Highlights
-
•
PBK models have helped to facilitate quantitative in vitro to in vivo extrapolation.
-
•
PBK modelling has the potential to play a significant role in reducing animal testing.
-
•
It is critical to assess the validity of PBK models built using non-animal data.
-
•
A framework is needed for communicating characteristics and results of PBK modelling.
Keywords: Physiologically based kinetic models, PBPK, PBTK, Toxicokinetics, In vitro, In silico
Abstract
The fields of toxicology and chemical risk assessment seek to reduce, and eventually replace, the use of animals for the prediction of toxicity in humans. In this context, physiologically based kinetic (PBK) modelling based on in vitro and in silico kinetic data has the potential to a play significant role in reducing animal testing, by providing a methodology capable of incorporating in vitro human data to facilitate the development of in vitro to in vivo extrapolation of hazard information. In the present article, we discuss the challenges in: 1) applying PBK modelling to support regulatory decision making under the toxicology and risk-assessment paradigm shift towards animal replacement; 2) constructing PBK models without in vivo animal kinetic data, while relying solely on in vitro or in silico methods for model parameterization; and 3) assessing the validity and credibility of PBK models built largely using non-animal data. The strengths, uncertainties, and limitations of PBK models developed using in vitro or in silico data are discussed in an effort to establish a higher degree of confidence in the application of such models in a regulatory context. The article summarises the outcome of an expert workshop hosted by the European Commission Joint Research Centre (EC-JRC) – European Union Reference Laboratory for Alternatives to Animal Testing (EURL ECVAM), on “Physiologically-Based Kinetic modelling in risk assessment – reaching a whole new level in regulatory decision-making” held in Ispra, Italy, in November 2016, along with results from an international survey conducted in 2017 and recently reported activities occurring within the PBK modelling field. The discussions presented herein highlight the potential applications of next generation (NG)-PBK modelling, based on new data streams.
1. Introduction
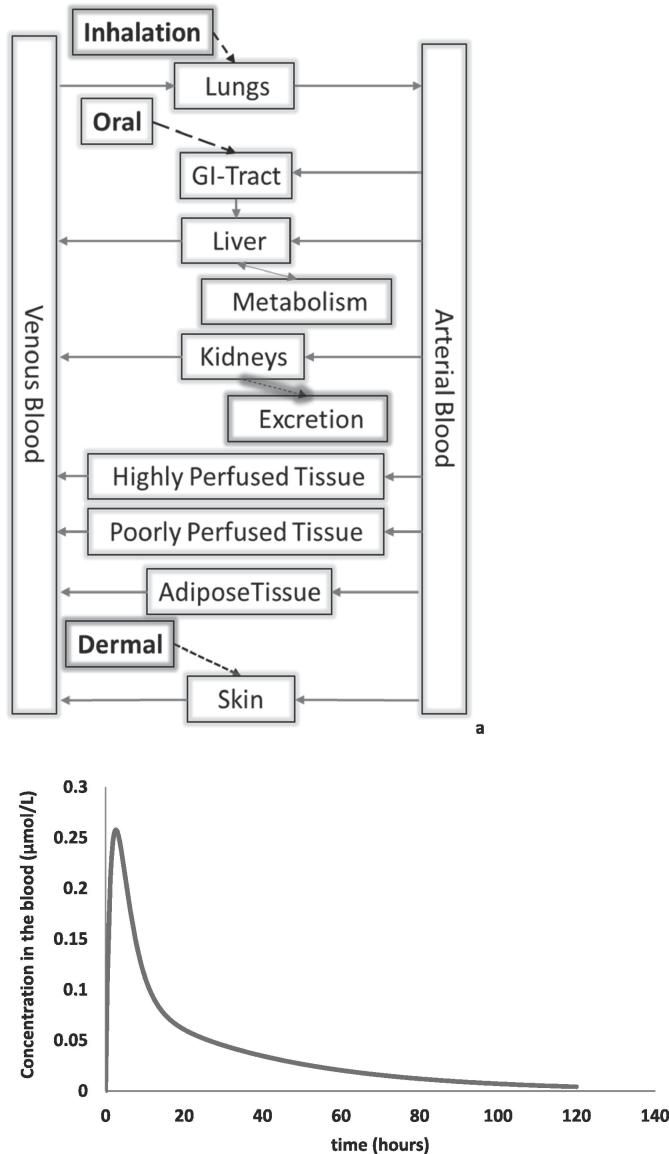
Modelling and simulation based approaches are gradually gaining interest as critical tools for safety and risk assessment of a variety of compounds including drugs, chemicals, consumer products, and food ingredients. These modelling approaches are recognised for the crucial role they play in, for example, predicting the biokinetics of drugs and chemicals in the organism without the need to conduct in vivo experiments. For more than 40 years, physiologically-based kinetic (PBK) models have been used to simulate biokinetics [2], [41], [28], [11]. In PBK models, the body is represented as a series of interconnected compartments linked via blood flow, as depicted in the schematic below (Fig. 1a), to simulate concentration-time curves in target organs or their surrogates, such as in blood (Fig. 1b). PBK models use differential equations to describe the absorption, distribution, metabolism, and elimination (ADME) processes that govern the fate and transport of the chemical among these interconnected compartments. Proper use of PBK models helps to reduce uncertainties and to identify data gaps inherent in hazard characterisation approaches that rely upon default extrapolation factors (e.g., a multiplication factor of 10 for inter-species extrapolation) to derive health-based guidance values from animal toxicity studies. PBK models provide a sound scientific basis to extrapolate across species, routes of exposure, and exposure scenarios, based on physiology and (physico-)chemical properties [32], [6]. As PBK models can be developed for specific individuals within the human population, they provide a means for quantifying inter-individual differences in kinetics, allowing for the determination of extrapolation factors across age groups or across populations of varying susceptibilities. With this information, safe chemical intake levels can be derived for individuals and populations. Most recently, PBK models have helped to facilitate quantitative in vitro to in vivo extrapolation (QIVIVE) approaches [70], [71], [72], [67], [35], enabling the use of in vitro toxicity data for the setting of safe intake levels. QIVIVE is an essential process in linking an in vitro measured biological (adverse) readout to a potential in vivo outcome [23]. QIVIVE provides a means of considering exposure and dosimetry, and enables the use of in vitro toxicity data for risk-based assessments beyond hazard-based assessments [5]. Once an in vitro concentration-response has been generated, the benchmark dose approach can be applied to the predicted dose-response data, to obtain an in vitro-based point of departure (PoD) or Reference Point (RfP) [34], [35].
Fig. 1.
(a) Schematic representation of a physiologically based kinetic (PBK) model, (b) with an example of a typical PBK model-output (time-dependent chemical concentration in blood).
1.1. Nomenclature
“Physiologically based pharmacokinetic” (PBPK) model is the most widely used term and was developed by the pharmaceutical field to simulate the kinetics of drugs. Despite the popular use of the term, “PBPK” is not entirely correct in the context of general chemical risk assessment. Another term preferred in the European Union (EU) and related to chemical risk assessment is “PBTK”, where TK is the abbreviation for “toxicokinetics”. However, this term is not entirely appropriate either [17]. Rather, a more general nomenclature, such as physiologically based biokinetic (PBBK) or the aforementioned PBK, might be seen as more appropriate. Regardless of the terminology used, PBK, PBPK, PBBK and PBTK can all be considered synonyms, and so throughout this document we will consistently use the more general terms of PBK model or PBK modelling. It is noted that the ever-increasing advancements in in vitro and in silico methodologies in the field of toxicology can be used in combination with PBK models to support regulatory decisions on the use of chemical substances. In the present manuscript the term next generation PBK (NG-PBK) model will be used to name these models. This term, NG-PBK, refers to PBK models that are developed without the provision of newly produced (i.e., without animal sacrifice) animal TK data for parametrisation and validation of those models, but rather through supporting in vitro, in silico, -omics, and micro-scale applications. NG-PBK models representing the human body should be parameterized and validated using in vitro, in silico, -omics data, micro-scale systems, and human in vivo data, when available. This stands also for PBK models built to represent animals (e.g. livestock, fish, bees), which should be parameterized and validated using in vitro, in silico, -omics, micro-scale systems and historical or (bio)-monitoring animal data of the species of interest, to avoid the need for animal sacrifice.
1.2. Milestones in the history of PBK modelling
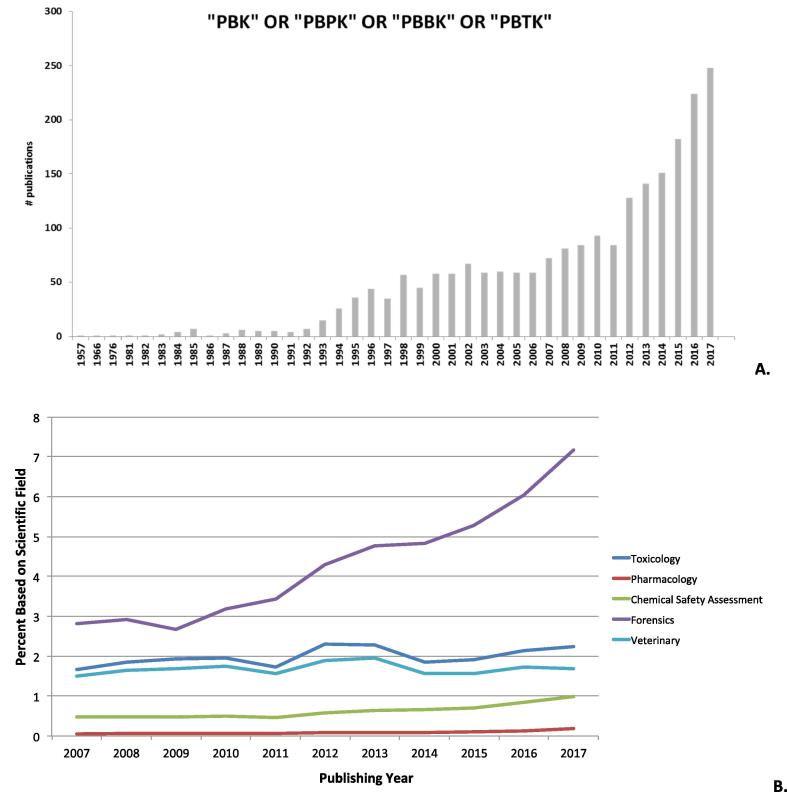
The principles behind PBK modelling were first reported in 1937 by Teorell, in a publication entitled “Kinetics of distribution of substances administered to the body” [58]. Although Teorell’s work was the first attempt to describe the body as a series of equations, the complexity of the mathematics, lack of data, and lack of computing power rendered his concepts incomplete until the 1960s. Between the 1960s and 1970s, several PBK models were developed for pharmaceutical drugs to target cancers [7], [8]. These publications paved the way for more than 2000 articles written on the topic of PB(P/T)K modelling within the last forty years (Fig. 2a). Over the past decade, there has been an increase in the development of PBK models for use in a variety of scientific fields, such as pharmacology, forensic sciences, and chemical risk assessment (Fig. 2b), although such an increase was not seen for toxicology and veterinary medicine. Many risk assessors remain reluctant to apply these models within their work [43], [49], [50], as PBK models are not often included in current hazard characterization and risk assessment protocols. In addition, some regulatory agencies may often have limited experience in using PBK models, and the complexity associated with the evaluation of model performance has also contributed to this reluctance.
Fig. 2.
(a) Number of papers published per year within the last 60 years. The search was conducted using the online repository PubMed on the 7th of March 2018, with key words string including “PBPK OR PBBK OR PBTK OR PBK”. (b) The number of papers (Fig. 2 A) published with key words string including “PBPK OR PBBK OR PBTK OR PBK” were normalized to the following terms: Toxicology; Pharmacology; Chemical Safety OR Risk assessment; Forensic Sciences and Veterinary.
Over the past 20 years, several workshops have been held to promote the applicability of PBK models in the academic, industrial, and regulatory sectors. For example, a 1995 European Centre for the Validation of Alternative Methods (ECVAM) workshop discussing the use of biokinetic and in vitro methods resulted in 15 recommendations that were submitted to support and guide future work in the PBK modelling field [9]. This workshop was followed by many others to better define the potential role of PBK modelling in science and risk assessment following a Three R (replacement, reduction and refinement) strategy [10]. In the same year, a workshop to address uncertainty and variability analysis in PBK modeling was held [4]. Loizou et al. [32] reported the need for clear descriptions of good modelling practices (GMP) for: 1) model development; 2) model characterisation; 3) model documentation; and 4) model evaluation. A subsequent thematic workshop aimed to critically appraise PBK modelling software platforms and to provide a more detailed state-of-the-art overview of non-animal based PBK parameterisation tools [6]. A CEN (European Committee for Standardization) workshop in 2014 strived for agreement upon the minimum requirements for the amount and type of information to be provided for exposure models, such as PBK models, along with documentation and guidelines for the structure and reporting of such information. The resulting CEN workshop agreement was expected to provide a more rigorous means of describing exposure models and to aid users in better understanding them [13], [1]. The following year, a workshop assessed the state of knowledge in the application of PBK models in regulatory decision-making, in addition to sharing and discussing best practices in the use of PBK modelling to inform dose selection in specific patient populations [61]. In 2017, a workshop organized by the National Centre for the Replacement, Refinement, and Reduction of Animals in Research (NC3Rs) encouraged experts in exposure science to consider the role of PBK models in the extrapolation of external exposure data to internal concentrations to promote the application of non-animal data in efficacy and safety testing (Burden et al., 2017; https://www.nc3rs.org.uk/applying-exposure-science-increase-utility-non-animal-data-efficacy-and-safety-testing). A Lorentz Center workshop entitled “Non-animal Methods for Toxicokinetics: Meeting New Paradigms in Toxicology” was held at the end of 2017 and emphasized the role of PBK models (https://www.lorentzcenter.nl/lc/web/2017/943/info.php3?wsid=943&venue=Oort; https://www.lorentzcenter.nl/lc/web/2017/943/report.pdf). The first European Partnership for Alternative Approaches to Animal Testing (EPAA) partners’ forum, held at the end of 2017, aimed to provide an overview on toxicokinetics and read-across with insight into the role of PBK models [30].
1.3. Framing the problem
The EURL ECVAM Strategy on Toxicokinetics1, as published in 2015, outlines opportunities for generating and making better use of TK data. The central feature of the strategy focuses on the use of PBK modelling to integrate data from in vitro and in silico methods for prediction of human whole-body biokinetic behavior, and enables QIVIVE to obtain safety guidance values expressed as external doses [5]. In the past, in vivo tissue/blood concentration-time data were a prerequisite for calibrating and evaluating the predictive capability of a PBK model [6]. The common practice was to start with an animal PBK model, calibrating it with animal in vivo data, and then re-parameterizing it based on in vitro biotransformation measurements or allometric scaling to develop a human PBK model. As the field of risk assessment evolves towards the goal of reducing, and eventually replacing, the use of animals for predicting human toxicity, PBK model development has seen a shift towards increased use of non-animal data for parameterization, along with increased use of the models for IVIVE. Efforts in this area should be directed towards developing standards that will increase the acceptance of in vitro methods for characterizing human-relevant ADME properties. To enhance the acceptance of PBK models at an international level, good modelling practice is required to guide the use of the in vitro and in silico methodologies in developing PBK models. As the first step, to initiate a dialogue on such a topic, the European Commission’s Joint Research Centre (JRC), EURL ECVAM, hosted a workshop on “Physiologically-Based Kinetic modelling in risk assessment – reaching a whole new level in regulatory decision-making” (Ispra, Italy, November 16–17, 2016). The workshop participants discussed challenges in: 1) applying NG-PBK modelling to support regulatory decision making; 2) constructing PBK models for safety assessment without animal in vivo data, relying solely on in vitro or in silico methods; and 3 assessing the validity of PBK models that rely only upon non-animal data. A portion of this current article summarizes the outcome of the workshop; detailed information on the workshop outcomes can be found in the workshop report [42].
In addition to the EURL ECVAM workshop, an international survey was conducted in 2017 to understand the applications of PBK modelling in the broader scientific and regulatory communities. An aggregate summary, including analysis of the results, has been published [43], while results presented per individual country are available online at http://apps.klimeto.com/pbk/. The survey provides insight into the current state of knowledge throughout the PBK modelling and user community, as well as a cursory volunteer contact list of modellers available for peer reviewing models. The main findings of the survey showed that though continuous expansion of the modelling community has allowed PBK models to gain ground for use in various scientific and regulatory risk assessment applications, this remains a slow process, due to a lack of guidance, data, and expertise, which continue to limit widespread acceptance of those models in such applications [43]. Here, we also discuss recently reported activities in the field, (subsequent to the 2016 EURL ECVAM workshop) that demonstrate both ongoing developments and the continued hesitancy within public health agencies to apply PBK modelling in their decisions. In addition, we will introduce as a new challenge the integration of NG-PBK modelling with toxicodynamic endpoints, as this will be essential for implementation of NG-PBK models.
2. Salient Features: Applying NG-PBK modelling to support regulatory decision making
As concluded from the 2017 survey [43], training, guidance, and dialogue are three main factors that will facilitate the successful acceptance of NG-PBK modelling in regulatory decision-making.
2.1. Dialogue and communication
While training and guidance are both essential, their maximum benefits cannot be achieved without frequent dialogue between regulators, modellers, and model proponents (chemical registrants). Such frequent dialogue not only allows the proposers to better understand the needs of the regulators, but also allows the regulators to provide modellers with feedback throughout the development, evaluation, and application processes. For example, risk assessors present at the 2016 EURL ECVAM workshop indicated that they prefer to use the simplest model possible, as finding sufficient input data is rather challenging, but would be willing to use more complex models if necessity dictates and sufficient input data are available. Thus, dialogue can help regulators to convey their needs for specific training and for model features, and help proponents to understand the criteria necessary for regulatory acceptance. Conversely, the regulators can learn what is technically or scientifically feasible and what is not. As such dialogue may prove to be time-consuming, establishing a harmonized template for model construction and evaluation would facilitate the process. The template should be flexible enough for any regulatory agency or country to use, and would ideally incorporate an agreed-upon ontology. To efficiently develop a PBK model to support regulatory risk assessment, modellers and end users (proponents and regulators) need to clearly define their goals of model use and related model requirements at an early stage. For example, if a read-across approach is likely to be applied by the end users, biokinetic data for a pre-determined set of relevant chemicals (target and source chemicals) will constitute important supporting material and should be included in the submission package. In situations where safety assessment is conducted for a new chemical on the market, the following criteria may be used to facilitate regulatory acceptance of a PBK model for this substance: 1) the model should be transparent, with a usable code; 2) model uncertainty should consider biological plausibility, and be clearly described and quantified when possible; 3) uncertainty in exposure scenarios should be characterised, because this uncertainty will propagate to PBK model results; 4) user-friendly platforms should be used where possible; 5) the model should be fit-for-purpose with no unnecessary additional complexity, and with all required parameters measurable; and 6) the model should consider sufficient coverage of chemical space, to allow for read-across approaches if desired. In cases where the model performance needs to be evaluated using human in vivo data, regulators may consider using data that are generated from human trials, such as micro-dosing. It should be emphasized that clinical studies would only be conducted once the safety of the chemical has been established and the clinical investigation represents de minimis risk to the subjects.
2.2. Training
Within the current climate of desire to reduce, refine, and replace animal testing through ongoing scientific and technological advancements, it would be beneficial to risk assessors/managers and other workers in safety assessment to be kept abreast of the development of NG-PBK models. In order to achieve this goal, information on a number of novel emerging technologies, in addition to PBK modelling, should be made more accessible. These include -omics, organ- on-a -chip, high-throughput screening methods, read-across, Adverse Outcome Pathways (AOPs), and IVIVE. Additionally, it would be helpful if guidance were available indicating how these different approaches are integrated in support of chemical safety assessment. On the other hand, it is not necessary for risk assessors/managers, etc. to have detailed knowledge related to all the diverse aspects of PBK modelling; rather, it may be sufficient to provide tailored training that focuses only on the specific needs of each regulatory sector and, where applicable, cross-sector needs. For example, some risk assessors may need or wish to run a model, and so they would require knowledge of the relevant software and expertise to review and run model codes. Other risk assessors may rely on a model peer review system to check the implementation and reliability of new model codes, and in this case, may only require sufficient knowledge to allow for interpretation of the data and to enable modelling predictions to be put in context. One option is for risk assessors to assemble technical committees that consist of members possessing a range of expertise, to review the model code and interpret model results. The training content/format should also be tailored to achieve maximum effectiveness in understanding the application of models. In addition to the traditional classroom setting, training formats could include webinars, ad hoc short courses, and more refined or specialised graduate-level courses. Further, online training could potentially generate a larger audience that would also allow the modelling and user community to continue to expand. Finally, since risk assessors generally place higher confidence in in vivo data, there is a need to make courses on alternative in vitro and in silico methods more accessible, to provide a path forward to acceptance of these NG-PBK model applications in regulatory decision making.
2.3. Guidance
While training is essential, establishing guidance and GMP on PBK model applications intended for regulatory purposes is also critical [32]. The GMP should include clear documentation on how to report a model’s scope and purposes, details of model development and evaluation, interpretation of results, and applications of the model in risk assessment [32]. It is recommended that the individual(s) or community network(s) responsible for each specific step in the development, evaluation, and application process be clearly identified, to increase transparency and allow end users to identify where targeted training may be required, if necessary, for a specific topic. The context in which the model is to be used, and thus the scope of the model development or amendment(s), should be clearly documented. This is especially important to avoid misuse of a reliable model, such as when results of the simulations are applied for the wrong purpose or when the model is applied outside of its applicability domain.
The WHO-IPCS published, in 2010, a guidance document on the characterisation and application of PBK models in risk assessment [68]. Nevertheless, no comprehensive guidance documentation is currently available for reporting and evaluating NG-PBK models without use of animal in vivo TK data, or for interpreting and applying outputs from these models for human safety assessment. Recently, several efforts have been made to produce such documentation. For example, the Scientific Committee for Consumer Safety (SCCS) considers all available scientific data in their safety evaluation of cosmetic substances, including data generated from PBK modelling. In the most recent Notes of Guidance for the Testing of Cosmetic Ingredients and Their Safety Evaluation (SCCS/1564/15)2, the SCCS defines the conditions for the use of PBK models submitted for risk assessment purposes. PBK modelling has already been accepted as a tool for risk assessment or for use as supporting information in some of the chemical-specific dossiers evaluated by the SCCS, EFSA, and US-EPA. The SCCS document could act as a starting point or as a template for a new general guidance document. Additionally, the new reporting guidelines from the US Food and Drug Administration (FDA) and European Medicine Agency (EMA) [60], [20], on harmonization of reporting and on qualification of PBK modelling and simulation, can also apply to NG-PBK models. To extend this concept, a working group at the Organisation for Economic Co-operation and Development (OECD), comprised of more than 45 scientists from different areas of scientific expertise, are drafting a guidance document for characterizing, validating, and reporting uncertainties in NG-PBK model applications.
3. Salient Features: Constructing PBK models for safety assessment without animal in vivo data
PBK models are built using three sets of parameters: i) physiological and anatomical parameters, with representative reference parameters taken from the species under study (animal or human); ii) biokinetic/ADME properties, which can be gathered using in vitro methods or by fitting the model to an in vivo data set; and iii) physico-chemical parameters, which are experimentally derived or obtained using in silico approaches such as quantitative activity relationship (QSAR) models [51]. For GMP, the PBK model construction should consider the compound exposure situation/dosing strategy to be simulated (problem formulation). The exposure descriptions should include route of administration, timeframe of the simulation (i.e. exposure duration), and exposure frequency. In the cases of complex models that include inter-individual variability among some physiological values, the number of individuals that should be incorporated into the simulation for sufficient statistical power analysis should also be considered.
In the case of NG-PBK models, assuming there is no possibility of generating in vivo animal data for the model calibration, there are two key pre-requisites to build the model:
-
•
Availability of in vitro and in silico alternatives to generate ADME properties (including prediction of metabolism) of sufficient quality
-
•
Availability and accessibility of modelling platforms.
3.1. Availability of in vitro and in silico data for ADME properties
Without in vivo data, the values of parameters in a PBK model will need to be derived from the results of in silico or in vitro experiments. Clearly, the accuracy of PBK models will be heavily reliant upon the quality of the model parameters, which often are not only tissue dependent but also chemical dependent.
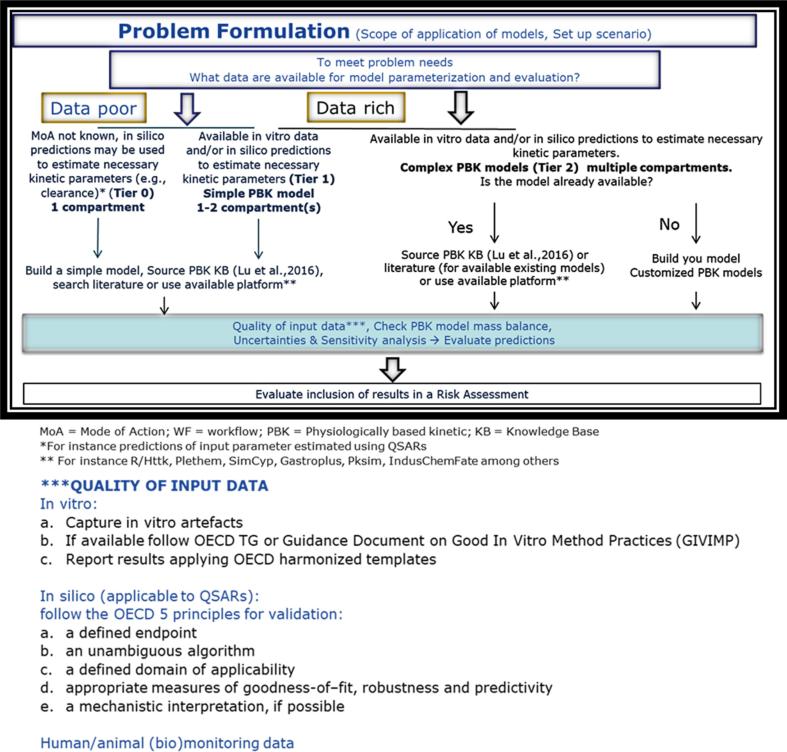
As it is useful to determine the minimum requirements for PBK models (with respect to data-poor and data-rich chemicals), a decision tree indicating requirements for different scenarios is presented here (Fig. 3). The most minimalistic model type, one-compartment models, parameterised with only protein binding and clearance data, have been developed and used to support chemical screening and prioritization [52], [64], [65], [66], [59], [71], [62]. Depending on the exposure route, a compartment representing the skin, intestine, or lung may need to be included in a model. If a compound is highly lipophilic, a fat compartment is required, and it may also be necessary for the model to describe uptake into the lymphatic system. Finally, depending on the hazard data available, additional compartments and biological processes may need to be added to the PBK model. Throughout development of the model, as more specific information is obtained on the chemical’s properties and mode of action (MoA), confidence is increased in the applicability of the models. A good strategy would be to begin with a generic model structure, then move to more specific models once knowledge is gained that indicates a unique biokinetic behavior of the compound in question. In using a simple model, it is possible that a key kinetic pathway specific to a given target chemical will not be taken into consideration. To address this issue, a database of all known ADME/TK processes, such as cell uptake (capturing the role of transporters), metabolism, and efflux, could be developed to help modellers identify which processes may need to be included for a specific chemical / purpose.
Fig. 3.
An example of a schematic decision tree to decide what tier of PBK model to apply when encountering data-poor or data-rich chemicals during model parameterization and based on problem formulation.
Membrane transporters influence the ADME processes of various endogenous and exogenous compounds [25], [54]. In recent decades, the pharmaceutical field has placed considerable effort into the study of transporters affecting drug disposition, therapeutic efficacy, and adverse outcomes, but little is known in regards to transporter effects on environmental chemicals [15]. Transporters can play a significant role in chemical distribution. As such, integration of membrane transporter-based experimental data during parameterization of several types of computational models (e.g., QSAR, pharmacophore, and PBK models), through use of platforms like SimCyp, PKSim, or GastroPlus, will enable better understanding of chemical/drug disposition [15].
Protein binding in plasma influences the partitioning of endogenous and exogenous compounds from the blood into the tissues. The plasma protein binding property is, among other things, related to lipophilicity, as binding becomes greater with more lipophilic chemicals, thus sequestering such chemicals in blood and limiting the systemic availability and distribution of unbound fraction of the chemical. A common and widely used method for estimating plasma protein binding in vitro is the rapid equilibrium method, which involves measurement of chemical transport across a dialysis membrane with a high surface area-to-volume ration within a Teflon-lined plate well [63].
Metabolism is an important feature to consider in a model, especially when a metabolite is assumed or known to be the toxic moiety. Both in vitro and in silico methods can be informative in providing predictions for metabolism and clearance. Kirchmair et al [24] reviewed software for predicting a range of features associated with metabolism (e.g. identification of labile moieties, enzyme interactions and metabolite prediction). The focus of these in silico tools is mainly the estimation of the qualitative nature of the metabolites (i.e., which metabolites are formed based on the parent compound’s molecular structure) and seldom allows for estimation of rate constants. A common criticism of software for predicting metabolites is the tendency for over-prediction: theoretically possible metabolites are not differentiated from those that occur experimentally. Some software platforms have attempted to address this issue through inclusion of filtering rules. For example, in order to reduce over-prediction within the Meteor Nexus software (Lhasa Ltd, Leeds), Marchant et al [37] describe a process whereby k-nearest neighbor analysis is combined with expert knowledge of biotransformation to reduce the over-prediction of metabolites. Such in silico models do not predict efflux of metabolites.
In vitro data for metabolism may be generated using tissue slices, organ (e.g., liver) homogenates, cell lines, spheroids, or (sub)cellular fractions (such as microsomes, baculosomes, S9, and cytosol), where metabolism is measured as loss of the parent compound or production of metabolite(s). It should be noted that if metabolism occurs very slowly, it may not be detected in a short-term in vitro assay. If a chemical is known to be predominantly excreted unchanged in urine, then metabolism is less relevant to the model. However, if metabolism of a parent compound is thought to be metabolized to undergo biliary excretion or to be excreted via the bile, then a model including such elimination pathways is necessary, first by determining which pathways of elimination are most relevant to the target chemical. In silico and in vitro models have also been developed for predicting different processes involved in elimination. These include in silico models for total clearance [77] and metabolism [47] and in vitro models for biliary excretion [22]. However, more work is required to develop models for elimination, and the applicability domain for existing models should be carefully considered before application to a wider range of chemicals. A current limitation is that there are no (OECD) guideline(s) addressing in vitro methods to determine kinetic parameters, except for the guideline on Skin Absorption (OECD TG 428). In the absence of standardised methods for generating in vitro parameters to calibrate PBK models, it is important that in vitro metabolism data or data regarding transporters are produced according to the new OECD good in vitro method practice (GIVIMP)3. The GIVIMP document is meant to serve as technical guidance on generating and applying quality data through good scientific and quality practices, to support the regulatory human safety assessment of chemicals using in vitro methods.
Bessems et al. [6] provides a general overview of the currently available in vitro and in silico methods for characterizing human ADME and the gaps and challenges faced. Mostrag-Szlichtyng et al [40] provide an extensive review specifically of in silico tools (i.e., QSAR models and software) for prediction of ADME properties that are relevant to PBK model building. More recently, Patel et al [44] have collated and assessed the quality of over 80 models for 31 absorption-, distribution-, and excretion-related endpoints [44].
Finally, toxicodynamic data derived from in vitro toxicity tests are typically based on nominal concentrations of the substances, which may contain significant errors due to the loss of biological, physical, and toxicological chemical processes in such tests. An in vitro biokinetic study plays a significant role in translating a nominal concentration used in in vitro systems to the actual level of cell exposure producing the effect. Several methodologies can be applied to address such a relationship, such as in vitro fate and transport mass balance models recently developed by several research teams [26], [27], [3], [21], [73].
3.2. Availability of modelling platforms
Currently, several open source modelling platforms, such as IndusChemFAte (Cefic LRI, http://cefic-lri.org/toolbox/induschemfate/), High-Throughput Toxicokinetics (httk)-r package ([74] https://cran.r-project.org/web/packages/httk/index.html), MEGEN-RVis ([33]; https://megen.useconnect.co.uk/), PLETHEM (http://www.scitovation.com/plethem.html), MERLIN-EXPO ([14], [55]; https://merlin-expo.eu/), and PK-Sim (www.systems-biology.com), and license-based platforms such as GastroPlus (www.simulations-plus.com) and SimCyp (https://www.certara.com), are available to individuals possessing varying degrees of expertise in PBK modeling. These platforms provide different computational tools that allow non-programmers to develop and run model simulations with varying options to gain a better understanding of model behavior, which is essential for interpretation of model output. The PBK models run from these platforms can be parameterised using in vitro or in silico data. However, programmers or users with modeling skills can also use R, MATLAB, and Berkeley Madonna software to develop customised PBK models, and to support the generation of innovative modeling components, which might otherwise not be generated through use of the more-structured commercial platforms.
A concern for the use of open source modelling platforms, as compared to use of their proprietary counterparts, is the lack of sustainable resources and funding that are needed for further development and maintenance of those platforms. While most of these platforms are initiated by a research grant, upon completion of the project, the developers are often unable to find other funding sources to maintain them. In order for a modelling platform to remain sustainable, it is essential to maintain access to the model’s equations, so that these can be easily coded later. Sustainability also depends on the ability of model updates to be communicated to end-users. Establishment of an open source library as a repository for all available model information, including a peer review process, is strongly recommended.
3.3. Integrating NG-PBK modelling with toxicodynamic endpoints
There is high value in the use of PBK models to predict internal target tissue doses for risk assessment applications, based on the assumption that a similar tissue response arises from an equivalent target tissue dose, rather than the external dose, across different exposure conditions. In addition, toxicodynamic processes that are interpreted in a high-throughput context from in vitro dose–response data can be integrated with PBK models, to link external exposure concentrations to target tissue doses to adverse endpoints. Such integration allows for support of several risk assessment extrapolations, such as QIVIVE and reverse dosimetry approaches. Examples of PBK/TD models are reported in table 3 of Punt et al. [75]. However, the application of PBK/TD models in risk assessment requires proper evaluation of model purpose, model assumptions and structure, mathematical representation, parameter estimation, computer implementation, and predictive capacity.
The topic of model evaluation will be captured in the next chapter.
4. Salient Features: Model evaluation- assessing the validity of PBK models that rely only upon non-animal data
A question that often arises is “How can we trust a PBK model prediction if there are no in vivo data to evaluate the simulation; how can the model gain credibility then?”
The following approaches could be applied and are described in further detail below: 1) read-across; 2) micro-scale systems; 3) pragmatic conservative scenario approach; 4) “credibility matrix”; 5) the reliability of dose metric predictions provided with uncertainty and sensitivity analyses (WHO 2010); and 6) population characteristics and virtual population libraries.
4.1. Read-across
For those cases in which in vivo data exist for one chemical, a read-across approach4 may be applied to parameterize models for other chemicals [76]. For example, if a valid PBK model exists for chemical A (source chemical), and chemical B (target chemical) lacks any in vivo data and has been shown to be similar in structure to chemical A, then the same parameterised PBK model structure/code and in vivo data for chemical A can be used for chemical B. This read-across approach has been demonstrated by case studies applying the PBK Knowledgebase developed by Lu et al. [36]. Alternatively, if parameterisation of the PBK model using available in vitro or in silico data for chemical B is possible, predictions can be compared to output from the model for chemical A based on in vivo data, in order to evaluate the PBK model for chemical B. When using such a model based on similarity between different chemicals, the influence of chemical-specific properties mediating ADME behaviour (e.g., log P, specific functional groups) should be carefully considered.
4.2. Micro-scale systems
Microscale systems, such as human-on-a-chip technology, could potentially be applied to measure and predict kinetics and whole body response to substances [56], thus aiding in evaluation and increased confidence in NG-PBK models. However, the limitations of these novel microscale systems should be carefully considered. For example, flow rates from model systems are often not scaled down in a similar manner as tissue volumes, thus rendering interpretation of the data difficult for PBK model applications.
4.3. Pragmatic conservative scenario approach
When in vivo data are lacking for model evaluation, a pragmatic conservative scenario could be followed in order to derive the most conservative estimate for risk assessment. For NG-PBK modeling, such an approach needs to be designed in such a way that the structure and input of the model is likely to lead to an overestimation of the internal concentration. This can be achieved by including uncertainty factors in the input parameters of the model. A worst-case estimate for absorption can for example be set to 100%. Other input parameters, such as metabolic clearance can be set to a value that is a certain extent lower than that measured for in vitro rates. To define the conservative boundaries around each input parameter, the uncertainties of each in vitro or in silico input method need to be identified.
4.4. Credibility matrix
There is a need to develop a framework for supporting the credibility of PBK models in support of risk assessment applications. As a first requirement for credibility, PBK models should be biologically plausible. Often, modellers or mathematicians exclude a number of biologically-relevant processes because these processes are considered to have no bearing on the model results and because models should be kept as simple as possible and created following the required purpose/problem formulation. However, such assumptions must always be discussed and agreed upon with biologists and toxicologists, to prevent the omission of critical biological and toxicological steps or key events. Good documentation of model assumptions is critical for modelers to demonstrate the validity of their models to reviewers and users, and visualization is a key feature when dealing with communication of these models. The recent EFSA uncertainty guidance document provides a reporting table for listing and evaluating model uncertainties [19].
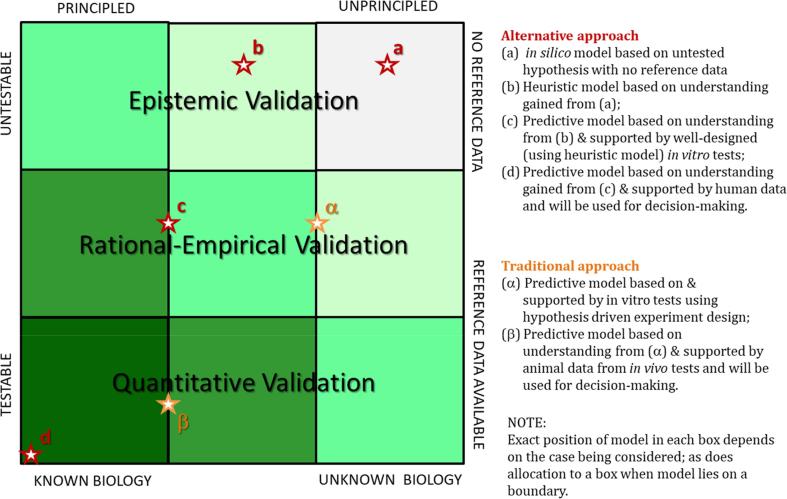
From the 2016 EURL ECVAM workshop, the following graphical representation and application of the “credibility matrix by Patterson & Whelan” has been proposed. The matrix (Fig. 4) allows for locating a specific model type based on the information available, i.e whether a model is principled and testable, as well as knowledge of the biology and the availability of data, which should aid in systematically establishing model credibility via a process of social epistemology [46]. If a model falls in the bottom left region (testable and with full knowledge), confidence in the model is likely high. However, if a model falls in the top right region of the matrix (not testable and without any knowledge of the system biology), confidence in the model is likely low due to the uncertainties associated with it. In other words, regulators are unlikely to trust model types found in the top right region of the matrix when making decisions. The question is, to what degree a PBK model would need to be placed towards the bottom left corner to attain sufficient credibility for regulators. In some sense, testable models do not really predict, but provide an estimate to compare against available data in a retrospective fashion.
Fig. 4.
Credibility matrix showing comparative loci for a model based on traditional in vivo data-based approaches and for a model based on an alternative approach (i.e., in vitro, in silico methods, and/or micro-scale systems). The rationale for the locations of the model types, indicated by stars and letters, are given in the side-bar legend. For example, in silico models placed at the top right, might consist of a simple model ‘a’ based on a limited set of data, for instance in a QSAR. This leads to a more sophisticated, but still heuristic, model ‘b’ based on the understanding gained from model ‘a’. The predictions from models ‘a’ and ‘b’ are used to design in vitro tests that enable the development of model ‘c’, which can be validated using the rational-empirical approach, thus enhancing its credibility. Finally, this leads to the development of clinical studies and model ‘d’, supported by its predecessors and quantitatively validated or confirmed using clinical data. This places model ‘d’ in the bottom left corner, as a model whose predictions stakeholders, including regulators, practitioners, and patients, will likely use to make decisions (adapted from [42], proposed by [46]).
The proposed framework should lay out the requirements for validating models with different degrees of knowledge and testability (e.g., quantitative validation), which could aid in quantifying the uncertainty currently existing with animal models, and which can help regulators assess whether models developed through in vitro and in silico methodologies can be equally reliable, or even more so, compared to current risk assessment approaches. Biological systems, by nature, are complex networks operating under simple rules that can be described by non-linear dynamic processes, and which exhibit non-trivial emergent and self-organizing behavior. As a result, a measured value might represent a particular, and perhaps unknown, state of a system, which makes its use, as a comparator for a predicted value, challenging. To handle such issues, approaches that operate on experience-based validation are required. Ideally, these, approaches would capture the diversity of experiences to establish generic digital twins, which are couplings of validated models with their real-world datasets (see [45]).
There is disagreement amongst modellers as to the meaning of the terms model evaluation, verification, and validation; for instance, EMA has shifted to use of the word “qualification”. Regardless of which term is more appropriate, the analytical purpose is to ensure that the model is appropriate for the task at hand, and that its predictions are a reasonable representation of reality. Once confirming that the model is a reasonable representation of reality for the intended purpose, several analyses may be used to “validate” a model, including sensitivity analysis, robustness analysis5, assumption justification, model argumentation, structured calibration, predictive performance, proper scoring rules, and relation to reality. To “verify” a model, the model scope should be revisited and the model equations and code reviewed. The following key elements were suggested by the 2016 EURL ECVAM workshop participants to achieve model credibility [42]:
-
•
Understand the model;
-
•
Understand the data underpinning the model;
-
•
State clearly the assumptions and hypothesis encoded;
-
•
Consider the gap between the model and reality, based on available observations.
This last item can be a description of what is lacking in the model. The outcomes of sensitivity analyses can be used to explain some model deficits. One possible approach, as opposed to the statement in the introduction regarding developing the simplest model, would be to start with a more complex model and then remove parameters to which the predictions are not sensitive. The potential problem with this approach is that when there are many parameters with large uncertainties, they may introduce a great deal of variation into the uncertainty analysis.
4.5. Reliability of dose metric predictions (model testing, uncertainty, and sensitivity)
In 2010, the World Health Organization (WHO) reported the level of confidence needed to gain credibility in a PBK model intended for risk assessment (WHO, 2010). The degree of confidence in a PBK model’s predictions depends upon how well the model has been tested against real data and whether adequate sensitivity and uncertainty analyses have been conducted, in order to support the reliability of predictions (WHO, 2010). In the case of NG-PBK models, the lack of “real data” (e.g. in vivo human data) that are required to evaluate model predictions for the purpose of validation render such validation nearly impossible. However, reporting of adequate sensitivity and uncertainty is certainly relevant and encouraged. Tables providing guidance in reporting results of uncertainty and sensitivity analyses have been provided in the WHO 2010 article, as a tool to better document the evaluation of model predictions (from WHO 2010; [39]). There are several areas that are considered to present current challenges in accepting model-informed drug development, which can also provide insight into necessary acceptance criteria for PBK model-based drug development. Among those criteria, most noteworthy is that the adequacy of submitted PBK models is to be based on their intended purposes at different stages of drug development [42]. That is, determination of whether a model is fit-for-purpose and the need to identify and transparently communicate the knowledge gaps. EMA and US FDA published a draft document in 2016 as guidance on the qualification and reporting of physiologically based pharmacokinetic (PBPK) modelling and simulations [20], [60]. The aim of this guideline is to describe the expected content that should be included in PBK modelling and simulation reports during regulatory submission, including applications for authorization of medicinal products, pediatric investigation plans, and clinical trial applications. This also includes the documentation needed to support the qualification of a PBK platform for an intended use, such as results of sensitivity and uncertainty analyses.
4.6. Population characteristics and virtual population libraries
This chapter reports information on population characteristics as virtual population libraries for the in silico medicine field. However we believe that this information could be also relevant for chemical risk assessment. Efforts undertaken to better capture the heterogeneity in the human species can certainly be applied to environmental chemical risks, as different population cohorts may be more at risk to specific chemical exposures than are other cohorts. Important aspects of human heterogeneity include inter-individual variations in lifestyle, health status (immunosuppressed, disease patient) genetic polymorphism (gene expression), physiology (uptake rate), biochemistry and molecular biology [38], all with respect to age. These factors will interact and influence the chemical ADME and biokinetic behaviors and toxicodynamics within the body. Parameters in a PBK model have a direct biological correspondence, providing a useful framework for determining the impact of observed variations in physiological and biochemical factors on the population variability in the achieved target of a particular chemical [16], [48], [38]. In addition, integration of genetic information from –omics studies will enhance predictions for precise and personalized medicine. Applications for predicting the kinetics of substances within specific populations, such as in the field of pediatrics, have been increasing in their development and use [31]. In the pharmaceutical field, population-specific PBK models can simulate untestable clinical outcomes, allowing for evaluating the effects of intrinsic (e.g., organ dysfunction, age, genetics, etc.) and extrinsic (e.g., drug-drug interactions) factors, alone or in combination, on drug target concentrations.
5. Next steps and future perspectives
With an increasing demand for application of alternative methods within the risk assessment framework, the need for the development of higher throughput NG-PBK models has also increased. A guidance document for GMP for PBK modelling could also be extended to other types of in silico biokinetic models, such as in vitro mass balance models [3], [73]. Existing guidance documents [68], [57], and those documents of EFSA [18], and CEN [12], that are less PBK-specific, require updating with respect to the current trends, due to the continuous evolution in science and risk assessment. The recent FDA [60] and EMA [20] guidelines are the first that open up the possibility to submit non-animal PBK model results for drug dossier submission and provide excellent examples that other agencies could follow. At the same time, the OECD is working on a guidance document for the characterization, validation, and reporting of physiologically based models for regulatory applications that should be ready in 2019, and which attempts to set principles for NG-PBK model validation.
However, the challenge remains in making appropriate use of in vitro data and/or in silico predictions when 1) building these models; 2) interpreting model outputs and integrating the outputs with other sources of information for risk assessment purposes; and 3) attempting to gain model credibility by underlining all uncertainties and assumptions when in vivo human data are unavailable for proper model evaluation. The uncertainty and variability associated with PBK models, and the proposed GMP [32], should be further developed and should include guidance for PBK models built using in vitro and in silico methodologies to estimate ADME properties. The use of a matrix in the new risk assessment paradigm, to underline and quantify the uncertainty associated with NG-PBK models, compared to models based on in vivo animal data, would be desirable.
Several standardised decision trees could be developed to guide modellers in their construction of a PBK model in the absence of in vivo data for calibration, and to guide risk assessors in application and interpretation of PBK models. For instance, PBK-predicted internal dose metrics vs. in vitro PoD from toxicity testing could be taken into account, along with in vitro results linking to in vivo adverse outcomes for a tiered assessment, perhaps through application of the traditional and internal threshold of toxicological concern approach [29], [69]. With the need for several international working groups to further develop such documentation, communication is required among these groups to ensure compatibility of in vitro kinetic and dynamic methods with PBK models, in addition to communication with regulators to fit the total risk-assessment framework. It should be noted that for such communication to be achieved, funding would be necessary.
There remains a need to create a community to address issues with human ADME/TK and NG-PBK models, such as the development of criteria for model construction and model evaluation. A group of scientists across the academic, industrial, and governmental landscapes should be available and willing to establish a peer review system for PBK models. Criteria should exist to select those individuals that will review the models, and templates and check lists should be provided to assist in the review process. A public repository is needed for PBK models that have been built and/or peer reviewed, and once this repository is developed, relevant documentation can be introduced from an independent peer review to support model credibility. Such a repository is in line with the work reported in Lu et al., [36] and will allow for the curation of more case studies and the creation of libraries of ad hoc PBK models that could be used for training purposes. Additionally, this repository will facilitate risk assessment approaches applying PBK models and IVIVE, and communicate to decision makers more efficiently the current state of science regarding the use of animal-free models in regulatory applications. Perspectives from the various industrial stakeholders (e.g. pharmaceutical, food safety, agricultural, and personal care product industries) also need to be communicated, to provide greater insight of current practice and understanding of future needs of these sectors, to enable promotion of best practices.
Application of NG-PBK models, in the context of exposure in specific population of patients, would be extremely valuable in the generation of virtual population/patient libraries. These libraries would enable clinical trials to entail populations with a greater number of “virtual” individuals, which might not otherwise be possible to conduct with a limited number of real persons/patients. Additionally, these libraries would introduce populations more rarely encountered, such as those possessing enzyme polymorphisms that exert a greater influence on drug-drug interactions or those with rare genetic diseases or health abnormalities. Such libraries would also prove useful in chemical risk assessment when evaluating interindividual variability in relation to chemical exposures and toxicological outcomes.
Finally, it is recommended that a means for training new modellers and risk assessors be established. Such training, which can be provided with specific courses or as a continuing education course at scientific conferences, will focus on PBK model development, evaluation, and application. Though several challenges still remain, the suggestions and steps presented in this work provide a path towards gaining acceptance of NG-PBK models in regulatory practices.
In summary, to facilitate the development and use of NG-PBK models, which do not rely on animal in vivo data, and their acceptance in the regulatory domain, the following are recommended:
-
i)
development of more transparent, accessible, and user-friendly software platforms that facilitate development and application of PBK models by a community of users, and which allow specific populations to be modelled or population variability to be evaluated;
-
ii)
development of resources to inform new developments in in silico and in vitro approaches that may be used to provide data for model development;
-
iii)
development and refinement of existing web applications and PBK model platforms that have the ability to conduct QIVIVE and reverse dosimetry in an automated manner;
-
iv)
knowledge sharing initiatives that allow members of the regulatory community, such as risk assessors and risk managers, to become familiar with relevant PBK model information, while model developers gain a better understanding of regulatory needs;
-
v)
GMPs and harmonised guidelines for reporting the steps taken during model development, evaluation, and application, with respect to NG- PBK models. This would include the use of a clear and common terminologies.
Acknowledgments
Acknowledgements
The authors would like to provide a special thanks to E. Ahs and G. Tosiou for logistical and practical support during the workshop. The authors thank S. Belz, R. Corvi, P. Prieto-Peraita, A. Richarz, and M. Whelan of the JRC for contributing to discussions during the workshop.
Funding information
This work was supported by the European Union Reference Laboratory for Alternatives to Animal Testing (EURL ECVAM) of the European Commission’s Joint Research Centre (JRC), Ispra, Italy. Funding for Dr. Leonard was provided by the Oak Ridge Institute for Science and Education Research Participation Program at the US EPA.
Disclaimer
The views expressed in this paper are those of the authors and do not necessarily reflect the views of their institutions. Authors declare no conflicts of interest.
Footnotes
https://ec.europa.eu/jrc/en/publication/eur-scientific-and-technical-research-reports/eurl-ecvam-strategy achieving-3rs-impact-assessment-toxicokinetics-and-systemic-toxicity?search
Quotation: “The underlining philosophy of read-across is that substances which are similar in chemical structure will have similar properties and thereby, have similar toxicokinetics and toxicodynamics. Experimental derived toxicological proprieties from one substance, often referred to as source chemical, can be read across to fill the data gap for a second substance, the target chemical, which has a similar molecular structure but is lacking data” [76].
Quotation from Saltelli et al 2000 Sensitivity Analysis – What is Sensitivity Analysis? “For a software engineer, SA could be related to the robustness and reliability of the software with respect to different assumptions “ … “For a statistician, involved in statistical modelling, SA is mostly known and practice under the heading of “robustness analysis” [53].
Supplementary data to this article can be found online at https://doi.org/10.1016/j.comtox.2018.11.002.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Altenpohl A., Ciffroy P., Paini A., Radovnkovic A., Suciu N.A., Tanaka T., Tediosi A., Verdonck F. Standard documentation of exposure models: Merlin-Expo case study. Handbook Environ. Chem. 2018;57:59–76. [Google Scholar]
- 2.Andersen, Krishnan, Quantitative Modeling in Toxicology: An Introduction Book Editor(s): Dr. Kannan Krishnan Dr Melvin E. Andersen. Wiley Online Library. First published: 30 March 2010, 2010. https://doi.org/10.1002/9780470686263.ch1.
- 3.Armitage J.M., Wania F., Arnot J.A. Application of mass balance models and the chemical activity concept to facilitate the use of in vitro toxicity data for risk assessment. Environ. Sci. Technol. 2014;48(16):9770–9779. doi: 10.1021/es501955g. [DOI] [PubMed] [Google Scholar]
- 4.Barton H.A., Chiu W.A., Setzer R.W., Andersen M.E., Bailer A.J., Bois F.Y. Characterizing uncertainty and variability in physiologically-based pharmacokinetic (PBPK) models: state of the science and needs for research and implementation. Toxicol. Sci. 2007;99(2):395–402. doi: 10.1093/toxsci/kfm100. [DOI] [PubMed] [Google Scholar]
- 5.Bell S.M., Chang X., Wambaugh J.F., Allen D.G., Bartels M., Brouwer K.L.R., Casey W.M., Choksi N., Ferguson S.S., Fraczkiewicz G., Jarabek A.M., Ke A., Lumen A., Lynn S.G., Paini A., Price P.S., Ring C., Simon T.W., Sipes N.S., Sprankle C.S., Strickland J., Troutman J., Wetmore B.A., Kleinstreuer N.C. In vitro to in vivo extrapolation for high throughput prioritization and decision making. Toxicol. In Vitro. 2018 Mar;47:213–227. doi: 10.1016/j.tiv.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bessems J.G., Loizou G., Krishnan K., Clewell H.J., Bernasconi C., Bois F.Y., Coecke S., Collnot E.M., Diembeck W., Farcal L.R. PBTK modelling platforms and parameter estimation tools to enable animal-free risk assessment: recommendations from a joint EPAA–EURL ECVAM ADME workshop. Regul. Toxicol. Pharmacol. 2014;68(1):119–139. doi: 10.1016/j.yrtph.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 7.Bischoff K.B., Dedrick R.L. Thiopental pharmacokinetics. J. Pharm. Sci. 1968;57(8):1346–1351. doi: 10.1002/jps.2600570814. [DOI] [PubMed] [Google Scholar]
- 8.Bischoff K.B., Dedrick R.L., Zaharko D.S., Longstreth J.A. Methotrexate pharmacokinetics. J. Pharm. Sci. 1971;60(8):1128–1133. doi: 10.1002/jps.2600600803. [DOI] [PubMed] [Google Scholar]
- 9.Blaauboer B., Bayliss M.K., Castell J., Evelo C.T.A., Frazier J.M., Groen K., Gulden M., Guillouzo A., Hissink A.M., Houston B., Johanson G., de Jongh J., Kedderis G.L., Reinhardt C.A., van de Sandt J.J.M., Semino G. The use of biokinetics and in vitro methods in toxicological risk evaluation. The report and recommendations of ECVAM Workshop 15. ATLA. 1996;24:473–497. [Google Scholar]
- 10.Bouvier d'Yvoire M., Prieto P., Blaauboer B.J., Bois F.Y., Boobis A., Brochot C., Coecke S., Freidig A., Gundert-Remy U., Hartung T. Physiologically-based Kinetic Modelling (PBK Modelling): meeting the 3Rs agenda. The report and recommendations of ECVAM Workshop 63. ATLA. 2007;35(6):661–671. doi: 10.1177/026119290703500606. [DOI] [PubMed] [Google Scholar]
- 11.Bois F.Y., Ochoa J.G.D., Gajewska M., Kovarich S., Mauch K., Paini A., Péry A., Benito J.V.S., Teng S., Worth A. Multiscale modelling approaches for assessing cosmetic ingredients safety. Toxicology. 2017;392:130–139. doi: 10.1016/j.tox.2016.05.026. [DOI] [PubMed] [Google Scholar]
- 12.CEN, European committee for standardization, CEN Workshop on Standard documentation of large chemical exposure models (WS MERLIN-EXPO); CWA 16938 Brussels, 2015. https://www.cen.eu/work/areas/chemical/Pages/WS-MerlinExpo.aspx.
- 13.Ciffroy P., Altenpohl A., Fait G., Fransman W., Paini A., Radovnikovic A., Simon-Cornu M., Suciu N., Verdonck F. Development of a standard documentation protocol for communicating exposure models. Sci. Total Environ. 2016;568:557–565. doi: 10.1016/j.scitotenv.2016.01.134. [DOI] [PubMed] [Google Scholar]
- 14.Ciffroy P., Alfonso B., Altenpohl A., Banjac Z., Bierkens J., Brochot C., Critto A., De Wilde T., Fait G., Tediosi A., Fierens T., Garratt J., Giubilato E., Grange E., Johansson E., Radomyski A., Reschwann K., Suciu N., Van Holderbeke M., Verdonck F., Vlajic A. Modelling the exposure to chemicals for risk assessment: a comprehensive library of multimedia and PBPK models for integration, prediction, uncertainty and sensitivity analysis — the MERLIN-expo tool. Sci. Total Environ. 2016;568:770–784. doi: 10.1016/j.scitotenv.2016.03.191. [DOI] [PubMed] [Google Scholar]
- 15.Clerbaux L.A., Coecke S., Lumen A., Kliment T., Worth A.P., Paini A. Capturing the applicability of in vitro-in silico membrane transporter data in chemical risk assessment and biomedical research. Sci Total Environ. 2018;15(645):97–108. doi: 10.1016/j.scitotenv.2018.07.122. Epub 2018 Jul 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clewell H.J., III, Andersen M.E. Use of physiologically based pharmacokinetic modeling to investigate individual versus population risk. Toxicology. 1996;111:315–329. doi: 10.1016/0300-483x(96)03385-9. [DOI] [PubMed] [Google Scholar]
- 17.Clewell H.J., III, Andersen M.E., Blaauboer B.J. On the incorporation of chemical-specific information in risk assessment. Toxicol. Lett. 2008;180(2008):100–109. doi: 10.1016/j.toxlet.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 18.EFSA Scientific opinion on good modelling practice in the context of mechanistic effect models for risk assessment of plant protection products. EFSA J. 2014;12(3):3589. [Google Scholar]
- 19.EFSA, Guidance on Uncertainty in EFSA Scientific Assessment, 2018, http://www.efsa.europa.eu/en/efsajournal/pub/5123. [DOI] [PMC free article] [PubMed]
- 20.EMA European Medicine Agency, Draft “Guideline on the qualification and reporting of physiologically based pharmacokinetic (PBPK) modelling and simulation.” 2016. http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2016/07/WC500211315.pdf.
- 21.Fischer F.C., Henneberger L., König M., Bittermann K., Linden L., Goss K.U., Escher B.I. Modeling Exposure in the Tox21 in vitro bioassays Chem. Res. Toxicol. 2017;5:1197–1208. doi: 10.1021/acs.chemrestox.7b00023. [DOI] [PubMed] [Google Scholar]
- 22.Ghibellini G., Leslie E.M., Brouwer K.L.R. Methods to Evaluate Biliary Excretion of Drugs in Humans: an Updated Review. Mol. Pharm. 2006;3(3):198–211. doi: 10.1021/mp060011k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groothuis F.A., Heringa M.B., Nicol B., Hermens J.L., Blaauboer B.J., Kramer N.I. Dose metric considerations in in vitro assays to improve quantitative in vitro-in vivo dose extrapolations. Toxicology. 2015;332:30–40. doi: 10.1016/j.tox.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 24.Kirchmair J., Göller A.H., Lang D., Kunze J., Testa B., Wilson I.D. Predicting drug metabolism: experiment and/or computation? (2015) Nat. Rev. Drug Discovery. 2015;14(6):387–404. doi: 10.1038/nrd4581. [DOI] [PubMed] [Google Scholar]
- 25.Klaassen C.D., Aleksunes L.M. Xenobiotic, bile acid, and cholesterol transporters. Pharmacol. Rev. 2010;62(1):1–96. doi: 10.1124/pr.109.002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kramer N.I., Busser F.J., Oosterwijk M.T., Schirmer K., Escher B.I., Hermens J.L. Development of a partition-controlled dosing system for cell assays. Chem. Res. Toxicol. 2010;23(11):1806–1814. doi: 10.1021/tx1002595. [DOI] [PubMed] [Google Scholar]
- 27.Kramer, N.I., Measuring, modeling, and increasing the free concentration of test chemicals in cell assays. Utrecht University, 2010b.
- 28.Krishnan, Peyret, Physiologically Based Toxicokinetic (PBTK) Modeling in Ecotoxicology, in: J. Devillers (ed.), Ecotoxicology Modeling, Emerging Topics in Ecotoxicology: Principles, Approaches and Perspectives 2, DOI 10.1007/978-1-4419-0197-2 6, Springer Science+Business Media, LLC, 2009.
- 29.Kroes R., Renwick A.G., Feron V., Galli C.L., Gibney M., Greim H., Guy R.H., Lhuguenot J.C., van de Sandt J.J.M. Application of the threshold of toxicological concern (TTC) to the safety evaluation of cosmetic ingredients. Food Chem. Toxicol. 2007;45:2533–2562. doi: 10.1016/j.fct.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 30.C. Laroche, M. Aggarwal, H. Bender, P. Benndorf, B. Birk, J. Crozier, G. Dal Negro, F. De Gaetano, C. Desaintes, I. Gardner, B. Hubesch, A. Irizar, D. John, V. Kumar, A. Lostia, I. Manou, M. Monshouwer, B.P. Müller, A. Paini, K. Reid, T. Rowan, M. Sachana, K. Schutte, C. Stirling, R. Taalman, L. van Aerts, Weissenhorn, R8, Sauer UG25, Finding Synergies for 3Rs – Toxicokinetics and Read-Across: Report from an EPAA Partners’ Forum Regul Toxicol Pharmacol. 2018 Aug 23;99:5–21. [DOI] [PubMed]
- 31.Leong R., Vieira M.L.T., Zhao P., Mulugeta Y., Lee C.S., Huang S.-M., Burckart G.J. Regulatory Experience With Physiologically Based Pharmacokinetic Modeling for Pediatric Drug Trials. Clin. Pharmacol. Ther. 2012;91(5):926–931. doi: 10.1038/clpt.2012.19. [DOI] [PubMed] [Google Scholar]
- 32.Loizou G.D., Spendiff M., Barton H.A., Bessems J., Bois F.Y., Bouvier M. Development of Good Modelling Practice for Physiologically Based Pharmacokinetic Models for Use in Risk Assessment: The First Steps. Reg. Toxicol. Pharmacol. 2008;50(3):400–411. doi: 10.1016/j.yrtph.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 33.Loizou G.D., Hogg A. MEGen: A Physiologically Based Pharmacokinetic Model Generator. Front. Pharmacol. 2011;2(56):1–14. doi: 10.3389/fphar.2011.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Louisse J., Bosgra S., Blaauboer B.J., Rietjens I.M., Verwei M. Prediction of in vivo developmental toxicity of all-trans-retinoic acid based on in vitro toxicity data and in silico physiologically based kinetic modeling. Arch. Toxicol. 2015;89(7):1135–1148. doi: 10.1007/s00204-014-1289-4. [DOI] [PubMed] [Google Scholar]
- 35.Louisse J., Beekmann K., Rietjens I.M. Use of Physiologically Based Kinetic Modeling-Based Reverse Dosimetry to Predict in Vivo Toxicity from in Vitro Data. Chem. Res. Toxicol. 2017;30(1):114–125. doi: 10.1021/acs.chemrestox.6b00302. [DOI] [PubMed] [Google Scholar]
- 36.Lu J., Goldsmith M.R., Grulke C.M., Chang D.T., Brooks R.D., Leonard J.A., Johnson J. Developing a physiologically-based pharmacokinetic model knowledgebase in support of provisional model construction. PLoS Comput. Biol. 2016;12:(2). doi: 10.1371/journal.pcbi.1004495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marchant C.A., Rosser E.M., Vessey J.D. A k-Nearest Neighbours Approach Using Metabolism-related Fingerprints to Improve In Silico Metabolite Ranking. Mol. Inf. 2017;36(3) doi: 10.1002/minf.201600105. [DOI] [PubMed] [Google Scholar]
- 38.McLanahan Physiologically based pharmacokinetic model use in risk assessment–Why being published is not enough. Tox. Sci. 2012;126:5–15. doi: 10.1093/toxsci/kfr295. [DOI] [PubMed] [Google Scholar]
- 39.Meek M.E., Barton H.A., Bessems J.G., Lipscomb J.C., Krishnan K. Case study illustrating the WHO IPCS guidance on characterization and application of physiologically based pharmacokinetic models in risk assessment. Regul. Toxicol. Pharmacol. 2013;66(1):116–129. doi: 10.1016/j.yrtph.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 40.A. Mostrag-Szlichtyng, A. Worth, In silico modelling of microbial and human metabolism: a case study with the fungicide carbendazim. JRC Technical Report EUR 24377 EN.
- 41.M. Mumtaz, J. Fisher, B. Blount, P. Ruiz, Application of Physiologically Based Pharmacokinetic Models in Chemical Risk Assessment, J. Toxicol. 2012, Article ID 904603, 11. [DOI] [PMC free article] [PubMed]
- 42.Paini A., Joossens E., Bessems J., Desalegn A., Dorne J.L., Gosling J.P., Heringa M., Klaric M., Kramer N., Loizou G., Louisse J., Lumen A., Madden J., Patterson E., Duarte Proenca S., Punt A., Setzer W.S., Suciu N., Troutman J., Tan Y.M. EURL ECVAM Workshop On New Generation of Physiologically-Based Kinetic Models In Risk Assessment. 2017 doi: 10.1016/j.comtox.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paini A., Leonard J.A., Kliment T., Tan Y.M., Worth A. Investigating the state of physiologically based kinetic modelling practices and challenges associated with gaining regulatory acceptance of model applications. Regul. Toxicol. Pharmacol. 2017;90:104–115. doi: 10.1016/j.yrtph.2017.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patel M., Chilton M.L., Sartini A., Gibson L., Barber C., Covy-Crump L., Przybylak K.R., Cronin M.T.D., Madden J.C. Assessment and reproducibility of quantitative structure-activity relationship models by the non-expert. J. Chem. Inf. Modell. 2018;58(3):673–682. doi: 10.1021/acs.jcim.7b00523. [DOI] [PubMed] [Google Scholar]
- 45.Patterson E.A., Taylor R.J., Bankhead M. A framework for an integrated nuclear digital environment. Prog. Nucl. Energy. 2016;87:97–103. [Google Scholar]
- 46.Patterson E.A., Whelan M.P. A framework to establish credibility of computational models in biology. Prog. Biophys. Mol. Biol. 2017;129:13–19. doi: 10.1016/j.pbiomolbio.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 47.Pirovano A., Brandmaier S., Huijbregts M.A.J., Ragas A.M.J., Veltman K., Hendriks A.J. The utilisation of structural descriptors to predict metabolic constants of xenobiotics in mammals. Environ. Toxicol. Pharmacol. 2015;39(1):247–258. doi: 10.1016/j.etap.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 48.Price P.S., Conolly R.B., Chaisson C.F., Gross E.A., Young J.S., Mathis E.T., Tedder D.R. Modeling interindividual variation in physiological factors used in PBPK models of humans. Crit. Rev. Toxicol. 2003;33:469–503. [PubMed] [Google Scholar]
- 49.Punt A., Bouwmeester H., Peijnenburg A.A.C.M. Non-animal approaches for kinetics in risk evaluations of food chemicals. ALTEX. 2017;34:501–514. doi: 10.14573/altex.1702211. [DOI] [PubMed] [Google Scholar]
- 50.Punt A., Bouwmeester H., Schiffelers M.J.W.A., Peijnenburg A.A.C.M. Expert opinions on the acceptance of alternative methods in food safety evaluations: Formulating recommendations to increase acceptance of non-animal methods for kinetics. Regul. Toxicol. Pharmacol. 2018;92:145–151. doi: 10.1016/j.yrtph.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 51.Rietjens I.M.C.M., Louisse J., Punt A. Tutorial on physiologically based kinetic modeling in molecular nutrition and food research. Mol Nutr Food Res. 2011;55:941–956. doi: 10.1002/mnfr.201000655. [DOI] [PubMed] [Google Scholar]
- 52.Rotroff D.M., Wetmore B.A., Dix D.J., Ferguson S.S., Clewell H.J., Houck K.A., Lecluyse E.L., Andersen M.E., Judson R.S., Smith C.M. Incorporating human dosimetry and exposure into high-throughput in vitro toxicity screening. Toxicol. Sci. 2010;117:348–358. doi: 10.1093/toxsci/kfq220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.A. Saltelli, What is Sensitivity Analysis? in: Sensitivity Analysis Edited by Saltelli et al., John Wiley & Sons Ltd., 2000. pp. 3–13.
- 54.SOLVO, Biotechnology, SOLVO The Transporter Book (3rd Edition), Produced and Published by SOLVO Biotechnology, 2017.
- 55.Suciu N., Tediosi A., Ciffroy P., Altenpohl A., Brochot C., Verdonck F. Potential for MERLIN-Expo, an advanced tool for higher tier exposure assessment, within the EU chemical legislative frameworks. Sci. Total Environ. 2016;562:474–479. doi: 10.1016/j.scitotenv.2016.04.072. [DOI] [PubMed] [Google Scholar]
- 56.Sung J.H., Srinivasan B., Esch M.B., McLamb W.T., Bernabini C., Shuler M.L., Hickman J.J. Using physiologically-based pharmacokinetic-guided “body-on-a-chip” systems to predict mammalian response to drug and chemical exposure. Exp. Biol. Med. (Maywood) 2014;239(9):1225–1239. doi: 10.1177/1535370214529397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.US EPA (U.S. Environmental Protection Agency), Approaches for the Application of Physiologically Based Pharmacokinetic (PBPK) Models and Supporting Data in Risk Assessment (Final Report). National Center for Environmental Assessment, Washington, DC, 2006. EPA/600/R- 05/043F.
- 58.Teorell T. Kinetics of the distribution of substances administered to the body II. The extravascular mode of administration. Arch. Int. Pharm. Ther. 1937;57:205–240. [Google Scholar]
- 59.Tonnelier A., Coecke S., Zaldívar J.M. Screening of chemicals for human bioaccumulative potential with a physiologically based toxicokinetic model. Arch. Toxicol. 2012;86:393–403. doi: 10.1007/s00204-011-0768-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.US FDA (U.S. Food and Drug Administration), Draft “Physiologically Based Pharmacokinetic Analyses — Format and Content Guidance for Industry”, 2018. https://www.fda.gov/ucm/groups/fdagov-public/@fdagov-drugs-gen/documents/document/ucm531207.pdf.
- 61.Wagner C., Zhao P., Pan Y., Hsu V., Grillo J., Huang S., Sinha V. Application of Physiologically Based Pharmacokinetic (PBPK) Modeling to Support Dose Selection: Report of an FDA Public Workshop on PBPK. CPT: Pharmacometrics Syst. Pharmacol. 2015;4(4):226–230. doi: 10.1002/psp4.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wambaugh J.F., Wetmore B.A., Pearce R., Strope C., Goldsmith R., Sluka J.P., Sedykh A., Tropsha A., Bosgra S., Shah I., Judson R., Thomas R.S., Setzer R.W. Toxicokinetic triage for environmental chemicals. Toxicol. Sci. 2015;147:55–67. doi: 10.1093/toxsci/kfv118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Waters N.J., Jones R., Williams G., Sohal B. (2008) Validation of a rapid equilibrium dialysis approach for the measurement of plasma protein binding. J Pharm Sci. 2008;97(10):4586–4595. doi: 10.1002/jps.21317. [DOI] [PubMed] [Google Scholar]
- 64.Wetmore B.A., Wambaugh J.F., Ferguson S.S., Sochaski M.A., Rotroff D.M., Freeman K., Clewell H.J., III, Dix D.J., Andersen M.E., Houck K.A. Integration of dosimetry, exposure, and high-throughput screening data in chemical toxicity assessment. Toxicol. Sci. 2012;125:157–174. doi: 10.1093/toxsci/kfr254. [DOI] [PubMed] [Google Scholar]
- 65.Wetmore B.A., Wambaugh J.F., Ferguson S.S., Li L., Clewell H.J., Judson R.S., Freeman K., Bao W., Sochaski M.A., Chu T.M. Relative impact of incorporating pharmacokinetics on predicting in vivo hazard and mode of action from high-throughput in vitro toxicity assays. Toxicol. Sci. 2013;132:327–346. doi: 10.1093/toxsci/kft012. [DOI] [PubMed] [Google Scholar]
- 66.Wetmore B.A., Allen B., Clewell H.J., Parker T., Wambaugh J.F., Almond L.M., Sochaski M.A., Thomas R.S. Incorporating population variability and susceptible subpopulations into dosimetry for high-throughput toxicity testing. Toxicol. Sci. 2014;142:210–224. doi: 10.1093/toxsci/kfu169. [DOI] [PubMed] [Google Scholar]
- 67.Wetmore B.A. Quantitative in vitro-to-in vivo extrapolation in a high-throughput environment. Toxicology. 2015;332:94–101. doi: 10.1016/j.tox.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 68.WHO/IPCS (World Health Organization. International Programme on Chemical Safety), Characterization and Application of Physiologically Based Pharmacokinetic Models in Risk Assessment. Harmonization Project Document No. 9. 2010. Geneva, Switzerland.
- 69.A. Worth, M. Cronin, S. Enoch, E. Fioravanzo, M. Fuart-Gatnik, M. Pavan, C. Yang, Applicability of the Threshold of Toxicological Concern (TTC) approach to cosmetics – preliminary analysis. JRC report EUR 25162 EN. Publications Office of the European Union, Luxembourg, 2012, available at: http://publications.jrc.ec.europa.eu/ repository/.
- 70.Yoon M., Campbell J.L., Andersen M.E., Clewell H.J. Quantitative in vitro to in vivo extrapolation of cell-based toxicity assay results. Crit. Rev. Toxicol. 2012;42(8):633–652. doi: 10.3109/10408444.2012.692115. [DOI] [PubMed] [Google Scholar]
- 71.Yoon M., Efremenko A., Blaauboer B.J., Clewell H.J. Evaluation of simple in vitro to in vivo extrapolation approaches for environmental compounds. Toxicol. in vitro. 2014;28:164–170. doi: 10.1016/j.tiv.2013.10.023. [DOI] [PubMed] [Google Scholar]
- 72.Yoon M., Kedderis G.L., Yan G.Z., Clewell H.J., 3rd. Use of in vitro data in developing a physiologically based pharmacokinetic model: Carbaryl as a case study. Toxicology. 2015;5(332):52–66. doi: 10.1016/j.tox.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 73.Zaldivar Comenges J.M., Joossens E., Sala Benito J.V., Worth A., Paini A. Theoretical and mathematical foundation of the virtual cell based assay – a review. Toxicol. In Vitro. 2017;45 Part 2:209–221. doi: 10.1016/j.tiv.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 74.Pearce Robert G., Woodrow Setzer R., Strope Cory L., Sipes Nisha S., Wambaugh John F. httk: R package for high-throughput toxicokinetics. J. Stat. Software. 2017;79(4):1–25. doi: 10.18637/jss.v079.i04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Punt A., Schiffelers M.-J., Horbach GJ W.A., van de Sandt J.M., Groothuis G.M.M., Rietjens I.M.C.M., Blaauboer B.J. Evaluation of research activities and research needs to increase the impact and applicability of alternative testing strategies in risk assessment practice. Regul. Toxicol. Pharmacol. 2011;61:105–114. doi: 10.1016/j.yrtph.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 76.Schultz T.W., Amcoff P., Berggren E., Gautier F., Klaric M., Knight D.J., Mahony C., Schwarz M., White A., Cronin M.T.D. A strategy for structuring and reporting a read-across prediction of toxicity. Regul. Toxicol. Pharm. 2015;72(3):586–601. doi: 10.1016/j.yrtph.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 77.Lombardo Franco, Scott Obach R., Varma Manthena V., Stringer Rowan, Berellini Giuliano. Clearance mechanism assignment and total clearance prediction in human based upon in silico models. J. Med. Chem. 2014;57(10):4397–4405. doi: 10.1021/jm500436v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.