Abstract
Cassava brown streak disease (CBSD) caused by Cassava brown streak virus (CBSV) and Ugandan cassava brown streak virus (UCBSV) is the main constraint to cassava (Manihot esculenta Crantz) production in Mozambique. Using RT-PCR to amplify partial coat protein nucleotide sequences, we detected for the first time the occurrence of CBSV in two non-cassava perennial wild plant species: Zanha africana (Radlk.) Exell. and Trichodesma zeylanicum (Burm.f.) R.Br., that occur widely within and near cassava fields in Nampula, Zambezia, Niassa and Cabo Delgado provinces. In addition, we also detected CBSV and UCBSV in Manihot carthaginensis subsp. glaziovii (Müell-Arg.) Allem., a wild cassava relative. These findings were verified in biological assays through mechanical inoculation of CBSV to T. zeylanicum, albeit at low rates of infection. Phylogenetic analysis clustered the CBSV isolates from the non-cassava plant species with those from cultivated cassava, with high sequence homology among CBSV (91.0–99.6%) and with UCBSV (84–92%) isolates. These results provide definitive evidence of a wider host range for CBSV and UCBSV in Mozambique, indicating that these viruses are not restricted to cultivated cassava. Our findings are key to understanding the epidemiology of CBSD and will aid in the development of sustainable management strategies for the disease.
Keywords: Cassava brown streak viruses, Wild host plants, Mechanical inoculation, Alternative host, Mozambique
Highlights
-
•
Alternative host plants of Cassava brown streak viruses were determined.
-
•
There was evidence of natural infection of CBSV in non-cassava relative plants.
-
•
CBSV was successfully transmitted in T. zeylanicum by mechanical inoculation.
-
•
CBSV and UCBSV, associated with CBSD in cassava, naturally infect M. glaziovii.
1. Introduction
Cassava (Manihot esculenta Crantz, family Euphorbiaceae) is the second most important crop after maize in Mozambique [1]. More than 80% of cassava production in Mozambique occurs in the north and central regions. Currently, production in these regions is severely constrained by two cassava brown streak viruses, Cassava brown streak virus (CBSV) and Ugandan cassava brown streak virus (UCBSV) [[2], [3], [4]], which cause Cassava brown streak disease (CBSD) [2,5]. The disease was first reported to be transmitted with very low efficiency by whitefly, Bemisia tabaci (Gennadius) [6,7], but [8] recently confirmed generally moderate rate of transmission of CBSV, ranging from 30 to 53% using 20 to 100 whiteflies. Recently, the presence of the DAG motif in CBSV sequences suggests that aphids could be potential vectors of CBSV as observed in Squash vein yellowing virus (SqVYV) and Coccinia mottle virus (COCMOV) [9]. Work to confirm aphid transmission of CBSVs is ongoing.
A virus disease survey of cassava was undertaken in 1999 in Zambezia and Nampula provinces, which are the main areas of production in Mozambique in which CBSD was identified for the first time in Mozambique. Disease incidences in some fields reached 80–100% and many of the main cassava cultivars were affected [10]. In subsequent country-wide surveys in 2010 and 2012, CBSD was found in Zambezia, Nampula and a third province, Cabo Delgado, all in northern Mozambique. The disease was highest in Zambezia (61.3% and 82.2%) and lowest in Cabo Delgado (23.6% and 35.1%) in 2010 and 2012, respectively. The local cultivars ‘Cadri’ and ‘Robero’ were the most affected, while ‘Likonde’ and ‘Amwalikampiche’ had low incidences and symptom severity, indicating some tolerance to the disease [11]. When compared to previous surveys conducted in 1999 and 2003, the increasing incidence and symptom severity suggests that farmers were replanting new fields with disease-affected cuttings. Recently, the 2015 and 2017 country-wide surveys indicated a reduction in CBSD incidence and severity, attributed mainly to wide adoption of improved cassava cultivars with increased tolerance to CBSD in Nampula and Zambezia (Nurbibi Cossa, unpublished and Cassava Disease Diagnostic annual reports).
The natural occurrence of Cassava brown streak viruses in M. carthaginensis subsp. glaziovii (Müell-Arg.) Allem. has been reported [12]. In addition, Nicotiana tabacum, N. benthamiana, N. debneyi, N. rustica, N. glutinosa, N. hesperis, N. occidentalis, Datura stramonium, Petunia hybrida, Chenopodium quinoa and C. amaranticolor were used as experimental hosts for CBSV [13,14]; [15]. Of these plant species, N. debneyi and N. benthamiana have proved the most useful for virus infection assays [3,4,16]. Pathogens can have highly variable host ranges: in natural conditions some infect only one or a few related species (i.e., specialist pathogens), but others can infect a wide range of hosts. For example, Tobacco rattle virus reportedly infects over 400 plant species belonging to 50 different families [17] and Cucumber mosaic virus infects 1200 plant species belonging to 100 families [18]. The Cassava mosaic begomoviruses (CMBs) that cause Cassava mosaic disease (CMD) naturally occur in cassava, but also infect Jatropha curcas under experimental and natural conditions [19]. [20] reported African cassava mosaic virus (ACMV) and East African cassava mosaic virus (EACMV) in M. carthaginensis subsp. glaziovii (Müell-Arg.) Allem., Senna occidentalis L. and the weed Combretum confertum Benth. Therefore, given these findings of alternative hosts for several crop-infecting viruses, including some important in cassava, it is plausible that CBSV or UCBSV could have additional, yet undiscovered alternative hosts. There is limited information on alternative hosts and their potential role in the spread of CBSV and UCBSV in sub-Saharan Africa. The lack of knowledge of the alternative hosts of CBSV and UCBSV is a key knowledge gap in the epidemiology and management of CBSD, especially in the endemic countries such as Mozambique. Available information on the natural host range of Cassava brown streak viruses indicates that they are largely restricted to cassava and wild relatives such as M. carthaginensis subsp. glaziovii (Müell-Arg.) Allem. This study aimed to identify alternative host plants for Cassava brown streak viruses in Mozambique.
2. Materials and methods
2.1. Areas surveyed and sample collection
To determine and identify alternative hosts for CBSV, leaf samples were collected in 2014 from four major cassava production areas namely Nampula, Zambezia, Niassa and Cabo Delgado. A total of 120 leaf samples showing virus-like disease symptoms such as chlorosis, yellow spotting, deformation, mosaic, wilting, leaf curling and necrotic lesions were collected from 15 plant species: M. carthaginensis subsp. glaziovii (Müell-Arg.) Allem., Mucuna pruriens, Cajanus cajan (L.) Millsp., Trichodesma zeylanicum (Burm.f.) R.Br., Paederia bojeriana (A.Rich.) Drake subsp. foetens (Hiern), Commelina benghalensis, Ageratum conyzoides (L.), Vernonia petersii Oliv. & Hiern ex Oliv., Zanha africana (Radlk.) Exell., Brachistegia spiciform Benth, Ocimum africanum Lour., Senna obtusifolia (L.) H.S.Irwin & Barneby, Ipomea tenuipes Verdc., Vernonia cinerea (L.) Less. and Nidorela sp. (Table 1) growing within or nearby (5–10 m away) cassava fields. The wild plant species were identified using a working list of all plant species website (http://www.theplantlist.org). Additionally, wild plant species collected in the fields were taken to the Botany Department at Mozambique Agricultural Research Institute for identification and further confirmation of the identity/taxonomy by a Botanist. The samples were labeled and kept in herbarium field kits to preserve their integrity until laboratory analysis.
Table 1.
Occurrence and ecology of non-cassava plant species sampled for Cassava brown streak viruses in Mozambique 2014.
| Local name | Botanical name | Plant family | CBSV/UCBSV Testing results |
Disease severity (1–5 scale) | Collection environment and ecology | Sample location | Frequency occurrence and distribution |
|---|---|---|---|---|---|---|---|
| Tree cassava | Manihot carthaginensis subsp. glaziovii (Müell-Arg.) Allem. | Euphorbiaceae (perennial shrub tree) | CBSV and UCBSV | 4 | Nearby cassava fields along boundaries and homestead | Zambezia, Nampula, Niassa |
High |
| Velvet-fruited zanha | Zanha africana (Radlk.) Exell. | Sapindaceae (perennial shrub tree) | CBSV | 3 | Within cassava fields and uncultivated areas | Nampula | High |
| Camel bush | Trichodesma zeylanicum (Burm.f.) R.Br. | Boraginaceae (annual/perennial weed) | CBSV | 3 | Within cassava fields and uncultivated areas | Nampula, Niassa | Very high |
| Velvet bean | Mucuna pruriens | Fabaceae (creeping vine legume) | – | 4 | Within cassava fields and uncultivated areas | Nampula | Low |
| Pigeon pea | Cajanus cajan (L.) Millsp. | Fabaceae (annual/perennial legume) | – | 2 | In the cassava fields | Zambezia | High |
| Paederia bojeriana | Paederia bojeriana (A.Rich.) Drake subsp. foetens (Hiern) | Rubiaceae | – | 3 | In cassava fields and within cassava fields | Zambezia | Low |
| Benghal dayflower | Commelina benghalensis | Commelinaceae (annual/perennial herb) | – | 2 | In cassava fields and within cassava fields | Nampula | High |
| Billygoat weed | Ageratum conyzoides (L.) | Asteraceae (perennial weed) | – | 3 | In cassava fields and within cassava fields | Zambezia, Niassa, C.Delgado |
High |
| – | Vernonia petersii Oliv. & Hiern ex Oliv. | Compositae (annual weed) | – | 3 | Within cassava fields and uncultivated areas | Zambezia, Nampula | Low |
| Zebrawood or Msasa | Brachistegia spiciform Benth | Fabaceae (perennial shrub tree) | – | 3 | Within cassava fields and uncultivated areas | Nampula | Very high |
| Lemon basil | Ocimum africanum Lour. | Lamiaceae (annual weed) | – | 4 | Nearby cassava fields and in the cassava field | Nampula | Very high |
| Cofeeweed/cassia | Senna obtusifolia (L.) H.S.Irwin & Barneby | Caesalpinioideae (annual/perennial herb) | – | 3 | Nearby cassava fields and in the cassava field | Nampula, Zambezia | Very high |
| Morning glory | Ipomea tenuipes Verdc. | Convolvulaceae (perennial) | – | 2 | Nearby cassava fields | Zambezia, Cabo Delgado | Low |
| Dandotapala | Vernonia cinerea (L.) Less | Asteraceae (annual shrub) | – | 3 | Within cassava fields and uncultivated areas | Zambezia, Nampula | Low |
| – | Nidorela sp. | – | 2 | Nearby cassava fields | Niassa | Low |
2.2. CBSD symptoms severity
To score the CBSD symptoms severity in M. carthaginensis subsp. glaziovii (Müell-Arg.), we used more comprehensive descriptions based on 1–5 scale of foliar CBSD symptom described by Refs. [21] and [22]: 1 = no visible symptoms, 2 = mild vein yellowing or chlorotic blotches on some leaves, 3 = pronounced/extensive vein yellowing or chlorotic blotches on leaves, but no lesions or streaks on stems, 4 = pronounced/extensive vein yellowing or chlorotic blotches on leaves and mild lesions or streaks on stems, and 5 = pronounced/extensive vein yellowing or chlorotic blotches on leaves and severe lesions or streaks on stems, defoliation and dieback.
2.3. RNA extraction
Total RNA was extracted from the leaf samples using a modified CTAB protocol as described previously [23,24]. The yield of RNA was quantified using a Thermo Scientific NanoDrop 2000/2000c (Thermo Scientific, Waltham, MA, USA) (full spectrum UV–Vis) at A260/280 ratio.
2.4. Reverse transcription
Total RNA (4 μg) was used to synthesize cDNA in two steps using an ImProm-II™ reverse transcriptase Kit (Promega, Madison, WI, USA) following the manufacturer's instructions. RT was performed with cycling conditions of 42 °C for 60 min and 70 °C for 10 min and the resulting cDNA was used for PCR.
2.5. PCR amplification
To screen for the presence of CBSV in the samples, PCR was conducted using the primers CBSDDF and CBSDDR, which are designed to amplify the partial coat protein (CP) gene and 3′-untranslated region (UTR) [12] – with expected fragment sizes of 344 bp (CBSV) and 430–440 bp (UCBSV). The PCR reaction mix of 25 μL consisted of 12.9 μL of sterile de-ionized water, 3.0 μL of 10 × PCR buffer +20 mM MgCl2, 1.0 μL of primers CBSDDF/CBSDDR (10 mM), 0.3 μL of Pfu DNA polymerase, 2.8 μL of dNTPs (2.5 mM) and 4.0 μL of cDNA template. The PCR cycling conditions were 94 °C for 2 min, followed by 35 cycles of 94 °C for 30 s, 51 °C for 30 s and 72 °C for 30 s for denaturation, annealing and extension, respectively. PCR products were analyzed by electrophoresis in 1 × TAE buffer on a 2% agarose gel stained with 0.5 μg/mL of ethidium bromide.
2.6. Cloning and sequencing
Samples with the expected product size (344 bp for CBSV and 440 bp UCBSV) from PCR were cloned separately using a Thermo Scientific CloneJET PCR Cloning Kit and transformed into E. coli JM109 (Thermo Scientific), following the manufacturer's instructions. Samples with two amplified bands were cut from the gel and purified using a GeneJET Gel Extraction Kit (Thermo Scientific) following the manufacturer's instructions and cloned as for the samples with one band. Recombinant DNA was extracted using a GeneJET Plasmid Miniprep Kit (Thermo Scientific), and sent for sequencing by Inqaba Biotech (Pretoria, South Africa).
2.7. Phylogenetic analysis of CBSV sequences
The resulting sequences were trimmed and edited using FinchTV 1.4.0 (http://jblseqdat.bioc.cam.ac.uk/gnmweb/download/soft/FinchTV_1.4/doc/) and multiple alignments representing partial CP and 3′-UTR sequences were performed using MEGA 5.02. Nucleotide sequences of isolates obtained from cassava relatives and non-relatives were aligned and compared with all available GenBank CBSV and UCBSV sequences from eastern and southern Africa as well as CBSV sequences from cassava collected in Mozambique during this study. Phylogenetic analysis was performed using the maximum likelihood method as implemented in MEGA 5.02 [25]. All phylogenetic analyses were performed using the best-fit substitution model for nucleotides (GTR + I + G) with 1000 bootstrap replicates.
2.8. Mechanical transmission of CBSV
2.8.1. Establishment of test plants
Infection assays of CBSV were established using T. zeylanicum, which was easier to grow than the shrub tree Z. africana. The plants were raised using seeds established in Hygromix growth medium (Hygrotech Pty Ltd, South Africa) and maintained under natural light in a screen house. Cypermethrin insecticide was applied weekly to the plants to control infestation by insects and possible transmission of viruses, and the plants maintained in an insect-proof net cage until inoculation.
2.8.2. Virus sources and mechanical transmission
A bioassay experiment for CBSV transmission was conducted using classical virology methods for mechanical inoculation as described by Refs. [26] and [27]. Thirty plants of T. zeylanicum were used for the infection assays, among which five were included as controls. Extracts of CBSD-symptomatic cassava leaves confirmed to be positive for CBSV in RT-PCR (Fig. 1A) were used as sources of virus inoculum and were rubbed onto the expanded leaf surfaces of 25 T. zeylanicum plants with aid of carborundum dust (Fig. 1B). For negative control plants, only buffer (0.02 M Phosphate, PH = 7.0) was applied to the leaves. The inoculated plants were covered with transparent plastic and maintained in a controlled environment in the laboratory for 48 h at 25 °C. The plants were transferred to the greenhouse where they were monitored for symptom development. Plants were inspected daily for symptom development for one month, and the leaves tested for the presence of Cassava brown streak viruses using RT-PCR.
Fig. 1.

Array of viral disease symptoms on wild non-cassava plant species detected with Cassava brown streak viruses: (A) spotted yellowing along secondary veins, feathery chlorosis and yellow mosaic on leaves of Zanha africana (Radlk.) Exell, (B) yellowing, feathery chlorosis and leaf curling on leaves of Trichodesma zeylanicum (Burm.f.) R.Br. and (C & D) chlorosis and yellowing on leaves of Manihot carthaginensis subsp. glaziovii (Müell-Arg.) Allem, in Mozambique, 2014.
3. Results
3.1. Viral disease symptoms on alternative host plants
Viral disease symptoms on Velvet-fruited zanha (Z. africana (Radlk.) Exell) and Camel bush (T. zeylanicum (Burm.f.) R.Br.) included: spotted yellowing along secondary veins, feathery chlorosis, yellow mosaic and leaf curling (Fig. 1A and B). In comparison, the cassava relative M. carthaginensis subsp. glaziovii (Müell-Arg.) Allem had typical severe chlorosis with severity scale of 4, on the 1–5 severity scale described by Refs. [21] and [22] on the leaves and necrosis on the stems (Fig. 1C and D). The symptoms were similar to those observed on cultivated cassava. The incidence of plants with virus-like disease symptoms was moderate (45–55%) to high (80–90%) in the study locations (data not provided), and this formed the basis for sampling the plant species reported here.
3.2. PCR amplification of Cassava brown streak viruses in non-cassava plants
A total of 120 plant samples comprising of weeds, shrubs, trees and cassava relatives were screened for presence of CBSV and UCBSV using species-specific primers. PCR analysis produced the expected bands of 344 bp and 440 bp for CBSV and UCBSV, respectively. CBSV was detected in six plant samples: four of cassava relative M. carthaginensis subsp. glaziovii (Müell-Arg.) Allem. and two non-cassava plant species, T. zeylanicum (Burm.f.) R.Br. and Z. africana (Radlk.) Exell. UCBSV was detected in one M. carthaginensis subsp. glaziovii (Müell-Arg.) Allem. sample. The rest of the samples that did not test positive with Cassava brown streak viruses were kept for future study to determine the causal viruses for the virus-like symptoms and establish their importance to agriculture.
3.3. Phylogenetic analysis
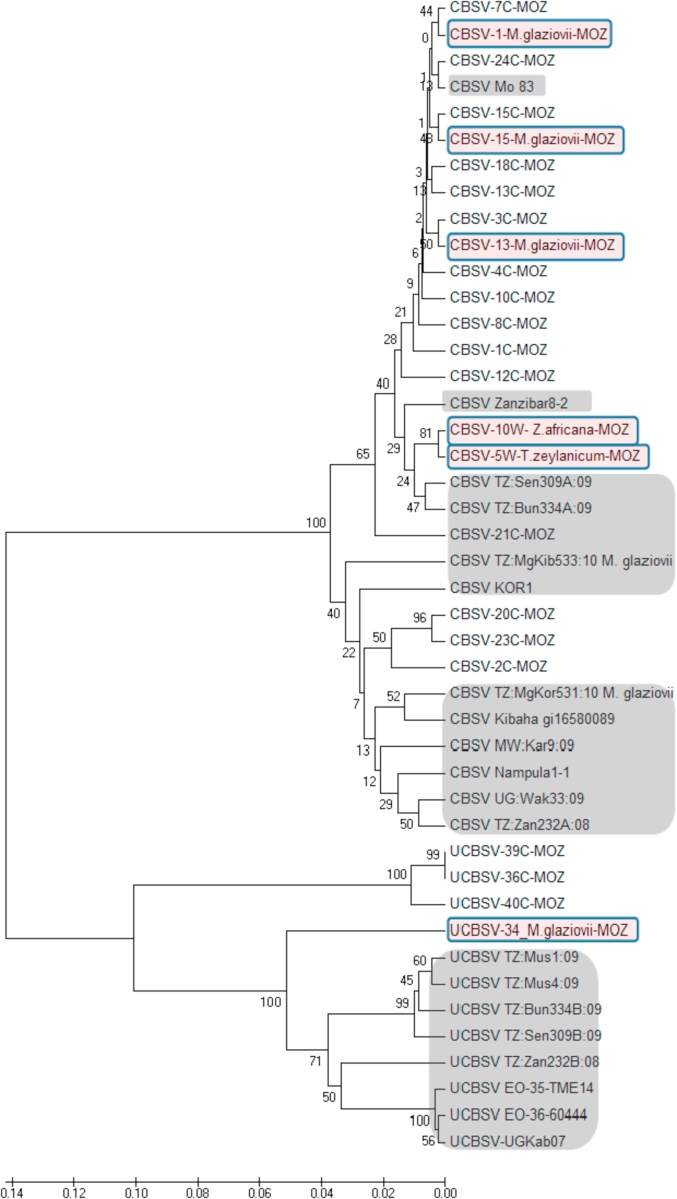
Phylogenetic analysis was carried out to determine the genetic relationships among the six CBSV isolates obtained from the non-cassava samples using partial sequences of the core region of CP and 3′-UTR. The partial sequences were aligned with 20 reference nucleotide sequences (11 of CBSV and eight of UCBSV) from GenBank (Table 2) using MEGA 5.02 [25] with a best-fit model. As expected, comparisons based on nucleotide sequences revealed the existence of two major groups: CBSV and UCBSV. Five out of six sequences clustered with CBSV sequences from Mozambique (Fig. 2), while one of the sequences obtained from M. carthaginensis subsp. glaziovii (Müell-Arg.) Allem. clustered with UCBSV (Fig. 2). Isolates obtained from T. zeylanicum (Burm.f.) R.Br., Z. africana (Radlk.) Exell and M. carthaginensis subsp. glaziovii (Müell-Arg.) Allem. shared 91.0–99.6% sequence similarity with CBSV affecting cassava in East Africa and Mozambique. However, the UCBSV isolate from M. carthaginensis subsp. glaziovii (Müell-Arg.) Allem. had lower sequence homology (84–92%) with isolates from cultivated cassava.
Table 2.
Cassava brown streak viruses isolates sequences used in the phylogenetic analysis in this study.
| Isolate name | Host | Accession number | Reference |
|---|---|---|---|
| UCBSV TZ:Mus1:09 | M. esculenta Crantz | HM453037 | [28] |
| UCBSV TZ:Mus4:09 | M. esculenta Crantz | HM453038 | [28] |
| UCBSV TZ:Sen309B:09 | M. esculenta Crantz | HM453036 | [28] |
| UCBSV EO-36-60444 | M. esculenta Crantz | KJ606231 | [29] |
| UCBSV-UGKab07 | M. esculenta Crantz | HG965222 | [28] |
| UCBSV TZ:Bun334B:09 | M. esculenta Crantz | HM453039 | [28] |
| UCBSV TZ:Zan232B:08 | M. esculenta Crantz | HM453040 | [28] |
| CBSV TZ:Sen309A:09 | M. esculenta Crantz | HM453033 | [28] |
| UCBSV EO-35-TME14 | M. esculenta Crantz | KJ606230 | [29] |
| CBSV Nampula1-1 | M. esculenta Crantz | HM346953 | [28] |
| CBSV TZ:MgKor531:10 M. glaziovii | M. carthaginensis subsp. glaziovii (Müell-Arg.) | HM453032 | [12] |
| CBSV KOR1 | M. esculenta Crantz | GU563327 | [12] |
| CBSV Mo 83 | M. esculenta Crantz | FN434436 | [4] |
| CBSV MW:Kar9:09 | M. esculenta Crantz | HM171296 | [28] |
| CBSV UG:Wak33:09 | M. esculenta Crantz | HM171312 | [28] |
| CBSV TZ:MgKib533:10 M. glaziovii | M. carthaginensis subsp. glaziovii (Müell-Arg.) | HM453031 | [12] |
| CBSV TZ:Zan232A:08 | M. esculenta Crantz | GU563325 | [12] |
| CBSV TZ:Bun334A:09 | M. esculenta Crantz | HM450034 | [28] |
| CBSV Zanzibar8-2 | M. esculenta Crantz | HM346957 | [28] |
| CBSV-10W-_Z.africana-MOZ | Zanha africana | Yet to be received | This study |
| CBSV-10C-MOZ | M. esculenta Crantz | Yet to be received | This study |
| CBSV-18C-MOZ | M. esculenta Crantz | Yet to be received | This study |
| CBSV-1C-MOZ | M. esculenta Crantz | Yet to be received | This study |
| CBSV-13C-MOZ | M. esculenta Crantz | Yet to be received | This study |
| CBSV-15C-MOZ | M. esculenta Crantz | Yet to be received | This study |
| CBSV-2C-MOZ | M. esculenta Crantz | Yet to be received | This study |
| CBSV-3C-MOZ | M. esculenta Crantz | Yet to be received | This study |
| CBSV-4C-MOZ | M. esculenta Crantz | Yet to be received | This study |
| CBSV-5W-T.zeylanicum-MOZ | Trichodesma zeylanicum | Yet to be received | This study |
| CBSV-7C-MOZ | M. esculenta Crantz | Yet to be received | This study |
| CBSV-8C-MOZ | M. esculenta Crantz | Yet to be received | This study |
| CBSV-12C-MOZ | M. esculenta Crantz | Yet to be received | This study |
| CBSV-20C-MOZ | M. esculenta Crantz | Yet to be received | This study |
| CBSV-21C-MOZ | M. esculenta Crantz | Yet to be received | This study |
| CBSV-23C-MOZ | M. esculenta Crantz | Yet to be received | This study |
| CBSV-24C-MOZ | M. esculenta Crantz | Yet to be received | This study |
| CBSV-13-Glaziovii-MOZ | M. carthaginensis subsp. glaziovii (Müell-Arg.) | Yet to be received | This study |
| CBSV-15-Glaziovii-MOZ | M. carthaginensis subsp. glaziovii (Müell-Arg.) | Yet to be received | This study |
| CBSV-1-Glaziovii-MOZ | M. carthaginensis subsp. glaziovii (Müell-Arg.) | Yet to be received | This study |
Fig. 2.
Phylogenetic tree constructed using the neighbor-joining method with MEGA5.2. The phylogenetic tree was generated based on partial CP-encoding nucleotide sequences of CBSV and UCBSV isolates collected in Nampula, Zambezia and Niassa Provinces. CBSV and UCBSV sequences from cassava relatives and non-relatives are indicated with pink shading, the reference isolates from GenBank are indicated with gray and the remaining are sequences from isolates collected during this study from cultivated cassava plants in Mozambique (isolates with terminal MOZ). The number at each branch represents the bootstrap value (1000 replicates).
3.4. Koch's postulates and virus infection assays
Out of the 25 T. zeylanicum (Burm.f.) R.Br. plants mechanically inoculated with CBSV, only three successfully developed viral disease symptoms. The first symptoms were recorded at 32 days after inoculation. The symptoms included chlorotic spots, leaf yellowing and wilting (Fig. 3A–C), and were similar to those observed on T. zeylanicum (Burm.f.) R.Br. in the field, except for the wilting. The presence of CBSV in the infected plants was confirmed with RT-PCR.
Fig. 3.

Symptoms induced by CBSV isolate (CBSV-8C-MOZ) after 5 weeks; 3 out of 25 inoculated Trichodesma zeylanicum (Burm.f.) R.Br. plants displayed viral disease symptoms, including (A) leaf yellowing, (B) wilting and (C) chlorotic spots.
3.5. Occurrence and distribution of the alternative host plants
Occurrence and distribution of M. carthaginensis subsp. glaziovii (Müell-Arg.) Allem., the wild cassava relative and the two non-cassava plant species T. zeylanicum (Burm.f.) R.Br.and Z. africana (Radlk.) Exell in Nampula, Zambezia, Niassa and Cabo Delgado provinces were assessed in general terms as either low, high or very high. The M. carthaginensis subsp. glaziovii (Müell-Arg.) Allem. occurred with high frequency as shrubs along boundaries of the sampled cassava fields, in homesteads and in uncultivated areas (Fig. 4). Zanha africana (Radlk.) Exell plants occurred with low frequency as short shrubs and/or stumps within and near the sampled cassava fields. In uncultivated areas, Z. africana (Radlk.) Exell plants occurred with high frequency mainly as trees (Fig. 5). However, T. zeylanicum (Burm.f.) R.Br. plants were among the predominant weeds with very high frequency in cassava fields (Fig. 6). Due to their ease of growth through seed dispersal, this species is considered a major weed in agricultural fields (Table 1).
Fig. 4.
Occurrence of Manihot carthaginensis subsp. glaziovii (Müell-Arg.) Allem., plants in (A) homesteads, with typical CBSD symptoms on (B–D) leaves and (E) stems in the sampled areas in Mozambique, 2014.
Fig. 5.
Symptomless (A) and viral disease symptomatic (B) plants of Zanha africana (Radlk.) Exell and (C) shrub/trees with viral disease symptoms growing in uncultivated areas next to cassava fields in the sampled areas in Mozambique, 2014.
Fig. 6.
Occurrence of (A) symptomless and (B) viral disease symptomatic plants of Trichodesma zeylanicum (Burm.f.) R.Br. in a cassava field and (C) weed plants growing around cassava plants showing typical CBSD symptoms on leaves in Mozambique, 2014.
4. Discussion
We report here, for the first time, the occurrence of CBSV in two non-cassava perennial wild plant species, Velvet-fruited zanha (Z. africana (Radlk.) Exell) and Camel bush (T. zeylanicum (Burm.f.) R.Br.), and UCBSV in M. carthaginensis subsp. glaziovii (Müell-Arg.) Allem., a wild cassava relative in Mozambique, based on results obtained in PCR using virus species-specific primers [12] and phylogenetic analyses of the partial CP sequences of the isolates. Pairwise nucleotide sequence comparisons revealed high sequence homology among CBSVs (91.0–99.6%) and UCBSV (84–92%) isolates. The viral disease symptoms recorded on Z. africana (Radlk.) Exell and T. zeylanicum (Burm.f.) R.Br.) in the field included spotted yellowing along secondary veins, feathery chlorosis, yellow mosaic and leaf curling. In comparison, M. carthaginensis subsp. glaziovii (Müell-Arg.) Allem. had severe chlorosis on leaves and necrosis on stems, symptoms typical of CBSD on cultivated cassava. CBSV was detected in more samples, including M. carthaginensis subsp. glaziovii (Müell-Arg.) Allem., Z. africana (Radlk.) Exell and T. zeylanicum (Burm.f.) R.Br.), than UCBSV which occurred only in M. carthaginensis subsp. glaziovii (Müell-Arg.) Allem. A recent study by Ref. [31] reported CBSV to have a more rapid rate of evolution, and to be the predominant virus associated with severe CBSD compared with UCBSV in Uganda. In Mozambique, [11] showed that CBSV was widely distributed and the most important species causing CBSD. In contrast, this study observed that UCBSV was confined to Zambezia Province in M. carthaginensis subsp. glaziovii (Müell-Arg.) Allem, tree cassava, which is a glabrous shrub or tree that grows to 6 m high, and occasionally taller (10–20 m). This perennial plant was introduced to Africa as a plantation crop for rubber production in the 19th century and quickly established as common flora in uncultivated areas. In the study areas of Mozambique, tree cassava occurred mainly as a boundary plant along farms and homesteads and was abundant in uncultivated areas. In many homesteads, a few plants were maintained as sources of leafy vegetables, the majority bearing clear viral disease symptoms. Zanha africana (Radlk.) Exell is a perennial tropical African savanna tree [[32], [33], [34], [35]]. In the current study, Z. africana (Radlk.) Exell occurred as short shrubs and/or stumps in and near the sampled cassava fields. Trichodesma zeylanicum (Burm.f.) R.Br.) is an annual/perennial weed species that is abundant in agricultural and unused fields. It is highly competitive, a quick grower and covers many areas. Of the three wild non-cassava host plant species, T. zeylanicum (Burm.f.) R.Br.) was the most abundant in the sampled cassava farmers’ fields.
We tested infection assays of CBSV isolated from cassava plants to T. zeylanicum (Burm.f.) R.Br.) raised from seed, and ably demonstrated the mechanical transmission of the virus from cassava to a non-cassava plant species, albeit at low rates of infection. We do not know the reasons for the low infection rates, but mechanical transmission of plant viruses can be very delicate even between herbaceous hosts. For example, plants with high levels of phenolic compounds, such as T. zeylanicum (Burm.f.) R.Br., were found to have high antibacterial and antiphytoviral activities [36,37], which inhibit disease development through inhibition of extracellular enzymes and antioxidant activity in plant tissue [38]. Similarly, resistance to mechanical viral infection in chili was attributed to increased quantity of phenolics [39]. Regarding transmission of cassava brown streak viruses [40], indicated that mechanical transmission could not be achieved by using a simple buffer in infection assays, and suggested the use of antioxidants in buffers to enhance mechanical inoculation. We suggest that future investigations could include grafting and/or vector mediated transmission in infection assays. However, notwithstanding the low infection rates in our study, mechanical transmission successfully confirmed T. zeylanicum (Burm.f.) R.Br. as a natural host for CBSV. Interestingly, the incidence of M. carthaginensis subsp. glaziovii (Müell-Arg.) Allem., Z. Africana (Radlk.) Exell and T. zeylanicum (Burm.f.) R.Br.) plants with viral disease symptoms that tested positive for CBSVs was moderate to high in the sampled areas. In this study, we did not investigate the vectors associated with transmission of the Cassava brown streak viruses detected in the non-cassava plant species and suggest this to be a focus for future research.
The high abundance and widespread distribution of M. carthaginensis subsp. glaziovii (Müell-Arg.) Allem., Z. africana (Radlk.) Exell and T. zeylanicum (Burm.f.) R.Br. plants in the CBSD-affected areas in Nampula and Zambezia suggests that these plants serve as important inoculum sources for Cassava brown streak viruses that infect cassava crops both during the season and off-season. We propose that a survey be conducted to further establish the incidence of CBSV infections in the three wild host plant species described in this study. In addition, awareness campaigns should be carried out to educate farmers, agricultural extension officers, scientists (plant breeders, entomologists and virologists) and other cassava stakeholders on the importance of wild non-cassava plant hosts in the spread and management of CBSD. Emphasis should be placed on disease symptom identification, scouting and roguing of suspected plants in cassava fields. Attempts should be made to plant cassava crops away from uncultivated areas with suspected viral disease symptomatic weeds, shrubs and trees, including the three wild plant hosts identified in this study, although this may be a challenge to achieve in areas with limited arable land and/or a lack of community participation.
Declaration of conflict of interest
The authors had no conflict of interest.
Acknowledgments
This study was funded by the Bill and Melinda Gates Foundation (Grant no. 51466) through a sub-grant to the Mozambique Agricultural Research Institute Maputo, Mozambique, under the auspices of Mikocheni Agricultural Research Institute through the “Disease Diagnostics for Sustainable Cassava Productivity in Africa” project. The authors acknowledge the financial support and smallholder farmers in Mozambique for allowing us to collect samples from their farms.
Footnotes
This article is part of a Special Issue entitled ‘Crop Pathology in Africa’ published at the journal Physiological and Molecular Plant Pathology 105C, 2019.
References
- 1.MINAG . Seminário Nacional de Extensao- Investigaçao; Moçambique: 2005. Ministerio da Agricultura; p. 45. [Google Scholar]
- 2.Mbanzibwa D.R., Tian Y.P., Tugume A.K., Mukasa S.B., Tairo F., Kyamanywa S., Kullaya A., Valkonen J.P.T. Genetically distinct strains of cassava brown streak virus in the lake victoria basin and the indian ocean coastal area of East Africa. Arch. Virol. 2009;154:353–359. doi: 10.1007/s00705-008-0301-9. [DOI] [PubMed] [Google Scholar]
- 3.Monger W.A., Seal S., Cotton S., Foster G.D. The identification of different isolates of cassava brown streak virus and development of a diagnostic test. Plant Pathol. 2001;50:768–775. [Google Scholar]
- 4.Winter S., Koerbler M., Stein B., Pietruszka A., Paape M., Anja B. The analysis of Cassava brown streak viruses reveals the presence of distinct virus species causing cassava brown streak disease in East Africa. J. Gen. Virol. 2010;91:1365–1372. doi: 10.1099/vir.0.014688-0. [DOI] [PubMed] [Google Scholar]
- 5.Storey H.H., Nichols R.F.W. Virus diseases of East African plants VII – a field experiment in the transmission of cassava mosaic virus. East Afr. Agric. 1938;3:446–449. [Google Scholar]
- 6.Maruthi M.N., Hillocks R.J., Mtunda K., Raya M.D., Muhanna M., Kiozia H., Thresh J.M. Transmission of cassava brown streak virus by Bemisia tabaci (Gennadius) J. Phytopathol. 2005;153:307–312. [Google Scholar]
- 7.Mware B., Narla R., Amata R., Olubayo F., Songa J., Kyamanyua S., Ateka E.M. Efficiency of cassava brown streak virus transmission by two whitefly species in coastal Kenya. J. Gen. Mol. Virol. 2009;1(4):40–45. [Google Scholar]
- 8.Maruthi M.N., Jeremiah S.C., Mohammed I.U., Legg J.P. The role of the whitefly, Bemisia tabaci (Gennadius), and farmer practices in the spread of cassava brown streak ipomoviruses. J. Phytopathol. 2017:1–11. doi: 10.1111/jph.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ateka E., Alicai T., Ndunguru J., Tairo F., Sseruwagi P., Kiarie S. Unusual occurrence of a DAG motif in the Ipomovirus Cassava brown streak virus and implications for its vector transmission. PloS One. 2017;12(11) doi: 10.1371/journal.pone.0187883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hillocks R.J., Thresh J.M., Tomas J., Botaos M., Macia R., Zavier R. Cassava brown streak disease in northern Mozambique. Int. J. Pest Manag. 2002;48:179–182. [Google Scholar]
- 11.Amisse J.J.G. Thesis submitted for MSc degree at University of Witwatersrand; Johannesburg, South Africa: 2013. Molecular Characterization of Cassava Brown Streak Viruses in Mozambique. [Google Scholar]
- 12.Mbanzibwa D.R., Tian Y.P., Tugume A.K., Mukasa S.B., Tairo F., Kyamanywa S., Kullaya A., Valkonen J.P.T. Simultaneous virus-specific detection of the two-cassava brown streak-associated viruses by RT-PCR reveals wide distribution in East Africa, mixed infection, and infections in Manihot glaziovii. J. Virol. Methods. 2011;171:394–400. doi: 10.1016/j.jviromet.2010.09.024. [DOI] [PubMed] [Google Scholar]
- 13.Bua B., Namara J. 9th African Crop Science, Conference Proceedings, Cape Town, South Africa, 28 September – 2 October 2009. 2009. Reaction of Nicotiana species to cassava brown streak virus from Uganda; pp. 647–650. [Google Scholar]
- 14.Lister R.M. Mechanical transmission of cassava brown streak virus. Nat. London. 1959;183:1588–1589. doi: 10.1038/1831588b0. [DOI] [PubMed] [Google Scholar]
- 15.Thresh J.M., Fargette D., Otim-Nape G.W. The viruses and virus diseases of cassava in Africa. Afr. Crop Sci. J. 1994;2:459–478. [Google Scholar]
- 16.Bock K.R. Studies on cassava brown streak virus disease in Kenya. Trop. Sci. 1994;34:134–145. [Google Scholar]
- 17.Schmelzer K. Untersuchungen iiber den Wirtspflanzenkreis des Tabakmauche Virus. Phytopathol. Z. 1957;30:281–314. [Google Scholar]
- 18.Zitter T.A., Murphy J.F. Cucumber mosaic. Plant Health Instr. 2009 [Google Scholar]
- 19.Appiah A.S., Amoatey H.M., Klu G.Y.P., Affu N.T., Azu E., Owusu G.K. Spread of African cassava mosaic virus from cassava (Manihot esculenta Crantz) to physic nut (Jatropha curcas L.) in Ghana. J. Phytol. 2012;4:31–37. [Google Scholar]
- 20.Ogbe F.O., Dixon A.G.O., Hughes J.d’A., Alabi O.J., Okechukwu R. Status of cassava begomoviruses and their new natural hosts in Nigeria. Plant Dis. 2006;90:548–553. doi: 10.1094/PD-90-0548. [DOI] [PubMed] [Google Scholar]
- 21.Hillocks R.J., Raya M.D., Thresh J.M. The association between root necrosis and above ground symptoms of brown streak virus infection of cassava in southern Tanzania. Int. J. Pest Manag. 1996;42:285–289. [Google Scholar]
- 22.Hillocks R.J., Thresh J.M. Natural Resources Institute; UK: 1998. Cassava Mosaic and Cassava Brown Streak Virus Diseases in Africa: a Comparative Guide to Symptoms and Aetiologies; p. 10. [Google Scholar]
- 23.Lodhi M.A., Ye G.N., Weeden N.F., Reisch B.I. A simple and efficient method for DNA extraction from grapevine cultivars and Vitis species. Plant Mol. Biol. Rep. 1994;12:6–13. [Google Scholar]
- 24.Xu Q.H., Zhang Z., Tong G., Gao Z.H., Qu S.C., Qiao Y.S. Effect of sorbitol on total RNA isolation from plum fruit flesh. Jiangsu J. Agric. Sc. 2010;26:390–394. [Google Scholar]
- 25.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holmes F.O. Local lesions in tobacco mosaic. Bot. Gaz. 1929;87:39–55. [Google Scholar]
- 27.Walkey D.G.A. Applied Plant Virology. Springer; Dordrecht: 1991. Mechanical transmission and virus isolation. [Google Scholar]
- 28.Mbanzibwa D.R., Tian Y.P., Tugume A.K., Patil B.L., Yadav J.S., Bagewadi B., Abarshi M.M., Alicai T., Changadeya W., Mkumbira J., Muli M.B., Mukasa S.B., Tairo F., Baguma Y., Kyamanywa S., Kullaya A., Maruthi M.N., Fauquet C.M., Valkonen J.P. Evolution of cassava brown streak disease-associated viruses. J. Gen. Virol. 2011;92:974–987. doi: 10.1099/vir.0.026922-0. [DOI] [PubMed] [Google Scholar]
- 29.Ogwok E., Alicai T., Rey M.E.C., Beyene G., Taylor N.J. Distribution and accumulation of cassava brown streak viruses within infected cassava (Manihot esculenta) plants. Plant Pathol. 2015;64:1235–1246. [Google Scholar]
- 31.Alicai T., Ndunguru J., Sseruwagi P., Tairo F., Okao-Okuja G., Nanvubya R., Kiiza L., Kubatko L., Kehoe M.A., Boykin L.M. Cassava brown streak virus has a rapidly evolving genome: implications for virus speciation, variability, diagnosis and host resistance. Science Report. 2016;6:36164. doi: 10.1038/srep36164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Archer R.H. Sapindaceae. In: Germishuizen G., Meyer N.L., editors. Plants of Southern Africa: an Annotated Checklist. National Botanical Institute; Pretoria: 2003. Strelitzia 14. [Google Scholar]
- 33.Beentje H.J. National Museums of Kenya; Nairobi: 1994. Kenya Trees, Shrubs and Lianas. [Google Scholar]
- 34.Exell A.W. Sapindaceae. In: W Exell A., Fernandes A., Wild H., editors. Flora Zambesiaca 2,2: 537–539. Crown Agents for Overseas Governments and Administrations; London: 1966. [Google Scholar]
- 35.Van Wyk B.[A.E.], Van Den Berg E., Coates Palgrave M., Jordaan M. Briza Publications; Pretoria: 2011. Dictionary of Names for Southern African Trees. [Google Scholar]
- 36.Cowan M.M. Plant products as antimicrobial agents. Clin. Microbiol. Rev. 1999;12:564–582. doi: 10.1128/cmr.12.4.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dunkic V., Bezic N., Vuko E., Cukrov D. Antiphytoviral activity of Satureja montana L. ssp. variegata (host) P. W. Ball essential oil and phenol compounds on CMV and TMV. Molecules. 2010;15:6713–6721. [Google Scholar]
- 38.Scalbert A. Antimicrobial properties of tannins. Phytochemistry. 1991;30:3875–3883. [Google Scholar]
- 39.Meena R.K., Patni V., Arora D.K. Study on phenolics and their oxidative enzyme in Capsicum annuum L, infected with geminivirus. Asian J. Exp. Sci. 2008;22:307–310. [Google Scholar]
- 40.Ogwok E., Patil B.L., Alicai T., Fauquet C.M. Transmission studies with Cassava brown streak Uganda virus (Potyviridae: Ipomovirus) and its interaction with abiotic and biotic factors in Nicotiana benthamiana. J. Virol Methods. 2010;169:296–304. doi: 10.1016/j.jviromet.2010.07.030. [DOI] [PubMed] [Google Scholar]