Abstract
Metabolism is generally considered as a neatly organised system of modular pathways, shaped by evolution under selection for optimal cellular growth. This view falls short of explaining and predicting a number of key observations about the structure and dynamics of metabolism. We highlight these limitations of a pathway-centric view on metabolism and summarise studies suggesting how these could be overcome by viewing metabolism as a thermodynamically and kinetically constrained, dynamical flow system. Such a systems-level, first-principles based view of metabolism can open up new avenues of metabolic engineering and cures for metabolic diseases and allow better insights to a myriad of physiological processes that are ultimately linked to metabolism. Towards further developing this view, we call for a closer interaction among physical and biological disciplines and an increased use of electrochemical and biophysical approaches to interrogate cellular metabolism together with the microenvironment in which it exists.
Keywords: Electrobiology, Metabolic organisation, Evolution of metabolism, Redox potential, Thermodynamics, Cellular physiology, Cellular trade-offs
Highlights
-
•
Cell metabolism represents an interconnected network rather than isolated pathways.
-
•
Capturing the interconnected nature of metabolism makes experimental and theoretical analysis of metabolism very challenging.
-
•
We highlight kinetic and thermodynamic constraints arising from a view of metabolism as an electron flow as a fundamental approach to study and understand metabolism.
-
•
We call for the development of experimental tools and models that focus on capturing redox and ion fluxes connected to electron flows through metabolism.
Metabolism refers to the collective set of biochemical reactions that occur within a cell. Early pioneering studies, crowned by several Nobel prizes, have mapped the so-called core metabolism that includes glycolytic and pentose phosphate pathways (PPP), Calvin–Benson cycle, gluconeogenesis and the tricarboxylic acid (TCA) cycle 1, 2. In parallel, the chemiosmotic theory of energy generation through the respiratory electron transport chain has been formulated and proven correct [3] (even though the mechanistic details are still a matter of debate 4∗∗, 5). These productive lines of research elucidating individual enzymatic reactions continue to make important contributions to our understanding of metabolism [6]. In this opinion article, we argue that it is now time to use this accumulating knowledge to develop an explanatory theoretical framework based on electron flows for interrogating cellular metabolic structure and dynamics at systems level.
Pathway-centric views of metabolism and its shortcomings
Following the early pioneering studies on the biochemistry of enzymes, a pathway-centric framework has dominated the description of metabolism 7∗, 8, 9. This textbook view organizes cellular metabolism into modular pathways that link sugars to pyruvate (i.e. the glycolysis pathways of pentose-phosphate (PP), Entner–Doudoroff (ET), and Embden–Meyerhof–Parnas (EMP)), and those that link pyruvate to the generation of the key reductive equivalents via the TCA (a.k.a. Krebs) cycle and associated glyoxylate and glutamate cycles. These pathways and their interactions with the biosynthesis pathways of lipids and amino acids are considered to give rise to a higher organisation of metabolism as catabolic (energy generating) and anabolic (energy consuming) reactions 7∗, 8, 9. Such organisational view is further enforced by graph-theory analyses that suggest a hierarchical and modular organisation of metabolism 10, 11 (although it is important to note that identifying appropriate null models is challenging when studying metabolism as a graph 12, 13 and different graph representations can give different results [14]).
This pathway-centric framework, thus, draws a picture of cellular metabolism as consisting of well-defined flows through modularly organised pathways. The pathways themselves are suggested to represent historical contingencies in evolution, where a key set of metabolites and metabolic conversions emerged as first forms of “life” [15] and subsequently maintained and expanded through an evolutionary process that is commonly seen as driven by cellular growth under diverse conditions. While this adaptive view of metabolism gives us one possible way of rationalising it, it ignores other possible driving forces and constraints in the evolution of metabolism (e.g. osmotic or toxic effects, trade-offs, cell stability) 16, 17, and falls short in explaining and predicting many of the key structural and dynamical features of metabolism. We list below some of these as open questions, and argue for the development of a new explanatory framework, which can offer starting points towards addressing them.
Why are metabolic systems diverse across different species? Pathways that offer alternative routes for glucose consumption are present or absent in different microbes [18]. Biosynthesis pathways for essential co-factors, such as thiamine or vitamin B12, are lacking in different algae and fungi species 19, 20, and even the TCA cycle structure can be varied in different organisms [21]. While the pathway-centric view tends to explain such structural diversity as “adaptations to different conditions”, we need a more predictive theory that can directly link specific environmental factors to specific metabolic structures (e.g. lack of a given enzyme predicted under a certain environment). For example, loss of metabolic pathways in some organisms is explained as an adaptive process under the provision of specific metabolites by other organisms [22], but it is left unclear why and under what conditions any organism would excrete any such metabolites. Similarly, functionally equivalent pathways are suggested to be adaptive under different conditions due to their different energetic costs 7∗, 18∗, but it is left unclear what those conditions might be. An ideal theoretical framework for metabolism should provide experimentally testable predictions about where and when to find which structural variants of metabolism.
What are the design principles of common metabolic system structures? While metabolic systems show some diversity, there are common structures that are conserved across many species, and that can be presented as key metabolic pathways. It is still not clear why these common pathways are structured in the way that they are. Several studies have tried to answer this question based on biophysical and energetic aspects 18∗, 23, 24∗, but did so while treating individual pathways as isolated units. These approaches should be further developed, given the fact that no pathway (and its reactions and metabolites) exists in isolation in a cellular environment, but instead operates under high interconnectedness through metabolites and common electron shuttles (Figure 1). Developing a system-level framework that could naturally explain the emergence of interconnected pathways as a whole remains to be an open challenge.
Figure 1.
Common metabolic pathways, shown in their broader context, and highlighting their interconnected nature. Reactions are compiled from Ref. [7] and do not represent the full list of known metabolic reactions in the cell. Metabolites are shown using their common names, while reactions are indicated with arrows. Yellow and green arrows indicate oxidation and reduction reactions, respectively, with arrow type indicating the involved electron shuttle; solid, dashed, and double dashed lines for NADH, NADPH, and FADH2. Reactions releasing and consuming ATP are shown in red and blue, respectively. Where multiple reactions are involved when going from one metabolite to another, this is indicated by multiple arrows. Involvement of co-factors and release of small molecules (such as carbon dioxide) are indicated partly (with smaller arrows). Metabolites involved in multiple reactions, and therefore forming additional metabolic cycles, are indicated with a grey backdrop.
Why do cells excrete metabolites and enzymes to their environment? Many metabolites are readily excreted from cells despite still carrying energy value, and many enzymes are known to operate outside of the cell. A pathway-centric view focused on optimization of growth does not provide any explanations for these observations, nor does it allow any predictions about which metabolites or enzymes might be secreted under which conditions. Adaptive arguments have been made about some metabolic excretions (i.e. ethanol) providing a mechanism to inhibit or kill competing species [25]. This particular example also highlights a broader issue with adaptive arguments in that they can readily be formulated for many different observations, but lack a clear sequential evolutionary path; in this case, for example, the killing argument does not explain how small amounts of metabolic excretions, that would not yet function as a toxic agent, could have emerged and stabilised. A series of recent studies indicate that metabolic excretions, in particular those involving organic acids, can arise from cellular trade-offs linked to biophysical constraints on enzymatic reactions such as space (i.e. cytosolic vs. membrane–bound reactions) 26∗∗, 27, 28 and protein capacity 29, 30. These studies highlight the importance of considering the biophysical basis and limits as directing evolutionary processes when explaining and predicting specific features of metabolic systems.
How does metabolism exhibit non-linear dynamics as a whole? The pathway-centric view of metabolism does not consider the dynamics of fluxes across pathways. This dynamic nature is evident from the interconnectedness of key metabolic pathways through common electron and energy shuttles such as NADH and ATP (Figure 1), and the fact that many of their constituting reactions are close to thermodynamic equilibrium under standard conditions [31] (Figure 2). These reactions could thus be operating reversibly with changing cellular conditions, such as pH and redox potential, as shown for some pathways 32, 33∗∗, 34∗, and lead to non-intuitive flux dynamics. In addition, the sharing of (or competition for) metabolites and enzymes across many reactions can lead to complex temporal dynamics such as oscillations and multi-stability, as theoretically shown for even simplified biochemical reaction systems 35∗, 36, 37∗, 38∗∗. Indeed, where measured, periodic oscillations are found to be present in central metabolism 39, 40, 41∗∗, 42. These oscillations are well studied in continuous yeast cultures, in which they have been shown for virtually all metabolites and coincide with a separation of oxidative and reductive phases [43]. Similarly, combined levels of NADH and NADPH in Escherichia coli, measured fluorimetrically, were found to oscillate in line with the cell division cycle [41]. It has been furthermore suggested that these oscillations are crucially linked to other biological oscillations, such as cell cycle and circadian rhythms 44, 45. While some models 46, 47, 48 and metabolic mediators 39, 49 have been proposed to explain these oscillations, a detailed and predictive understanding of metabolic oscillations still needs to be developed. Besides oscillations, bistable dynamics have been predicted and experimentally shown for isolated enzymatic reactions and pathways 36, 50, 51, 52, 53. These dynamics are still challenging to analyse at the single-cell level, but emerging fluorescence approaches offer a promising route [54]. Finally, the dynamics arising from (seemingly) futile enzymatic cycles have been discussed theoretically as a mechanism for achieving robustness against perturbations [37], but have not been studied in the context of temporal dynamics. Theoretical models of futile cycles have shown that these can give rise to bistability and oscillations under certain parameter regimes [50], but these ideas have not been explored experimentally. Overall, the dynamics of metabolism either at the whole system level or at the level of commonly observed dynamical modules (like futile cycles) is understudied.
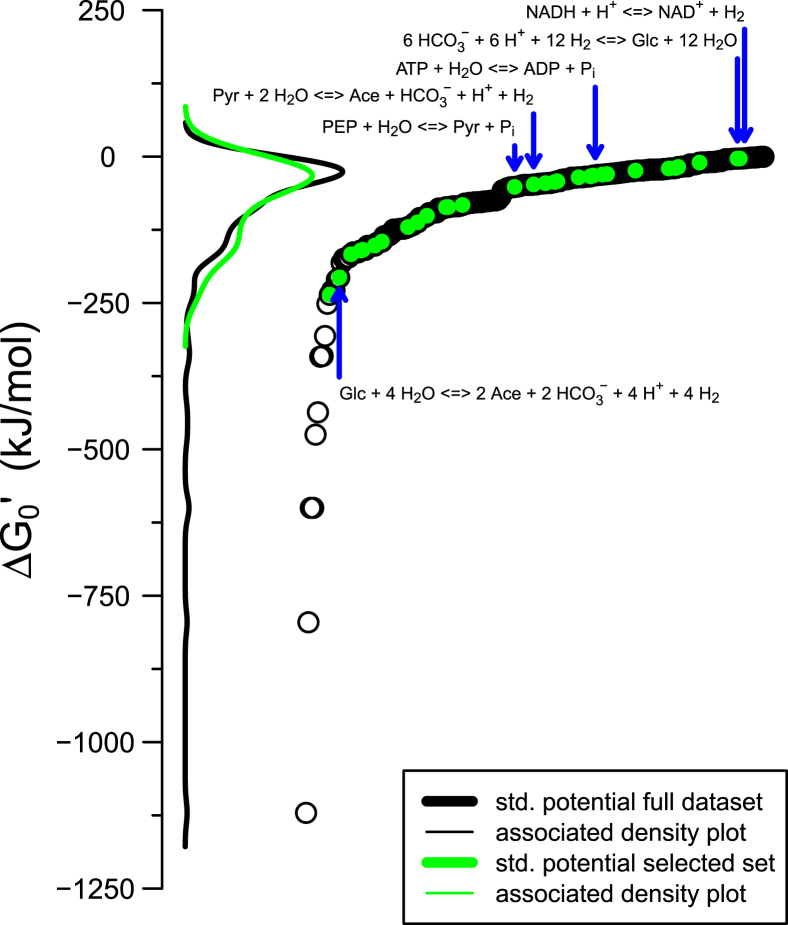
Figure 2.
Thermodynamics of common metabolic conversions as collated from references 31∗, 73∗. The y-axis shows the standard Gibbs free energy at physiological conditions (ΔG0′), while the x-axis is used for reaction index (ordering them from lowest to highest ΔG0′). In calculating the ΔG0′, reactions are considered in their spontaneous direction and the H+/H2 pair is used for electron balancing (in the case of redox reactions). Each reaction is shown as a single data point, while lines are for the density distribution (as indicated in the inset). Reactions that were included in Fig. 1 are shown as green filled points and their distribution by the green line. This figure suggests that most metabolic conversions have a ΔG0′ value close to zero (see distributions), but calls for a further mapping of metabolites and reactions in order to allow more general conclusions to be drawn (with regards to how these distributions compare to those of all possible reactions). We note that reactions with the most negative ΔG0′ (<-250 kJ/mol, left side in plot) are those involving the reduction of nitrate, oxygen, and sulphite; common terminal electron acceptors.
Metabolism as a thermodynamically driven electron flow system
Life depends on free energy gradients [55]. Organisms, or cells, act as “disequilibrium converters” [56] that exploit these energy gradients and couple thermodynamically spontaneous and nonspontaneous reactions through a defined path, the metabolic reaction network. The ratio of the forward and reverse kinetic constants of a reaction equals to the equilibrium constant Keq, which in turn determines its standard Gibbs free energy at a dynamical steady state (ΔG0 = —R⋅T⋅ln(Keq), where R is universal gas constant and T is temperature in Kelvin) 56, 57, 58. The actual reaction free energy (and hence the spontaneous direction of a reaction) at a given condition is based on ΔG0 (Figure 2), which relates to the concentrations of substrates and products under that condition. Thus, changing concentrations through one reaction can make another reaction feasible, allowing metabolism to couple external gradients to internal ones in a turnstile-like mechanism [56]. For example, light-energy drives photosynthetic reactions forward (i.e. water-oxidation with carbon dioxide reduction), and once a pool of metabolites is generated, the reverse reaction (e.g. glucose oxidation with oxygen reduction) becomes spontaneous (i.e. energy releasing), allowing excess energy for other non-spontaneous reactions.
These dynamics of metabolic flows can be formalised through non-equilibrium thermodynamics, which is well developed for simplified enzymatic reaction networks 58, 59. The use of this formalism as a general theoretical framework for studying the entire cellular metabolism, however, has not been fully explored. This is mainly due to a lack of information about thermodynamic properties (i.e. standard Gibbs energies) and intracellular concentrations of many metabolites. The former issue can be approached computationally using statistical (e.g. group contribution) or quantum chemical methods, however, these are currently limited and can only cover a fraction of cellular metabolites 60∗, 61. The intracellular metabolite concentrations can be increasingly determined using technological developments in high-throughput mass spectrometry and ion chromatography [33], hence, with knowledge of a reaction's ΔG0, allowing direct assessment of the thermodynamic state of the reaction.
Development of a dynamical thermodynamic framework for cellular metabolism can benefit from a special focus on redox reactions. These reactions hold a key interconnecting role among different pathways and offer electrochemical interrogation of their dynamics 62, 63. Redox reactions interlink for example glucose oxidation via PP, ET, and EMP pathways, pyruvate oxidation through the TCA cycle, and biosynthesis of many of the amino acids (Figure 1), through the shared use of conserved moieties acting as electron shuttles, such as NAD+/NADH, NADP+/NADPH, FAD/FADH2, ferredoxins, and quinones. This, crucially, makes the ratio of the concentration of the oxidised and reduced forms of these molecules a key factor in determining the thermodynamics of (and flows through) different metabolic paths.
Considering this interconnected set of redox reactions can crucially allow us to consider electronic circuits as an analogy to metabolic systems [64] on one hand, and to attempt their manipulation by electrochemical means on the other 65, 66, 67. Indeed, it has been shown that influencing the ratio of key electron shuttles (e.g. NAD+/NADH) through enzymatic and electrochemical means can directly influence metabolic pathway fluxes and the dynamics of metabolic excretions 63, 68, 69, 70∗.
Potential explanatory powers of viewing metabolism as electron flows
We argued so far that considering metabolism as a thermodynamically driven flow system, rather than isolated pathways, can provide an overarching theoretical framework and that focusing this framework to redox reactions can further allow the formulation of new explanatory and predictive theories. We list below some of the key areas where this electrical view of metabolism can make immediate impact in our understanding of metabolism.
Energetic barriers in electron flow driving metabolic pathway diversity and utilization. When strong electron acceptors are not available in the environment, cells have to use weaker electron acceptors such as organic acids or inorganic molecules (such as H+) as electron sinks (i.e. to maintain electron flow). The result is a smaller free energy available from the overall redox reaction that the cell implements, and an increased risk of the system reaching equilibrium by product accumulation (i.e. thermodynamic inhibition). Recently, it has been shown theoretically that this type of thermodynamic inhibition due to product accumulation can lead to diversity of microbial growth-supporting metabolic reactions and possibly internal metabolic pathways [71]. This theory aligns well with the observation that organisms, such as methanogens and sulfate reducers, adapted to weak or fluctuating electron acceptors show diverse respiratory and fermentative pathways and enzymes 55∗, 72, 73∗. It would be interesting to see if theories relating to thermodynamic inhibition could be developed further to link the diversity of broader organization of metabolic systems (in particular redox shuttle and respiratory enzyme usage) to environmental conditions, and in particular availability and ecological dynamics of common electron acceptors.
Metabolic excretions to maintain electron flows. Thermodynamic inhibition could also be a driving force beyond observed metabolic excretions. For example, oxidation of certain metabolites can become thermodynamically inhibited if the reduced forms of their paired electron shuttle have reached a low concentration. Such an inhibition would be lifted if another metabolite could be reduced using the oxidized form of the same shuttle molecule. This coupling can be further facilitated if one (or some) of the metabolites can be made to act as a sink, e.g. by excreting them from the cell. The increased rate of formation and excretion of organic and amino acids through reductive reactions can be understood within this view, as a mechanism to combat shifts in the NAD+/NADH ratio. Indeed, experimental manipulation of the NAD+/NADH ratio in E. coli and yeast were found to directly influence the dynamics of metabolic excretions through fermentative pathways (i.e. acetate and ethanol excretion) 69, 70∗. Thus, furthering a theoretical framework based on thermodynamically driven redox paths might allow us to predict metabolic excretions under different conditions.
Enzymatic excretions combatting toxic effects arising from redox reactions involving electron sinks (i.e. respiration). Redox reactions with compounds that act as final electron acceptors (i.e. electron sinks), should ideally involve strong electron acceptors so to provide a significant energy gradient (see Figure 2). This, however, creates an additional constraint on metabolic system structure and dynamics in that strong terminal redox reactions can also result in the generation of even stronger redox active compounds [74]. The respiration (i.e. reduction) of O2, for example, is found to lead to a generation of reactive oxygen species (ROS) at a rate up to 20% [75], while sulfate and nitrate respiration results in ‘toxic’ sulfide and nitrite, respectively. To avoid additional, uncontrolled redox reactions by these strong oxidizing agents, cells must have evolved ways to generate enzymes and redox metabolites that can act as effective neutralizers against strong redox agents. While catalases and peroxidases are known enzymes that can combat ROS toxicity [76], the generation of ROS from respiration could have been a strong driver also for the overall metabolic structure and dynamics. Indeed, connections between resistance to ROS and metabolic flux changes have been shown 77, 78, 79, 80, 81, and ROS mitigation is suggested as an explanation for excretions of metal oxidizing enzymes in bacteria 82, 83.
Compartmentalization of electron flows in space or time. Spatial compartmentalization of metabolism can be seen in form of specialized organelles within an individual cell 84, 85 (e.g. mitochondria and chloroplasts) and in form of subpopulations within an isogenic cell population 54, 86. Such compartmentalization could be understood in the context of thermodynamic inhibition due to product accumulation and the associated issue of maintaining electron flows: local microenvironments that can maintain a disequilibrium through separation or flow of products from their reactions can allow overcoming thermodynamic inhibition. A similar effect could also be achieved by separation of processes over time, i.e. by implementing oscillatory dynamics that can balance the impact of opposing processes (e.g. on the NAD+/NADH ratio). Indeed, theoretical analyses suggest that oscillations, as observed for example in the NAD+/NADH ratio in yeast and eukaryotic cells 39, 44, 49, may enhance the thermodynamic efficiency of glycolysis [87] or, more broadly, coupled chemical reaction systems [88]. This is in line with experimental observations of periodic NADH/NADPH concentration oscillations correlating with cell cycle phases 41∗∗, 43, which could be an adaptation to alternately maximize thermodynamic driving force through pathways of interest with temporal separation.
Testing and establishing the electrical view of metabolism – the road ahead
It has been long-recognized that metabolism is a prime example of a system obeying non-equilibrium thermodynamics [89]. Bringing this view to practical study and engineering of metabolism, however, remains a challenge. We believe that the presented framework highlighting a view of metabolism that is based on maintenance of electron flows, under constraints arising from thermodynamics, kinetics, and interconnectedness of pathways through common electron and energy shuttles, can initiate further studies towards overcoming this challenge. This framework calls for more interaction between experimental and modelling disciplines and the development of new theoretical and experimental tools.
On the theoretical side, we note that efforts have been made to expand the stoichiometric, constraint-based optimization models of metabolism (i.e. flux balance analysis, FBA) with thermodynamics 90, 91, 92, 93 and also with overall constraints that can mimic some of the biophysical constraints arising from resource and space limitations 27, 94, 95, 96. It would be important to continue these developments, and also consider coupling FBA with the modelling of cell environment dynamics 95, 97 towards incorporating possible thermodynamic inhibitions and electron acceptor availabilities in these environments. Inevitably, however, capturing the full dynamics of metabolism as electron flows will require kinetic and even spatial models that account for physiological parameters such as pH and membrane potential. To achieve this, current kinetic models, which usually assume fixed ratios for common electron and energy shuttles, and tend to consider pathways in isolation, need to be further developed. Dynamics of electron shuttles and the possibility of reaction reversibility will need to be incorporated to account for thermodynamics. Statistical thermodynamics simulations, as used recently to simulate TCA cycle dynamics [98], can be useful in this context but would need to be expanded and further developed to account for the larger parts and interconnected nature of metabolism. Similarly, emerging thermodynamic models for modelling overall metabolic conversions embedded through metabolism (i.e. thermodynamic microbial growth models) [99] can be possibly adapted to model cellular metabolism. As kinetic models incorporate more biochemical realism through reaction reversibility, electron and energy shuttle dynamics, and pathway interconnectedness, the major challenge will be to maintain models as tractable and ‘simple enough’ so that they can still generate experimentally testable insights and predictions.
On the experimental side, approaches integrating physiological, metabolomic, and electrochemical techniques will need to be developed to better understand electron flows in metabolism and constraints on these flows. In particular, we note that combined application of fluorescent reporters (as being increasingly used to interrogate cellular redox states and physiology (e.g. 41∗∗, 100, 101, 102, 103, 104)) and emerging nano-scale electrochemical probing methods [105] can provide powerful insights into the electron flow dynamics at cellular and population levels. These methods can be particularly suited to link metabolic dynamics to higher–level complex physiological processes such as cellular differentiations. Intriguingly, cellular differentiation is often associated with specialization of metabolism (e.g. bacterial spores 106, 107, T cells [108], cancer cells [109]). We envision that development of the electron-flow based view to metabolism may provide a novel insight to these cellular differentiation processes as arising from thermodynamic limits and imbalances in metabolism. This, in turn, can open up novel means of controlling these processes using electrochemistry, and opening up new medical intervention methods.
Conflict of interest statement
Nothing declared.
Acknowledgments
This work is funded by The University of Warwick and by the Biotechnological and Biological Sciences Research Council (BBSRC), with grant IDs: BB/K003240/2 (to OSS) and BB/M017982/1 (to the Warwick Integrative Synthetic Biology Centre, WISB).
This review comes from a themed issue on Systems ecology and evolution
Edited by Eörs Szathmáry and Ferenc Jordán
References
- 1.Kresge N., Simoni R.D., Hill R.L. Otto Fritz, Meyerhof and the elucidation of the glycolytic pathway. J Biol Chem. 2005;280:e3. [PubMed] [Google Scholar]
- 2.Krebs H.A., Johnson W.A. Metabolism of ketonic acids in animal tissues. Biochem J. 1937;31:645–660. doi: 10.1042/bj0310645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- Mulkidjanian A.Y., Heberle J., Cherepanov D.A. Protons @ interfaces: implications for biological energy conversion. Biochim Biophys Acta – Bioenerg. 2006;1757:913–930. doi: 10.1016/j.bbabio.2006.02.015. [DOI] [PubMed] [Google Scholar]; This review highlights what is still not understood about chemiosmosis, and how this affects bacterial energy metabolism in different pH environments.
- 5.Nath S. Analysis of molecular mechanisms of ATP synthesis from the standpoint of the principle of electrical neutrality. Biophys Chem. 2017;224:49–58. doi: 10.1016/j.bpc.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Prosser G.A., Larrouy-Maumus G., de Carvalho L.P.S. Metabolomic strategies for the identification of new enzyme functions and metabolic pathways. EMBO Rep. 2014;15:657–669. doi: 10.15252/embr.201338283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk G. Springer New York; 1986. Bacterial metabolism. [Google Scholar]; An important and comprehensive reference for metabolic pathway structure and energetics.
- 8.Voet D., Voet J.G. Wiley; 2011. Biochemistry. [Google Scholar]
- 9.Berg J.M., Tymoczko J.L., Stryer L. W. H. Freeman & Co.; 2002. Biochemistry. [Google Scholar]
- 10.Ravasz E., Somera A.L., Mongru D.A., Oltvai Z.N., Barabási A.L. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- 11.Jeong H., Tombor B., Albert R., Oltvai Z.N., Barabási A.L. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 12.Stumpf M.P.H., Wiuf C., May R.M. Subnets of scale-free networks are not scale-free: sampling properties of networks. Proc Natl Acad Sci. 2005;102:4221–4224. doi: 10.1073/pnas.0501179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox Keller E. Revisiting “scale-free” networks. Bioessays. 2005;27:1060–1068. doi: 10.1002/bies.20294. [DOI] [PubMed] [Google Scholar]
- 14.Arita M. The metabolic world of Escherichia coli is not small. Proc Natl Acad Sci. 2004;101:1543–1547. doi: 10.1073/pnas.0306458101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin W., Russell M.J. On the origin of biochemistry at an alkaline hydrothermal vent. Philos Trans R Soc B Biol Sci. 2007;362:1887–1926. doi: 10.1098/rstb.2006.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schuster S., Pfeiffer T., Fell D.A. Is maximization of molar yield in metabolic networks favoured by evolution? J Theor Biol. 2008;252:497–504. doi: 10.1016/j.jtbi.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 17.Papp B., Teusink B., Notebaart R.A. A critical view of metabolic network adaptations. HFSP J. 2009;3:24–35. doi: 10.2976/1.3020599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamholz A., Noor E., Bar-Even A., Liebermeister W., Milo R. Glycolytic strategy as a tradeoff between energy yield and protein cost. Proc Natl Acad Sci. 2013;110:10039–10044. doi: 10.1073/pnas.1215283110. [DOI] [PMC free article] [PubMed] [Google Scholar]; An important contribution that formalizes the different enzyme investments required to achieve the same net forward flux among functionally equivalent pathways due to thermodynamic differences.
- 19.Helliwell K.E., Wheeler G.L., Smith A.G. Widespread decay of vitamin-related pathways: coincidence or consequence? Trends Genet. 2013;29:469–478. doi: 10.1016/j.tig.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Jiang X., Zerfaß C., Feng S., Eichmann R., Asally M., Schäfer P., Soyer O.S. Impact of spatial organization on a novel auxotrophic interaction among soil microbes. ISME J. 2018;12:1443–1456. doi: 10.1038/s41396-018-0095-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steinhauser D., Fernie A.R., Araújo W.L. Unusual cyanobacterial TCA cycles: not broken just different. Trends Plant Sci. 2012;17:503–509. doi: 10.1016/j.tplants.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 22.Morris J.J., Lenski R.E., Zinser E.R. The black queen hypothesis: evolution of dependencies through adaptive gene loss. mBio. 2012;3 doi: 10.1128/mBio.00036-12. e00036-12-e00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Court S.J., Waclaw B., Allen R.J. Lower glycolysis carries a higher flux than any biochemically possible alternative. Nat Commun. 2015;6:8427. doi: 10.1038/ncomms9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Even A., Flamholz A., Noor E., Milo R. Rethinking glycolysis: on the biochemical logic of metabolic pathways. Nat Chem Biol. 2012;8:509–517. doi: 10.1038/nchembio.971. [DOI] [PubMed] [Google Scholar]; An important contribution, highlighting the role of multitude of biophysical factors that might have influenced evolution of key pathways.
- 25.Piskur J., Rozpedowska E., Polakova S., Merico A., Compagno C. How did Saccharomyces evolve to become a good brewer? Trends Genet. 2006;22:183–186. doi: 10.1016/j.tig.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Szenk M., Dill K.A., de Graff A.M.R. Why Do Fast-Growing Bacteria Enter Overflow Metabolism? Testing the Membrane Real Estate Hypothesis. Cell Syst. 2017;5:95–104. doi: 10.1016/j.cels.2017.06.005. [DOI] [PubMed] [Google Scholar]; A recent and important study of biophysical limits and constraints on the metabolic fluxes among respiratory and fermentative pathways.
- 27.Beg Q.K., Vazquez A., Ernst J., de Menezes M.A., Bar-Joseph Z., Barabasi A.-L., Oltvai Z.N. Intracellular crowding defines the mode and sequence of substrate uptake by Escherichia coli and constrains its metabolic activity. Proc Natl Acad Sci. 2007;104:12663–12668. doi: 10.1073/pnas.0609845104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Majewski R.A., Domach M.M. Simple constrained-optimization view of acetate overflow in E. coli. Biotechnol Bioeng. 1990;35:732–738. doi: 10.1002/bit.260350711. [DOI] [PubMed] [Google Scholar]
- 29.Molenaar D., van Berlo R., de Ridder D., Teusink B. Shifts in growth strategies reflect tradeoffs in cellular economics. Mol Syst Biol. 2009;5:323. doi: 10.1038/msb.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basan M., Hui S., Okano H., Zhang Z., Shen Y., Williamson J.R., Hwa T. Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature. 2015;528:99–104. doi: 10.1038/nature15765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S.L., Smith-Magowan D. The thermodynamics of the krebs cycle and related compounds. J Phys Chem Ref Data. 1990;19:1049–1073. [Google Scholar]; A key reference paper, listing thermodynamics data for reactions forming the TCA cycle.
- 32.Timmers P.H.A., Welte C.U., Koehorst J.J., Plugge C.M., Jetten M.S.M., Stams A.J.M. Reverse methanogenesis and respiration in methanotrophic archaea. Archaea. 2017;2017:1–22. doi: 10.1155/2017/1654237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett B.D., Kimball E.H., Gao M., Osterhout R., Van Dien S.J., Rabinowitz J.D. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]; An example of how quantitation of metabolites allows direct interpretations of the thermodynamics of metabolic pathways.
- Noor E., Bar-Even A., Flamholz A., Reznik E., Liebermeister W., Milo R. Pathway thermodynamics highlights kinetic obstacles in central metabolism. PLoS Comput Biol. 2014;10 doi: 10.1371/journal.pcbi.1003483. [DOI] [PMC free article] [PubMed] [Google Scholar]; An important contribution formalising the relation between reaction forward flux and thermodynamics.
- Feng S., Sáez M., Wiuf C., Feliu E., Soyer O.S. Core signalling motif displaying multistability through multi-state enzymes. J R Soc Interface. 2016;13:20160524. doi: 10.1098/rsif.2016.0524. [DOI] [PMC free article] [PubMed] [Google Scholar]; A theoretical study suggesting that enyzme or substrate sharing among reactions, which is also common in metabolism, can result in multistability in system dynamics.
- 36.Cimino A., Hervagault J.-F. Irreversible transitions in a model substrate cycle. An experimental illustration. FEBS Lett. 1990;263:199–205. doi: 10.1016/0014-5793(90)81373-v. [DOI] [PubMed] [Google Scholar]
- Reich J.G., Selkov E. Academic Press; 1981. Energy metabolism of the cell. [Google Scholar]; An important reference point for the understanding of kinetics and nonlinar dynamics of enzymatic reaction systems
- Liu J., Prindle A., Humphries J., Gabalda-Sagarra M., Asally M., Lee D.D., Ly S., Garcia-Ojalvo J., Süel G.M. Metabolic co-dependence gives rise to collective oscillations within biofilms. Nature. 2015;523:550–554. doi: 10.1038/nature14660. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first demonstration of metabolism related oscillations in a spatial context, discovered through the application of redox sensitive fluorescent dyes.
- 39.Sohn H.-Y., Murray D.B., Kuriyama H. Ultradian oscillation of Saccharomyces cerevisiae during aerobic continuous culture: hydrogen sulphide mediates population synchrony. Yeast. 2000;16:1185–1190. doi: 10.1002/1097-0061(20000930)16:13<1185::AID-YEA619>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 40.Iotti S., Borsari M., Bendahan D. Oscillations in energy metabolism. Biochim Biophys Acta – Bioenerg. 2010;1797:1353–1361. doi: 10.1016/j.bbabio.2010.02.019. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Milias-Argeitis A., Heinemann M. Dynamic single-cell NAD(P)H measurement reveals oscillatory metabolism throughout the E. coli cell division cycle. Sci Rep. 2018;8:2162. doi: 10.1038/s41598-018-20550-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; An important study, attempting to develop fluorescent dyes for the measurement of redox state in single bacterial cells.
- 42.Ahn E., Kumar P., Mukha D., Tzur A., Shlomi T. Temporal fluxomics reveals oscillations in TCA cycle flux throughout the mammalian cell cycle. Mol Syst Biol. 2017;13:953. doi: 10.15252/msb.20177763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murray D.B., Amariei C., Sasidharan K., Machné R., Aon M.A., Lloyd D. Temporal partitioning of the yeast cellular network. In: Aon M., Saks V., editors. Systems Biology of metabolic and signaling networks. Springer series in biophysics. Schlattner U. Springer Berlin Heidelberg; 2014. pp. 323–349. [Google Scholar]
- 44.Papagiannakis A., Niebel B., Wit E.C., Heinemann M. Autonomous metabolic oscillations robustly gate the early and late cell cycle. Mol Cell. 2017;65:285–295. doi: 10.1016/j.molcel.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 45.O'Neill J.S., Feeney K.A. Circadian redox and metabolic oscillations in mammalian systems. Antioxid Redox Signal. 2014;20:2966–2981. doi: 10.1089/ars.2013.5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolf J., Heinrich R. Effect of cellular interaction on glycolytic oscillations in yeast: a theoretical investigation. Biochem J. 2000;345:321. [PMC free article] [PubMed] [Google Scholar]
- 47.Smallbone K., Messiha H.L., Carroll K.M., Winder C.L., Malys N., Dunn W.B., Murabito E., Swainston N., Dada J.O., Khan F. A model of yeast glycolysis based on a consistent kinetic characterisation of all its enzymes. FEBS Lett. 2013;587:2832–2841. doi: 10.1016/j.febslet.2013.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolf J., Sohn H.-Y., Heinrich R., Kuriyama H. Mathematical analysis of a mechanism for autonomous metabolic oscillations in continuous culture of Saccharomyces cerevisiae. FEBS Lett. 2001;499:230–234. doi: 10.1016/s0014-5793(01)02562-5. [DOI] [PubMed] [Google Scholar]
- 49.Richard P., Bakker B.M., Teusink B., Dam K., Westerhoff H.V. Acetaldehyde mediates the synchronization of sustained glycolytic oscillations in populations of yeast cells. Eur J Biochem. 1996;235:238–241. doi: 10.1111/j.1432-1033.1996.00238.x. [DOI] [PubMed] [Google Scholar]
- 50.Guidi G.M., Goldbeter A. From bistability to oscillations in a model for the isocitrate dehydrogenase reaction. Biophys Chem. 1998;72:201–210. doi: 10.1016/s0301-4622(98)00134-3. [DOI] [PubMed] [Google Scholar]
- 51.Kotte O., Volkmer B., Radzikowski J.L., Heinemann M. Phenotypic bistability in Escherichia coli's central carbon metabolism. Mol Syst Biol. 2014;10:736. doi: 10.15252/msb.20135022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mulukutla B.C., Yongky A., Grimm S., Daoutidis P., Hu W.-S. Multiplicity of steady states in glycolysis and shift of metabolic state in cultured mammalian cells. PloS One. 2015;10 doi: 10.1371/journal.pone.0121561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Heerden J.H., Wortel M.T., Bruggeman F.J., Heijnen J.J., Bollen Y.J.M., Planqué R., Hulshof J., O'Toole T.G., Wahl S.A., Teusink B. Lost in transition: start-up of glycolysis yields subpopulations of nongrowing cells. Science. 2014;343:1245114. doi: 10.1126/science.1245114. [DOI] [PubMed] [Google Scholar]
- 54.Rosenthal A.Z., Qi Y., Hormoz S., Park J., Li S.H.-J., Elowitz M.B. Metabolic interactions between dynamic bacterial subpopulations. Elife. 2018;7 doi: 10.7554/eLife.33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoepp-Cothenet B., van Lis R., Atteia A., Baymann F., Capowiez L., Ducluzeau A.-L., Duval S., ten Brink F., Russell M.J., Nitschke W. On the universal core of bioenergetics. Biochim Biophys Acta - Bioenerg. 2013;1827:79–93. doi: 10.1016/j.bbabio.2012.09.005. [DOI] [PubMed] [Google Scholar]; An important contribution highlighting and surveying the central role of redox reactions in metabolism.
- 56.Branscomb E., Russell M.J. Turnstiles and bifurcators: the disequilibrium converting engines that put metabolism on the road. Biochim Biophys Acta – Bioenerg. 2013;1827:62–78. doi: 10.1016/j.bbabio.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 57.Bergethon P.R. Springer; New York: 2010. The physical basis of biochemistry. [Google Scholar]
- 58.Ederer M., Gilles E.D. Thermodynamically feasible kinetic models of reaction networks. Biophys J. 2007;92:1846–1857. doi: 10.1529/biophysj.106.094094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qian H., Reluga T.C. Nonequilibrium thermodynamics and nonlinear kinetics in a cellular signaling switch. Phys Rev Lett. 2005;94:028101. doi: 10.1103/PhysRevLett.94.028101. [DOI] [PubMed] [Google Scholar]
- Jinich A., Sanchez-Lengeling B., Ren H., Harman R., Aspuru-Guzik A. A mixed quantum chemistry/machine learning approach for the fast and accurate prediction of biochemical redox potentials and its large-scale application to 315,000 redox reactions. bioRxiv. 2018 doi: 10.1021/acscentsci.9b00297. [DOI] [PMC free article] [PubMed] [Google Scholar]; An alternative approach towards trying to fill the gap in the knowledge (estimation) of reaction thermodynamics.
- 61.Du B., Zhang Z., Grubner S., Yurkovich J.T., Palsson B.O., Zielinski D.C. Temperature-dependent estimation of Gibbs energies using an updated group-contribution method. Biophys J. 2018;114:2691–2702. doi: 10.1016/j.bpj.2018.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaneko M., Ishikawa M., Hashimoto K., Nakanishi S. Molecular design of cytocompatible amphiphilic redox-active polymers for efficient extracellular electron transfer. Bioelectrochemistry. 2017;114:8–12. doi: 10.1016/j.bioelechem.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 63.Rawson F.J., Downard A.J., Baronian K.H. Electrochemical detection of intracellular and cell membrane redox systems in Saccharomyces cerevisiae. Sci Rep. 2015;4:5216. doi: 10.1038/srep05216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berry M.N. An electrochemical interpretation of metabolism. FEBS Lett. 1981;134:133–138. doi: 10.1016/0014-5793(81)80585-6. [DOI] [PubMed] [Google Scholar]
- 65.Zerfaß C., Chen J., Soyer O.S. Engineering microbial communities using thermodynamic principles and electrical interfaces. Curr Opin Biotechnol. 2018;50:121–127. doi: 10.1016/j.copbio.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 66.Kato S. Biotechnological aspects of microbial extracellular electron transfer. Microb Environ. 2015;30:133–139. doi: 10.1264/jsme2.ME15028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lovley D.R. Electromicrobiology. Annu Rev Microbiol. 2012;66:391–409. doi: 10.1146/annurev-micro-092611-150104. [DOI] [PubMed] [Google Scholar]
- 68.Vemuri G.N., Altman E., Sangurdekar D.P., Khodursky A.B., Eiteman M.A. Overflow metabolism in Escherichia coli during steady-state growth: transcriptional regulation and effect of the redox ratio. Appl Environ Microbiol. 2006;72:3653–3661. doi: 10.1128/AEM.72.5.3653-3661.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hou J., Scalcinati G., Oldiges M., Vemuri G.N. Metabolic impact of increased NADH availability in Saccharomyces cerevisiae. Appl Environ Microbiol. 2010;76:851–859. doi: 10.1128/AEM.02040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri G.N., Eiteman M.A., McEwen J.E., Olsson L., Nielsen J. Increasing NADH oxidation reduces overflow metabolism in Saccharomyces cerevisiae. Proc Natl Acad Sci. 2007;104:2402–2407. doi: 10.1073/pnas.0607469104. [DOI] [PMC free article] [PubMed] [Google Scholar]; An important contribution demonstrating the relation between metabolic excretion dynamics and cellular redox balances.
- Großkopf T., Soyer O.S. Microbial diversity arising from thermodynamic constraints. ISME J. 2016;10:2725–2733. doi: 10.1038/ismej.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]; A theoretical demonstration that thermodynamic inhibition can result in diversity of metabolic routes from a single compound.
- 72.Thauer R.K., Kaster A.-K., Seedorf H., Buckel W., Hedderich R. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol. 2008;6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- Thauer R.K., Jungermann K., Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]; An important and early compilation of reaction thermodynamics discussed in the context of microbial metabolism.
- 74.Davies K.J. Oxidative stress: the paradox of aerobic life. Biochem Soc Symp. 1995;61:1–31. doi: 10.1042/bss0610001. [DOI] [PubMed] [Google Scholar]
- 75.Imlay J.A. Pathways of oxidative damage. Annu Rev Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 76.Mishra S., Imlay J. Why do bacteria use so many enzymes to scavenge hydrogen peroxide? Arch Biochem Biophys. 2012;525:145–160. doi: 10.1016/j.abb.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ralser M., Wamelink M.M., Kowald A., Gerisch B., Heeren G., Struys E.A., Klipp E., Jakobs C., Breitenbach M., Lehrach H. Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J Biol. 2007;6:10. doi: 10.1186/jbiol61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mailloux R.J., Lemire J., Appanna V.D. Metabolic networks to combat oxidative stress in Pseudomonas fluorescens. Antonie Leeuwenhoek. 2011;99:433–442. doi: 10.1007/s10482-010-9538-x. [DOI] [PubMed] [Google Scholar]
- 79.Alhasawi A., Leblanc M., Appanna N.D., Auger C., Appanna V.D. Aspartate metabolism and pyruvate homeostasis triggered by oxidative stress in Pseudomonas fluorescens: a functional metabolomic study. Metabolomics. 2015;11:1792–1801. [Google Scholar]
- 80.Tharmalingam S., Alhasawi A., Appanna V.P., Lemire J., Appanna V.D. Reactive nitrogen species (RNS)-resistant microbes: adaptation and medical implications. Biol Chem. 2017;398:1193–1208. doi: 10.1515/hsz-2017-0152. [DOI] [PubMed] [Google Scholar]
- 81.Lemire J., Alhasawi A., Appanna V.P., Tharmalingam S., Appanna V.D. Metabolic defence against oxidative stress: the road less travelled so far. J Appl Microbiol. 2017;123:798–809. doi: 10.1111/jam.13509. [DOI] [PubMed] [Google Scholar]
- 82.Zerfaß C., Christie-Oleza J.A., Soyer O.S. Manganese oxide biomineralization is a social trait protecting against nitrite toxicity. bioRxiv. 2018 [Google Scholar]
- 83.Banh A., Chavez V., Doi J., Nguyen A., Hernandez S., Ha V., Jimenez P., Espinoza F., Johnson H.A. Manganese (Mn) oxidation increases intracellular Mn in Pseudomonas putida GB-1. PloS One. 2013;8 doi: 10.1371/journal.pone.0077835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zecchin A., Stapor P.C., Goveia J., Carmeliet P. Metabolic pathway compartmentalization: an underappreciated opportunity? Curr Opin Biotechnol. 2015;34:73–81. doi: 10.1016/j.copbio.2014.11.022. [DOI] [PubMed] [Google Scholar]
- 85.Brandriss M.C., Magasanik B. Subcellular compartmentation in control of converging pathways for proline and arginine metabolism in Saccharomyces cerevisiae. J Bacteriol. 1981;145:1359–1364. doi: 10.1128/jb.145.3.1359-1364.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Campbell K., Vowinckel J., Ralser M. Cell-to-cell heterogeneity emerges as consequence of metabolic cooperation in a synthetic yeast community. Biotechnol J. 2016;11:1169–1178. doi: 10.1002/biot.201500301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Richter P., Ross J. Concentration oscillations and efficiency: glycolysis. Science. 1981;211:715–717. doi: 10.1126/science.6450447. [DOI] [PubMed] [Google Scholar]
- 88.Himeoka Y., Kaneko K. Enzyme oscillation can enhance the thermodynamic efficiency of cellular metabolism: consequence of anti-phase coupling between reaction flux and affinity. Phys Biol. 2016;13:026002. doi: 10.1088/1478-3975/13/2/026002. [DOI] [PubMed] [Google Scholar]
- 89.Gräber P. Free energy transduction and biochemical cycle kinetics. Bioelectrochem Bioenerg. 1990;24:69. [Google Scholar]
- 90.Beard D.A., Babson E., Curtis E., Qian H. Thermodynamic constraints for biochemical networks. J Theor Biol. 2004;228:327–333. doi: 10.1016/j.jtbi.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 91.Hoppe A., Hoffmann S., Holzhütter H.-G. Including metabolite concentrations into flux balance analysis: thermodynamic realizability as a constraint on flux distributions in metabolic networks. BMC Syst Biol. 2007;1:23. doi: 10.1186/1752-0509-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Feist A.M., Henry C.S., Reed J.L., Krummenacker M., Joyce A.R., Karp P.D., Broadbelt L.J., Hatzimanikatis V., Palsson B.Ø. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol. 2007;3:121. doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Henry C.S., Broadbelt L.J., Hatzimanikatis V. Thermodynamics-based metabolic flux analysis. Biophys J. 2007;92:1792–1805. doi: 10.1529/biophysj.106.093138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhuang K., Vemuri G.N., Mahadevan R. Economics of membrane occupancy and respiro-fermentation. Mol Syst Biol. 2014;7 doi: 10.1038/msb.2011.34. 500–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Großkopf T., Consuegra J., Gaffé J., Willison J.C., Lenski R.E., Soyer O.S., Schneider D. Metabolic modelling in a dynamic evolutionary framework predicts adaptive diversification of bacteria in a long-term evolution experiment. BMC Evol Biol. 2016;16:163. doi: 10.1186/s12862-016-0733-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mori M., Hwa T., Martin O.C., De Martino A., Marinari E. Constrained allocation flux balance analysis. PLoS Comput Biol. 2016;12 doi: 10.1371/journal.pcbi.1004913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Harcombe W.R., Riehl W.J., Dukovski I., Granger B.R., Betts A., Lang A.H., Bonilla G., Kar A., Leiby N., Mehta P. Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell Rep. 2014;7:1104–1115. doi: 10.1016/j.celrep.2014.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cannon W.R. Simulating metabolism with statistical thermodynamics. PloS One. 2014;9 doi: 10.1371/journal.pone.0103582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Desmond-Le Quéméner E., Bouchez T. A thermodynamic theory of microbial growth. ISME J. 2014;8:1747–1751. doi: 10.1038/ismej.2014.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.te Winkel J.D., Gray D.A., Seistrup K.H., Hamoen L.W., Strahl H. Analysis of antimicrobial-triggered membrane depolarization using voltage sensitive dyes. Front Cell Dev Biol. 2016;4 doi: 10.3389/fcell.2016.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Arlt J., Martinez V.A., Dawson A., Pilizota T., Poon W.C.K. Painting with light-powered bacteria. Nat Commun. 2018;9:768. doi: 10.1038/s41467-018-03161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Prindle A., Liu J., Asally M., Ly S., Garcia-Ojalvo J., Süel G.M. Ion channels enable electrical communication in bacterial communities. Nature. 2015;527:59–63. doi: 10.1038/nature15709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lin M.Z., Schnitzer M.J. Genetically encoded indicators of neuronal activity. Nat Neurosci. 2016;19:1142–1153. doi: 10.1038/nn.4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.San Martín A., Sotelo-Hitschfeld T., Lerchundi R., Fernández-Moncada I., Ceballo S., Valdebenito R., Baeza-Lehnert F., Alegría K., Contreras-Baeza Y., Garrido-Gerter P. Single-cell imaging tools for brain energy metabolism: a review. Neurophotonics. 2014;1:011004. doi: 10.1117/1.NPh.1.1.011004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bentley C.L., Kang M., Unwin P.R. Scanning electrochemical cell microscopy: new perspectives on electrode processes in action. Curr Opin Electrochem. 2017;6:23–30. [Google Scholar]
- 106.Sonenshein A.L. Control of key metabolic intersections in Bacillus subtilis. Nat Rev Microbiol. 2007;5:917–927. doi: 10.1038/nrmicro1772. [DOI] [PubMed] [Google Scholar]
- 107.Sporer A.J., Kahl L.J., Price-Whelan A., Dietrich L.E.P. Redox-based regulation of bacterial development and behavior. Annu Rev Biochem. 2017;86:777–797. doi: 10.1146/annurev-biochem-061516-044453. [DOI] [PubMed] [Google Scholar]
- 108.Peng M., Yin N., Chhangawala S., Xu K., Leslie C.S., Li M.O. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science. 2016;354:481–484. doi: 10.1126/science.aaf6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jain M., Nilsson R., Sharma S., Madhusudhan N., Kitami T., Souza A.L., Kafri R., Kirschner M.W., Clish C.B., Mootha V.K. Metabolite profiling identifies a key role for Glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]