Abstract
Disrupting protein-protein interaction for molecularly targeted cancer therapeutics can be a challenging but promising strategy. Compounds that disrupt the interaction between Menin, a chromatin-binding protein, and oncogenic MLL fusion proteins (FPs) have shown significant promise in pre-clinical models of leukemia and show a high degree of selectivity for leukemia versus normal hematopoietic cells. Biochemical and structural studies demonstrate that, in addition to disrupting Menin-MLL-FP interaction, such compounds also inhibit Menin-MLL1, Menin-MLL2, and other Menin interacting proteins. Here we address the degree to which disruption of Menin-MLL-FP interactions or Menin-MLL1/MLL2 interactions contribute to the anti-leukemia effect of Menin inhibition. We show that Men1 deletion in MLL-AF9-transformed leukemia cells produces distinct cellular and molecular consequences as compared to Mll1;Mll2 co-deletion, and that compounds disrupting Menin-MLL-N terminal interactions largely phenocopy Menin loss. Moreover, we show that Mll1;Mll2-deficient leukemia cells exhibit enhanced sensitivity to Menin-interaction inhibitors consistent with each regulating complementary genetic pathways. These data illustrate the heightened dependency of MLL-FPs on Menin, as compared to wild-type MLL1/MLL2 for regulation of downstream target genes, and argue that the predominant action of Menin inhibitory compounds is through direct inhibition of MLL-FPs without significant contribution from MLL1/MLL2 inhibition.
Introduction
Patients with chromosomal translocations involving the Mixed Lineage Leukemia 1 gene (MLL, MLL1, KMT2A) represent an exception to overall favorable outcomes for children with acute leukemia.1 Menin, encoded by the Men1 gene, is a tumor suppressor in neuroendocrine tissues but is essential for MLL1 fusion oncoprotein (MLL-FP)-mediated leukemogenesis. MLL-FP binding to Menin bridges an interaction with Lens Epithelium-Derived Growth Factor (LEDGF), which in turn binds histone H3 dimethyl lysine 36 (H3K36me2) modified chromatin.2,3 Menin also interacts with endogenous wild-type MLL1 and MLL2 2,4,5 with quantitative proteomics indicating nearly 1:1 stoichiometry of Menin with MLL1 and MLL2 complexes.6 Because of the essential nature of the Menin/LEDGF interaction for MLL-FPs to target to chromatin, small-molecule inhibitors have been developed that disrupt Menin binding to the N-terminus of MLL-FPs.7–11 Menin binds to MLL-FPs, MLL1, MLL2 and other proteins using the same pocket, hence small-molecule inhibitors may disrupt all of these interactions in cells. To clarify through which pathways inhibitors of the Menin-MLL N-terminus act, we compared cellular and molecular alterations in in MLL-AF9 transformed leukemia cells using genetic and pharmacologic manipulation of Menin, Mll1 and Mll2.
Methods
Mice and generation of MLL-AF9-transformed cells.
Lin-/Sca-1+/c-Kit+ (LSK) or c-Kit+ cells were sorted from Cre:ERT2;Mll1F/F;Mll2F/F and Cre:ERT2;Men1F/F mice and transduced with MSCV-MLL-AF9-YFP or MSCV-MLL-AF9-GFP (gifts from Drs. Scott Armstrong and Mick Milsom) as described.12 Transduced cells were replated in M3434 medium (StemCell Technologies) > 4 rounds to generate transformed cells.
Quantitative Real-Time PCR (qRT-PCR).
Procedures were as described12 using TaqMan Gene Expression Master Mix (Applied Biosystems) assays or primers as follows: Magohb: Applied Biosystems Mm01200054_m1.; Mef2c: Applied Biosystems Mm01340842_m1; Gapdh: Applied Biosystems 4308313; Meis1: GAGCAAGGTGATGGCTTGGA and TGTCCTTATCAGGGTCATCATCG; Meis1 Probe: AACAGTGTAGCTTCCCCCAGCACAGGT. The following primer pairs were fused with SYBR Green Supermix (Biorad): Pigp: TGCCCGTCTACCTCCTTATC and ATGGGGACATCTCTCAATGC. Jmjd1c: CACATTCTTGGATCTGTGACCA and ATGCTGTCTTTGCAGTTGAGG. Cdkn2c: AACCATCCCAGTCCTTCTGTCA and CCCCTTTCCTT TGCTCCTAATC. Il3ra: CTGGCATCCCACTCTTCAGAT and GGTCCCAGC TCAGTGTGTA.
RNA-sequencing (RNA-seq) and genomic analyses.
RNA-seq and Gene Set Enrichment Analysis (GSEA) were performed as described12 and the gene lists represented in the Venn diagrams and supplemental files represent data filtered for greater than 2-fold change and p<0.05. Diagrams and overlap lists were generated using BioVenn.13 The RNA-seq data reported in this article have been deposited at the NCBI Gene Expression Omnibus with the accession code GSE117933.
Cell proliferation and viability assays.
Proliferation was assessed by BrdU incorporation according to the manufacturer’s protocol using the APC BrdU Flow Kit (BD Biosciences) using a 30-minute incubation with BrdU. Cell viability was determined by propidium iodide (PI, Sigma-Aldrich) and Annexin V-APC (Biolegend) staining. Cre induction in leukemia cells was initiated in culture medium supplemented with 100 nM 4-OHT (Sigma-Aldrich). After 24 hours, 4-OHT was removed by exchanging the medium and cells were cultured for the time indicated in the Figure Legends. Both MI-2 (Cayman) and MI-2–2 were dissolved in DMSO. Three thousand cells in 200 μL were treated with MI-2 or MI-2–2 for 3 days.
Chemistry.
Chemical synthesis and chemical characterization of MI-2 and MI-2–2 compounds have been described previously.9,10
Results and Discussion
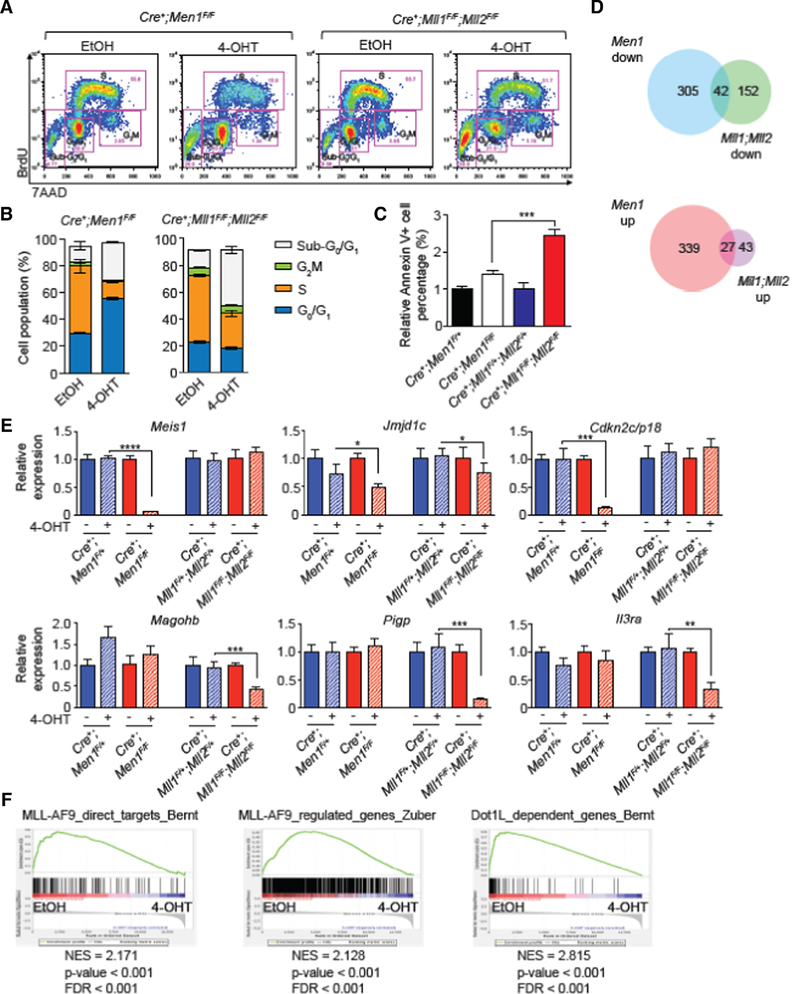
We recently showed that co-deletion of Mll1 and Mll2 in MLL-FP-transformed leukemia inhibited cell growth through modulation of several leukemia survival pathways.12 MLL-FPs have also been shown to directly regulate anti-apoptotic survival pathways.14 To deconvolute the contributions of MLL-FP versus MLL1/MLL2 inhibition that would occur with Menin inhibitors, we compared the effect of deleting Men1 to that of co-deletion of Mll1 and Mll2. We selected the time points for analysis based on complete gene deletion and loss of the corresponding transcript (observed by 48 hours, supplemental Figure 1A). A severe reduction in S-phase cells was observed 5 days after initiating Men1 deletion (from 56% to 16%) concomitant with a G0/G1 accumulation (Figure 1A-B). In contrast, Mll1;Mll2 deletion resulted in milder reduction in S-phase cells (from 56% to 32%) and much larger accumulation of sub-G0/G1 cells. The selective effect on cell cycle may be due to the fact that Men1 deletion affects expression of the cyclin-dependent kinases CDK4 and CDK6 and Mll1;Mll2 deletion does not (Figure 1A-B, supplemental Figure 1B). Mll1;Mll2-deleted cells exhibited increased Annexin V binding and PI permeability at day 3 which accumulated over time (Figure 1C and data not shown), suggesting that cell death plays a larger role in the growth inhibition observed upon co-deletion of Mll1 and Mll2.12 To broadly compare the molecular characteristics of Men1 versus Mll1;Mll2 deficiency, we performed side-by-side RNA-seq analysis in MLL-AF9-transformed cells at day 3 prior to the execution of cell cycle/cell death phenotypes. Men1 deletion resulted in both up- and down-regulated genes (366 and 347 genes, respectively, Figure 1D and Supplemental Table 1), while Mll1;Mll2 deletion resulted in fewer changes using the same criteria for data analysis (70 up-regulated and 194 down-regulated, Figure 1D). Comparison of Men1- versus Mll1;Mll2-deregulated genes showed minimal overlap of differentially expressed genes; only 12% of the down- and 7% of the up-regulated genes in Men1-deficient leukemia cells were shared with Mll1;Mll2-deficient cells (Figure 1D). We performed qRT-PCR validation in MLL-AF9 cells focusing on MLL-FP-regulated genes or those unique to the MLL1/MLL2 regulated pathways.12,15 The MLL-FP targets Meis1 and Jmjd1c were significantly and reproducibly down-regulated upon Men1 deletion, as was the Menin-regulated gene Cdkn2c/p18, whereas expression of these genes changed minimally or not at all upon Mll1;Mll2 deletion (Figure 1E and ref. 12). In contrast, MLL2-regulated genes Magohb, Pigp and Il3ra were down-regulated by Mll1;Mll2 deletion and not Men1 deletion (Figure 1E). Men1 deletion specifically reduced expression of MLL-AF9-bound, MLL-AF9 up-regulated, and Dot1L-dependent genes, consistent with its role as requisite binding partner of MLL-FPs (Figure 1F). In contrast, Mll1;Mll2 deletion did not significantly affect the same gene sets (see Figure 2A). These results illustrate that Men1 deletion specifically affects MLL-FP activity rather than the combined activity of MLL-FPs and endogenous MLL1/MLL2.15,16 Although loss of MLL1/MLL2 effectively kills MLL-AF9 cells, it apparently does so through distinct mechanisms and pathways as compared to Men1 deletion. MLL-FPs lack the C-terminal chromatin-targeting motifs of wild-type MLL1 and MLL2,17–19 which may result in a stronger dependency on the N-terminal Menin-LEDGF complex than wild-type MLL1 or MLL2.
Figure 1. Loss of Men1 versus Mll1;Mll2 shows different cellular and molecular effects in MLL-AF9-transformed cells.
A-B) Representative FACS plots (A) and quantification (B) showing BrdU incorporation and DNA content in Men1- and Mll1;Mll2-deficient MLL-AF9 cells. MLL-AF9-transformed cells were treated with ethanol or 4-OHT for 24 hours to induce the deletion of Men1 or Mll1;Mll2. After an additional 4 days, cells were incubated with BrdU for 30 minutes and were processed to detect BrdU and DNA content by flow cytometry. The apoptotic sub-G0/G1, G2M, S and G0/G1 gates are shown. C) Relative Annexin V+ cell percentage compared to control cells 3 days after initiating gene deletion. D) Venn diagram showing the overlap of down-regulated and up-regulated genes with Men1 and Mll1;Mll2 deletion. Differentially expressed genes (Table S1) were identified as described in Methods. E) qRT-PCR of select genes after Men1 or Mll1;Mll2 deletion in MLL-AF9-transformed cells. Gene expression was determined 3 days after initiating gene deletion. Data are represented as averages ± standard deviation (SD); *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. One representative experiment of five is shown. F) GSEA plots showing enrichment of MLL-AF9 direct binding targets or Dot1L-dependent genes16 and MLL-AF9-regulated genes15 in Men1-deficient (4-OHT) MLL-AF9-transformed cells.
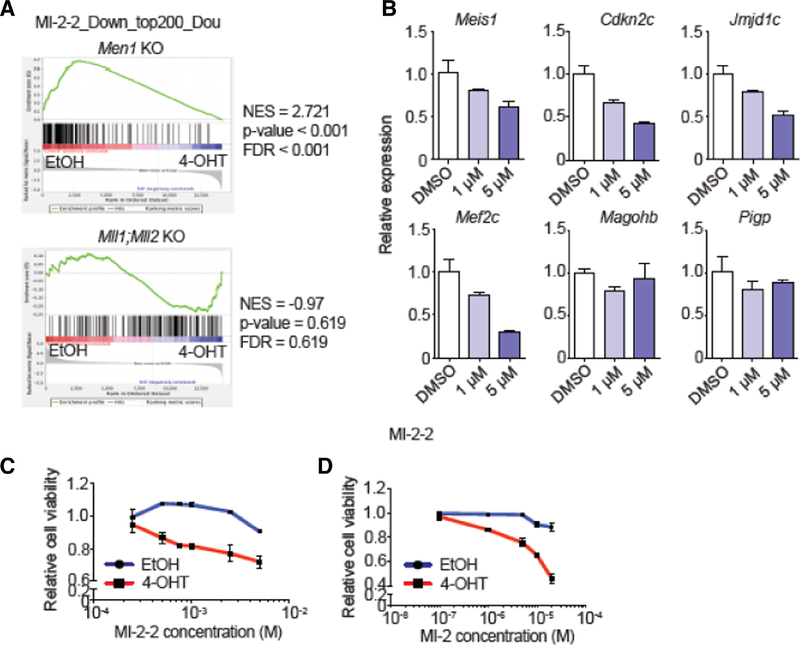
Figure 2. Menin inhibitors phenocopy Men1 deletion and collaborate with Mll1;Mll2 loss to kill leukemia cells.
A) GSEA plots showing enrichment of MI-2–2 regulated genes20 in Men1-deleted MLL-AF9 cells and in Mll1;Mll2-deleted MLL-AF9 cells. B) qRT-PCR of selected target genes after MI-2–2 treatment in MLL-AF9-transformed cells. Gene expression was determined at 72 hours post treatment. Data are represented as averages ± SD. C-D) Relative cell viability after and MI-2–2 (C) and MI-2 (D) treatment in MLL-AF9-transformed cells with or without Mll1;Mll2. MLL-AF9-transformed cells with the genotype of Cre+; Mll1F/F; Mll2F/F were treated with ethanol or 4-OHT for 24 hours, washed, then treated with compound or vehicle for 3 additional days. Cell viabilities were all normalized to corresponding vehicle (ethanol) control. Data are represented as mean ± SD. One representative of two independent experiments is shown.
Since Menin and MLL1/MLL2 regulate distinct pathways, we hypothesized that Mll1;Mll2 co-deletion would be complimentary to inhibition of the Menin-MLL-FP interaction. Thus, we tested the combined effect of Mll1;Mll2 deletion with the Menin-MLL1 small-molecule inhibitor MI-2 and its higher affinity variant MI-2–2.9,10 Comparing Men1 deletion to the published effect of MI-2–2 on murine MLL-AF9 cells,20 MI-2–2 down-regulated genes were significantly enriched in our Men1−/− AML data using GSEA (NES > 2.7, p < 0.001) whereas Mll1;Mll2 double knockout genes were not (NES < 1.5, p-value > 0.1; Figure 2A. Validation experiments using independently transformed MLL-AF9 cells showed that MI-2–2 inhibited expression of the direct MLL-FP target genes Meis1, Cdkn2c, Jmjd1c and Mef2c, but not MLL2 target genes (Figure 2B). We therefore tested the sensitivity of Mll1;Mll2-deleted, MLL-AF9 transformed cells to Menin inhibitors relative to the parental cells to determine whether inhibition of both pathways provided additional cell killing. Strikingly, Mll1;Mll2-deficient MLL-AF9-transformed cells showed over 10-fold increased sensitivity to both MI-2 and MI-2–2 (Figure 2C-D).9,10 Thus combined inhibition of Menin and MLL1/MLL2 more effectively kills MLL-rearranged leukemia cells, likely due to their effects on distinct genetic networks. These data also predict that targeting MLL1/MLL2 in combination with other MLL-FP directed strategies (eg. DOT1L inhibitors) may result in synergistic killing of leukemia cells, similar to the demonstration that targeting MLL-FPs with two different molecular inhibitors can produce synergistic effects.21
The observation that MLL-FPs are much more strongly dependent upon Menin genetically or upon Menin interaction (based on use of MI-2–2) demonstrates an interesting and perhaps unanticipated gained dependency by the fusion oncoprotein. This phenomenon may be attributed to the distinct multivalent chromatin interactions between MLL1 and is fusion oncoprotein derivatives, which may also be related to the observed spreading of both MLL-FPs and Menin into gene bodies in MLL-FP expressing cell lines.14,22 Our data begin to unravel the nature of this gained dependency, but further molecular characterization will be required to completely understand the basis for the apparently selective effect of Menin inhibitors on MLL-FPs, and how Menin inhibition can be combined with additional strategies to maximize the specificity and selectivity of its anti-leukemia activity.
Supplementary Material
Highlights:
Loss of Men1 has a greater impact on cell cycle progression than Mll1;Mll2 loss
Loss of Men1 in AML predominantly affects the MLL-AF9 driven leukemogenic program
Mll1;Mll2 deletion increases AML sensitivity to Menin inhibitors
Acknowledgments
We thank Kathrin Bernt for critical comments and our laboratory members for discussion and critical comments. This work was supported by N.I.H. grants HL090036, CA224436 to PE and CA046934, CA160467 to JG.
Footnotes
Disclosure of potential conflicts of interest
P.E. owns Amgen stock and has consulted for Servier Oncology. J.G. receives research support, has equity ownership and consults for Kura Oncology, Inc. Y.C. is employed by Seattle Genetics, Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pui CH et al. Childhood Acute Lymphoblastic Leukemia: Progress Through Collaboration. J Clin Oncol 33, 2938–2948, doi: 10.1200/JCO.2014.59.1636 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yokoyama A et al. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell 123, 207–218, doi:S0092-8674(05)00979-7 [pii] 10.1016/j.cell.2005.09.025 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Zhu L et al. ASH1L Links Histone H3 Lysine 36 Dimethylation to MLL Leukemia. Cancer Discov 6, 770–783, doi: 10.1158/2159-8290.CD-16-0058 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hughes CM et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell 13, 587–597 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Milne TA et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell 10, 1107–1117, doi:S1097276502007414 [pii] (2002). [DOI] [PubMed] [Google Scholar]
- 6.van Nuland R et al. Quantitative dissection and stoichiometry determination of the human SET1/MLL histone methyltransferase complexes. Mol Cell Biol 33, 2067–2077, doi: 10.1128/MCB.01742-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murai MJ et al. The same site on the integrase-binding domain of lens epithelium-derived growth factor is a therapeutic target for MLL leukemia and HIV. Blood 124, 3730–3737, doi: 10.1182/blood-2014-01-550079 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borkin D et al. Pharmacologic inhibition of the Menin-MLL interaction blocks progression of MLL leukemia in vivo. Cancer Cell 27, 589–602, doi: 10.1016/j.ccell.2015.02.016 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grembecka J et al. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat Chem Biol 8, 277–284, doi: 10.1038/nchembio.773 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi A et al. Structural insights into inhibition of the bivalent menin-MLL interaction by small molecules in leukemia. Blood 120, 4461–4469, doi: 10.1182/blood-2012-05-429274 blood-2012-05-429274 [pii] (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S et al. Design of the First-in-Class, Highly Potent Irreversible Inhibitor Targeting the Menin-MLL Protein-Protein Interaction. Angew Chem Int Ed Engl, doi: 10.1002/anie.201711828 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Chen Y et al. MLL2, Not MLL1, Plays a Major Role in Sustaining MLL-Rearranged Acute Myeloid Leukemia. Cancer Cell 31, 755–770 e756, doi: 10.1016/j.ccell.2017.05.002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hulsen T, de Vlieg J & Alkema W BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9, 488, doi: 10.1186/1471-2164-9-488 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerry J et al. MLL-AF4 Spreading Identifies Binding Sites that Are Distinct from Super-Enhancers and that Govern Sensitivity to DOT1L Inhibition in Leukemia. Cell Rep 18, 482–495, doi: 10.1016/j.celrep.2016.12.054 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuber J et al. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev 25, 1628–1640, doi: 10.1101/gad.17269211 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernt KM et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 20, 66–78, doi: 10.1016/j.ccr.2011.06.010 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ali M, Hom RA, Blakeslee W, Ikenouye L & Kutateladze TG Diverse functions of PHD fingers of the MLL/KMT2 subfamily. Biochim Biophys Acta 1843, 366–371, doi: 10.1016/j.bbamcr.2013.11.016 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milne TA et al. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis. Mol Cell 38, 853–863, doi: 10.1016/j.molcel.2010.05.011 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Couture JF, Collazo E & Trievel RC Molecular recognition of histone H3 by the WD40 protein WDR5. Nat Struct Mol Biol 13, 698–703, doi: 10.1038/nsmb1116 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Xu J et al. MLL1 and MLL1 fusion proteins have distinct functions in regulating leukemic transcription program. Cell Discov 2, 16008, doi: 10.1038/celldisc.2016.8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dafflon C et al. Complementary activities of DOT1L and Menin inhibitors in MLL-rearranged leukemia. Leukemia 31, 1269–1277, doi: 10.1038/leu.2016.327 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Ruthenburg AJ, Li H, Patel DJ & Allis CD Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol 8, 983–994, doi: 10.1038/nrm2298 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.