Abstract
Extended spectrum β-lactamase (ESBL)-producing Klebsiella pneumoniae remain a critical clinical concern worldwide. The aim of this study was to characterize ESBL-producing K. pneumoniae detected within and between two hospitals in uMgungundlovu district, South Africa, using whole genome sequencing (WGS). An observational period prevalence study on antibiotic-resistant ESKAPE (i.e. Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp.) bacteria was carried out in hospitalized patients during a two-month period in 2017. Rectal swabs and clinical specimens were collected from patients hospitalized and were screened for ESBL-producing, Gram-negative ESKAPE bacteria using cefotaxime-containing MacConkey agar and ESBL combination disk tests. Nine confirmed ESBL-K. pneumoniae isolated from six patients and two hospitals were whole genome sequenced using an Illumina MiSeq platform. Genome sequences were screened for presence of integrons, insertion sequences, plasmid replicons, CRISPR regions, resistance genes and virulence genes using different software tools. Of the 159 resistant Gram-negative isolates collected, 31 (19.50%) were ESBL-producers, of which, nine (29.03%) were ESBL-K. pneumoniae. The nine K. pneumoniae isolates harboured several β-lactamase genes, including blaCTX-M-15, blaTEM-1b, blaSHV-1, blaOXA-1 concomitantly with many other resistance genes e.g. acc(6′)-lb-cr, aadAI6, oqxA and oqxB that confer resistance to aminoglycosides and/or fluoroquinolones, respectively. Three replicon plasmid types were detected in both clinical and carriage isolates, namely ColRNAI, IncFIB(K), IncF(II). Sequence type ST152 was confirmed in two patients (one carriage isolate detected on admission and one isolate implicated in infection) in one hospital. In contrast, ST983 was confirmed in a clinical and a carriage isolate of two patients in two different hospitals. Our data indicate introduction of ESBL-producing K. pneumoniae isolates into hospitals from the community. We also found evidence of nosocomial transmission within a hospital and transmission between different hospitals. The Clustered Regularly Interspaced Palindromic Repeats (CRISPR)-associated cas3 genes were further detected in two of the nine ESBL-KP isolates. This study showed that both district and tertiary hospital in uMgungundlovu District were reservoirs for several resistance determinants and highlighted the necessity to efficiently and routinely screen patients, particularly those receiving extensive antibiotic treatment and long-term hospitalization stay. It also reinforced the importance of infection, prevention and control measures to reduce the dissemination of antibiotic resistance within the hospital referral system in this district.
Subject terms: Microbial genetics, Microbial genetics, Clinical microbiology, Clinical microbiology
Introduction
The emergence of extended-spectrum-β-lactamases (ESBLs)-producing Klebsiella pneumoniae (ESBL-KP) represent a serious clinical concern in both healthcare and community settings1–3. ESBL-producing Enterobacteriaceae including K. pneumoniae were listed as pathogens of critical priority for research and development of antibiotics by the World Health Organization in 20174.
ESBL enzymes confer resistance to a large spectrum of β-lactam antibiotics including penicillins and third-generation cephalosporins, thus limiting effective therapeutic options, increasing morbidity, mortality and hospital costs2,5,6. Among the four molecular classes of beta-lactamases (A, B, C, and D) defined according to the Ambler classification, the most prevalent ESBLs belong to class A, namely CTX-M, TEM and SHV families. CTX-M enzymes are predominant worldwide; and five different groups of CTX-M including CTX-M-1-2-8-9-25 groups have been described in K. pneumoniae strains1,2,7.
A group of clinical bacteria, termed “ESKAPE” (Enterococcus spp., Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter spp.) pathogens encompass bacteria that can escape the activity of and develop high levels of resistance to multiple antibiotics8. A study on the complexity and diversity of 25 K. pneumoniae strains isolated in clinical samples (sputum, endotracheal aspiration, blood, pleural cavity tap, urine) showed different resistance genes with 100% prevalence of blaTEM and blaSHV. Additionally, various TEM and SHV types (SHV-19, 20, 21 and 22, and TEM-1, 53 and 63) were observed in K. pneumoniae isolates in KwaZulu-Natal9. Similarly, several studies conducted in the Western Cape region of South Africa have shown that various TEM-, SHV-, and CTX-M types were commonly detected in South African clinical isolates of K. pneumoniae10,11. The plasticity of K. pneumoniae related to the acquisition of resistance genes coupled with its propensity to act as nosocomial pathogen underscore the necessity for further investigation in different South African settings. This study undertook the molecular characterization of antibiotic resistance genes, virulence factors and mobile genetic elements associated with ESBL-KP isolated from carriage and clinical samples from patients in the public hospitals in the uMgungundlovu district, KwaZulu-Natal, South Africa using whole genome sequencing (WGS).
Results
Bacterial strains and phenotypic analyses
Of 159 cefotaxime-resistant Gram-negative ESKAPE bacteria isolated, 31 (19.5%) were ESBL-producers including K. pneumoniae (n = 9, 29.03%), Acinetobacter baumannii (n = 8, 25.81%), Pseudomonas aeruginosa (n = 8, 25.81%), Enterobacter aerogenes (n = 4, 12.90%) and Enterobacter cloacae (n = 2, 6.45%). Only the nine K. pneumoniae isolated from six patients were further analyzed. Amongst them, three (33%) were clinical isolates identified in the tertiary hospital and the remaining six (67%) isolates were identified in rectal swabs in both district (n = 3) and tertiary (n = 3) hospitals. Table 1 shows antibiotic resistance profiles of these isolates. All nine ESBL K. pneumoniae isolates were resistant to penicillins and cephalosporins, aminoglycosides, fluoroquinolones and trimethoprim (Table 1). The resistance observed was corroborated with WGS analyses which revealed the presence of several antimicrobial resistance determinants.
Table 1.
Antibiotic resistance profiles of carriage and clinical ESBL-producing K. pneumoniae isolates.
| Isolate ID | Patient ID (date of isolation) | MLST | Hospital | MIC values (µg/ml) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ampicillin | Cefoxitin | Cefotaxime | Ceftazidime | Imipenem | Meropenem | Gentamicin | Amikacin | Ciprofloxacin | Ofloxacin | Trimethoprim | ||||
| Carriage isolates | ||||||||||||||
| A105R2B2 | A105 (10/06/2017) | ST607 | District | ≥512 | ≥512 | ≥512 | ≥512 | 16 | 16 | 16 | 32 | 64 | 128 | ≥512 |
| A105R1B5 | ST983 | District | ≥512 | ≥512 | ≥512 | ≥512 | 32 | 16 | 8 | 8 | 64 | 128 | ≥512 | |
| A111R1B2 | A111 (10/06/2017) | ST17 | District | ≥512 | ≥512 | ≥512 | 32 | 2 | 0.25 | 4 | 8 | 32 | 64 | ≥512 |
| G702R1B5 | G702 (01/07/2017) | ST152 | Tertiary | ≥512 | 64 | ≥512 | ≥512 | 8 | 0.5 | 128 | 32 | ≥512 | ≥512 | ≥512 |
| G702R2B5 | G702 (03/07/2017) | ST152 | Tertiary | ≥512 | ≥512 | ≥512 | ≥512 | 4 | 2 | ≥512 | 128 | ≥512 | ≥512 | ≥512 |
| G702R3B2 | G702 (04/07/2017) | ST152 | Tertiary | ≥512 | 8 | ≥512 | ≥512 | 64 | 0.5 | 512 | 8 | 64 | 64 | ≥512 |
| Clinical isolates | ||||||||||||||
| ED01500733 | ED01500733 (13/07/2017) | ST983 | Tertiary | ≥32 | ≤4 | ≥64 | 16 | ≤0.25 | ≤0.25 | ≥16 | ≤2 | 2 | 2 | ≥512 |
| ED01502268 | ED01502268 (17/07/2017) | ST432 | Tertiary | ≥32 | ≤4 | ≥64 | ≥64 | ≤0.25 | ≤0.25 | 16 | 8 | 4 | 8 | ≥512 |
| ED01503757 | ED01503757 (23/07/2017) | ST152 | Tertiary | ≥32 | ≤4 | ≥64 | ≥64 | ≤0.25 | ≤0.25 | ≥16 | 8 | 4 | 4 | ≥512 |
Moreover, three isolates from three different patients showed imipenem resistance while meropenem resistance was observed in two isolates and elevated MICs for imipenem were evident in several isolates. The MICs of carbapenems varied across the type of specimen but all carriage isolates except A111R1B2 were resistant to imipenem according to the EUCAST breakpoints. In addition, two isolates (A105R1B5 and A105R2B2) out of the six carriage K. pneumoniae were resistant to meropenem. Carbapenemase genes were not found in the nine K. pneumoniae isolates although several isolates were resistant to imipenem and meropenem or evidenced elevated carbapenem MICs. Previous studies showed that deficiency or alteration of OmpK35 and OmpK36 confers increased resistance to carbapenems K. pneumoniae12,13. OMP analysis subsequently revealed the absence of OmpK35 and OmpK36 genes in all K. pneumoniae isolates although the wild type OmpK37 gene was present.
Genotypic analyses and antimicrobial resistance determinants
The nine ESBL-K. pneumoniae isolates harbored several resistance genes conferring resistance to aminoglycosides [aadAI6, aac(6′)Ib-cr, aac(3′)II-a, aph(6)Id, aph(3′)-Ib], sulphamides (sul1, sul2), tetracyclines (tetA, tetB, tetD), trimethroprim (dfrA14, dfrA27), phenicols (catA1, catB4) and fluoroquinolones (oqxA, oqxB). The most common β-lactam resistance genes were blaTEM-1-B (n = 9; 100%), blaCTX-M-15 (n = 9; 100%), blaSHV-1 (n = 8; 89%) and blaOXA-1 (n = 4; 44.5%). In addition, fosA (n = 9; 100%) and ARR-3 (n = 6; 66.7%) encoding for resistance to fosfomycin and rifampicin, respectively, were also detected. Alterations in the fluoroquinolone drug target due to modification in the quinolone-resistance-determining region (QRDR) including DNA gyrase subunit A (gyrA) and topoisomerase IV subunit C (parC) were observed in four isolates (ED01503757, G702R1B5, G702R2B5 and G702R3B2). In gyrA, mutation encoding the amino-acid substitution serine at codon 83 to phenyalanine (Ser83F) was observed in all four isolates whilst in parC the amino-acid substitution concerned serine at codon 80 to leucine (Ser80L). All K. pneumoniae strains carried an integron integrase gene IntIPac.
Multi-drug resistant (MDR) efflux pumps
All K. pneumoniae isolates harbored numerous MDR efflux pump genes including CmeA, CmeB, MATE, MFS, MacA, MarcB, MarA, OML, RND, AcrB and AcrAB. These MDR efflux pumps encode for resistance to several families of antibiotics including tetracyclines, fluoroquinolones, macrolides, tigecycline and β-lactams.
Multi-locus sequence type analysis (MLST) and core genome multi-locus sequence type analysis (cgMLST)
Analyses of MLST profiles has shown high variation among the seven housekeeping genes and identified five different sequence types (STs) including ST152 (n = 4), ST983 (n = 2), and three singleton ST432, ST607 and ST17 (Table 2). The four K. pneumoniae ST152 strains isolated from two patients were detected in clinical (n = 1) and carriage (n = 3) samples in the tertiary hospital while the two K. pneumoniae ST983 were each identified in carriage and clinical sample of patients admitted in the district and tertiary hospital, respectively (Table 2). The single-locus variants ST432, ST607 and ST17 were isolated from tertiary (n = 1) and district (n = 2) hospital, respectively.
Table 2.
Summary of phenotypic and genotypic characteristics of ESBL-producing K. pneumoniae isolates.
| Isolate | Patient ID (date of isolation) | Hospital levels | Sample types | *Antibiotic resistance genes | Plasmid types | pMLST | MLST | Integrons |
|---|---|---|---|---|---|---|---|---|
| A105R2B2 | A105 (10/06/2017) | District | Carriage | aadAI6, aadAI6/aadA10, aac(6′)Ib-cr, aac(3)-IIa, aac(3)-IId, aph(6)Id, blaSHV-1, blaSHV-1, blaTEM-1B, blaCTXM-15, oqxA, oqxB, fosA, qnrB6, ARR-3, sul1, sul2, dfrA27 | ColRNAI, FIA(pBK30683), IncFIB(K), FII(pBK30683), IncFII(K), IncR | IncF [K13:A13:B-] | ST607 | IntIPac |
| A111R1B2 | A111 (10/06/2017) | District | Carriage | aadAI6, aadA6/aadA10, aac(6′)Ib-cr, aph(6)Id, aph(3′)Ib, aac(3)-IIa, blaTEM-1B, blaSHV-11, blaCTX-M-15, blaSCO-1, oqxA, oqxB, qnrB6, qnrB54, fosA2, ARR-3, sul1, sul2, dfrA27 | ColRNAI, FIA(pBK30683), IncFIB(K), IncFII(K), IncR | IncF [K2-like:A13:B-] | ST17 | IntIPac |
| ED01502268 | ED01502268 (17/07/2017) | Tertiary | Clinical | aph(6)Id, aph(3’)Ib, blaTEM-1B, blaSHV-60, blaCTX-M-14, oqxA, oqxB, fosA, sul1, sul2, tet(A), tet(C), dfrA1 | IncFIB(K), IncFII(K) | IncF [K9:A-:B-] | ST432 | IntIPac |
| ED01500733 | ED01500733 (13/07/2017) | Tertiary | Clinical | aph(6)Id, aph(3′)Ib, blaTEM-1B, blaSHV-38, blaSHV-168, blaCTX-M-15, qnrB6, oqxA, oqxB, fosA, sul2, tet(A), tet(C), dfrA14, qnrB1 | IncFIB(K), IncFII(K) | IncF [K7:A-:B-] | ST983 | IntIPac |
| A105R1B5 | A105 (10/06/2017) | District | Carriage | aph(6)Id, aph(3′)Ib, blaTEM-1B, blaSHV-38, blaSHV-168, blaCTX-M-15, qnrB6, qnrB1, fosA2, oqxA, oqxB, fosA, sul2, tet(A), tet(C), dfrA14 | IncFIB(K), IncFII(K) | IncF [K7:A-:B-] | ST983 | IntIPac |
| G702R3B2 | G702 (04/07/2017) | Tertiary | Carriage | aadAI6, aac(6′)Ib-cr, aac(3)-IIa, aph(6)-Id, aph(3′)-Ib,blaTEM-1B, blaSHV-1, blaCTX-M-15, blaOXA-1, oqxA, oqxB, fosA, mph(A), catB4, catA1, ARR-3, sul1, sul2, tet(D), dfrA27 | ColRNAI, IncFIB(K), IncF(II) | IncF [K12:A-:B-] | ST152 | IntIPac |
| ED01503757 | ED01503757 (23/07/2017) | Tertiary | Clinical | aadAI, aadA2, aadAI6, aadAI6/aadA10, aac(6′)Ib-cr, aac(3)-IIa, aph(6)-Id, aph(3′)-Ib,blaTEM-1B, blaSHV-1, blaCTX-M-15, blaOXA-1, oqxA, oqxB, fosA, mph(A), catB4, catA1, ARR-3, sul1, sul2, tet(D), tet(C), dfrA27 | ColRNAI, IncFIB(K), IncF(II) | IncF [K12:A-:B-] | ST152 | IntIPac |
| G702R1B5 | G702 (01/07/2017) | Tertiary | Carriage | aadAI6, aadAI6/aadA10, aac(6′)Ib-cr, aac(3)-IIa, aph(6)-Id, aph(3′)-Ib, aac(6′)Ib8, blaTEM-1B, blaSHV-1, blaCTX-M-15, blaOXA-1, oqxA, oqxB, fosA, mph(A), catI, catA1, catB3, ARR-3, sul1, sul2, dfrA27, tet(C) | ColRNAI, IncFIB(K), IncF(II) | IncF [K12:A-:B-] | ST152 | IntIPac |
| G702R2B5 | G702 (03/07/2017) | Tertiary | Carriage | aadA5, aadAI6, aadAI6/aadA10, aac(6′)Ib8, aac(6′)Ib-cr, aac(3)-IIa, aph(6)-Id, aph(3′)-Ib, blaTEM-1B, blaSHV-1, blaCTX-M-15, blaOXA-1, oqxA, oqxB, qnrB6, fosA, mph(A), catA1, catI, catB3, catB4, ARR-3, sul1, sul2, tet(D), tet(A), dfrA27, dfrA14, tet(C) | ColRNAI, IncFIB(K), IncF(II), ColpVC, IncFIB(pKPHS1), IncFII(K), IncN, IncQ1 | IncN ST-5IncF [K12:A-:B-] | ST152 | IntIPac |
*Altogether, 2605 antibiotic resistance genes were investigated; pMLST: Plasmid multi-locus sequence type.
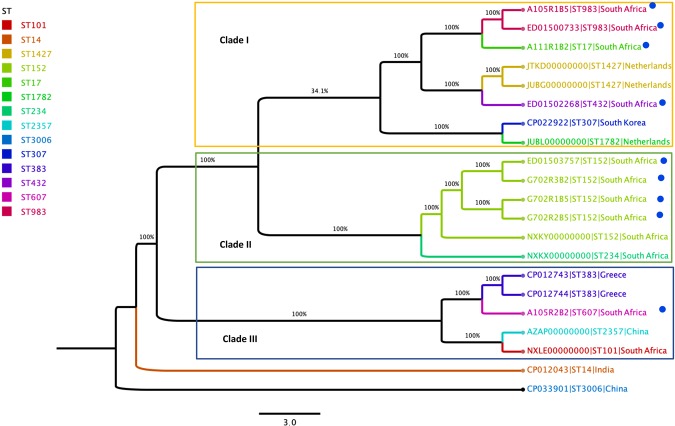
The cgMLST K. pneumoniae scheme was defined with NCBI data using K. pneumoniae K069 as the reference genome. The close relatedness between a batch of carriage (A105R1B5) and clinical (ED01500733) ST 983 strains isolated from the district and tertiary hospital, respectively was evident, with 100% identity and an allelic distance of zero (Fig. 1). Similarly, high genetic similarity was observed between carriage (G702R3B2, G702R1B5, G702R2B5) and clinical K. pneumoniae (ED01503757) ST152 strains originating from the tertiary hospital with 99% identity and an allelic distance of zero (Fig. 1).
Figure 1.
Phylogeny based on core genome multilocus sequence typing genes of 21 K. pneumoniae genomes. The following information is provided for each isolate: name/reference, MLST types (STs), and country. STs are highlighted as indicated in the legend and isolates present in the study are marked with a blue dot.
Mobile genetic elements (MGEs) analysis
PlasmidFinder showed that all strains harboured at least two plasmids of the incompatibility group F (IncF), namely the IncFII and IncFIB (Table 2). It further revealed that the IncF plasmid detected in all ESBL-KP ST152 were associated with the same plasmid replicons ST, K12:A-:B-, while the ST of the IncF plasmid replicons detected in ESBL-KP ST983 isolates was K7:A-:B-. The carriage strains, A111R1B2 (ST17) and A105R1B2 (ST607) isolated in the district hospital, additionally carried an IncR plasmid. In silico plasmid analyses also revealed that one ESBL-KP ST152 (G702R2B5) isolated in a carriage sample harboured eight plasmids including ColpVC, ColRNAI, IncFIB(K), IncFIB(pKPHS1), IncFII, IncFII(K), IncN (ST-5) and IncQ1. In addition, an integron integrase IntIPac closely to IntI-1 was detected through ISFinder in all ESBL-KP strains (Table 2).
PHAST algorithm demonstrated that 8 out of 9 strains (88.9%) hosted at least one intact bacteriophage (Supplementary Table 1). Entero P88 (n = 5; 62.5%) was the most prevalent intact phage followed by Salmon 118970 (n = 3; 37.5%), Entero mEp 235 (n = 2, 25%), and Klebsi phiKO2 (n = 2, 25%). Five and four phage regions were identified in K. pneumoniae ST152 (G702R2B5) and K. pneumoniae ST607 (A105R2B2) isolated in district hospital and carriage samples, respectively. Figure 2 shows the genomic organization of the phage regions in the K. pneumoniae ST152 (G702R2B5) while Fig. 3 represents its complete genomic structure.
Figure 2.
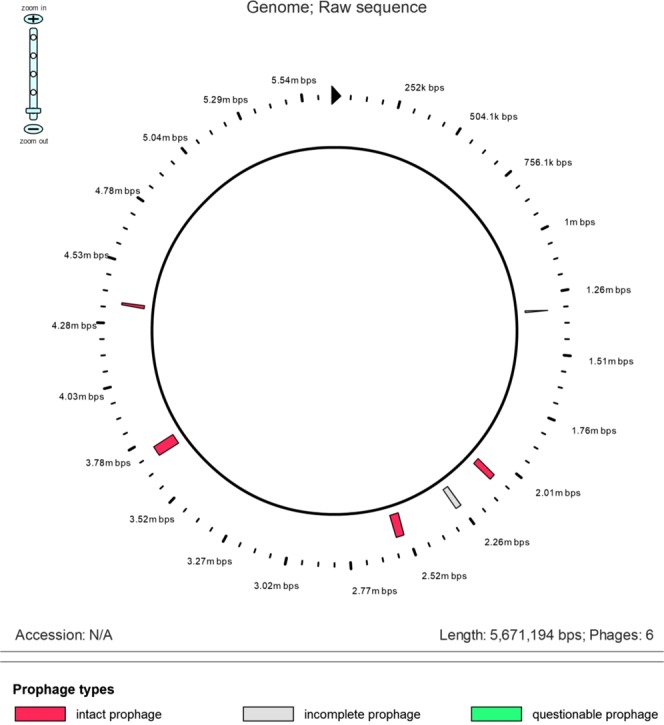
Genomic organization circular view of the phage regions in the K. pneumoniae ST152 (G702R2B5). The raw sequence presents the intact, incomplete and questionable prophage of K. pneumoniae G702R2B5.
Figure 3.
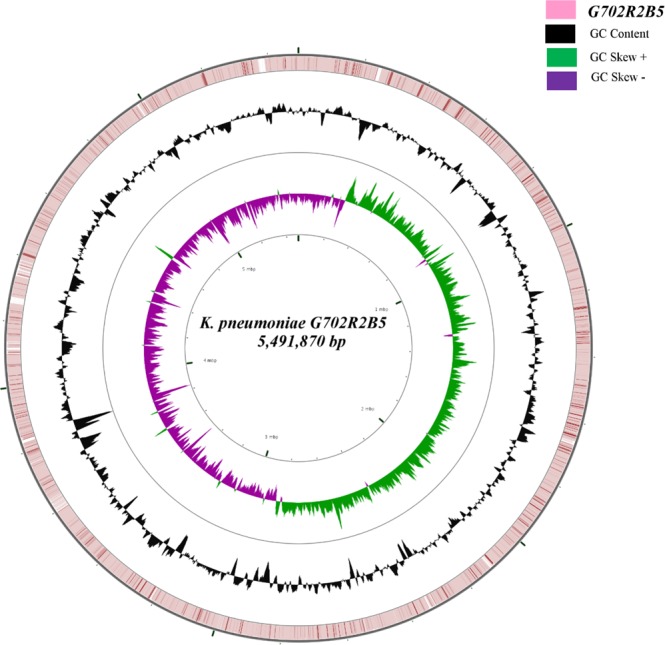
ESBL-producing K. pneumoniae G702R2B5 ring representation using CGView Server V 1.041. The inner ring displays the percent of identity comparing the complete genome of K. pneumoniae U25(CP012043) and K. pneumoniae G702R2B5. The two next (inner) rings display the GC skew and GC content, respectively. The outer most ring indicates the complete genome K. pneumoniae G702R2B5.
CRISPRFinder identified CRISPR (Clustered Regularly Interspaced Palindromic Repeats) regions in only 2 out of 9 ESBL-KP namely ESBL-KP ST607 (A105R2B2) and ST152 (G702R2B5). At least one CRISPR array including CRISPR1 or CRISPR2 was detected in these isolates. These two were both identified as CRISPR-associated cas3 gene (Fig. 4). ESBL-KP ST607 harboured six plasmids [ColRNAI, FIA(pBK30683), IncFIB(K), FII(pBK30683), IncFII(K) and IncR], four phages (Salmon SEN5, Entero P4, Pseudo JBD44, Entero fiAA91_ss) and both two CRISPRs (Supplementary Table 1). CRISPR1 and CRISPR2 were located at nucleotides 194435 to 195133 with 11 spacers and 203887 to 205203 with 21 spacers, respectively. In contrast, ESBL-KP ST152 (G702R2B5) carried eight plasmids [ColRNAI, IncFIB(K), IncFII, ColpVC, IncFIB(pKPHS1), IncFII(K), IncN and IncQ1], four phages (Klebsi PKP126, Salmon SSU5, Salmon SSU5, Salmon Fels 2) and the CRISPR1, located at nucleotides 11181 to 11759 with nine spacers.
Figure 4.
CRISP arrays detected in the K. pneumoniae G702R2B5 (3A) and A105R2B2 (3B). Two different Characterization of CRISPR arrays detected including (CRISPR 2) and (CRISPR 1 and 2) in K. pneumoniae G702R2B5 strain. The first CRISPR2 array composed of six direct repeated sequences and nine spacer sequences was located at nucleotides 11242 to 11731. CRISPR1 array composed twelve direct repeated sequences and eleven spacer sequences was located at nucleotides 194435 to 195045, CRISPR2 composed twenty-two direct repeated sequences and twenty-one spacers was located at nucleotides 203887 to 205176.
Discussion
The increasing prevalence of ESBL-KP remains a major clinical concern worldwide. To understand the molecular epidemiology of ESBL-KP, we studied using WGS, antibiotic resistance genes, MGEs and genetic lineages associated with circulating ESBL-KP isolated from carriage and clinical samples of hospitalized patients in uMgungundlovu district, South Africa.
The increasing prevalence of ESBL-KP has been associated with high mortality in developing country14. However, a 5% prevalence of ESBL-KP was detected in faecal carriage and clinical samples in our study. This is consistent with the report of Jallad et al.15, which shown 9.7% of ESBL-KP from faecal carriage among healthy patients in nursing homes in Lebanon15. In contrast, this finding is lower than that described by Perovic et al.16, where a 68.9% prevalence of ESBL-KP was detected in bloodstream infections in the public healthcare sector in Free State, Gauteng, Limpopo, KwaZulu-Natal, and Western Capes provinces in South Africa16. Similarly, Rashid et al.5 reported a 32.43% prevalence of ESBL-KP from faecal carriage in healthy patients hospitalized in tertiary hospital in India5. The discrepancies observed in the ESBL-KP prevalence could be attributed to variation of the geographic location, level of exposure to healthcare settings, hospital levels, antibiotic stewardship programs and antibiotic use.
The molecular characterization of diverse resistance determinants associated with the circulating ESBL-KP strains was undertaken following the health referral system. High level of resistance was detected in the tertiary hospital with one isolate, G702R2B5 (ST152) harboring 48 resistance genes in contrast to the district hospital where a maximum of 36 resistance genes were identified in the isolate A111R1B2 (ST17). The presence of genes encoding resistance to β-lactams, aminoglycosides, fluoroquinolones, fosfomycin, rifampicin, sulphonamide were reported in both clinical and carriage samples in the tertiary hospital. This is consonant to that reported in the literature where blaCTX-M-15, blaSHV-28, and blaTEM-1B and fosA3 were the common genes implicated in the resistance of cephalosporins, monobactams and fosfomycin identified in carriage and clinical K. pneumoniae isolate in Lebanon17 and China18. This suggest that ESBL-KP either in clinical or carriage sample, could be a probable reservoir of resistance genes for other bacterial species and be responsible for genetic transfer to other species. The dissemination of ESBL-KP in these healthcare settings could probably be attributed to a lack of effective infection, prevention and control (IPC) measures for their containment.
An interesting finding of this study was the detection of the clonal lineage ST152 (n = 4; 44.5%) circulating in both carriage (at admission, n = 3) and clinical sample (n = 1) of the tertiary hospital. These isolates were characterized by their multidrug resistance which was confirmed by the concomitant presence of several β-lactam (blaCTX-M-15, blaSHV-11, blaTEM-1B and blaOXA-1) resistance genes. This is consistent with the literature which confirmed that the blaSHV gene is a normal chromosomal gene in K. pneumoniae and that CTX-M-15 is the most predominant CTX-M enzyme worldwide19. In addition, resistance genes for aminoglycoside [aadAI6, aac(6′)Ib-cr, aac(3)-IIa, aph(6)-Id and aph(3′)-Ib], fluoroquinolone (oqxA, oqxB, and qnrB6), fosfomycin (fosA), macrolide [mph(A)], rifampicin (ARR-3), phenicol (catB4, catA1), tetracycline [tet(D)], trimethoprim (dfrA27) and sulphonamide (sul1, sul2) were also identified in these isolates. It is acknowledged that aac(6′)-Ib-cr is a variant of the aac(6′)-Ib gene which acetylates fluoroquinolones and has a low-level resistance to aminoglycosides. Mutation in gyrA (Ser83F) and parC (Ser80L) have further been detected in these four ST 152 isolates. Our MICs corroborate these findings since all these isolates exhibited high level resistance to fluoroquinolones, except for K. pneumoniae ED01503757 where moderate fluoroquinolone resistance was observed. ESBL-KP harboring similar resistance genes have been reported in Italia20 and Lebanon hospital8. Tokajian et al.17, showed that CTX-M-15 was associated with MDR-K. pneumoniae and revealed that qnrB6 was frequently observed in African countries17. Several studies showed that ESBL-producing K. pneumoniae ST152 is associated with resistance to carbapenems21–23.
Taken all together, the fact that ESBL-KP ST152 strains isolated in the tertiary hospital harbored the same resistance genes and mobile genetic elements including plasmids [ColRNAI, IncFIB(K), IncF(II)] and integrons (IntIPac) suggests that this clone could be associated with intra- and/or inter-hospital dissemination. This clonal spread was corroborated in our cgMLST analysis, where in clade II a high genetic relationship (99.90% identity and allelic distance of zero), was observed in our collection of carriage (G702R3B2, G702R1B5, G702R2B5) and clinical K. pneumoniae (ED01503757) ST152 strains, all originating from the tertiary hospital. In addition, a close relationship was observed with the K. pneumoniae ST152 strain K069 (NXKY0000000) detected in a clinical sample in Pretoria, South Africa. Of further interest is that one of the K. pneumoniae ST152 (ED01503757) isolates was carbapenem susceptible whereas the other ST152 isolates showed reduced susceptibility to carbapenems. Although the contribution of efflux mechanisms was neither determined by a MIC reduction assay nor gene expression assay, we hypothesized that the overexpression or repression of the numerous multidrug resistance efflux pumps detected in these isolates could be associated with the various MICs of imipenem and meropenem as described24.
Another interesting finding of this study, was the detection of two ESBL-KP ST 983 harboring similar resistant determinants [aph(6)Id, aph(3’)Ib, blaTEM-1B, blaSHV-38, blaCTX-M-15, qnrB66, oqxA, oqxB, fosA, sul2, tet(A), dfrA14], plasmids [IncFIB(K), IncFII(K)] and plasmid MLST [IncF (K7:A-:B-)]. They were isolated in two patients hospitalized in the district (A105R1B5) and tertiary (ED01500733) hospital, suggesting the probable clonal spread of the ESBL-KP ST983 inter-hospital in uMgungundlovu district as a result of the health referral system. The cgMLST analysis confirms that the two clinical (ED01500733) and carriage (A105R1B5) K. pneumoniae ST 983 in clade I were closely genetically related with 100% identity, an allelic distance of zero and an allele difference of one. Meanwhile, they shared a common ancestor with the K. pneumoniae ST 17 (A111R1B2) detected in the carriage sample of the district hospital. The two ST983 isolates ED01500733 and A105R1B5 were as such closely related to this ST17 isolate with an allelic distance of zero, a 99.41% percent identity and allele differences of 16640 and 16639 between A111R1B2 and A105R1B5, and between A111R1B2 and ED01500733, respectively. Our findings thus suggest that these ST 152 and ST 983 lineages could spread between patients in the same or different wards, within and/or across hospitals reflective of the referral system between hospitals in the public healthcare system. This spread further intimates sub-optimal infection prevention and control practices and provide motivation for the pro-active surveillance and routine screening of admitted patients in uMgungundlovu district in South Africa, in order to prevent the emergence and subsequent spread of new highly resistant clones.
Whilst Ambler classes A, B, C and D carbapenemase-producing K. pneumoniae strains gained worldwide attention due to the high resistance conferred to carbapenems, K. pneumoniae has evolved to become resistant to almost all β-lactams without harbouring carbapenemase genes13. This phenomenon has been possible with the concomitant use of multiple resistance mechanisms such as acquisition of an Ambler class A or C β-lactamases, with the loss of the OmpK35 and OmpK36 porins and/or overexpression of MDR efflux pumps13. In fact, some studies25,26 have established the impact of MDR-efflux pumps and porin losses on the membrane permeability of K. pneumoniae. The carbapenem resistance detected phenotypically (meropenem 16 mg/L; MIC imipenem 8–64 mg/L) in some isolates was not corroborated genotypically with the detection of specific carbapenemase encoding genes by WGS. This could hence be explained by the fact that resistance was not mediated by specific carbapenemase genes but rather by porin loss and/or MDR efflux pumps present in all isolates. Efflux systems have been reported in several clinically important bacteria and the overexpression of MDR efflux pumps can lead to high-level multi-drug resistance25,26. All the isolates harbored several MDR efflux pumps including CmeA, CmeB, MATE, MFS, MacA, MarcB, AcrB, MarA, OML, RND and AcrAB. We postulate that MDR efflux pumps were implicated in multi-drug resistance observed in the majority of isolates. K. pneumoniae contains three well-known porins including the two major porins OmpK35 and OmpK36 that are homologous to the OmpF and OmpC of Escherichia coli respectively, as well as the small porin OmpK37. Given that OmpK35 and OmpK36 porins play a critical role in the cell penetration of antibiotics, their loss can lead to reduce susceptibility or resistance to cephalosporins and carbapenems, especially in strains harboring Ambler class A, B, C or D β-lactamase12,13. The detection of OmpK37 porin that allows penetration by carbapenems but not other β-lactams, may explain why all isolates expressed high level resistance to cefoxitin (except for clinical isolates), cefotaxime and ceftazidime, and null or moderate resistance to carbapenems. We thus hypothesized that the deficiency in OmpK35 and OmpK36 coupled with the presence of blaCTX-M-15, and MDR efflux pumps, could play a significant role in conferring K. pneumoniae resistance to carbapenems and third generation cephalosporins in our study.
ABR is generally mediated by intrinsic or acquired resistance genes located on chromosome or MGEs, respectively. The CRISPR-Cas system was demonstrated to cleave plasmid DNA, thereby protecting bacteria from transduction (phage infection) and other horizontal gene transfers. They were supposed to be a defense mechanism against infection by diverse extra-chromosomal agents26. The correlation between the presence of CRISPR-Cas system and antibiotic resistance has already been studied and an inverse correlation between its presence and acquisition of antibiotic resistance was described in 48 Enterococcus faecalis strains26. In K. pneumoniae, CRISPR-Cas system has been detected in a very few strains worldwide. Apart from our isolates A105R2B2 and G702R2B5, only two complete K. pneumoniae genomes (NC_018522, NC_012731) and five draft genomes sequences (NC_012731, NZ_ANGH02000012, NZ_APGM01000001, NZ_JH930419, NZ_JH930428) harbor it. Even though CRISPR-Cas system serves to protect bacteria against phage infections and horizontal gene transfer, their presence among ESBL-KP ST607 (A105R2B2) and ST152 (G702R2B5) that were the most resistant isolates, suggest their probable implication in the acquisition of resistance genes. This is consistent with a report which demonstrated that in Klebsiella genomes, CRISPR-Cas systems are located among genes encoding for proteins that are likely involved in metabolism as well as resistance to antibiotics22. Additionally, the detection of several phages in these two highly resistant strains along with CRISPR-associated cas3 genes shed light on areas for further investigation on the emergence of ABR and transmission of antibiotic resistance genes.
In summary, our findings reveal the dissemination of ESBL-producing K. pneumoniae within and between wards and hospitals in uMgungundlovu district, South Africa. It shows that hospital is a reservoir for several resistance determinants and highlights the necessity to efficiently and routinely screen patients, particularly those receiving extensive antibiotic treatment and long-term hospitalization stay. It also reinforces the need for infection, prevention and control measures to reduce the dissemination of ABR in this district.
Methods
Ethical approval
Ethical approval was obtained from the Biomedical Research Ethics committee (BREC) (No. BF512/16, sub-study of BCA444/16) of the University of KwaZulu-Natal, South Africa. Permission to conduct the research was also granted from the Department of Health, uMgungundlovu District and hospital managers. All methods were performed in accordance with the relevant guidelines and regulations.
Study design and bacterial isolates
This study took place in two hospitals at different level of care (district and tertiary), from May to July 2017 in uMgungundlovu district, South Africa. The district and tertiary hospitals were approximately 70 Km apart. Oral and written informed consent were obtained from all study participants after explanation of the procedure and purpose of the study. Rectal swabs were collected aseptically with Amies swabs from all admitted in-patients >18 years old to form the carriage sample. Isolates routinely processed in the microbiological laboratory during the sampling period formed the clinical sample. Every patient included in this study was screened for the presence of Gram-negative ESKAPE bacteria. All samples were cultured onto MacConkey agar with and without cefotaxime (2 mg/L). After incubation for 18–24 h at 37 °C, each morphotype growing on MacConkey with cefotaxime (MCA + CTX) was subjected to Gram staining, catalase and oxidase tests, followed by biochemical identification with API 20E (bioMérieux, Marcy l’Etoile, France) and the Vitek® 2 System (bioMérieux, Marcy l’Etoile, France).
The strains sequenced in this study were isolated from carriage (A105R2B2, A105R1B2, A111R1B2, G702R1B5, G702R2B5, G702R3B2) and clinical samples including sputum (ED01503757, ED01502268) and urine (ED01500733) of six patients hospitalised in the district or tertiary hospital. The isolates A105R2B2, A105R1B2, A111R1B2 were detected in rectal swabs of two patients (A105 and A111) admitted in the medical ward of the district hospital while the isolates G702R1B5, G702R2B5, G702R3B2 were recovered from rectal swabs of a single patient (G702) but at different time-point (admission, after 48 h and at discharge) in the tertiary hospital. The clinical isolates ED01503757, ED01502268, ED01500733 were identified from three patients in the tertiary hospital. These isolates were closely related on enterobacterial-repetitive-polymerase chain reaction (ERIC-PCR)27 analysis28. Given that we aimed to evidence clonal spread of ESBL-KP, within each ERIC-cluster, representative isolates of related strains originating from different level of care were considered for WGS. These isolates were more representative because they belonged to the same ERIC-PCR cluster and antibiotic resistant patterns.
Phenotypic screening of ESBL-production and antimicrobial susceptibility testing
All isolates were phenotypically screened for ESBL, AmpC, KPC and MBL production using combination disk test sets (ROSCO DIAGNOSTICA, Taastrup, Denmark). Minimum inhibitory concentrations (MICs) were determined broth microdilution for all selected isolates. Ampicillin, cefoxitin, cefuroxime, cefotaxime, ceftriaxone, imipenem, meropenem, amikacin, gentamicin, trimethoprim, ciprofloxacin, moxifloxacin, nitrofurantoin, tetracycline, were tested and interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints29. E. coli ATCC 25922, K. pneumoniae ATCC 700603 and K. pneumoniae ATCC 51503 were used as controls.
DNA Extraction
Genomic DNA (gDNA) was extracted with the GenElute® Bacterial Genomic DNA Kit (Sigma-Aldrich, St. Louis, MO, USA) according the manufacturer’s instructions. The purity and concentration of the extracted gDNA were determined by fluorometric analysis (Qubit®) and agarose gel electrophoresis.
Genomic DNA Sequencing and assembly
Multiplexed paired-end libraries (2 × 300 bp) were prepared with the Nextera XT DNA sample preparation kit (Illumina, San Diego, CA, USA) and sequencing performed on an Illumina MiSeq instrument with 100 × coverage by the National Institute of Communicable Diseases Sequencing Core Facility, South Africa. The generated reads were quality trimmed and de novo assembled using CLC Genomics Workbench version 10 (CLC, Bio-QIAGEN, Aarhus, Denmark) and SPAdes version 3.530 to nullify any gaps.
Genome analysis
The assembled contigs were uploaded to the prokaryotic genome annotation pipeline server (https://www.ncbi.nlm.nih.gov/genome/annotation_prok/) and RAST server (http://rast.nmpdr.org/)31 for annotation.
Identification of the resistome, virulome and mobile genetic elements
The GoSeqIt tool was used to annotate and determine known antimicrobial resistance genes, virulence factors and plasmids using ResFinder32, VirulenceFinder33 and PlasmidFinder34, respectively. The RAST SEED viewer aided the identification of integrons and transposases flanking the β-lactamase genes35. The identification, annotation and visualization of prophage associated regions were performed using PHAge Search Tool (PHAST) server36. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and insertion sequence elements were investigated with the CRISPRFinder server (http://crispr.i2bc.paris-saclay.fr/Server/) and ISFinder (https://www-is.biotoul.fr/)37, respectively. Outer membrane porin genes were analyzed with the Sequence Search Antibiotic Resistance Tool (SSTAR, version 1.1.01, https://github.com/tomdeman-bio/Sequence-Search-Tool-for-Antimicrobial-Resistance-SSTAR-)38 software that used a standalone Basic Local Alignment Search Tool (BLAST) and a database combining ARG-ANNOT and ResFinder to identify known antibiotic resistance genes, detect putative new variants, modification and/or truncated genes. In addition, the Comprehensive Antibiotic Resistance Database (CARD; https://card.mcmaster.ca) was used to corroborate the results. Finally, the contigs of the K. pneumoniae G702R2B5 were mapped against the complete genome of K. pneumoniae U25 (CP012043) for visualization of the genomic organization39.
Multilocus Sequence typing (MLST) and core genome multi-locus sequence type analysis (cgMLST)
The scheme of Diancourt et al.38, which considers the allelic variation amongst seven housekeeping genes (gapa, infb, mdh, pgi, phoe, rpob and tonb) to assign STs was used for in silico multi-locus sequence type (MLST)-analyses and WGS data were used for the MLST assignment of K. pneumoniae isolates40.
A genome-wide gene-by-gene comparison approach was used to assess the clonal relatedness between isolates within and across wards and hospitals. The core genes were determined from the annotated genome assemblies, predicted coding regions were extracted and converted into protein sequences. A phylogeny was drawn for K. pneumoniae using Rapid large-scale prokaryote pangenome analysis (Roary; https://sanger-pathogens.github.io/Roary/) to estimate the tree for the core genome. The genome of K. pneumoniae strain K069 (Accession number NXKY01000005.1) served as reference genome and the following 12 query international K. pneumoniae genomes (Accession numbers JUBG00000000, JTKD00000000, JUBL00000000, JUBM00000000, AZAP00000000, CP012743, CP012744, CP012043, NXLE01000020.1, NXKY01000005.1, NXKX01000020.1, CP022922.1, CP033901.1) obtained from NCBI database were used to assess the cgMLST target genes. Altogether, 2944 core genes were extracted with an alignment length of 2,852,207 bp shared by the nine K. pneumoniae genomes. The allelic distance from the cgMLST was visualized using Figtree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/) in a maximum likelihood phylogenetic tree including isolate name, ST type and country.
Supplementary information
Acknowledgements
We are grateful to the NCBI GENBANK submission staff for help with genome upload, decontamination and deposition procedures. This work was supported by the Antimicrobial Research Unit (ARU) and College of Health Sciences (CHS) of the University of KwaZulu-Natal. The National Research Foundation funded this study through the NRF Incentive Funding for Rated Researchers (Grant No. 85595), the NRF Competitive Grant for Rated Researchers (Grant no. 106063) and the DST/NRF South African Research Chair in Antibiotic Resistance and One Health (Grant No. 98342) awarded to professor S.Y.Essack. The South African Medical Research Council through the Self-Initiated Research (Sir) Grant also funded the WGS aspect of study. Any opinions, findings and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the organizations or agencies that provided support for the project. The funders had no role in the study design, nor the decision to submit the work for publication.
Author Contributions
R.C.F. co-conceptualized the study, undertook sample collection, microbiological laboratory and data analyses, prepared tables, interpreted results, contributed to bioinformatics analysis, and drafted the manuscript. L.L.F. undertook sample collection, microbiological laboratory analyses, contributed to data analysis and vetting of the results. M.S. undertook bioinformatics analyses. A.I. performed whole genome sequencing analysis. S.Y.E. co-conceptualized the study and undertook critical revision of the manuscript. All authors read and approve the final manuscript.
Data Availability
This whole-genome shotgun project PRJNA429538 of K. pneumoniae strains ED01500733, ED01502268, A105R2B2, A111R1B2, G702R3B2, ED01503757, A105R1B5, G702R1B5, G702R2B5 has been deposited at DDBJ/EMBL/GenBank under accession numbers POWS00000000, POTV00000000, POWR00000000, POTU00000000, POWQ00000000, POTT00000000, POTS00000000, POWP00000000 and POWO00000000, respectively.
Competing Interests
Professor Essack is a member of the Global Respiratory Infection Partnership sponsored by unrestricted educational grant from Reckitt and Benckiser, UK. All other authors declare that there is no competing financial and non-financial interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-42672-2.
References
- 1.Founou L, Founou R, Essack S. Antibiotic Resistance in the Food Chain: A Developing Country-Perspective. Front Microbiol. 2016;7:1881. doi: 10.3389/fmicb.2016.01881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kluytmans-van den Bergh M, et al. Whole-Genome Multilocus Sequence Typing of Extended-Spectrum-Beta-Lactamase-Producing Enterobacteriaceae. J Clin Microbiol, 2016;54:12. doi: 10.1128/JCM.01648-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lei L, et al. mcr-1 in Enterobacteriaceae from Companion Animals, Beijing, China, 2012–2016. Emerg Infect Dis J. 2017;23:4. doi: 10.3201/eid2304.161732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization . Research, Discovery, and Development of New Antibiotics. Geneva: World Health Organization; 2017. Global Priority List of Antibiotic-Resistant Bacteria to Guide. [Google Scholar]
- 5.Rashid, M. et al. Carriage of ESBL and AmpC-Positive Enterobacteriaceae in Gastrointestinal Tract of Healthy Community Subjects and Hospitalized Patients and Detection of blaCTX-M Gene in ESBL Positive Isolates. Int J Curr Microbiol App Sci (2015).
- 6.Mohammed Y, Gadzama GB, Zailani SB, Aboderin AO. Characterization of Extended-Spectrum Beta-lactamase from Escherichia coli and Klebsiella Species from North Eastern Nigeria. J Clin Diagn Res. 2016;10:2. doi: 10.7860/JCDR/2016/16330.7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woerther PL, Burdet C, Chachaty E, Andremont A. Trends in Human Fecal Carriage of Extended-Spectrum β-Lactamases in the Community: Toward the Globalization of CTX-M. Clin Microbiol Rev. 2013;26:4. doi: 10.1128/CMR.00023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Founou RC, Founou LL, Essack SY. Extended spectrum beta-lactamase mediated resistance in carriage and clinical gram-negative ESKAPE bacteria: a comparative study between a district and tertiary hospital in South Africa. Antimicrob Res. Infect Control. 2018;7:134. doi: 10.1186/s13756-018-0423-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Essack SY, Hall LM, Pillay DG, McFadyen ML, Livermore DM. Complexity and diversity of Klebsiella pneumoniae strains with extended-spectrum beta-lactamases isolated in 1994 and 1996 at a teaching hospital in Durban, South Africa. Antimicrob Agents Chemother. 2001;45:1. doi: 10.1128/AAC.45.1.88-95.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buys H, Muloiwa R, Bamford C, Eley B. Klebsiella pneumoniae bloodstream infections at a South African children’s hospital 2006-2011, a cross-sectional study. BMC Infect Dis. 2016;16:570. doi: 10.1186/s12879-016-1919-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandeep. V., Larry. O., Isaac. M., Mary B.J. Molecular characteristics and antibiotic resistance profiles of Klebsiella isolates in Mthatha, Eastern Cape Province, South Africa. Int J Microbiol, 7 (2017). [DOI] [PMC free article] [PubMed]
- 12.Shi W, et al. Carbapenem and cefoxitin resistance of Klebsiella pneumoniae strains associated with porin OmpK36 loss and DHA-1 beta-lactamase production. Braz J Microbiol. 2013;44:2. doi: 10.1590/S1517-83822013000200015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaczmarek FM, Dib-Hajj F, Shang W, Gootz TD. High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of bla(ACT-1) beta-lactamase production, porin OmpK35/36 insertional inactivation, and down-regulation of the phosphate transport porin phoe. Antimicrob Agents Chemother. 2006;50:10. doi: 10.1128/AAC.00285-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Founou, R. C., Founou, L. L., Essack, S. Y. Clinical and economic impact of antibiotic resistance in developing countries: A systematic review and meta-analysis. Plos One12(12) (2017). [DOI] [PMC free article] [PubMed]
- 15.Jallad, M., Naoufal, R., Irani, J., Azar, E. Extended Spectrum Beta-Lactamase Carriage State among Elderly Nursing Home Residents in Beirut. The Scientific World Journal (2015). [DOI] [PMC free article] [PubMed]
- 16.Perovic O, et al. National sentinel site surveillance for antimicrobial resistance in Klebsiella pneumoniae isolates in South Africa, 2010-2012. S Afr Med J. 2014;104(8):563. doi: 10.7196/SAMJ.7617. [DOI] [PubMed] [Google Scholar]
- 17.Tokajian S, Eisen JA, Jospin G, Farra A, Coil DA. Whole genome sequencing of extended-spectrum β-lactamase producing Klebsiella pneumoniae isolated from a patient in Lebanon. Front Cell Infect Microbiol. 2015;5:32. doi: 10.3389/fcimb.2015.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mei, Y. f. et al. Virulence and Genomic Feature of a Virulent Klebsiella pneumoniae Sequence Type 14 Strain of Serotype K2 Harboring blaNDM–5 in China. Front Microbiol8(335) (2017). [DOI] [PMC free article] [PubMed]
- 19.Aniela W, et al. Porin alterations present in non-carbapenemase producing Enterobacteriaceae with high and intermediate levels of carbapenem resistance in Chile. J Med Microbiol. 2012;61:1270–1279. doi: 10.1099/jmm.0.045799-0. [DOI] [PubMed] [Google Scholar]
- 20.Becker L, et al. Whole Genome Sequence Analysis of CTX-M-15 Producing Klebsiella Isolates Allowed Dissecting a Polyclonal Outbreak Scenario. Front. Microbiol. 2018;9:322. doi: 10.3389/fmicb.2018.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou K, et al. Characterization of a CTX-M-15 Producing Klebsiella Pneumoniae Outbreak Strain Assigned to a Novel Sequence Type (1427) Front Microbiol. 2015;6:1250. doi: 10.3389/fmicb.2015.01250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaman TU, et al. Clonal diversity and genetic profiling of antibiotic resistance among multidrug/carbapenem-resistant Klebsiella pneumoniae isolates from a tertiary care hospital in Saudi Arabia. BMC Infect Dis. 2018;18:1. doi: 10.1186/s12879-018-3114-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dianelys Q, et al. High Clonal Diversity in a Non-Outbreak Situation of Clinical ESBL-Producing Klebsiella pneumoniae Isolates in the First National Surveillance Program in Cuba. Microb Drug Resist. 2014;20:1. doi: 10.1089/mdr.2013.0024. [DOI] [PubMed] [Google Scholar]
- 24.Filgona J, Banerjee T, Anupurba S. Role of efflux pumps inhibitor in decreasing antibiotic resistance of Klebsiella pneumoniae in a tertiary hospital in North India. J Infect Dev Ctries. 2015;9:815–820. doi: 10.3855/jidc.6216. [DOI] [PubMed] [Google Scholar]
- 25.Rimoldi SG, et al. Whole genome sequencing for the molecular characterization of carbapenem-resistant Klebsiella pneumoniae strains isolated at the Italian ASST Fatebenefratelli Sacco Hospital, 2012–2014. BMC Infect Dis. 2017;17:666. doi: 10.1186/s12879-017-2760-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin TL, et al. Imipenem represses CRISPR-Cas interference of DNA acquisition through H-NS stimulation in Klebsiella pneumoniae. Scientific Reports. 2016;6:31644. doi: 10.1038/srep31644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Founou RC, Founou LL, Essack SY. Extended spectrum beta-lactamase mediated resistance in carriage and clinical gram-negative ESKAPE bacteria: a comparative study between a district and tertiary hospital in South Africa. Antimicrobial Resistance & Infection Control. 2018;7(1):134. doi: 10.1186/s13756-018-0423-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.EUCAST. Breakpoint tables for interpretation of MICs and zone diameters. Available from http://www.eucast.org. 2017.
- 30.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aziz RK, et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics. 2008;9(1):75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zankari E, et al. Identification of acquired antimicrobial resistance genes. JAC. 2012;67(11):2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joensen KG, et al. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014;52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carattoli A, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Overbeek, R. et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res, 42(Database issue), D206–D214 (2014). [DOI] [PMC free article] [PubMed]
- 36.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39(Suppl. 2):W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ostria-Hernandez ML, Sanchez-Vallejo CJ, Ibarra JA, Castro-Escarpulli G. Survey of clustered regularly interspaced short palindromic repeats and their associated Cas proteins (CRISPR/Cas) systems in multiple sequenced strains of Klebsiella pneumoniae. BMC research notes. 2015;8:332. doi: 10.1186/s13104-015-1285-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Man T, Limbago B. SSTAR, a stand-alone easy-to-use antimicrobial resistance gene predictor. mSphere. 2016;1(1):e00050–00015. doi: 10.1128/mSphere.00050-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grant JR, Arantes AS, Stothard P. Comparing thousands of circular genomes using the CGView comparison tool. BMC Genomics. 2012;13:202. doi: 10.1186/1471-2164-13-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This whole-genome shotgun project PRJNA429538 of K. pneumoniae strains ED01500733, ED01502268, A105R2B2, A111R1B2, G702R3B2, ED01503757, A105R1B5, G702R1B5, G702R2B5 has been deposited at DDBJ/EMBL/GenBank under accession numbers POWS00000000, POTV00000000, POWR00000000, POTU00000000, POWQ00000000, POTT00000000, POTS00000000, POWP00000000 and POWO00000000, respectively.