Abstract
Introduction
The extracellular matrix (ECM) in the tumor microenvironment contains high densities of collagen that are highly aligned, resulting in directional migration called contact guidance that facilitates efficient migration out of the tumor. Cancer cells can remodel the ECM through traction force controlled by myosin contractility or proteolytic activity controlled by matrix metalloproteinase (MMP) activity, leading to either enhanced or diminished contact guidance.
Methods
Recently, we have leveraged the ability of mica to epitaxially grow aligned collagen fibrils in order to assess contact guidance. In this article, we probe the mechanisms of remodeling of aligned collagen fibrils on mica by breast cancer cells.
Results
We show that cells that contact guide with high fidelity (MDA-MB-231 cells) exert more force on the underlying collagen fibrils than do cells that contact guide with low fidelity (MTLn3 cells). These high traction cells (MDA-MB-231 cells) remodel collagen fibrils over hours, pulling so hard that the collagen fibrils detach from the surface, effectively delaminating the entire contact guidance cue. Myosin or MMP inhibition decreases this effect. Interestingly, blocking MMP appears to increase the alignment of cells on these substrates, potentially allowing the alignment through myosin contractility to be uninhibited. Finally, amplification or dampening of contact guidance with respect to a particular collagen fibril organization is seen under different conditions.
Conclusions
Both myosin II contractility and MMP activity allow MDA-MB-231 cells to remodel and eventually destroy epitaxially grown aligned collagen fibrils.
Electronic supplementary material
The online version of this article (10.1007/s12195-018-0547-6) contains supplementary material, which is available to authorized users.
Keywords: Directed migration, Second harmonic generation, MT1-MMP, MMP-14, Function blocking antibody
Introduction
The migration of cells plays an important role in many pathological and physiological processes. Cancer cells are able to degrade basement membrane barriers and invade surrounding tissue and metastasize to other organs. Once cancer cells have penetrated the basement membrane, the tumor microenvironment (TME) presents directional cues including aligned collagen fibers, allowing for more efficient cell migration from the basement membrane towards blood and lymph vessels. Second harmonic generation (SHG) imaging and other imaging approaches have demonstrated that collagen is well aligned in the TME and is a critical marker of the invasive phenotype and is a negative prognostic indicator.9,40 Aligned collagenous extracellular matrix (ECM) directs migration during a process called contact guidance. In vitro environments that mimic the collagen organization of the TME are powerful tools to understand cell-ECM interactions in the TME. There have been numerous approaches to fabricate contact guidance cues in vitro. Collagen fibers have been oriented in 3D using flow,26,46 using magnetic fields,7,11 by contractile cells,43,48 through rotational alignment21,22 or tension44 and by electrospinning.15,18,31 However, these environments can be difficult to control and imaging cell migration within them can be challenging, particularly if subcellular information is sought. Consequently, 2D contact guidance environments that are easier to control and image still hold value. In 2D environments, common techniques include, flow-induce alignment,54 electrospinning,15,18,31 fiber drawing,32,34 microcontact printing,41,45 and generating grooves or gratings.3,5,42,50 However, many of these techniques suffer from either not presenting natural fibers (many but not all electrospinning techniques and fiber drawing) or not presenting fibers at all (microcontact printing and gratings). Natural fibers54 recapitulate the nanotopography and spatial ligand presentation characteristics of fibers in vivo. Epitaxial growth of aligned collagen fibrils on mica20 provides an approach by which to assemble aligned collagen fibrils on a 2D surface. Cells engage in contact guidance on epitaxially grown collagen fibrils.39 However, the robustness of the contact guidance differs among cells.52 This diversity in response to this 2D contact guidance cue mimics behavior seen in 3D contact guidance cues38 and in mouse models for invasion and metastasis58 and has been used as a substrate by which to assess the role of adhesion and contractility in tuning contact guidance fidelity.53
Many of the contact guidance systems mentioned above are immovable because the contact guidance cue is attached to or constitutes a stiff substrate. Consequently, the contact guidance cue cannot be remodeled. However, cell-mediated ECM remodeling is at the heart of tissue morphogenesis and is a key feature of cells within the TME, where both cancer and stromal cells cooperate to remodel the ECM. During contact guidance, cells adhere to ECM through integrins and generate traction force on ECM by coupling myosin II-mediated contractility to adhesion through different F-actin structures. Some of this force transmission is used to overcome adhesion to the substrate and pull up the tail of the cell. However, force transmission is also used to deform surrounding ECM, commonly resulting in ECM alignment. Different cells in the TME exert a different amount of force on the ECM leading to remodeling. For example, MTLn3 breast cancer cells generate less traction force than MDA-MB-231 breast cancer cells.27,33,55 When the ECM in 2D contact guidance systems cannot be altered by the cell, there is a complete decoupling of contact guidance from ECM remodeling. This provides for a clean experiment whereby contact guidance can be assessed for a particular contact guidance cue. However, this shields the effects associated with ECM remodeling that may act to either amplify or degrade the contact guidance response to aligned collagen. Epitaxial growth of collagen fibrils proceeds through a modest attachment to mica governed by electrostatic interactions between collagen and negative charges tuned by potassium ion binding and left after mica cleaving29,37. Collagen fibril alignment is achieved due to an atomic ridge that runs along one axis of the hexagonal packed crystal structure of mica.49 Consequently, while aligned collagen fibrils can be stably grown on mica, they can also be rearranged by cells.25 In addition, they may reveal differences in contact guidance due to collagen fiber deformation that are not revealed by other contact guidance systems (electrospinning, fiber drawing, microcontact printing, and generating grooves or gratings).53
Not only do cells use myosin II-mediated contractile force to remodel the ECM, but they also degrade ECM through matrix metalloproteinase (MMP) activity.30 ECM degradation facilitates tumor cell invasion and metastasis, thus, MMPs are regarded as important regulators. MMP activity has traditionally been thought to be dispensable for cell migration on 2D surfaces, because the role of MMPs in 3D is to open ECM pores, allowing cells to more easily move through dense ECM networks, particularly those that are cross-linked. However, MMPs can also process a number of surface-bound or extracellular proteins including cell surface receptors and growth factors and cytokines. This has led to observations that MMP activity even affects cell behavior on 2D substrates. Indeed, we have shown that tissue inhibitors of matrix metalloproteinase activity (TIMPs) are able to modulate contact guidance in both 2D and 3D environments.28 One of the most important MMPs due to is requirement for cell migration in 3D ECM is MMP-14 (MT1-MMP). MMP-14 is a collagenase that acts to degrade ECM local to the cell and likely cooperates with myosin II-mediated contractility to degrade ECM. Indeed, it appears that contractile force generation is important in activating MMP-14, other MMPs or degradative structures like invadopodia.1,12,13,19 While the local structure of collagen on epitaxially grown collagen fibril substrates has been assessed in response to perturbations in contractility and proteinase activity,25 the global structure of collagen fibrils over time and during contact guidance has not been determined. Furthermore, it is not known whether myosin II-mediated contractility or MMP-mediated proteinase activity acts to either amplify or dampen the contact guidance response.
In this paper, we find that high traction exerting cells (MDA-MB-231 cells) deform and remodel collagen fibril structures more dramatically than low traction force exerting cells (MTLn3 cells) by tracking fluorescent nanospheres attached to epitaxially grown collagen fibrils on mica substrates. MDA-MB-231 cells stained for F-actin at different times showed that these cells detach from substrate, forming large aggregates, resulting in a decreased cell density. MTLn3 cells did not detach in his way. Furthermore, if MDA-MB-231 cells were treated with a contractility inhibitor (blebbistatin), MMP inhibitors (marimastat or 3A2 (MT1-MMP function blocking antibody)) or both, detachment was blocked, implicating cellular contractility and proteinase activity in the detachment phenotype. Blocking MMPs generally and MT1-MMP, specifically, appears to increase the alignment of cells on these substrates, whereas blocking contractility appears to decrease alignment. Collagen fibril orientation was assessed using second harmonic generation (SHG) imaging. MMPs partially degrade the collagen signal, but contractility is needed for a dramatic large scale change in collagen fibril coverage. Finally, we compared cell alignment versus collagen fibril alignment and show that under control conditions, contact guidance is amplified by the cell, whereas blocking contractility or MMP activity acts to dampen the contact guidance signal, resulting in lower than expected cell alignment. This indicates that contractility and MMP activity both directly affect contact guidance and indirectly affect contact guidance through ECM remodeling.
Materials and Methods
Cell Culture
A human mammary basal/claudin low carcinoma cell line (MDA-MB-231, ATCC, Manassas, VA, USA) was cultured in Dulbecco’s modified Eagles medium (DMEM) (Sigma Aldrich, St. Louis, MO, USA) containing 10% fetal bovine serum (FBS) (Gibco, Grand Island, New York, USA) and 1% penicillin–streptomycin (pen-strep) (Gibco) at 37 °C in 5% CO2. A rat mammary basal adenocarcinoma cell line (MTLn3, Jeffrey E. Segall, Albert Einstein College of Medicine) was authenticated using IDEXX BioResearch (Westbrook, Maine, USA) and cultured in MEMα (Gibco) supplemented with 5% FBS (Gibco) and 1% pen-strep (Gibco) at 37 °C in 5% CO2. Imaging media for MDA-MB-231 and MTLn3 cells was the same as the subculturing media, with the exception that no phenol red was included and that 12 mM HEPES (Sigma Aldrich) was included. Serum free conditions produced much less migratory cells, so FBS was including for migration experiments.
Contractility and Proteinase Inhibitors
Blebbistatin (Sigma Aldrich) was used to inhibit myosin II activity and marimastat (R&D Systems, Minneapolis, MN, USA) was used to broadly inhibit MMPs. The 3A2 was isolated from a customized synthetic antibody library using phage panning.36 Its gene was cloned in a Fab expression vector containing the PhoA promoter, a STII leader peptide, and a His tag at the C terminus of the VH.10 Transformed E. coli BL21cells were grown overnight at 30 °C in 2 × YT supplemented with 100 µg mL−1 ampicillin. Cells were harvested by centrifugation and treated with osmotic shock to prepare periplasmic fractions. The 3A2 Fab was purified by nickel-nitrilotriacetic acid chromatography (Qiagen), dialyzed against 50 mM HEPES and 150 mM NaCl (pH 6.8), and analyzed by SDS-PAGE. Its concentration was measured with a NanoDrop 2000 instrument (Thermo Scientific).
Assembling Collagen Substrates
Collagen fibrils were epitaxially grown on 15 mm × 15 mm pieces of muscovite mica (highest grade VI, Ted Pella, Redding, CA, USA) that were freshly cleaved using tape.52 Rat tail collagen type I was diluted (BD Bioscience, 10 μg mL−1) in the buffer solution consisted of 50 mM Tris–HCl (Fisher Scientific) and 200 mM KCl (Fisher Scientific) at pH 9.2. After incubation of 18 h the collagen solution was washed with deionized water, the mica was laid against the edge of a tissue culture dish and the mica was allowed to dry overnight and was used the next day.
Imaging Deformations on Aligned Collagen Fibrils using Nanosphere Tracking
Nanospheres were added to substrates to mark deformations. Carboxylate-modified nanospheres (580 nm excitation max, 605 nm emission max, 40 nm O.D., ThermoFisher Scientific, Waltham, MA USA) were sonicated to fully suspend nanospheres and disrupt aggregates. Then nanospheres suspended in PBS were added to collagen fibrils assembled on mica for 45 min. Collagen fibrils with attached nanospheres were washed 3×, the last time with distilled water and dried before use in deformation assays. Cells were plated at 30,000 cells mL−1 in 2 mL of media in 35 mm dishes with bead-labeled collagen fibrils. Imaging media used for live-cell experiments was conditioned in a dish containing 960,000 cell mL−1 of MDA-MB-231 cells or MTLn3 cells, the day prior to use. Conditioned media was centrifuged to clarify any cells or cell debris. MDA-MB-231 cells (± 30 μM blebbistatin) and MTLn3 cells were incubated in the conditioned imaging medium for 2–3 or 11–12 h, respectively, before imaging. Substrates with attached cells were inverted onto two strips of double sided tape attached to a microscope slide to generate a flow chamber. The chamber was filled with conditioned imaging media and sealed with a 1:1:1 mixture of vasoline, lanolin and paraffin wax. Chambers were imaged by phase contrast microscopy on a heated stage at 37 °C every 30 s for 6 h. Phase contrast and fluorescence images were captured at 20× (NA 0.50, Nikon) with a charge-coupled device (CoolSNAP HQ2, Photometrics) attached to an inverted microscope (Eclipse Ti, Nikon).
Matlab was used to detect microspheres positions in each image and then we applied an intensity threshold to find each microsphere centroid and track each from image sequences using a standard particle-tracking algorithm (http://site.physics.georgetown.edu/matlab/tutorial.html)6 and measure the mean displacement of microspheres. The mean displacement () of microsphere at any time point, i, was defined as:
| 1 |
Where xi is an x-axis position of a single microsphere at time point, i, yi is a y-axis position of a single nanosphere at time point, i, and and are average x and y positions over the entire timelapse of a single nanosphere.
MMP Activity Assays
The MMP quenched cleavage peptide used for MMP activity assays was a broad-spectrum MMP-substrate (Suc-Ala-Ala-Pro-Phe-2,4-difluoroanilide, M-2305, Bachem, Bubendorf, Switzerland). The MMP-14 catalytic subunit was constructed, expressed and refolded as previously described.35 To conduct the MMP activity assay, cells were plated at 1.5 × 106 in a 60 mm tissue culture dish in the same media used for migration experiments. After 24 h the media was replaced with media lacking FBS due to interference with the fluorescence signal. After 24 h the media was removed and put on ice, and the cells were trypsinized and counted. Fluorescence was measured from the collected media after addition of 10 µM MMP-substrate and normalized to the number of cells. Additionally, the activity of the MMP-14 catalytic domain in α-MEM and DMEM was assessed by adding 23 nM MMP-14 to 10 µM MMP-substrate.
Fluorescence Imaging
MDA-MB-231 cells were plated at 40,000–60,000 cells mL−1 in 2 mL of media in 35 mm dishes for 4, 12 and 24 h on highly aligned type I collagen fibrils. Cells that were treated with 30 μM blebbistatin, 10 μM marimastat or marimastat and blebbistatin (10 and 30 μM) or 500 μM 3A2 antibody were incubated for 12 and 24 h on highly aligned type I collagen fibrils. Aliquots of 16% paraformaldehyde (Fisher), which were premade and frozen, were heated to 37 °C and diluted to 4% in cytoskeleton buffer. The cytoskeleton buffer included 10 mM MES pH 6.1, 3 mM MgCl, 138 mM KCl, and 2 mM EGTA. Cells were then washed with Tris base buffered saline (TBS) containing 150 mM NaCl and 20 mM Tris–Cl pH 7.4 3× for 5 min each. Fixing was quenched using 100 mM glycine and permeabilized using 0.5% triton-X solution in cytoskeletal buffer for 10 min. Cells were washed 3× for 5 min each in TBS and were treated with 0.1% tween-20 v/v, 2% BSA w/v, and alexa 488-phalloidin in TBS for 1 h. After treatment, mica samples were mounted using ProLong Gold mounting media onto microscope slides face down for imaging. Fixed and stained cells were imaged by epifluorescence using the same microscope as the fluorescent nanospheres.
Assessing Cell Orientation
A 4 × 4 matrix of images was taken for each sample within a particular condition. After the images were taken, ImageJ was utilized to analyze cell orientation angle and aspect ratio. The fluorescence cell image was background subtracted with a 50 pixel (16 μm) rolling ball algorithm, thresholded and converted to a binary image. The binary image was eroded, dilated, opened, closed, eroded, and finally dilated. The cell regions of interest (ROIs) were then analyzed for any ROI areas greater than 500 pixels. The analyzed outlines were compared to the original phase contrast image to ensure the size and shape was accurate. Overlapping cells were outlined manually to ensure an accurate cell count and area. Only single cells were used in analysis of orientation and aspect ratio. ImageJ was used to analyze mean greyscale value, area, and ellipse properties for each ROI. The ellipse fit in ImageJ gives the major and minor axes lengths as well as the angle of the major axis with respect to a horizontal vector. The aspect ratio of the ellipse was used to determine whether the orientation angle of the cell was used. Cells with an aspect ratio greater than 2 were considered oriented and included in the orientation analysis. Distributions of orientation angles were generated. These distributions were fit to the sum of three von Mises distributions offset by 60° each. The equation used is
| 2 |
where ai is the fraction of cells in peak i, κ determines the breadth of the distribution where large κ results in narrower peaks, μ is the location of each peak (offset by 60°), I0 is the modified Bessel function of the first kind.
The angle distribution for a particular sample was determined to have a peak if the fit to the von Mises distribution resulted in at least one peak amplitude with a 95% confidence interval that did not include zero. Samples with peaks were reoriented so the major peak was at 30° and the second peak was at 90°. The non-aligned samples were not reoriented. Fractional distributions were then averaged across samples for a given condition. These averaged distributions were then fitted to Eq. (2). The aligned cell fraction was the area underneath the major peak in each condition’s average von Mises fit. In addition, the κ was calculated for each condition based on the fit.
Secondary Harmonic Generation Microscopy Imaging
The fixed cell samples with different contractility or proteinase perturbations were imaged using a mode-locked Ti:Sapphire laser (100 fs pulse width, 1 kHz repetition rate, Libra, Coherent, Santa Clara, CA) that produces an 800 nm fundamental. The average power at the sample image plane was controlled using a combination of a half-wave plate and a Glan–Thompson polarizer (Thorlabs, Newton, NJ). Second harmonic signal was collected in the transmission mode. For this setup, an inverted microscope (AmScope, Irvine, CA) and Nikon plan fluorite objective (20×, NA = 0.5, Nikon, Melville, NY) was used to focus the beam and the SHG transmission was collected with a Nikon water immersion objective (40x, 0.8 NA, Nikon, Melville, NY). The transmitted SHG signal was reflected by a dichroic mirror (DMLP425T, Thorlabs, Newton, NJ) and separated from the fundamental beam with two short pass filters < 450 nm (FGB37 M, Thorlabs, Newton, NJ) and 808 nm notch filter (NF-808.0-E-25.0 M, Melles Griot, Rochester, NY), before detection by an intensified charge coupled device (iCCD, iStar 334T, Andor, Belfast, UK). Polarized SHG imaging was conducted using a Glan–Thompson polarizer and a half-wave plate mounted on a motor driven rotational stage (Thorlabs Newton, NJ) to achieve linear polarization. Images of the samples were collected every 10° from 0° to 350°. A minimum of three images for each experimental condition was taken. From this collection of images, ROIs were fit using the following equation:
| 3 |
where and are second-order susceptibility tensor element ratios, θe and θo are the incident polarization angle and collagen fiber angle, respectively, and c is a normalization constant. The orientation angle of collagen in each ROI was calculated and a histogram of orientations was generated. Collagen organization was observed by fitting the orientation angle histogram with Eq. [2] as were the cell distributions. Over small length scales, the different ROIs were classified as ordered, non-ordered and non-collagen, to evaluate the small scale ordering or presence of the collagen fibrils locally. The non-ordered ROI fraction was determined through the deviation of the data from the predicted von Mises distribution in one bin (10°). The non-collagen ROI fraction was determined by identifying ROIs that were below a set threshold intensity as done previously. As the total number of ROIs were kept constant throughout the analysis, the remaining ROIs were calculated as the ordered fraction.
Statistics
The number of experiments and cells were stated in all the figure legends. At least three independent experiments were conducted. In general, means were calculated with error bars representing 95% confidence intervals. Statistical significance was determined by a Tukey’s HSD test in JMP (SAS, Cary, North Carolina).
Results
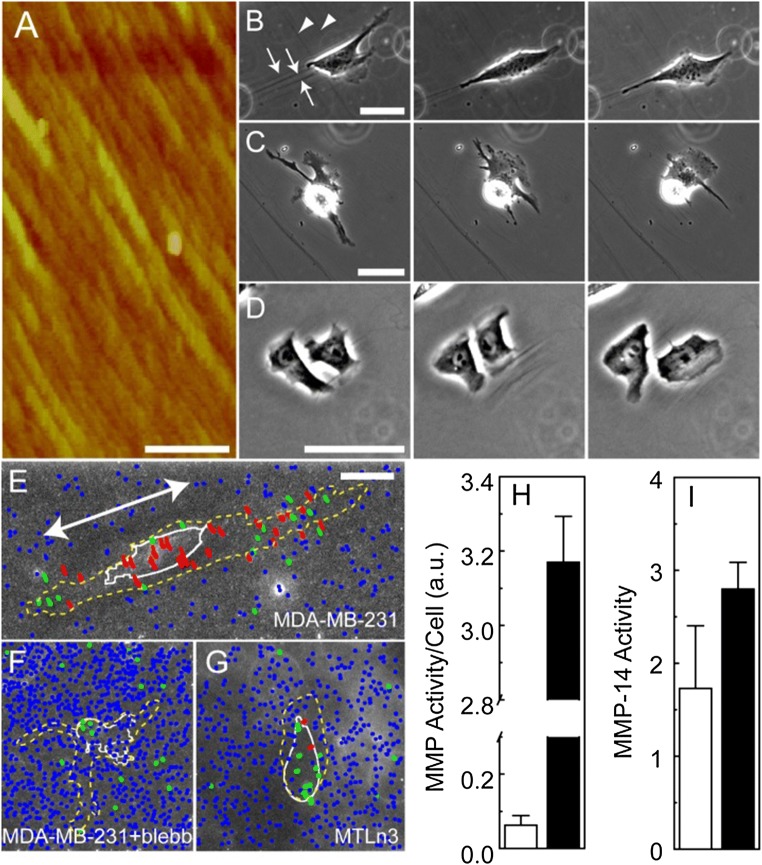
Aligned collagen fibrils that are epitaxially grown on mica have been shown to form a tunable substrate for the study of 2D contact guidance.39,52 As has been previously shown, this system consists of fibrils aligned on a surface and that form a thin (3 nm) and relatively continuous layer of aligned collagen fibrils (Fig. 1a). Previously, we showed that contact guidance efficiency on these substrates as measured by the propensity to make migration steps in the direction of the long axis of the fibrils was different between cells that exert vastly different traction forces. MDA-MB-231 sense the contact guidance cue and migrate directionally, but MTLn3 cells do not.52 In addition, turning cell contractility either up or down can correspondingly alter contact guidance efficiency in a dose dependent manner.53 MDA-MB-231 cells move on these substrates along the collagen fibrils (Fig. 1b). MDA-MB-231 cells whose contractility has been inhibited with blebbistatin frequently extended protrusions orthogonal to the long axis of the collagen fibrils and did not dramatically alter collagen fibrils (Fig. 1c). Similarly, MTLn3 cells frequently extended in directions orthogonal to the fibril alignment and did not dramatically deform fibrils (Fig. 1d). Collagen fibrils can be seen to deform using phase contrast microscopy, but are frequently difficult to image and quantify (Figs. 1b–1d). Consequently, we used fluorescently labeled negatively charged carboxylate-modified nanospheres that electrostatically attach to positively charged collagen fibrils assembled on the mica surfaces as fiduciary marks and tracked fiber deformations over time. Three different colors (red, green and blue) were used to represent large, intermediate and small sums of the mean displacements. MDA-MB-231 cells, whose polarity and migration alignment match the long axis of the collagen fibrils, pull on collagen fibrils in the direction orthogonal to the direction of migration, resulting in trajectories that run orthogonal to the collagen fibrils (Fig. 1e). Nanospheres that were close, but outside the cell edge or underneath the MDA-MB-231 cell displayed large displacements. When contractility is blocked using blebbistatin, the magnitude of the deformations is much smaller (Fig. 1f). Additionally, MTLn3 cells deform the substrate much less than MDA-MB-231 cells (Fig. 1g). Furthermore, the MMP activity was measured in both cell lines. Surprisingly, MDA-MB-231 cells expressed active MMPs, but at a much lower activity per unit cell as compared to MTLn3 cells (Fig. 1h). However, little difference was seen when the activity of MMP-14 catalytic domain was assayed in media in the absence of cells (Fig. 1i).
Figure 1.
Cell alignment and traction on epitaxially grown aligned collagen fibrils. (a) Atomic force microscopy image of epitaxially grown, aligned collagen fibrils. The scale bar is 1 μm. (b–d) Timelapse phase contrast images of an (a) MDA-MB-231 cell, (b) MDA-MB-231 cell treated with blebbistatin and (c) MTLn3 cell on aligned collagen fibrils. (a) Collagen fibrils are marked with arrows and move between 0 min (left), 200 min (center) and 400 min (right). Arrow head marks scratches in the mica that do not deform over time. The scale bar length is 30 μm. Trajectories of fluorescent nanospheres adsorbed on aligned collagen fibrils assembled on mica for (e) MDA-MB-231 cells, (f) MDA-MB-231 cells treated with blebbistatin (30 μM) and (g) MTLn3 cells. Beads are colored differently depending on their mean displacement. Red indicates mean displacements larger than 600 nm, green indicates mean displacements between 300 and 600 nm and blue represents mean displacement smaller than 300 nm. The cell outline from one time point is shown in white and the region over which the cell migrates over the entire time lapse is shown as the yellow dashed line. The primary migration direction for (e) is shown as the arrow. (h) MMP activity in MDA-MB-231 (white) and MTLn3 (black) cells MMP activity on a per cell basis. Note the break in the axis. (i) Activity of the head domain of MMP-14 in the media used for MDA-MB-231 (white) and MTLn3 (black) cell migration experiments. Error bars are 95% confidence intervals.
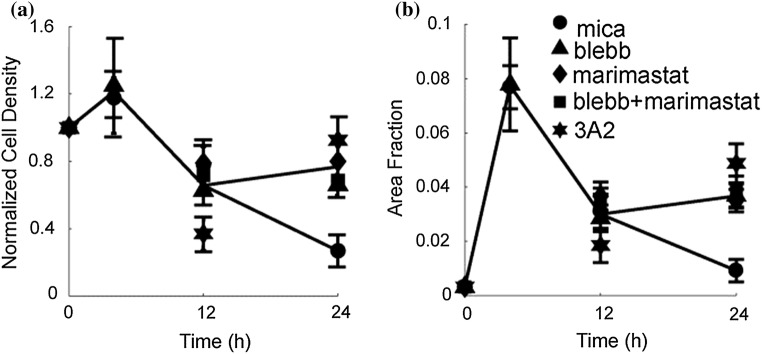
MDA-MB-231 cells robustly engage in contact guidance and take migration steps primarily in directions along the long axis of the collagen fibrils over the first 10 h.52,53 However, we observed that for longer times (12–24 h), cells pull (using contractility) and potentially degrade (using MMP activity) the collagen fibrils to such a large extent that the cells detach. This detachment phenotype is robust and is not seen in MTLn3 cells. Consequently, we quantified detachment for MDA-MB-231 cells. MDA-MB-231 cells were fixed and stained for F-actin at 4, 12 and 24 h after plating and images of cells were acquired. When the cells detach from the substrate they form large aggregates that are not frequently imaged in the samples that we take, thus cells that appear in the images correspond to cells still attached to the substrate. Cell density was normalized to the expected cell density at time 0, for given cell concentration and substrate size. Under control conditions, normalized cell density does not change over the first 4 h, but the area fraction that the cells occupy increases, due to cell spreading (Figs. 2a and 2b). Both normalized cell density and area fraction decrease at 12 h and continue decreasing through 24 h (Figs. 2a and 2b). Contractility and proteinase inhibitors were then used to probe the effects of force transmission and proteolytic cleavage in modulating this response. Contractility was blocked using blebbistatin, which inhibits the myosin II motor domain. Proteinase activity was inhibited generally using marimastat, which is a small molecule inhibitor that binds well to many different MMP catalytic sites, or specifically with an MMP-14 function blocking antibody (3A2), which specifically recognizes MMP-14 through its stalk domain, but blocks its catalytic function. Interestingly, if cells are treated with a contractility blocker (blebbistatin), a pan-MMP blocker (marimastat), both a contractility blocker and pan-MMP blocker (blebbistatin and marimastat) or an MMP-14 function blocking antibody (3A2), the decrease in cell density and area fraction seen between 12 and 24 h is inhibited (Figs. 2a and 2b). This suggests that both contractility and MMP activity (MMP-14 activity in particular) are responsible for this decrease in cell density. We have previously shown that MDA-MB-231 cells robustly respond to the contact guidance cue as quantified by a directionality index, which is the average over all migration steps of the cosine of twice the angle between the cell migration direction and the direction of the long axis of the collagen fibrils.52 Consequently, we wanted to assess the alignment over the same timescale that cell detachment was occurring.
Figure 2.
(a) Normalized cell density and (b) area fraction occupied by MDA-MB-231 cells change with time on aligned collagen fibrils assembled on mica (circles). The decrease seen at 24 h is blocked when treated with a myosin II-mediated contractility inhibitor (blebbistatin), a pan-MMP inhibitor (marimastat) or an MMP-14 function blocking antibody (3A2). Blebbistatin (30 μM, triangle), marimastat (10 μM, diamond), marimastat and blebbistatin (10 and 30 μM, squares) or 3A2 antibody (500 μM, star). Error bars represent 95% confidence intervals. Nexperiments > 3 and Ncell > 160.
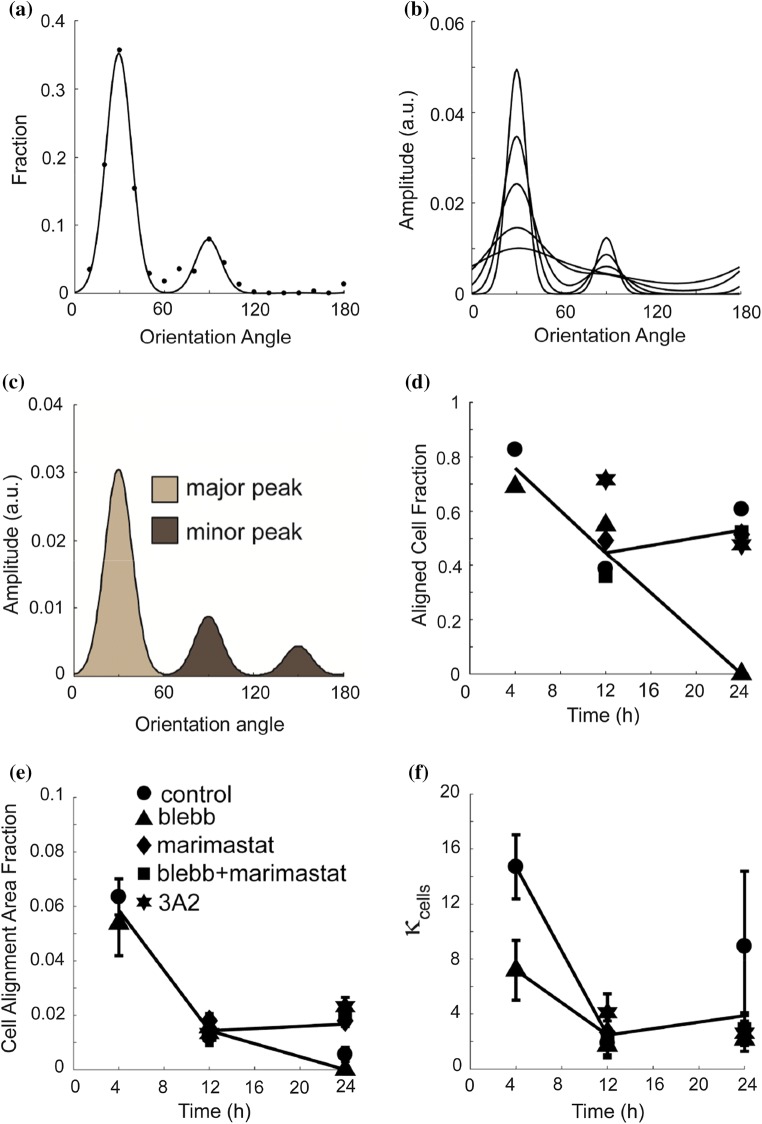
We assessed cell alignment on the collagen fibrils over time and under different inhibitor treatments by examining the orientation distribution. This is similar to what we have done previously,52 however instead of using curvilinear Gaussian distributions, which assume an infinite independent variable; we have chosen spherical von Mises distributions, which assume a repeating independent variable. Three von Mises distributions shifted at 60° angles were summed and fit to the experimental fraction of cells oriented in a particular direction (Figs. 3a and S1). The 60° is set and is based on the hexagonal structure of the underlying mica crystal.29,49,56 One dominant collagen alignment direction is favored. However, additional directions will appear due to incomplete mica cleavage, particularly on samples with small numbers of cells. We assume that the parameter that describes spread of each peak is identical for a given condition. Consequently, the only adjustable parameters are the parameter that describes the spread (κcells) and two of three fraction maxima (the final maximum is set by constraining the integrated function fraction to be equal to 1) (Eq. (2)). As κcells increases, the peak widths (full-width half maximum, FWHM) narrow and more alignment is seen (Fig. 3b). Each peak is tested to determine if it is statistically considered a peak (see “Materials and Methods”). Consequently, one measure of alignment is the fraction of cells that falls under the major peak, the aligned cell fraction (Fig. 3d). Only the major peaks are included in the integration (Fig. 3c). When cells were plated on aligned collagen fibrils for 4 h, they adhered, spread on the surface and roughly 80% of them aligned (Fig. 3d). When the cells were treated with proteinase inhibitors, no change in alignment was observed over the initial 24 h. In fact, the alignment marginally increased at 12 h when cells were treated with 3A2, an MMP-14 specific inhibitor. Blebbistatin treated cells on the other hand completely degraded the alignment by 24 h. Interestingly, the combination of blebbistatin and marimastat resulted in a retained orientation after 24 h. Because the area fraction occupied by cells changes over time (Fig. 2b), we calculated the product of the area fraction and the aligned cell fraction, which yields the cell alignment area fraction (Fig. 3e). This indicates that few cells are aligned at 24 h for both control and blebbistatin treatment. However, this is due to different reasons. Control cells pull themselves off the substrate, whereas blebbistatin blocks cell detachment, but results in cells that are unable to align. Another measure of alignment is the κcells for the fit of the cell orientation distribution (Fig. 3f). The κcells for control cells is high and decreases after 4 h. After blebbistatin treatment, cell alignment is lower at 4 h. In agreement with the aligned cell fraction, 3A2 treatment, blocking MT1-MMP appears to increase the alignment of cells on these substrates. Given that blocking contractility appears to both block cell detachment and alignment at 24 h and that 3A2 increases cell alignment at 12 h, we were interested in assessing collagen fibril alignment changes under the same perturbations. This would allow us to determine if cell alignment changes were due to changes in the alignment of the collagen fibrils themselves or in the cells ability to interpret aligned structures.
Figure 3.
Cell alignment of MDA-MB-231 cells treated with a myosin II-mediated contractility inhibitor (blebbistatin), a pan-MMP inhibitor (marimastat) or an MMP-14 function blocking antibody (3A2). (a) The angle distribution of cells on aligned collagen fibrils assembled on mica at 4 h (circles). The sum of three von Mises distributions separated by 60 degrees was used to fit the data. (b) The sum of three von Mises distributions with different values for the distribution spread parameter (κcells=1, 2, 5, 10 and 20). As κcells increases, the spread of the distribution descreases. κcells is inversely proportional to the full width half max. (c) Schematic of cell angle distribution for the one major and two minor peaks. The cell alignment fraction is calculated as fraction of the total area that is under the major peak. (d) Cell alignment fraction, (e) cell alignment area fraction and (f) distribution spread parameter, κcells as a function of time and exposed to different inhibitors. Cells plated on aligned collagen fibrils assembled on mica (control, circle) treated with blebbistatin (30 μM, triangle), marimastat (10 μM, diamond), marimastat and blebbistatin (30 and 10 μM, squares) or 3A2 antibody (500 nM, star). Error bars represent 95% confidence intervals. Nexperiments > 3 and Ncell > 160.
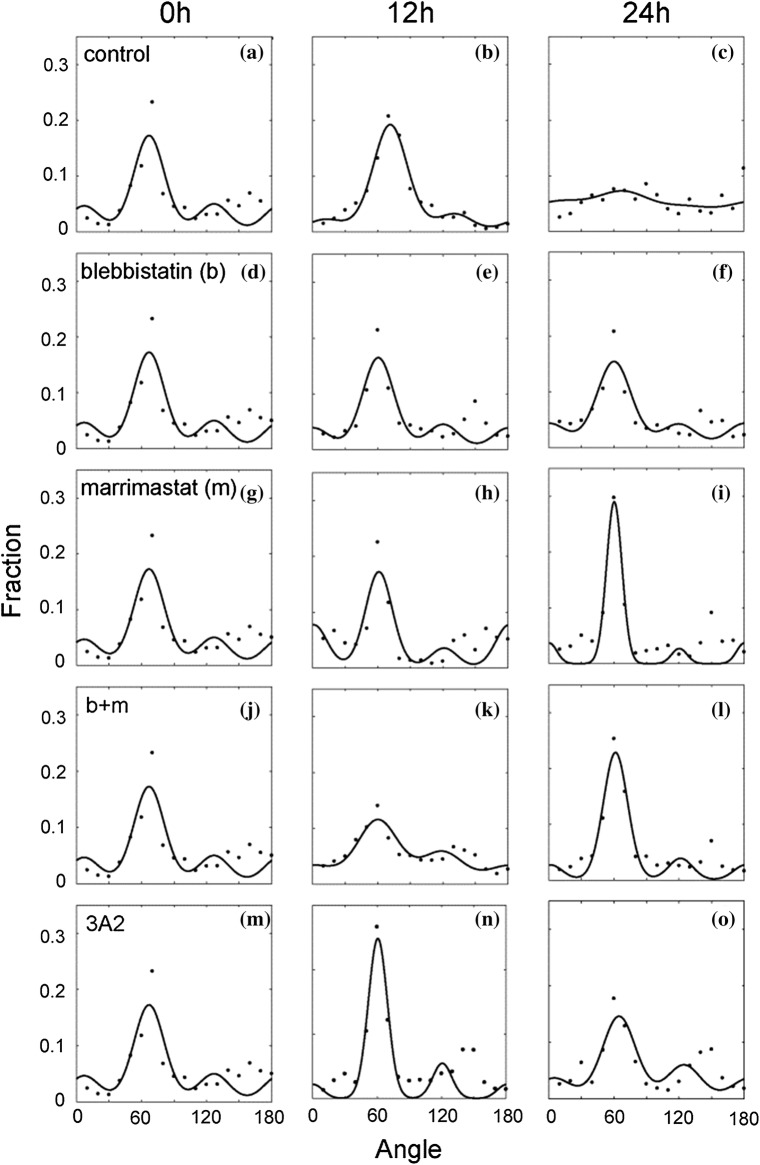
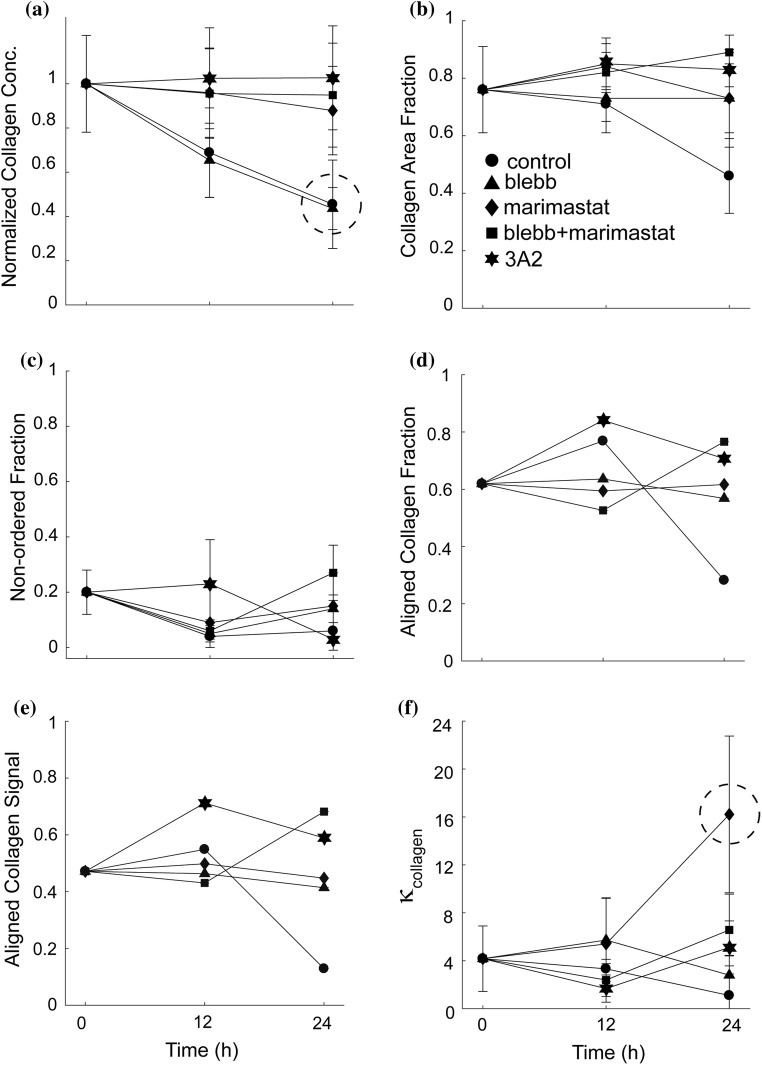
The collagen fibrils were imaged using SHG imaging and the distribution of collagen fibril orientations was determined and fitted to the same distribution as used above. In control samples collagen orientation did not differ between substrates that had no cells and substrates plated with cells for 12 h (Figs. 4a and 4b). However, collagen fiber alignment decreased dramatically between 12 and 24 h (Figs. 4b and 4 c). In order to study whether contractility plays an important role in rearranging aligned fibrils, MDA-MB-231 cells were treated with blebbistatin. This resulted in blocking changes in collagen orientation distribution (Figs. 4d–4f). When MMP activity was blocked using marimastat, the alignment appeared to improve over time (Figs. 4g–4i). This response was somewhat tempered when blebbistatin was added (Figs. 4j–4l). Interestingly, blocking MT1-MMP with 3A2 had distinct effects compared to marimastat. A peak in alignment appeared at 12 h and marginally decreased after that (Figs. 4m–4o). Since this distribution indicates orientation and not density, we quantified the collagen concentration under the same conditions by using the intensity of the SHG images. Aligned collagen assembled on mica in the presence of cells showed a marked decrease in collagen density over time (Fig. 5a). This decrease was not dependent on contractility as blebbistatin does not block the response (Fig. 5a). However, blocking either MT1-MMP alone (3A2), all MMPs (marimastat) or contractility and MMPs (blebbistatin + marimastat) does inhibit the decrease in collagen density (Fig. 5a). The images obtained through SHG microscopy were processed to identify the fraction of area that was ordered, non-ordered and contained no collagen (see “Materials and Methods”). This processing occurred over one ROI (~ 1 μm). The fraction of the area that was occupied by collagen under the different perturbations was calculated (Fig. 5b). Surprisingly, while blebbistatin did not inhibit the decrease in collagen intensity (Fig. 5a), it did inhibit the decrease in area fraction that was covered by collagen (Fig. 5b). There were no significant differences in the non-ordered collagen fractions across the samples (Fig. 5c) and aligned collagen fraction roughly correlated with collagen area fraction (Fig. 5d) and matched what was seen in Fig. 4. Since the collagen density changes across conditions, we calculated the product of the collagen area fraction and the aligned collagen fraction, giving the amount of collagen that is organized (Fig. 5d). The fitting parameter κcollagen, showed no significant difference across the samples, apart from the marimastat treatment sample at 24 h, where its collagen alignment increased in line with Fig. 4i.
Figure 4.
Collagen alignment on mica substrates seeded with MDA-MB-231 cells treated with a myosin II-mediated contractility inhibitor (blebbistatin), a pan-MMP inhibitor (marimastat) or an MMP-14 function blocking antibody (3A2). The angular distribution of collagen of MDA-MB-231 cells at 0, 12, and 24 h treated with (a–c) no inhibitor (d–f) blebbistatin (g–i) marimastat (j–l) both marimastat and blebbistatin, and (m–o) 3A2. Circles are data points and the lines are the von Mises fits to the data. Nexperiments > 3 and Nimages > 9.
Figure 5.
Collagen fiber alignment on mica substrates seeded with MDA-MB-231 cells treated with a myosin II-mediated contractility inhibitor (blebbistatin), a pan-MMP inhibitor (marimastat) or an MMP-14 function blocking antibody (3A2). (a) The relative intensity of the collagen fibers on the mica-based substrates over 24 h, normalized to the initial intensity of the control at 0 h. The mica and blebbistatin substrates are circled. (b) The collagen area fraction of the sum of both ordered and non-ordered collagen fractions. (c) The non-ordered collagen fractions of the collagen signal. (d) Collagen alignment fraction, (e) Aligned collagen signal fraction, as a product of the aligned collagen fraction and the normalized collagen intensity and (f) distribution spread parameter, κcollagen. Cells plated on aligned collagen fibrils assembled on mica (control, circle) treated with blebbistatin (30 μM, triangle), marimastat (10 μM, diamond), marimastat and blebbistatin (10 and 30 μM, squares) or 3A2 antibody (500 nM, star). Error bars represent 95% confidence intervals. Nexperiments > 3 and Nimages > 9.
Discussion
Contact guidance in response to aligned collagen fibers is an important facet of cancer invasion and metastasis.9,40 The degree to which cells engage in contact guidance is intimately related to the degree of collagen fiber alignment and consequently the noise in the contact guidance signal.17,51 However, collagen fiber alignment dynamically changes over time due to both contractility and MMP activity of stromal cells in the tumor microenvironment. Cancer cells can also remodel collagen fibers during the process of migration.4,8 This can either act to change mechanical properties close to the cell23 or locally disorganize or organize collagen fibers, thus amplifying2,14 or dampening the original aligned collagen fiber signal. This results in different degrees of contact guidance and different invasion or metastasis rates. In this paper, we examine the link between cell and collagen alignment by perturbing the myosin II and MMP activity of MDA-MB-231 cells migrating on aligned collagen fibrils. Epitaxially grown, aligned collagen fibrils are loosely attached to mica substrate due to non-covalent, electrostatic interactions between collagen and the surface. Because of the loose interaction between collagen and the mica surface, MDA-MB-231 cells robustly detach between 12 and 24 h, producing a sensitive phenotype that can be used to probe how perturbations in cell traction generation and collagen degradation affect cell and collagen fibril presence and alignment. MTLn3 cells do not detach and appear to deform the collagen fibrils less. It is possible that this difference is due to the different amounts of serum in the media used for each cell line. Serum contains fibronectin16 that can bind collagen. However, given the prior work outlining the differences in traction forces, the differences in deformations that we observe are likely cell-line intrinsic. Furthermore, MTLn3 cells appear to generate more MMP activity per unit cell. However, for technical reasons, this measurement was made in serum free conditions and only assessed secreted MMPs. Surface-bound MMP-14 activity was not assessed and is likely important in collagen degradation.25 Indeed, MDA-MB-231 cells express MMP-14.57
When myosin II was inhibited, MDA-MB-231 cells did not detach from the surface. Several possibilities for this result exist. First, it is possible that detachment requires contractility-mediated traction force generation to pull the collagen fibrils off the surface. Indeed, collagen area fraction appears to remain constant as others have seen on these substrates in different cells.25 However, collagen concentration on the surface appears to decrease when myosin II is inhibited. We will discuss potential mechanisms to explain this discrepancy below. Second, it is possible that detachment requires MMP activity and blocking myosin II acts to subdue or decrease MMP activity. Indeed, we13 and others19 have shown that contractility is required for maximum ECM degradation and MMP activity and contractility inhibitors can block MMP activity almost as potently as MMP inhibitors. This would seem to agree with the fact that myosin II inhibition erases a decrease in collagen area fraction, although this piece of evidence also agrees with the first mechanism. The decrease in collagen density in the presence of myosin II inhibition would argue against a large drop in MMP activity. Taken together, it is likely these mechanisms are acting jointly to cause contractility-mediated decreases in cell remodeling.
MMP inhibition acts somewhat differently. Both pan-MMP and MMP-14 inhibitors block cell detachment. Furthermore, both seem to block the decrease in both collagen intensity and area fraction covered with collagen. This agrees with data indicating that MMP-14 is a primary collagenase related to collagen fiber remodeling and aligned collagen fibrils specifically.25 Interestingly, the alignment of collagen goes up when MMPs in general or MMP-14 is inhibited. Contractility is still at work and cells may be able to better align fibers close to the cell. The disorganization of the collagen fibrils may initially proceed through MMP-14 with other secreted MMPs following, hence the different kinetics of enhanced collagen fibril alignment in response to the MMP-14 function blocking antibody with respect to marimastat. While we certainly see cooperation between contractility and degradation in remodeling the collagen fibrils and affecting cell detachment and alignment, we did not notice any synergy. Conditions, where both blebbistatin and marimastat are added appeared to produce phenotypes from one or the other of the single perturbation experiment.
Interestingly, although collagen alignment was not changed between 0-24 h under blebbistatin treatment, normalized collagen concentration decreased to the same extent as the control condition, while fraction of area occupied by collagen did not change. This might suggest that MMPs can thin or partially degrade the collagen signal, but contractility is needed for the dramatic large-scale changes in collagen fibril coverage. It is unlikely that these changes are due to changes in collagen secretion as MDA-MB-231 cells do not secrete collagen.24 These collagen fibril fields formed from many laterally interacting collagen fibrils are thin, measuring on the order of nanometers in thickness. The SHG signal is presumably collected as the square of the integrated average collagen density throughout the thickness of the fibrils, and no out-of-focus collagen exists. Consequently, if the fibrils were to thin out due to MMP activity, but not be completely degraded in particular areas, this would result in lower collagen intensity with roughly the same area fraction covered by collagen. The collagen alignment remained roughly the same after blebbistatin, but the cell alignment decreased. Indeed, there is indication that myosin II acts to mediate in part contact guidance and the presence of a contact guidance cue does not necessarily indicate that a cell will recognize and align its polarity with the long axis of the collagen fibrils.
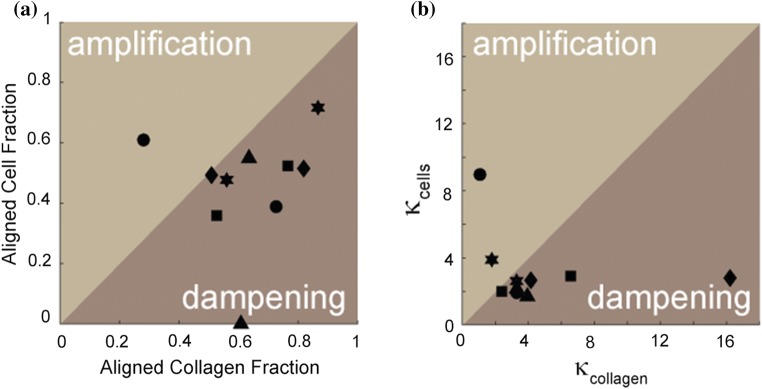
Finally, we wanted to assess how the alignment of cells corresponded to the alignment of collagen across all conditions. Consequently we plotted aligned cell fraction against aligned collagen fraction (Fig. 6a) and κcells against κcollagen (Fig. 6b). If both cell and collagen metrics are the same, we would predict no amplification or dampening of the contact guidance signal. However, if the cell metrics are larger than the collagen metrics, amplification occurs. Under the control condition, amplification is seen. When most of the collagen is ripped from the surface at 24 h and only a few aligned collagen fibrils remain, the existing cells on aligned collagen fibrils are still highly aligned (Fig. 6a). When myosin II or MMPs are inhibited, dampening occurs. Inhibition of contractility retains collagen alignment constant over time as it appears to block large changes in collagen density, but cells are disordered, presumably due to the direct affect that contractility inhibitors have on the cells ability to engage in contact guidance. Contractility appears to modulate contact guidance,53 but not spreading on contact guidance cues.47 However, inhibition of degradation appears to act differently. Blocking MMP activity actually enhances the collagen alignment, but the cell alignment stays roughly the same or decreases slightly. This suggests an indirect effect on contact guidance through modulation of the alignment of the collagen and likely not through direct changes in contact guidance. This suggests that myosin II-mediated contractility and MMP-mediated degradation cooperatively act during contact guidance, but may affect it through both direct and indirect mechanisms.
Figure 6.
(a) Aligned cell and collagen fraction and (b) distribution spread parameter (κcollagen) for both cells and collagen under control and inhibitor conditions. Cells plated on aligned collagen fibrils assembled on mica (circle) treated with blebbistatin (30 μM, triangle), marimastat (10 μM, diamond), marimastat and blebbistatin (10 and 30 μM, squares) or 3A2 antibody (500 nM, star). Cell amplification of the directional cue occurs when either the alignment fraction or the κcollagen is larger for cells than for collagen. Cell dampening of the directional cue occurs when either the alignment fraction or the κcollagen is smaller for cells than for collagen.
Conclusions
In this paper, we show that MDA-MB-231 cells deform epitaxially grown and aligned collagen fibrils more robustly than MTLn3 cells. During remodeling, both contractility and MMPs cooperate to degrade and detach collagen from the mica, leading to large scale cell detachment. However, the effects of myosin II-mediated contraction and MMP-mediated degradation are subtly different. Since myosin II regulates contractility-dependent alignment as well as collagen deformation leading to remodeling, cells that remain on the substrate are not well aligned. In addition, collagen fibrils are still degraded under myosin II inhibition, but not to the extent that leads to large scale cell detachment. MMP activity on the other hand acts to temper collagen fibril alignment by cells as blocking MMP activity leads to increased collagen alignment over time. Cooperation between contractility and degradation acts to amplify the response to the contact guidance cue. Cells are more aligned than what the collagen fibril field might suggest. By inhibiting contractility, most collagen fibrils keep the same alignment, but cell alignment becomes more disordered, dampening the sensing of the contact guidance cue. By inhibiting MMP degradation, collagen fibrils are more aligned than in the control condition, while cell alignment remains about the same, also dampening the response to the contract guidance cue, but for a different reason. This work suggests that contractility and degradation impact contact guidance through different mechanisms that both directly affect contact guidance at the level of cell motility and indirectly affect contact guidance at the level of collagen rearrangement.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We acknowledge Zhiqi Yao and Andrew Hillier with help on the AFM imaging and Jacob Nuhn for help with the MMP assays.
Funding
This work was supported by the National Institutes of Health/National Institute for General Medical Sciences [R01GM115672]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Conflict of interest
Juan Wang, Anuraag Boddupalli, Joseph Koelbl, Dong Hyun Nam, Xin Ge, Kaitlin M. Bratlie and Ian C. Schneider state they have no conflicts of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Abbreviations
- TME
Tumor microenvironment
- SHG
Second harmonic generation
- ECM
Extracellular matrix
- TIMP
Tissue inhibitor of matrix metalloproteinase
- MMP
Matrix metalloproteinase
- DMEM
Dulbecco’s modified Eagles medium
- FBS
Fetal bovine serum
- NA
Numerical aperture
- HSD
Honest significant difference
References
- 1.Alexander NR, Branch KM, Parekh A, Clark ES, Lwueke LC, Guelcher SA, Weaver AM. Extracellular matrix rigidity promotes invadopodia activity. Curr. Biol. 2008;18:1295–1299. doi: 10.1016/j.cub.2008.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barocas VH, Tranquillo RT. An anisotropic biphasic theory of tissue-equivalent mechanics: the interplay among cell traction, fibrillar network deformation, fibril alignment, and cell contact guidance. J. Biomech. Eng. Trans. ASME. 1997;119:137–145. doi: 10.1115/1.2796072. [DOI] [PubMed] [Google Scholar]
- 3.Biela SA, Su Y, Spatz JP, Kemkemer R. Different sensitivity of human endothelial cells, smooth muscle cells and fibroblasts to topography in the nano-micro range. Acta Biomater. 2009;5:2460–2466. doi: 10.1016/j.actbio.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Chiu C-L, Digman MA, Gratton E. Cell matrix remodeling ability shown by image spatial correlation. J. Biophys. 2013;2013:8. doi: 10.1155/2013/532030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark P, Connolly P, Curtis ASG, Dow JAT, Wilkinson CDW. Topographical control of cell behavior. 2. Multiple grooved substrata. Development. 1990;108:635–644. doi: 10.1242/dev.108.4.635. [DOI] [PubMed] [Google Scholar]
- 6.Crocker JC, Grier DG. Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci. 1996;179:298–310. doi: 10.1006/jcis.1996.0217. [DOI] [Google Scholar]
- 7.Dickinson RB, Guido S, Tranquillo RT. Biased cell-migration of fibroblasts exhibiting contact guidance in oriented collagen gels. Ann. Biomed. Eng. 1994;22:342–356. doi: 10.1007/BF02368241. [DOI] [PubMed] [Google Scholar]
- 8.Drifka CR, Loeffler AG, Esquibel CR, Weber SM, Eliceiri KW, Kao WJ. Human pancreatic stellate cells modulate 3D collagen alignment to promote the migration of pancreatic ductal adenocarcinoma cells. Biomed. Microdevices. 2016;18:105. doi: 10.1007/s10544-016-0128-1. [DOI] [PubMed] [Google Scholar]
- 9.Drifka CR, Loeffler AG, Mathewson K, Keikhosravi A, Eickhoff JC, Liu YM, Weber SM, Kao WJ, Eliceiri KW. Highly aligned stromal collagen is a negative prognostic factor following pancreatic ductal adenocarcinoma resection. Oncotarget. 2016;7:76197–76213. doi: 10.18632/oncotarget.12772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fellouse FA, Sidhu S. Making Antibodies in Bacteria. Baca Raton: CRC Press; 2013. pp. 151–172. [Google Scholar]
- 11.Guo C, Kaufman LJ. Flow and magnetic field induced collagen alignment. Biomaterials. 2007;28:1105–1114. doi: 10.1016/j.biomaterials.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 12.Haage A, Nam DH, Ge X, Schneider IC. Matrix metalloproteinase-14 is a mechanically regulated activator of secreted MMPs and invasion. Biochem. Biophys. Res. Commun. 2014;450:213–218. doi: 10.1016/j.bbrc.2014.05.086. [DOI] [PubMed] [Google Scholar]
- 13.Haage A, Schneider IC. Cellular contractility and extracellular matrix stiffness regulate matrix metalloproteinase activity in pancreatic cancer cells. Faseb J. 2014;28:3589–3599. doi: 10.1096/fj.13-245613. [DOI] [PubMed] [Google Scholar]
- 14.Hall MS, Alisafaei F, Ban E, Feng XZ, Hui CY, Shenoy VB, Wu MM. Fibrous nonlinear elasticity enables positive mechanical feedback between cells and ECMs. Proc. Natl. Acad. Sci. USA. 2016;113:14043–14048. doi: 10.1073/pnas.1613058113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartman O, Zhang C, Adams EL, Farach-Carson MC, Petrelli NJ, Chase BD, Rabolt JF. Microfabricated electrospun collagen membranes for 3-D cancer models and drug screening applications. Biomacromolecules. 2009;10:2019–2032. doi: 10.1021/bm8012764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayman EG, Ruoslahti E. Distribution of fetal bovine serum fibronectin and endogenous rat-cell fibronectin in extracellular matrix. J. Cell Biol. 1979;83:255–259. doi: 10.1083/jcb.83.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacchetti E, Di Rienzo C, Meucci S, Nocchi F, Beltram F, Cecchini M. Wharton’s Jelly human Mesenchymal Stem Cell contact guidance by noisy nanotopographies. Sci. Rep. 2014;4:3830. doi: 10.1038/srep03830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janani G, Pillai MM, Selvakumar R, Bhattacharyya A, Sabarinath C. An in vitro 3D model using collagen coated gelatin nanofibers for studying breast cancer metastasis. Biofabrication. 2017;9:015016. doi: 10.1088/1758-5090/aa5510. [DOI] [PubMed] [Google Scholar]
- 19.Jerrell RJ, Parekh A. Cellular traction stresses mediate extracellular matrix degradation by invadopodia. Acta Biomater. 2014;10:1886–1896. doi: 10.1016/j.actbio.2013.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang FZ, Horber H, Howard J, Muller DJ. Assembly of collagen into microribbons: effects of pH and electrolytes. J. Struct. Biol. 2004;148:268–278. doi: 10.1016/j.jsb.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 21.Julias M, Buettner HM, Shreiber DI. Varying assay geometry to emulate connective tissue planes in an in vitro model of acupuncture needling. Anat. Rec. 2011;294:243–252. doi: 10.1002/ar.21308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Julias M, Edgar LT, Buettner HM, Shreiber DI. An in vitro assay of collagen fiber alignment by acupuncture needle rotation. Biomed. Eng. Online. 2008;7:19. doi: 10.1186/1475-925X-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keating M, Kurup A, Alvarez-Elizondo M, Levine AJ, Botvinick E. Spatial distributions of pericellular stiffness in natural extracellular matrices are dependent on cell-mediated proteolysis and contractility. Acta Biomater. 2017;57:304–312. doi: 10.1016/j.actbio.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim SH, Lee HY, Jung SP, Kim S, Lee JE, Nam SJ, Bae JW. Role of secreted type I collagen derived from stromal cells in two breast cancer cell lines. Oncol. Lett. 2014;8:507–512. doi: 10.3892/ol.2014.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirmse R, Otto H, Ludwig T. Interdependency of cell adhesion, force generation and extracellular proteolysis in matrix remodeling. J. Cell Sci. 2011;124:1857–1866. doi: 10.1242/jcs.079343. [DOI] [PubMed] [Google Scholar]
- 26.Koster S, Leach JB, Struth B, Pfohl T, Wong JY. Visualization of flow-aligned type I collagen self-assembly in tunable pH gradients. Langmuir. 2007;23:357–359. doi: 10.1021/la062473a. [DOI] [PubMed] [Google Scholar]
- 27.Kraning-Rush CM, Carey SP, Califano JP, Smith BN, Reinhart-King CA. The role of the cytoskeleton in cellular force generation in 2D and 3D environments. Phys. Biol. 2011;8:015009. doi: 10.1088/1478-3975/8/1/015009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee KB, Nam DH, Nuhn JAM, Wang J, Schneider IC, Ge X. Direct expression of active human tissue inhibitors of metalloproteinases by periplasmic secretion in Escherichia coli. Microb. Cell. Fact. 2017;16:73. doi: 10.1186/s12934-017-0686-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leow WW, Hwang W. Epitaxially guided assembly of collagen layers on mica surfaces. Langmuir. 2011;27:10907–10913. doi: 10.1021/la2018055. [DOI] [PubMed] [Google Scholar]
- 30.Malik R, Lelkes PI, Cukierman E. Biomechanical and biochemical remodeling of stromal extracellular matrix in cancer. Trends Biotechnol. 2015;33:230–236. doi: 10.1016/j.tibtech.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matthews JA, Wnek GE, Simpson DG, Bowlin GL. Electrospinning of collagen nanofibers. Biomacromolecules. 2002;3:232–238. doi: 10.1021/bm015533u. [DOI] [PubMed] [Google Scholar]
- 32.Meehan S, Nain AS. Role of suspended fiber structural stiffness and curvature on single-cell migration, nucleus shape, and focal-adhesion-cluster length. Biophys. J. 2014;107:2604–2611. doi: 10.1016/j.bpj.2014.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mierke CT, Rosel D, Fabry B, Brabek J. Contractile forces in tumor cell migration. Eur. J. Cell Biol. 2008;87:669–676. doi: 10.1016/j.ejcb.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nain AS, Phillippi JA, Sitti M, MacKrell J, Campbell PG, Amon C. Control of cell behavior by aligned micro/nanofibrous biomaterial scaffolds fabricated by spinneret-based tunable engineered parameters (STEP) technique. Small. 2008;4:1153–1159. doi: 10.1002/smll.200800101. [DOI] [PubMed] [Google Scholar]
- 35.Nam DH, Ge X. Development of a periplasmic FRET screening method for protease inhibitory antibodies. Biotechnol. Bioeng. 2013;110:2856–2864. doi: 10.1002/bit.24964. [DOI] [PubMed] [Google Scholar]
- 36.Nam DH, Rodriguez C, Remacle AG, Strongin AY, Ge X. Active-site MMP-selective antibody inhibitors discovered from convex paratope synthetic libraries. Proc. Natl. Acad. Sci. USA. 2016;113:14970–14975. doi: 10.1073/pnas.1609375114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narayanan B, Gilmer GH, Tao JH, De Yoreo JJ, Ciobanu CV. Self-assembly of collagen on flat surfaces: the interplay of collagen-collagen and collagen-substrate interactions. Langmuir. 2014;30:1343–1350. doi: 10.1021/la4043364. [DOI] [PubMed] [Google Scholar]
- 38.Nuhn JAM, Perez AM, Schneider IC. Contact guidance diversity in rotationally aligned collagen matrices. Acta Biomater. 2018;66:248–257. doi: 10.1016/j.actbio.2017.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poole K, Khairy K, Friedrichs J, Franz C, Cisneros DA, Howard J, Mueller D. Molecular-scale topographic cues induce the orientation and directional movement of fibroblasts on two-dimensional collagen surfaces. J. Mol. Biol. 2005;349:380–386. doi: 10.1016/j.jmb.2005.03.064. [DOI] [PubMed] [Google Scholar]
- 40.Provenzano PP, Eliceiri KW, Campbell JM, Inman DR, White JG, Keely PJ. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006;4:38. doi: 10.1186/1741-7015-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramirez-San Juan GR, Oakes PW, Gardel ML. Contact guidance requires spatial control of leading-edge protrusion. Mol. Biol. Cell. 2017;28:1043–1053. doi: 10.1091/mbc.e16-11-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ray A, Lee O, Win Z, Edwards RM, Alford PW, Kim DH, Provenzano PP. Anisotropic forces from spatially constrained focal adhesions mediate contact guidance directed cell migration. Nat. Commun. 2017;8:14923. doi: 10.1038/ncomms14923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ray A, Slama ZM, Morford RK, Madden SA, Provenzano PP. Enhanced directional migration of cancer stem cells in 3D aligned collagen matrices. Biophys. J. 2017;112:1023–1036. doi: 10.1016/j.bpj.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riching KM, Cox BL, Salick MR, Pehlke C, Riching AS, Ponik SM, Bass BR, Crone WC, Jiang Y, Weaver AM, Eliceiri KW, Keely PJ. 3D collagen alignment limits protrusions to enhance breast cancer cell persistence. Biophys. J. 2014;107:2546–2558. doi: 10.1016/j.bpj.2014.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruiz SA, Chen CS. Microcontact printing: a tool to pattern. Soft Matter. 2007;3:168–177. doi: 10.1039/B613349E. [DOI] [PubMed] [Google Scholar]
- 46.Saeidi N, Sander EA, Ruberti JW. Dynamic shear-influenced collagen self-assembly. Biomaterials. 2009;30:6581–6592. doi: 10.1016/j.biomaterials.2009.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sales A, Holle AW, Kemkemer R. Initial contact guidance during cell spreading is contractility-independent. Soft Matter. 2017;13:5158–5167. doi: 10.1039/C6SM02685K. [DOI] [PubMed] [Google Scholar]
- 48.Singh S, Bandini SB, Donnelly PE, Schwartz J, Schwarzbauer JE. A cell-assembled, spatially aligned extracellular matrix to promote directed tissue development. J. Mater. Chem. B. 2014;2:1449–1453. doi: 10.1039/c3tb21864c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun M, Stetco A, Merschrod EF. Surface-templated formation of protein microfibril arrays. Langmuir. 2008;24:5418–5421. doi: 10.1021/la703292h. [DOI] [PubMed] [Google Scholar]
- 50.Teixeira AI, Abrams GA, Bertics PJ, Murphy CJ, Nealey PF. Epithelial contact guidance on well-defined micro- and nanostructured substrates. J. Cell Sci. 2003;116:1881–1892. doi: 10.1242/jcs.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tonazzini I, Meucci S, Faraci P, Beltram F, Cecchini M. Neuronal differentiation on anisotropic substrates and the influence of nanotopographical noise on neurite contact guidance. Biomaterials. 2013;34:6027–6036. doi: 10.1016/j.biomaterials.2013.04.039. [DOI] [PubMed] [Google Scholar]
- 52.Wang J, Petefish JW, Hillier AC, Schneider IC. Epitaxially grown collagen fibrils reveal diversity in contact guidance behavior among cancer cells. Langmuir. 2015;31:307–314. doi: 10.1021/la503254x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang J, Schneider IC. Myosin phosphorylation on stress fibers predicts contact guidance behavior across diverse breast cancer cells. Biomaterials. 2017;120:81–93. doi: 10.1016/j.biomaterials.2016.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Worthen DM, Cleveland PH, Slight JR, Abare J. Selective binding-affinity of human-plasma fibronectin for the collagens I-IV. Investig. Ophthalmol. Vis. Sci. 1985;26:1740–1744. [PubMed] [Google Scholar]
- 55.Wyckoff JB, Pinner SE, Gschmeissner S, Condeelis JS, Sahai E. ROCK- and myosin-dependent matrix deformation enables protease-independent tumor-cell invasion in vivo. Curr. Biol. 2006;16:1515–1523. doi: 10.1016/j.cub.2006.05.065. [DOI] [PubMed] [Google Scholar]
- 56.Yang DL, Zeng FX, Sun M, Gu WH, Li L. Investigation on properties of collagen nanowires quasiepitaxially grown on mica lattice plane. Chin. J. Anal. Chem. 2017;45:465–469. doi: 10.1016/S1872-2040(17)61003-X. [DOI] [Google Scholar]
- 57.Zarrabi K, Dufour A, Li J, Kuscu C, Pulkoski-Gross A, Zhi JZ, Hu YJ, Sampson NS, Zucker S, Cao J. Inhibition of matrix metalloproteinase 14 (MMP-14)-mediated cancer cell migration. J. Biol. Chem. 2011;286:33167–33177. doi: 10.1074/jbc.M111.256644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou ZN, Sharma VP, Beaty BT, Roh-Johnson M, Peterson EA, Van Rooijen N, Kenny PA, Wiley HS, Condeelis JS, Segall JE. Autocrine HBEGF expression promotes breast cancer intravasation, metastasis and macrophage-independent invasion in vivo. Oncogene. 2014;33:3784–3793. doi: 10.1038/onc.2013.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.