Abstract
Purpose
To clinically use the UCSF500, a pancancer, next-generation sequencing assay in uveal melanoma (UM) and to correlate results with gene expression profiling (GEP) and predictive factors for metastasis.
Methods
Cohort study. Tumor samples of adult UM patients were analyzed with the UCSF500 and GEP. Main outcomes were copy number changes in chromosomes 1, 3, 6, and 8 and mutations in GNAQ, GNA11, SF3B1, EIF1AX, BAP1, SRSF2, U2AF1, and PLCB4. Chromosome 3 loss (a metastasis predictor) was tested for correlation with GEP class, tumor characteristics (largest basal diameter, thickness, ciliary body involvement, and extraocular extension), and histology (presence of epithelioid cells, closed loops, and mitotic count).
Results
The 62 patients had a mean age of 59 years (range, 24–89 years). Chromosome 3 loss was detected in 30 patients and was associated with larger basal tumor diameter (Wilcoxon rank sum test, P = 0.015), greater thickness (Wilcoxon rank sum test, P = 0.016) and tumor, node, metastasis stage (Fisher test, P = 0.006), epithelioid cytology (Fisher test, P < 0.001), BAP1 mutation (Fisher test, P < 0.001), and chromosome 8q gain (Fisher test, P < 0.001). Class 2 tumors were much more likely to have chromosome 3 loss than class 1 (odds ratio, 121; P < 0.001). Eleven patients developed metastatic UM, of which five died during the study. All metastatic cases had chromosome 3 loss, 8 gain, BAP1 mutation, and class 2 GEP. Five class 1 tumors had chromosome 3 loss.
Conclusions
UCSF500 detects chromosomal copy number changes and missense mutations that correlate strongly with metastasis predictors, including GEP.
Translational Relevance
Next-generation sequencing of UM should enhance survival prognostication.
Keywords: uveal melanoma, choroidal melanoma, next generation sequencing, prognostication
Introduction
Approximately 50% of patients with uveal melanoma (UM) develop metastatic disease, which is almost always fatal. Using multivariable analysis including patient age, sex, clinical features incorporated in the tumor, node, metastasis (TNM) staging system of the American Joint Committee on Cancer, histopathology, and cytogenetics has become the gold standard for personalized survival prognostication.1–6
Of all predictive factors, tumor genetic information remains the most informative, with chromosome 3 loss and BAP1 mutation, as well as class 2 gene expression profile (GEP) correlating most strongly with metastatic death.7,8 Chromosome 8 gain also confers increased mortality risk, with some studies suggesting that chromosome 3 loss and 8 gain must both be present for metastasis to occur.9,10 Chromosome 6p gain is associated with a relatively good prognosis and is rare in tumors with chromosome 3 loss. When chromosome 6p gain coexists with chromosome 3 loss, survival is better than with monosomy 3 alone.10,11
More than 85% of UMs show GNAQ or GNA11 mutation, which are mutually exclusive, initiating oncogenic events, and are, therefore, useful in differentiating UM from other lesions such as choroidal metastases.12,13 Loss-of-function mutations in the BAP1 tumor suppressor gene on chromosome 3 are found in 49% of UM and are strongly correlated with metastasis.14 Mutations in EIF1AX and SF3B1, which also tend to occur in a mutually exclusive pattern, are associated with low and intermediate risk, respectively.15–19 Other less common UM mutations include SRSF2, U2AF1, PLCB4, and CYSLTR2.18,20,21 CYSLTR2 and PLCB4 occur mutually exclusively with GNAQ and GNA11 as initiating oncogenic events and do not influence metastatic behavior. SRSF2 and U2AF1 appear to function as splicing factors akin to SF3B1.18
The UCSF500 assay uses hybrid capture enrichment of target DNA to interrogate approximately 500 genes that are frequently mutated in cancers, including genes known to be altered in UM. This test also detects genome-wide chromosomal copy number changes using the off-target sequencing reads.22 It has been in clinical use at University of California, San Francisco (UCSF) since 2013 and is used for a wide variety of solid and blood cancers.23
The aims of this study were to determine the clinical use of the UCSF500 assay in UM patients and to correlate results with known genetic, clinical, and histopathologic predictors of metastasis. It was not our intention to identify novel prognostic markers but rather to reconfirm known correlations and to determine the clinical utility of the UCSF500 assay for UM. We compared genetic alterations including prognostically relevant copy number changes of chromosomes. Chromosome 3 loss (a metastasis predictor) was correlated with known genetic aberrations in UM, such as GNAQ, GNA11, SF3B1, EIF1AX, and BAP1 mutations, GEP class, and with established clinical and histopathologic parameters associated with metastatic risk. Results here are based on our experience using the UCSF500 for UM since 2013, with preliminary findings presented in 2016.24 To our knowledge, this is the first study to report both next-generation sequencing (NGS) and GEP results in a cohort of UM patients.
Methods
Patients
The 62 patients in this study were recruited prospectively from the UCSF Ocular Oncology Service, starting in August of 2013. Patients were included if treated with proton beam radiotherapy, plaque brachytherapy, enucleation, or endoresection. They were excluded if they had received prior radiotherapy, in case radiotherapy might have influenced genetic testing results. Four patients who were invited to participate in the study declined to have any genetic testing of their tumors. This study was conducted in accordance with the Declaration of Helsinki and Good Clinical Practice Guidelines, under the approval of the UCSF Institutional Review Board (CC number 14852). Consent for the use of tissues and data for research was obtained from all patients.
Clinical Methods
Clinical evaluation included full ophthalmologic examination and measurement of largest basal tumor diameter and tumor thickness by B-scan echography (Eyecubed; Ellex, Adelaide, Australia). Systemic clinical examination was performed in a routine manner with abdominal magnetic resonance imaging performed both with and without contrast at time of diagnosis and every 6 months thereafter. Postoperatively, patients were referred to a medical oncologist for systemic screening.
Tumors were categorized as involving ciliary body if they extended anterior to ora serrata. Extraocular extension was recorded as being present if this was noted clinically or histopathologically.
Tumor Sampling
Tumors treated by proton beam radiotherapy or brachytherapy were sampled by transscleral fine-needle aspiration or transretinal vitrector biopsy25 (25- or 27-gauge). All biopsies were performed prior to radiation at the time of tantalum marker or plaque insertion. For eyes undergoing enucleation or endoresection, a fresh tumor sample was obtained at the time of surgery. Tissue samples were provided to the UCSF Clinical Cancer Genomics Laboratory for DNA extraction, library preparation, and sequencing and to Castle Biosciences (Friendswood, TX) for GEP.
Histopathologic Examination
The diagnosis of melanoma was confirmed by light microscopy on sections stained with hematoxylin and eosin and, if necessary, by Melan A immunohistochemistry. Melanoma cells were categorized as spindle, epithelioid, or mixed, according to the modified Callender system.26 For statistical analysis, tumors were dichotomized as either having no epithelioid cells (spindle cell morphology) or having the presence of any epithelioid cells (mixed or epithelioid morphology).
Genetic Analysis
Next-Generation Sequencing
The UCSF500 assay was performed at the UCSF Clinical Cancer Genomics Laboratory. This assay uses solution phase hybrid capture using a custom bait library (NimblegenSeqCap)27 of 2.8 Mbp of genomic sequence and includes all exons of approximately 500 cancer-related genes and selected introns of 40 of these genes. In addition, the assay captures 2,000 unique sequences containing common single nucleotide polymorphisms within regions devoid of constitutional copy number variations to assist in genome-wide copy number and allelic imbalance analysis.
Barcoded samples were pooled, and captured libraries were sequenced with a read length of 100 nucleotides from both directions at >500-fold coverage on a single lane of an Illumina HiSeq 2500 instrument. Duplicate sequencing reads were removed computationally to allow for accurate allele frequency determination and copy number calling. The analysis was based on the human reference sequence UCSC build hg19 (NCBI build 37) by using the following software packages: BWA, 0.7.10-r789; Samtools, 1.1 (using htslib 1.1); Picard tools, 1.97 (1504); GATK, 2014.4-3.3.0-0-ga3711; CNVkit, 0.3.3; Pindel, 0.2.5a7; SATK, 2013.1-10-gd6fa6c3; Annovar, v2015Mar22; Freebayes, 0.9.20; and Delly, 0.5.9. Copy number changes were calculated from on- and off-target reads using CNVkit.22,28–37
Gene Expression Profiling
Tumor samples were immediately placed after biopsy or harvesting in provided fixative and packed with dry ice and sent to Castle Biosciences for gene-expression profiling, with methods as previously reported.8
Statistical Analysis
All data analyses were performed using the R statistical language.38 To test associations between chromosome 3 loss and other clinical and other genetic factors, the exact Wilcoxon rank sum test39 and the Fisher's exact test were used for continuous and categorical variables, respectively. The odds ratios and corresponding confidence intervals were estimated using the fisher.test function.
All categorical variables were dichotomized except for TNM stage, which was grouped into four levels: T1, T2, T3, and T4. The Cochrane-Armitage test38 was used to test for trend of TNM stage with chromosome 3 status. All tests were two-sided, with P values less than 0.05 regarded as significant. Kaplan-Meier curves were plotted. The log-rank test was performed to demonstrate associations between metastasis-free survival and chromosome 3 loss, chromosome 8q gain, BAP1 mutation, and GEP class.
Results
Patient Demographics
Of the 62 patients, 36 were male and 26 were female. Fifty-three patients were white, 6 were Asian, and 3 patients were Hispanic (Table 1). The mean age was 58.56 years (SD, 13.24; range, 24–89 years).
Table 1.
Patient Demographics, Tumor Clinical Features, TNM Stage, and Treatmenta
| Patient Information |
Value, N = 62 |
| Demographics | |
| Mean age (median, range), years | 58.65 (58, 24.00–89.00) |
| Sex | |
| Male | 35 (57) |
| Female | 26 (43) |
| Race | |
| White | 53 (85) |
| Asian | 6 (10) |
| Hispanic | 3 (5) |
| Tumor clinical features | |
| Affected eye | |
| Right | 38 (61) |
| Left | 24 (39) |
| Location | |
| Choroidal | 54 (87) |
| Ciliary body | 3 (5) |
| Ciliochoroidal | 5 (8) |
| Extraocular extension | 3 (5) |
| TNM stage (all N0M0) | |
| T1 | 18 (29) |
| T1a | 17 |
| T1b | 1 |
| T2 | 19 (31) |
| T2a | 18 |
| T2b | 1 |
| T3 | 19 (31) |
| T3a | 18 |
| T3b | 1 |
| T4 | 6 (10) |
| T4b | 3 |
| T4c | 3 |
| Ophthalmic treatment | |
| Proton beam radiotherapy | 34 (55) |
| Enucleation | 18 (29) |
| Local resection | 8 (13) |
| Plaque brachytherapy | 2 (3) |
Values shown are number (nearest %) except where indicated.
Baseline Study Eye Characteristics
The tumors involved the right eye in 38 patients and the left eye in 24, with ciliary body involvement in 8 patients, and extraocular extension in 3. The mean largest basal tumor diameter was 11.94 mm (range, 4.80–20.00) and the median tumor thickness was 5.65 mm (range, 0.69–16.00). The TNM stage was T1 in 18 patients, T2 in 19, T3 in 19, and T4 in 6 (Table 1). All were N0M0 at diagnosis.
Histopathologic and Genetic Results
The laboratory results are summarized in Figure 1. The tumors were classified as spindle cell in 25 patients, epithelioid in 15, and mixed in 19. Histology was inconclusive in three patients, because of pauci-cellular samples.
Figure 1.
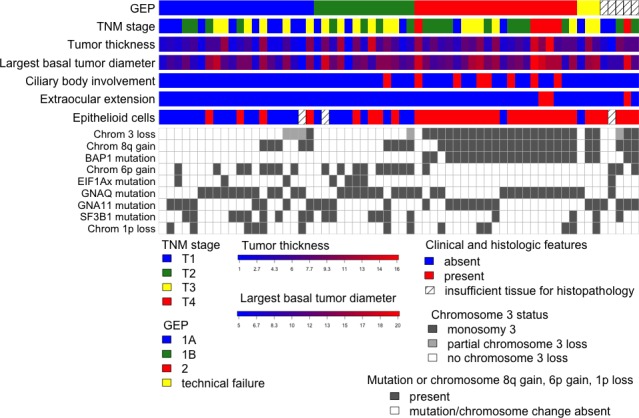
Tiling plot including clinical, histopathologic, and genetic features for all patients.
Next-Generation Sequencing
Chromosome 3 loss (monosomy 3) was seen in 30 (48%) tumors, with 5 of these showing partial loss of chromosome 3. Chromosome 8q gain and 6p gain were seen in 32 (52%) and 21 (34%) tumors, respectively. Genome-wide copy chromosomal copy number changes in two representative cases are shown in Figure 2. GNAQ and GNA11 mutations were present in 36 (58%) and 26 (42%) tumors, respectively, and were mutually exclusive. BAP1 inactivating mutations were present in 23 (37%) patients. SF3B1 mutations and EIF1AX mutations were detected in 19 (31%) and 8 (13%) tumors, respectively. No tumors had SRSF2, U2AF1, or PLCB4 mutations.
Figure 2.
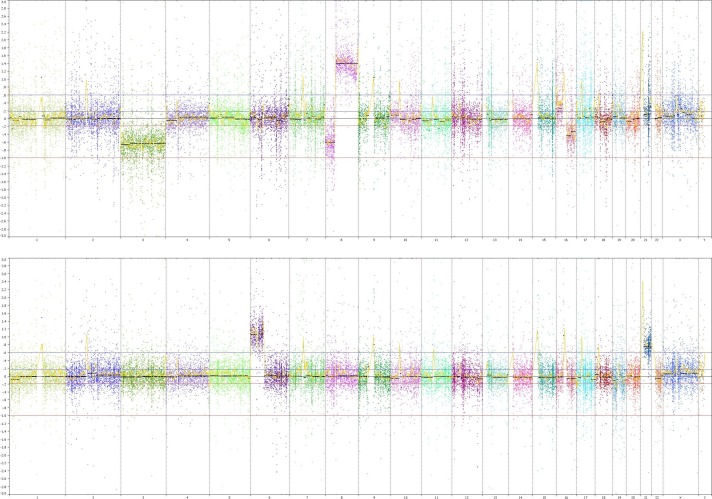
Genome-wide copy number profiles for two representative cases. Shown are the raw log2 ratios of the normalized bin counts from CNVKit (y-axis) along genomic coordinates (x-axis) with segments obtained by circular binary segmentation (black lines) Top: Copy number alterations include losses of chromosomes 3, 8p, and 16q and gain of 8q in a UM that also carried a GNAQ p.Q209P and a truncating BAP1 p.Q4* mutation. Bottom: Copy number gains of 6p and 21 in a tumor with an GNAQ p.Q209L mutation.
Gene Expression Profiling
Of the 62 tests performed by Castle Biosciences, three were categorized as “technical failure.” In five patients, GEP was not performed. This was because of transport problems in one case, with two patients declining Castle testing, and two samples (both from tumors <1 mm thick) considered too small for both types of genetic analysis so that only UCSF500 was performed.
Treatment
Treatment consisted of proton beam radiotherapy in 34 patients, enucleation in 18, endoresection in 8, and plaque brachytherapy in 2 patients (Table 1).
Biopsy Technique
Transretinal biopsy (either with vitrector after tantalum marker insertion or during endoresection) was performed in 36 patients and transscleral fine-needle aspiration in 8, with tumor harvesting after enucleation in 18 patients.
Correlation of Genetic Alterations With Predictors of Metastasis
UCSF500 results and clinical and histopathologic features are presented in Figure 1 and Table 2.
Table 2.
Tumor Histology and Genetics
| Variable |
Value, N (%) |
| Histology, N = 59 | |
| Spindle cell | 25 (42) |
| Epithelioid | 15 (25) |
| Mixed | 19 (32) |
| Genetics—UCSF500, N = 62 | |
| Mutations of relevance | |
| GNAQ | 36 (58) |
| GNA11 | 26 (42) |
| SF3B1 | 19 (31) |
| EIF1AX | 8 (13) |
| BAP1 | 23 (37) |
| Chromosomal aberrations | |
| Chromosome 1 loss | 14 (23) |
| Chromosome 3 loss (any) | 30 (48) |
| Chromosome 3 loss (partial) | 5 (8) |
| Chromosome 6 gain | 21 (34) |
| Chromosome 8 gain | 32 (52) |
| Genetics—gene expression profiling, N = 54 | |
| Class 1A | 21 (39) |
| Class 1B | 13 (24) |
| Class 2 | 20 (36) |
| Test result of technical failure | 3 (37) |
| Test not done (UCSF500 done only) | 5 |
The associations between chromosome 3 loss (partial or total) and clinical, histopathologic, and other genetic alterations are presented in Table 3. Chromosome 3 loss was more frequent in tumors with larger basal diameter (Wilcoxon rank sum test, P = 0.015), greater thickness (P = 0.016), more advanced TNM stage (P = 0.006), presence of any epithelioid cells (P < 0.001), BAP1 mutation (P < 0.001), and chromosome 8q gain (P < 0.001). There was no statistically significant association found between chromosome 3 loss and the mutational status of GNAQ (P = 0.200), GNA11 (P = 0.200), or SF3B1 (P = 0.280). An inverse association was seen between chromosome 3 loss and chromosome 6p gain (P < 0.001).
Table 3.
Association of Clinical and Genetic Variables With Chromosome 3 Lossa
| Variable |
P Value |
OR |
95% CI |
| Clinical features | |||
| Largest basal diameter | 0.015 | N/A | |
| Thickness | 0.016 | N/A | |
| TNM stage | 0.006 | N/A | |
| Epithelioid cells (any) | <0.001 | 19.07 | (4.30–123.20) |
| Ciliary body involvement | 0.131 | 3.96 | (0.63–43.55) |
| Extraocular extension | 0.097 | ∞ | (0.48–∞) |
| Genetics—UCSF500 | |||
| Mutations of relevance | |||
| GNAQ | 0.198 | 0.47 | (0.15–1.46) |
| GNA11 | 0.198 | 2.12 | (0.68–6.79) |
| SF3B1 | 0.409 | 0.56 | (0.16–1.91) |
| EIF1AX | 0.054 | 0.14 | (0.003–1.18) |
| BAP1 | <0.001 | ∞ | (21.75–∞) |
| Chromosomal aberrations | |||
| Chromosome 1 loss | 0.544 | 1.70 | (0.44–6.95) |
| Chromosome 6 gain | <0.001 | 0.15 | (0.03–0.59) |
| Chromosome 8 gain | <0.001 | 35.51 | (7.60–244.70) |
| Genetics—gene expression profiling | |||
| Class 2 vs. class 1 | <0.001 | 120.80 | (13.65–5,905.96) |
Significant associations are in bold. OR, odds ratio; CI, confidence interval; N/A, not applicable.
Class 2 GEP tumors were much more likely to have chromosome 3 loss than class 1 GEP tumors (odds ratio 121, P = 6.2 × 10−10).
Association With Metastasis-Free Survival
The median metastasis-free survival time was 27 months (range, 6–60). Metastatic disease developed in 11 (18%) patients, 5 (8%) of whom died. All patients with metastases had chromosome 3 loss, chromosome 8q gain, BAP1 mutation, and class 2 GEP. Chromosome 3 loss (P = 6.54 × 10−5) alone and combined chromosome 3 loss, chromosome 8q gain, and BAP1 mutation (P = 5 × 10−7) were associated with metastatic disease. Kaplan-Meier curves (Figure 3) show that the median survival was 37 months in patients whose tumors showed chromosome 3 loss and 35 months in patients whose tumor showed combined chromosome 3 loss, chromosome 8q gain and BAP1 mutation (log rank test, P < 0.001).
Figure 3.
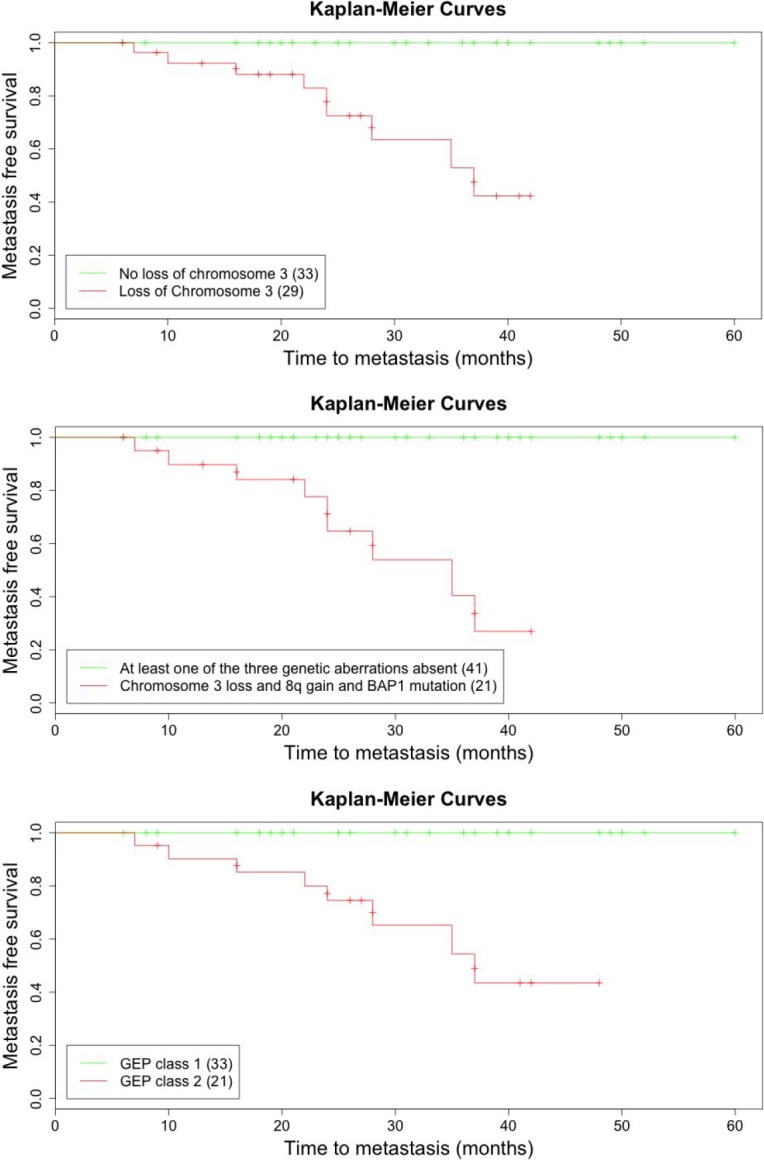
Kaplan-Meier curves demonstrating time to metastasis. Top: Time to metastasis in patients according to chromosome 3 loss, detected with the UCSF500 assay. Middle: Time to metastasis in patients with combination of chromosome 3 loss, 8 gain and BAP1 mutation, and time to metastasis with at least one of these three genetic features absent. Bottom: Time to metastasis in patients according to GEP class (class 1 vs. class 2).
Discussion
Main Findings
Genetic predictors for metastasis that were detected with the UCSF500 assay correlated closely with clinical and histologic risk factors, as well as GEP results. The UCSF500 assay had a low failure rate, even with small tumor biopsy samples, providing meaningful genetic results in all 62 patients tested. Its deployment was straightforward in routine clinical practice.
Discussion of Methods
Targeted sequencing of DNA can detect multiple types of genetic alterations, including single nucleotide variants, small insertions/deletions, structural rearrangements, and copy number alterations, thereby providing a richer view of relevant genetic alterations compared to fluorescence in situ hybridization and single gene sequencing.2 The method is quantitative and sensitive enough to detect melanoma mutations, even in the presence of a considerable fraction of stromal cells. The low complexity of the genetic landscape of UMs with highly recurrent mutations at hotspots of a few genes allows for accurate quantitation of tumor fractions, providing a potential advantage over gene expression analyses.
An additional advantage of UCSF500 is its ability to confirm that the biopsy specimen is of uveal melanocytic origin, by detecting GNAQ and GNA11 mutations. The richness of molecular data provided by the UCSF500 may assist in diagnostic accuracy for other tumors. In a recent study using the UCSF500 in pediatric neurooncologic patients, the diagnosis of six tumors (19%) was revised after genomic profiling, including high-grade glioma to pilocytic astrocytoma, medulloblastoma to pineoblastoma, ependymoma to high-grade glioma, and medulloblastoma to central nervous system high-grade neuroepithelial tumor with BCOR alteration.23
An advantage of this pancancer assay is also that genetic mutations of relevance to a variety of cancers can also be added to the assay as new discoveries are made. The version of the assay in this study investigated mutations in GNAQ, GNA11, SF3B1, EIF1AX, BAP1, and PCLB4. Since the study, U2AF1 and CYSLTR2 have been added. In addition, although all sequencing was performed on fresh tumor specimens, the UCSF500 assay can also be used to analyze formalin-fixed, paraffin-embedded tissue,23 although this was not needed in this study because fresh samples were available for all patients.
Discussion of NGS Results
The association of genetic alterations, as detected with NGS, with known predictors of metastasis, listed in Table 3, are in keeping with the published literature. Chromosome 3 loss was associated with BAP1 mutation, chromosome 8q gain, largest basal tumor diameter, tumor thickness, TNM stage, and presence of epithelioid cells.1,8,9,40,41 As anticipated, chromosome 3 loss was inversely associated with chromosome 6p gain.9,15,16,17,18 No PCLB4, SRF2, or U2AF1 mutations were identified. Although CYSLTR2 was not included in the UCSF500 at the time of the study, all tumors sequenced had GNAQ and GNA11 mutations. Because CYSLTR2 is mutually exclusive of these mutations, it is unlikely the lack of the gene in the assay affected study results.21
As expected, all 11 tumors that metastasized during the study period had chromosome 3 loss, chromosome 8q gain, BAP1 mutation, and class 2 GEP.9,11,41 Interestingly, the combination of chromosome 3 loss and chromosome 8 gain has also been shown to confer increased metastatic risk in oral cancer.42
No significant association between SF3B1 mutation and chromosome 3 loss was seen. There is a lack of consensus on the prognostic value of the SF3B1 mutation, with some reports indicating lower metastatic risk15,16,18,43 and another indicating potentially increased risk of late metastasis.19 Longer-term follow up will be needed to determine the prognostic relevance of SF3B1 mutations.
The tissue sample was sufficient for NGS in all 62 tumors. Six of these tumors were less than 2 mm in thickness, with two less than 1 mm in thickness (i.e., 0.69 mm and 0.74 mm, respectively). Although the histopathology was interpreted as inconclusive due to paucicellular sample in three of these cases, NGS identified GNAQ or GNA11 mutations in all of these, providing evidence that the neoplasms were of melanocytic lineage. In two of these tumors with inconclusive histology, NGS also confirmed malignancy by identifying chromosome 6p gain in one tumor and SF3B1 mutation, loss of chromosome 1p, partial chromosome 3 deletion, and chromosome 8q gain in the other tumor. The third tumor in which histology failed was 8.80 mm thick and sampled transsclerally by fine-needle aspiration biopsy. The specimen was inadequate for histology probably because too much was used for genetic studies. In any case, NGS provided a diagnosis of melanoma by demonstrating GNA11 and SF3B1 mutations as well as chromosome 6p gain.
NGS Versus GEP
Notably, there was concordance between GEP class and NGS sequencing result in all class 2 tumors, which showed loss of chromosome 3 (P = 6 × 10−10) and the majority (18 of 21 class 2 tumors) had chromosome 8 gain. Surprisingly, 4 of 20 (20%) class 1A tumors and 1 of 13 (8%) class 1B tumors showed chromosome 3 loss. Of these class 1 tumors with chromosome 3 loss, only one patient had true monosomy 3; the other four patient specimens demonstrated partial/focal loss of chromosome 3, sparing the BAP1 locus. Long-term follow up is needed to determine if partial chromosome 3 loss will lead to increased metastatic risk in these patients.
Strengths and Weaknesses of Study
Strengths of this study are the prospective data collection and the large number of patients. The main weakness is the short follow up, which resulted in few patients with metastatic disease and prediction of time to metastasis.
Comparison With Other Studies
The results obtained with NGS in this study are comparable to those reported with microsatellite analysis, fluorescence in situ hybridization, GEP, and other genetic methods used to analyze UM.1,2,8,9,43,44 A study by Smit et al.45 demonstrated that an NGS kit developed in Rotterdam, The Netherlands, specifically for UM showed concordant results with other methods of genetic analysis.45 This was done with a bank of carefully selected specimens and not in routine clinical practice, unlike our study. The Rotterdam panel analyzed 98 amplicons, which included 17 amplicons covering highly polymorphic regions in chromosomes 1 and 8, and 21 amplicons on chromosome 3, with the remainder covering all exons on the BAP1 gene and the exons containing known mutation hotspots on GNAQ, GNA11, EIF1AX, and SF3B1. In comparison, the UCSF500 assay provides substantially higher resolution of copy number changes; it uses hybrid capture rather than polymerase chain reaction amplification for enrichment, providing a more accurate assessment of copy number alterations.22 Although the preferred input for UCSF500 extracted from formalin-fixed paraffin-embedded tissue is 100 ng to yield a median coverage of 200 or more unique sequence reads, it can reliably detect UM typical alterations in unfixed samples with 10 ng of input material or less.
Conclusions
When used in routine clinical practice, the UCSF500 test successfully identifies known genetic predictors of metastasis in UM.
The strong concordance between UCSF500 results and known genetic, clinical, and histopathologic predictors suggests that the UCSF500 assay is a useful prognostic test in patients with UM. As with other genetic methods, the NGS results only indicate whether the UM is likely to have metastasized; multivariate analysis that includes anatomic, histopathologic, and genetic predictors is required to provide more accurate assessment of an individual patient's life expectancy.46
Acknowledgments
Supported in part by That Man May See, Inc., San Francisco, CA, an unrestricted grant and a Career Development Award from Research to Prevent Blindness, New York, NY (ARA); grants EY002162 (Core Grant for Vision Research) and 1K23EY027466 (K23 Mentored Patient-Oriented Research Career Development Award to ARA) from the National Eye Institute, Bethesda, MD; a National Cancer Institute Outstanding Investigatory Award (1R35CA220481) to BCB; and Cancer Center Support Grant 5P30CA082103 (RR and ABO) from the National Cancer Institute, Bethesda, MD.
Disclosure: A.R. Afshar, None; B.E. Damato, None; J.M. Stewart, None; L.B. Zablotska, None; R. Roy, None; A.B. Olshen, None; N.M. Joseph, None; B.C. Bastian, None
References
- 1.Damato B, Eleuteri A, Taktak AFG, Coupland SE. Estimating prognosis for survival after treatment of choroidal melanoma. Prog Retin Eye Res. 2011;30:285–295. doi: 10.1016/j.preteyeres.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Damato B, Duke C, Coupland SE, et al. Cytogenetics of uveal melanoma: a 7-year clinical experience. Ophthalmology. 2007;114:1925–1931. doi: 10.1016/j.ophtha.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 3.Damato B, Eleuteri A, Fisher AC, Coupland SE, Taktak AF. Artificial neural networks estimating survival probability after treatment of choroidal melanoma. Ophthalmology. 2008;115:1598–1607. doi: 10.1016/j.ophtha.2008.01.032. [DOI] [PubMed] [Google Scholar]
- 4.Cook SA, Damato B, Marshall E, Salmon P. Psychological aspects of cytogenetic testing of uveal melanoma: preliminary findings and directions for future research. Eye(Lond) 200923:581–585. doi: 10.1038/eye.2008.54. [DOI] [PubMed] [Google Scholar]
- 5.Dogrusoz M, Bagger M, van Duinen SG, et al. The prognostic value of AJCC staging in uveal melanoma is enhanced by adding chromosome 3 and 8q status. Invest Ophthalmol Vis Sci. 2017;58:833–842. doi: 10.1167/iovs.16-20212. [DOI] [PubMed] [Google Scholar]
- 6.Finger PT. The 7th edition AJCC staging system for eye cancer: an international language for ophthalmic oncology. Arch Pathol Lab Med. 2009;133:1197–1198. doi: 10.5858/133.8.1197. [DOI] [PubMed] [Google Scholar]
- 7.Prescher G, Bornfeld N, Hirche H, et al. Prognostic implications of monosomy 3 in uveal melanoma. Lancet. 1996;347:1222–1225. doi: 10.1016/s0140-6736(96)90736-9. [DOI] [PubMed] [Google Scholar]
- 8.Onken MD, Worley LA, Char DH, et al. Collaborative Ocular Oncology Group report number 1: prospective validation of a multi-gene prognostic assay in uveal melanoma. Ophthalmology. 2012;119:1596–1603. doi: 10.1016/j.ophtha.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White VA, Chambers J, Courtright PD, et al. Correlation of cytogenetic abnormalities with the outcome of patients with uveal melanoma. Cancer. 1998;83:354e359. [PubMed] [Google Scholar]
- 10.Damato B, Dopierala JA, Coupland SE. Genotypic profiling of 452 choroidal melanomas with multiplex ligation-dependent probe amplification. Clin Cancer Res. 2010;16:6083–6092. doi: 10.1158/1078-0432.CCR-10-2076. [DOI] [PubMed] [Google Scholar]
- 11.Shields CL, Say EAT, Hasanreisoglu M, et al. Personalized prognosis of uveal melanoma based on cytogenetic profile in 1059 patients over an 8 year period. Ophthalmology. 2017;124:1523–1531. doi: 10.1016/j.ophtha.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Van Raamsdonk CD, Bezrookove V, Green G, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue nevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Raamsdonk CD, Griewank KG, Crosby MB, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harbour JW, Onken MD, Roberson ED, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin M, Masshofer L, Temming P, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet. 2013;45:933–936. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harbour JW, Roberson ED, Anbunathan H, et al. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat Genet. 2013;45:133–135. doi: 10.1038/ng.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ewens KG, Kanetsky PA, Richards-Yutz J, et al. Chromosome 3 status combined with BAP1 and EIF1AX mutation profiles are associated with metastasis in uveal melanoma. Invest Ophthalmol Vis Sci. 2014;55:5160–5167. doi: 10.1167/iovs.14-14550. [DOI] [PubMed] [Google Scholar]
- 18.Robertson AG, Shih J, Yau C, et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2017;32:204–220.e15. doi: 10.1016/j.ccell.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yavuzyiqitoqlu S, Koopmans AE, Verdijk RM, et al. Uveal melanomas with SF3B1 mutations: a distinct subclass associated with late-onset metastases. Ophthalmology. 2016;123:1118–1128. doi: 10.1016/j.ophtha.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 20.Johansson P, Aoude LG, Wadt K, et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget. 2016;7:4624–4631. doi: 10.18632/oncotarget.6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore AM, Ceraudo E, Sher JJ, et al. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat Genet. 2016;48:675–680. doi: 10.1038/ng.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Talevich ES, Shain AH, Bastian BC. CNVkit: genome wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol 2016; 12:e1004873. doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kline CN, Joseph NM, Grenert JP, et al. Targeted next-generation sequencing of pediatric neuro-oncology patients improves diagnosis, identifies pathogenic germline mutations, and directs targeted therapy. Neuro Oncol. 2017;19:699–709. doi: 10.1093/neuonc/now254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Afshar AR. Presented at the American Society of Retinal Specialists Annual Meeting. San Francisco, CA: 2016. “Next-generation sequencing of uveal melanoma.”. August 12. [Google Scholar]
- 25.Sen J, Groenewald C, Hiscott PS, et al. Transretinal choroidal tumor biopsy with a 25-gauge vitrector. Ophthalmology. 2006;113:1028–1031. doi: 10.1016/j.ophtha.2006.02.048. [DOI] [PubMed] [Google Scholar]
- 26.McLean IW, Foster WD, Zimmerman LE, et al. Modifications of Callender's classification of uveal melanoma at the Armed Forces Institute of Pathology. Am J Ophthalmol. 1983;96:502–509. doi: 10.1016/s0002-9394(14)77914-0. [DOI] [PubMed] [Google Scholar]
- 27.Wagle N, Berger MF, Davis MJ, et al. High- throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. 2015;10:1556–1566. doi: 10.1038/nprot.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rausch T, Zichner T, Schlattl A, et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i9. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Broad Institute; Picard: A set of tools (in Java) for working with next generation sequencing data in the BAM. Available at: http://broadinstitute.github.io/picard. [Google Scholar]
- 33.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv. 2012. (1207.3907 [q-bio.GN])
- 34.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye K, Schulz MH, Long Q, et al. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2016 Available at: https://www.R-project.org.
- 39.Hothorn T, Hornik K, van de Wiel MA, Zeileis A. A lego system for conditional interference. Am Stat. 2006;60:257–263. [Google Scholar]
- 40.Damato B, Coupland SE. A reappraisal of the significance of largest basal diameter of posterior uveal melanoma. Eye (Lond) 2009;23:2152–2160. doi: 10.1038/eye.2009.235. quiz 2161–2162. [DOI] [PubMed] [Google Scholar]
- 41.Kalirai H, Dodson A, Faqir S, et al. Lack of BAP1 protein expression in uveal melanoma is associated with increased metastatic risk and has utility in routine prognostic testing. Br J Cancer. 2014;111:1373–1380. doi: 10.1038/bjc.2014.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhattacharya A, Roy R, Snijders AM, Hamilton G. Two distinct routes to oral cancer differing in genome instability and risk for cervical node metastasis. Clin Cancer Res. 17:7024–7034. doi: 10.1158/1078-0432.CCR-11-1944. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Furney SJ, Pedersen M, Gentien D. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 2013;3:1122–1129. doi: 10.1158/2159-8290.CD-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Field MG, Durante MA, Anbunathan H, et al. Punctuated evolution of canonical genomic aberrations in uveal melanoma. Nat Comm. 2018;9:116. doi: 10.1038/s41467-017-02428-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smit KN, van Poppelen NM, Vaarwater J, et al. Combined mutation and copy-number variation detection by targeted next-generation sequencing in uveal melanoma. Mod Path. 2018;31:763–771. doi: 10.1038/modpathol.2017.187. [DOI] [PubMed] [Google Scholar]
- 46.Eleuteri A, Damato B, Coupland SE, Taktak A. Enhancing survival prognostication in patients with choroidal melanoma by integrating pathologic clinical and genetic predictors of metastases. Int J Biomed Eng Technol. 2012;8:18–35. [Google Scholar]