Abstract
The discovery of interleukin-2 (IL-2) changed the molecular understanding of how the immune system is controlled. IL-2 is a pleiotropic cytokine, and dissecting the signaling pathways that allow IL-2 to control the differentiation and homeostasis of both pro- and anti-inflammatory T cells is fundamental to determining the molecular details of immune regulation. The IL-2 receptor couples to JAK tyrosine kinases and activates the STAT5 transcription factors. However, IL-2 does much more than control transcriptional programs; it is a key regulator of T cell metabolic programs. The development of global phosphoproteomic approaches has expanded the understanding of IL-2 signaling further, revealing the diversity of phosphoproteins that may be influenced by IL-2 in T cells. However, it is increasingly clear that within each T cell subset, IL-2 will signal within a framework of other signal transduction networks that together will shape the transcriptional and metabolic programs that determine T cell fate.
Keywords: interleukin-2, JAK1/3, PI3K, cytokine signaling
The History of IL-2
Interleukin-2 (IL-2), a 15.5-kDa variably glycosylated globular protein, was first described as T cell growth factor (TCGF), a potent mitogen and growth regulator of T cells in vitro (1–6). The discovery of IL-2 stemmed from work that documented that conditioned media from lymphocytes activated polyclonally with plant lectins, such as phytohemagglutinin, contained a protein that could support the proliferative expansion of cytotoxic T lymphocytes and the isolation of antigen-specific T cell clones (1). In seminal work, the group led by Kendall Smith used rigorous protein enrichment protocols to purify TCGF (IL-2) to homogeneity and generate bioactive, biosynthetically radiolabeled TCGF (7, 8). The availability of radiolabeled TCGF/IL-2 made it possible to explore how this cytokine interacted with T cells. This led to a fundamental discovery: the biological effects of TCGF were mediated by a high-affinity cytokine receptor that was expressed selectively on T cells activated via their T cell antigen receptor, thereby ensuring the immune specificity of IL-2 actions (5, 6).
During the 1980s, the generation of monoclonal antibodies against T cell populations accelerated many discoveries: In 1982, an antibody called anti-Tac was proposed to be a candidate IL-2 receptor antibody because it could block the binding of IL-2 to T cells. In addition, the expression of the Tac antigen on immune-activated T cells coincided with the expression of the high-affinity IL-2 receptor (9). However, there were discrepancies to be accounted for, such as the observation that activated T cells expressed a maximum of 4–8 × 103 high-affinity IL-2 binding sites, but much higher numbers of Tac antigen (40–100 × 103) binding sites (9). The cloning of the Tac antigen and its expression in heterologous cells then revealed that the Tac antigen was able to bind IL-2, but with low affinity (10, 11). The search was then on for other components that could confer the high-affinity binding of IL-2 to its receptor. After much work, it became clear that the high-affinity IL-2 receptor (IL-2R) comprised three polypeptides: Tac antigen (now known as CD25 or the IL-2 receptor α chain, IL-2Rα), an IL-2 receptor β chain (CD122 or IL-2Rβ), and a γ subunit (γc, or CD132) (12–15). The coexpression of all three subunits is needed to confer high-affinity IL-2 binding to a cell. The discrepancy in Tac antigen numbers and high-affinity IL-2 binding sites was explained by the limiting numbers of IL-2Rβ/γc subunits. Subsequently, the expression of IL-2α has been quantified and shown to exceed the levels of IL-2Rβ and/or γc using flow cytometry (16) and mass spectrometry (17).
The discovery of the IL-2/IL-2R system promoted an explosion of work to purify and identify other cytokines and their receptors. This work has exposed a complex system in which the γc from IL-2R functions as a common subunit for multiple cytokines, including IL-4, IL-7, IL-9, IL-15, and IL-21; and where the γc subunit and the IL-2Rβ subunit, in complex with the unique IL-15Rα chain, form the receptor for IL-15 (18). The discovery and biochemical characterization of IL-2, and the experiments to define the IL-2 receptor, were instrumental to effecting a change in the way immunologists viewed immunoregulation: They highlighted the fundamental importance of secreted cytokines for the control of T cell proliferation and differentiation. Indeed, it was quickly recognized that the loss of the γc was the molecular basis for X-linked severe combined immunodeficiency disease (XSCID) (19).
The discovery of IL-2 has had a significant impact on immunology research. Experiments to probe the molecular basis for IL-2 gene transcription led to the identification of key antigen receptor signal transduction pathways, including the calcium/calcineurin pathway that controls the nuclear translocation of the transcription factor nuclear factor of activated T cells (NFAT) (20). Additionally, the importance of IL-2 as an immunomodulatory cytokine, and as a valuable research tool that permits the proliferation of T cell populations for in vitro analysis and immunotherapy protocols, has resulted in the publication of thousands of studies. This review focuses on our current understanding of the biological role of IL-2 with an emphasis on the intracellular signaling pathways controlled by IL-2 in T cells, and factors that might influence the biological outcomes of IL-2 signaling.
The IL-2 Paradox
The powerful growth factor and mitogenic activities of IL-2 led to the hypothesis that IL-2 would be an essential cytokine that promoted T cell immune responses in vivo. However, this paradigm was challenged by the phenotype of mice with deleted IL-2 alleles: IL-2−/− mice developed normally during their first 3–4 weeks (21), but 50% of IL-2−/− mice died within 9 weeks of birth and the remaining mice had anemia, aberrant lymphoproliferation, and an inflammatory bowel disease resembling ulcerative colitis (22). A similar autoimmune phenotype was seen when CD25 alleles were deleted in mice (23), indicating that this important immunoregulatory role of IL-2 was mediated by the high-affinity IL-2 receptor. These data forced a reevaluation of the role of IL-2 as simply a positive regulator of effector T cell proliferation and rather suggested that the fundamental role of IL-2 is to suppress immune responses. This concept is reinforced by studies showing that polymorphisms that can alter IL-2 signaling in humans are associated with autoimmune diseases including type 1 diabetes, celiac disease, multiple sclerosis, Graves’ disease, and rheumatoid arthritis (24–26).
It was rapidly recognized that the severe immune autoreactivity of IL-2−/− mice is caused by a loss of immunosuppressive CD4+ FoxP3+ regulatory T cells (Tregs) (27, 28). IL-2 is critical for the development of Tregs in the thymus and the regulation, proliferation, and maintenance of Tregs in peripheral tissues, and is essential for maintaining the transcriptional program required for Treg function. This includes sustaining the expression of high levels of FoxP3, the key transcription factor that determines Treg identity (29). IL-2−/− mice have very low numbers of Tregs, and those Tregs that are present do not function effectively to control autoimmunity (30).
What about the role of IL-2 in effector T cells? Evaluation of the importance of IL-2 for effector T cell differentiation in vivo has been complicated by the lymphoproliferation induced by the loss of Tregs. However, it is clear that IL-2 influences effector T cell differentiation and is a critical determinant of the fate decisions of antigen receptor–activated T cells (31–38). CD4+ α/β T cells differentiate following immune activation to produce distinct effector subpopulations that can be distinguished based on their cytokine production, expression of lineage-specific transcriptional master regulators, and cytokine requirements (39). Th1 cells produce IFN-γ and depend on signals from IL-12 and activation of the transcription factors STAT4 and T-bet. The requirement for IL-12 signals renders Th1 cells dependent on the expression of the IL-12 receptor (IL-12R), which consists of IL-12Rβ1 and IL-12Rβ2. Th2 cells produce the cytokines IL-4, IL-5, and IL-13 and depend on IL-4 signals and the transcriptional activities of STAT6 and GATA3. Thus, Th2 cells depend on the expression of the IL-4 receptor, which consists of IL-4Rα and γc. Th17 helper cells, which control neutrophil function and epithelial barrier integrity, produce IL-17A, IL-17F, and IL-22 and depend on signals from IL-6, in combination with TGF-β, IL-23, and IL-21. These signals result in activation of STAT3 and the expression of the transcription factor RORγt to promote Th17 differentiation. Therefore, expression of the IL-6 receptor, which consists of IL-6Rα and gp130, is important for Th17 differentiation. Finally, T follicular helper (Tfh) cells support B cell–mediated immune responses and germinal center formation in the spleen. Tfh cells require IL-21 and IL-6 for their development, with BCL6 acting as a master regulator of the differentiation of Tfh cells (40).
One fundamental question is what directs CD4+ T cells to adopt these different fates. Here it is clear that IL-2 has a role. IL-2 can, thus, promote Th1 and Th2 fate decisions in antigen receptor–activated CD4+ T cells (31, 35, 36, 41) but suppress the differentiation of Th17 cells (32, 42) and Tfh cells (37, 43) (Figure 1). IL-2 controls CD4+ T cell fate by controlling the expression of the key cytokine receptors, transcription factors, chromatin regulators, and effector cytokines. For example, IL-2 can induce the expression of Eomesodermin (34, 44), T-bet (Tbx21) (36), and Blimp-1 (34, 45) while suppressing BCL6 (46, 47). Additionally, IL-2 can regulate how cells may respond to other proinflammatory cytokines by stimulating the expression of the IL-12R, IL-12Rβ1, and IL-12Rβ2 chains (36) and the IL-4Rα chain (41), and suppressing the expression of IL-6Rα and gp130 (36) and IL-7R (48).
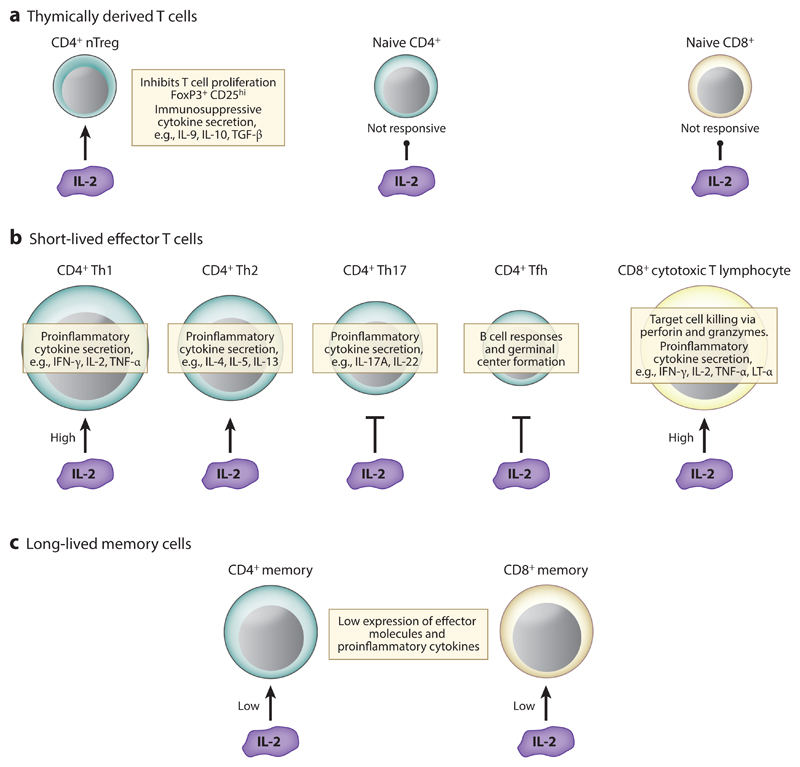
Figure 1.
IL-2 biology: IL-2 positively influences the homeostasis and development of a number of T cell lineages. Expression of IL-2R is restricted to nTregs (a) and antigen-activated T cells (b,c), thereby guaranteeing the specificity of IL-2 activities. IL-2-responsive T cell subsets have extremely diverse characteristics and proinflammatory and anti-inflammatory biological roles. Moreover, IL-2 inhibits the differentiation of Th17 and Tfh cells (b), thus making IL-2 an important regulator of T cell lineage commitment. In addition, the levels of IL-2 signaling can define the effector/memory fates of CD4+ and CD8+ T cells: High levels of IL-2 favor the development of short-lived effector cells (b), while low levels of IL-2 signaling promote the differentiation of memory T cells (c). IL-2 stimulation of T cell function is depicted by arrows, IL-2 inhibition of lineage development is represented by inhibitory lines, and no response to IL-2 is indicated by a line with a circle at the end. Abbreviations: IFN-γ, interferon gamma; IL-2R, interleukin-2 receptor; LT-α, lymphotoxin alpha; nTreg, thymically derived regulatory T cell; Tfh, follicular helper T; TNF-α, tumor necrosis factor alpha.
IL-2 is also important for CD8+ effector cells (Figure 1). In response to infection, naive antigen-specific CD8+ T cells proliferate and differentiate into effector cytotoxic T lymphocytes (CTLs) that make proinflammatory cytokines such as IFN-γ and acquire the ability to kill infected cells. IL-2 can influence these effector activities in CD8+ T cells by inducing the expression of IFN-γ (49, 50), TNF-α, and lymphotoxin α (51), by stimulating expression of the cytolytic effector molecules granzyme B (34, 44, 52) and perforin (34, 52), and by promoting effective target cell killing (34, 44). IL-2 also acts to control the size of CTLs by regulating amino acid uptake and protein synthesis (53–55). The majority of CTLs die after pathogen clearance; however, a small number do not terminally differentiate but switch off active effector functions and survive as long-lived, self-renewing memory T cells. An understanding of how IL-2 can control the balance in the effector/memory cell fates of CD8+ T cells has grown greatly in recent years. High IL-2 signaling drives T cells to become terminally differentiated, short-lived effector cells and promotes the expression of critical cytolytic effector molecules and cytokines by immune-activated CD8+ T cells (Figure 1); this is because IL-2 induces the expression of Blimp-1 while suppressing the expression of molecules associated with central memory effectors, such as BCL6, the IL-7Rα chain (CD127) and L-selectin (CD62L) (33, 34, 56). In contrast, low doses of IL-2 signaling can support a memory phenotype in activated CD8+ T cells (33, 34, 56) and Tfh-like or memory phenotype in CD4+ T cells (38) (Figure 1). Low-dose IL-2 allows BCL6, IL-7Rα, and L-selectin to be re-expressed and may not drive expression of high levels of cytolytic molecules, thus more readily allowing the maturation of memory cells. The low levels of Blimp-1 in cells differentiated in low levels of IL-2 correlate with a more efficient IL-2 production by these cells upon rechallenge (34). This may be important for potentiating the autocrine IL-2 signaling by CD8+ T cells that is important for CD8+ T cell memory responses (57, 58).
IL-2 Signal Transduction
IL-2 controls diverse T cell biology by binding with high affinity (Kd ≈ 10−11 M) to the trimeric IL-2R consisting of IL-2Rα, IL-2Rβ, and γc. The IL-2Rβ and γc dimer can bind IL-2 with lower affinity (Kd ≈ 10−9 M), but the phenotype of the CD25 knockout mouse indicates that most of the biological functions of IL-2 are mediated by the high-affinity IL-2 receptor. Moreover, in early experiments, it became clear that key signaling functions of the IL-2 receptor were mediated by the high-affinity receptor. For example, IL-2 receptor triggering causes the Ras GTPase to switch from the inactive GDP loaded to the active GTP bound state: The concentration of IL-2 that activated Ras coincided with those that bound to the high-affinity IL-2 receptor (59). GTP-bound Ras binds and activates the serine/threonine kinase RAF-1, which directs activity of the MAP kinases ERK1/2 (60). These studies were the first to indicate that some of the biological effects of IL-2 could be mediated by the activation of cytosolic serine/threonine kinases. This concept was reinforced by studies showing that IL-2 controlled the activity of the p70S6 ribosomal kinase (61–63) in a signaling pathway regulated by the immunosuppressive drug rapamycin. This latter result is now known to reflect IL-2 control of the activity of the nutrient-sensing serine/threonine kinase mammalian target of rapamycin complex 1 (mTORC1) (64).
The challenge, then, was to understand how IL-2 receptors could couple to cytosolic GTPases and serine/threonine kinases. The IL-2 receptor subunits lack intrinsic kinase or other enzymatic activity. However, it was quickly recognized that IL-2 controlled T cell function by regulating the activity of cytosolic tyrosine kinases. It was originally proposed that IL-2 signaled to T cells via SRC family kinases, such as LCK and FYN (65, 66). This hypothesis did not survive experimental tests, and it is now known that LCK and FYN have high constitutive activity prior to IL-2 receptor engagement and hence signal independently of IL-2 signals (55). A key breakthrough in understanding how the IL-2 receptor functions came from the discovery that the IL-2 receptor uses Janus family kinase (JAK) members to induce signal transduction (67–70). The IL-2Rβ chain binds JAK1, the γc binds to JAK3, and IL-2 receptor occupancy results in JAK1/3 activation (71). Consequently, JAK1 and/or JAK3 selective kinase inhibitors are powerful tools for blocking IL-2-mediated proliferative responses (72, 73). The importance of JAK3 for γc signaling is also clear because the phenotype of humans lacking expression/function of JAK3 is associated with the same type of severe combined immunodeficiency (SCID) found for humans with γc loss of expression/function (73, 74). There is, however, still some debate over the role of the kinase activity of JAK3 versus its role as a signaling scaffold in mediating IL-2–IL-2R transduction (75).
The discovery of the IL-2-JAK connection rapidly led to the discovery that IL-2 induced the tyrosine phosphorylation and DNA binding of a high-molecular-weight protein of the signal transducer and activator of transcription (STAT) family (76). This was subsequently mapped to key tyrosine residues close to the C terminus of STAT5 proteins, STAT5A Y694 and STAT5B Y699 (77, 78) (Figure 2a). In addition to the tyrosine phosphorylations, IL-2-induced serine phosphorylation of STAT5 has been proposed to regulate STAT5 transcriptional function (78). Serine residues increased in phosphorylation in response to IL-2 have been mapped to STAT5A S127 and/or S128 (55) (Figure 2a), sites that may influence the transcriptional activity of the protein (79). IL-2 can also regulate other STATs and induce phosphorylation of STAT3 on Y705 and S727 (77, 80) (Figure 2a) and STAT1 phosphorylation on Y701 (15). The model for STAT5 activation is that activation of JAK1 and JAK3 induces a number of tyrosine phosphorylations in the IL-2Rβ chain, including Y338, Y392, and Y510 in the human receptor (Y341, Y395, and Y498 in the mouse IL-2 receptor) (15) (Figure 2a). The phosphorylation of IL-2Rβ at Y395 and Y498 allows the recruitment of STAT5A and STAT5B transcription factors via their SH2 domains and their subsequent tyrosine phosphorylation (Figure 2a); following phosphorylation, STAT5 proteins dissociate from the IL-2β subunit and form dimers, through the head-to-tail association of the SH2 domains with the phosphorylated phosphotyrosine sites. This induces their transcriptional activation and translocation to the nucleus. These STAT dimers can bind to gamma interferon–activated sequence (GAS) DNA sequences (TTCN3GAA) to induce transcription. The STAT5 dimers can also form tetramers through interactions between residues (I28, F81, and L82) in their N-terminal regions. These STAT5 tetramers bind to pairs of GAS motifs separated by a linker of 6–22 nucleotides (51). Mutational studies demonstrate that STAT5 dimers and tetramers are both important for IL-2-induced gene expression (51).
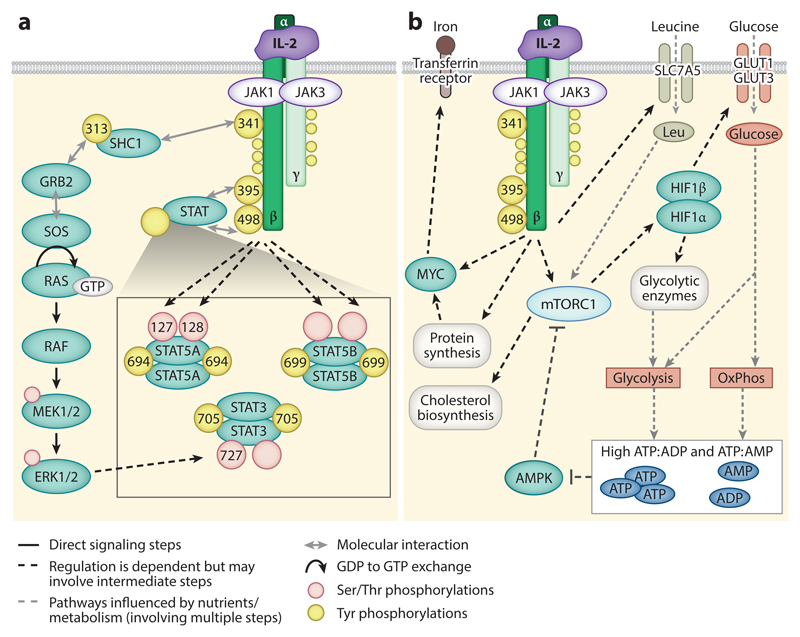
Figure 2.
Signaling downstream of the IL-2 receptor (IL-2R): (a) Janus kinase-signal transducer and activator of transcription (JAK-STAT) signal transduction. The IL-2Rβ chain couples to JAK1, and the γ chain (γc) couples to JAK3. IL-2R occupancy results in the activation of the JAKs and the tyrosine phosphorylation of IL-2Rβ and γc (yellow circles). The tyrosine phosphorylations in IL-2Rβ are best characterized in terms of the signaling complexes they coordinate; the phosphorylation of Y395 and Y498 (in murine IL-2Rβ) permits the recruitment of STAT5A, STAT5B, and STAT3, while the phosphorylation of Y341 allows recruitment of SHC1. Following their recruitment to the receptor, STAT5 proteins and SHC1 are tyrosine phosphorylated by JAKs. Tyrosine phosphorylation of STAT5 permits dimerization, nuclear translation, and STAT5-mediated transcription. Tyrosine phosphorylation of SHC1 allows the recruitment of GRB2 and SOS to facilitate GTP loading of Ras and activation of the classical Raf-ERK mitogen-activated protein kinase (MAP kinase) cascade. STAT proteins are also phosphorylated on serine residues (peach circles) in response to IL-2 signaling. While the STAT5 serine kinase is unknown, the STAT3 proteins are phosphorylated in response to MAP kinase signaling. The sites of IL-2-regulated phosphorylation in (murine) IL-2R, SHC1 (isoform 2), STAT5A, and STAT5B are annotated where positions have been mapped. (b) Metabolic pathways activated downstream of IL-2R. IL-2 sustains the expression of MYC and activation of the mammalian target of rapamycin complex 1 (mTORC1) and the hypoxia inducible factor 1 transcriptional complex (HIF1α/HIF1β) to maintain the uptake of nutrients such as iron and glucose. IL-2 also sustains the expression of the amino acid transporter SLC7A5, which transports many of the essential amino acids, such as leucine, into cells. Amino acids are required to sustain mTORC1 activity and fuel translation. Glucose metabolism, via glycolysis and oxidative phosphorylation (OxPhos), sustains ATP generation, and high ATP:ADP and ATP:AMP ratios, thus preventing the activation of AMP-activated protein kinase (AMPK), which inhibits mTORC1. Protein synthesis is essential for maintaining the expression of proteins, such as MYC, with high turnover rates. Through the mTORC1 pathway, IL-2 can also sustain glycolytic metabolism and other biosynthesis in cytotoxic T lymphocytes to support cell proliferation, growth, and the expression of effector molecules.
IL-2 activation of STAT5-mediated transcriptional programs is undoubtedly key to the biological actions of IL-2 (28, 73, 81–83). The critical importance of STAT5A/B-mediated transcriptional processes has been highlighted by studies of knockout mice. STAT5A/B-deficient mice fail to generate Tregs (84–86), demonstrating the crucial role for IL-2-JAK1/3-STAT5 in Treg development. In addition, immune-activated peripheral T cells from STAT5A/B knockout mice are unable to proliferate (81, 87). Moreover, in humans, loss of function in STAT5B is associated with a loss of Tregs (88, 89). STAT5A and STAT5B are thus critical mediators of IL-2 signals. Analyses of STAT5 binding sites using chromatin immunoprecipitation (ChIP) in CD4+ T cells have shown that a number of essential lineage-defining transcription factors are direct STAT5A and STAT5B target genes. These include FoxP3 (85, 90), Tbx21 (T-bet) (36), Maf (41, 91), and Gata3 (41). STAT5 can additionally bind to the Il2ra gene (90), Il12rb2 (36), and Il4ra (41), correlating with the ability of IL-2 to induce the expression of these genes, sustain its own signaling, and render T cells susceptible to inputs from other cytokines to promote the differentiation of Th1 and Th2 cells. Another STAT5 target gene is Socs3 (41, 51, 92)—which encodes a protein that negatively regulates IL-2 signals. However, not all STAT5-binding sites are associated with increases in gene expression: STAT5 binding to the Il17a-Il17f locus is repressive, thereby inhibiting STAT3-mediated transcription of the Il17 gene (42) and suppressing Th17 cell differentiation. Similarly, STAT5 has been shown to bind the Bcl6 gene in hematopoietic cell lines (93) and in CD4+ T cells (47, 94), correlating with repressed expression of the gene. The mechanisms by which STAT5 may regulate gene expression in T cells, such as recruitment of RNA polymerase II, coactivating or corepressing proteins, or chromatin-remodeling enzymes, are still to be elucidated. The amino acid sequences of STAT5A and STAT5B show over 90% similarity, and homodimers of each of these two proteins recognize very similar GAS motifs (81). While differences in STAT5A and STAT5B gene targets have been suggested in T cell subsets (41, 90), it is likely this reflects differences in their relative expression in vivo rather than functional differences (94).
IL-2 Signaling Beyond STAT5
Although activation of STAT5 is important for IL-2 signaling, experiments with constitutively active mutants have found active STAT5 is not sufficient to mimic the effects of IL-2 on T cell biology (95). It is, therefore, clear that IL-2 signaling mechanisms extend beyond STATs and include other signaling networks. As discussed, IL-2 drives the accumulation of active, GTP-bound Ras GTPases (59). The key pathway for IL-2 control of Ras is mediated by the adapter SHC, which is tyrosine phosphorylated on Y317 (Y313 in mouse) in response to IL-2 receptor engagement (96) (Figure 2a). Tyrosine-phosphorylated SHC then forms a complex with the SH2 domain of the adapter GRB2, which binds constitutively via its tandem SH3 domains to the Ras guanine nucleotide exchange protein SOS (Son of Sevenless) (97) (Figure 2a). This complex catalyzes the exchange of GDP for GTP on Ras, thereby allowing Ras to accumulate in its GTP-bound state and activate the Raf-ERK MAP kinase cascade (Figure 2a).
Another important serine/threonine kinase for IL-2 signal transduction is mammalian target of rapamycin (mTOR) (43, 64) (Figure 2b). The mTOR kinase is a constituent of two different kinase complexes, mTORC1 and mTORC2. The core subunits of mTORC1 are mTOR, RAPTOR, and mLST8, while the core subunits of mTORC2 are mTOR, RICTOR, and mLST8 (98). The significance of the mTOR complex for T cell biology is evidenced by the importance of the mTORC1 inhibitor rapamycin as a T cell immunosuppressant. Moreover, CD4+ T cell differentiation into Th1 cells is inhibited by T cell–specific knockout of the mTOR kinase (99), RHEB (100), and RICTOR (100, 101), thus implicating both mTORC1 and mTORC2 complexes in Th1 development. Interestingly, Th2 cells appear to depend on mTORC2 signaling, as Rheb−/− T cells were capable of differentiating into Th2 cells (100) but mTOR−/− and RICTOR-deficient T cells were not (99–102). In contrast, IL-2-mediated activation of mTORC1 has been shown to be involved in IL-2 suppression of Tfh differentiation (43).
In effector CD8+ cytotoxic T cells, IL-2 induces and maintains a robust activation of mTORC1 to control IL-2-induced metabolic and transcriptional programs (53, 54, 64). Hence mTORC1 activity appears to play an important role in shaping the CTL proteome to control energy-generating metabolic process (17, 64). Loss of mTORC1 signaling thus causes CTLs to decrease expression of glucose transporters (17, 64) and glycolytic enzymes (Figure 2b) and to decrease their rate of glycolysis while maintaining oxidative phosphorylation pathways (17). In addition, mTORC1 activity is required to sustain the expression of proteins required for cholesterol biosynthesis, such as 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) (17) (Figure 2b). Interestingly, mTORC1 activity may suppress some metabolic pathways in CTLs; rates of glutaminolysis are increased in mTORC1-inhibited (i.e., rapamycin-treated) CTLs (17).
One important role for mTORC1 activity in CTLs is to control the expression of effector molecules including perforin, granzymes, and IFN-γ (17). Moreover, a key differentiation step for effector CTLs is to switch off the expression of chemokine receptors and adhesion molecules, such as CD62L, CCR7, and S1P1, that direct naive and memory T cells to secondary lymphoid organs (103). This part of the CTL transcriptional program is essential to prevent CTLs entering secondary lymphoid tissue and hence promotes CTL trafficking to peripheral tissues, where they mediate destruction of pathogen-infected cells. The inhibition of mTORC1 in IL-2-maintained CTLs causes the cells to retain CD62L and CCR7 expression and the capacity to home to secondary lymphoid tissue (103).
How does IL-2-regulated mTORC1 control CTL function? A crucial molecule here is the transcription factor HIF1α. IL-2 sustains expression of HIF1α in CTLs via an mTORC1-dependent pathway (Figure 2b). HIF1α forms a transcriptional complex with ARNT/HIF1β to regulate gene expression. HIF1 controls a subset of mTORC1-regulated genes, including those for glucose transporters, genes involved in glycolysis and pyruvate metabolism, and the gene for the effector molecule perforin (64) (Figure 2b). Not all effector molecules are regulated by HIF1, as HIF1 knockout CTLs are still capable of producing IFN-γ and expressing FAS ligand and lymphotoxin α (64). However, the HIF1 pathway is critical for mTORC1 repression of the expression of the lymph node–homing molecules CD62L, CCR7, and S1P1 (64). Immune-activated HIF1 null CD8+ T cells thus retain the migratory pattern of naive T cells to secondary lymphoid tissue (64).
How is mTORC1 activity sustained by IL-2 in CTLs? It is known that IL-2 control of mTORC1 activity is dependent on JAK kinase activity (55). Additionally, mTORC1 activity in IL-2-stimulated T cells is regulated by nutrient availability. Indeed, a constant source of amino acid signaling via leucine, glutamine, and arginine is required for mTORC1 activity (98) and is a prerequisite for mTORC1 activation in T cells (54) (Figure 2b). mTORC1 activity in CTLs is also highly dependent on sustained availability of glucose (104). CTLs are able to sense glucose levels via the AMP-activated protein kinase (AMPK). If the glucose supply to T cells is limited, AMPK is activated, triggering a negative feedback pathway to restrict mTORC1 activity (105) (Figure 2b). The high sensitivity of mTORC1 activity to nutrients makes it probable that the ability of IL-2 to maintain amino acid uptake, and possibly glucose uptake (53, 54, 64), is a key driver of mTORC1 activity. In this respect, one key IL-2-JAK1/3-maintained molecule in this nutrient-sensitive signaling pathway is the system L amino acid transporter, SLC7A5 (Figure 2b). This transporter sustains the uptake of leucine and other large neutral amino acids (54) and is the most highly expressed IL-2 regulated leucine transporter in CTLs (17).
Since the advancement of quantitative mass spectrometry technologies, a number of studies have taken unbiased approaches to investigate the phosphorylation pathways induced in cells in response to IL-2 activation on a global scale. Studies in KIT225 cells (106–109), F15R-Kit cells (110), and primary mouse CTL cultures (55) have made it possible to evaluate the diversity and true extent of phosphorylation networks influenced by IL-2 signaling (Figure 3). The picture of signaling that emerged from the primary CTL phosphoproteomics study (55) exposed the striking number and range of protein phosphorylations that can be influenced by IL-2; out of 6,458 phosphorylation sites identified in CTLs, IL-2 increased the phosphorylation of 707 and decreased 224 phosphorylations. These data highlight how much of IL-2 signaling is underappreciated, particularly that IL-2 can both positively and negatively influence the phosphorylation status of proteins in a cell. The majority of the phosphorylations influenced by IL-2 were at phosphoserine and phosphothreonine sites, a number of which were inversely regulated by treating IL-2-maintained CTLs with the JAK1/3 inhibitor tofacitinib (55). Thus, these data give a perspective of the diverse phosphoserine and phosphothreonine signals that IL-2-JAK1/3 signaling can modulate. The phosphoproteins regulated by IL-2 included adaptor proteins, proteins involved in processes in the nucleus, the actin and microtubule cytoskeleton, and membrane trafficking (55, 106–110) (Figure 3). The web of phosphorylation networks uncovered by such global studies demonstrates how multiple aspects of T cell biology may be synchronized by IL-2. In addition, they reveal how IL-2 may control the differentiation and fate decisions in T cells. Of particular note was the ability of IL-2 to regulate the phosphorylation of histone modifiers and chromatin-remodeling enzymes (55, 109) (Figure 3), indicating how IL-2 can control gene expression and may influence memory cell recall responses of T cells. Such data also emphasize that IL-2 may direct protein expression in T cells separately from instigating transcriptional programs: IL-2 can regulate the phosphorylation of mRNA-binding proteins such as YBX1 and LARP1 (55), which can control the stability of mRNAs and direct which are translated into proteins (Figure 3). Additionally, IL-2 regulated the phosphorylation of components of the cellular translational machinery, correlating with the ability of IL-2 to control the rate of protein synthesis in CTLs (55) (Figure 3). These phosphoproteomic studies have given unparalleled insights into the proteins that are influenced by IL-2 signaling. Future phosphoproteomics and proteomics analyses will powerfully accelerate understanding of the molecules that are regulated by IL-2 in different cell types and permit in-depth dissection of how IL-2 can be tailored to regulate both pro- and anti-inflammatory T cells.
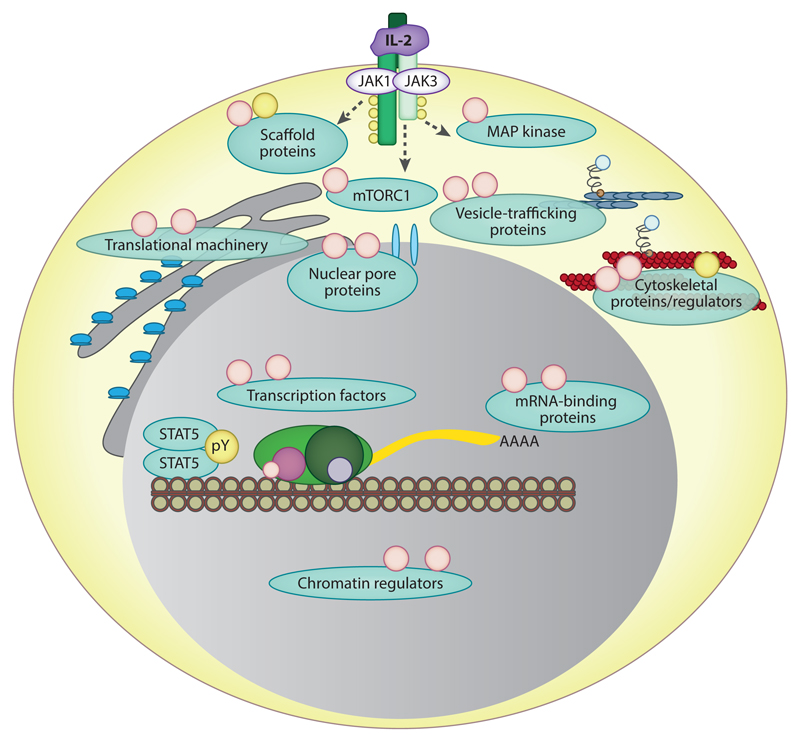
Figure 3.
The phosphorylation networks of IL-2 signaling in effector T lymphocytes. Global phosphoproteomic studies have revealed the range of tyrosine (yellow circles) and serine/threonine phosphorylations (peach circles) regulated following IL-2 stimulation of cytotoxic T lymphocytes. These phosphorylations are on diverse proteins involved in multiple aspects of T cell biology and include scaffold proteins, proteins that can direct gene expression, and cytoskeletal regulators. These comprehensive phosphoproteomic studies highlight that IL-2 signaling involves much more than JAK1/3-STAT5 and reinforce the importance of serine/threonine kinase pathways in IL-2 responses.
Indeed, an important aspect to consider is how IL-2 may downregulate or switch off signaling pathways in T cells. Some potential pathways linking IL-2R occupancy to tyrosine phosphatases have been characterized: SHC-GRB2 complexes associated with IL-2R may allow the recruitment of the adaptor, GAB2, which, in turn, may recruit the protein tyrosine phosphatase SHP-2 (PTPN11) to the IL-2 receptor (111). SHP-2 has been shown to be phosphorylated in response to IL-2 (112): This may lead to molecular rearrangements that induce the activation of its phosphatase activity (113). However, SHP-2 may additionally act as an adaptor molecule in IL-2 signal transduction, and despite its phosphatase activities, it has been implicated in positively regulating IL-2 signaling (114). Interestingly, SHP-1, a phosphatase related to SHP-2, has also been found to associate with the IL-2R complex and may influence the tyrosine phosphorylation of receptor components (115) in a negative feedback loop to terminate IL-2R signaling. IL-2 via STAT5 can also stimulate the expression of the SH2-domain-containing E3-ubiquitin ligases, CIS, SOCS1, and SOCS3 (51). These proteins can associate with IL-2Rβ to block interaction sites, directly inhibit JAK1 activity, and/or mediate proteasomal degradation of bound proteins (116). Therefore, the extent of activation of such inhibitory pathways may also affect IL-2 signaling outcomes in T cells.
IL-2 and PI3K Signaling
A key signaling pathway in T cells is mediated by the lipid phosphatidylinositol-3,4,5-trisphosphate [PI(3,4,5)P3]. Cellular levels of PI(3,4,5)P3 are controlled by the balanced activity of kinases and phosphatases. Class I phosphatidylinositol 3-kinases (PI3Ks) phosphorylate the 3 position of the inositol ring of phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2] to produce PI(3,4,5)P3. The class I PI3Ks comprise a 110-kDa catalytic subunit and an adapter regulatory subunit: There are four catalytic isoforms (α, β, γ, δ) and three adapter subunits (p85α, p85β, p55γ). In mature T cells, the majority of PI(3,4,5)P3 is produced by the actions of the p110δ catalytic subunit in a complex with p85α or p85β (117). PI(3,4,5)P3 can then be metabolized in one of two ways: either via the lipid phosphatase PTEN (phosphatase and tensin homolog deleted on chromosome 10), which removes the phosphate group from the 3 position of PI(3,4,5)P3 to produce PI(4,5)P2, or via the SHIP family phosphatases (SHIP-1 and SHIP-2), which dephosphorylate PI(3,4,5)P3 to PI(3,4)P2 (117).
PI(3,4,5)P3 can organize signaling transduction in cells by interacting with pleckstrin homology (PH) domains in proteins and localizing these proteins to membranes. Proteins that bind to PI(3,4,5)P3 in T cells include the serine/threonine kinases phosphoinositide-dependent kinase l (PDK1) and protein kinase B (PKB; also known as AKT), Tec family tyrosine kinases, and guanine nucleotide exchange proteins for Rho family GTPases (118). In addition to establishing the localization of proteins, the binding of PH domains to PI(3,4,5)P3 can initiate conformational changes in proteins: For example, the catalytic activity of AKT is allosterically regulated by PI(3,4,5)P3 (119). The activities of PI(3,4,5)P3-binding serine/threonine kinases play an important role in the differentiation of CTLs. PDK1 controls the activity of mTORC1 and T cell metabolic programs (64). Additionally, PDK1 controls the activity of AKT in IL-2-maintained CTLs: PI(3,4,5)P3 facilitates the colocalization of AKT and PDK1, allowing PDK1 to phosphorylate AKT on T308, thereby activating AKT (120). The PI3K/AKT axis is key for effector T cell differentiation because of the ability of AKT to phosphorylate and enforce the nuclear exclusion, and, consequently, repress the activities of the FOXO1/3 transcription factors. This regulation of FOXO1/3 is an important governor of T cell fate because FOXO1/3 proteins induce the expression of lymphoid homing trafficking molecules while simultaneously repressing the expression of effector molecules in CTLs, thus directing effector/memory CD8+ T cell differentiation (121–123). Therefore, the activation of PI(3,4,5)P3-AKT is pivotal for effector T cell differentiation (121, 124). However, although loss of PI3K-p110δ activity is detrimental to T cell development, constitutive activation of PI3K-p110δ also impairs immune function (117, 125–127). It is increasingly clear that the activation of PI3K-p110δ must be finely balanced: Determining how cells appropriately regulate this pathway is key to understanding the molecular mechanisms that control immune homeostasis. However, understanding the regulation of this pathway by IL-2 has been challenging.
In the 1990s, it was proposed that activation of the PI3K-AKT signaling pathway was regulated by IL-2 (62, 63, 128). This model stemmed from observations that p85/PI3K-p110 could be recruited into the IL-2R signaling complex (129–131). The p85 regulatory subunit of PI3K contains an SH2 domain that can bind with high affinity to phosphorylated tyrosines, normally within the motif YXXM. The pY392 (mouse Y395) in IL-2Rβ has been implicated in recruiting the p85 adaptor subunit of PI3K to the IL-2R complex (129). However, this site does not fit the consensus YXXM-binding motif for the p85 SH2 domain. Interestingly, other studies indicated that p85 may be recruited to the receptor by non–phosphotyrosine-mediated interactions with IL-2Rβ and JAK1 (130). An alternative mechanism by which p85 may be recruited to the receptor is via interactions with tyrosine-phosphorylated GAB2, which can associate with IL-2R via SHC-GRB2 complexes (131).
One approach to investigate the role of PI3K-AKT signaling in T cells examined the impact of overexpressing plasma membrane–localized PI3K-p110 or constitutively active AKT in the IL-2R-expressing T cell tumor line KIT225 (63). These experiments indicated that active PI3K or AKT could partially mimic IL-2 and regulate the transcription factor E2F, which is key for T cell cycle progression. Such results demonstrated what the PI3K-AKT signaling pathway was able to do in T cells, but they did not prove that the pathway was regulated by IL-2. Other experiments investigated the impact of overexpressing a mutated p85 subunit that was unable to interact with the p110 catalytic subunit on IL-2 signaling. The rationale for these experiments was that the mutated p85 subunit would associate with p85 docking sites in IL-2R or adaptor proteins but would fail to recruit p110δ to the plasma membrane and, hence, function as a dominant negative PI3K. Overexpression of this mutant p85 blocked IL-2 control of cell cycle progression (62); however, it is now clear that this p85 construct would not be selective for PI3K docking sites and when overexpressed would have the potential to block multiple signaling complexes, not just activation of PI3K.
Arguably, the most persuasive experiments to indicate that IL-2 activated PI3K in T cells came from studies using the PI3K inhibitors LY294002 and wortmannin. For example, LY294002 blocked IL-2-mediated cell cycle progression of KIT225 cells (62) and IL-2-induced cell growth of CD8+ T cells (53), thus supporting a role for PI3K signaling in the IL-2 regulatory program. Further analysis of the biochemical pathways regulated by LY294002 and wortmannin linked loss of PI3K-AKT signals to the activation of FOXO transcription factors and the inhibition of mTORC1 (132, 133). As a result, it has become a paradigm that IL-2 signaling activates the PI3K-AKT pathway to inhibit FOXO1/3 transcription factors and activate mTORC1 and its substrate, p70S6K (p70 ribosomal S6 kinase), in T cells. Consequently, many subsequent studies have used the output of p70S6K, as measured by the magnitude of the phosphorylation of the ribosomal protein S6, as a surrogate method to assess the accumulation of PI(3,4,5)P3 or activation of the PI3K-AKT signaling pathway in T cells. However, we now know that using LY294002 and wortmannin to identify downstream targets of the PI3K pathway is flawed, as these inhibitors have numerous off-target effects. Both inhibitors directly block mTOR catalytic activity, and LY294002 is also a potent inhibitor of the PIM family of serine/threonine kinases (134, 135). Therefore, decreased phosphorylation of mTORC1 substrates in response to LY294002 or wortmannin can occur via direct inhibition of mTOR activity and may not reflect any role for PI3K-AKT signaling. Moreover, any impact of LY294002 on T cell biology would reflect the impact of combined inhibition of PI3Ks, mTOR, and PIM kinases and should be interpreted cautiously.
A further complication in evaluating PI3K-AKT signaling downstream of IL-2R has been the assessment of AKT phosphorylation itself: Many studies use an antibody that detects AKT S473 phosphorylation, rather than an antibody that recognizes the activating T-loop phosphorylation site in AKT, phospho-T308. AKT S473 is phosphorylated by the mTORC2 complex and, unlike phospho-T308, is not essential for AKT activity (120). Moreover, loss of AKT S473 phosphorylation does not necessarily result in a decrease in AKT T308 phosphorylation or correlate with a decrease in cellular levels of PI(3,4,5)P3 (17, 136). Thus, monitoring AKT S473 does not inform on AKT activity or cellular levels of PI(3,4,5)P3. Therefore, to assess the PI3K-AKT signaling pathway accurately, PI(3,4,5)P3 must either be measured biochemically (137) or be monitored with imaging techniques that follow the membrane accumulation of fluorescently tagged, and PI(3,4,5)P3-specific, PH domain probes (138, 139). Any assessment of AKT activity should be made by quantifying AKT T308 phosphorylation. If a substitute measure of AKT activity is required, the phosphorylation and nuclear exclusion of the direct AKT substrates FOXO1/FOXO3 are likely to be more reliable than those downstream of mTORC1 signaling.
In view of these caveats, what is known about the role of PI3K-AKT in the context of T cell biology and IL-2 signaling? Here, key advances have been made following the development of small molecules with improved inhibitor specificities, and by looking at T cells that have wildtype p110δ alleles replaced with catalytically inactive p110δD910A alleles (64, 121). For example, IC87114 is a highly specific inhibitor of PI3K-p110δ and AKTi is an inhibitor of AKT that, rather than targeting its catalytic pocket, interferes with the PI(3,4,5)P3-mediated recruitment of AKT to the plasma membrane (140). Studies have shown that the IL-2-mediated proliferative expansion of effector CD8+ CTLs is normal in the presence of AKTi or IC87114 (121). Both IC87114 and AKTi block the phosphorylation of AKT T308 and the phosphorylation of the AKT substrates, FOXO1 and FOXO3, demonstrating that they inhibit the activation of AKT. However, a critical finding using these inhibitors was that neither compound blocked the phosphorylation of the mTORC1 substrate, p70S6K1, or phosphorylation of the p70S6K1 substrates, 4EBP1 and S6 (64). Thus, these data indicate that PI3K-AKT activity is not responsible for maintaining mTORC1 activity in CTLs. Further supporting this model, CTLs expressing the catalytically inactive p110δD910A protein did not activate AKT but showed normal proliferative responses and mTORC1-dependent phosphorylation of S6K1 and S6 (64, 121). Additionally, loss of p110δ or AKT activity had no impact on the ability of IL-2 to sustain the expression of the glucose metabolic pathways mediated by HIF1α downstream of mTORC1 (64, 121). Conversely, deletion of PTEN, which increases cellular PI(3,4,5)P3 and can drive the activation of AKT, is not sufficient to activate mTORC1 and mTORC1-dependent glucose metabolism (141). Together, these data demonstrate that IL-2 can activate and sustain mTORC1 activity without the need for activation of PI3K or AKT.
In terms of understanding the link between IL-2 signaling and the accumulation of PI(3,4,5)P3 levels in CTLs, direct measurements of cellular lipids has confirmed that PI(3,4,5)P3 levels decrease in IL-2-maintained CTLs following PI3K-p110δ inhibition (55). However, no measurable effect on PI(3,4,5)P3 levels was detected following treatment of IL-2-maintained CTLs with a JAK1/3 inhibitor or in response to IL-2 removal. It was also clear that IL-2-JAK1/3 activity did not regulate FOXO1 phosphorylation and nuclear exclusion in CTLs substantially (55). Rather, cellular levels of PI(3,4,5)P3, the activity of AKT, and the phosphorylation and nuclear exclusion of FOXO transcription factors are controlled by IL-2-independent signaling networks mediated by SRC family kinases LCK and FYN (55). Although LCK activity is associated with antigen receptor signaling in T cells, the activity of LCK is constitutive and not controlled by antigen receptor occupancy (142). Thus, IL-2-stimulated effector T cells have high levels of activity of the SRC family kinases p56LCK and FYN, but the activity of FYN and LCK is intrinsic and is not controlled by IL-2. Hence, while the original IL-2 signal transduction models placed PI(3,4,5)P3-AKT pathways downstream of IL-2R and JAKs, these models need to be revised to include our new understanding that the PI(3,4,5)P3-AKT pathway may be maintained in T cells independently from IL-2-JAK1/3 signaling (Figure 4). Indeed, such findings may explain why AKT activity is not maintained or induced in all IL-2-dependent T cell subsets (143). This key revision of IL-2 signal transduction highlights that IL-2 signals within a context of other “preorganized” signal transduction pathways (Figure 4). Indeed, the role of these preexisting pathways is increasingly appreciated. Another kinase that, like LCK and FYN, is constitutively active in CTLs is GSK3. The activity of GSK3 controls expression of MYC in IL-2-maintained CTLs (144). Similarly, the nuclear exclusion of the chromatin regulator of HDAC7 is determined by the constitutive activity of serine/threonine kinases (145). Thus, the output of IL-2 signaling in individual T cell subsets may be defined by extrinsic factors including the expression levels and activity of constitutively active kinases.
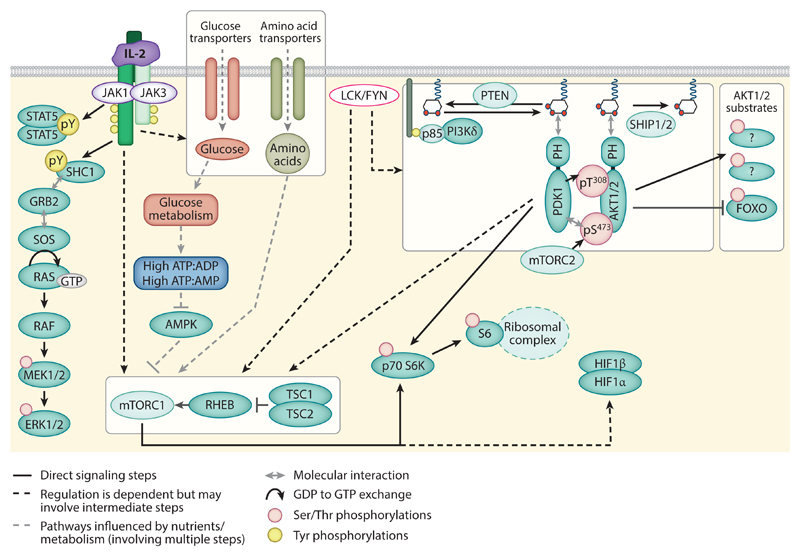
Figure 4.
Revised model for IL-2 signal transduction: IL-2 signaling outputs are likely to be modulated by other cellular signals, such as “preorganized” phosphorylation networks that do not depend on IL-2. Constitutive LCK/FYN activity has the potential to influence the phosphorylation of a number of signaling molecules in T cells and may be a key controller of the accumulation of PI(3,4,5)P3 in IL-2-dependent T cells. Levels of PI(3,4,5)P3 in cells depend on the balance in activity of phosphatidylinositol 3-kinase δ isoform (PI3Kδ or p110δ), which phosphorylates PI(4,5)P2 to generate PI(3,4,5)P3, and the PI(3,4,5)P3 phosphatases, PTEN and SHIP1/2. PI(3,4,5)P3 can coordinate the localization of a number of proteins, including the kinases phosphoinositide-dependent kinase 1 (PDK1) and AKT. Phosphorylation of T308 in the activation loop of AKT is induced by PDK1 and is coordinated by PI(3,4,5)P3 and mTORC2-mediated phosphorylation of AKT S473. Activated AKT may then go on to phosphorylate and inhibit FOXO family transcription factors, to influence transcriptional processes in T cells. However, PI3K-AKT activity in cytotoxic T lymphocytes is not a prerequisite for mTORC1 activity, and sustained activation of this complex is dependent on glucose metabolism and amino acid levels that can be regulated by IL-2 and environmental nutrient availability.
Defining IL-2 Signaling in Different T Cell Subsets
A defining characteristic of IL-2 is that its biological activities appear to be highly tailored to induce cell type–specific responses. Hence, a key role for IL-2 in Tregs is to support their differentiation and homeostasis, whereas in effector T cells IL-2 stimulates differentiation, proliferation, and cell growth. Moreover, these activities appear to be exclusive, as, for example, IL-2 does not induce rapid proliferation in Tregs (143). In this respect, there is much excitement around the concept of developing synthetically modified forms of IL-2 that can selectively regulate specific subsets of T cells, such as stimulating Tregs, but not effector cells, to treat autoimmunity.
Most studies that have dissected IL-2 signaling have been performed in antigen receptor–activated CD4+ or CD8+ T cells where IL-2 is a potent mitogen and growth factor. How, then, are the diverse signaling pathways that have been mapped to IL-2 signaling in these effector cells functioning in different T cell subsets? For example, as Tregs and effector cells have very different phenotypes, and since IL-2-activated signals, such as mTORC1, can have opposing impacts on their development, it is unlikely that IL-2 activates and sustains identical pathways in Tregs versus effector cells.
A possible answer is that IL-2 induces different signals in different T cell subsets. This could occur because different thresholds of IL-2 signals are received by cells depending on the concentrations of IL-2 in their environment and the duration for which cells are exposed to them. In vivo, the IL-2 gradients are extremely localized (146), so only certain cells will receive high concentrations of IL-2. This could dictate biological responses to IL-2, as early studies revealed that T cells can count the IL-2 signals that they receive. Thus, at least some IL-2 responses are quantal (all-or-none) rather than analog. For example, single-cell analysis of the DNA synthesis in response to a saturating dose of IL-2 compared to a half-saturating dose of IL-2 revealed that the higher concentration of IL-2 induced cell cycle progression more rapidly. Therefore, in situations where the IL-2 is limiting, it takes longer for the required number of activated receptors to accumulate (147–150).
Another likelihood is that signals induced by IL-2 will be modified by other signaling inputs. Many studies are now beginning to emphasize that IL-2 signaling does not act in isolation, but rather against a backdrop of other signaling pathways that can integrate with, and modulate, IL-2 responses. One factor that will influence this is the proteome of the T cell, such as the expression levels of kinases, phosphatases, adaptor proteins, and other signaling molecules that will influence signal transduction by IL-2. Examples of this include the levels of the lipid phosphatase PTEN, which has been shown to be a critical determinant of AKT signals in Tregs (143, 151), and the levels of LCK and FYN, which may influence how IL-2-dependent T cell subsets accumulate PI(3,4,5)P3. Similarly, other signals received by T cells may influence the IL-2 response. For example, Tregs require continuous signals from antigen receptors (152, 153). In addition, following immune challenge, the specific cytokine milieu induced by innate immune cells may influence the outcomes of IL-2 signaling responses.
One final consideration about IL-2 signaling is that it has largely been dissected based on understanding IL-2R-induced phosphorylation networks. However, other types of regulatory modifications on proteins may affect IL-2-mediated signaling pathways too. For example, recent studies in cell lines suggest that IL-2R occupancy can induce protein acetylation to regulate signaling (154). Additionally, the ability of IL-2 to potentiate glucose and glutamine uptake in certain T cell subsets may also permit or stimulate the modification of proteins by O-GlcNAcylation. This could be important in defining phosphorylation events, as O-GlcNAc modifies proteins on serine and threonine residues. For example, O-GlcNacylation of MYC on T58 blocks the phosphorylation that licenses MYC for degradation. Thus, O-GlcNAcylation was found to be important for stabilized MYC expression in CTLs (155). In this respect, another factor that may shape IL-2 signaling responses is microenvironmental nutrient levels, such as glucose and amino acids. Similarly, environmental oxygen levels, through modulating protein proline hydroxylation (156), may add a further level of regulation to IL-2 signals. Therefore, a key objective, and challenge, for further understanding IL-2 biology will be to determine how IL-2 signaling may be influenced by other immunoregulatory factors.
Acknowledgments
This work was supported by the Wellcome Trust (Principal Research Fellowship 097418/Z/11/Z and 205023/Z/16/Z to D.A.C.) and Tenovus Scotland (S.H.R.). We thank other members of the Cantrell group for critical reading of the manuscript.
Footnotes
Disclosure Statement
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Literature Cited
- 1.Morgan DA, Ruscetti FW, Gallo R. Selective in vitro growth of T lymphocytes from normal human bone marrows. Science. 1976;193(4257):1007–8. doi: 10.1126/science.181845. [DOI] [PubMed] [Google Scholar]
- 2.Gillis S, Smith KA. Long term culture of tumour-specific cytotoxic T cells. Nature. 1977;268(5616):154–56. doi: 10.1038/268154a0. [DOI] [PubMed] [Google Scholar]
- 3.Gillis S, Baker PE, Ruscetti FW, Smith KA. Long-term culture of human antigen-specific cytotoxic T-cell lines. J Exp Med. 1978;148(4):1093–98. doi: 10.1084/jem.148.4.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith KA. Interleukin-2: inception, impact, and implications. Science. 1988;240(4856):1169–76. doi: 10.1126/science.3131876. [DOI] [PubMed] [Google Scholar]
- 5.Cantrell DA, Smith KA. Transient expression of interleukin 2 receptors: consequences for T cell growth. J Exp Med. 1983;158(6):1895–911. doi: 10.1084/jem.158.6.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meuer SC, Hussey RE, Cantrell DA, Hodgdon JC, Schlossman SF, et al. Triggering of the T3-Ti antigen-receptor complex results in clonal T-cell proliferation through an interleukin 2-dependent autocrine pathway. PNAS. 1984;81(5):1509–13. doi: 10.1073/pnas.81.5.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robb RJ, Smith KA. Heterogeneity of human T-cell growth factor(s) due to variable glycosylation. Mol Immunol. 1981;18(12):1087–94. doi: 10.1016/0161-5890(81)90024-9. [DOI] [PubMed] [Google Scholar]
- 8.Robb RJ, Kutny RM, Chowdhry V. Purification and partial sequence analysis of human T-cell growth factor. PNAS. 1983;80(19):5990–94. doi: 10.1073/pnas.80.19.5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith KA, Cantrell DA. Interleukin 2 regulates its own receptors. PNAS. 1985;82(3):864–68. doi: 10.1073/pnas.82.3.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leonard WJ, Depper JM, Crabtree GR, Rudikoff S, Pumphrey J, et al. Molecular cloning and expression of cDNAs for the human interleukin-2 receptor. Nature. 1984;311(5987):626–31. doi: 10.1038/311626a0. [DOI] [PubMed] [Google Scholar]
- 11.Nikaido T, Shimizu A, Ishida N, Sabe H, Teshigawara K, et al. Molecular cloning of cDNA encoding human interleukin-2 receptor. Nature. 1984;311(5987):631–35. doi: 10.1038/311631a0. [DOI] [PubMed] [Google Scholar]
- 12.Sharon M, Klausner R, Cullen B, Chizzonite R, Leonard W. Novel interleukin-2 receptor subunit detected by cross-linking under high-affinity conditions. Science. 1986;234(4778):859–63. doi: 10.1126/science.3095922. [DOI] [PubMed] [Google Scholar]
- 13.Sugamura K, Takeshita T, Asao H, Kumaki S, Ohbo K, et al. The IL-2/IL-2 receptor system: involvement of a novel receptor subunit, gamma chain, in growth signal transduction. Tohoku J Exp Med. 1992;168(2):231–37. doi: 10.1620/tjem.168.231. [DOI] [PubMed] [Google Scholar]
- 14.Takeshita T, Asao H, Ohtani K, Ishii N, Kumaki S, et al. Cloning of the gamma chain of the human IL-2 receptor. Science. 1992;257(5068):379–82. doi: 10.1126/science.1631559. [DOI] [PubMed] [Google Scholar]
- 15.Liao W, Lin J-X, Leonard WJ. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity. 2013;38(1):13–25. doi: 10.1016/j.immuni.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feinerman O, Jentsch G, Tkach KE, Coward JW, Hathorn MM, et al. Single-cell quantification of IL-2 response by effector and regulatory T cells reveals critical plasticity in immune response. Mol Syst Biol. 2010;6:437. doi: 10.1038/msb.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hukelmann JL, Anderson KE, Sinclair LV, Grzes KM, Murillo AB, et al. The cytotoxic T cell proteome and its shaping by the kinase mTOR. Nat Immunol. 2016;17(1):104–12. doi: 10.1038/ni.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by γc family cytokines. Nat Rev Immunol. 2009;9(7):480–90. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noguchi M, Yi H, Rosenblatt HM, Filipovich AH, Adelstein S, et al. Interleukin-2 receptor gamma chain mutation results in X-linked severe combined immunodeficiency in humans. Cell. 1993;73(1):147–57. doi: 10.1016/0092-8674(93)90167-o. [DOI] [PubMed] [Google Scholar]
- 20.Macian F. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol. 2005;5(6):472–84. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- 21.Schorle H, Holtschke T, Hünig T, Schimpl A, Horak I. Development and function of T cells in mice rendered interleukin-2 deficient by gene targeting. Nature. 1991;352(6336):621–24. doi: 10.1038/352621a0. [DOI] [PubMed] [Google Scholar]
- 22.Sadlack B, Merz H, Schorle H, Schimpl A, Feller AC, Horak I. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell. 1993;75(2):253–61. doi: 10.1016/0092-8674(93)80067-o. [DOI] [PubMed] [Google Scholar]
- 23.Willerford DM, Chen J, Ferry JA, Davidson L, Ma A. Interleukin-2 receptor α chain regulates the size and content of the peripheral lymphoid compartment. Immunity. 1995;3(4):521–30. doi: 10.1016/1074-7613(95)90180-9. [DOI] [PubMed] [Google Scholar]
- 24.Sharfe N, Dadi HK, Shahar M, Roifman CM. Human immune disorder arising from mutation of the alpha chain of the interleukin-2 receptor. PNAS. 1997;94(7):3168–71. doi: 10.1073/pnas.94.7.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gregersen PK, Olsson LM. Recent advances in the genetics of autoimmune disease. Annu Rev Immunol. 2009;27:363–91. doi: 10.1146/annurev.immunol.021908.132653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Todd JA. Etiology of type 1 diabetes. Immunity. 2010;32(4):457–67. doi: 10.1016/j.immuni.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Fontenot JD, Rasmussen JP, Gavin MA, Rudensky AY. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat Immunol. 2005;6(11):1142–51. doi: 10.1038/ni1263. [DOI] [PubMed] [Google Scholar]
- 28.Cheng G, Yu A, Malek TR. T-cell tolerance and the multi-functional role of IL-2R signaling in T-regulatory cells. Immunol Rev. 2011;241(1):63–76. doi: 10.1111/j.1600-065X.2011.01004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rudensky AY. Regulatory T cells and Foxp3. Immunol Rev. 2011;241(1):260–68. doi: 10.1111/j.1600-065X.2011.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malek TR. The biology of interleukin-2. Annu Rev Immunol. 2008;26:453–79. doi: 10.1146/annurev.immunol.26.021607.090357. [DOI] [PubMed] [Google Scholar]
- 31.Cote-Sierra J, Foucras G, Guo L, Chiodetti L, Young HA, et al. Interleukin 2 plays a central role in Th2 differentiation. PNAS. 2004;101(11):3880–85. doi: 10.1073/pnas.0400339101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26(3):371–81. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 33.Kalia V, Sarkar S, Subramaniam S, Haining WN, Smith KA, Ahmed R. Prolonged interleukin-2Rα expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity. 2010;32(1):91–103. doi: 10.1016/j.immuni.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 34.Pipkin ME, Sacks JA, Cruz-Guilloty F, Lichtenheld MG, Bevan MJ, Rao A. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 2010;32(1):79–90. doi: 10.1016/j.immuni.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao W, Lin J-X, Leonard WJ. IL-2 family cytokines: new insights into the complex roles of IL-2 as a broad regulator of T helper cell differentiation. Curr Opin Immunol. 2011;23(5):598–604. doi: 10.1016/j.coi.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao W, Lin J-X, Wang L, Li P, Leonard WJ. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat Immunol. 2011;12(6):551–59. doi: 10.1038/ni.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ballesteros-Tato A, León B, Graf BA, Moquin A, Adams PS, et al. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity. 2012;36(5):847–56. doi: 10.1016/j.immuni.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyman O, Sprent J. The role of interleukin-2 during homeostasis and activation of the immune system. Nat Rev Immunol. 2012;12(3):180–90. doi: 10.1038/nri3156. [DOI] [PubMed] [Google Scholar]
- 39.O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327(5969):1098–102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325(5943):1006–10. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao W, Schones DE, Oh J, Cui Y, Cui K, et al. Priming for T helper type 2 differentiation by interleukin 2-mediated induction of interleukin 4 receptor α-chain expression. Nat Immunol. 2008;9(11):1288–96. doi: 10.1038/ni.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang X-P, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, et al. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol. 2011;12(3):247–54. doi: 10.1038/ni.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ray JP, Staron MM, Shyer JA, Ho P-C, Marshall HD, et al. The interleukin-2-mTORC1 kinase axis defines the signaling, differentiation, and metabolism of T helper 1 and follicular B helper T cells. Immunity. 2015;43(4):690–702. doi: 10.1016/j.immuni.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hinrichs CS, Spolski R, Paulos CM, Gattinoni L, Kerstann KW, et al. IL-2 and IL-21 confer opposing differentiation programs to CD8+ T cells for adoptive immunotherapy. Blood. 2008;111(11):5326–33. doi: 10.1182/blood-2007-09-113050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gong D, Malek TR. Cytokine-dependent Blimp-1 expression in activated T cells inhibits IL-2 production. J Immunol. 2007;178(1):242–52. doi: 10.4049/jimmunol.178.1.242. [DOI] [PubMed] [Google Scholar]
- 46.Oestreich KJ, Huang AC, Weinmann AS. The lineage-defining factors T-bet and Bcl-6 collaborate to regulate Th1 gene expression patterns. J Exp Med. 2011;208(5):1001–13. doi: 10.1084/jem.20102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oestreich KJ, Mohn SE, Weinmann AS. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat Immunol. 2012;13(4):405–11. doi: 10.1038/ni.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xue H-H, Kovanen PE, Pise-Masison CA, Berg M, Radovich MF, et al. IL-2 negatively regulates IL-7 receptor α chain expression in activated T lymphocytes. PNAS. 2002;99(21):13759–64. doi: 10.1073/pnas.212214999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reem GH, Yeh NH. Interleukin 2 regulates expression of its receptor and synthesis of gamma interferon by human T lymphocytes. Science. 1984;225(4660):429–30. doi: 10.1126/science.6429853. [DOI] [PubMed] [Google Scholar]
- 50.Kasahara T, Hooks JJ, Dougherty SF, Oppenheim JJ. Interleukin 2-mediated immune interferon (IFN-gamma) production by human T cells and T cell subsets. J Immunol. 1983;130(4):1784–89. [PubMed] [Google Scholar]
- 51.Lin J-X, Li P, Liu D, Jin H-T, He J, et al. Critical role of STAT5 transcription factor tetramerization for cytokine responses and normal immune function. Immunity. 2012;36(4):586–99. doi: 10.1016/j.immuni.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janas ML, Groves P, Kienzle N, Kelso A. IL-2 regulates perforin and granzyme gene expression in CD8+ T cells independently of its effects on survival and proliferation. J Immunol. 2005;175(12):8003–10. doi: 10.4049/jimmunol.175.12.8003. [DOI] [PubMed] [Google Scholar]
- 53.Cornish GH, Sinclair LV, Cantrell DA. Differential regulation of T-cell growth by IL-2 and IL-15. Blood. 2006;108(2):600–8. doi: 10.1182/blood-2005-12-4827. [DOI] [PubMed] [Google Scholar]
- 54.Sinclair LV, Rolf J, Emslie E, Shi Y-B, Taylor PM, Cantrell DA. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat Immunol. 2013;14(5):500–8. doi: 10.1038/ni.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ross SH, Rollings C, Anderson KE, Hawkins PT, Stephens LR, Cantrell DA. Phosphoproteomic analyses of interleukin 2 signaling reveal integrated JAK kinase-dependent and -independent networks in CD8+ T cells. Immunity. 2016;45(3):685–700. doi: 10.1016/j.immuni.2016.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manjunath N, Shankar P, Wan J, Weninger W, Crowley MA, et al. Effector differentiation is not prerequisite for generation of memory cytotoxic T lymphocytes. J Clin Investig. 2001;108(6):871–78. doi: 10.1172/JCI13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feau S, Arens R, Togher S, Schoenberger SP. Autocrine IL-2 is required for secondary population expansion of CD8+ memory T cells. Nat Immunol. 2011;12(9):908–13. doi: 10.1038/ni.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sa Q, Woodward J, Suzuki Y. IL-2 produced by CD8+ immune T cells can augment their IFN-γ production independently from their proliferation in the secondary response to an intracellular pathogen. J Immunol. 2013;190(5):2199–207. doi: 10.4049/jimmunol.1202256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Graves JD, Downward J, Izquierdo Pastor M, Rayter S, Warne PH, Cantrell DA. The growth factor IL-2 activates p21ras proteins in normal human T lymphocytes. J Immunol. 1992;148(8):2417–22. [PubMed] [Google Scholar]
- 60.Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nat Rev Mol Cell Biol. 2004;5(11):875–85. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- 61.Kuo CJ, Chung J, Fiorentino DF, Flanagan WM, Blenis J, Crabtree GR. Rapamycin selectively inhibits interleukin-2 activation of p70 S6 kinase. Nature. 1992;358(6381):70–73. doi: 10.1038/358070a0. [DOI] [PubMed] [Google Scholar]
- 62.Brennan P, Babbage JW, Burgering BM, Groner B, Reif K, Cantrell DA. Phosphatidylinositol 3-kinase couples the interleukin-2 receptor to the cell cycle regulator E2F. Immunity. 1997;7(5):679–89. doi: 10.1016/s1074-7613(00)80388-x. [DOI] [PubMed] [Google Scholar]
- 63.Reif K, Burgering BM, Cantrell DA. Phosphatidylinositol 3-kinase links the interleukin-2 receptor to protein kinase B and p70 S6 kinase. J Biol Chem. 1997;272(22):14426–33. doi: 10.1074/jbc.272.22.14426. [DOI] [PubMed] [Google Scholar]
- 64.Finlay DK, Rosenzweig E, Sinclair LV, Feijoo-Carnero C, Hukelmann JL, et al. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J Exp Med. 2012;209(13):2441–53. doi: 10.1084/jem.20112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Delespine-Carmagnat M, Bouvier G, Allée G, Fagard R, Bertoglio J. Biochemical analysis of interleukin-2 receptor β chain phosphorylation by p56lck. FEBS Lett. 1999;447(2–3):241–46. doi: 10.1016/s0014-5793(99)00301-4. [DOI] [PubMed] [Google Scholar]
- 66.Hatakeyama M, Kono T, Kobayashi N, Kawahara A, Levin SD, et al. Interaction of the IL-2 receptor with the src-family kinase p56lck: identification of novel intermolecular association. Science. 1991;252(5012):1523–28. doi: 10.1126/science.2047859. [DOI] [PubMed] [Google Scholar]
- 67.Johnston JA, Kawamura M, Kirken RA, Chen YQ, Blake TB, et al. Phosphorylation and activation of the Jak-3 Janus kinase in response to interleukin-2. Nature. 1994;370(6485):151–53. doi: 10.1038/370151a0. [DOI] [PubMed] [Google Scholar]
- 68.Russell SM, Johnston JA, Noguchi M, Kawamura M, Bacon CM, et al. Interaction of IL-2R beta and gamma c chains with Jak1 and Jak3: implications for XSCID and XCID. Science. 1994;266(5187):1042–45. doi: 10.1126/science.7973658. [DOI] [PubMed] [Google Scholar]
- 69.Witthuhn BA, Silvennoinen O, Miura O, Lai KS, Cwik C, et al. Involvement of the Jak-3 Janus kinase in signalling by interleukins 2 and 4 in lymphoid and myeloid cells. Nature. 1994;370(6485):153–57. doi: 10.1038/370153a0. [DOI] [PubMed] [Google Scholar]
- 70.Suzuki K. Janus kinase 3 (Jak3) is essential for common cytokine receptor γ chain (γc)-dependent signaling: comparative analysis of γc, Jak3, and γc and Jak3 double-deficient mice. Int Immunol. 2000;12(2):123–32. doi: 10.1093/intimm/12.2.123. [DOI] [PubMed] [Google Scholar]
- 71.Miyazaki T, Kawahara A, Fujii H, Nakagawa Y, Minami Y, et al. Functional activation of Jak1 and Jak3 by selective association with IL-2 receptor subunits. Science. 1994;266(5187):1045–47. doi: 10.1126/science.7973659. [DOI] [PubMed] [Google Scholar]
- 72.O’Shea JJ, Kontzias A, Yamaoka K, Tanaka Y, Laurence A. Janus kinase inhibitors in autoimmune diseases. Ann Rheum Dis. 2013;72(Suppl. 2):ii111–15. doi: 10.1136/annrheumdis-2012-202576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.O’Shea JJ, Schwartz DM, Villarino AV, Gadina M, McInnes IB, Laurence A. The JAK-STAT pathway: impact on human disease and therapeutic intervention. Annu Rev Med. 2015;66:311–28. doi: 10.1146/annurev-med-051113-024537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Macchi P, Villa A, Giliani S, Sacco MG, Frattini A, et al. Mutations of Jak-3 gene in patients with autosomal severe combined immune deficiency (SCID) Nature. 1995;377(6544):65–68. doi: 10.1038/377065a0. [DOI] [PubMed] [Google Scholar]
- 75.Haan C, Rolvering C, Raulf F, Kapp M, Drückes P, et al. Jak1 has a dominant role over Jak3 in signal transduction through γc-containing cytokine receptors. Chem Biol. 2011;18(3):314–23. doi: 10.1016/j.chembiol.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 76.Beadling C, Guschin D, Witthuhn BA, Ziemiecki A, Ihle JN, et al. Activation of JAK kinases and STAT proteins by interleukin-2 and interferon alpha, but not the T cell antigen receptor, in human T lymphocytes. EMBO J. 1994;13(23):5605–15. doi: 10.1002/j.1460-2075.1994.tb06898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Johnston JA, Bacon CM, Finbloom DS, Rees RC, Kaplan D, et al. Tyrosine phosphorylation and activation of STAT5, STAT3, and Janus kinases by interleukins 2 and 15. PNAS. 1995;92(19):8705–9. doi: 10.1073/pnas.92.19.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beadling C, Ng J, Babbage JW, Cantrell DA. Interleukin-2 activation of STAT5 requires the convergent action of tyrosine kinases and a serine/threonine kinase pathway distinct from the Raf1/ERK2 MAP kinase pathway. EMBO J. 1996;15(8):1902–13. [PMC free article] [PubMed] [Google Scholar]
- 79.Clark DE, Williams CC, Duplessis TT, Moring KL, Notwick AR, et al. ERBB4/HER4 potentiates STAT5A transcriptional activity by regulating novel STAT5A serine phosphorylation events. J Biol Chem. 2005;280(25):24175–80. doi: 10.1074/jbc.M414044200. [DOI] [PubMed] [Google Scholar]
- 80.Ng J, Cantrell D. STAT3 is a serine kinase target in T lymphocytes: Interleukin 2 and T cell antigen receptor signals converge upon serine 727. J Biol Chem. 1997;272(39):24542–49. doi: 10.1074/jbc.272.39.24542. [DOI] [PubMed] [Google Scholar]
- 81.Lin JX, Leonard WJ. The role of Stat5a and Stat5b in signaling by IL-2 family cytokines. Oncogene. 2000;19(21):2566–76. doi: 10.1038/sj.onc.1203523. [DOI] [PubMed] [Google Scholar]
- 82.Malek TR, Castro I. Interleukin-2 receptor signaling: at the interface between tolerance and immunity. Immunity. 2010;33(2):153–65. doi: 10.1016/j.immuni.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Owen DL, Farrar MA. STAT5 and CD4+ T cell immunity. F1000Research. 2017;6:32. doi: 10.12688/f1000research.9838.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Snow JW, Abraham N, Ma MC, Herndier BG, Pastuszak AW, Goldsmith MA. Loss of tolerance and autoimmunity affecting multiple organs in STAT5A/5B-deficient mice. J Immunol. 2003;171(10):5042–50. doi: 10.4049/jimmunol.171.10.5042. [DOI] [PubMed] [Google Scholar]
- 85.Yao Z, Kanno Y, Kerenyi M, Stephens G, Durant L, et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109(10):4368–75. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA. IL-2 receptor β-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J Immunol. 2007;178(1):280–90. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- 87.Moriggl R, Topham DJ, Teglund S, Sexl V, McKay C, et al. Stat5 is required for IL-2-induced cell cycle progression of peripheral T cells. Immunity. 1999;10(2):249–59. doi: 10.1016/s1074-7613(00)80025-4. [DOI] [PubMed] [Google Scholar]
- 88.Cleary A, Nadeau K, Tu W, Hwa V, Dionis KY, et al. Decreased generation and function of CD4+CD25hi T regulatory cells in human STAT5b deficiency. Blood. 2005;106(11):768. (Abstr.) [Google Scholar]
- 89.Cohen AC, Nadeau KC, Tu W, Hwa V, Dionis K, et al. Cutting edge: Decreased accumulation and regulatory function of CD4+ CD25high T cells in human STAT5b deficiency. J Immunol. 2006;177(5):2770–74. doi: 10.4049/jimmunol.177.5.2770. [DOI] [PubMed] [Google Scholar]
- 90.Kanai T, Seki S, Jenks JA, Kohli A, Kawli T, et al. Identification of STAT5A and STAT5B target genes in human T cells. PLOS ONE. 2014;9(1):e86790. doi: 10.1371/journal.pone.0086790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rani A, Afzali B, Kelly A, Tewolde-Berhan L, Hackett M, et al. IL-2 regulates expression of C-MAF in human CD4 T cells. J Immunol. 2011;187(7):3721–29. doi: 10.4049/jimmunol.1002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Basham B, Sathe M, Grein J, McClanahan T, D’Andrea A, et al. In vivo identification of novel STAT5 target genes. Nucleic Acids Res. 2008;36(11):3802–18. doi: 10.1093/nar/gkn271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Walker SR, Nelson EA, Frank DA. STAT5 represses BCL6 expression by binding to a regulatory region frequently mutated in lymphomas. Oncogene. 2007;26(2):224–33. doi: 10.1038/sj.onc.1209775. [DOI] [PubMed] [Google Scholar]
- 94.Villarino A, Laurence A, Robinson GW, Bonelli M, Dema B, et al. Signal transducer and activator of transcription 5 (STAT5) paralog dose governs T cell effector and regulatory functions. eLife. 2016;5:e08384. doi: 10.7554/eLife.08384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hand TW, Cui W, Jung YW, Sefik E, Joshi NS, et al. Differential effects of STAT5 and PI3K/AKT signaling on effector and memory CD8 T-cell survival. PNAS. 2010;107(38):16601–6. doi: 10.1073/pnas.1003457107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ravichandran KS, Burakoff SJ. The adapter protein Shc interacts with the interleukin-2 (IL-2) receptor upon IL-2 stimulation. J Biol Chem. 1994;269(3):1599–1602. [PubMed] [Google Scholar]
- 97.Reif K, Buday L, Downward J, Cantrell DA. SH3 domains of the adapter molecule Grb2 complex with two proteins in T cells: the guanine nucleotide exchange protein Sos and a 75-kDa protein that is a substrate for T cell antigen receptor-activated tyrosine kinases. J Biol Chem. 1994;269(19):14081–87. [PubMed] [Google Scholar]
- 98.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168(6):960–76. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, et al. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30(6):832–44. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12(4):295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee K, Gudapati P, Dragovic S, Spencer C, Joyce S, et al. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity. 2010;32(6):743–53. doi: 10.1016/j.immuni.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Powell JD, Pollizzi KN, Heikamp EB, Horton MR. Regulation of immune responses by mTOR. Annu Rev Immunol. 2012;30(1):39–68. doi: 10.1146/annurev-immunol-020711-075024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sinclair LV, Finlay D, Feijoo C, Cornish GH, Gray A, et al. Phosphatidylinositol-3-OH kinase and nutrient-sensing mTOR pathways control T lymphocyte trafficking. Nat Immunol. 2008;9(5):513–21. doi: 10.1038/ni.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rolf J, Zarrouk M, Finlay DK, Foretz M, Viollet B, Cantrell DA. AMPKα1: a glucose sensor that controls CD8 T-cell memory. Eur J Immunol. 2013;43(4):889–96. doi: 10.1002/eji.201243008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hardie DG. AMPK—sensing energy while talking to other signaling pathways. Cell Metab. 2014;20(6):939–52. doi: 10.1016/j.cmet.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Osinalde N, Moss H, Arrizabalaga O, Omaetxebarria MJ, Blagoev B, et al. Interleukin-2 signaling pathway analysis by quantitative phosphoproteomics. J Proteom. 2011;75(1):177–91. doi: 10.1016/j.jprot.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 107.Osinalde N, Sanchez-Quiles V, Akimov V, Blagoev B, Kratchmarova I. SILAC-based quantification of changes in protein tyrosine phosphorylation induced by interleukin-2 (IL-2) and IL-15 in T-lymphocytes. Data Brief. 2015;5:53–58. doi: 10.1016/j.dib.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Osinalde N, Sanchez-Quiles V, Akimov V, Guerra B, Blagoev B, Kratchmarova I. Simultaneous dissection and comparison of IL-2 and IL-15 signaling pathways by global quantitative phosphoproteomics. Proteomics. 2015;15(2–3):520–31. doi: 10.1002/pmic.201400194. [DOI] [PubMed] [Google Scholar]
- 109.Osinalde N, Mitxelena J, Sanchez-Quiles V, Akimov V, Aloria K, et al. Nuclear phosphoproteomic screen uncovers ACLY as mediator of IL-2-induced proliferation of CD4+ T lymphocytes. Mol Cell Proteom. 2016;15(6):2076–92. doi: 10.1074/mcp.M115.057158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Arneja A, Johnson H, Gabrovsek L, Lauffenburger DA, White FM. Qualitatively different T cell phenotypic responses to IL-2 versus IL-15 are unified by identical dependences on receptor signal strength and duration. J Immunol. 2014;192(1):123–35. doi: 10.4049/jimmunol.1302291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Arnaud M, Mzali R, Gesbert F, Crouin C, Guenzi C, et al. Interaction of the tyrosine phosphatase SHP-2 with Gab2 regulates Rho-dependent activation of the c-fos serum response element by interleukin-2. Biochem J. 2004;382(Pt 2):545–56. doi: 10.1042/BJ20040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Adachi M, Ishino M, Torigoe T, Minami Y, Matozaki T, et al. Interleukin-2 induces tyrosine phosphorylation of SHP-2 through IL-2 receptor beta chain. Oncogene. 1997;14(13):1629–33. doi: 10.1038/sj.onc.1200981. [DOI] [PubMed] [Google Scholar]
- 113.Lu W, Gong D, Bar-Sagi D, Cole PA. Site-specific incorporation of a phosphotyrosine mimetic reveals a role for tyrosine phosphorylation of SHP-2 in cell signaling. Mol Cell. 2001;8(4):759–69. doi: 10.1016/s1097-2765(01)00369-0. [DOI] [PubMed] [Google Scholar]
- 114.Gadina M, Stancato LM, Bacon CM, Larner AC, O’Shea JJ. Involvement of SHP-2 in multiple aspects of IL-2 signaling: evidence for a positive regulatory role. J Immunol. 1998;160(10):4657–61. [PubMed] [Google Scholar]
- 115.Migone TS, Cacalano NA, Taylor N, Yi T, Waldmann TA, Johnston JA. Recruitment of SH2-containing protein tyrosine phosphatase SHP-1 to the interleukin 2 receptor; loss of SHP-1 expression in human T-lymphotropic virus type I-transformed T cells. PNAS. 1998;95(7):3845–50. doi: 10.1073/pnas.95.7.3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Linossi EM, Babon JJ, Hilton DJ, Nicholson SE. Suppression of cytokine signaling: the SOCS perspective. Cytokine Growth Factor Rev. 2013;24(3):241–48. doi: 10.1016/j.cytogfr.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lucas CL, Chandra A, Nejentsev S, Condliffe AM, Okkenhaug K. PI3Kδ and primary immuno-deficiencies. Nat Rev Immunol. 2016;16(11):702–14. doi: 10.1038/nri.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cantrell DA. Phosphoinositide 3-kinase signalling pathways. J Cell Sci. 2001;114(Part 8):1439–45. doi: 10.1242/jcs.114.8.1439. [DOI] [PubMed] [Google Scholar]
- 119.Milburn CC, Deak M, Kelly SM, Price NC, Alessi DR, Van Aalten DMF. Binding of phosphatidylinositol 3,4,5-trisphosphate to the pleckstrin homology domain of protein kinase B induces a conformational change. Biochem J. 2003;375(Part 3):531–38. doi: 10.1042/BJ20031229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol. 2010;11(1):9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 121.Macintyre AN, Finlay D, Preston G, Sinclair LV, Waugh CM, et al. Protein kinase B controls transcriptional programs that direct cytotoxic T cell fate but is dispensable for T cell metabolism. Immunity. 2011;34(2):224–36. doi: 10.1016/j.immuni.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hedrick SM, Hess Michelini R, Doedens AL, Goldrath AW, Stone EL. FOXO transcription factors throughout T cell biology. Nat Rev Immunol. 2012;12(9):649–61. doi: 10.1038/nri3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hess Michelini R, Doedens AL, Goldrath AW, Hedrick SM. Differentiation of CD8 memory T cells depends on Foxo1. J Exp Med. 2013;210(6):1189–1200. doi: 10.1084/jem.20130392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Waugh C, Sinclair L, Finlay D, Bayascas JR, Cantrell D. Phosphoinositide (3,4,5)-triphosphate binding to phosphoinositide-dependent kinase 1 regulates a protein kinase B/Akt signaling threshold that dictates T-cell migration, not proliferation. Mol Cell Biol. 2009;29(21):5952–62. doi: 10.1128/MCB.00585-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lucas CL, Kuehn HS, Zhao F, Niemela JE, Deenick EK, et al. Dominant-activating germline mutations in the gene encoding the PI(3)K catalytic subunit p110δ result in T cell senescence and human immunodeficiency. Nat Immunol. 2014;15(1):88–97. doi: 10.1038/ni.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Angulo I, Vadas O, Garçon F, Banham-Hall E, Plagnol V, et al. Phosphoinositide 3-kinase δ gene mutation predisposes to respiratory infection and airway damage. Science. 2013;342(6160):866–71. doi: 10.1126/science.1243292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Okkenhaug K, Ali K, Vanhaesebroeck B. Antigen receptor signalling: a distinctive role for the p110δ isoform of PI3K. Trends Immunol. 2007;28(2):80–87. doi: 10.1016/j.it.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ward SG, Cantrell DA. Phosphoinositide 3-kinases in T lymphocyte activation. Curr Opin Immunol. 2001;13(3):332–38. doi: 10.1016/s0952-7915(00)00223-5. [DOI] [PubMed] [Google Scholar]
- 129.Truitt KE, Mills GB, Turck CW, Imboden JB. SH2-dependent association of phosphatidylinositol 3′-kinase 85-kDa regulatory subunit with the interleukin-2 receptor β chain. J Biol Chem. 1994;269(8):5937–43. [PubMed] [Google Scholar]
- 130.Migone TS, Rodig S, Cacalano NA, Berg M, Schreiber RD, Leonard WJ. Functional cooperation of the interleukin-2 receptor beta chain and Jak1 in phosphatidylinositol 3-kinase recruitment and phosphorylation. Mol Cell Biol. 1998;18(11):6416–22. doi: 10.1128/mcb.18.11.6416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gu H, Maeda H, Moon JJ, Lord JD, Yoakim M, et al. New role for Shc in activation of the phosphatidylinositol 3-kinase/Akt pathway. Mol Cell Biol. 2000;20(19):7109–20. doi: 10.1128/mcb.20.19.7109-7120.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Monfar M, Lemon KP, Grammer TC, Cheatham L, Chung J, et al. Activation of pp70/85 S6 kinases in interleukin-2-responsive lymphoid cells is mediated by phosphatidylinositol 3-kinase and inhibited by cyclic AMP. Mol Cell Biol. 1995;15(1):326–37. doi: 10.1128/mcb.15.1.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Stahl M, Dijkers PF, Kops GJPL, Lens SMA, Coffer PJ, et al. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J Immunol. 2002;168(10):5024–31. doi: 10.4049/jimmunol.168.10.5024. [DOI] [PubMed] [Google Scholar]
- 134.Brunn GJ, Williams J, Sabers C, Wiederrecht G, Lawrence JC, Abraham RT. Direct inhibition of the signaling functions of the mammalian target of rapamycin by the phosphoinositide 3-kinase inhibitors, wortmannin and LY294002. EMBO J. 1996;15(19):5256–67. [PMC free article] [PubMed] [Google Scholar]
- 135.Jacobs MD, Black J, Futer O, Swenson L, Hare B, et al. Pim-1 ligand-bound structures reveal the mechanism of serine/threonine kinase inhibition by LY294002. J Biol Chem. 2005;280(14):13728–34. doi: 10.1074/jbc.M413155200. [DOI] [PubMed] [Google Scholar]
- 136.Najafov A, Shpiro N, Alessi DR. Akt is efficiently activated by PIF-pocket- and PtdIns(3,4,5)P3-dependent mechanisms leading to resistance to PDK1 inhibitors. Biochem J. 2012;448(2):285–95. doi: 10.1042/BJ20121287. [DOI] [PubMed] [Google Scholar]
- 137.Clark J, Anderson KE, Juvin V, Smith TS, Karpe F, et al. Quantification of PtdInsP3 molecular species in cells and tissues by mass spectrometry. Nat Methods. 2011;8(3):267–72. doi: 10.1038/nmeth.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Costello PS, Gallagher M, Cantrell DA. Sustained and dynamic inositol lipid metabolism inside and outside the immunological synapse. Nat Immunol. 2002;3(11):1082–89. doi: 10.1038/ni848. [DOI] [PubMed] [Google Scholar]
- 139.Fabre S, Lang V, Harriague J, Jobart A, Unterman TG, et al. Stable activation of phosphatidylinositol 3-kinase in the T cell immunological synapse stimulates Akt signaling to FoxO1 nuclear exclusion and cell growth control. J Immunol. 2005;174(7):4161–71. doi: 10.4049/jimmunol.174.7.4161. [DOI] [PubMed] [Google Scholar]
- 140.Wu W-I, Voegtli WC, Sturgis HL, Dizon FP, Vigers GPA, Brandhuber BJ. Crystal structure of human AKT1 with an allosteric inhibitor reveals a new mode of kinase inhibition. PLOS ONE. 2010;5(9):e12913. doi: 10.1371/journal.pone.0012913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Grzes KM, Swamy M, Hukelmann JL, Emslie E, Sinclair LV, Cantrell DA. Control of amino acid transport coordinates metabolic reprogramming in T-cell malignancy. Leukemia. 2017;31:2771–79. doi: 10.1038/leu.2017.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nika K, Soldani C, Salek M, Paster W, Gray A, et al. Constitutively active Lck kinase in T cells drives antigen receptor signal transduction. Immunity. 2010;32(6):766–77. doi: 10.1016/j.immuni.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bensinger SJ, Walsh PT, Zhang J, Carroll M, Parsons R, et al. Distinct IL-2 receptor signaling pattern in CD4+CD25+ regulatory T cells. J Immunol. 2004;172(9):5287–96. doi: 10.4049/jimmunol.172.9.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Preston GC, Sinclair LV, Kaskar A, Hukelmann JL, Navarro MN, et al. Single cell tuning of Myc expression by antigen receptor signal strength and interleukin-2 in T lymphocytes. EMBO J. 2015;34(15):2008–24. doi: 10.15252/embj.201490252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Navarro MN, Goebel J, Feijoo-Carnero C, Morrice N, Cantrell DA. Phosphoproteomic analysis reveals an intrinsic pathway for the regulation of histone deacetylase 7 that controls the function of cytotoxic T lymphocytes. Nat Immunol. 2011;12(4):352–61. doi: 10.1038/ni.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Oyler-Yaniv A, Oyler-Yaniv J, Whitlock BM, Liu Z, Germain RN, et al. A tunable diffusion-consumption mechanism of cytokine propagation enables plasticity in cell-to-cell communication in the immune system. Immunity. 2017;46(4):609–20. doi: 10.1016/j.immuni.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cantrell DA, Smith KA. The interleukin-2 T-cell system: a new cell growth model. Science. 1984;224(4655):1312–16. doi: 10.1126/science.6427923. [DOI] [PubMed] [Google Scholar]
- 148.Smith KA. The quantal theory of how the immune system discriminates between “self and non-self”. Med Immunol. 2004;3(1):3. doi: 10.1186/1476-9433-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Smith KA. The structure of IL2 bound to the three chains of the IL2 receptor and how signaling occurs. Med Immunol. 2006;5:3. doi: 10.1186/1476-9433-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Smith KA. The quantal theory of immunity. Cell Res. 2006;16(1):11–19. doi: 10.1038/sj.cr.7310003. [DOI] [PubMed] [Google Scholar]
- 151.Zeiser R, Leveson-Gower DB, Zambricki EA, Kambham N, Beilhack A, et al. Differential impact of mammalian target of rapamycin inhibition on CD4+CD25+Foxp3+ regulatory T cells compared with conventional CD4+ T cells. Blood. 2007;111(1):453–62. doi: 10.1182/blood-2007-06-094482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Levine AG, Arvey A, Jin W, Rudensky AY. Continuous requirement for the TCR in regulatory T cell function. Nat Immunol. 2014;15(11):1070–78. doi: 10.1038/ni.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Vahl JC, Drees C, Heger K, Heink S, Fischer JC, et al. Continuous T cell receptor signals maintain a functional regulatory T cell pool. Immunity. 2014;41(5):722–36. doi: 10.1016/j.immuni.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 154.Kuwabara T, Kasai H, Kondo M. Acetylation modulates IL-2 receptor signaling in T cells. J Immunol. 2016;197(11):4334–43. doi: 10.4049/jimmunol.1601174. [DOI] [PubMed] [Google Scholar]
- 155.Swamy M, Pathak S, Grzes KM, Damerow S, Sinclair LV, et al. Glucose and glutamine fuel protein O-GlcNAcylation to control T cell self-renewal and malignancy. Nat Immunol. 2016;17(6):712–20. doi: 10.1038/ni.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ivan M, Kaelin WG. The EGLN-HIF O2-sensing system: multiple inputs and feedbacks. Mol Cell. 2017;66(6):772–79. doi: 10.1016/j.molcel.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]