Abstract
Background
Advances in genomics have greatly improved the survival rate in cancer patients. However, due to genetic heterogeneity, pancreatic ductal adenocarcinoma (PDAC) is still difficult to diagnose early, and its survival rate is extremely low. Therefore, we identified biomarkers that predict the prognosis of PDAC patients using independent cohort data.
Materials and methods
To develop a novel prognostic biomarker, we used the gene expression and clinical data from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO). Kaplan–Meier survival curve using median values of genes as cutoff showed that EIF4G1 was the only statistically significant gene in the 3 cohorts. We analyzed the prognostic significance of EIF4G1 using the time-dependent area under the curve (AUC) of Uno’s C-index, the AUC value of the receiver operating characteristics (ROC) at 3 years, and multivariate Cox analysis. We also compared EIF4G1 levels between tumors and matched non-tumor tissues.
Results
EIF4G1 is the only prognostic gene in patients with PDAC, which was selected by Kaplan–Meier survival analysis. The survival curve showed that high expression of EIF4G1 was associated with poor prognosis of PDAC with a good discriminative ability in 3 independent cohorts. The risk stratifying ability of EIF4G1 was demonstrated by analyzing C-indices and AUC values. Multivariate Cox regression confirmed its prognostic significance. EIF4G1 expression was significantly higher in PDAC tissues than in the matched normal tissues.
Conclusion
EIF4G1 could be used as a novel prognostic marker for PDAC and to determine suitable treatment options.
Keywords: EIF4GI, pancreatic ductal adenocarcinoma, prognosis, GEO, TCGA
Introduction
Pancreatic cancer has a very poor prognosis and is difficult to detect early.1 Of pancreatic cancer cases, 90% are pancreatic ductal adenocarcinoma (PDAC).2 Only surgical treatment is known to be effective in patients with PDAC. Surgical resection is performed only in 10%–20% of the cases,3 because most cases are at an advanced stage at the time of diagnosis.4,5 Moreover, the 5-year survival rate is less than 10% because most patients show relapse or metastasis even if they undergo complete surgical resection.6 Therefore, biomarkers for PDAC that could predict the prognosis accurately and facilitate early diagnosis are indispensable.
As the importance of precision medicine has been emphasized recently, genomic research is active and its use is expanding from the bench to the bedside, in the actual diagnosis and treatment process.7,8 Through these efforts, public databases including The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), and others related to patients with various cancer types and their genomes have been developed, and studies have been actively conducted with these data. Using the data sets in these databases and our novel statistical methods, we can report a single gene or set of genes in a specific cancer that can predict prognosis.9,10
In the present study, we investigated whether a gene could be used as a biomarker to predict the prognosis of patients with PDAC based on 3 cohorts from TCGA and GEO. Finally, we found the only gene that could predict prognosis in PDAC. Furthermore, the prognosis was found to be stratified according to the gene expression level.
Materials and methods
Patients’ data and study design
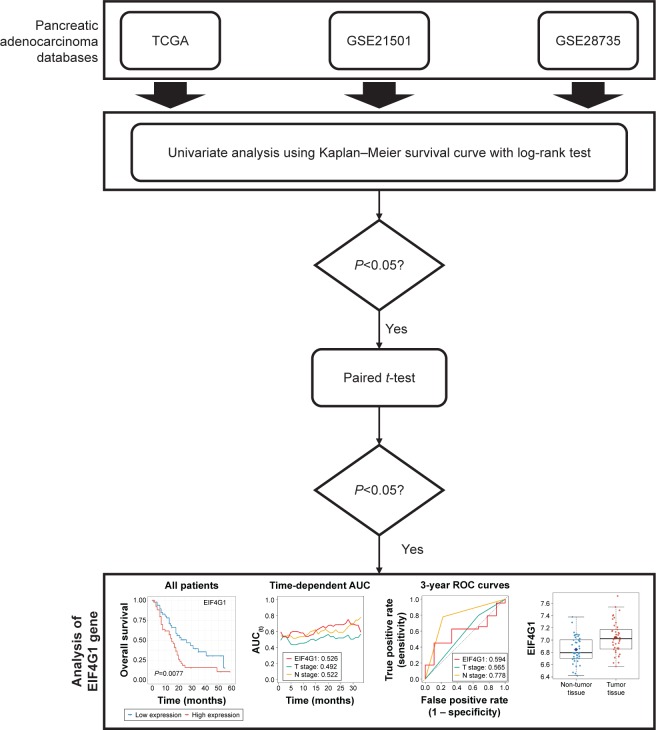
We investigated all pancreatic cancer cohorts in the GEO database and included only the GSE21501 and GSE28735 data sets in this study, because they contained survival information. The RNA-seq and microarray data and clinical data of PDAC were downloaded from TCGA,11,12 GSE21501,13 and GSE2873514 in March 2018. Patients lacking clinical information were excluded. We identified the prognostic significances of mRNAs in 3 independent cohorts. We then performed paired t-test or unpaired (Wilcoxon rank sum test) test to determine whether the statistically significant genes in all cohorts were increased in cancer tissues compared with those in normal tissues using the TCGA or GSE28735 cohorts. The overall process is described in Figure 1. These processes were performed in R software version 3.5.0 (The R Foundation for Statistical Computing, 2018) using the “cgdsr”, “TCGAbiolinks”, and “GEOquery” R packages.
Figure 1.
Study protocol.
Abbreviations: AUC, area under the curve; ROC, receiver operating characteristics; TCGA, The Cancer Genome Atlas.
Statistical analysis
Kaplan–Meier survival curves were used to identify the discriminatory power of EIF4G1. We determined the optimal cutoff value of the survival curve as described previously.4,15,16 Furthermore, we used 2 methods to evaluate biomarker performance as follows: 1) Uno’s C-index in the time-dependent area under the curve (AUC) analysis and 2) AUC values in receiver operating characteristic (ROC) curves at the 3-year mark as described in our previous studies.17,18 These values were calculated using the R packages “survival” and “survAUC”. The paired t-test in the GSE28735 or unpaired test in the TCGA was performed to analyze the EIF4G1 expression values between the matched tumor and non-tumor tissue samples using the “coin” package. We used uni- and multivariate Cox regression analyses to compare the effect of EIF4G1 on prognosis along with several clinical variables. Additionally, to identify the prognostic significance of miR-NAs in TCGA, we used the Oncomir (http://www.oncomir.org), which can analyze survival outcome about miRNAs.19
Results
To select the gene that could predict the prognosis of PDAC using public databases, the clinical and genetic information of 316 patients with PDAC from 3 independent cohorts (TCGA, n=172; GSE21501, n=102 and GSE28735, n=42) were downloaded and analyzed. The patient information used in the present study is detailed in Table 1. The patients in TCGA were almost diagnosed at an early stage, whereas the patients in GSE21501 were diagnosed almost at a late stage.
Table 1.
| Group | TCGA | GSE21501 | GSE28735 | |
|---|---|---|---|---|
| EIF4G1 | All patients | 172 | 102 | 42 |
| High expression (event) | 58 (40) | 54 (39) | 24 (16) | |
| Low expression (event) | 114 (52) | 48 (27) | 18 (13) | |
| Patients’ information | Male | 94 | – | – |
| Female | 78 | – | – | |
| Stages I and II | 164 | – | – | |
| Stages III and IV | 8 | – | – | |
| T1 and T2 | – | 18 | – | |
| T3 and T4 | – | 80 | – | |
| N0 | – | 28 | – | |
| N1 | – | 73 | – |
Abbreviation: TCGA, The Cancer Genome Atlas.
Prognostic performance of EIF4G1 in PDAC
We obtained the median value of gene expression for all genes in each cohort. Each cohort was divided into 2 groups based on the median value of each gene. The survival of the 2 groups was compared using Kaplan–Meier survival analysis, and statistically significant genes were extracted for each cohort. Among the commonly extracted genes in all 3 cohorts, only EIF4G1 showed prognostic significance in all 3 cohorts.
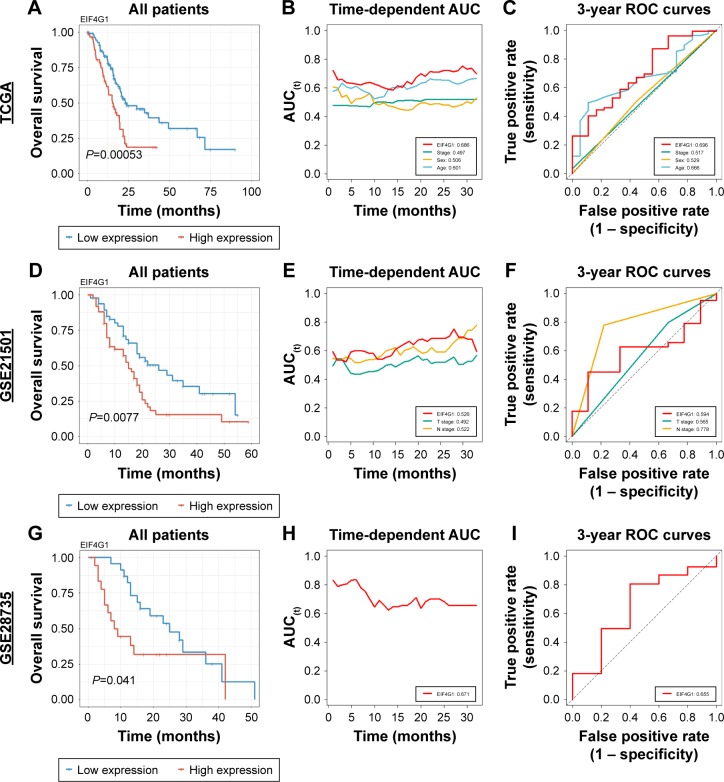
We analyzed the Kaplan–Meier curves for survival according to the EIF4G1 expression level to demonstrate the prognostic performance of EIF4G1 in PDAC. Intriguingly, lower expression of EIF4G1 was significantly associated with good prognosis in all 3 cohorts (TCGA, P=0.00053; GSE21501, P=0.0077 and GSE28735, P=0.041) (Figure 2A, D, and G). The results of univariate analysis of overall survival in each cohort suggested that the EIF4G1 expression level was statistically significant in all cohorts (Table 2). Furthermore, multivariate analysis of the TCGA and GSE21501 demonstrated a significant prognostic performance of EIF4G1 in PDCA, which was consistent with the abovementioned survival analysis (TCGA, P=0.00132, GSE21501, P=0.025, Table 2). The HR of EIF4G1 is particularly high when compared to other variables (Table 2). In addition, age in TCGA was a significant variable that could stratify prognosis (P=0.02121, Table 2).
Figure 2.
Survival analyses of EIF4G1 in 3 independent cohorts.
Notes: Kaplan–Meier estimates of all patients in TCGA (A), GSE21501 (D), and GSE28735 (G) according to EIF4G1 expression. Time-dependent AUC of EIF4G1 with clinical variables in TCGA (red, EIF4G1; green, stage; yellow, sex; and blue, age) (B), GSE21501 (red, EIF4G1; green, T stage; and yellow, N stage) (E), and GSE28735 (red, EIF4G1) (H). The 3-year ROC of EIF4G1 with clinical variables in TCGA (red, EIF4G1; green, stage; yellow, sex; and blue, age) (C), GSE21501 (red, EIF4G1; green, T stage; and yellow, N stage) (F), and GSE28735 (red, EIF4G1) (I).
Abbreviations: AUC, area under the curve; ROC, receiver operating characteristic; TCGA, The Cancer Genome Atlas.
Table 2.
Univariate and multivariate analyses of overall survival in each cohort
| Parameters | Univariate analysis | Multivariate analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| P-value | HR | 95% Cl | P-value | HR | 95% Cl | |||
| TCGA | ||||||||
| EIF4G1 | <0.0001*** | 2.072 | 1.359 | 3.157 | 0.00132** | 1.9974 | 1.3093 | 3.047 |
| Stage | 0.716 | 0.8072 | 0.2545 | 2.561 | 0.7005 | 0.7968 | 0.2504 | 2.535 |
| Sex | 0.39 | 0.8354 | 0.5543 | 1.259 | 0.2903 | 0.8010 | 0.5308 | 1.209 |
| Age | 0.0136* | 1.0261 | 1.005 | 1.047 | 0.02121* | 1.024 | 1.0036 | 1.045 |
| GSE21501 | ||||||||
| EIF4G1 | 0.00896** | 1.964 | 1.184 | 3.258 | 0.025* | 1.8076 | 1.077 | 3.034 |
| T stage (T1 and T2 vs T3 and T4) | 0.73 | 0.8977 | 0.4862 | 1.657 | 0.6047 | 0.8424 | 0.4400 | 1.613 |
| N stage (N0 vs N1) | 0.0425* | 1.8399 | 1.021 | 3.316 | 0.0709 | 1.7773 | 0.9523 | 3.317 |
| GSE28735 | ||||||||
| EIF4G1 | 0.045* | 2.185 | 1.018 | 4.691 | ||||
Notes: Statistically significant values are expressed in bold. *, ** and *** indicate significance at the <0.05, <0.01, and <0.001 respectively.
Abbreviation: TCGA, The Cancer Genome Atlas.
To compare the prognostic significance between miR-NAs and EIF4G1, we identified the log-rank test results of miRNAs by using Oncomir. There are 216 miRNAs significantly associated with survival in TCGA (Table S1). Unfortunately, because there is no miRNA information in GSE21501 and GSE28735 cohorts, we cannot compare miRNAs and EIF4G1.
Biomarker ability of EIF4G1 in PDAC
We compared Uno’s C-index values and AUC values at 3 years for the expression level of EIF4G1 with other variables such as tumor staging, sex, and age, which could be obtained from the clinical information of each cohort to examine the ability of EIF4G1 as a biomarker. EIF4G1 showed the highest C-index values in 3 independent cohorts (TCGA, 0.686; GSE21501, 0.526; and GSE28735, 0.671; Figure 2B, E, and H). Consistent with the results of Uno’s C-index, the 3-year AUC value was slightly less than 0.6 in GSE21501 (0.594; Figure 2F) and nearly 0.7 in the other 2 cohorts (TCGA, 0.696 and GSE28735, 0.655; Figure 2C and I).
Overexpression of EIF4G1 in PDAC
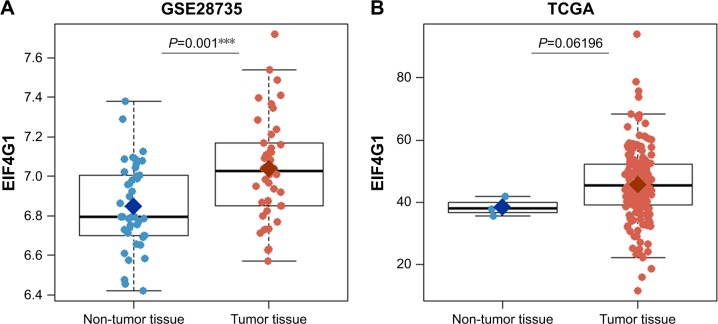
In order to confirm that EIF4G1 could predict prognosis as a tumor biomarker, the expression level of EIF4G1 was analyzed in tumor tissues and in non-tumor normal tissues using the GSE28735 or TCGA. Expression level of EIF4G1 was significantly higher in the PDAC tissues than in the matched normal tissues in GSE28735 (P<0.0001, Figure 3A). In TCGA, EIF4G1 expression seems to be higher than normal tissues, but it is not statistically significant (P=0.06196, Figure 3B).
Figure 3.
Comparison of EIF4G1 gene expression between matched non-tumor (blue) and tumor tissues (red) in GSE28735 (A) or TCGA (B).
Note: ***indicates significance at <0.001.
Abbreviation: TCGA, The Cancer Genome Atlas.
Discussion
EIF4G1 encodes a scaffold protein upon which ribosomes and the eukaryotic initiation factor (EIF) 4F complex assemble.20,21 The EIF4F complex regulates the key step of initiation in the translation of almost all genes in eukaryotes.21,22 Increased E1F4G1 expression has been found in inflammatory breast cancer, lung cancer, hypo-pharyngeal cancer, and nasopharyngeal cancer, which are consistent with our findings.20,23 Higher expression of EIF4G1 is also associated with shorter overall survival in various cancers.20,22–25 EIF4G1 may thus play a tumorigenic role by enhancing the translation of IRES-containing p120 mRNA, which contributes to the survival of breast tumor cells.22 However, the process by which EIF4G1 has been identified in tumorigenesis has not been fully elucidated in many cancers, including PDAC.
Despite the development of precision medicine, the only prognostic/diagnostic marker for PDAC is CA19-9.26 Although CA19-9 has been used widely, it is not useful for screening because of its low positive predictive value (<1%). Furthermore, increased false positivity of CA19-9 has been shown in the presence of obstructive jaundice (10%–60%).27 Recently, given the importance of molecular markers, many studies have been performed to identify novel biomarkers for pancreatic cancer using multi-omics data.19,28–30 We also investigated novel prognostic markers in patients with PDAC using 3 independent cohorts from TCGA and GEO databases. As described in Table 1, the patients’ information from TCGA and GSE21501 is quite different. EIF4G1, which is associated with survival in both cohorts, is likely to be a universal prognostic predictor applied to patients of all stages. Additionally, other clinical variables except for age were not statistically related to survival. These results may be due to the fact that the patient composition of each cohort (TCGA, GSE21501) is biased toward one side.
Conclusion
We demonstrated the prognostic significance of EIF4G1 in patients with PDAC using public databases. EIF4G1 is known to contribute to tumorigenesis as well as to tumor progression in several cancers. EIF4G1 is more expressed in cancer tissues and is associated with poor prognosis as its expression increases. We suggest that EIF4G1 could act as a prognostic biomarker to help determine the precise treatment strategy for PDAC.
Supplementary Materials
Acknowledgments
This study was supported by Biomedical Research Institute Grant (2018B032), Pusan National University Hospital.
Footnotes
Author contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59(4):225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Kwon HJ, Kim SG. Surgical treatment for advanced pancreatic cancer. Korean J Hepatobiliary Pancreat Surg. 2012;16(3):89–92. doi: 10.14701/kjhbps.2012.16.3.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li CP, Chao Y, Chi KH, et al. Concurrent chemoradiotherapy treatment of locally advanced pancreatic cancer: gemcitabine versus 5-fluorouracil, a randomized controlled study. Int J Radiat Oncol Biol Phys. 2003;57(1):98–104. doi: 10.1016/s0360-3016(03)00435-8. [DOI] [PubMed] [Google Scholar]
- 4.Kim YH, Jeong DC, Pak K, et al. Gene network inherent in genomic big data improves the accuracy of prognostic prediction for cancer patients. Oncotarget. 2017;8(44):77515–77526. doi: 10.18632/oncotarget.20548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eto S, Ishikawa M, Asanoma M, Tashiro Y, Matsuyama K, Oshio T. A long-term survival case of advanced biliary cancer with repeated resection due to recurrence in the pancreaticogastrostomy site after pancreaticoduodenectomy. Ann Hepatobiliary Pancreat Surg. 2018;22(2):173–177. doi: 10.14701/ahbps.2018.22.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zammarchi F, Morelli M, Menicagli M, et al. KLF4 is a novel candidate tumor suppressor gene in pancreatic ductal carcinoma. Am J Pathol. 2011;178(1):361–372. doi: 10.1016/j.ajpath.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman AA, Letai A, Fisher DE, Flaherty KT. Precision medicine for cancer with next-generation functional diagnostics. Nat Rev Cancer. 2015;15(12):747–756. doi: 10.1038/nrc4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnedos M, Vicier C, Loi S, et al. Precision medicine for metastatic breast cancer–limitations and solutions. Nat Rev Clin Oncol. 2015;12(12):693–704. doi: 10.1038/nrclinonc.2015.123. [DOI] [PubMed] [Google Scholar]
- 9.Kim YH, Jeong DC, Pak K, et al. SLC2A2 (GLUT2) as a novel prognostic factor for hepatocellular carcinoma. Oncotarget. 2017;8(40):68381–68392. doi: 10.18632/oncotarget.20266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suh S, Kim YH, Goh TS, et al. mRNA expression of SLC5A5 and SLC2A family genes in papillary thyroid cancer: an analysis of the cancer genome atlas. Yonsei Med J. 2018;59(6):746–753. doi: 10.3349/ymj.2018.59.6.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stratford JK, Bentrem DJ, Anderson JM, et al. A six-gene signature predicts survival of patients with localized pancreatic ductal adenocarcinoma. PLoS Med. 2010;7(7):e1000307. doi: 10.1371/journal.pmed.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang G, Schetter A, He P, et al. DPEP1 inhibits tumor cell invasiveness, enhances chemosensitivity and predicts clinical outcome in pancreatic ductal adenocarcinoma. PLoS One. 2012;7(2):e31507. doi: 10.1371/journal.pone.0031507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ha M, Han ME, Kim JY, Jeong DC, Oh SO, Kim YH. Prognostic role of TPD52 in acute myeloid leukemia: a retrospective multicohort analysis. J Cell Biochem. 2019;120(3):3672–3678. doi: 10.1002/jcb.27645. [DOI] [PubMed] [Google Scholar]
- 16.Han ME, Kim JY, Kim GH, Park SY, Kim YH, Oh SO. SAC3D1: a novel prognostic marker in hepatocellular carcinoma. Sci Rep. 2018;8(1):15608. doi: 10.1038/s41598-018-34129-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho SH, Pak K, Jeong DC, Han ME, Oh SO, Kim YH. The AP2M1 gene expression is a promising biomarker for predicting survival of patients with hepatocellular carcinoma. J Cell Biochem. 2019;120(3):4140–4146. doi: 10.1002/jcb.27699. [DOI] [PubMed] [Google Scholar]
- 18.Goh TS, Lee JS, Il Kim J, et al. Prognostic scoring system for osteosarcoma using network-regularized high-dimensional Cox-regression analysis and potential therapeutic targets. J Cell Physiol. 2019 doi: 10.1002/jcp.28065. [DOI] [PubMed] [Google Scholar]
- 19.Wong NW, Chen Y, Chen S, Wang X. OncomiR: an online resource for exploring pan-cancer microRNA dysregulation. Bioinformatics. 2018;34(4):713–715. doi: 10.1093/bioinformatics/btx627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tu L, Liu Z, He X, et al. Over-expression of eukaryotic translation initiation factor 4 gamma 1 correlates with tumor progression and poor prognosis in nasopharyngeal carcinoma. Mol Cancer. 2010;9:78. doi: 10.1186/1476-4598-9-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Badura M, Braunstein S, Zavadil J, Schneider RJ. DNA damage and eIF4G1 in breast cancer cells reprogram translation for survival and DNA repair mRNAs. Proc Natl Acad Sci U S A. 2012;109(46):18767–18772. doi: 10.1073/pnas.1203853109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silvera D, Arju R, Darvishian F, et al. Essential role for eIF4GI overexpression in the pathogenesis of inflammatory breast cancer. Nat Cell Biol. 2009;11(7):903–908. doi: 10.1038/ncb1900. [DOI] [PubMed] [Google Scholar]
- 23.Cao Y, Wei M, Li B, et al. Functional role of eukaryotic translation initiation factor 4 gamma 1 (EIF4G1) in NSCLC. Oncotarget. 2016;7(17):24242–24251. doi: 10.18632/oncotarget.8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaiswal PK, Koul S, Shanmugam PST, Koul HK. Eukaryotic translation initiation factor 4 gamma 1 (eIF4G1) is upregulated during prostate cancer progression and modulates cell growth and metastasis. Sci Rep. 2018;8(1):7459. doi: 10.1038/s41598-018-25798-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang S, Zhou Y, Chen Y, Ke G, Wen H, Wu X. Decreased expression of EIF4A1 after preoperative brachytherapy predicts better tumor-specific survival in cervical cancer. Int J Gynecol Cancer. 2014;24(5):908–915. doi: 10.1097/IGC.0000000000000152. [DOI] [PubMed] [Google Scholar]
- 26.Goonetilleke KS, Siriwardena AK. Systematic review of carbohydrate antigen (CA 19-9) as a biochemical marker in the diagnosis of pancreatic cancer. Eur J Surg Oncol. 2007;33(3):266–270. doi: 10.1016/j.ejso.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Ballehaninna UK, Chamberlain RS. The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: an evidence based appraisal. J Gastrointest Oncol. 2012;3(2):105–119. doi: 10.3978/j.issn.2078-6891.2011.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, Xu H, Zhu XX. Abnormal expression levels of sMICA and NKG2D are correlated with poor prognosis in pancreatic cancer. Ther Clin Risk Manag. 2016;12:11–18. doi: 10.2147/TCRM.S96869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding M, Li Y, Yang Y, et al. Elevated expression of Tiam1 is associated with poor prognosis and promotes tumor progression in pancreatic cancer. Onco Targets Ther. 2018;11:4367–4375. doi: 10.2147/OTT.S171425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou X, Lu Z, Wang T, Huang Z, Zhu W, Miao Y. Plasma miRNAs in diagnosis and prognosis of pancreatic cancer: a miRNA expression analysis. Gene. 2018;673:181–193. doi: 10.1016/j.gene.2018.06.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.