Passage Human Dermal Papilla Cell Secretome in Relation to Inducing Hair Follicle Regeneration. PLoS One. 2016;11:e0167474. doi: 10.1371/journal.pone.0167474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyachi S, Iwasaki I, Shiraiwa Y. Historical perspective on microalgal and cyanobacterial acclimation to low- and extremely high-CO2 conditions. Photosynth. Res. 2003;77:139–153. doi: 10.1023/A:1025817616865. [DOI] [PubMed] [Google Scholar]
- 18.Solovchenko A, Khozin-Goldberg I. High-CO2 tolerance in microalgae: possible mechanisms and implications for biotechnology and bioremediation. Biotechnol. Lett. 2013;35:1745–1752. doi: 10.1007/s10529-013-1274-7. [DOI] [PubMed] [Google Scholar]
- 19.Nishimura T, et al. Mechanism of low CO2-induced activation of the cmp bicarbonate transporter operon by a LysR family protein in the cyanobacterium Synechococcus elongatus strain PCC 7942. Mol. Microbiol. 2008;68:98–109. doi: 10.1111/j.1365-2958.2008.06137.x. [DOI] [PubMed] [Google Scholar]
- 20.Price GD. Inorganic carbon transporters of the cyanobacterial CO2 concentrating mechanism. Photosynth. Res. 2011;109:47–57. doi: 10.1007/s11120-010-9608-y. [DOI] [PubMed] [Google Scholar]
- 21.Omata T, et al. Identification of an ATP-binding cassette transporter involved in bicarbonate uptake in the cyanobacterium Synechococcus sp. strain PCC 7942. Proc. Natl. Acad. Sci. USA. 1999;96:13571–13576. doi: 10.1073/pnas.96.23.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shibata M, et al. Genes essential to sodium-dependent bicarbonate transport in cyanobacteria: function and phylogenetic analysis. J. Biol. Chem. 2002;277:18658–18664. doi: 10.1074/jbc.M112468200. [DOI] [PubMed] [Google Scholar]
- 23.Price GD, Woodger FJ, Badger MR, Howitt SM, Tucker L. Identification of a SulP-type bicarbonate transporter in marine cyanobacteria. Proc. Natl. Acad. Sci. USA. 2004;101:18228–18233. doi: 10.1073/pnas.0405211101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shibata M, et al. Distinct constitutive and low-CO2-induced CO2 uptake systems in cyanobacteria: genes involved and their phylogenetic relationship with homologous genes in other organisms. Proc. Natl. Acad. Sci. USA. 2001;98:11789–11794. doi: 10.1073/pnas.191258298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maeda S, Badger MR, Price GD. Novel gene products associated with NdhD3/D4-containing NDH-1 complexes are involved in photosynthetic CO2 hydration in the cyanobacterium, Synechococcus sp. PCC7942. Mol. Microbiol. 2002;43:425–435. doi: 10.1046/j.1365-2958.2002.02753.x. [DOI] [PubMed] [Google Scholar]
- 26.Battchikova N, et al. Dynamic changes in the proteome of Synechocystis 6803 in response to CO2 limitation revealed by quantitative proteomics. J. Proteome Res. 2010;9:5896–5912. doi: 10.1021/pr100651w. [DOI] [PubMed] [Google Scholar]
- 27.Schwarz D, et al. Metabolic and transcriptomic phenotyping of inorganic carbon acclimation in the Cyanobacterium Synechococcus elongatus PCC 7942. Plant Physiol. 2011;155:1640–1655. doi: 10.1104/pp.110.170225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang HL, Postier BL, Burnap RL. Alterations in global patterns of gene expression in Synechocystis sp. PCC 6803 in response to inorganic carbon limitation and the inactivation of ndhR, a LysR family regulator. J. Biol. Chem. 2004;279:5739–5751. doi: 10.1074/jbc.M311336200. [DOI] [PubMed] [Google Scholar]
- 29.Eisenhut M, et al. Long-term response toward inorganic carbon limitation in wild type and glycolate turnover mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Plant. Physiol. 2007;144:1946–1959. doi: 10.1104/pp.107.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Omata T, Gohta S, Takahashi Y, Harano Y, Maeda S. Involvement of a CbbR homolog in low CO2-induced activation of the bicarbonate transporter operon in cyanobacteria. J. Bacteriol. 2001;183:1891–1898. doi: 10.1128/JB.183.6.1891-1898.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Badger MR, Price GD. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J. Exp. Bot. 2003;54:609–622. doi: 10.1093/jxb/erg076. [DOI] [PubMed] [Google Scholar]
- 32.Price GD, Badger MR, Woodger FJ, Long BM. Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J. Exp. Bot. 2008;59:1441–1461. doi: 10.1093/jxb/erm112. [DOI] [PubMed] [Google Scholar]
- 33.Slaymaker DH, et al. The tobacco salicylic acid-binding protein 3 (SABP3) is the chloroplast carbonic anhydrase, which exhibits antioxidant activity and plays a role in the hypersensitive defense response. Proc. Natl. Acad. Sci. USA. 2002;99:11640–11645. doi: 10.1073/pnas.182427699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirokawa Y, Matsuo S, Hamada H, Matsuda F, Hanai T. Metabolic engineering of Synechococcus elongatus PCC 7942 for improvement of 1,3-propanediol and glycerol production based on in silico simulation of metabolic flux distribution. Microb. Cell Fact. 2017;16:212. doi: 10.1186/s12934-017-0824-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rippka R. Isolation and purification of cyanobacteria. Methods Enzymol. 1988;167:3–27. doi: 10.1016/0076-6879(88)67004-2. [DOI] [PubMed] [Google Scholar]
- 36.Omata T, Ohmori M, Arai N, Ogawa T. Genetically engineered mutant of the cyanobacterium Synechococcus PCC 7942 defective in nitrate transport. Proc. Natl. Acad. Sci. USA. 1989;86:6612–6616. doi: 10.1073/pnas.86.17.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maeda S, Omata T. Substrate-binding lipoprotein of the cyanobacterium Synechococcus sp. strain PCC 7942 involved in the transport of nitrate and nitrite. J. Biol. Chem. 1997;272:3036–3041. doi: 10.1074/jbc.272.5.3036. [DOI] [PubMed] [Google Scholar]
- 38.Herrero A, Muro-Pastor AM, Flores E. Nitrogen control in cyanobacteria. J. Bacteriol. 2001;183:411–425. doi: 10.1128/JB.183.2.411-425.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luque I, Flores E, Herrero A. Nitrite reductase gene from Synechococcus sp. PCC 7942: homology between cyanobacterial and higher-plant nitrite reductases. Plant Mol. Biol. 1993;21:1201–1205. doi: 10.1007/BF00023618. [DOI] [PubMed] [Google Scholar]
- 40.Rubio LM, Herrero A, Flores E. A cyanobacterial narB gene encodes a ferredoxin-dependent nitrate reductase. Plant Mol. Biol. 1996;30:845–850. doi: 10.1007/BF00019017. [DOI] [PubMed] [Google Scholar]
- 41.Meeks JC, et al. Pathways of assimilation of [13N]N2 and 13NH4+ by cyanobacteria with and without heterocysts. J. Bacteriol. 1978;134:125–130. doi: 10.1128/jb.134.1.125-130.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Omata T, Andriesse X, Hirano A. Identification and characterization of a gene cluster involved in nitrate transport in the cyanobacterium Synechococcus sp. PCC7942. Mol. Gen. Genet. 1993;236:193–202. doi: 10.1007/BF00277112. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki I, Sugiyama T, Omata T. Primary structure and transcriptional regulation of the gene for nitrite reductase from the cyanobacterium Synechococcus PCC 7942. Plant Cell Physiol. 1993;34:1311–1320. [Google Scholar]
- 44.Vazquez-Bermudez MF, Herrero A, Flores E. Carbon supply and 2-oxoglutarate effects on expression of nitrate reductase and nitrogen-regulated genes in Synechococcus sp. strain PCC 7942. FEMS Microbiol. Lett. 2003;221:155–159. doi: 10.1016/S0378-1097(03)00208-8. [DOI] [PubMed] [Google Scholar]
- 45.Zou D. Effects of elevated atmospheric CO2 on growth, photosynthesis and nitrogen metabolism in the economic brown seaweed, Hizikia fusiforme (Sargassaceae, Phaeophyta) Aquaculture. 2005;250:726–735. doi: 10.1016/j.aquaculture.2005.05.014. [DOI] [Google Scholar]
- 46.Helling RB. Why does Escherichia coli have two primary pathways for synthesis of glutamate? J. Bacteriol. 1994;176:4664–4668. doi: 10.1128/jb.176.15.4664-4668.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Helling RB. Pathway choice in glutamate synthesis in. Escherichia coli. J. Bacteriol. 1998;180:4571–4575. doi: 10.1128/jb.180.17.4571-4575.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klimov VV, Allakhverdiev SI, Feyziev Ya M, Baranov SV. Bicarbonate requirement for the donor side of photosystem II. FEBS Lett. 1995;363:251–255. doi: 10.1016/0014-5793(95)00327-6. [DOI] [PubMed] [Google Scholar]
- 49.Govindjee Bicarbonate-Reversible Inhibition of Plastoquinone Reductase in Photosystem II. Z. Naturforsch. 1993;48c:251–258. doi: 10.1515/znc-1993-3-422. [DOI] [Google Scholar]
- 50.Lorimer GH, Badger MR, Andrews TJ. The activation of ribulose-1,5-bisphosphate carboxylase by carbon dioxide and magnesium ions. Equilibria, kinetics, a suggested mechanism, and physiological implications. Biochemistry. 1976;15:529–536. doi: 10.1021/bi00648a012. [DOI] [PubMed] [Google Scholar]
- 51.Bahr JT, Jensen RG. Activation of ribulose bisphosphate carboxylase in intact chloroplasts by CO2 and light. Arch. Biochem. Biophys. 1978;185:39–48. doi: 10.1016/0003-9861(78)90141-8. [DOI] [PubMed] [Google Scholar]
- 52.Lorimer GH. Evidence for the existence of discrete activator and substrate sites for CO2 on ribulose-1,5-bisphosphate carboxylase. J. Biol. Chem. 1979;254:5599–5601. [PubMed] [Google Scholar]
- 53.Makino A, Mae T. Photosynthesis and plant growth at elevated levels of CO2. Plant Cell Physiol. 1999;40:999–1006. doi: 10.1093/oxfordjournals.pcp.a029493. [DOI] [Google Scholar]
- 54.Raines CA. The Calvin cycle revisited. Photosynth. Res. 2003;75:1–10. doi: 10.1023/A:1022421515027. [DOI] [PubMed] [Google Scholar]
- 55.Ding F, Wang M, Zhang S, Ai X. Changes in SBPase activity influence photosynthetic capacity. growth, and tolerance to chilling stress in transgenic tomato plants. Sci. Rep. 2016;6:32741. doi: 10.1038/srep32741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lefebvre S, et al. Increased sedoheptulose-1,7-bisphosphatase activity in transgenic tobacco plants stimulates photosynthesis and growth from an early stage in development. Plant Physiol. 2005;138:451–460. doi: 10.1104/pp.104.055046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harrison EP, Willingham NM, Lloyd JC, Raines CA. Reduced sedoheptulose-1,7-bisphosphatase levels in transgenic tobacco lead to decreased photosynthetic capacity and altered carbohydrate accumulation. Planta. 1998;204:27–36. doi: 10.1007/s004250050226. [DOI] [Google Scholar]
- 58.Raines CA, Harrison EP, Olcer H, Lloyd JC. Investigating the role of the thiol-regulated enzyme sedoheptulose-1,7-bisphosphatase in the control of photosynthesis. Physiol. Plant. 2000;110:303–308. doi: 10.1034/j.1399-3054.2000.1100303.x. [DOI] [Google Scholar]
- 59.Hartman FC, Harpel MR. Structure, function, regulation, and assembly of D-ribulose-1,5-bisphosphate carboxylase/oxygenase. Annu. Rev. Biochem. 1994;63:197–234. doi: 10.1146/annurev.bi.63.070194.001213. [DOI] [PubMed] [Google Scholar]
- 60.Spreitzer RJ. Genetic dissection of rubisco structure and function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1993;44:411–434. doi: 10.1146/annurev.pp.44.060193.002211. [DOI] [Google Scholar]
- 61.Martin W, Brinkmann H, Savonna C, Cerff R. Evidence for a chimeric nature of nuclear genomes: eubacterial origin of eukaryotic glyceraldehyde-3-phosphate dehydrogenase genes. Proc. Natl. Acad. Sci. USA. 1993;90:8692–8696. doi: 10.1073/pnas.90.18.8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koksharova O, Schubert M, Shestakov S, Cerff R. Genetic and biochemical evidence for distinct key functions of two highly divergent GAPDH genes in catabolic and anabolic carbon flow of the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 1998;36:183–194. doi: 10.1023/A:1005925732743. [DOI] [PubMed] [Google Scholar]
- 63.Liang F, Lindblad P. Effects of overexpressing photosynthetic carbon flux control enzymes in the cyanobacterium Synechocystis PCC 6803. Metab. Eng. 2016;38:56–64. doi: 10.1016/j.ymben.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 64.Keller Y, Bouvier F, D’Harlingue A, Camara B. Metabolic compartmentation of plastid prenyllipid biosynthesis–evidence for the involvement of a multifunctional geranylgeranyl reductase. Eur. J. Biochem. 1998;251:413–417. doi: 10.1046/j.1432-1327.1998.2510413.x. [DOI] [PubMed] [Google Scholar]
- 65.Shpilyov AV, Zinchenko VV, Shestakov SV, Grimm B, Lokstein H. Inactivation of the geranylgeranyl reductase (ChlP) gene in the cyanobacterium Synechocystis sp. PCC 6803. Biochim. Biophys. Acta. 2005;1706:195–203. doi: 10.1016/j.bbabio.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 66.Helman Y, et al. Genes encoding A-type flavoproteins are essential for photoreduction of O2 in cyanobacteria. Curr. Biol. 2003;13:230–235. doi: 10.1016/S0960-9822(03)00046-0. [DOI] [PubMed] [Google Scholar]
- 67.Clarke DJ, et al. Subdivision of the bacterioferritin comigratory protein family of bacterial peroxiredoxins based on catalytic activity. Biochemistry. 2010;49:1319–1330. doi: 10.1021/bi901703m. [DOI] [PubMed] [Google Scholar]
- 68.Stork T, Michel KP, Pistorius EK, Dietz KJ. Bioinformatic analysis of the genomes of the cyanobacteria Synechocystis sp. PCC 6803 and Synechococcus elongatus PCC 7942 for the presence of peroxiredoxins and their transcript regulation under stress. J. Exp. Bot. 2005;56:3193–3206. doi: 10.1093/jxb/eri316. [DOI] [PubMed] [Google Scholar]
- 69.Das PK, Bagchi SN. Role of bacterioferritin comigratory protein and glutathione peroxidase-reductase system in promoting bentazone tolerance in a mutant of Synechococcus elongatus PCC7942. Protoplasma. 2012;249:65–74. doi: 10.1007/s00709-011-0262-9. [DOI] [PubMed] [Google Scholar]
- 70.Yamamoto H, et al. Thioredoxin peroxidase in the Cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 1999;447:269–273. doi: 10.1016/S0014-5793(99)00309-9. [DOI] [PubMed] [Google Scholar]
- 71.Gao Y, et al. Identification of the proteomic changes in Synechocystis sp. PCC 6803 following prolonged UV-B irradiation. J. Exp. Bot. 2009;60:1141–1154. doi: 10.1093/jxb/ern356. [DOI] [PubMed] [Google Scholar]
- 72.Mishra S, Jha AB, Dubey RS. Arsenite treatment induces oxidative stress, upregulates antioxidant system, and causes phytochelatin synthesis in rice seedlings. Protoplasma. 2011;248:565–577. doi: 10.1007/s00709-010-0210-0. [DOI] [PubMed] [Google Scholar]
- 73.Sharma P, Jha AB, Dubey RS, Pessarakli M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012;2012:26. [Google Scholar]
- 74.el-Mansi EMT, Holms WH. Control of carbon flux to acetate excretion during growth of Escherichia coli in batch and continuous cultures. J. Gen. Microbiol. 1989;135:2875–2883. doi: 10.1099/00221287-135-11-2875. [DOI] [PubMed] [Google Scholar]
- 75.Wolfe AJ. The acetate switch. Microbiol. Mol. Biol. Rev. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jose S, Suraishkumar GK. High carbon (CO2) supply leads to elevated intracellular acetyl CoA levels and increased lipid accumulation in Chlorella vulgaris. Algal Res. 2016;19:307–315. doi: 10.1016/j.algal.2016.08.011. [DOI] [Google Scholar]
- 77.Sun Z, Chen YF, Du J. Elevated CO2 improves lipid accumulation by increasing carbon metabolism in Chlorella sorokiniana. Plant Biotechnol. J. 2016;14:557–566. doi: 10.1111/pbi.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sandrini G, et al. Rapid adaptation of harmful cyanobacteria to rising CO2. Proc. Natl. Acad. Sci. USA. 2016;113:9315–9320. doi: 10.1073/pnas.1602435113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ungerer J, Wendt KE, Hendry JI, Maranas CD, Pakrasi HB. Comparative genomics reveals the molecular determinants of rapid growth of the cyanobacterium Synechococcus elongatus UTEX 2973. Proc. Natl. Acad. Sci. USA. 2018;115:E11761–E11770. doi: 10.1073/pnas.1814912115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin PC, Pakrasi HB. Engineering cyanobacteria for production of terpenoids. Planta. 2018 doi: 10.1007/s00425-018-3047-y. [DOI] [PubMed] [Google Scholar]
- 81.Jazmin LJ, et al. Isotopically nonstationary 13C flux analysis of cyanobacterial isobutyraldehyde production. Metab. Eng. 2017;42:9–18. doi: 10.1016/j.ymben.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guerreiro AC, et al. Daily rhythms in the cyanobacterium Synechococcus elongatus probed by high-resolution mass spectrometry-based proteomics reveals a small defined set of cyclic proteins. Mol. Cell. Proteomics. 2014;13:2042–2055. doi: 10.1074/mcp.M113.035840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shilov IV, et al. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics. 2007;6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- 84.Vizcaino JA, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zavřel T, Sinetova MA, Červený J. Measurement of Chlorophyll a and Carotenoids Concentration in Cyanobacteria. Bio-protocol. 2015;5:e1467. doi: 10.21769/BioProtoc.1467. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.