Introduction
The myelin sheath provides an electrical insulation through the multilayers of nonconductive plasma membrane. Insulating myelin sheath allows rapid transduction of nerve impulses [1]. There are two major components of the myelin sheath, a protein component that is geared towards maintaining structural integrity and a lipid component that is asymmetrically distributed across the membrane [2]. Myelin basic protein (MBP) is one of the major structural proteins, which is also the second most abundant protein in the central nervous system (CNS) myelin. MBP has been associated with various neurological diseases, underscoring its importance in health and disease [3-6].
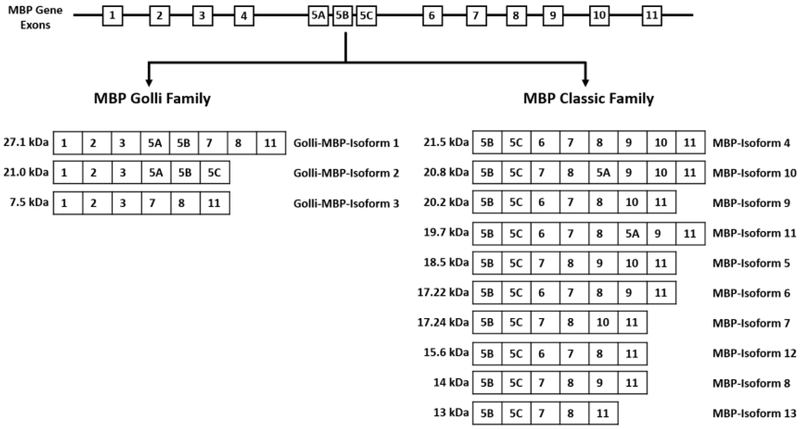
MBP is a multifunctional protein critically involved in the maintenance of the myelin sheath compact structure [7,8]. There are 6 distinct MBP isoforms reported in humans and 13 distinct MBP isoforms reported in mice, all of which arise from alternative splicing of a single gene [8]. These transcript variants are categorized into two main families: 1) the Golli (Genes of Oligodendrocyte Lineage) family ubiquitously expressed throughout the immune and nervous system and 2) the classic family that is primarily expressed in oligodendrocytes [9,10]. There are 11 exons that comprise the murine MBP gene, which give rise to MBP isoforms 1 to 3 in the Golli family, and MBP isoforms 4 to 13 in the classic family [9,7].
The versatility of MBP gives rise to a broad functional spectrum which encompasses many biological processes, all of which remain to be fully understood [7,8,11]. The Golli family appears to be involved in calcium regulation in T-cells and oligodendrocytes, suggesting their involvement in T-cell activation, oligodendrocyte development and calcium dependent biological processes [7]. On the other hand, the classic MBP family plays a major role within oligodendrocytes, which is primarily associated with stability of the myelin sheath [8,9,5]. However, certain members of the classic MBP family have been associated with other functions including cell signaling, nuclear translocation, lipid interactions, interaction with cytoskeletal elements, and regulation of gene expression [12-21]. Given the wide range of functions that MBP seems to be involved in and the numerous isoforms it has, the present report explored the various transcripts that are naturally found in the murine CNS.
Materials and Methods
Neuronal tissue acquisition, RNA extraction, and cDNA synthesis
All animal protocols were approved by the Institutional Animal Care and Use Committee of the University of Miami (IACUC protocol number: 16-235). Neuronal tissue (brain) from five C57BL6/J mice at 2 months of age was dissected and 30 mg of neuronal tissue was utilized for RNA extraction. The Qiagen RNeasy Mini Kit (74104) was used for RNA extraction and all surfaces and tools for dissection were cleaned utilizing an RNase decontamination solution (Thermo, AM9780). Extracted RNA was purified from contaminant DNA using the TURBO DNA-free Kit (Ambion, AM1907). cDNA was made from DNA-free RNA utilizing oligo (dT) to specifically target the reverse transcription of mRNA transcripts (Invitrogen Superscript III, 18080-051).
Primer design
Primers were designed specifically for exon 5B (forward), exon 11 (reverse) and exon 1 (forward) of the MBP gene. Exon 5B forward primer: 5’-TGGCATCACAGAAGAGACCC-3’, exon 11 reverse primer: 5’-AGCGGCTGTCTCTTCCTCC-3’, and exon 1 forward primer 5’-ATGGGAAACCACTCTGGAAAGA-3’.
PCR and product purification
The Amresco PCR Kit (N555) was utilized for amplification of transcript and optimization of primer annealing temperature (65.5°C). PCR products were separated in a 3% agarose gel and bands of interest were excised under UV lamp visualization. The PCR product was extracted from the excised band utilizing the Qiagen QIAquick Gel Extraction Kit (28704) and the eluted product was send for sequencing (Genewiz, South Plainfield, NJ).
Enzymatic digestion and mass spectrometry analysis
Neuronal tissue was separated using the XCell SureLock Mini-Cell system (Thermo, EI0001). Gel was stained using SimplyBlue (Invitrogen, LC6060). Band excised at 32.5 kDa was denature using 6 M urea, reduced in 10 mM Dithiothreitol, alkylated in 15 mM Iodoacetamide, and quenched in 20 mM Dithiothreitol. Samples were digested with either Trypsin (Promega, V511A), Chymotrypsin (Promega, V106A), or Proteinase K (Thermo, 17916). Mass spectrometry was performed on a Q-Exactive instrument after fractionation on a coupled Easy nLC 1000 nano-liquid chromatography system (Thermo Fisher Scientific, Waltham, MA) as described in our other published reports [22].
Results and Discussion
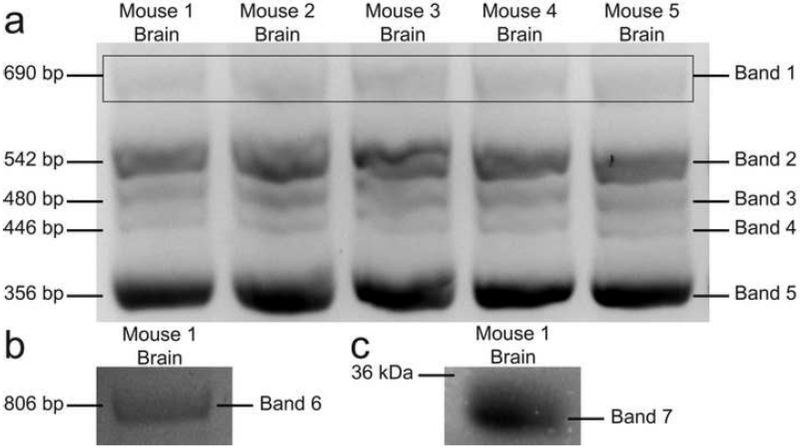
Primers for MBP transcripts were designed to encompass the first and last exons that are common in all the classic MBP transcripts (exon 5B and exon 11, respectively) in order to capture the transcripts present in the CNS (Fig. 1). Agarose DNA gel analysis demonstrated the presence of five major bands, which could be accounted for by the molecular weight of known MBP transcripts (Fig. 2a). These encompassed classic-MBP isoform 9 (band 2, 542 bp), classic-MBP isoform 7 (band 3, 480 bp), classic-MBP isoform 6 (band 4, 446 bp), and classic-MBP isoform 13 (band 5, 356 bp). However, band 1 corresponded to a transcript of about 690 bp, which could not be accounted for by any of the known MBP transcripts. The highest transcript molecular weight corresponds to Golli-MBP isoform 1 (MBP-1, 750 bp) followed by the classic-MBP isoform 4 (MBP-4, 588 bp), demonstrating the lack of an intermediate transcript that could account for the 690 bp unknown transcript. The sequence for MBP-1 (NM_010777.3) is the canonical sequence that is used as a reference for the comparison of other MBP transcript variants. MBP-1 is composed of exons 1, 2, 3, 5A, 5B, 7, 8, and 11, which gives rise to the possibility that the 690 bp unknown transcript is part of the larger MBP-1 transcript. However, MBP-1 transcript analysis demonstrated that the primer designed for exon 5B is localized 400 bp downstream of the start site. This indicates that the unknown transcript corresponding to 690 bp could not be part of the region detected by primers against exon 5B and exon 11 of the MBP-1 transcript, since it would result in a transcript of 350 bp.
Fig. 1.
Diagram representation of the different MBP family members and their individual exon composition. The different MBP isoforms are the result of alternative splicing of a single gene
Fig. 2.
Representative DNA Agarose gel of transcript amplification. (a) Transcripts obtained from using primers for exon 5B and exon 11. Five different C57BL6/J mouse brains. Band 1 – Unknown transcript (690 bp), Band 2 – MBP-9 (542 bp), Band 3 – MBP-7 (480 bp), Band 4 – MBP-6 (446 bp), and Band 5 – MBP-13 (356 bp). Bands on the rectangle were excised, purified, and sent for sequencing. (b) Unknown transcript obtained from using primers for exon 1 and exon 11. Band 6 – Unknown transcript (806 bp). (c) Coomassie blue stain of brain homogenate and excise band for mass spectrometry protein analysis. Band 7 – excised band
To verify the nature of this unknown transcript variant, band 1 was excised, purified, and sent for sequencing (Table 1). Sequence analysis demonstrated that the unknown transcript could be part of a larger transcript. This larger transcript corresponds to a 32.5 kDa MBP isoform (XM_006526456.2) which has not been reported in the literature. Subsequent transcript analysis utilizing a primer for exon 1, revealed the presence of an 806 bp unknown transcript (band 6, Fig. 2b). Given that the highest molecular weight MBP isoform reported is the Golli-MBP isoform 1 (750 bp), this finding further supported the possibility that the 806 bp unknown transcript corresponds to the transcript for the 32.5 kDa MBP isoform.
Table 1.
DNA sequence and composite protein sequence for excised bands. The composite sequence is composed of underlined sequence corresponding to band 6 and non-underlined sequence corresponds to band 1. Lowercase protein sequence correspond to the partial sequence of exon 11 not detected by DNA sequence analysis
| Band 1 | TGGCATCACAGAAGAGACCCTCACAGCGATCCAAGTACCTGGCCACAGCAAGTACCATGGACCATGCC AGGCATGGCTTCCTCCCAAGGCACAGAGACACGGGCATCCTTGACTCCATCGGGCGCTTCTTTAGCGG TGACAGGGGTGCGCCCAAGCGGGGCTCTGGCAAGGACTCACACACGAGAACTACCCATTATGGCTCC CTGCCCCAGAAGTCGCAGCACGGCCGGACCCAAGATGAAAACCCAGTAGTCCATTTCTTCAAGAACAT TGTGACACCTCGAACACCACCTCCATCCCAAGGGAAGGGGAGAGGCCTGTCCCTCAGCAGATTTAGCT GGGGGGCCGAGGGGCAGAAGCCAGGATTTGGCTACGGAGGCAGAGCTTCCGACTATAAATCGGCTCA CAAGGGATTCAAGGGGGCCTACGACGCCCAGGGCACGCTTTCCAAAATCTTTAAGCTGGGAGGAAGAG ACAGCCGCT |
| Band 6 | ATGGGAAACCACTCTGGAAAGAGAGAATTATCTGCTGAGAAGGCCAGTAAGGATGGAGAGATTCACCG AGGAGAGGCTGGAAAGAAGAGAAGCGTGGGCAAGCTTTCTCAGACGGCCTCAGAGGACAGTGATGTG TTTGGGGAGGCAGATGCGATCCAGAACAATGGGACCTCGGCTGAGGACACGGCGGTGACAGACTCCA AGCACACAGCAGACCCAAAGAATAACTGGCAAGGCGCCCACCCAGCTGACCCAGGGAACCGCCCCCA CTTGATCCGCCTCTTTTCCCGAGATGCCCCGGGAAGGGAGGACAACACCTTCAAAGACAGGCCCTCAG AGTCCGACGAGCTTCAGACCATCCAAGAAGACCCCACAGCAGCTTCCGGAGGCCTGGATGTGATGGCA TCACAGAAGAGACCCTCACAGCGATCCAAGTACCTGGCCACAGCAAGTACCATGGACCATGCCAGGCA TGGCTTCCTCCCAAGGCACAGAGACACGGGCATCCTTGACTCCATCGGGCGCTTCTTTAGCGGTGACA GGGGTGCGCCCAAGCGGGGCTCTGGCAAGGACTCACACACGAGAACTACCCATTATGGCTCCCTGCC CCAGAAGTCGCAGCACGGCCGGACCCAAGATGAAAACCCAGTAGTCCATTTCTTCAAGAACATTGTGA CACCTCGAACACCACCTCCATCCCAAGGGAAGGG |
| Composite sequence |
ATGGGAAACCACTCTGGAAAGAGAGAATTATCTGCTGAGAAGGCCAGTAAGGATGGAGAGATTCACCG AGGAGAGGCTGGAAAGAAGAGAAGCGTGGGCAAGCTTTCTCAGACGGCCTCAGAGGACAGTGATGTG TTTGGGGAGGCAGATGCGATCCAGAACAATGGGACCTCGGCTGAGGACACGGCGGTGACAGACTCCA AGCACACAGCAGACCCAAAGAATAACTGGCAAGGCGCCCACCCAGCTGACCCAGGGAACCGCCCCCA CTTGATCCGCCTCTTTTCCCGAGATGCCCCGGGAAGGGAGGACAACACCTTCAAAGACAGGCCCTCAG AGTCCGACGAGCTTCAGACCATCCAAGAAGACCCCACAGCAGCTTCCGGAGGCCTGGATGTGATGGCA TCACAGAAGAGACCCTCACAGCGATCCAAGTACCTGGCCACAGCAAGTACCATGGACCATGCCAGGCA TGGCTTCCTCCCAAGGCACAGAGACACGGGCATCCTTGACTCCATCGGGCGCTTCTTTAGCGGTGACA GGGGTGCGCCCAAGCGGGGCTCTGGCAAGGACTCACACACGAGAACTACCCATTATGGCTCCCTGCC CCAGAAGTCGCAGCACGGCCGGACCCAAGATGAAAACCCAGTAGTCCATTTCTTCAAGAACATTGTGA CACCTCGAACACCACCTCCATCCCAAGGGAAGGGGAGAGGCCTGTCCCTCAGCAGATTTAGCTGGGG GGCCGAGGGGCAGAAGCCAGGATTTGGCTACGGAGGCAGAGCTTCCGACTATAAATCGGCTCACAAG GGATTCAAGGGGGCCTACGACGCCCAGGGCACGCTTTCCAAAATCTTTAAGCTGGGAGGAAGAGACAG CCGCT |
| Predicted composite protein sequence | MGNHSGKRELSAEKASKDGEIHRGEAGKKRSVGKLSQTASEDSDVFGEADAIQNNGTSAEDTAVTDSKHT ADPKNNWQGAHPADPGNRPHLIRLFSRDAPGREDNTFKDRPSESDELQTIQEDPTAASGGLDVMASQKRP SQRSKYLATASTMDHARHGFLPRHRDTGILDSIGRFFSGDRGAPKRGSGKDSHTRTTHYGSLPQKSQHGR TQDENPVVHFFKNIVTPRTPPPSQGKGRGLSLSRFSWGAEGQKPGFGYGGRASDYKSAHKGFKGAYDAQ GTLSKIFKLGGRDSRsgspmarr |
Composite sequence GeneBank Accession Number: MH926013
The band corresponding to this 806 bp unknown transcript was excised, purified and sent for sequencing, which matched with the predicted sequence for the 32.5 kDa MBP isoform (Table 1). The sequence was submitted to the GeneBank database (accession number MH926013). Bioinformatics analysis revealed the protein sequence for this 806 bp unknown transcript and comparison with the sequence from MBP-1 demonstrated that the 806 bp unknown transcript shares the same exon composition with the addition of exon 9 and 10 (Table 1, Fig. 3). To determine the presence of this novel MBP protein, CNS tissue from the same animal was separated via SDS-PAGE gel electrophoresis and the band corresponding to the molecular weight of 32.5 kDa was excised and analyzed by mass spectrometry (band 7, Fig. 2c). Mass spectrometry analysis of three enzymatic digestions (Trypsin, Chymotrypsin, and Proteinase K) demonstrated a 99% coverage of the predicted protein sequence (Table 2).
Fig. 3.
Novel MBP variant bioinformatics analysis. (a) Exon composition of Golli-MBP isoform 1 and novel MBP variant. (b) Protein sequence alignment of Golli-MBP isoform 1 and novel MBP variant. Score = 485 bits (1248), Expect = 2e-180, Method = Compositional matrix adjust, Identities = 250/302 (83%), Positives = 250/302 (82%), Gaps = 52/302 (17%)
Table 2.
Peptides identified from mass spectrometric analysis using enzymatic digestions. Identification of peptide sequence from digestions with Trypsin, Chymotrypsin, and Proteinase K
| Enzyme | Exp. MH+ * | Peptide |
|---|---|---|
| Trypsin | 1046.5 | DTGILDSIGRF |
| 728.3 | FFSGDRG | |
| 1800.8 | FSWGAEGQKPGFGYGGRA | |
| 726.4 | HGFLPRH | |
| 1339.7 | HRDTGILDSIGRF | |
| 1460.7 | TQDENPVVHFFKN | |
| 4015.9 | DRPSESDELQTIQEDPTAASGGLDVMASQKRPSQRSK | |
| 3473.5 | LSQTASEDSDVFGEADAIQN NGTSAEDTAVTDSK | |
| 2653.4 | NNWQGAHPADPGNRPHLIRL FSR | |
| 2044.0 | YLATASTMDHARHGFLPR | |
| 1727.9 | DSHTRTTHYGSLPQK | |
| 1006.5 | ASDYKSAHK | |
| 832.4 | RELSAEK | |
| 774.4 | SAHKGFK | |
| 753.3 | EDNTFK | |
| Chymotrypsin | 841.4 | DSIGRFF |
| 1801.9 | FSGDRGAPKRGSGKVPW | |
| 2508.2 | GSLPQKSQHGRTQDENPVVHFF | |
| 1498.7 | KGAYDAQGTLSKIF | |
| 1733.8 | KLGGRDSRSGSPMARR | |
| 2003.1 | KNIVTPRTPPPSQGKGRGL | |
| 2596.4 | KNIVTPRTPPPSQGKGRGLSLSRF | |
| 1654.8 | SGDRGAPKRGSGKVPW | |
| 882.4 | SLSRFSW | |
| 1383.6 | SWGAEGQKPGFGY | |
| Proteinase K | 1128.5 | MGNHSGKREL |
| 648.3 | SKDGEI | |
| 976.4 | EKASKDGEI | |
| 1513.7 | TDSKHTADPKNNW | |
| 1224.6 | HRGEAGKKRSV | |
| 1107.4 | QNNGTSAEDTA | |
| 1364.6 | SRDAPGREDNTF | |
| 651.2 | SEDSDV | |
| Experimentally determined composite protein sequence |
MGNHSGKRELSAEKASKDGEIHRGEAGKKRSVGKLSQTASEDSDVFGEADAIQNN GTSAEDTAVTDSKHTADPKNNWQGAHPADPGNRPHLIRLFSRDAPGREDNTFKDR PSESDELQTIQEDPTAASGGLDVMASQKRPSQRSKYLATASTMDHARHGFLPRHRD TGILDSIGRFFSGDRGAPKRGSGKDSHTRTTHYGSLPQKSQHGRTQDENPVVHFFK NIVTPRTPPPSQGKGRGLSLSRFSWGAEGQKPGFGYGGRASDYKSAHKGFKGAYD AQGTLSKIFKLGGRDSRSGSPMARR |
|
Mass spectrometric peptide sequence identification showed 99% coverage of predicted protein sequence. Trypsin (red), Chymotrypsin (blue) and Proteinase K (purple) digestion identified peptide sequences are shown in the predicted protein sequence.
Experimental MH+ determined by Q-Exactive was found to be in complete agreement with Theoretical MH+.
These findings expand the number of murine myelin basic protein transcript variants that are found in nature, which is crucial for understanding the different factors contributing to myelin associated diseases. This is crucial for clinical significance given that it provides an understanding of the natural state of the central nervous system myelin, which is often perturbed during disease. Given that this novel MBP variant contains exons from the Golli family (exons 1 to 3), this classifies the novel MBP variant as a member of the MBP Golli family and we propose for this novel MBP variant to be named Golli-MBP isoform 14. As it is the case with other members of the MBP Golli family their functions are not completely understood and the scope of this study limits the conclusions that can be made regarding this novel MBP’s biological function. However, the similarity it holds with the Golli family members suggests that this novel MBP variant could share similar functions in oligodendrocyte development and calcium dependent biological processes. Future experiments will seek to understand the biological role it may potentially have in the murine CNS.
Acknowledgements
This work was supported by an unrestricted grant to the University of Miami from Research to Prevent Blindness (RPB), Department of Defense grant WHX81-16-0715 and NIH grant EY027257.
References
- 1.Hartline DK, Colman DR (2007) Rapid conduction and the evolution of giant axons and myelinated fibers. Current biology : CB 17 (1):R29–35. doi: 10.1016/j.cub.2006.11.042 [DOI] [PubMed] [Google Scholar]
- 2.Jahn O, Tenzer S, Werner H (2009) Myelin Proteomics: Molecular Anatomy of an Insulating Sheath. Molecular Neurobiology 40 (1):55–72. doi: 10.1007/s12035-009-8071-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumann N, Pham-Dinh D (2001) Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiological reviews 81 (2):871–927. doi: 10.1152/physrev.2001.81.2.871 [DOI] [PubMed] [Google Scholar]
- 4.Kramer EM, Schardt A, Nave KA (2001) Membrane traffic in myelinating oligodendrocytes. Microscopy research and technique 52 (6):656–671. doi: 10.1002/jemt.1050 [DOI] [PubMed] [Google Scholar]
- 5.Moscarello MA, Wood DD, Boulias C, Ackerley C (1994) Myelin in multiple sclerosis is developmentally immature. Journal of Clinical Investigation 94 (1):146–154. doi: 10.1172/JCI117300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kursula P (2008) Structural properties of proteins specific to the myelin sheath. Amino acids 34 (2):175–185. doi: 10.1007/s00726-006-0479-7 [DOI] [PubMed] [Google Scholar]
- 7.Boggs JM (2008) Myelin Basic Protein. Nova Science Publishers, Incorporated, New York, UNITED STATES [Google Scholar]
- 8.Boggs JM (2006) Myelin basic protein: a multifunctional protein. Cellular and Molecular Life Sciences CMLS 63 (17):1945–1961. doi: 10.1007/s00018-006-6094-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harauz G, Ishiyama N, Hill CM, Bates IR, Libich DS, Fares C (2004) Myelin basic protein-diverse conformational states of an intrinsically unstructured protein and its roles in myelin assembly and multiple sclerosis. Micron (Oxford, England : 1993) 35 (7):503–542. doi: 10.1016/j.micron.2004.04.005 [DOI] [PubMed] [Google Scholar]
- 10.Feng JM, Fernandes AO, Campagnoni CW, Hu YH, Campagnoni AT (2004) The golli-myelin basic protein negatively regulates signal transduction in T lymphocytes. Journal of neuroimmunology 152 (1–2):57–66. doi: 10.1016/j.jneuroim.2004.03.021 [DOI] [PubMed] [Google Scholar]
- 11.Min Y, Kristiansen K, Boggs JM, Husted C, Zasadzinski JA, Israelachvili J (2009) Interaction forces and adhesion of supported myelin lipid bilayers modulated by myelin basic protein. Proceedings of the National Academy of Sciences of the United States of America 106 (9):3154. doi: 10.1073/pnas.0813110106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pedraza L, Fidler L, Staugaitis SM, Colman DR (1997) The active transport of myelin basic protein into the nucleus suggests a regulatory role in myelination. Neuron 18 (4):579–589 [DOI] [PubMed] [Google Scholar]
- 13.Staugaitis SM, Colman DR, Pedraza L (1996) Membrane adhesion and other functions for the myelin basic proteins. Bioessays 18 (1):13–18. doi: 10.1002/bies.950180106 [DOI] [PubMed] [Google Scholar]
- 14.Dyer CA, Philibotte TM, Billings-Gagliardi S, Wolf MK (1995) Cytoskeleton in myelin-basic-protein-deficient shiverer oligodendrocytes. Developmental neuroscience 17 (1):53–62. doi: 10.1159/000111273 [DOI] [PubMed] [Google Scholar]
- 15.Dyer CA (2002) The structure and function of myelin: from inert membrane to perfusion pump. Neurochem Res 27 (11):1279–1292 [DOI] [PubMed] [Google Scholar]
- 16.Boggs JM, Rangaraj G (2000) Interaction of lipid-bound myelin basic protein with actin filaments and calmodulin. Biochemistry 39 (26):7799–7806 [DOI] [PubMed] [Google Scholar]
- 17.Libich DS, Hill CM, Bates IR, Hallett FR, Armstrong S, Siemiarczuk A, Harauz G (2003) Interaction of the 18.5-kD isoform of myelin basic protein with Ca2+ -calmodulin: effects of deimination assessed by intrinsic Trp fluorescence spectroscopy, dynamic light scattering, and circular dichroism. Protein science : a publication of the Protein Society 12 (7):1507–1521. doi: 10.1110/ps.0303603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boggs JM, Rangaraj G, Hill CM, Bates IR, Heng YM, Harauz G (2005) Effect of arginine loss in myelin basic protein, as occurs in its deiminated charge isoform, on mediation of actin polymerization and actin binding to a lipid membrane in vitro. Biochemistry 44 (9):3524–3534. doi: 10.1021/bi0473760 [DOI] [PubMed] [Google Scholar]
- 19.Boggs JM, Rangaraj G, Gao W, Heng YM (2006) Effect of phosphorylation of myelin basic protein by MAPK on its interactions with actin and actin binding to a lipid membrane in vitro. Biochemistry 45 (2):391–401. doi: 10.1021/bi0519194 [DOI] [PubMed] [Google Scholar]
- 20.Hardy RJ, Lazzarini RA, Colman DR, Friedrich VL Jr. (1996) Cytoplasmic and nuclear localization of myelin basic proteins reveals heterogeneity among oligodendrocytes. J Neurosci Res 46 (2):246–257. doi: [DOI] [PubMed] [Google Scholar]
- 21.Carre JL, Goetz BD, O'Connor LT, Bremer Q, Duncan ID (2002) Mutations in the rat myelin basic protein gene are associated with specific alterations in other myelin gene expression. Neurosci Lett 330 (1):17–20 [DOI] [PubMed] [Google Scholar]
- 22.Travers TS, Harlow L, Rosas IO, Gochuico BR, Mikuls TR, Bhattacharya SK, Camacho CJ, Ascherman DP (2016) Extensive Citrullination Promotes Immunogenicity of HSP90 through Protein Unfolding and Exposure of Cryptic Epitopes. Journal of immunology (Baltimore, Md : 1950) 197 (5):1926–1936. doi: 10.4049/jimmunol.1600162 [DOI] [PMC free article] [PubMed] [Google Scholar]