Abstract
A common focus of fermentation process optimization is the product titer. Different strategies to boost fermentation titer target whole‐cell biocatalyst selection, process control, and medium composition. Working at higher product concentrations reduces the water that needs to be removed in the case of aqueous systems and, therefore, lowers the cost of downstream separation and purification. Different approaches to achieve higher titer in fermentation are examined. Energy and water consumption data collected from different Cargill fermentation plants, i.e., ethanol, lactic acid, and 2‐keto‐L‐gulonic acid, confirm that improvements in fermentation titer play a decisive role in downstream economics and environmental footprint.
Keywords: Downstream processing, Energy saving, Microbial fermentation, Titer increase, Water reduction
1. Aqueous Fermentation Systems
In nature, the different organic molecules that correspond to commodity chemicals, i.e., alcohols, polyols, acids, amino acids, polysaccharides etc., exhibit excellent solubility in water and the bioconversion reactions they derive from primarily occur in hydrophilic systems, with the exception of long‐chain fatty acids 1. Therefore, industrial processes designed to produce the respective biomolecules in large volumes involving renewable resources and natural reaction systems, in most cases are carried out in water suspension 2, 3, 4. Currently, due to the use of renewable resources as fermentation substrates for the production of value‐added compounds 5, the manufacturing processes result in rather complex aqueous systems, wherefrom the products need to be separated 6. Thus, these technological approaches require the implementation of advanced downstream processing methods, which contribute to higher production costs 7.
Additionally, to facilitate greater yields and overcome inhibition phenomena or mass transfer issues, highly diluted systems are employed. For further processing, especially for but not limited to platform molecules serving as intermediates within different value chains of the chemical industry, it is crucial that the water is reduced to a minimum or even removed 7, 8. Likewise, logistic considerations also dictate the concentration of the products for efficient storage and transportation to end customers.
With regard to downstream processing and chemical work‐up, high concentrations significantly contribute to lowering capital investment cost (CAPEX) and operational expenditures (OPEX), which constitute key elements in designing competitive processes. In general, for industrial commodity fermentations based on renewable feedstock the largest costs are allocated with downstream processing, e.g., separation and purification 9, 10. The perspective of optimizing all parameters associated with the use of whole‐cell biocatalysts, i.e., strain performance, medium composition, and operating conditions, with an emphasis on achieving high titer can reduce CAPEX, energy costs, and water consumption throughout the process.
In the optimization of fermentation processes, increasing the titer of the fermentation directly impacts the energy requirements. For dilute solutions, increasing the titer disproportionally reduces the requirement as about 0.9 kg of water per kg fermentation product need to be removed to raise the concentration from 10 to 11 %. However, only 0.09 kg of water per kg of fermentation product need to be removed to increase the concentration from 33 to 34 %. So, while there are economic benefits in reduced energy use, there are also environmental benefits. Greenhouse gas emissions will also be decreased with reduced energy use, with the exact decline depending upon the evaporation technology (multiple effect evaporation, mechanical vapor recompression, combined heat and power) and the primary energy sources to drive the evaporation process.
2. Strategies for Titer Increase in Biotechnological Processes
Increasing the fermentation titer during the production of any molecule depends upon a number of aspects related to the microorganism and the process design (Fig. 1). Initially, the selection of a suitable microorganism that will fit the type of process exhibiting high productivity and substrate conversion rate are considered primordial for any economically competitive, large‐scale process 11, 12. The possibility to improve the phenotype of a given strain, as well as the use of recombinant microorganisms engineered for dedicated purposes are reviewed in the current study. Subsequently, constant monitoring of crucial processing parameters, such as inoculum quality, medium composition, feeding strategy, pH, agitation, aeration, and process temperature are required 12, 13.
Figure 1.
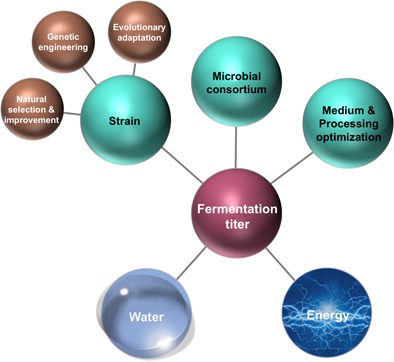
Schematic overview of strategies to improve the upstream part of fermentation processes, associated with whole‐cell biocatalyst selection, operation setup, and medium composition to elevate product titers. Consequently, high product titers impact the downstream efforts for separation and purification, economizing water and energy consumption.
Fermentation process development can focus on multiple targets including productivity (gram product per liter and hour), metabolic yield, and titer. Each of these impacts the economics of commodity‐based fermentation processes. While optimization of these parameters post‐commercialization can still achieve reductions in energy and water use, the greatest effect on CAPEX relies on optimization prior to the build. Therefore, it is the titer that can drive the best return on investment with respect to overall CAPEX and OPEX.
3. Strain Development
3.1. Natural Selection and Strain Improvement
In 1928, Alexander Fleming isolated a Penicillium chrysogenum strain producing a hitherto unknown antimicrobial substance, namely, the antibiotic penicillin. Subsequently, more research was performed during the later decades leading to the discovery of another fungal strain producing 100‐fold more antibiotic under submerged conditions. Further improvements through mutagenesis resulted in 100 000‐fold higher titer compared to the original fungal strain 14. Random mutagenesis mediated by chemical mutagens, UV irradiation, and high‐throughput screening of strains has traditionally been deployed for increasing the titers of fermentation processes 15.
For instance, the production of micafungin, a semisynthetic lipopeptide used worldwide against invasive fungal infections including candidemia and abscesses, involves an enzymatic deacylation step performed via an acylase produced by a Streptomyces sp. strain. Through consecutive mutagenesis steps and selection in small‐scale assays, enhanced acylase activity was achieved. When combined with an optimized fermentation setup the hyper‐production of the targeted enzyme reached 65‐fold increase 16.
For the discovery of hyper‐butanol‐producing solventogenic Clostridium strains, a rapid and high‐throughput spectrophotometric assay was designed in 96‐well plates to enable identification of strains with desirable phenotype of improved butanol production 17. Similarly, in an approach to select a potent lipid‐producing yeast, a miniaturized assay was designed using flowerplates mimicking the agitation applied in bioreactors, allowing for real‐time monitoring of growth and lipid production. In this way, different strains of Yarrowia lipolytica were evaluated with respect to storage of lipids in microtiter‐scale but under conditions close to those of bench‐top bioreactors 18.
In general, despite the tremendous increase in production rates that can be achieved based on reported cases, these methods have certain drawbacks. Strain improvement constitutes a very time‐consuming approach since several rounds of mutagenesis are normally needed at the risk of causing accumulation of undesirable mutations that eventually lead to crippled strains 14. Moreover, such approaches with random outcomes may not always allow the overview of the obtained phenotype without intense testing. Likewise, the high‐throughput screenings are cumbersome and in some instances do not represent industrial conditions 18. Therefore, the great potential of using metabolic engineering has propelled the efforts to improve cell‐catalysts 12, 13.
3.2. Genetic Modifications through Strain Engineering
Nowadays, with the advances in molecular biology and bioinformatics, coupled with the boom of sequencing and omic techniques, it is possible to express natural products in bacterial or yeast cells. The most frequently encountered species are Escherichia coli 19 and Saccharomyces cerevisiae 20, 21. The competitive advantages that render these species ideal hosts are their relatively rapid growth, their deeply dissected physiology and well‐known behavior in fermenters as well as the wide accessibility to genetic tools amenable to these organisms. The knowledge around the genetics of these species and the requirement of inexpensive cultivation media to achieve high cell densities in bioreactors are also key factors 21. The approaches are not limited to the production of industrially relevant commodity chemicals but also to the production of recombinant proteins 22. For example, E. coli has been implemented for the manufacturing of nearly 30 % of the approved recombinant therapeutics 19.
Another industrially important production process of bulk biochemicals is that of amino acids, i.e., L‐lysine and L‐glutamate, which serve among others as flavor enhancers 11. The fermentation process was initiated making use of wild strains of Corynebacterium glutamicum and has recently become one of the most promising processes for commercial production of amino acids because genetic engineering routes facilitated a substantial yield increase for the targeted compounds of up to 50 mass %, along with higher specificity and productivity. By applying site‐specific mutations, pathway modifications, and transcriptional attenuation regulations on E. coli, the production of aromatic amino acids, i.e., L‐phenylalanine, L‐tyrosine, and L‐tryptophan, and branched chain amino acids, i.e., L‐valine, L‐leucine, and L‐isoleucine, was successful 11. These compounds are valuable for use as feed supplements, cosmetic and pharmaceutical applications, and due to engineered E. coli strains a wider range of simple carbohydrates, e.g., glucose, sucrose, mannose, xylose, arabinose, galactose, and fructose, can serve as substrates, as well as alternative, low‐cost feedstocks, consisting of industrial by‐products containing glycerol 11.
The most abundant component of mammalian collagen is trans‐4‐hydroxy‐L‐proline (Hyp) which is widely used as an additive in personal care applications. Currently, Hyp is generated through collagen hydrolysis, which incurs heavy environmental pollution. Metabolic engineering on C. glutaricum targeting the reconstruction of the TCA cycle and redirecting the carbon flux towards Hyp synthesis led to a final concentration of 21.7 g L−1, opening the way for a pioneering production of Hyp in glucose‐minimal medium 23. A different type of metabolic engineering, employed in a Bacillus subtilis strain, resulted in the coproduction pathway for uridine and acetoin for the first time. By using glycerol as carbon source, a simultaneous accumulation of 40 g L−1 uridine and 60 g L−1 acetoin was achieved improving the economics of the fermentation and generating two value‐added compounds in a single fed‐batch process 24.
Production of fermentative butanol as an alternative fuel from plant biomass has reached improved titers employing recombination or modification of Clostridium strains, targeting the overexpression or disruption of genes 25. Likewise, a Clostridium tyrobutyricum strain has been metabolically engineered for improved butyric acid production (+61 %) and high butyrate/acetate ratio (+32 %) resulting in an overall more economic process performance 26. Concerning the secretory production of recombinant proteins from microbial hosts, several applications have been described with medical or industrial interest.
Yeasts like S. cerevisiae and others, e.g., Pichia pastoris, Hansenula polymorpha, and Kluyveromyces lactis, are thermotolerant, halotolerant, and utilize unusual carbon sources providing an inventory of versatile and cost‐effective expression systems 27. The recombinant protein therapeutics approved by food and drug authorities in Europe and USA are almost exclusively produced by S. cerevisiae, e.g., insulin, serum albumin, and hepatitis antigen 21. S. cerevisiae, which is most widely employed for bioethanol production from agro‐food residues is engineered to deplete pentoses, apart from hexoses so as to increase the titers 15, 20.
Nonetheless, shortcomings can be encountered as the optimization process can be complicated and strenuous. E. coli hyper‐expression of eukaryotic proteins in some cases results in formation of inclusion bodies due to higher transcription rates, adversely impacting the protein functionality and final titers 19. Apart from this, the issue of protein degradation needs to be addressed carefully as it can significantly impede the production of high amounts of stable protein with the correct folding. To avoid such issues, an integrated approach is followed 22.
In the future, genome editing will allow the engineering of strains with desired features in a simpler and more amenable manner. Currently, the emergence of an innovative approach for targeted genome editing mediated by an adaptive immunity system present in Streptococcus pyogenes, which incorporates foreign DNA (bacteriophage or plasmid) into the bacterial genome to memorize and later cleave it, could serve as a tool for integration of foreign genomic elements in a microorganism. This system, namely CRISPR/Cas9, could facilitate rapid and precise metabolic pathway modifications without the shortcomings of strain engineering involving auxotrophic markers and multiple‐step processes 27. This highly sophisticated genome‐editing tool has been used in anaerobic clostridial acetogens to develop an efficient microbial chassis to ferment C1 gases, such as CO and CO2, into biofuels and chemicals. A Clostridum ljungdahlii mutant was used as a paradigm for the selective redirection of carbon‐to‐ethanol production in an efficient way using CRISPR/Cas9, avoiding the addition of antibiotic resistance genes. Such applications revolutionize the field of Synthetic Biology, opening new paths to sustainable solutions for fuels from non‐petrochemical sources and reduction of greenhouse gas emissions 28.
3.3. Evolutionary Adaptation of Strains
Several attempts to construct evolved bioethanol‐producing S. cerevisiae yeast strains have been reported with respect to mixed sugar fermentation, i.e., hexoses and pentoses. Recombinant and wild‐type strains have been subjected to consecutive cultivation steps or batch fermentations to trigger adaptation to and utilization of non‐preferable substrates 29. Evolutionary engineering was applied on an industrial strain of S. cerevisiae by continuous fermentation on xylose and arabinose. Subsequently, the fermentation was carried out under increasing concentration of pentoses and eventually selection of strains was performed after 70 generations. The evolved strains showed improved growth rate and final cell mass when growing on pentoses and achieved greater ethanol yields under both aerobic and anaerobic conditions 30. In the same context, improvement through evolution was deployed in a S. cerevisiae strain to valorize a stream of red‐algal biomass for production of bioethanol. The strain had reduced catabolic repression at all growth phases towards galactose, and a coordinated carbon and amino acid metabolism was investigated through a multi‐omic approach. Similar applications open wide horizons for sustainable biotechnological solutions mediated by pioneering fermentation routes 31.
Overall, such efforts have already been reviewed concerning improved stress tolerance (e.g., ethanol, acetic acid, 3‐hydroxypropionic acid, high temperature), vitamin prototrophy, higher rates of substrate consumption (e.g., glycerol, xylose, arabinose), and product formation (e.g., enhanced aromatic amino acid flux). This approach constitutes a promising path for strain improvement especially when genetic engineering is combined with automated serial‐transfer or sequential batch experiments that facilitate massive parallelization 32.
4. Implementation of a Microbial Consortium
In several biotechnological processes, the implementation of cocultures has an advantageous impact on production yields, cost‐effectiveness of media formulations, and ensuring control of the process. Therefore, the production of enzyme molecules, numerous chemicals, food or pharmaceutical additives as well as antimicrobial substances rely on a beneficial interaction between multiple microorganisms 33.
For the industrial production of acetic acid destined for food and pharmaceutical applications, the cultivation of two microorganisms is usually employed in a two‐step fermentation setup or in a single bioreactor process, wherein conversion of glucose into acetic acid occurs. The yeast Zymomonas mobilis and an acetic acid bacterium (AAB) member of genera Acetobacter, Gluconobacter or Gluconacetobacter are used in coculture to firstly oxidize glucose into ethanol and subsequently oxidize ethanol into acetic acid, respectively. The latter oxidative route is carried out through primary dehydrogenase enzymes anchored in the periplasmic side of the cytoplasmic membrane and associated with the terminal ubiquinol oxidase via ubiquinone during respiration of the respective AAB strain. The fermentation process is faster and more economical, reducing the time and water usage by combining the two steps in one biofermentor 33.
In the case of bioethanol production from plant material with high cellulose, hemicellulose, and lignin content, a combination of cultures is implemented. The strategy is straightforward deploying yeast strains that apart from the high hydrolytic potential and the tolerance to inhibitor molecules formed during fractionation/pretreatment of the substrate also possess complementary carbohydrate uptake specificities as both pentose and hexose molecules are generated during the chemical steps prior to fermentation 4, 20. In addition, faster utilization of cleaved monomers prevents product inhibition of the cellulolytic enzymes, which accelerates the bioethanol production process. S. cerevisiae is usually combined with Pichia fermentans or Scheffersomyces stipitis to target glucose and xylose, respectively, leading to a more efficient carbohydrate utilization pattern and elevated bioethanol yields 34.
In the case of the industrial production of 2‐keto‐L‐gulonic acid (KGA), the precursor molecule of vitamin C, the fermentation process is often conducted with a coculture 35, 36. Ketogulonicigenium vulgare is the microorganism converting sorbose into KGA and variations are widely encountered in this biotechnological process 37. The enzymes involved in this bioconversion are encoded by genes which have been located in different positions and multiple copies in the chromosome or the plasmids of K. vulgare strains 38, 39. The genomic variance though, is poorly correlated with the yield of tested strains. To optimize the performance of K. vulgare with respect to conversion of sorbose into KGA, the presence of an allochthonous strain belonging to the genus Bacillus is required. This process is essentially implementing a microbial consortium of two bacteria, of which the former is the producing and the latter is the helper strain 40. In this coculture, the establishment of microbial cooperation is rather unclear and requires substantial investigation. However, there has been evidence that a member of the genus Bacillus boosts the yield and productivity of K. vulgare through mutualistic interactions based on the exchange of amino acids, growth cofactors, i.e., purines and nucleosides, and adaptation to environmental stressors 41, 42.
Different species have been described in association with K. vulgare such as Bacillus cereus, Bacillus endophyticus, Bacillus megaterium, and Bacillus thuringiensis, and the enhanced growth and metabolite kinetics, as well as the improved adaptation mechanism for both strains after numerous consecutive cultivation cycles have been demonstrated 43, 44, 45. Comparative proteomic analysis before and after the evolutionary adaptation of the coculture indicated the underlying basis of this mutualistic relationship 43.
5. Optimization of Medium Composition and Processing Conditions
Previously, medium optimization methods relied on classical approaches, which were cumbersome, cost‐demanding, time‐consuming, and mainly required numerous experimental repetitions to evaluate parameter contribution, nonetheless with low accuracy 46. Currently, for the fine‐tuning of fermentation conditions and the identification of essential medium components, modern mathematical/statistical techniques are employed, facilitating a faster and more reliable definition of a fermentation setup. An increase in productivity reduces the overall cost of the product, as well as the production cost; hence, it is one of the important topics for research.
Nutrients that constitute a source of carbon, nitrogen, and phosphorous are essential elements for the buildup of all biomolecules. Glucose and glycerol are widely used carbon sources in the fermentation industry as they combine broad availability with competitive price. Microorganisms can assimilate them easily leading to increased growth rates; however, for the production of secondary metabolites, like antibiotics, such carbon sources can impede the efficient product formation 14. In these complex scenarios, the carbon sources can negatively regulate secondary metabolite gene expression or post‐transcriptional processing. Numerous studies corroborate the dependence of achievable titer on nutrient type or concentration, as well as the formation and accumulation of precursors. A range of simple carbohydrates, i.e., glucose and glycerol, cause carbon catabolite repression, suppressing the formation of antibiotics, unlike other substrates such as lactose and galactose 14. On the other hand, ammonium serves as the nitrogen source in most cases due to the rapid utilization, whereas phosphorus is supplied in the form of phosphate salt which at low concentration facilitates the initiation of antibiotic production.
In the case of amino acid production by C. glutamicum, a wide spectrum of simple carbohydrates can be used, albeit when glucose concentration > 50 g L−1 coincides with an L‐glutamic acid concentration > 12 g L−1, the growth decreases significantly 11. The specifications mentioned in the aforementioned examples prove that medium limitations cannot be predicted on a genetic basis only but require an iterative optimization of the medium. Nowadays, non‐statistical approaches, such as evolutionary computational methods and artificial neural networks, have been employed for medium design like in the case of polyols and the production of xylitols 47.
Nonetheless, apart from the composition of a production medium, the fermentation conditions and processing configurations are equally crucial. Selection of batch, fed‐batch or continuous operation mode have different specificities. Also, parameters related to the bioreactor, such as type and scale of the fermentor, applied shear stress due to agitation, aeration intensity, and dissolved oxygen, can affect titer and productivity 12. Although processing conditions are extensively reviewed, they remain process‐dependent and adapted to the available infrastructure and capabilities of production plants. For instance, an engineered Pichia pastoris was evaluated under different agitation, aeration, and pH conditions for production of 2,3‐butanediol using glucose as feedstock in 5‐L fermentors. Low agitation and moderate aeration resulted in higher production of 2,3‐butanediol compared to acetoin, and a modified medium formulation was implemented through statistical medium optimization 48.
The production of nemadectin, a broad‐spectrum insecticide synthesized by Streptomyces cyaneogriseus ssp. noncyanogenus was optimized with regard to dissolved oxygen and shear rate 49. The product is one of the most widely applied and biggest selling acaricides and anthelmintics currently available. Studying the effect of these two parameters in 5‐L bioreactors helped define the agitation rate to achieve the optimal dissolved oxygen level and the shear stress that triggered the highest nemadectin titer. Overall, fine‐tuning of processing conditions requires multiple parallel experiments by maintaining all factors constant except for one variable. Subsequent modeling of the obtained results can facilitate the optimal setup for a process that will again be assessed at industrial scale.
6. Impact of Product Titer on Energy and Water Consumption
Cargill plants across the globe have been implementing the aforementioned approaches in a very wide range of commodity fermentations. The primary goal apart from developing competitive and cost‐efficient processes is also reducing the environmental footprint concerning energy demand and water consumption. In Figs. 2–4, the significant improvement from both aspects is reported using data collected throughout the continuous monitoring of production facility performances. In Fig. 2, ethanol production from yeast fermentation using renewable resources results in a complex aqueous system, wherefrom ethanol needs to be separated. The analysis performed in two distinct locations underpinned that under normal operation conditions small increases in titer of ethanol (+ 6 %) during the fermentation step can reduce downstream steam usage by 4–6 %. Further improvement in ethanol titer coinciding with an increase of around 20 % resulted in approximately 10 % less steam.
Figure 2.
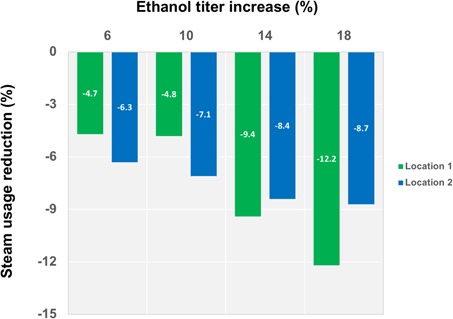
Graph correlating ethanol titer increase and steam usage reduction data, collected from two Cargill plants.
Figure 4.
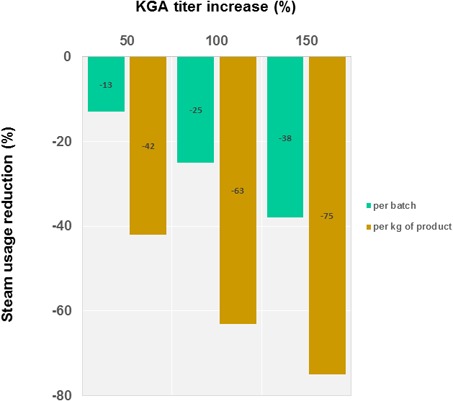
Graph correlating KGA titer increase with steam usage reduction throughout the historical improvement of the process.
In Fig. 3, the improvement of the lactic acid titer is correlated with water consumption data from Cargill's lactic acid production facility. The water consumption attained an overall reduction of 21 %. Stepwise water reduction was concomitant with sequential reduction in energy usage for downstream processing.
Figure 3.
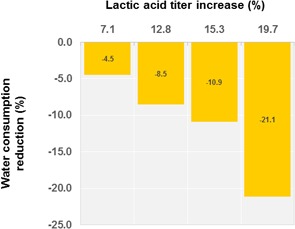
Graphs correlating lactic acid titer increase with water consumption reduction.
During the EU‐funded consortium project Prodias (grant agreement no. 637077), Cargill focused on optimization of the fermentation titer in KGA as a means of reducing its water and energy footprint in this process. This work builds on a historic approach to fermentation optimization, which has achieved a cumulative 150 % increase in titer. Consequently, the steam required to concentrate the fermentation mass and purify KGA has decreased by 38 and 75 %, respectively (Fig. 4).
Cargill continues to invest to improve its water and energy efficiency and reduce greenhouse gas emissions. In February 2018, Cargill announced a commitment to reduce absolute greenhouse gas emissions in our operations by 10 % by 2025. Improvements in fermentation titer contribute to this goal.
7. Conclusions
The improvements in fermentation processes associated with the whole‐cell biocatalyst, operating parameters, and medium composition can contribute to elevated product titers. Working with high concentrations of product in the fermentation mass requires less downstream CAPEX and OPEX for separation and purification, as well as lowering the environmental impact with respect to water and energy consumption. The data collected from different Cargill plants confirm that developing strategies to boost fermentation titer can result in more competitive processes and also reduce the environmental footprint of biotechnological applications.
Abbreviations
- AAB
acetic acid bacterium
- CAPEX
capital investment cost
- Hyp
trans‐4‐hydroxy‐L‐proline
- KGA
2‐keto‐L‐gulonic acid
- OPEX
operational expenditures
Acknowledgements
The research project receives funding from the European Union's (EU's) framework program for research and innovation Horizon 2020 (2014–2020) under grant agreement no. 637077.
The authors have declared no conflict of interest.
References
- 1. Ball P., Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 13327–13335. DOI: 10.1073/pnas.1703781114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liew F., Martin M. E., Tappel R. C., Heijstra B. D., Mihalcea C., Köpke M., Front. Microbiol. 2016, 7, 694 DOI: 10.3389/fmicb.2016.00694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mohapatra S., Mishra C., Behera S. S., Thatoi H., Renewable Sustainable Energy Rev. 2017, 78, 1007–1032. DOI: 10.1016/j.rser.2017.05.026 [DOI] [Google Scholar]
- 4. Kumar D., Murthy G. S., Biotechnol. Biofuels 2011, 4, 27 DOI: 10.1186/1754-6834-4-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Diaz A. B., Blandino A., Caro I., Trends Food Sci. Technol. 2018, 71, 52–64. DOI: 10.1016/j.tifs.2017.10.016 [DOI] [Google Scholar]
- 6. Banik R. M., Santhiagu A., Kanari B., Sabarinath C., Upadhyay S. N., World J. Microbiol. Biotechnol. 2003, 19, 337–348. DOI: 10.1023/A:1023940809095 [DOI] [Google Scholar]
- 7. Gu B. H., Zheng P., Yan Q., Liu W., Sep. Purif. Technol. 2014, 138, 47–54. DOI: 10.1016/j.seppur.2014.09.034 [DOI] [Google Scholar]
- 8. Soccol C. R., da Costa E. S. F., Letti L. A. J., Karp S. G., Woiciechowski A. L., Porto de Souza Vandenberghe L., Biotechnol. Res. Innovation 2017, 1, 52–71. DOI: 10.1016/j.biori.2017.01.002 [DOI] [Google Scholar]
- 9. Bekatorou A., Dima A., Tsafrakidou P., Boura K., Lappa K., Kandylis P., Pissaridi K., Kanellaki M., Koutinas A. A., Bioresour. Technol. 2016, 220, 34–37. DOI: 10.1016/j.biortech.2016.08.039 [DOI] [PubMed] [Google Scholar]
- 10. Phanthumchinda N., Thitiprasert S., Tanasupawat S., Assabumrungrat S., Thongchul N., Process Biochem. 2018, 68, 205–213. DOI: 10.1016/j.procbio.2018.02.013 [DOI] [Google Scholar]
- 11. D'Este M., Alvarado‐Morales M., Angelidaki I., Biotechnol. Adv. 2017, 36, 14–25. DOI: 10.1016/j.biotechadv.2017.09.001 [DOI] [PubMed] [Google Scholar]
- 12. Gronemeyer P., Ditz R., Strube J., J. Bioeng. 2014, 1, 188–212. DOI: 10.3390/bioengineering1040188 [DOI] [PubMed] [Google Scholar]
- 13. Saha B. C., Ferment. Biotechnol. 2003, 862, 3–17. DOI: 10.1021/bk-2003-0862.ch001 [DOI] [Google Scholar]
- 14. Rokem J. S., Lantz A. E., Nielsen J., Nat. Prod. Rep. 2007, 24, 1262 DOI: 10.1039/b617765b [DOI] [PubMed] [Google Scholar]
- 15. Kim S. R., Park Y. C., Jin Y. S., Seo J. H., Biotechnol. Adv. 2013, 31, 851–861. DOI: 10.1016/j.biotechadv.2013.03.004 [DOI] [PubMed] [Google Scholar]
- 16. Ueda S., Kinoshita M., Tanaka F., Tsuboi M., Shimizu S., N. Oohata , Hino M., Yamada M., Isogai Y., Hashimoto S., J. Biosci. Bioeng. 2011, 112, 409–414. DOI: 10.1016/j.jbiosc.2011.06.002 [DOI] [PubMed] [Google Scholar]
- 17. Agu C. V., Lai S. M., Ujor A. V., Biswas P. K., Jones A., Gopalan V., Ezeji T. C., Sci. Rep. 2018, 8, 3379 DOI: 10.1038/s41598-017-18074-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Back A., Rossignol T., Krier F., Nicaud J. M., Dhulster P., Microb. Cell Fact. 2016, 15, 147 DOI: 10.1186/s12934-016-0546-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Singha T. K., Gulati P., Mohanty A., Khasa Y. P., Kapoor R. K., Kumar S., Process Biochem. 2017, 55, 17–31. DOI: 10.1016/j.procbio.2017.01.026 [DOI] [Google Scholar]
- 20. Chu B. C. H., Hug L., Biotechnol. Adv. 2007, 25, 425–41. DOI: 10.1016/j.biotechadv.2007.04.001 [DOI] [PubMed] [Google Scholar]
- 21. Çelik E., Çalik P., Biotechnol. Adv. 2012, 30, 1108–1118. DOI: 10.1016/j.biotechadv.2011.09.011 [DOI] [PubMed] [Google Scholar]
- 22. Mergulhão F. J. M., Summers D. K., Monteiro G. A., Biotechnol. Adv. 2005, 23, 177–202. DOI: 10.1016/j.biotechadv.2004.11.003 [DOI] [PubMed] [Google Scholar]
- 23. Zhang Y., Zhang Y., Shang X., Wang B., Hu Q., Liu S., T. Wen , Biotechnol. Bioeng., in press. DOI: 10.1002/bit.26818 [DOI] [PubMed] [Google Scholar]
- 24. Fan X., Wu H., Jia Z., Li G., Li Q., Chen N., Xie X., Appl. Microbiol. Biotechnol., in press. DOI: 10.1007/s00253-018-9316-7 [DOI] [PubMed] [Google Scholar]
- 25. Zheng J., Tashiro Y., Wang Q., Sonomoto K., J. Biosci. Bioeng. 2015, 119, 1–9. DOI: 10.1016/j.jbiosc.2014.05.023 [DOI] [PubMed] [Google Scholar]
- 26. Luo H., Yang R., Zhao Y., Wang Z., Liu Z., Huang M., Q. Zeng , Bioresour. Technol. 2018, 253, 343–354. DOI: 10.1016/j.biortech.2018.01.007 [DOI] [PubMed] [Google Scholar]
- 27. Löbs A.‐K., Schwartz C., Wheeldon I., Synth. Syst. Biotechnol. 2017, 2, 198–207. DOI: 10.1016/j.synbio.2017.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Humphreys C. M., Minton N. P., Curr. Opin. Biotechnol. 2018, 50, 174–181. DOI: 10.1016/j.copbio.2017.12.023 [DOI] [PubMed] [Google Scholar]
- 29. Wisselink H. W., Toirkens M. J., Wu Q., Pronk J. T., van Maris A. J. A., Appl. Environ. Microbiol. 2009, 75, 907–914. DOI: 10.1128/AEM.02268-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garcia Sanchez R., Karhumaa K., Fonseca C., Sànchez Nogué V., Almeida J. R., Larsson C. U., Bengtsson O., Bettiga M., Hahn‐Hägerdal B., Gorwa‐Grauslund M. F., Biotechnol. Biofuels 2010, 3, 13 DOI: 10.1186/1754-6834-3-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim S. J., Lee J. E., Lee D. Y., Park H., Kim K. H., Park Y. C., Appl. Microbiol. Biotechnol., in press. DOI: 10.1007/s00253-018-9306-9 [DOI] [Google Scholar]
- 32. Mans R., Daran J. M. G., Pronk J. T., Curr. Opin. Biotechnol. 2018, 50, 47–56. DOI: 10.1016/j.copbio.2017.10.011 [DOI] [PubMed] [Google Scholar]
- 33. Bader J., Mast‐Gerlach E., Popović M. K., Bajpai R., Stahl U., J. Appl. Microbiol. 2010, 109, 371–387. DOI: 10.1111/j.1365-2672.2009.04659.x [DOI] [PubMed] [Google Scholar]
- 34. Tesfaw A., Assefa F., Int. Scholarly Res. Not. 2014, 2014, 532852 DOI: 10.1155/2014/532852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang E. X., Ding M. Z., Ma Q., Dong X. T., Yuan Y. J., Microb. Cell Fact. 2016, 15, 21 DOI: 10.1186/s12934-016-0418-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jia N., Ding M. Z., Zou Y., Gao F., Yuan Y. J., Sci. Rep. 2017, 7, 46759 DOI: 10.1038/srep46759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jia N., Ding M., Du Y., Feng S., Gao F., Yuan Y., Genome Accounce. 2016, 4, e01426‐16. DOI: 10.1128/genomeA.01426-16 [DOI] [Google Scholar]
- 38. Liu L., Li Y., Zhang J., Zhou Z., Liu J., Li X., Zhou J., Du G., Wang L., Chen J., J. Bacteriol. 2011, 193, 6108–6109. DOI: 10.1128/JB.06007-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xiong X. H., Han S., Wang J. H., Jiang Z. H., Chen W., N. Jia , Wei H. L., Cheng H., Yang Y. X., Zhu B., You S., He J. Y., Hou W., Chen M. X., Yu C. J., Jiao Y. H., Zhang W. C., J. Bacteriol. 2011, 193, 315–316. DOI: 10.1128/JB.01189-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jia N., Du J., Ding M. Z., Gao F., Yuan Y. J., PLoS One 2015, 10, e0135104. DOI: 10.1371/journal.pone.0135104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ma Q., Zhou J., Zhang W., Meng X., Sun J., Yuan Y. J., PLoS One 2011, 6, e26108. DOI: 10.1371/journal.pone.0026108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ma Q., Zhang W., Zhang L., Qiao B., Pan C., Yi H., Wang L., Yuan Y. J., PLoS One 2012, 7, e32156. DOI: 10.1371/journal.pone.0032156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ding M. Z., Zou Y., Song H., Yuan Y. J., PLoS One 2014, 9, e94889. DOI: 10.1371/journal.pone.0094889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jia N., Ding M. Z., Gao F., Yuan Y. J., Sci. Rep. 2016, 6, 28794 DOI: 10.1038/srep28794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ma Q., Zou Y., Lv Y., Song H., Yuan Y. J., PLoS One 2014, 9, e91789. DOI: 10.1371/journal.pone.0091789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Singh V., Haque S., Niwas R., Srivastava A., Pasupuleti M., Tripathi C. K. M., Front. Microbiol. 2017, 7, 2087 DOI: 10.3389/fmicb.2016.02087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nagata Y., Chu K. H., Biotechnol. Lett. 2003, 25, 1837–1842. DOI: 10.1023/A:1026225526558 [DOI] [PubMed] [Google Scholar]
- 48. Yang Z., Zhang Z., Biotechnol. Biofuels 2018, 11, 35 DOI: 10.1186/s13068-018-1031-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Song X., Zhang Y., Xue J., Li C., Wang Z., Wang Y., Bioresour. Technol. 2018, 255, 180–188. DOI: 10.1016/j.biortech.2017.09.033 [DOI] [PubMed] [Google Scholar]


