Abstract
Phylogenies recurrently demonstrate that oceanic island systems have been home to rapid clade diversification and adaptive radiations. The existence of adaptive radiations posits a central role of natural selection causing ecological divergence and speciation, and some plant radiations have been highlighted as paradigmatic examples of such radiations. However, neutral processes may also drive speciation during clade radiations, with ecological divergence occurring following speciation. Here, we document an exceptionally rapid and unique radiation of Lamiaceae within the New Caledonian biodiversity hotspot. Specifically, we investigated various biological, ecological, and geographical drivers of species diversification within the genus Oxera. We found that Oxera underwent an initial process of rapid cladogenesis likely triggered by a dramatic period of aridity during the early Pliocene. This early diversification of Oxera was associated with an important phase of ecological diversification triggered by significant shifts of pollination syndromes, dispersal modes, and life forms. Finally, recent diversification of Oxera appears to have been further driven by the interplay of allopatry and habitat shifts likely related to climatic oscillations. This suggests that Oxera could be regarded as an adaptive radiation at an early evolutionary stage that has been obscured by more recent joint habitat diversification and neutral geographical processes. Diversification within Oxera has perhaps been triggered by varied ecological and biological drivers acting in a leapfrog pattern, but geographic processes may have been an equally important driver. We suspect that strictly adaptive radiations may be rare in plants and that most events of rapid clade diversification may have involved a mixture of geographical and ecological divergence.
Keywords: Adaptive radiation, allopatry, leapfrog radiation, Lamiaceae, New Caledonia, niche shifts, Oxera
Oceanic islands are widely regarded as laboratories of evolution mainly owing to their isolated past biogeographical history (Losos and Ricklefs 2009). Their geographical isolation limits dispersal events from outside, resulting in ecological niches being filled by species diversification rather than colonization. Consequently, islands are expected to have sheltered many adaptive radiations, where single lineages have diversified rapidly into several distinct niches, resulting in disproportionately high phenotypic diversity (Schluter 2000) when compared with their continental relatives (Silvertown et al. 2005). This phenomenon can be particularly apparent on large and high-elevation islands that exhibit diverse and sharp environmental gradients (Whittaker et al. 2008). Although adaptive radiations may not necessarily result from an acceleration of speciation rates (see Givnish 2015 for a discussion), all adaptive radiations are spurred by ecological opportunities; that is, the release of vacant niches due to unused resources, species extinctions or the acquisition of new traits (Losos 2010). The existence of adaptive radiations thus posits a central role of natural selection causing ecological divergence and speciation, and some plant radiations have been highlighted repeatedly as paradigmatic examples of this scenario, such as the silversword alliance and lobeliads in Hawaii (Carlquist et al. 2003; Givnish et al. 2009), the Macaronesian Aeonium and Echium (Jorgensen and Olesen 2001), and Veronica (Hebe) in New Zealand (Wagstaff and Garnock-Jones 1998).
However, adaptive radiations sensu stricto appear to be quite uncommon in the plant kingdom, with the aforementioned well-documented cases probably representing rare exceptions. In fact, many radiations likely occur, at least in part, in a non-adaptive way (Givnish 1997, Rundell and Price 2009). Under such a scenario, speciation is not primarily driven by ecological divergence, but mainly by neutral divergence in allopatry. Ecological differentiation may occur following non-ecological speciation, through random trait divergence, adaptation to different climatic regimes or habitats, or due to trait divergence favoring local coexistence if secondary sympatry occurs (e.g., Tobias et al. 2014). While molecular phylogenies will often provide little power to discriminate between different scenarios, it remains possible to assess the relative importance of ecological differentiation and geographic divergence in rapid species diversifications. Considering that theoretical evidence suggests that most rapid evolutionary radiations may have occurred through a mixture of ecological speciation and neutral divergence in allopatry (Aguilée et al. 2012, 2011), we can legitimately wonder whether purely adaptive radiations have ever occurred in plants.
The New Caledonian biodiversity hotspot is a remote archipelago in the southwest Pacific, composed of an old-, large-, and high-elevation main island (= Grande Terre, ca. 37 Ma, ca. 16,600 km , 1600 m max. elevation) exhibiting exceptional levels of species richness and endemism (ca. 3400 species, 75% endemic; Morat et al. 2012; Munzinger et al. 2016). The singular geological and climatic New Caledonian history led to the implementation of complex and abrupt environmental gradients, resulting in a mosaic of highly distinct habitats. The archipelago topography is especially complex on Grande Terre, with many relatively steep valleys and high summits (Bonvallot 2012) presenting both physical barriers to dispersal and strong selection gradients. The archipelago also has a suite of diverse bedrock types, in particular volcano-sedimentary, metamorphic and ultramafic (metal-rich soils with chemical and physical properties that constrain plant growth). These latter bedrock types, which act as strong environmental filters and exhibit a patchy distribution, have probably played a key role in plant speciation and flora evolution (Pillon et al. 2010). In addition, Pliocene and Pleistocene climatic fluctuations considerably affected the dynamic of New Caledonian biotas, leading to the origination of new habitats such as the unique shrubby sclerophyllous vegetation (i.e., maquis; Jaffré 1980), during periods of forest contraction (Hope and Pask 1998; Stevenson 2004; Stevenson and Hope 2005). This likely triggered speciation in palms (Pintaud et al. 2001), and has contributed to the persistence of old angiosperm lineages in forest refugia (Pouteau et al. 2015). All the aforementioned characteristics have contributed to the tremendous plant diversity of New Caledonia (Jaffré 1993), and have also driven numerous plant radiations (e.g., Psychotria; Pycnandra; Munzinger et al. 2016). Though plant radiations are increasingly highlighted in the archipelago through molecular investigations, few studies have focused on the relative effect of drivers implied in their in situ diversification (e.g., Barrabé et al. 2014; Pillon et al. 2014; Paun et al. 2016).
, 1600 m max. elevation) exhibiting exceptional levels of species richness and endemism (ca. 3400 species, 75% endemic; Morat et al. 2012; Munzinger et al. 2016). The singular geological and climatic New Caledonian history led to the implementation of complex and abrupt environmental gradients, resulting in a mosaic of highly distinct habitats. The archipelago topography is especially complex on Grande Terre, with many relatively steep valleys and high summits (Bonvallot 2012) presenting both physical barriers to dispersal and strong selection gradients. The archipelago also has a suite of diverse bedrock types, in particular volcano-sedimentary, metamorphic and ultramafic (metal-rich soils with chemical and physical properties that constrain plant growth). These latter bedrock types, which act as strong environmental filters and exhibit a patchy distribution, have probably played a key role in plant speciation and flora evolution (Pillon et al. 2010). In addition, Pliocene and Pleistocene climatic fluctuations considerably affected the dynamic of New Caledonian biotas, leading to the origination of new habitats such as the unique shrubby sclerophyllous vegetation (i.e., maquis; Jaffré 1980), during periods of forest contraction (Hope and Pask 1998; Stevenson 2004; Stevenson and Hope 2005). This likely triggered speciation in palms (Pintaud et al. 2001), and has contributed to the persistence of old angiosperm lineages in forest refugia (Pouteau et al. 2015). All the aforementioned characteristics have contributed to the tremendous plant diversity of New Caledonia (Jaffré 1993), and have also driven numerous plant radiations (e.g., Psychotria; Pycnandra; Munzinger et al. 2016). Though plant radiations are increasingly highlighted in the archipelago through molecular investigations, few studies have focused on the relative effect of drivers implied in their in situ diversification (e.g., Barrabé et al. 2014; Pillon et al. 2014; Paun et al. 2016).
The old age, geographic isolation, and geologic and topographic complexity of New Caledonia suggest that adaptive radiations may be common among many genera endemic to New Caledonia. However, few of these genera exhibit the joint signatures of morphological and ecological divergence expected from adaptive radiations, and rather seem to be relictual lineages (Pillon et al. 2017). The woody genus Diospyros (Ebenaceae) was the first purported clear case of an adaptive radiation of a plant group within New Caledonia due to its wide ecological diversity (Paun et al. 2016), although the radiation was not related to obvious morphological and/or physiological differences. The tree genus Geissois (Cunoniaceae) was described as a cryptic adaptive radiation, in which species exhibit notable differences in leaf element composition likely linked to the occupation of varied soils (Pillon et al. 2014), but does not fully satisfy the criteria of an adaptive radiation as outlined by Givnish (2015), as no physiological adaptation has yet been highlighted. It thus seems that New Caledonia has harbored few, if any, adaptive radiations in a strict sense (sensu Givnish 2015 and Schluter 2000). This has been explained as a consequence of the relatively old age of the island, the prevalence of woody species, the paucity of potential pollinator species, and/or a reduced number of ecological opportunities as compared with other island systems (Pillon et al. 2017).
In this work, we report on a molecular systematic investigation of the genus Oxera (Lamiaceae), which has been hypothesized to have diversified in New Caledonia through adaptive radiation (Pillon et al. 2014). The genus is composed of 33 species endemic to New Caledonia (Gâteblé unpublished data, available at http://endemia.nc/flore/fiche588), and was recently enlarged to include five other Australasian and Pacific species (Barrabé et al. 2015). It was demonstrated that the entire New Caledonian clade originated from a single colonization and constitutes the tenth largest plant radiation in the archipelago, which is puzzling given that Lamiaceae are an under-represented family within the flora of New Caledonia (Pillon et al. 2010). Distinct subclades were partially circumscribed within Oxera and can arguably be considered as several independent micro-radiations within the genus (Barrabé et al. 2015). Oxera exhibits strongly divergent morphology in terms of life form, flowers and fruits, and occupies a vast diversity of distinct habitats. In relation to this remarkable morphological diversity, we hypothesize that shifts of functional, reproductive (pollination and dispersal), and environmental niches may have played a key role during the diversification of Oxera. This role can be two-fold: 1) niche shifts may have triggered species differentiation and speciation, and 2) certain niche types may have accelerated the rate of genetic divergence and speciation.
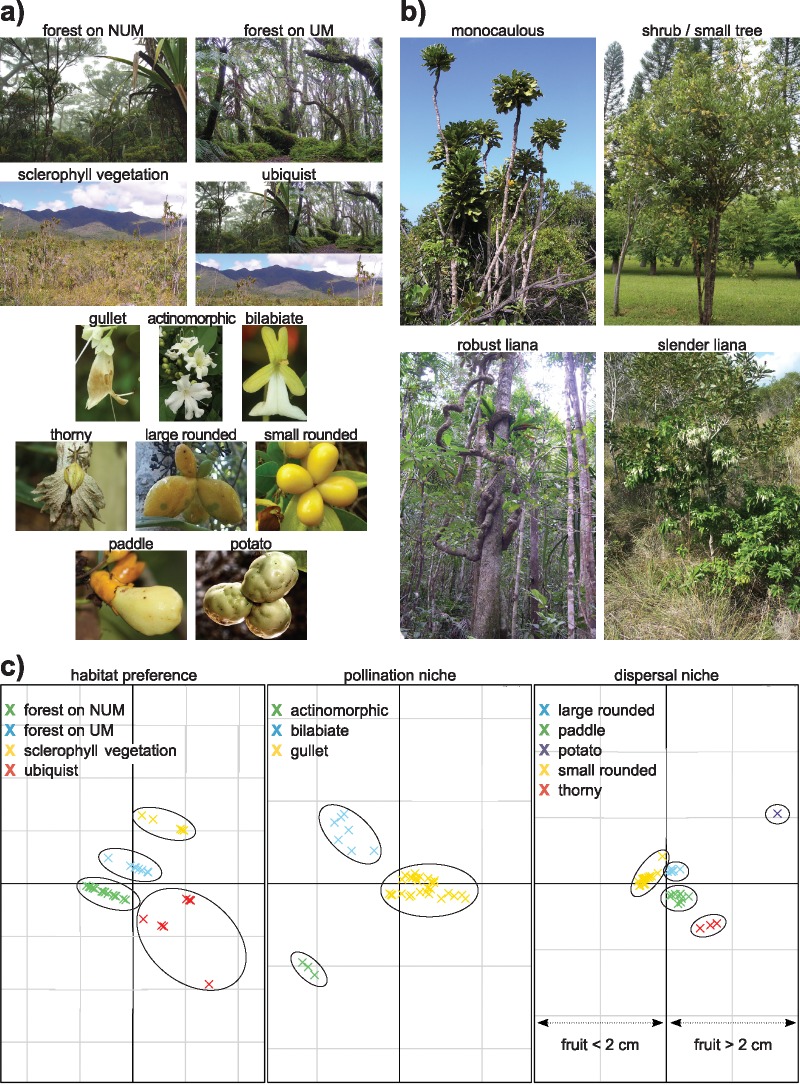
The dazzling morphological and ecological diversity of New Caledonian Oxera could reflect adaptations to different ecological factors (see Givnish 2010 for a discussion of life form adaptations). First, the great array of life forms observed in Oxera could be associated with different environment preferences, as observed in the silversword alliance (Carpenter et al. 2003), and this may have created conditions that facilitated parapatric isolation and speciation. Robust lianas mainly grow in closed rainforests with leaves usually reaching the canopy, while slender lianas are mostly encountered in open sclerophyllous vegetation and forest edges; monocaulous trees (i.e., single-stemmed) are mainly confined to rainforest understories; and finally, shrub species are often able to establish in both closed and more open vegetation (Gâteblé, pers. obs.; see Fig. 1a and b). Such contrasting life forms clearly reflect functional strategies regarding growth under different light conditions (Givnish 1995; Santiago and Wright 2007; Selaya and Anten 2008).
Figure 1.
Biological and ecological data sets of Oxera. a) Illustration of habitat preferences and biological syndromes, from top to bottom: habitat preferences (UM = ultramafic soils; NUM  non-ultramafic soils), pollination niche, and dispersal niche. b) Illustration of life forms. c) Niche syndromes inferred from the Hill and Smith principal component method, according to the first two axes of multivariate analyses.
non-ultramafic soils), pollination niche, and dispersal niche. b) Illustration of life forms. c) Niche syndromes inferred from the Hill and Smith principal component method, according to the first two axes of multivariate analyses.
Second, most species of Oxera are confined to narrow environmental conditions within New Caledonia. They occur either in rainforests, dry forests or maquis, or occur at different elevations and/or on different bedrock types (limestone, ultramafic, and volcano-sedimentary rocks). Although some species of Oxera are clearly specialized toward particular habitats or have very limited geographic ranges in narrow valleys or on remote summits, others are encountered throughout the archipelago due to wider environmental tolerances. These contrasting habitat and geographic patterns may be hypothesized as the signature of parapatric or allopatric speciation processes, and provide an interesting basis for testing whether habitat or geography (or both) have been major drivers of clade diversification.
Third, the floral diversity of Oxera was previously linked to distinct flower pollinators (de Kok 1998) based upon field observations of animal visits (Table 1).
Table 1.
In situ direct observations of flower visitors and fruit consumers of Oxera
| Flowers | Fruits | ||||||
|---|---|---|---|---|---|---|---|
| Subclade | Species | Flower visitor | Flower type (multivariate analyses) | Fruit consumer | Fruit type (multivariate analyses) | Maximum mericarp length | References |
| Australasian Oxera | Oxera splendida | Actinomorphic | Cassowaries, flying foxes, rats | Potato | 62.5 | Richards (1990), Hutson et al. (1992), Cooper and Cooper (2004) | |
| Pacific Oxera | Oxera amicorum | Honey eater (Myzomela cardinalis, Gymnomyza viridis) | Actinomorphic | Columbidae (Ducula pacifica, Didunculus strigirostris), flying foxes | Rounded | 41.7 | Belcher and Sibson (1982), de Kok (1997), Beichle (1987), Trail (1994), Pillon (pers. obs.) |
| Pacific Oxera | Oxera lehuntei | Honey eater | Actinomorphic | Rounded | 30.5 | de Kok (1997) | |
| “baladica” subclade | Oxera aff. nuda | Paddle | 40.6 | ||||
| “baladica” subclade | Oxera baladica | Honey eater (Glycifohia undulata), Zosteropidae (Zosterops sp.) | Gullet | Paddle | 26.3 | de Kok (1997), Fleurot (pers. obs.) | |
| “baladica” subclade | Oxera doubetiae sp.nov.ined. | Thorny | 40.6 | ||||
| “baladica” subclade | Oxera garoensis sp.nov.ined. | ||||||
| “baladica” subclade | Oxera ounemoae sp.nov.ined. | Thorny | 30.6 | ||||
| “baladica” subclade | Oxera papineaui sp.nov.ined. | Paddle | 25.9 | ||||
| “baladica” subclade | Oxera sessilifolia | Thorny | 40.4 | ||||
| “baladica” subclade | Oxera vanuatuensis | Birds | Gullet | Paddle | 35.0 | de Kok (1997) | |
| “neriifolia” subclade | Oxera neriifolia | Pieridae (Anaphaeis java, Eurema hecabe), Sphingidae (Gnathothlibus erotus, Hippotion scrofa, Hippotion celerio, Psilogramma lifuensis, Compsulyx cochereaui, etc.) | Bilabiate | Rounded | 14.8 | de Kok (1997), Gâteblé and Ouriet (pers. obs.) | |
| “neriifolia” subclade | Oxera ovata | Pieridae (Anaphaeis java), Sphingidae (Gnathothlibus salesnei) | Bilabiate | Rounded | 15.5 | Haxaire and Salesne (2016), de Kok 574 (in herb.) | |
| “neriifolia” subclade | Oxera sororia | Rounded | 13.5 | ||||
| “oblongifolia” subclade | Oxera crassifolia | Rounded | 18.1 | ||||
| “oblongifolia” subclade | Oxera glandulosa | Rounded | 11.4 | ||||
| “oblongifolia” subclade | Oxera oblongifolia | Bilabiate | Sturnidae (Aplonis striatus) | Rounded | 14.6 | de Kok 568 (in herb.) | |
| “oblongifolia” subclade | Oxera oreophila | Lepidoptera | Bilabiate | Rounded | 11.9 | de Kok (1997) | |
| Oxera morierei lineage | Oxera morierei | Honey eater (Glycifohia undulata) | Gullet | Rounded | 16.2 | de Kok (1997) | |
| “pulchella” subclade | Oxera aff. balansae | Rounded | 12.2 | ||||
| “pulchella” subclade | Oxera balansae | Birds | Gullet | Sturnidae (Aplonis striatus) | Rounded | 8.4 | de Kok (1997), Desmoulins (pers. obs.) |
| “pulchella” subclade | Oxera brevicalyx | Honey eater (Glycifohia undulata) | Gullet | Sturnidae (Aplonis striatus) | Rounded | 7.9 | de Kok (1997), Desmoulins (pers. obs.) |
| “pulchella” subclade | Oxera pulchella | Honey eater (Myzolema caledonica), Zosteropidae (Zosterops sp.) | Gullet | Sturnidae (Aplonis striatus) | Rounded | 11.3 | Desmoulins (pers. obs.), Fleurot (pers. obs.) |
| “robusta” subclade | Oxera coriacea | Honey eater (Glycifohia undulata) | Gullet | Rounded | 40.8 | de Kok (1997) | |
| “robusta” subclade | Oxera coronata | Rounded | 29.9 | ||||
| “robusta” subclade | Oxera longifolia | Honey eater (Lichmera incana) | Gullet | Rounded | 37.2 | de Kok (1997) | |
| “robusta” subclade | Oxera palmatinervia | Honey eater (Glycifohia undulata, Philemon diemenensis) | Gullet | Columbidae (Drepanoptila holosericea, Ducula Goliath) | Rounded | 34.0 | de Kok (1997), Bachy (pers. obs.), Desmoulins (pers. obs.) |
| “robusta” subclade | Oxera robusta | Rounded | 35.7 | ||||
| “subverticillata” subclade | Oxera aureocalyx sp.nov.ined. | Paddle | 40.7 | ||||
| “subverticillata” subclade | Oxera merytifolia | Paddle | 32.6 | ||||
| “subverticillata” subclade | Oxera subverticillata | Honey eater (Philemon diemenensis) | Gullet | Paddle | 30.9 | de Kok (1997) | |
| “subverticillata” subclade | Oxera tiwaeana sp.nov.ined. | Paddle | 31.9 | ||||
| “sulfurea” subclade | Oxera gmelinoides | Rounded | 13.8 | ||||
| “sulfurea” subclade | Oxera microcalyx | Honey eater (Glycifohia undulata), Zosteropidae (Zosterops xanthochroa) | Gullet | Rounded | 13.5 | de Kok (1997) | |
| “sulfurea” subclade | Oxera ouameniensis sp.nov.ined. | Rounded | 12.7 | ||||
| “sulfurea” subclade | Oxera pancheri | Rounded | 13.5 | ||||
| “sulfurea” subclade | Oxera rugosa | Rounded | 18.4 | ||||
| “sulfurea” subclade | Oxera sulfurea | Honey eater (Lichmera incana, Myzomela caledonica, Glycifohia undulata) | Gullet | Sturnidae (Aplonis striatus) | Rounded | 11.7 | de Kok (1997), Desmoulins (pers. obs.), Gâteblé (pers. obs.) |
These pollinator preferences could potentially be drivers of evolution of reproductive isolation within Oxera. Honey-eaters (Meliphagidae) and white-eyes (Zosteropidae) have been observed visiting Oxera species displaying showy and curved flowers, profusely producing nectar, and bearing arched anthers typically extending out of the corolla (Table 1). Long-tubular white and/or green bilabiate flowers, sometimes releasing a strong fragrance, have been observed to be visited by butterflies and moths (Pieridae and Sphingidae; Table 1; de Kok 1997; Haxaire and Salesne 2016). Despite the lack of recorded visitor observation, the flower attributes of the Australasian O. splendida, which bears large, flared, white flowers that open nocturnally and release a strong fragrance, are very similar to that of Fagraea, renowned for its bat-pollination (Momose et al. 1998).
Fourth, the large variety of fruit morphology exhibited by Oxera suggests different dispersal agents, although these mechanisms have not been fully identified in the field (Table 1). We assume that the different behaviors and flight abilities of animal dispersers would have affected the spatial scale of gene flow, and thus produced different rates of genetic divergence between Oxera lineages, due to varied dispersal abilities and habitat selection of dispersers (Givnish 2010, Theim et al. 2014). The largest white fruits encountered in Australasia are consumed by bats and cassowaries (Richards 1990; Hutson et al. 1992; Cooper and Cooper 2004). Other large colored fruits (ca.  2 cm) could be exclusively eaten by imperial pigeons (Ducula, Columbidae), which is the only bird able to swallow and effectively disperse such large fruits (Meehan et al. 2002; Carpenter et al. 2003; Cibois et al. 2017; P. Bachy pers. comm. for O. palmatinervia). It is noteworthy that the sedentary behavior of pigeons confines them mostly to forest understories (Colombo 2008; Wotton and Kelly 2012). Smaller fruits (ca.
2 cm) could be exclusively eaten by imperial pigeons (Ducula, Columbidae), which is the only bird able to swallow and effectively disperse such large fruits (Meehan et al. 2002; Carpenter et al. 2003; Cibois et al. 2017; P. Bachy pers. comm. for O. palmatinervia). It is noteworthy that the sedentary behavior of pigeons confines them mostly to forest understories (Colombo 2008; Wotton and Kelly 2012). Smaller fruits (ca.  2 cm) are consumed by more diminutive birds such as Sturnidae (Table 1; Tassin et al. 2010), which probably move over short distances, but are able to shift between different vegetation types (Barré et al. 2006). Finally, large grey or brown fruits having armed appendages are considered to be consumed by large geckos (Rhacodactylus, Mniarogekko, and Correlophus genera) because they are not discouraged by the fruits’ repelling structures (Whitaker and Bauer, pers. comm.). Large geckos also have limited spatial movements (Clark et al. 2009), which may have constrained gene flow in Oxera.
2 cm) are consumed by more diminutive birds such as Sturnidae (Table 1; Tassin et al. 2010), which probably move over short distances, but are able to shift between different vegetation types (Barré et al. 2006). Finally, large grey or brown fruits having armed appendages are considered to be consumed by large geckos (Rhacodactylus, Mniarogekko, and Correlophus genera) because they are not discouraged by the fruits’ repelling structures (Whitaker and Bauer, pers. comm.). Large geckos also have limited spatial movements (Clark et al. 2009), which may have constrained gene flow in Oxera.
Given the large diversity of morphological traits, ecological attributes, and geographical ranges exhibited by Oxera, we suspect that species diversification within subclades of this genus has been driven by several ecological and biological factors acting at different time periods, which is in a leapfrog pattern (as originally defined by Chase and Palmer 1997). Available information on species coexistence within similar regions or habitats may also help test whether sympatric speciation has occurred in relation to different pollination and dispersal niches. Indeed, such a scenario suggests that sister species with distinct biotic niches should co-occur either in the same region or habitat. Sister species divergence based only on ecological preference or geographic range would otherwise suggest that speciation mainly occurred through parapatric or allopatric speciation, respectively. We thus posit that phylogenetic patterns of species ranges and niches bear the signature of both selective and neutral forces as engines of clade diversification. Here, we report the first detailed phylogenetic reconstruction of this exceptional radiation of Lamiaceae, the genus Oxera, within the New Caledonian biodiversity hotspot, and attempt to identify the major factors that have shaped its current species diversity. We used a combination of phylogenetics, molecular dating, and comparative analyses of species diversification and niche evolution in order to 1) reconstruct the tempo and rate of species diversification in Oxera, 2) extricate and identify the different drivers of its diversification, and 3) appraise their respective effects. This study specifically asks a pivotal question: Is Oxera the single clear adaptive radiation in New Caledonia?
Materials and Methods
Species Sampling and Molecular Data Sets
In order to both establish species relationships and to estimate divergence times within the genus Oxera, the focal ingroup was composed of the 38 recognized species according to the most recent systematic investigations in New Caledonia (Gâteblé unpublished data, available at http://endemia.nc/flore/fiche588), and taxonomical revisions by de Kok and Mabberley (1999a, 1999b) for Australasia and Pacific. We added to the data set the sister lineage Hosea, as well as 10 species belonging to the closely related Clerodendrum clade identified in Yuan et al. (2010). Three more divergent species of Lamiaceae were used as external outgroups (i.e., Ajuga chamaepitys, Teucrium pyrenaicum, Rotheca sp. Wen 9487; Supplementary Appendix S1a; available on Dryad at https://doi.org/10.5061/dryad.mm1ft40). A total of 51 species were used in this sampling, which we refer to as the Oxera data set, including 22 species with previously published sequences (downloaded from GenBank), and 29 species with new sequences generated for this study. This molecular data set consisted of the 12 DNA loci sequenced in Barrabé et al. (2015). This allowed us to improve 1) the phylogenetic resolution and 2) branch length estimates relative to Barrabé et al. (2015). All new DNA sequences were generated using the DNA extraction, amplification, and sequencing protocols described in Barrabé et al. (2015). All accessions used in this study are listed in the Supplementary Appendix S1a available on Dryad.
For divergence time estimation, we also used two additional alignments composed of four DNA plastid regions (matK, rps16, trnL-F, and trnL-trnF; Supplementary Appendix S2a available on Dryad) from two data sets, with taxon sampling that spanned the Lamiales and Lamiaceae. The Lamiales data set included: 1) a subset of the Oxera data set (i.e., five species; Supplementary Appendix S1c available on Dryad); and 2) 177 representatives of various lineages within Lamiales, whose sequences were downloaded from GenBank (Supplementary Appendix S1c available on Dryad). The Lamiaceae data set was thus composed of 1) the aforementioned Oxera subset and 2) 59 taxa representing all major Lamiaceae lineages (downloaded likewise from GenBank; Supplementary Appendix S1b available on Dryad). This large-scale sampling allowed us to incorporate Lamiales and Lamiaceae fossils as calibration points (see below).
All DNA alignment matrices used in this study are available on Dryad.
Phylogenetic Inference
For the three molecular data sets, phylogenetic inferences were conducted using Bayesian Markov Chain Monte Carlo (MCMC) as implemented in MrBayes v3.2.1 (Ronquist et al. 2012) based on single-locus and concatenated data sets. Best-fit nucleotide sequence evolution models were identified using jModelTest (Posada 2008) based on the Akaike criterion (more details on settings are provided in Supplementary Appendix S2a available on Dryad). The combined data sets were partitioned to allow each locus to possess specific model parameters (Nylander et al. 2004). The methodological approach of the Bayesian MCMC analyses followed that described in Barrabé et al. (2015) and the details of parameter settings are provided in Supplementary Appendix S2b available on Dryad. All analyses were run on the Cyber Infrastructure for Phylogenetic Research cluster (CIPRES; http://www.phylo.org/; last accessed 18 October 2017). The post-burn-in trees resulting from the MCMC stationary phase were used to construct a majority-rule consensus tree and calculate Bayesian posterior probabilities (PPs); clades were considered well-supported when PP values were  0.95). The three resulting consensus trees are available in TreeBASE (ID 23353) and Dryad.
0.95). The three resulting consensus trees are available in TreeBASE (ID 23353) and Dryad.
In order to check for phylogenetic incongruence between nuclear and plastid loci, we also ran two phylogenetic reconstructions separately for all six concatenated nuclear regions and all six concatenated plastid regions, using the same MrBayes parameter settings and models of nucleotide evolution as above. These two analyses were only applied to the Oxera data set. We then assessed whether there was any supported (PP  0.95) incongruence between two trees.
0.95) incongruence between two trees.
Fossil Dating and Divergence Time Estimation
We performed a two-step approach to estimate the temporal evolution of Oxera. This allowed us to test the impact of different dating calibrations and taxonomic sampling on divergence time estimates. In the first step, we performed a fossil calibration on the Lamiales and Lamiaceae data sets using fossils described in a recent Lamiaceae molecular dating study (Yao et al. 2016). We incorporated three Lamiaceae (Melissa, Ocimum, and Stachys) and three Lamiales fossils (Bignoniaceae, Acanthaceae, and Oleaceae) for dating the phylogeny inferred with the Lamiales data set, and two Lamiaceae fossils (Melissa and Ocimum) for dating the phylogeny inferred with the Lamiaceae data set (for further details on the design and parameters of these calibrations, see Supplementary Appendix S2c, available on Dryad and explanations provided inYao et al. 2016). For root calibrations we used divergence times estimated in Magallón et al. (2015): 1) the divergence between Lamiales and Gentianales—Solanales for the Lamiales data set and 2) the divergence between Plocospermataceae and other Lamiales for the Lamiaceae data set. In the second step, the divergence time estimate between Rotheca (sp. Wen 9487) and other Lamiaceae recovered during the first step (i.e., the dating of the Lamiaceae data set) was used as a secondary root calibration point (see Results section) for the Oxera data set.
For the three molecular data sets, molecular divergence times were estimated using the Bayesian MCMC approach implemented in BEAST v.1.8.0 (Drummond and Rambaut 2007). DNA regions were combined and partitions were set as in the above Bayesian MCMC analyses (Supplementary Appendix S2c available on Dryad). An uncorrelated relaxed molecular clock model was selected following a lognormal distribution, and the Birth–Death process implemented for the tree prior. Other details on analyses, calibration and parameter settings are provided in Supplementary Appendix S2c available on Dryad. The post burn-in trees were summarized, and Bayesian PPs, median height (= age estimate), and the 95% highest posterior density heights interval of each node (95% HPD) assessed, using a Maximum Clade Credibility target tree (named as MCCT tree for the Oxera data set) in Treeannotator v.1.8.0 (Drummond and Rambaut 2007). For the Oxera data set a subset of 100 dated trees (RD trees hereafter) were also randomly sampled from the stationary phase to integrate phylogenetic uncertainty in some of the following analyses (see below). All divergence time analyses were conducted using CIPRES. The three resulting Maximum Clade Credibility trees are available in TreeBASE (ID 23353) and Dryad.
Comparative Analyses of Species Diversification
To obtain an overall view of the Oxera diversification, we first generated a lineage through time diagram, based on a pruned version of the MCCT tree including only Oxera species and a single sample per species (pMCCT tree), and also on the RD trees (pruned as in the pMCCT tree). We also calculated net diversification rates, following the conservative approaches implemented in Pillon (2012) and Barrabé et al. (2014), to allow comparisons with other oceanic insular plant radiations, using median crown ages and their 95% HPD estimated from the molecular dating, and two extreme values for relative extinction (null and equal to 0.9). Net diversification rates were likewise estimated per unit of area and log(area).
To assess more precisely the rate and tempo of species diversification in Oxera through time, we carried out diversification analyses with maximum likelihood (ML) and Bayesian modeling approaches. The ML analyses were conducted in the TreePar R package (Stadler 2011), using an optimization algorithm to infer past rates of speciation and extinction and their temporal variation. We fitted various diversification models: a pure birth model, a constant birth–death model, several more complex models allowing one to several rate shifts through time (up to 10 shifts), and finally several diversity-dependent models. All models were fitted on the pMCCT tree (see Supplementary Appendix S2d, available on Dryad for further details on parameter settings) and on the RD trees to account for phylogenetic uncertainty. The best-fit diversification model was identified using the corrected Akaike criterion, and a likelihood-ratio test was used for comparing nested pure birth, birth–death, and rate shift models.
The Bayesian diversification analyses were computed using BAMM (Rabosky et al. 2014). This allows modelling complex dynamics of speciation and extinction on phylogenetic trees using a reversible jump MCMC algorithm. We pruned the MCCT tree in the same manner as the pMCCT tree, but retained species belonging to the Clerodendrum clade to assess early variation in Oxera diversification rates. The sampling fraction was completed with an estimate of species richness, especially for the Clerodendrum clade (retrieved from the Kew Checklist website: http://wcsp.science.kew.org; last accessed 18 October 2017). As we expected two rate shifts to have taken place during the evolution of Oxera and its relatives (considering the large sizes of the genera Clerodendrum and Oxera as compared with the other small genera), the prior on the number of diversification shifts was set to 2. All other priors were estimated in the BAMMtools R package (Rabosky et al. 2014), with the default settings used for all other parameters. The MCMC was run for 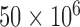 generations and sampled every 10,000 generations. The analysis was conducted with three independent Markov chains. The first 10% generations of each run were discarded after checking for chain convergence and adequate MCMC sampling. The remaining generations were summarized to generate the 95% credibility interval for rate shift configurations, the marginal shift probability tree (where branch lengths are proportional to the probability that a shift occurred on a given branch), the best rate shift configuration with the highest maximum a posteriori probability, the phylorate plot (showing variation of mean diversification rates with a colored gradient) and finally the rates-through time plots (only built for the Oxera focal group).
generations and sampled every 10,000 generations. The analysis was conducted with three independent Markov chains. The first 10% generations of each run were discarded after checking for chain convergence and adequate MCMC sampling. The remaining generations were summarized to generate the 95% credibility interval for rate shift configurations, the marginal shift probability tree (where branch lengths are proportional to the probability that a shift occurred on a given branch), the best rate shift configuration with the highest maximum a posteriori probability, the phylorate plot (showing variation of mean diversification rates with a colored gradient) and finally the rates-through time plots (only built for the Oxera focal group).
Biological, Ecological, and Geographic Range Data sets
To identify and disentangle suspected drivers of species diversification in Oxera, we built a data set by compiling data on five major life traits: 1) flower morphology, 2) fruit morphology, 3) life forms, 4) geographical occurrences, and 5) environmental preferences. The flower and fruit morphology (i.e., organ dimensions, colors, shapes, and textures) were compiled from herbarium and fieldwork observations on organ dimensions, colors, shapes, and textures. Those traits were coded as 13 discrete and three continuous characters for flower morphology, and three discrete and three continuous traits for fruits (Supplementary Appendix S3a, d, and e available on Dryad). The life form data set was composed of a single discrete trait corresponding to the four life forms usually ascribed to Oxera, namely monocaulous, robust liana, slender liana, and shrub/small tree (Supplementary Appendix S3a and b available on Dryad). Note that no functional or physiological traits related to light requirements or nutrient use strategy were available for Oxera.
The geographical distributions of New Caledonian species were retrieved from the databases of the herbaria of Nouméa (NOU, VIROT), the Museum national d’Histoire naturelle of Paris (P, SONNERAT), the University of Zurich (Z), and additional field observations. The geographical coordinates of herbarium specimen records were then databased, error-corrected for distribution, and finally incorporated into a Geographical Information System (GIS) under QGIS 2.8.1. For Australasian and Pacific species distributions were assessed from de Kok and Mabberley (1999b) and manually added into the same GIS layer.
The environmental data set was composed of three variables that best explain plant distributions across New Caledonia (Jaffré 1993; Morat 1993; Supplementary Appendix S3a and c available on Dryad): geological substrates, vegetation types, and elevation. Species’ rock types were coded as occurring on ultramafic rocks, on volcano-sedimentary and metamorphic rocks, or ubiquists. These geological attributes were determined by coupling geographical occurrences with a layer of geological substrate provided by the Direction de l’Industrie, des Mines et de l’Energie de la Nouvelle-Calédonie (New Caledonia). Species’ vegetation types were coded as occurring in wet forests, in sclerophyllous vegetation types (i.e., dry forests and/or maquis vegetation), and ubiquists, based on herbarium and field observations (Gâteblé, pers. obs.). Species’ elevation ranges were extracted from the GIS layer generated by a digital elevation model (resolution of 10 m) provided by the Direction des Infrastructures de la Topographie et des Transports Terrestres (New Caledonia), and then averaged across geographical occurrences of each species.
To identify ecological syndromes among Oxera species, we performed multivariate analyses on the environmental, flower, and fruit data sets. As we used both continuous and discrete traits, we conducted the PCA analyses by using the Hill and Smith (1976) principal component method under the ade4 R package (Chessel et al. 2004). This analysis allowed clustering species among pollinator syndromes, dispersal syndromes, and habitat preferences, which we subsequently treated as new discrete traits according to the species clustering (i.e., the niche data sets; Supplementary Appendix S3f available on Dryad). We posit that the ecological syndrome of a species reflects the ecological niche it occupies (Johnson 2010, Pauw 2013). No data on species growth and architecture were available, we therefore, could not evaluate life forms in a similar manner; this trait was treated as a discrete variable without any step of ordination analysis.
Comparative Analyses of Niche Evolution
To assess the mode and tempo of evolution of each niche during the Oxera radiation (i.e., of pollination and dispersal syndromes or habitat preferences), we measured their respective amount of phylogenetic signal by estimating 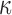 and
and 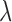 Pagel statistics (Pagel 1999). Values of
Pagel statistics (Pagel 1999). Values of 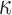 and
and 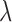 close to 1 indicate a strong phylogenetic signal and gradual trait evolution (i.e., according to a model of Brownian motion), and deviation from this expectation provides insight into temporal patterns of trait evolution. Values of
close to 1 indicate a strong phylogenetic signal and gradual trait evolution (i.e., according to a model of Brownian motion), and deviation from this expectation provides insight into temporal patterns of trait evolution. Values of 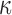 close to 0 depicts punctual evolution, where trait divergence occurs independently from branch lengths in the phylogeny. Values of
close to 0 depicts punctual evolution, where trait divergence occurs independently from branch lengths in the phylogeny. Values of 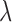 close to 0 reveal a low phylogenetic signal, where most trait change occurred late in evolutionary history and closely related species thus share very little trait similarity. For the habitat, pollination, and dispersal niches
close to 0 reveal a low phylogenetic signal, where most trait change occurred late in evolutionary history and closely related species thus share very little trait similarity. For the habitat, pollination, and dispersal niches 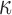 and
and 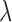 statistics were estimated using the “fitContinuous” and “phylosig” functions of the geiger and phytools R packages, respectively (Harmon et al. 2008; Revell 2012). Estimations were performed on the RD trees, and by averaging estimated values for each of the first n axes of each PCA explaining 90% of the cumulative variance. For life form, each statistic was simply averaged through the RD trees using the “fitdiscrete” function (geiger) and the single discrete trait.
statistics were estimated using the “fitContinuous” and “phylosig” functions of the geiger and phytools R packages, respectively (Harmon et al. 2008; Revell 2012). Estimations were performed on the RD trees, and by averaging estimated values for each of the first n axes of each PCA explaining 90% of the cumulative variance. For life form, each statistic was simply averaged through the RD trees using the “fitdiscrete” function (geiger) and the single discrete trait.
To reconstruct the evolutionary history of habitat preference, pollination, dispersal niches, and life form, we conducted ancestral state reconstruction analyses. We first assessed ancestral niche states using the “ace” function in the ape R package (Paradis et al. 2004) applied to the pMCCT tree, and the niche and life form data sets. We fitted three ML evolution models: the “ER” (with equal state transition rates), “SYM” (symmetrical), and “ARD” (with all rates unequal) models, identifying the best-fitting model using comparisons performed on the corrected Akaike criterion, and likelihood ratio test. We then estimated their respective absolute shift number and timing by performing stochastic character mappings (Huelsenbeck et al. 2003), using the “make.simmap” function in phytools. We launched 500 simulations using an estimated prior distribution on the root node, the best-fit ML model recovered in the previous analyses and applied first to the pMCCT tree, and then to the RD trees to obtain most likely envelopes of past shift number for habitat preference, pollination, dispersal niches and life-forms. For 11 of the 100 RD trees, we encountered intractable optimization issues when using the “make.simmap” function. We subsequently removed the 11 trees from these analyses and used only 89 RD trees. Absolute shift timings were extracted using a custom R-script from all simulations and summarized by plotting the median number of state changes in different time bins of a 0.5 Myr time-grid.
To investigate whether a particular niche state (pollination, dispersal, or habitat) or life-form would have affected clade diversification rates of Oxera, we conducted multi-state trait-based analyses that estimated simultaneously trait-dependent speciation, extinction, and transition rates on a phylogeny and character distribution (Fitzjohn et al. 2009). These were performed using the “make.musse” function under the diversitree R package (Fitzjohn et al. 2009), applied to the pMCCT tree, and the niche and life form data sets. We first fitted 17 models from a null model (rates independent of trait states) to a full model (all rate components dependent of trait states; Supplementary Appendix S4 available on Dryad), conserving the best-fitting using the corrected Akaike criterion. We performed subsequent Bayesian analyses through a MCMC, run for 10,000 generations, seeding them with rate estimates recovered from the previous ML analyses as priors, and discarding the first 1000 generations as burn-in. PP distributions of all parameters were summarized using the diversitree R-package. We also fitted the best model and compared it to the second best model across all RD trees to check that model selection was robust to phylogenetic uncertainty.
Geographical Evolution versus Habitat Evolution
To investigate the relative roles of geographical and environmental divergence during the diversification of Oxera, we computed age-range correlations (Fitzpatrick and Turelli 2006; Warren et al. 2008) where metrics of range overlaps, range asymmetries, and habitat distances computed between species pairs are compared with their estimated time of divergence (from the phylogeny). These metrics have been recurrently used to diagnose different speciation and post-speciation scenarios (Anacker and Strauss 2014; Grossenbacher et al. 2014).
Species geographical ranges were retrieved from the geographical data set by creating for each species a single convex polygon enclosing its locations using the “convex hull” tool under QGIS. For narrow endemic species, with one or two occurrences, we applied a buffer of 0.5 km around each location, and used a range of 3.141592 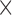 0.5
0.5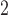 km
km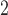 for the former, and of 6.283184
for the former, and of 6.283184 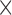 0.5
0.5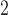 km
km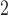 for the latter. From these distribution data, range overlaps, range asymmetries, and habitat distances metrics were computed between all possible pairs of species. Range overlap was defined as the area occupied by two species divided by the area of the more narrowly ranging species, ranging from 0 (no co-occurrence) to 1 (complete co-occurrence). Range asymmetry was calculated as the area of the wider-ranging species divided by that of the smaller (ranging from 1 to infinity). The habitat similarity between two species was assessed by calculating their Euclidian distance (habitat distance) based on the eigenvector values of the first
for the latter. From these distribution data, range overlaps, range asymmetries, and habitat distances metrics were computed between all possible pairs of species. Range overlap was defined as the area occupied by two species divided by the area of the more narrowly ranging species, ranging from 0 (no co-occurrence) to 1 (complete co-occurrence). Range asymmetry was calculated as the area of the wider-ranging species divided by that of the smaller (ranging from 1 to infinity). The habitat similarity between two species was assessed by calculating their Euclidian distance (habitat distance) based on the eigenvector values of the first 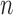 axes explaining 90% of the cumulate variance retrieved from the environmental PCA (see above). These geographical variables (areas of all Oxera species, range asymmetries and range overlaps of all Oxera species pairs) are available on Dryad.
axes explaining 90% of the cumulate variance retrieved from the environmental PCA (see above). These geographical variables (areas of all Oxera species, range asymmetries and range overlaps of all Oxera species pairs) are available on Dryad.
Age-range correlation analyses were performed using an extended version of the “age.range.correlation” function of the phyloclim R package (Heibl and Calenge 2013; https://github.com/danlwarren/arc-extensions/blob/master/age.range.correlation.2.R; last accessed 18 October 2017), applied to our pMCCT and the RD trees. We launched 1000 iterations for the Monte Carlo resampling procedure to create the null hypothesis of no relationship between phylogenetic relatedness and range overlap, range asymmetry, or habitat distance, and tested it. We calculated linear regressions, whose slopes and intercepts indicate speciation mode (allopatric vs. sympatric) or habitat evolution (conservatism vs. divergence) through time. For each metric a “super-p-value” was calculated, corresponding to the tree proportion across the RD trees for which the linear regression was significant. We also assessed the frequency of range overlaps across all random species pairs and across all sister species pairs using a kernel density plot.
Finally, we estimated the respective importance of geographical versus habitat divergence and whether local co-existence is possible between closely related Oxera species. To do so, we superimposed species geographical overlaps to their habitat preferences (summarized into diagrams combining information on their geological, elevation, and vegetation attributes) within each Oxera subclade (as delimited in Barrabé et al. 2015) to determine whether closely related species could possibly occur in sympatry.
Results
Phylogenetic Inference
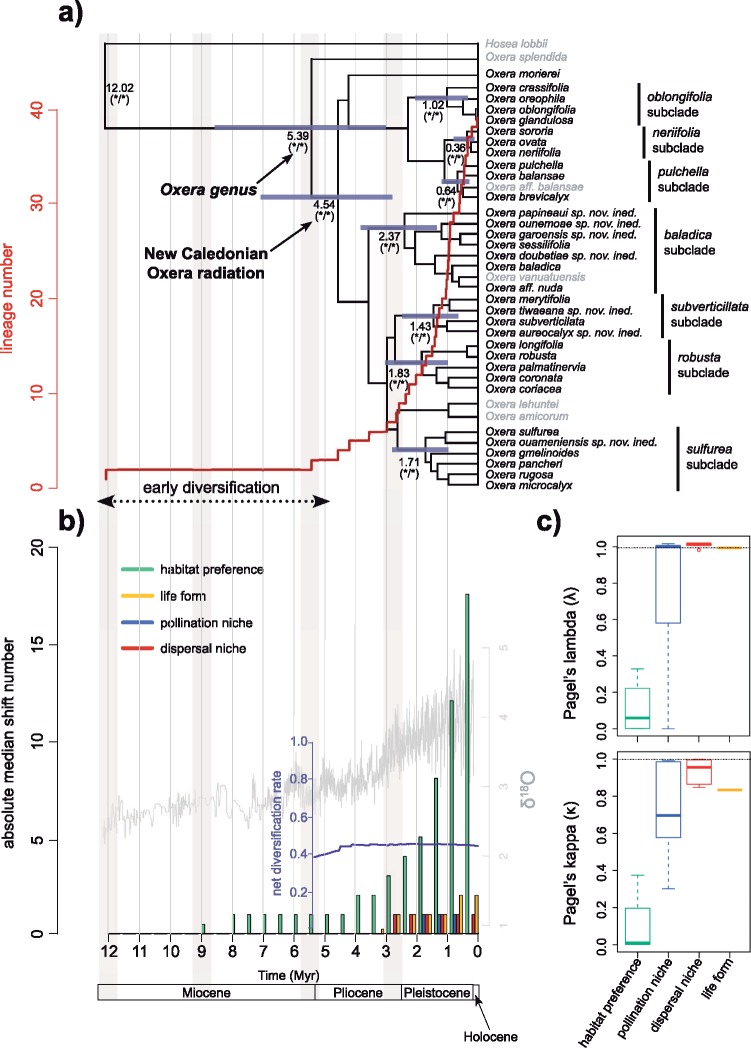
The Bayesian MCMC analyses of the three species sampling (Lamiales, Lamiaceae, and Oxera data sets) recovered the same following robust phylogenetic relationships (Supplementary Appendices S5–S7 available on Dryad). The sister genera Hosea and Oxera formed a well-supported clade that was sister to the Clerodendrum clade (PP 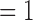 ). Analyses based on the Oxera data set provided more resolution and detail on the internal Oxera relationships, which are congruent with the ones established in Barrabé et al. (2015) (Supplementary Appendix S5 available on Dryad). The Australasian O. splendida was placed as sister to the New Caledonian radiation (PP
). Analyses based on the Oxera data set provided more resolution and detail on the internal Oxera relationships, which are congruent with the ones established in Barrabé et al. (2015) (Supplementary Appendix S5 available on Dryad). The Australasian O. splendida was placed as sister to the New Caledonian radiation (PP 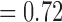 ), which included all New Caledonian and nested Pacific subclades. We retrieved seven well-supported New Caledonian subclades (PP
), which included all New Caledonian and nested Pacific subclades. We retrieved seven well-supported New Caledonian subclades (PP 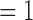 ; Fig. 2a), namely the oblongifolia, neriifolia, pulchella, baladica, subverticillata, robusta, and sulfurea subclades.
; Fig. 2a), namely the oblongifolia, neriifolia, pulchella, baladica, subverticillata, robusta, and sulfurea subclades.
Figure 2.
Evolutionary history of Oxera in New Caledonia, and of its habitat preferences, pollination and dispersal niches, and life forms. a) BEAST chronogram of the Oxera data set with superimposed lineage through time plot. Median age estimates and blue node bars (corresponding to the 95% highest posterior densities) are indicated for each node of interest. Bayesian PP from the BEAST analyses (on the right) and MCMC Bayesian analyses (on the left) are indicated with an asterisk for each node of interest when 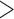 0.95, and with a hyphen when
0.95, and with a hyphen when 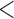 0.95. Vertical light grey rectangles indicate periods of intense weathering as inferred from Chevillotte et al. (2006). b) Absolute shift timings of the three niches and life forms inferred from stochastic character mappings, with global delta
0.95. Vertical light grey rectangles indicate periods of intense weathering as inferred from Chevillotte et al. (2006). b) Absolute shift timings of the three niches and life forms inferred from stochastic character mappings, with global delta 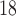 O variation through time (in grey; from Zachos et al. 2001) and net diversification rate variation through time of Oxera, as inferred from BAMM analyses (in dark blue), superimposed. c) Phylogenetic signal for habitat preferences, pollination and dispersal niches, and life forms inferred from estimations of the
O variation through time (in grey; from Zachos et al. 2001) and net diversification rate variation through time of Oxera, as inferred from BAMM analyses (in dark blue), superimposed. c) Phylogenetic signal for habitat preferences, pollination and dispersal niches, and life forms inferred from estimations of the 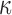 and
and 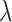 Pagel statistics.
Pagel statistics.
A few supported incongruences (PP 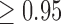 ) were observed between the phylogenetic trees reconstructed separately from the six nuclear loci and from the six plastid loci (Supplementary Appendix S8 available on Dryad), but these incongruencies were all located near the tips (i.e., between close species). This discordance did not affect the robustness of the seven New Caledonian subclades previously recovered. These minor incongruences underscore the importance of accounting for phylogenetic uncertainty by using the 100 RD trees in most of our evolution and geographical analyses (see Material and Methods section).
) were observed between the phylogenetic trees reconstructed separately from the six nuclear loci and from the six plastid loci (Supplementary Appendix S8 available on Dryad), but these incongruencies were all located near the tips (i.e., between close species). This discordance did not affect the robustness of the seven New Caledonian subclades previously recovered. These minor incongruences underscore the importance of accounting for phylogenetic uncertainty by using the 100 RD trees in most of our evolution and geographical analyses (see Material and Methods section).
Molecular Dating
The three molecular BEAST dating analyses provided congruent divergence times for Oxera and relatives (Fig. 2a, Supplementary Appendices S9–S11 available on Dryad), which can thus be considered as a reliable appraisal of colonization dates and tempo of species diversification. Oxera likely emerged during the late Miocene, and the New Caledonian radiation sensu stricto is estimated to have initiated during the early Pliocene. The median divergence time estimates for the stem node of Oxera were 11.46 Ma (95% HPD = 5.65–19.68), 11.2 Ma (95% HPD  5.61–19.06), and 12.02 Ma (95% HPD
5.61–19.06), and 12.02 Ma (95% HPD  6.21–19.16), as recovered with the Lamiales, Lamiaceae, and Oxera data sets, respectively (Fig. 2a, Supplementary Appendices S9–S11 available on Dryad). For the crown node, age estimates were 6.34 Ma (95% HPD
6.21–19.16), as recovered with the Lamiales, Lamiaceae, and Oxera data sets, respectively (Fig. 2a, Supplementary Appendices S9–S11 available on Dryad). For the crown node, age estimates were 6.34 Ma (95% HPD  2.89–11.48), 6.28 Ma (95% HPD
2.89–11.48), 6.28 Ma (95% HPD  3.07–10.73), and 5.39 Ma (95% HPD
3.07–10.73), and 5.39 Ma (95% HPD  2.97–8.42), respectively. The Oxera data set provided an estimate of 4.54 Ma (95% HPD
2.97–8.42), respectively. The Oxera data set provided an estimate of 4.54 Ma (95% HPD  2.7–6.92) for the crown node of the New Caledonian radiation (Fig. 2a).
2.7–6.92) for the crown node of the New Caledonian radiation (Fig. 2a).
Oxera Diversification
The lineage through time plot highlighted a gradual and exponential increase of the number of Oxera lineages through time (Fig. 2a). The net conservative diversification rates are summarized in Tables 2 and 3. For the New Caledonian radiation, these rates were 0.2 species/Myr for an extinction of 0.9 and 1.04 species/Myr for a null extinction. With respect to per-unit-area and per-unit-log(area) they were estimated between 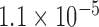 and
and 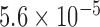 species/Myr/km
species/Myr/km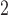 , and between 0.02 and 0.106 species/Myr/log(km
, and between 0.02 and 0.106 species/Myr/log(km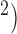 , respectively. The BAMM analyses inferred two early instances of changes in the diversification rate. The phylorate plot showed an increase in mean diversification rates at the crown node of Oxera and the second deepest node of the Clerodendrum clade, whereas Hosea demonstrated a decrease in these rates (Supplementary Appendix S12a available on Dryad). Five of the six most probable 95% credible rate shift configurations (with a cumulative PP of 0.378; Supplementary Appendix S13 available on Dryad) showed significant rate shifts mainly located on the two consecutive branches leading to the New Caledonian Oxera radiation, and those leading to most of the Clerodendrum clade (Supplementary Appendix S13 available on Dryad). Both shifts were depicted in the marginal shift probability tree, where these latter branches were significantly longer than most others, indicating a high probability that these shifts occurred on their respective branches (Supplementary Appendix S12b available on Dryad). The Oxera rates through time plots highlighted that speciation rates remained constant (with a mean value of ca. 0.71 species/Myr). Extinction rates decreased slightly at the radiation onset but remained constant after (mean value of ca. 0.31 species/Myr). The resulting net diversification rates increased early in the radiation onset and later remained quite constant (mean value of ca. 0.4 species/Myr; Fig. 2b, Supplementary Appendix S12c available on Dryad). The ML diversification analyses selected the pure birth model as best-fitting our data for the genus Oxera, with a net diversification rate of 0.575 species/Myr (Supplementary Appendix S14 available on Dryad). Accordingly, the pure birth model was the best selected model on 93% of the RD trees.
, respectively. The BAMM analyses inferred two early instances of changes in the diversification rate. The phylorate plot showed an increase in mean diversification rates at the crown node of Oxera and the second deepest node of the Clerodendrum clade, whereas Hosea demonstrated a decrease in these rates (Supplementary Appendix S12a available on Dryad). Five of the six most probable 95% credible rate shift configurations (with a cumulative PP of 0.378; Supplementary Appendix S13 available on Dryad) showed significant rate shifts mainly located on the two consecutive branches leading to the New Caledonian Oxera radiation, and those leading to most of the Clerodendrum clade (Supplementary Appendix S13 available on Dryad). Both shifts were depicted in the marginal shift probability tree, where these latter branches were significantly longer than most others, indicating a high probability that these shifts occurred on their respective branches (Supplementary Appendix S12b available on Dryad). The Oxera rates through time plots highlighted that speciation rates remained constant (with a mean value of ca. 0.71 species/Myr). Extinction rates decreased slightly at the radiation onset but remained constant after (mean value of ca. 0.31 species/Myr). The resulting net diversification rates increased early in the radiation onset and later remained quite constant (mean value of ca. 0.4 species/Myr; Fig. 2b, Supplementary Appendix S12c available on Dryad). The ML diversification analyses selected the pure birth model as best-fitting our data for the genus Oxera, with a net diversification rate of 0.575 species/Myr (Supplementary Appendix S14 available on Dryad). Accordingly, the pure birth model was the best selected model on 93% of the RD trees.
Table 2.
Comparison of net diversification rates of Oxera with other New Caledonian plant lineages/radiations
| Family | Radiation | Stem age (Ma) | Crown age (Ma) | Number of New Caledonian species retrieved from Munzinger et al. (2016) | Species number restricted to sclerophyll vegetations | Species number able to growth in sclerophyll vegetations | Proportion of species strictly growing in sclerophyll vegetations (%) | Proportion of species able to grow in sclerophyll vegetations | Net diversification rate (based on the equation of Magallón and Sanderson (2001)) for an extinction null, and using crown ages (in species/Myr) | Works where age estimates have been published |
|---|---|---|---|---|---|---|---|---|---|---|
| Sapotaceae | Planchonella clade III | 18 | 15 | 3 | 1 | 2 | 33.3 | 66.7 | 0.03 | Swenson et al. (2013) |
| Proteaceae | Beauprea clade I (“filipes” clade) | 78.2 | 28.3 | 7 | 1 | 5 | 14.3 | 71.4 | 0.04 | He et al. (2016) |
| Nothofagaceae | Nothofagus | 26.1 | 16.4 | 5 | 0 | 0 | 0 | 0 | 0.06 | Sauquet et al. (2012) |
| Sapotaceae | Planchonella clade I | 15.3 | 12.05 | 4 | 0 | 3 | 0 | 75 | 0.06 | Swenson et al. (2013) |
| Proteaceae | Kermadecia - Sleumerodendron | 13.6 | 12.3 | 5 | 0 | 0 | 0 | 0.07 | Sauquet et al. (2009) | |
| Myrtaceae | Metrosideros clade B | 21.25 | 11 | 5 | 1 | 3 | 20 | 60 | 0.08 | Papadopulos et al. (2011) |
| Myrtaceae | Metrosideros clade A | 16.25 | 13.6 | 7 | 4 | 7 | 57.1 | 100 | 0.09 | Papadopulos et al. (2011) |
| Arecaceae | Clinosperma—Cyphokentia | 21.5 | 10.5 | 6 | 0 | 0 | 0 | 0 | 0.1 | Baker and Couvreur (2012) |
| Myrtaceae | Metrosideros clade C | 29.1 | 8.25 | 5 | 2 | 4 | 40 | 80 | 0.11 | Papadopulos et al. (2011) |
| Arecaceae | Chambeyronia—Kentiopsis | 9 | 4.5 | 6 | 0 | 0 | 0 | 0 | 0.12 | Baker and Couvreur (2012) |
| Sapotaceae | Pichonia | 19 | 10.1 | 7 | 5 | 5 | 71.4 | 57.1 | 0.12 | Swenson et al. (2013) |
| Sapotaceae | Planchonella clade II | 33 | 17.4 | 18 | 7 | 7 | 38.9 | 38.9 | 0.13 | Swenson et al. (2013) |
| Rubiaceae | Margaritopsis | 8.47 | 5.21 | 4 | 1 | 4 | 25 | 100 | 0.13 | Barrabé et al. (2014) |
| Rubiaceae | Psychotria clade NC1 | 17.06 | 4.78 | 4 | 4 | 4 | 100 | 100 | 0.15 | Barrabé et al. (2014) |
| Rutaceae | Oxanthera | max 5.8 | 5.8 | 5 | 5 | 5 | 100 | 100 | 0.16 | Pfeil and Crisp (2008) |
| Sapotaceae | Pleioluma clade I | 10 | 4.9 | 5 | 0 | 2 | 0 | 40 | 0.19 | Swenson et al. (2013) |
| Arecaceae | Burretiokentia—Cyphophoenix | 11.5 | 7.5 | 9 | 0 | 0 | 0 | 0 | 0.2 | Baker and Couvreur (2012) |
| Sapotaceae | Pycnandra | 29.8 | 16.2 | 57 | 17 | 20 | 29.8 | 35.1 | 0.21 | Swenson et al. (2013) |
| Araucariaceae | Araucaria | 16 | 8 | 13 | 0 | 7 | 0 | 53.8 | 0.23 | Kranitz et al. (2014) |
| Loganiaceae | Geniostoma | 9 | 6.5 | 10 | 1 | 4 | 10 | 40 | 0.25 | Foster et al. (2014) |
| Podocarpaceae | Dacrydium | 5 | 3.05 | 5 | 0 | 3 | 0 | 60 | 0.3 | Keppel et al. (2011) |
| Podocarpaceae | Podocarpus | 6.9 | 4.2 | 8 | 2 | 2 | 25 | 25 | 0.33 | Quiroga et al. (2016) |
| Sapotaceae | Pleioluma clade II | 7.5 | 4.2 | 8 | 6 | 8 | 75 | 100 | 0.33 | Swenson et al. (2013) |
| Anacardiaceae | Euroschinus | 11.85 | 3.7 | 7 | 0 | 2 | 0 | 28.6 | 0.34 | Weeks et al. (2014) |
| Ebenaceae | Diospyros clade III | 9.1 | 7.2 | 25 | 8 | 16 | 32 | 64 | 0.35 | Turner et al. (2013) |
| Pandanaceae | NC Pandanus subgen. Lophostigma | 8.5 | 6.5 | 24 | 2 | 2 | 8.3 | 8.3 | 0.38 | Gallaher et al. (2015) |
| Ericaceae | Dracophyllum | 5.2 | 3.5 | 8 | 7 | 6 | 87.5 | 75 | 0.4 | Wagstaff et al. (2010) |
| Rubiaceae | Psychotria clade NC2 | 7.63 | 6.9 | 78 | 32 | 40 | 41 | 51.3 | 0.53 | Barrabé et al. (2014) |
| Lamiaceae | NC Oxera | 5.39 | 4.54 | 33 | 5 | 11 | 15.2 | 29.7 | 0.62 | This study |
| Rubiaceae | Thiollierea | 6.5 | 3 | 13 | 13 | 13 | 100 | 100 | 0.62 | Manns et al. (2012) |
Note: In grey, sclerophyllous clades.
Table 3.
Comparison of net diversification rates of Oxera with other rapid island plant lineages/radiations
| Family | Radiation | Archipelago | Number of species | Area (km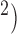
|
Median crown age (95% HPD in Ma) | Net diversification rate and using crown ages (species/Myr) | Net diversification rate at a area unit (species/ Myr/km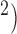
|
Net diversification rate at a log(area) unit (species/Myr/ log(km
|
Works where age estimates and species number have been published | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Extinction = 0 | Extinction = 0.9 | ||||||||||
| Lamiaceae | New Caledonian Oxera | New Caledonia | 33 | 18,600 | 2.7 | 6.92 | 0.41–1.04 | 0.2–0.51 | 0.000011–0.000056 | 0.02–0.106 | This study |
| Rubiaceae | Psychotria clade NC2 | New Caledonia | 78 | 18,600 | 4.62 | 9.82 | 0.37–0.79 | 0.22–0.46 | 0.000012–0.000043 | 0.022–0.081 | Barrabé et al. (2014) |
| Asteraceae | Cheilorophus | Macaronesia | 20 | 10,372 | 5.98 | 15.35 | 0.15–0.39 | 0.07–0.17 | 0.000006–0.000037 | 0.007–0.042 | Vitales et al. (2014) |
| Euphorbiaceae | Euphorbia | Hawaii | 16 | 16,644 | 0.72 | 3.97 | 0.52–2.89 | 0.22–1.2 | 0.000013–0.000174 | 0.022–0.297 | Yang et al. (2018) |
| Asteraceae | Bidens | Hawaii | 19 | 16,644 | 1.3 | 3.1 | 0.73–1.73 | 0.32–0.75 | 0.000019–0.000104 | 0.032–0.178 | Knope et al. (2012) |
| Asteraceae | Echium | Macaronesia | 19 | 10,372 | 2.7 | 3.9 | 0.58–0.83 | 0.25–0.36 | 0.000024–0.00008 | 0.027–0.09 | Knope et al. (2012) |
| Gesneriaceae | Cyrtandra | Hawaii | 58 | 16,644 | 5.2 | 0.65 | 0.36 | 0.000021–0.000039 | 0.037–0.067 | Clark et al. (2008, 2009) | |
| Campanulaceae | Lobeliads | Hawaii | 126 | 16,644 | 10.49 | 16.71 | 0.25–0.39 | 0.15–0.24 | 0.000009–0.000024 | 0.016–0.041 | Givnish et al. (2009) |
| Crassulaceae | Aeonium | Macaronesia | 63 | 10,372 | 13.25 | 17.15 | 0.2–0.26 | 0.11–0.15 | 0.000011–0.000025 | 0.012–0.028 | Kim et al. (2008) |
| Plantaginaceae | Veronica | New Zealand | 120 | 268,680 | 5.7 | 0.72 | 0.44 | 0.000002–0.000003 | 0.035–0.057 | Ogutcen (2016), Wagstaff et al. (2002) | |
Niche Ordination
The multivariate analyses allowed the separation of four, three, and five clearly distinct ecological syndromes from the species’ environmental, flower, and fruit data sets, respectively (Fig. 1c). The first four, nine, and six axes of the PCA analyses (explaining 90 % of the cumulate variance), respectively, were retained for subsequent comparative analyses. The three major habitats occupied by Oxera were forests on non-ultramafic rocks, forests on ultramafic rocks, sclerophyllous vegetation, and a fourth class was defined for ubiquist species. The three pollination syndromes characterized species with actinomorphic, gullet, and bilabiate floral morphs each adapted to bat, bird, moth/butterfly pollination, respectively. For dispersal syndromes the PCA axis markedly separated species with small fruits 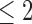 cm (on the left) from those with large fruits
cm (on the left) from those with large fruits 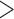 2 cm (on the right; Fig. 1c). The five dispersal syndromes discriminated species with potato-like (consumed by cassowaries and bats), paddle-like (by Columbidae), small rounded (by Columbidae and/or small birds such as Sturnidae), large rounded (by Columbidae), and thorny fruit morphs (presumably by large geckos).
2 cm (on the right; Fig. 1c). The five dispersal syndromes discriminated species with potato-like (consumed by cassowaries and bats), paddle-like (by Columbidae), small rounded (by Columbidae and/or small birds such as Sturnidae), large rounded (by Columbidae), and thorny fruit morphs (presumably by large geckos).
Niche Evolution
Our estimates of both Pagel statistics showed distinct evolutionary patterns (Fig. 2c; Supplementary Appendix S15 available on Dryad). We found low values of 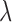 , indicating low phylogenetic signal (i.e., a recent accelerated evolution), for the habitat preference (Fig. 2c), and high phylogenetic signal for the dispersal niche and life form, suggesting that these niche characteristics have diversified early. For the pollination niche, estimates of
, indicating low phylogenetic signal (i.e., a recent accelerated evolution), for the habitat preference (Fig. 2c), and high phylogenetic signal for the dispersal niche and life form, suggesting that these niche characteristics have diversified early. For the pollination niche, estimates of 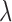 were more spread out, with a lower 5% quantile of 0.2, a higher 95% quantile of 1.01, and a median value of 0.99; this also indicated a relatively high phylogenetic signal (Fig. 2c). Estimates of the
were more spread out, with a lower 5% quantile of 0.2, a higher 95% quantile of 1.01, and a median value of 0.99; this also indicated a relatively high phylogenetic signal (Fig. 2c). Estimates of the 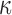 statistic were close to 0 for the habitat preference (indicating a punctual evolution), and close to 1 for the dispersal niche and life form, respectively (indicating more gradual evolution). For the pollination niche,
statistic were close to 0 for the habitat preference (indicating a punctual evolution), and close to 1 for the dispersal niche and life form, respectively (indicating more gradual evolution). For the pollination niche, 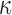 scaled between 0.35 (5% quantile) and 0.99 (95% quantile), with a median value of 0.69.
scaled between 0.35 (5% quantile) and 0.99 (95% quantile), with a median value of 0.69.
The best-fit evolutionary model identified in our ancestral state reconstruction analyses was the “ER” model for each niche/trait data set (Supplementary Appendix S16 available on Dryad). The reconstructed ancestral habitat preference of Oxera was ambiguous: forests on both ultramafic and non-ultramafic rocks were together reconstructed with a high likelihood (forested vegetation was recovered as ancestral for this node with cumulated likelihoods of both forest types of ca. 0.57). However, the ancestral pollination, dispersal, and life form were inferred with much less ambiguity; the Oxera ancestor was very likely a robust liana with gullet flowers producing small rounded fruits (Supplementary Appendix S17 available on Dryad). Analyses of stochastic character mapping allowed us to estimate that very few pollination, dispersal, and life form shifts occurred during the evolution of Oxera (up to two shifts per time bin of 0.5 Myr; Fig. 2b), and that most of these trait shifts occurred between 2 Ma and 0.5 Ma for pollination, and following 3 Ma and 3.5 Ma for dispersal and life form, respectively. Habitat shifts occurred between 9 Ma and present, but from 4 Ma onward their number increased markedly, finally reaching 17 shifts between 0.5 Ma and the present.
The best-fitting model of trait-dependent species diversification identified for habitats had speciation and transition rates all equal, a null extinction rate (Supplementary Appendix S4 available on Dryad), and no discernible effect of any habitat type. For the pollination syndromes, dispersal types and life forms, the best-fitting model was that with a null extinction, transition rates all equal, and a particular character state in which speciation rates were significantly different from other states (Supplementary Appendix S4 available on Dryad). These results were very robust to phylogenetic uncertainty, as 100% of the RD trees consistently yielded the same best models. Slender lianas had on average significantly higher speciation rates comparing to other life forms, actinomorphic flowers, and potato-like fruits had on average significantly lower speciation rates comparing to other floral and fruit types (Supplementary Appendix S18 available on Dryad).
Geographical Evolution
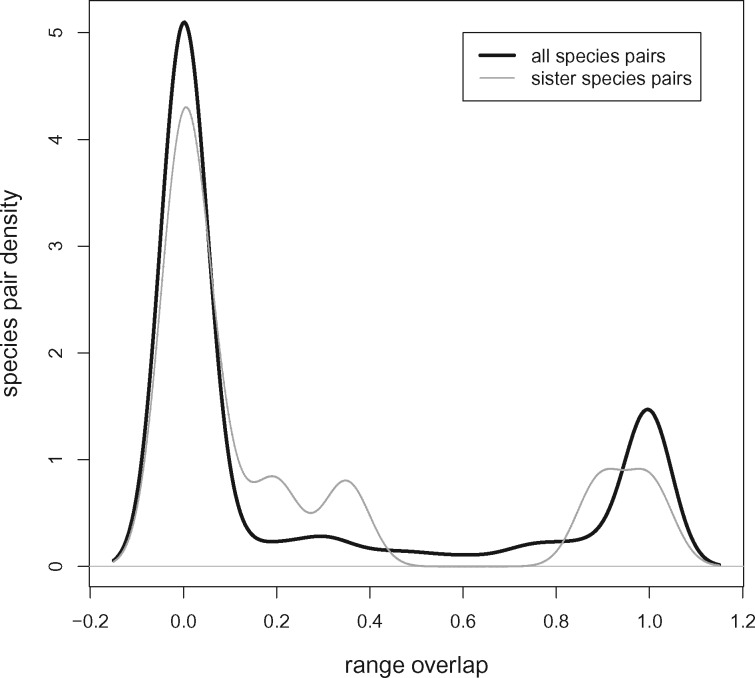
The range overlap between all species pairs was generally low (mean value of 0.187, and a standard deviation of 0.357), suggesting a high level of allopatry between all species. The kernel density plot highlighted two range overlap frequency peaks, the highest located for a range overlap of 0 and the shortest to 1 (Fig. 3). Age range correlation analyses showed that the slope of range asymmetry through time was significantly positive for linear regressions (all 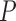 -values
-values  0.05 across RD trees, Supplementary Appendix S19 available on Dryad). For range overlap and habitat distance, linear regression slopes with divergence times were negative in both cases but not significant.
0.05 across RD trees, Supplementary Appendix S19 available on Dryad). For range overlap and habitat distance, linear regression slopes with divergence times were negative in both cases but not significant.
Figure 3.
The Kernel density plot of range overlap through all possible species pairs (in black) and all possible sister species pairs (in grey) in New Caledonian Oxera.
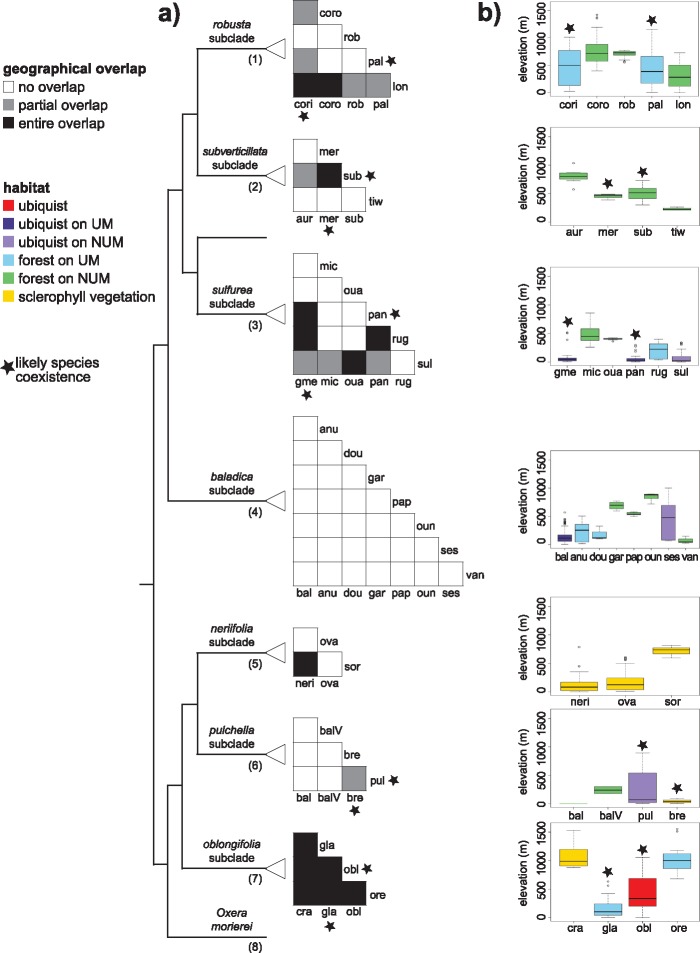
Different Oxera subclades highlighted distinct patterns of geographical overlap (Fig. 4a and b). The baladica subclade was entirely and strictly allopatric since none of its species overlapped geographically. The oblongifolia subclade was completely sympatric as all its species overlapped. The other subclades (i.e., “neriifolia” “pulchella” “robusta” “subverticillata” and “sulfurea” subclades) exhibited more complex patterns (Fig. 4a). Overall, of the 96 possible pairs of species encountered within the seven New Caledonian subclades, we recorded 23 cases of partial or entire geographical overlap (Fig. 4a and b), but most of them involved species occurring in markedly distinct habitats. In fact, among all 96 species pairs, only five pairs involved geographically overlapping pairs of species growing in the same habitat, suggesting possible local co-existence (i.e., O. oblongifolia and O. glandulosa, O. coriacea, and O. palmatinervia, O. merytifolia, and O. subverticillata, O. gmelinoides, and O. pancheri). However, none of these species pairs have been observed co-occurring in the field (Gâteblé, pers. obs.).
Figure 4.
Species coexistence within Oxera, indicated by subclade, with stars indicating likely coexistence; mapped on the BEAST maximum credibility clade chronogram from the Oxera data set. a) Geographical overlap between all species of each subclade. b) Ecological ranges of all species indicated per subclade, plotted according to their altitudinal ranges and their habitat preferences (UM  ultramafic soils, NUM
ultramafic soils, NUM  non-ultramafic soils). Species names are indicated with the following abbreviations, listed according to subclade: robusta subclade (cori
non-ultramafic soils). Species names are indicated with the following abbreviations, listed according to subclade: robusta subclade (cori  O. coriacea, coro
O. coriacea, coro  O. coronata, lon
O. coronata, lon  O. longifolia, pal
O. longifolia, pal  O. palmatinervia, rob
O. palmatinervia, rob  O. robusta), subverticillata subclade (aur
O. robusta), subverticillata subclade (aur  O. aureocalyx sp. nov. ined., mer
O. aureocalyx sp. nov. ined., mer  O. merytifolia, sub
O. merytifolia, sub  O. subverticillata, tiw
O. subverticillata, tiw  O. tiwaeana sp. nov. ined.), sulfurea subclade (gme
O. tiwaeana sp. nov. ined.), sulfurea subclade (gme  O. gmelinoides, mic
O. gmelinoides, mic  O. microcalyx, oua
O. microcalyx, oua  O. ouameniensis sp. nov. ined., pan
O. ouameniensis sp. nov. ined., pan  O. pancheri, rug
O. pancheri, rug  O. rugosa, sul
O. rugosa, sul  O. sulfurea), baladica subclade (bal
O. sulfurea), baladica subclade (bal  O. baladica, anu
O. baladica, anu 
 . aff. nuda, dou
. aff. nuda, dou  O. doubetiae sp. nov. ined., gar
O. doubetiae sp. nov. ined., gar  O. garoensis sp. nov. ined., oun
O. garoensis sp. nov. ined., oun  O. ounemoae sp. nov. ined., pap
O. ounemoae sp. nov. ined., pap  O. papineaui sp. nov. ined., ses
O. papineaui sp. nov. ined., ses  O. sessilifolia, van
O. sessilifolia, van  O. vanuatuensis), neriifolia subclade (neri
O. vanuatuensis), neriifolia subclade (neri  O. neriifolia, ova
O. neriifolia, ova  O. ovata, sor
O. ovata, sor  O. sororia), pulchella subclade (bal
O. sororia), pulchella subclade (bal  O. balansae, balV
O. balansae, balV 
 aff. balansae, bre
aff. balansae, bre  O. brevicalyx, pul
O. brevicalyx, pul  O. pulchella), and oblongifolia subclade (cra
O. pulchella), and oblongifolia subclade (cra  O. crassifolia, gla
O. crassifolia, gla  O. glandulosa, obl
O. glandulosa, obl  O. oblongifolia, ore
O. oblongifolia, ore  O. oreophila).
O. oreophila).
Discussion
An Early Burst of Diversification
Our study shows that the genus Oxera is a quite recent and rapid plant radiation that initiated during the late Miocene, and that most of the New Caledonian subclades within Oxera began to diversify in the early Pliocene (Table 2, Fig. 2). It is noteworthy that this New Caledonian radiation occurred long after the emergence of Grande Terre about 37 Ma (Cluzel et al. 2012). Thus, the radiation of Oxera originated in the archipelago via long-distance or stepping-stone dispersal, but was not triggered by Gondwanan fragmentation. This continental dispersal to the New Caledonian archipelago was probably aided by the ancestral dispersal type (i.e., small rounded fleshy fruits; Supplementary Appendix S17 available on Dryad), which may be the most efficient dispersal mode in the group. Following this dispersal to New Caledonia, the successful establishment of ancestral Oxera may have been facilitated by the climbing habit of its ancestor (robust woody liana identified as the ancestral state; Supplementary Appendix S17 available on Dryad), which is believed to allow the occupation of a broad range of local environments (Gianoli 2004). The finding that the ancestral colonizer of extant New Caledonian Oxera was reconstructed as having a liana life-form with fleshy fruits makes it a quite unique case since the liana form is generally associated with wind dispersal (Givnish 2010), implying dispersal limitations over water, and that the liana niche in islands remains often empty.
Diversification analyses identified an early burst of diversification (BAMM analyses; Fig. 2a and b, Supplementary Appendix S12 available on Dryad), likely located between the divergence of the Oxera ingroup and its sister lineage Hosea (ca. 12 Ma) and the crown node of Oxera within New Caledonia (ca. 4.5 Ma). This late Miocene/early Pliocene burst of species diversification indicates that Oxera underwent an initial period of rapid cladogenesis, especially in New Caledonia, concordant with an initial decreasing of extinction rates; Figure 2b, Supplementary Appendix S12 available on Dryad. This coincides with the origination and early diversification of other major New Caledonian radiations (e.g., Psychotria clade NC2), and especially lineages typical of sclerophyllous vegetation types (e.g., Thiollierea; Table 2).
This period of intense species diversification is concordant with the global onset of Pliocene glaciations (Zachos et al. 2001) as well as the temperature declines and severe aridification recorded in the South Pacific during the Pliocene (Karas et al. 2011). It also coincides with the timing of intense soil morphogenesis and weathering which is estimated to have occurred around 5.5 Ma in New Caledonia (Fig. 2a and b; Chevillotte et al. 2006). Such climatic events have been suggested to have impacted the evolutionary history of many plant lineages by leading to the extinction of certain species (thus alleviating competitive effects), and the emergence of new biotas (e.g., more sclerophyllous and open vegetation), especially since the Miocene (Donoghue and Edwards 2014). It is important to note that some of these novel habitats exhibit a patchy spatial distribution, which has likely increased genetic divergence. This ostensibly has generated vacant niches, created ecological opportunities, increased spatial divergence and thus triggered speciation in many Australasian lineages, as highlighted in Australia for Banksia, Eucalyptus, and Allocasuarina (Crisp et al. 2004; Gallagher et al. 2003). The early rapid diversification of Oxera suggests that similar climatic and physical events fostered diversification within Oxera shortly after its arrival into New Caledonia.
An Explosive Species Diversification, with Early Divergence in Biotic Niches
The rate of species diversification estimated for Oxera is exceptionally high, and represents the fastest known New Caledonian plant radiation, rivalled only by Thiollierea (Manns et al. 2012; Table 2). Estimated speciation rates within Oxera are comparable to the fastest island plant radiations documented in previous studies (e.g., Bidens in Hawaii or Echium in Macaronesia; Table 3). The detected early burst of diversification has been followed by a rather constant diversification, as inferred from BAMM and ML Treepar analyses even under weak divergence origination scenarios (Supplementary Appendices 12 and 14 available on Dryad); this may reflect the ongoing challenge within the evolutionary biology community to realistically model diversification processes, especially extinction (Stadler 2011; Morlon 2014). Nevertheless, it seems that since its origination Oxera has continued to diversify under constant speciation and rather null extinction, as exemplified by the exponential increase of species lineages through time (Fig. 2a).
Trait-dependent diversification models suggest that no specific adaptation has disproportionately favored speciation in Oxera (all biotic traits had equal speciation rates; Supplementary Appendices S4 and S18 available on Dryad), excepting perhaps the slender liana life form (acquired with the emergence of the neriifolia, pulchella, and oblongifolia subclades; see Table 4, Fig. 2a, Supplementary Appendix S18 available on Dryad), which is found in open sclerophyllous vegetation and exhibits high speciation rates probably because open habitats have increased in concert with Pliocene and Pleistocene cooling while remaining spatially fragmented (see below). However, the proliferation of species within Oxera might be related to the capacity of the genus to adopt different growth forms and floral and fruit morphologies, as hypothesized for angiosperms in general (Ricklefs and Renner 1994), probably in response to varied ecological opportunities available after its colonization of New Caledonia.
Table 4.
Combinations of ancestral node states (or character state for the single-species morierei lineage) of life forms, pollination and dispersal niches for each Oxera subclade, inferred from ancestral state reconstruction analyses
| Subclade/lineage | Life form | pollination niche | dispersal niche |
|---|---|---|---|
| robusta (1) | Robust liana | Gullet | Large rounded |
| subverticillata (2) | Robust liana | Gullet | Paddle |
| sulfurea (3) | Shrub | Gullet | Small rounded |
| baladica (4) | Monocaulous | Gullet | Paddle |
| neriifolia (5) | Slender liana | Bilabiate | Small rounded |
| pulchella (6) | Slender liana | Gullet | Small rounded |
| oblongifolia (7) | Slender liana | Bilabiate | Small rounded |
| morierei (8) | Slender liana | Gullet | Small rounded |
Numbers in brackets refer to tree node numbers in Figure 4. Clades with similar combinations are highlighted in grey.
The early diversification of Oxera seems to have been mainly associated with niche shifts driven by pollination syndromes, dispersal strategies, and life-form variety (Fig. 2b). This suggests that the early diversification burst within the clade coincides with ecological diversification, particularly in regards to pollen vectors, dispersal agents, and light requirements. Although it is not possible to determine whether these niche shifts were directly involved in the process of lineage divergence (ecological speciation), or have rather been caused by post-speciation adaptation to diverse ecological opportunities, these results suggest that diverse ecological opportunities have likely facilitated the diversification of Oxera within New Caledonia. The decelerated diversification of pollination syndromes, dispersal niches, and life-forms (revealed by a rather high phylogenetic signal; Fig. 2c) likely indicates that all or most of these niches previously vacant were ultimately filled during the early history of the clade. Our comparative analyses indicate that from the late Pliocene to early Pleistocene, the major Oxera lineages (i.e., the seven New Caledonian subclades plus O. morierei) had already originated (Fig. 2a), most of them occupying a unique niche combination. It is important to note that all recovered Oxera subclades were robust to contrasting phylogenetic signal between nuclear and plastid genes (Supplementary Appendix S8 available on Dryad). Hence, each Oxera subclade robustly holds a unique trait combination (Table 4), excepting perhaps the neriifolia and oblongifolia subclades, which similarly exhibit slender lianas, bilabiate flowers and small rounded fruits. This evolutionary divergence in life forms seems not to be correlated with habitat divergence, since these two characteristics have evolved largely independently across Oxera (Fig. 2b and c), suggesting that different growth forms occupy distinct functional niches related to light requirements within habitats.
Our study thus detected strong ecological segregation among Oxera subclades and species (Fig. 1c), but the precision of our morphological data may not be sufficient to discriminate finer ecological syndromes encountered in Oxera. Compared with our results, the genus may exploit an even larger diversity in pollinators and dispersers than highlighted in this study. For example, the actinomorphic flowers of Pacific species are sometimes visited by honeyeaters (Belcher and Sibson 1982; de Kok 1997). Gullet flowers with small and narrow tubes, such as the ones of O. sessilifolia, O. garoensis sp. nov. ined., and O. doubetiae sp. nov. ined., may be more efficiently pollinated by Diptera or Coleoptera rather than birds (Jourdan, pers. comm.). It would be illuminating to conduct detailed investigations on the pollination biology of different Oxera species using more precise measurements of floral morphology characters. This could reveal that pollination syndromes have in fact spurred diversification during the evolution of Oxera.
Recent Diversification Driven by the Interplay of Habitat Shifts and Allopatry
Our comparative analyses further suggest that since the end of Pliocene and the onset of the Pleistocene, the diversification of Oxera has been impacted by several punctual shifts in habitat preference (Fig. 2b and c). These events coincided with another intense period of soil morphogenesis and weathering events in New Caledonia which occurred between 3 and 2.5 Ma (Fig. 2a and b; Chevillotte et al. 2006), as well as several climatic oscillations documented in the south-western Pacific (Dodson and Macphail 2004). These events have likely caused myriad spatial rearrangements of New Caledonian biotas (Hope and Pask 1998, Stevenson et al. 2001, Stevenson 2004; Stevenson and Hope 2005), thus generating novel ecological opportunities, and hence the recent diversification of Oxera.
Phylogenetic patterns of geographic range overlap and asymmetry between pairs of species show that geographical isolation seems to have played a prominent role during the diversification of Oxera. This suggests that neutral divergence may have been at least as important as ecological speciation. Most species pairs within Oxera have clear allopatric distributions (null range overlap more frequent through all species pairs; Fig. 3). This is exemplified by the baladica subclade, which appears to have diversified almost entirely in allopatry (Fig. 4). Few species pairs clearly originated from sympatric speciation, as suggested by complete range overlaps and trait divergence (Figs. 3 and 4). Except for the oblongifolia subclade, the geographical range overlap between species remains remarkably low ( 0.4; Supplementary Appendix S19 available on Dryad) indicating that given the recent radiations of Oxera, most sister species have remained in allopatry, and that secondary sympatry may not have yet occurred in most sister species pairs.
0.4; Supplementary Appendix S19 available on Dryad) indicating that given the recent radiations of Oxera, most sister species have remained in allopatry, and that secondary sympatry may not have yet occurred in most sister species pairs.
The examination of geographic and ecological overlap between species pairs suggests that local species coexistence may in fact be very rare in Oxera. Among the 96 putative pairs of species (within the seven New Caledonian subclades), only five pairs potentially coexist locally, although this has never been noted after years of field observations (Gâteblé, pers. obs.). For example in the oblongifolia subclade (which exhibits largely sympatric species), each species grows in a specific set of environmental conditions, excepting the ubiquist O. oblongifolia (Fig. 4). Contemporary species coexistence is probably prevented by the recent interplay of allopatry and parapatry due to habitat shifts and environmental oscillations that have caused cycles of forest expansion and retraction. With Oxera being mainly a forest-adapted group (25 of 38 species confined to forests, and forest habitat identified as ancestral; Supplementary Appendix S17 available on Dryad), it is likely that vegetation cycles along the complex topographical gradients of New Caledonia have triggered spatial divergence and probably speciation. This process has been further amplified by the limited dispersal ability of Oxera since its dispersers are relatively sedentary. Such processes of vegetation oscillations along steep physical gradients, sometimes creating barriers to dispersal, is now considered to be a major driver of clade diversification, as demonstrated in recent theoretical simulations (Aguilée et al. 2012).
Conclusion: Is OXERA a Clear Adaptive Radiation from New Caledonia?
The genus Oxera constitutes a remarkable example of a rapid diversifying lineage that originated from a single ancestor. We observed that the group experienced an early burst of species diversification, followed by an exceptionally high and constant species diversification rate (Fig. 2b). This signature is considered typical of adaptive radiations for some authors (Guyer and Slowinski 1993; Slowinski and Guyer 1993; Soulebeau et al. 2015) but not all (Givnish 1997, 2015), mainly because it is usually thought that early diversification bursts are driven by the exploitation of new ecological opportunities, which allows lineages to enter new adaptive zones. The early history of Oxera seems to have been paved by niche shifts toward novel pollination, dispersal and life-form traits. These niche shifts later decelerated, perhaps due to a saturation of available niche space. However, our analyses do not highlight any diversity dependent diversification processes, which are often considered as another hallmark of adaptive radiations (Rabosky 2009). This is likely because other ecological opportunities have helped further diversification of the group after its initial phase of biotic niche exploration. More recent opportunities likely emerged through landscape formation and environmental oscillations which may have triggered allopatric and parapatric speciation processes. In fact, the explosive diversification of Oxera within New Caledonia appears to have been triggered by varied ecological opportunities during the course of evolution. Adaptive radiation should not be narrowly defined through quantitative tests or by quantifying diversification tempos, since some well-known adaptive radiations have little or no effect on speciation, or even a negative effect (Givnish 2015), and are therefore more identified by high rates of morphological and ecological diversification (Givnish 1997; Sanderson 1998); this remains to be investigated for the Oxera diversification.
The explosive radiation experienced by Oxera thus constitutes a remarkable model for studying the drivers involved in the evolution of the New Caledonian flora, especially for recent epochs. The genus is an unclassifiable, multifaceted radiation seemingly triggered by both adaptive and neutral processes (adaptive, geographical, climatic, etc.), and driven by several classes of biotic and abiotic factors. Oxera could be therefore regarded as a partial adaptive radiation since changing ecological opportunities have contributed to its radiation, with an early diversification of biotic traits (life-form, pollination, dispersal), but more recently driven by the joint action of habitat changes and allopatric speciation leading to scant local coexistence between species. Although different diversification drivers may have been overlapping in time, the early stage of Oxera diversification could be then viewed as a leapfrog radiation (sensu Chase and Palmer 1997), where diversification drivers have relayed to one another at different time periods, but geographic processes may have later been an equally important driver of speciation. Indeed, past climatic history of New Caledonia has also acted as a major evolutionary force, with two pivotal events, a dramatic aridity at the early Pliocene leading to the release of newly vacant niches, and the accentuation of glacial-interglacial cycles from the early Pleistocene, causing repeated biota rearrangements. From our study, it is legitimate to ask whether any other case of adaptive radiation can be clearly demonstrated in old islands system such as New Caledonia, since the distinctive hallmark of past adaptive divergence is progressively blurred by more recent environmental fluctuations creating new ecological opportunities.
Acknowledgements
The authors would like to thank all the people who provided plant material, access to specimens or bibliography: Laurence Ramon, Chanel Sam, and Stéphanie Stephens (PVNH); Felix Forest, Edith Kapinos, and Helen Fortune Hopkins (K); Vicki Funk and Carol Kelloff (US); Darren Crayn (CNS); Josephine Milne and Wayne Gebert (MEL); David Middleton and Nura Abdul Karim (SING); Jérôme Munzinger and Jacqueline Fambart-Tinel (NOU); Christiane Anderson (MICH); and the IAC’s team, especially Jacqueline Ounémoa, Virginie Lemay, Donny Wamytan, Laurent Demaret, Michel Moenteapo, Edmond Poitchili, Nicolas Anger, Hervé Vandrot, and Hélène Udo, for having collected, propagated and grown all New Caledonian Oxera species over the past 10 years. Jim Leebens-Mack kindly allowed us to use the 1Kp data for the Oxera species. We thank Daniel and Irène Létocart, Neil Sleddon, Druce Horton, and Kartuz Greenhouses for providing us with pictures of Oxera and habitats. Armelle Tardivel is also thanked for arranging the figures of this manuscript. We are also grateful to Daniel and Irène Létocart and Dominique Fleurot for their unconditional assistance in the field. We also thank the relevant New Caledonian authorities (Direction de l’Environnement de la Province Sud, Direction du Développement Economique et de l’Environnement de la Province Nord and Direction du Développement Economique de la Province des îles Loyauté) and the Vanuatuan Department of Environmental Protection and Conservation for collecting permits. The authors are very grateful to Julien Renaud, Nicolas Fernandez, and Damien Georges, who greatly helped on the SIG development, and the analyses computations under R. Some computations presented in this paper were performed using the CIMENT infrastructure (https://ciment.ujf-grenoble.fr), which is supported by the Rhône-Alpes region (GRANT CPER07_13 CIRA: http://www.ci-ra.org) and France-Grille (http://www.france-grilles.fr). Finally, we thank Yohan Pillon (IRD) for his corrections and useful comments on the manuscript, as well as Vincent Savolainen, James Rosindell, and Tom Givnish for their constructive evaluation of our manuscript.
Supplementary Material
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.mm1ft40 for DNA alignment matrices and geographical variables. The resulting trees are available from TreeBASE (ID 23353).
References
- Aguilée R.,, Claessen D.,, Lambert A. 2012. Adaptive radiation driven by the interplay of eco-evolutionary and landscape dynamics. Evolution. 67:1291–1306. [DOI] [PubMed] [Google Scholar]
- Aguilée R.,, Lambert A.,, Claessen D. 2011. Ecological speciation in dynamic landscapes. J. Evol. Biol. 24:2663–2677. [DOI] [PubMed] [Google Scholar]
- Anacker B.L.,, Strauss S.Y. 2014. The geography and ecology of plant speciation: range overlap and niche divergence in sister species. Proc. R. Soc. B Biol. Sci. 281:20132980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker W.J.,, Couvreur T.L.P. 2012. Global biogeography and diversification of palms sheds light on the evolution of tropical lineages. I. Historical biogeography. J. Biogeogr. 40:286–298. [Google Scholar]
- Barrabé L.,, Karnadi-Abdelkader G.,, Ounémoa J.,, de Kok R.P.J.,, Robert N.,, Gâteblé G. 2015. Recircumscription of Oxera (Lamiaceae: Ajugoideae) to include Faradaya based on molecular and anatomical data. Bot. J. Linn. Soc. 179:693–711. [Google Scholar]
- Barrabé L.,, Maggia L.,, Pillon Y.,, Rigault F.,, Mouly A.,, Davis A.P.,, Buerki S. 2014. New Caledonian lineages of Psychotria (Rubiaceae) reveal different evolutionary histories and the largest documented plant radiation for the archipelago. Mol. Phylogenet. Evol. 71:15–35. [DOI] [PubMed] [Google Scholar]
- Barré N.,, Villard P.,, Manceau N.,, Monimeau L.,, Ménard C. 2006. Les oiseaux de l’archipel des Loyauté (Nouvelle-Calédonie): inventaire et éléments d’écologie et de biogéographie. Revue d’Écologie 61:175–194. [Google Scholar]
- von Beichle U. 1987. Lebensraum, Bestand und Nahrungsaufnahme der Zahntaube, Didunculus strigirostris. J. Orn. 128:75–89. [Google Scholar]
- Belcher W.J.,, Sibson R.B. 1982. Birds of Fiji in colour. Auckand: Collins Publishers. [Google Scholar]
- Bonvallot J. 2012. L’orohydrographie. In: Bonvallot J.,, Gay J.-C.,, Habert E., editors. Atlas de la Nouvelle-Calédonie. Marseille-Nouméa: IRD-Congrès de la Nouvelle-Calédonie p. 25–28. [Google Scholar]
- Carlquist S.J.,, Baldwin B.G.,, Carr G.D. 2003. Tarweeds & Silverswords: evolution of the Madiinae (Asteraceae). St Louis, MO: Missouri Botanical Garden Press. [Google Scholar]
- Carpenter R.J.,, Read J.,, Jaffré T. 2003. Reproductive traits of tropical rain-forest trees in New Caledonia. J. Trop. Ecol. 19:351–365. [Google Scholar]
- Chase M.W.,, Palmer J.D. 1997. Leapfrog radiation in floral and vegetative traits among twig epiphytes in the orchid subtribe Oncidiinae. In: Givnish T.J.,, Sytsma K.J., editors. Molecular evolution and adaptive radiation. Cambridge: Cambridge University Press; p. 331–350. [Google Scholar]
- Chessel D.,, Dufour A.B.,, Thioulouse J. 2004. The ade4 package—I: one-table methods. R News. 4:5–10. [Google Scholar]
- Chevillotte V.,, Chardon D.,, Beauvais A.,, Maurizot P.,, Colin F. 2006. Long-term tropical morphogenesis of New Caledonia (Southwest Pacific): importance of positive epeirogeny and climate change. Geomorphology. 81:361–375. [Google Scholar]
- Cibois A.,, Thibault J.-C.,, Pasquet E. 2017. Uniform phenotype conceals double colonization by reed-warblers of a remote Pacific archipelago. J. Biogeogr. 34:1150–1166. [Google Scholar]
- Clark J.R.,, Ree R.H.,, Alfaro M.E.,, King M.G.,, Wagner W.L.,, Roalson E.H. 2008. A comparative study in ancestral range reconstruction methods: retracing the uncertain histories of insular lineages. Syst. Biol. 57:693–707. [DOI] [PubMed] [Google Scholar]
- Clark J.R.,, Wagner W.L.,, Roalson E.H. 2009. Patterns of diversification and ancestral range reconstruction in the southeast Asian-Pacific angiosperm lineage Cyrtandra (Gesneriaceae). Mol. Phylogenet. Evol. 53:982–994. [DOI] [PubMed] [Google Scholar]
- Cluzel D.,, Maurizot P.,, Collot J.,, Sevin B. 2012. An outline of the geology of New Caledonia; from Permian-Mesozoic Southeast Gondwanaland active margin to Cenozoic obduction and supergene evolution. Episodes. 35:72–86. [Google Scholar]
- Colombo R. 2008. Stratégies de dispersion chez quelques espèces de Sapotaceae de Nouvelle-Calédonie. Implications pour la conservation des forêts humides. Nouméa: Institut de Recherche pour le Développement, Université de Montpellier II. [Google Scholar]
- Cooper W.,, Cooper W.T. 2004. Fruits of the Australian tropical rainforest. Clifton Hill: Nokomis Publications. [Google Scholar]
- Crisp M.D.,, Cook L.G.,, Steane D. 2004. Radiation of the Australian flora: what can comparisons of molecular phylogenies across multiple taxa tell us about the evolution of diversity in present-day communities? Phil. Trans. R. Soc. B. 359:1551–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kok R.P.J. 1997. The biology and systematics of Oxera, Faradaya and Hosea (Labiatae) [D. Phil. Thesis]. [Oxford]: University of Oxford. [Google Scholar]
- de Kok R.P.J. 1998. The systematics and pollination biology of Oxera Labill., Faradaya F. Muell. and Hosea Ridl. (Labiatae). In: Saw L.G.,, Chua L.S.L,, Khoo K.C., editors. 4th International Flora Malesiana Symposium. Kuala Lumpur: Forest Research Institute Malaysia; p. 87–91. [Google Scholar]
- de Kok R.P.J.,, Mabberley D.J. 1999a. Generic and intra-generic delimitation of Oxera Labill. (Labiatae). Kew Bulletin. 54:257–264. [Google Scholar]
- de Kok R.P.J.,, Mabberley D.J. 1999b. The genus Faradaya F. Muell. (Labiatae). Blumea. 44:321–342. [Google Scholar]
- Dodson J.R.,, Macphail M.K. 2004. Palynological evidence for aridity events and vegetation change during the Middle Pliocene, a warm period in Southwestern Australia. Glob. Planet. Change. 41:285–307. [Google Scholar]
- Donoghue M.J.,, Edwards E.J. 2014. Biome shifts and niche evolution in plants. Annu. Rev. Ecol. Evol. Syst. 45:547–572. [Google Scholar]
- Drummond A.J.,, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzjohn R.G.,, Maddison W.P.,, Otto S.P. 2009. Estimating trait-dependent speciation and extinction rates from incompletely resolved phylogenies. Syst. Biol. 58:595–611. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick B.M.,, Turelli M. 2006. The geography of mammalian speciation: mixed signals from phylogenies and range maps. Evolution. 60:601–615. [PubMed] [Google Scholar]
- Foster C.S.P.,, Ho S.Y.W.,, Conn B.J.,, Henwood M.J. 2014. Molecular systematics and biogeography of Logania R.Br. (Loganiaceae). Mol. Phylogenet. Evol. 78:324–333 [DOI] [PubMed] [Google Scholar]
- Gallagher S.J.,, Greenwood D.R.,, Taylor D.,, Smith A.J.,, Wallace M.W.,, Holdgate G.R. 2003. The Pliocene climatic and environmental evolution of southeastern Australia: evidence from the marine and terrestrial realm. Palaeogeogr. Palaeoclimatol. Palaeoecol. 193:349–382. [Google Scholar]
- Gallaher T.,, Callmander M.W.,, Buerki S.,, Keeley S.C. 2015. A long distance dispersal hypothesis for the Pandanaceae and the origins of the Pandanus tectorius complex. Mol. Phylogenet. Evol. 83:20–32. [DOI] [PubMed] [Google Scholar]
- Gianoli E. 2004. Evolution of a climbing habit promotes diversification in flowering plants. Proc. Biol. Sci. 271:2011–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givnish T.J. 1995. Plant stems: biomechanical adaptation for energy capture and influence on species distributions. In: Gartner B.L., editor. Plant stems: physiology and functional morphology. San Diego, CA: Academic Press; p. 3–49. [Google Scholar]
- Givnish T.J. 1997. Adaptive radiation and molecular systematics: issues and approaches. In: Givnish T.J.,, Systma K.J., editors. Molecular evolution and adaptive radiation. Cambridge: Cambridge University Press; p. 1–54. [Google Scholar]
- Givnish T.J. 2010. Ecology of plant speciation. Taxon. 59:1326–1366. [Google Scholar]
- Givnish T.J. 2015. Adaptive radiation versus ‘radiation’ and ‘explosive diversification’: why conceptual distinctions are fundamental to understanding evolution. New Phytol. 207:297–303. [DOI] [PubMed] [Google Scholar]
- Givnish T.J.,, Millam K.C.,, Mast A.R.,, Paterson T.B.,, Theim T.J.,, Hipp A.L.,, Henss J.M.,, Smith J.F.,, Wood K.R.,, Sytsma K.J. 2009. Origin, adaptive radiation and diversification of the Hawaiian lobeliads (Asterales: Campanulaceae). Proc. Biol. Sci. 276:407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossenbacher D.L.,, Veloz S.D.,, Sexton, J.P. 2014. Niche and range size patterns suggest that speciation begins in small, ecologically diverged populations in North American monkeyflowers (Mimulus spp.). Evolution. 68:1270–1280. [DOI] [PubMed] [Google Scholar]
- Guyer C.,, Slowinski J.B. 1993. Adaptive radiation and the topology of large phylogenies. Evolution. 47:253–263. [DOI] [PubMed] [Google Scholar]
- Harmon L.J.,, Weir J.T.,, Brock C.D.,, Glor R.E.,, Challenger W. 2008. GEIGER: investigating evolutionary radiations. Bioinformatics. 24:129–131. [DOI] [PubMed] [Google Scholar]
- Haxaire J.,, Salesne T. 2016. Description d’un nouveau Sphingidae du genre Gnathothlibus Wallengren, 1858 de Nouvelle-Calédonie (Lepidoptera, Sphingidae). The Eur. Entomol. 8:179–208. [Google Scholar]
- He T.,, Lamont B.B.,, Fogliani B. 2016. Pre-Gondwanan-breakup origin of Beauprea (Proteaceae) explains its historical presence in New Caledonia and New Zealand. Sci. Adv. 2:e1501648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heibl C.,, Calenge C. 2013. Phyloclim: integrating phylogenetics and climatic niche modelling. Available from: https://CRAN.R-project.org/package=phyloclim. [Google Scholar]
- Hill M.O.,, Smith A.J.E. 1976. Principal component analysis of taxonomic data with multi-state discrete characters. Taxon. 25:249–255. [Google Scholar]
- Hope G.,, Pask J. 1998. Tropical vegetational changes in the late Pleistocene of New Caledonia. Palaeogeogr. Palaeoclimatol. Palaeoecol. 142:1–21. [Google Scholar]
- Huelsenbeck J.P.,, Nielsen R.,, Bollback J.P. 2003. Stochastic mapping of morphological characters. Syst. Biol. 52:131–158. [DOI] [PubMed] [Google Scholar]
- Hutson A.M.,, Mickelburgh S.P.,, Racey P.A. 1992. Old world fruit bats: an action plan for their conservation. Gland: IUCN. [Google Scholar]
- Jaffré T. 1980. Etude écologique du peuplement végétal des sols dérivés de roches ultrabasiques en Nouvelle-Calédonie. Paris: ORSTOM. [Google Scholar]
- Jaffré T. 1993. The relationship between ecological diversity and floristic diversity in New Caledonia. Biodiv. Lett. 1:82–87. [Google Scholar]
- Johnson S.D. 2010. The pollination niche and its role in the diversification and maintenance of the Southern African flora. Phil. Trans. R. Soc. B. 365:499–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen T.H.,, Olesen J.M. 2001. Adaptive radiation of island plants: evidence from Aeonium (Crassulaceae) of the Canary Islands. Perspect. Plant Ecol. Syst. 4:29–42. [Google Scholar]
- Karas C.,, Nurnberg D.,, Tiedemann R.,, Garbe-Schonberg D. 2011. Pliocene climate change of the Southwest Pacific and the impact of ocean gateways. Earth Planet. Sci. Lett. 301:117–124. [Google Scholar]
- Keppel G.,, Prentis P.,, Biffin E.,, Hodgskiss P.,, Tuisese S.,, Tuiwawa M.V.,, Lowe A.J. 2011. Diversification history and hybridisation of Dacrydium (Podocarpaceae) in remote Oceania. Aust. J. Bot. 59:262–273. [Google Scholar]
- Kim S.-C.,, McGowen M.R.,, Lubinsky P.,, Barber J.C.,, Mort ME,, Santos-Guerra A. 2008. Timing and tempo of early and successive adaptive radiations in Macaronesia. PLoS One. 3:e2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knope M.L.,, Morden C.W.,, Funk V.A.,, Fukami, T. 2012. Area and the rapid radiation of Hawaiian Bidens (Asteraceae). J. Biogeogr. 39:1206–1216. [Google Scholar]
- Kranitz M.L.,, Biffin E.,, Clark A.,, Hollingsworth M.L.,, Ruhsam M.,, Gardner M.F.,, Thomas P.,, Mill R.R.,, Ennos R.A.,, Gaudeul M.,, Lowe A.J.,, Hollingsworth P.M. 2014. Evolutionary diversification of new Caledonian Araucaria. PLoS One. 9:e110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losos J.B. 2010. Adaptive radiation, ecological opportunity, and evolutionary determinism. Am. Nat. 175:623–639. [DOI] [PubMed] [Google Scholar]
- Losos J.B.,, Ricklefs R.E. 2009. Adaptation and diversification on islands. Nature. 457:830–836. [DOI] [PubMed] [Google Scholar]
- Magallón S.,, Sanderson M.J. 2001. Absolute diversification rates in Angiosperm clades. Evolution 55:1762–1780. [DOI] [PubMed] [Google Scholar]
- Magallón S.,, Gómez-Acevedo S.,, Sánchez-Revees L.L.,, Hernández-Hernández T. 2015. A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol. 207:437–453. [DOI] [PubMed] [Google Scholar]
- Manns U.,, Wikström N.,, Taylor C.M.,, Bremer B. 2012. Historical biogeography of the predominantly Neotropical subfamily Cinchonoideae (Rubiaceae): into or out of America. Int. J. Plant Sci. 173:261–286. [Google Scholar]
- Meehan H.J.,, McConkey K.R.,, Drake D.R. 2002. Potential disruptions to seed dispersal mutualisms in Tonga, Western Polynesia. J. Biogeogr. 29:695–712. [Google Scholar]
- Momose K.,, Nagamitsu T.,, Inoue T. 1998. Thrips cross-pollination of Popowia pisocarpa (Annonaceae) in a lowland dipterocarp forest in Sarawak. Biotropica. 30:444–448. [Google Scholar]
- Morat P. 1993. Our knowledge of the flora of New Caledonia: endemism and diversity in relation to vegetation types and substrates. Biodiv. Lett. 1:72–81. [Google Scholar]
- Morat P.,, Jaffré T.,, Tronchet F.,, Munzinger J.,, Pillon Y.,, Veillon J.-M.,, Chalopin M. 2012. Le référentiel taxonomique Florical et les caractéristiques de la flore vasculaire indigène de la Nouvelle-Calédonie. Adansonia. 34:179–221. [Google Scholar]
- Morlon H. 2014. Phylogenetic approaches for studying diversification. Ecol. Lett. 17:508–525. [DOI] [PubMed] [Google Scholar]
- Munzinger J.,, Morat P.,, Jaffré T.,, Gâteblé G.,, Pillon Y.,, Tronchet F.,, Veillon J.-M.,, Chalopin M. 2016. FLORICAL: checklist of the vascular indigenous flora of New Caledonia. Version 22.IV.2016. Available from: http://www.botanique.nc/herbier/florical. [Google Scholar]
- Nylander J.A.A.,, Ronquist F.,, Huelsenbeck J.P.,, Nieves-Aldrey J.L. 2004. Bayesian phylogenetic analysis of combined data. Syst. Biol. 53:47–67. [DOI] [PubMed] [Google Scholar]
- Ogutcen E. 2016. The effects of dispersal and pollination on Plantaginaceae diversification [D. Phil. Thesis]. [Calgary: ]: University of Calgary. [Google Scholar]
- Pagel M. 1999. Inferring the historical patterns of biological evolution. Nature. 401:877–884. [DOI] [PubMed] [Google Scholar]
- Papadopulos A.S.T.,, Baker W.J.,, Crayn D.,, Butlin R.,, Kynast R.G.,, Hutton I.,, Savolainen V. 2011. Speciation with gene flow on Lord Howe Island. PNAS. 108:13188–13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E.,, Claude J.,, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 20:289–290. [DOI] [PubMed] [Google Scholar]
- Paun O.,, Turner B.,, Trucchi E.,, Munzinger J.,, Chase M.W.,, Samuel R. 2016. Processes driving the adaptive radiation of a tropical tree (Diospyros, Ebenaceae) in New Caledonia, a biodiversity hotspot. Syst. Biol. 65:212–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauw A. 2013. Can pollination niches facilitate plant coexistence? Trends Ecol. Evol. 28:30–37. [DOI] [PubMed] [Google Scholar]
- Pfeil B.E.,, Crisp M.D. 2008. The age and biogeography of Citrus and the orange subfamily (Rutaceae: Aurantioideae) in Australasia and New Caledonia. Am. J. Bot. 95:1621–1631. [DOI] [PubMed] [Google Scholar]
- Pillon Y. 2012. Time and tempo of diversification in the flora of New Caledonia. Bot. J. Linn. Soc. 170:288–298. [Google Scholar]
- Pillon Y.,, Barrabé L.,, Buerki S. 2017. How many genera of vascular plants are endemic to New Caledonia? A critical review based on phylogenetic evidence. Bot. J. Linn. Soc. 183:177–198. [Google Scholar]
- Pillon Y.,, Hopkins M.S.,, Rigault F.,, Jaffré T.,, Stacy E. 2014. A. Cryptic adaptive radiation in tropical forest trees in New Caledonia. New Phytol. 202:521–530. [DOI] [PubMed] [Google Scholar]
- Pillon Y.,, Munzinger J.,, Amir H.,, Lebrun M. 2010. Ultramafic soils and species sorting in the flora of New Caledonia. J. Ecol. 98:1108–1116. [Google Scholar]
- Pintaud J.-C.,, Jaffré T.,, Puig H. 2001. Chorology of New Caledonian palms and possible evidence of Pleistocene rain forest refugia. C. R. Acad. Sci. III. 324:453–463. [DOI] [PubMed] [Google Scholar]
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25:1253–1256. [DOI] [PubMed] [Google Scholar]
- Pouteau R.,, Trueba S.,, Feild T.S.,, Isnard S. 2015. New Caledonia: a Pleistocene refugium for rain forest lineages of relict angiosperms. J. Biogeogr. 42:2062–2077. [Google Scholar]
- Quiroga M.P.,, Mathiasen P.,, Iglesias A.,, Mill R.R.,, Premoli A.C. 2016. Molecular and fossil evidence disentangle the biogeographical history of Podocarpus, a key genus in plant geography. J. Biogeogr. 43:372–383. [Google Scholar]
- Rabosky D.L. 2009. Ecological limits and diversification rate: alternative paradigms to explain the variation in species richness among clades and regions. Ecol. Lett. 12:735–743 [DOI] [PubMed] [Google Scholar]
- Rabosky D.L.,, Grundler M.,, Anderson C.,, Title P.,, Shi J.J.,, Brown J.W.,, Huang H.,, Larson J.G. 2014. BAMMtools: an R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods Ecol. Evol. 5:701–707. [Google Scholar]
- Revell L.J. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3:217–223. [Google Scholar]
- Ricklefs R.E.,, Renner S.S. 1994. Species richness within families of flowering plants. Evolution. 48:1619–1636 . [DOI] [PubMed] [Google Scholar]
- Richards G.C. 1990. The spectacled flying-fox, Pteropus conspicillatus (Chiroptera: Pteropodidae), in north Queensland. 2. Diet, seed dispersal and feeding ecology. Aust. Mammal. 13:25–31. [Google Scholar]
- Ronquist F.,, Teslenko M.,, van der Mark P.,, Ayres D.L.,, Darling A.,, Höhna S.,, Larget B.,, Liu L.,, Suchard M.A.,, Huelsenbeck J.P. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundell R.J.,, Price T.D. 2009. Adaptive radiation, nonadaptive radiation, ecological speciation and nonecological speciation. Trends Ecol. Evol. 24:394–399. [DOI] [PubMed] [Google Scholar]
- Sanderson M.J. 1998. Reappraising adaptive radiation. Am. J. Bot. 85:1650–1655. [Google Scholar]
- Santiago L.S.,, Wright S.J. 2007. Leaf functional traits of tropical forest plants in relation to growth form. Funct. Ecol. 21:19–27. [Google Scholar]
- Sauquet H.,, Ho S.Y.,, Gandolfo M.A.,, Jordan G.J.,, Wilf P.,, Cantrill D.J.,, Baily M.J.,, Bromham G.K.,, Carpenter R.J.,, Lee D.M.,, Murphy D.J.,, Sniderman J.M.,, Udovicic F. 2012. Testing the impact of calibration on molecular divergence times using a fossil-rich group: the case of Nothofagus (Fagales). Syst. Biol. 61:289–313. [DOI] [PubMed] [Google Scholar]
- Sauquet H.,, Weston P.H.,, Anderson C.L.,, Barker N.P.,, Cantrill D.J.,, Mast A.R.,, Savolainen V. 2009. Contrasted patterns of hyperdiversification in Mediterranean hotspots. PNAS. 106:221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D. 2000. The ecology of adaptive radiation. Oxford: Oxford University Press. [Google Scholar]
- Selaya N.G.,, Anten N.P.R. 2008. Differences in biomass allocation, light interception and mechanical stability between lianas and trees in early secondary tropical forest. Funct. Ecol. 22:30–39. [Google Scholar]
- Silvertown J.,, Francisco-Ortega J.,, Carine M. 2005. The monophyly of island radiations: an evaluation of niche pre-emption and some alternative explanations. J. Ecol. 93:653–657. [Google Scholar]
- Slowinski J.B.,, Guyer C. 1993. Testing whether certain traits have caused amplified diversification—an improved method based on a model random speciation and extinction. Am. Nat. 142:1019–1024. [DOI] [PubMed] [Google Scholar]
- Soulebeau A.,, Aubriot X.,, Gaudeul M.,, Rouhan G.,, Hennequin S.,, Haevermans T.,, Dubuisson J.-Y.,, Jabbour F. 2015. The hypothesis of adaptive radiation in evolutionary biology: hard facts about a hazy concept. Org. Divers. Evol. 15:747–761. [Google Scholar]
- Stadler T. 2011. Inferring speciation and extinction processes from extant species data. PNAS. 108:16145–16146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson J. 2004. A late-Holocene record of human impact from the southwest coast of New Caledonia. Holocene. 14:888–898. [Google Scholar]
- Stevenson J.,, Dodson J.R.,, Prosser I.P. 2001. A late Quaternary record of environmental change and human impact from New Caledonia. Palaeogeogr. Palaeoclimatol. Palaeoecol. 168:97–123. [Google Scholar]
- Stevenson J.,, Hope G.A. 2005. Comparison of late Quaternary forest changes in New Caledonia and northeastern Australia. Quaternary Res. 64:372–383. [Google Scholar]
- Swenson U.,, Nylinder S.,, Munzinger J. 2013. Towards a natural classification of Sapotaceae subfamily Chrysophylloideae in Oceania and Southeast Asia based on nuclear sequence data. Taxon. 62:746–770. [Google Scholar]
- Tassin J.,, Boissenin M.,, Barré, N. 2010. Can Ptilinopus greyii (Columbidae) disperse seeds in New Caledonia’s dry forests? Pac. Sci. 64:527–532. [Google Scholar]
- Theim T.J.,, Shirk R.Y.,, Givnish T.J. 2014. Spatial genetic structure in four understory Psychotria species (Rubiaceae) and implications for tropical forest diversity. Am. J. Bot. 101:1189–1199. [DOI] [PubMed] [Google Scholar]
- Tobias J.A.,, Cornwallis C.K.,, Derryberry E.P.,, Claramunt S.,, Brumfield R.T.,, Seddon N. 2014. Species coexistence and the dynamics of phenotypic evolution in adaptive radiation. Nature. 506: 359–363. [DOI] [PubMed] [Google Scholar]
- Trail P.W. 1994. The phenology of rainforest plants in Tutuila, American Samoa. Biological Report Series 58 American Samoa Department of Marine & Wildlife Resources. [Google Scholar]
- Turner B.,, Munzinger J.,, Duangjai S.,, Temsch E.M.,, Stockenhuber R.,, Barfuss M.H.J.,, Chase M.W.,, Samuel R. 2013. Molecular phylogenetics of New Caledonian Diospyros (Ebenaceae) using plastid and nuclear markers. Mol. Phylogenet. Evol. 69:740–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitales D.,, Garnatje T.,, Pellicer J.,, Vallès J.,, Santos-Guerra A.,, Sanmartín I. 2014. The explosive radiation of Cheirolophus (Asteraceae, Cardueae) in Macaronesia. BMC Evol. Biol. 14:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff S.J.,, Bayly M.J.,, Garnock-Jones, P.J.,, Albach D.C. 2002. Classification, origin, and diversification of the New Zealand hebes (Scrophulariaceae). Ann. Mo. Bot. Gard. 89:38–63. [Google Scholar]
- Wagstaff S.J.,, Dawson M.I.,, Venter S.,, Munzinger J.,, Crayn D.,, Steane D.A.,, Lemson K.L. 2010. Origin, diversification, and classification of the Australasian genus Dracophyllum (Richeeae, Ericaceae). Ann. Mo. Bot. Gard. 97:235–258. [Google Scholar]
- Wagstaff S.J.,, Garnock-Jones P.J. 1998. Evolution and biogeography of the Hebe complex (Scrophulariaceae) inferred from ITS sequences. N. Z. J. Bot. 36:425–437. [Google Scholar]
- Warren D.L.,, Glor R.E.,, Turelli M. 2008. Environmental niche equivalency versus conservatism: quantitative approaches to niche evolution. Evolution. 62:2868–2883. [DOI] [PubMed] [Google Scholar]
- Weeks A.,, Zapata F.,, Pell S.K.,, Daly D.C.,, Mitchell J.D.,, Fine P.V.A. 2014. To move or to evolve: contrasting patterns of intercontinental connectivity and climatic niche evolution in “Terebinthaceae” (Anacardiaceae and Burseraceae). Front. Genet. 5:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker R.J.,, Triantis K.A.,, Ladle R.J. 2008. A general dynamic theory of oceanic island biogeography. J. Biogeogr. 35:977–994. [Google Scholar]
- Wotton D.M.,, Drake D.R.,, Powlesland R.G.,, Ladley J.J. 2016. The role of lizards as seed dispersers in New Zealand. J. R. Soc. N. Z. 46:40–65. [Google Scholar]
- Wotton D.M.,, Kelly D. 2012. Do larger frugivores move seeds further? Body size, seed dispersal distance, and a case study of a large, sedentary pigeon. J. Biogeogr. 39:1973–1983. [Google Scholar]
- Yang Y.,, Morden C.W.,, Sporck-Koehler M.J.,, Sack L.,, Wagner W.L.,, Berry P.E. 2018. Repeated range expansion and niche shift in a volcanic hotspot archipelago: Radiation of C4 Hawaiian Euphorbia subgenus Chamaesyce (Euphorbiaceae). Ecol. Evol. 8:8523–8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao G.,, Drew B.T.,, Yi T.S.,, Yan H.F.,, Yuan Y.M.,, Ge X.J. 2016. Phylogenetic relationships, character evolution and biogeographic diversification of Pogostemon s.l. (Lamiaceae). Mol. Phylogenet. Evol. 98:184–200. [DOI] [PubMed] [Google Scholar]
- Yuan Y.-W.,, Mabberley D.J.,, Steane D.A.,, Olmstead R.G. 2010. Further disintegration and redefinition of Clerodendrum (Lamiaceae): implications for the understanding of the evolution of an intriguing breeding strategy. Taxon. 59:125–133. [Google Scholar]
- Zachos J.,, Pagani M.,, Sloan L.,, Thomas E.,, Billups K. 2001. Trends, rhytms, and aberrations in global climate 65 Ma to present. Science. 292:686–693. [DOI] [PubMed] [Google Scholar]