Abstract
Cell penetrating peptides (CPPs) have garnered significant attention as a method to introduce reporters and therapeutics into intact cells. While numerous studies have been performed identifying new CPP sequences, relatively little is known about their uptake efficiency at the single-cell level. Here, a droplet microfluidic trapping array was used to characterize CPP uptake across a population of single intact cells. The microfluidic device allowed for facile and rapid isolation and analysis of single cell fluorescence in a 787-member overhead trapping array with >99% droplet trapping efficiency. The permeability efficiencies of four different CPPs were studied and compared in HeLa cells. Population analysis was performed using linkage hierarchical cluster analysis by R programming to bin cells into subpopulations expressing very low to very high peptide uptake efficiencies. CPP uptake was observed to be heterogeneous across the population of cells with peptide concentration and sequence both playing important roles in the diversity of CPP uptake, the overall peptide uptake efficiency, and the intracellular homogeneity of peptide distribution. This microfluidic-based analytical approach finds application in personalized medicine and provides new insight in the heterogeneity of CPP uptake which has the potential to affect both biosensor and drug internalization in intact cells.
Keywords: Peptides, High-throughput screening, Single-cell analysis, Microfluidics
Grphical Abstract
Introduction
Due to the abundance of pharmaceuticals designed to target intracellular proteins, efficient methods to deliver drugs, vectors, and biosensors into intact cells has gained tremendous attention in recent years [1]. Examples include small molecule inhibitors used as chemotherapeutics for treating cancer and other human diseases, silencing RNA, and biosensors for measuring the activity of intracellular enzymes [2–4]. In most cases, population heterogeneity manifests itself in non-normal distributions in which case the mean value of the blended responses is a poor representative of the entire population, and therefore fails to accurately describe the population behavior [5]. For this reason, there have been substantial efforts to analyze a large number of individual cells and obtain population distributions to better describe and predict the behavior of cell populations and address the heterogeneity associated with them [6, 7]. Traditionally, the gold standard for performing single cell analysis has been flow cytometry; however, it is limited to static measurements [5, 8]. This limitation has been overcome by using laser scanning cytometry which allows for scanning a single cell over time, but its throughput is lower than flow cytometry and it is still limited to whole cell responses, and therefore fails to characterize the intracellular events [5].
In recent years, microfluidics have revolutionized the state of single cell analysis by facilitating high-throughput screening of therapeutic-level quantities of cells using minute sample sizes which is made possible due to the micron level sizes of flow channels in microfluidic devices [9]. Droplet microfluidics, which operate through generating picolitre-sized droplets in an immiscible continuous oil flow, have allowed researchers to perform high-throughput dynamic analysis of single cells while simultaneously monitoring the dynamics of the intracellular environment which is of critical importance for many biochemical applications [8, 10, 7, 11]. Droplet microfluidic devices provide several advantages compared to competing technologies including overcoming the limitations of flow cytometry to whole cell responses by facilitating precise dynamic measurements in the sub-cellular level and facilitating the analysis of suspension cell lines that cannot be assayed using microtiter plates and microfluidic cell trappers. Moreover, droplet microfluidic devices contribute to system miniaturization by reducing the sample volumes from tens of microliters to in standard microtiter plates to the nano- or picoliter scale. Droplet microfluidics coupled with increasingly sensitive molecular biology tools has opened the venue for transformational progress in clinical diagnostics and prognostics in the past few years [9, 12]. Several reports have demonstrated the application of droplet microfluidics for performing biological assays such as PCR based biomarker discovery [13] enzyme kinetics analysis [14], cell-cell communication [15] and several other applications [11]. Meanwhile, fluorescent assays provide the most sensitive and robust methods for observing biological processes [16]. Traditionally, the analysis of biochemical fluorescent substrates in single cells using droplet microfluidics was performed using a two-step process. First, a device was used to encapsulate individual cells in droplets followed by off-chip collection and incubation. These droplets were then transferred to a second device for screening and analysis [17]. Alternatively, the analysis can be performed by encapsulating single cells and imaging under continuous flow in a large downstream channel; however, in this method, indexing the droplets to keep track single cells is challenging. Recent efforts with droplet generators have incorporated a droplet trapping array downstream of the flow channel to facilitate dynamic high-throughput screening of single cells on-chip while minimizing the chance for contamination and decreasing operator influence [18]. Interesting recent examples include a microfluidic trap array developed by Jackson-Holmes and colleagues for analyzing individual stem cell aggregates [19] and a static droplet array developed by Jin and colleagues for analysis of cell-cell communication in a confined microenvironment [15]. Trapping arrays are highly beneficial due to enabling process integration and increasing the overall analytical throughput by confining cells in pre-defined patterns which significantly reduces the computational burden and the time required for both image collection and automated image processing. As such, microfluidic droplet trapping arrays make an ideal platform for performing single cell screening of intracellular fluorescence by enhancing system miniaturization, integration, throughput, and automation; all of which are major areas of focus in current directions of microfluidic developments.
This paper describes the use of one such microfluidic droplet trapping arrays to perform single cell analysis of peptide uptake in intact cancer cells. In this work, a 787-membered trapping array was incorporated with a series of fluorescent cell penetrating peptides (CPPs): two commercially available peptides (TAT and ARG) and two novel CPPs, (RWRWR and OWRWR) to perform sequence-, concentration- and population-based uptake and intracellular distribution studies on single cancer cells. This microfluidic droplet trapping array enabled easy encapsulation and compartmentalization of single cells for fluorescent imaging. These images were processed by identifying regions of interest (ROIs) around individual cells to quantify the uptake signals measured by fluorescent microscopy. The measured fluorescent signals were then analyzed by R programming to perform hierarchical cluster analysis and bin the cells into underlying subpopulations based on peptide uptake. This analysis confirmed previous findings that CPP uptake is strongly dependent upon concentration. The intracellular distribution of peptide uptake in single cells was also characterized to reveal a correlation with the overall uptake efficiency. Results indicated that increasing peptide concentrations caused the CPPs to distribute more homogeneously in the cytoplasm of the cells and resulted a stronger uptake signal. Numerical simulations on single cell scatter plots and population histograms allowed for predictive analysis of cell populations and identification of common trends. Finally, a comparison between the uptake of a select CPP (ARG) at a 50 μM concentration in two different model cell lines with different sizes, HeLa (average cross-sectional area of 500 μm2) and OPM-2 (average cross-sectional area of 90 μm2) revealed that although the mean fluorescent intensities measured for the different cell lines were significantly different, their corresponding fluorescent densities (fluorescence/area) were not. This observation identified cell size as a major predictor of the fluorescent outputs measured in different cell lines. Similarly, this finding demonstrated that the uptake potential of CPPs was independent of the cell line being an adherent or a suspension line. These robust single cell analyses shine light on the heterogeneity of CPP uptake and its intracellular distribution in cancer cells, thus providing better understanding on biosensor and drug internalization in intact cells.
Materials and Methods
Device design
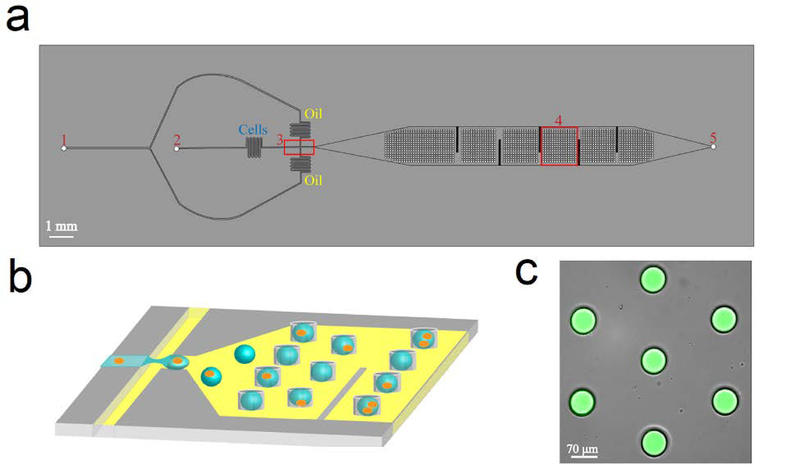
The features of the microfluidic droplet trapping array consist of two layers, each of which has a height of 40 μm. The bottom layer consists of the flow focusing junction followed by a large open channel. The features of the flow focusing junction and oil and water inlets in the bottom layer are similar to those presented by Mazutis et al.[22] The top layer consists of the trapping array which is located overhead of the larger channel, downstream of the flow focusing junction (Fig. 1). The oil and water phase inlet ports are 750 and 470 μm in diameter. The oil inlet channel is split into two channels which converge at the flow focusing junction leading to a channel that is 20 μm wide. The aqueous inlet stream enters the junction in a channel that is 34 μm wide. After formation, droplets flow through a 700 μm narrow channel (to allow for droplet stabilization) and into the trapping array channel. Here, an array of 787 microwells, 70 μm in diameter and 40 μm in height are imprinted into the overhead PDMS in a hexagonal pattern to facilitate optimal trapping and prevent droplet ejection. The trapping array is similar to one presented by Khorshidi et al.[18] In order to assure high droplet trapping efficiency, vertical fins were incorporated in the bottom fluidic layer to increase the residence time of the droplets and prevent them from flowing adjacent to the walls and bypassing the trapping array. At the end of the trapping array channel, an outlet port was incorporated for the flow to exit the channel.
Fig 1.
Schematic of the microfluidic droplet trapping platform. (a) Top view of the device illustrating the (1) oil inlet, (2) water inlet, (3) flow-focusing junction, (4) droplet trapping array, and (5) outlet. (b) Schematic of the generation and trapping of droplets containing cells into 70 μm circular microwells imprinted 40 μm into the PDMS directly above the downstream channel. (c) Overlaid microscope image of aqueous droplet containing 4mM 5(6)-carboxyfluorescein isolated in the trapping array.
Device fabrication
The droplet microfluidic trapping array was fabricated using a combination of soft lithography and polydimethylsiloxane (PDMS) replication techniques as previously reported [22]. AutoCAD (2015 version, AutoDesk) was used to create geometries for the microchannels which were printed onto iron oxide/chrome masks (Front Range). A silicon master was fabricated using a two-step lithography process to generate the bottom fluidic layer and top trapping array. SU-8 2025 (MicroChem) was spun onto a clean 4” silicon wafer (University Wafer) using a spin coater (WS-650 Series Spin Processor, Laurell Technologies Corp) at 3000 rpm for 30 s to achieve a thickness of 40 μm for the bottom fluidic layer. The wafer was pre-baked at 65°C for 5 min and then baked at 95°C for 25 min followed by a gradual cooldown to 25 °C. UV exposure was performed in a custom-built UV exposure system with a Blak-Ray B-100 series UV lamp (UVP, LLC) with 1 mW/cm2 power intensity for 60 s. A post exposure bake was performed at 65°C for 5 min and at 95°C for 25 min. A second 40 μm layer of SU-8 2025 was spun onto the wafer to generate the overhead trapping array followed by the same pre-exposure bake, UV exposure, and post-exposure bake steps. Following the second post exposure bake step, the wafer was immersed in an SU-8 developer solution (MicroChem) for ~5 min followed by a rinse with isopropyl alcohol (VWR) to remove all uncrosslinked SU-8. The wafer was dried with compressed nitrogen and then hard baked at 150°C for 30 min to stabilize the patterns.
PDMS replication was performed using traditional methods. Briefly, Sylgard 184 (Dow Corning, Ellsworth Adhesives) was poured onto the silicon master at a ratio of 10:1 (base: curing agent). The PDMS was cured on a hot plate at 65°C for a minimum of 6 hours (but no longer than 24 hours). The PDMS replica was cut and the inlet and outlet holes were created using a blunted 18-gauge needle (BD Biosciences), after which the replica was permanently bonded to a glass coverslip (75mm x 25 mm, Corning) using a Harrick Plasma Cleaner PDC-32G (Harrick Plasma) for 30 s. These microfluidic devices were allowed a minimum waiting period of 3 h to yield optimal bonding between the PDMS and the glass. The fluidic channels were rendered hydrophobic by Aquapel treatment to prevent channel wetting and the formation of a continuous aqueous phase in the device. After treatment, the device was flushed with Novec 7500 (3M) carrier oil and dried by compressed nitrogen. Prior to experimentation, the device was placed in a vacuum chamber for 30 min to prevent the diffusion of gas bubbles from the PDMS into the fluidic channels.
Droplet generation and trapping was facilitated by pressure drive flow from two syringe pumps (Pico Plus Elite, Harvard Apparatus) connected to the microfluidic device using 0.022” polytetrafluoroethylene (PTFE) tubing (Cole Parmer). The PTFE tubing was connected to 1 cc syringes (BD Biosciences) with 23-gauge luek lok needles (BD Biosciences) which were placed in the syringe pumps. The oil phase was injected into the device at a rate of 230 μL/h and contained Novec 7500 oil supplemented with 3 wt% 008-Neat Fluorosurfactant (Ran Biotechnologies) to prevent droplet accumulation in the device. The aqueous phase was injected into the device at a rate of 90 μL/h and contained either deionized water, extracellular buffer (ECB; 5.036 mM HEPES pH 7.4, 136.89 mM NaCl, 2.68 mM KCl, 2.066 mM MgCl2•6H2O, 1.8 mM CaCl2•2H2O, and 5.55 mM glucose), or ECB with cells. Droplet generation and trapping was observed using an inverted fluorescent microscope (DMi8, Lecia Microsystems) with a digital CMOS camera C11440 (Hamamatsu Photonics) equipped with LASX software 3.3.0 to image and analyse droplets and cells. After the droplet trapping array was filled, the device was flushed with only the oil phase to remove any remaining droplets from the bottom layer.
Peptide synthesis and purification
The RWRWR and OWRWR peptides were synthesized and purified as described by Safa et al. in their previous work[20].
Cell culture and reagents
HeLa cells (LSU AgCenter Tissue Culture Facility) and green fluorescent protein (GFP)-expressing HeLa cells (GFP-HeLa) were maintained in Dulbecco’s modified eagle medium (DMEM) with 10% v/v fetal bovine serum (FBS, Seradigm). OPM-2 cells were maintained in RPMI 1640 media supplemented with 12% FBS, 21.8 mM glucose, 8.6 mM HEPES (pH 7.4) and 1.0 mM sodium pyruvate. All media components were from Corning unless otherwise noted. FAM-tagged TAT and FAM-tagged ARG peptides were purchased from Anaspec.
Cell permeability assay
The quantification of peptide uptake was performed using an adapted protocol from Qian et al[23]. All peptides were reconstituted in sodium phosphate buffer (2.26 mM NaH2PO4•H2O and 8.43 mM Na2HPO4•7H2O, pH 7.4). Stock peptide concentration was determined using a NanoDrop (Thermo Scientific) at the wavelength of 492 nm using the UV-Vis function and diluted in ECB to reach the desired concentration. For experiments with HeLa cells, the cells were washed with phosphate buffered saline (PBS: 137 mM NaCl, 10 mM Na2HPO4, 27 mM KCl, and 1.75 mM KH2PO4 at pH 7.4), trypsinized, and re-suspended in ECB to a final density of 6×106 cells/mL. The cells were washed an additional time with ECB and then re-suspended in 2 mL of the peptide solutions. OPM-2 cells were pelleted (1800 rcf, 2.5 min at 25°C), washed twice with ECB, and re-suspended to final density of 6×106 cells/mL. These cells were then pelleted and re-suspended in 2 mL of the peptide solutions. Cells were incubated with the peptides for 60 min at 37°C in the dark in a CO2 incubator to internalize CPPs into intact cells. Peptide incubation was the only temperature-sensitive step, so all subsequent steps were performed at room temperature. After incubation, the cell suspension was centrifuged (1800 rcf, 2.5 min at 25°C) to remove the peptide solution followed by two additional washes with ECB. Finally, the cells were re-suspended in 2 mL ECB, placed into a 5 ml syringe (BD Biosciences) that was wrapped in foil and were immediately injected into the aqueous inlet of the microfluidic device. Experiments with GFP-HeLa cells followed a similar protocol except for the peptide incubation step.
Image processing and statistical analysis
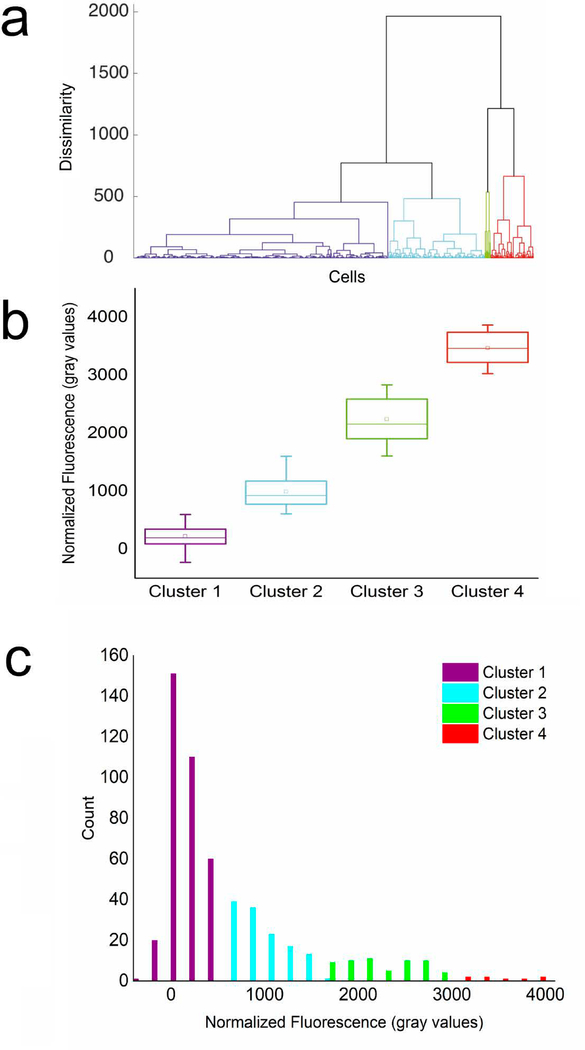
The DMi8 inverted fluorescent microscope was used to collect images of trapped droplets across the entire trapping array using the FITC filter set (λex: 440–520 nm and λem = 497–557 nm) from Chroma Tech. All experiments with HeLa cell were performed using a 10X (N PLAN 10×/0.25 DRY) objective. All experiments with OPM-2 cells were performed using a 20X (HC PL FLUOTAR L 20×/0.4 DRY) objective due to their smaller size. A Hamamatsu-Flash4-CL-880338 camera used to collect images using a digitization of 16 bits and quality mode of 209.8 MHz in all experiments. The fluorescent intensities of individual cells were quantified in gray values using thresholding in the LAS X software by defining ROIs around each individual cell. Random ROIs were also defined in free space away from the trapped droplets to measure the droplet background noise for each image. It was confirmed that the background noise measured using the FITC filter set fit within a very small range for all images. The noise was found to be slightly different across different experiments depending on the peptide studied. Peptides with higher solubilities demonstrated a lower median droplet background noise. The fluorescent value for each individual cell was normalized against the noise measured for the same image by subtracting the noise from the signal. A minimum of 100 HeLa cells were analyzed per experiment. The normalized fluorescent signals for each population of single cells were clustered in subpopulations using average linkage hierarchical cluster analysis by R programming. Through hierarchical clustering, cells with similar fluorescence signals are combined into groups called clusters. These clusters are distinct from each other, while the cells within each cluster are similar to each other thus, resulting in the formation of a tree-structured hierarchy of clusters, called a dendrogram, depicting the difference and proximity of each cluster[24, 25]. As an illustration of the analysis method, the clustering dendrogram obtained for HeLa cells incubated with a 10 μM OWRWR peptide solution is illustrated in Fig. 3a. In this dendrogram, the horizontal axis represents the datapoints obtained for individual cells, and the vertical axis represents dissimilarity. The dissimilarity was calculated using Euclidean distance measure. Finally, the dendrogram was cut at an appropriate dissimilarity threshold to produce the clusters. The optimal number of clusters for binning each dataset were determined exclusively for that dataset. An established R function named NbClust as reported by Charrad, et al. was used to determine the optimal number of clusters[25]. This function calculated 30 defined deterministic indices by varying the number of clusters to report the optimal cluster number by maximizing the number of favorable indices. Another established R function, named clValid as reported by Brock, et al. was then used to assess the quality of clustering using three indices. Each index in this function reflected the compactness, connectedness, and separation of the cluster partitions[24]. This assigned a cluster to each group of data points. These clusters which are presented in Fig. 3b were then marked on the histogram (Fig. 3c). Population histograms, simulation functions, and statistical analysis presented in Fig. 5 and in Electronic Supplementary Material (ESM) Figs. S1-S4 were performed using the statistical analysis package in Origin Pro.
Fig 3.
Identification of distinct subpopulations of HeLa cells based on peptide uptake using linkage hierarchical cluster analysis. A population of HeLa cells incubated with a 10 μM solution of OWRWR and imaged using the microfluidic droplet trapping array. (a) Agglomerative dendrogram tree for hierarchical cluster analysis. The horizontal axis represents the fluorescent signal from each individual cell analyzed and the vertical axis represents dissimilarity measured using the Euclidean distance measure. (b) The cells were binned into four clusters as the subpopulations present in the sample. (c) Distribution histogram of data for n=537 cells demonstrating a logarithmic normal trend and the four identified subpopulations.
Fig. 5.
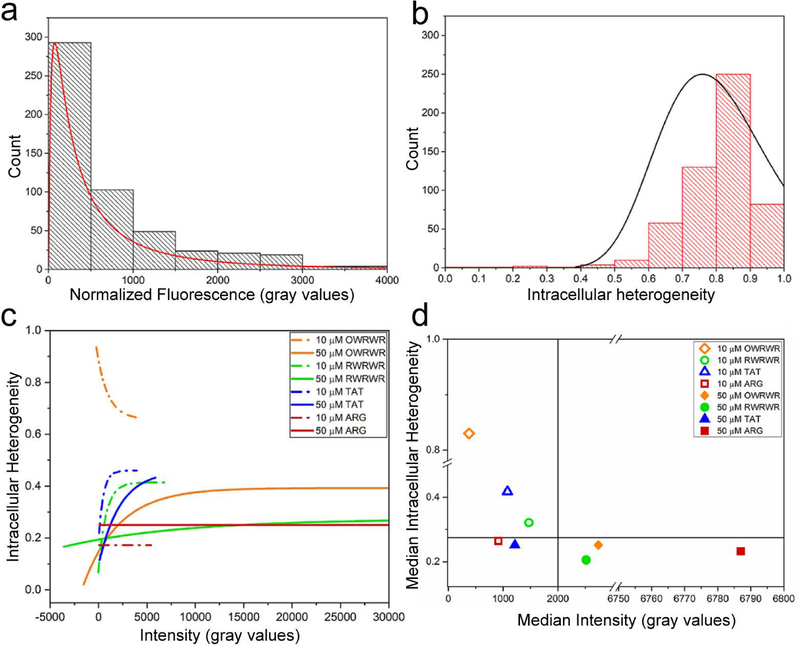
Concentration-dependent correlation between intracellular peptide distribution and CPP uptake efficiency. A coefficient of variance (COV) was calculated for each individual cell analyzed as a metric of intracellular heterogeneity to determine the effect of peptide concentration on intracellular distribution. (a) Population of fluorescent intensity signals for HeLa cells incubated with 10 μM OWRWR demonstrating a logarithmic normal distribution (n=539, r2=0.80, p<0.05) (b) Observed intracellular heterogeneity (COV) of 10 μM OWRWR peptide solution in HeLa cells demonstrating normal distribution (n=539, r2=0.65, p<0.05) (c) Correlation between intracellular heterogeneity and fluorescent for the entire population of HeLa cells incubated with four peptides at the two concentrations numerically simulated with exponential associations. (d) Comparison of the median values of the intracellular heterogeneity and the median cellular fluorescence intensity for all conditions.
Results and Discussion
Single cell fluorescence imaging of CPP uptake in intact cells using a microfluidic droplet trapping array.
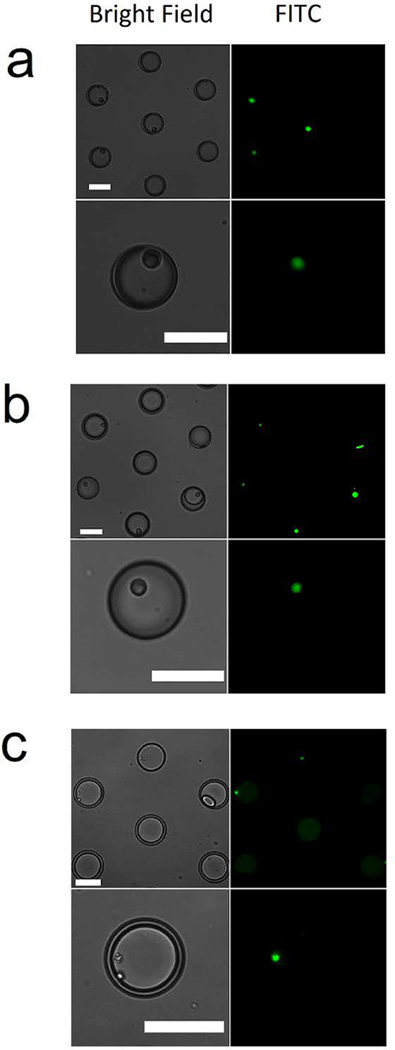
The droplet microfluidic trapping array allowed for the rapid and facile encapsulation of single cells incubated with CPPs to directly quantify peptide uptake using high-resolution fluorescent microscopy. The microfluidic device was confirmed to generate ~180 pL aqueous droplets at a rate of 200 Hz coupled with a ~99% droplet trapping efficiency in the trapping array. Single cell encapsulation from the flow focusing junction followed a Poisson distribution as previously described[26]. An example analysis from triplicate GFP-expressing HeLa encapsulation experiments (n=787 droplets from each experiment), yielded an average of 284±31 (35%) single cells, 146±13 (17%) multiple cells and 356±33 (48%) empty droplets. However, in some cases it was observed that ~55–65% of the trapped droplets contained single or multiple cells. It was suspected that the higher observed single cell encapsulation in the trapped droplets in the overhead array was due to a greater propensity for cell containing droplets to be trapped overhead compared to empty droplets. Submerging the device in water allowed for maintaining droplet stability for at least 14 hours. Cellular viability was assessed in cells encapsulated in the aqueous droplets using Calcein AM (live) and Ethidium homodimer-1 (dead) after the cells had been encapsulated and trapped in the device for 5 hours, well beyond the time needed to perform the analysis described here. All encapsulated cells were confirmed to be alive in the trapped droplets indicating that cellular manipulation using the droplet microfluidic trapping array did not affect cellular viability (data not shown). Two different cell lines were analyzed in terms of CPP uptake, and the effect of cell size and cell type on peptide uptake efficiency was studied. HeLa cells (~24 μm in diameter) are a larger, adherent cervical cancer cell line and OPM-2 (~10 μm in diameter) are a smaller, non-adherent (suspension) multiple myeloma-derived cell line. The CPPs studied here include two commercially available CPPs [TAT (FAM-YGRKKRRQRRR) and ARG (FAM-RRRRRRRRR)] and novel CPPs previously identified and described by Safa et al. [RWRWR (Ac-RWVRVpGO(FAM)WIRQ-NH2), and OWRWR (Ac-OWVRVpGO(FAM)WIRQ-NH2)][20]. All four of these peptides contain a fluorescent tag [5(6) carboxyfluorescein or FAM] in their sequence to visualize peptide internalization and allow for the direct quantification of peptide uptake in bulk samples as well as in intact cells following established fluorescent labeling, standard fluorometry, and fluorescent microscopy techniques[20, 21, 23]. TAT and ARG are commercially-available, well-studied fluorescent CPPs. RWRWR and OWRWR are novel fluorescent CPPs previously described by Safa et al [20]. RWRWR and OWRWR were fully characterized in terms of uptake dynamics in addition to the dependence on temperature and concentration on peptide internalization using bulk, off-chip studies. Multiple control experiments were reported in which cells were incubated with FAM alone, buffer only (blank control), a non-permeable peptide (negative control), and positive controls using both TAT and ARG[20]. Results demonstrated that the negative control peptide and the FAM alone experiments did not produce any fluorescent output in the cells which confirmed that the measured intracellular fluorescence was exclusively due to peptide uptake and verified the cell permeability of these peptides [20]. The OWRWR and RWRWR peptides showed enhanced stability under intracellular conditions compared to TAT and ARG, while TAT and ARG demonstrated higher permeability efficiencies compared to OWRWR and RWRWR on average. In this current study, the uptake efficiencies of these four peptides were assayed on single-cell level to characterize population heterogeneity. By injecting CPP-treated cells into the aqueous inlet of the device, single cells were isolated in aqueous droplets and collected in the overhead trapping array. Selective bright-field and FITC images of CPP uptake in single intact cells are illustrated in Fig. 2. Experiments were repeated to quantify peptide uptake for the four peptides at two different concentrations (10 μM and 50 μM) one at a time and provide an in-depth analysis of each population. The microfluidic droplet trapping array was able to encapsulate both HeLa cells (Fig. 2a–b) and OPM-2 cells (Fig. 2c) incubated with the RWRWR peptide (Fig. 2a), TAT peptide (Fig. 2b), and the ARG peptide (Fig. 2c). These images are representative of all experiments for the four peptides at the two concentrations with the two different cell lines. The high resolution of microscopy images facilitated precise measurement of fluorescent outputs in single intact cells and allowed for parametrization of intracellular peptide distributions by providing pixel-by-pixel detail of the cross-sectional area of each individual cell. The droplet microfluidic platform presented here coupled with high-resolution fluorescent microscopy enables this type of analysis in suspension cell lines which cannot be performed using competing technologies such as microtiter plates due to their incompatibility with suspension cell lines. This approach also overcomes the limitation of classic flow cytometry to whole cell responses by facilitating intracellular measurements.
Fig 2.
Visualization of CPP uptake in single intact HeLa cells encapsulated in droplets. Brightfield (left) and fluorescent (right) images of HeLa cells incubated with a (a) 50 μM RWRWR peptide solution and a (b) 50 μM TAT peptide solution at two magnifications. (c) OPM2 cells incubated with a 50 μM ARG peptide solution. Scale bar is 70 μm in all images.
Quantification of CPP uptake in intact single cells characterizes population heterogeneity.
The fluorescent signals produced due to FAM-tagged CPP uptake in individual cells were quantified, normalized, and used to identify subpopulations of cells based on the degree of peptide uptake. The following analysis is for the uptake of 10 μM OWRWR in HeLa cells (Fig. 3); however, the approach and results were similar in nature for the other three peptides at both concentrations. For this experiment, the optimal number of clusters was identified to be four by maximizing the number of favorable deterministic indices among the 30 indices defined in the NbClust function[25]. To assess the goodness of the clustering in terms of compactness, connectedness, and separation of the cluster partitions, 3 indices (Connectivity[27], Dunn index[28], and Silhouette Width[29]) were calculated using the clValid function[24]. Connectivity is a number in the range between [0,∞), to be minimized, the Dunn index is a number in the range between [0,∞), to be maximized, and Silhouette is a number in a range between [–1, 1] to be maximized. The calculated values for these indices for all the experiments are reported in Table S1 (see ESM). For the cells incubated with 10 μM OWRWR, these values were found to be 12.02 for Connectivity, 0.01 for Dunn, and 0.65 for Silhouette. The relatively low connectivity value coupled with the relatively high Silhouette value suggest adequate clustering results corresponding to population heterogeneity. Interestingly, the Dunn index as defined to be the ratio of the smallest distance between observations not in the same cluster to the largest intra-cluster distance was found to be 0.01. The small value calculated for the Dunn index suggested that the subpopulations identified are not completely distinct or well-separated. This result is not unexpected given that all the subpopulations identified belong to a population of seemingly identical immortal HeLa cells exposed to the exact same experimental conditions. The subtle difference observed among the subpopulations can only be attributed to cell to cell heterogeneity. This analysis was repeated for each experimental condition (peptide and concentration) for the total population of cells analyzed to identify three to five underlying subpopulations. The negative normalized fluorescence values calculated in this section correspond to cells demonstrating little to no peptide uptake result from cells with intracellular fluorescence values darker than the background noise. A detectable signal corresponding to background noise occurs due to the presence of residual free-floating peptides in the aqueous droplets. In all cases, peptide uptake in HeLa cells was found to be heterogenous with the distribution of the normalized fluorescent signals following a logarithmic trend with r-squared values above 0.95 (Fig. 3c). Similarly, a study by Wang et al.[7] had previously reported a logarithmic normal population distribution of TAT-quantum dot endocytosis in Chinese hamster ovary (CHO) cells. The identification of such behavioral patterns in biological events through single cell analysis can provide important insight to predict cell behavior as triggered by certain environmental stimuli.
Clustering of cells into distinct subpopulations based on peptide uptake identifies a potential bias in comparing population means to overall peptide uptake.
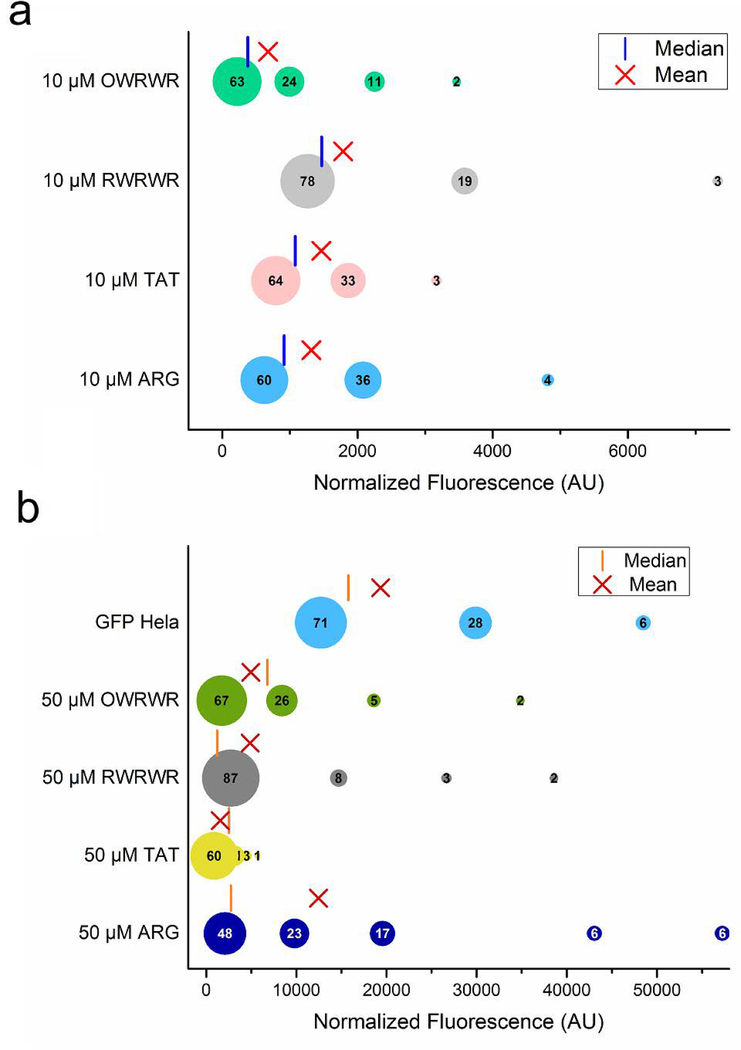
To visualize the distribution and size of each of the subpopulations, the clusters were grouped using a bubble plot (Fig. 4). As expected, the findings confirm an approximate 10-fold increase in the normalized signals when increasing the peptide concentration from 10 μM to 50 μM. This is consistent with previous data presented by Safa et al. using off-chip, bulk fluorescent studies[20]. Furthermore, this analysis found that the largest cluster for all four peptides at both concentrations was the cluster representing the lowest grouping of normalized fluorescent signals. This implies that there is a significant portion of the cellular population with a lower degree of peptide uptake, which can have dramatic results if the CPPs are conjugated to biosensors or therapeutics. Conversely, each peptide at each concentration was observed to have a small population of cells with extremely high normalized fluorescent signals which correspond to substantial uptake of the peptides. The presence of these high performers has the potential to bias results when using bulk population metrics like fluorometry or only looking at 3–5 cells with fluorescent microscopy, the two most common approaches utilized to assess CPP uptake. For example, the ARG peptide is widely accepted to be an ideal CPP; however, the finding here implies that the global uptake of ARG across a net population is biased by a small subpopulation with very high uptake. This can be observed when comparing the population mean and median values to the values for each of the subpopulations incubated with 50 μM peptide (Fig. 4b). The mean value for 50 μM ARG is 12462 compared to values of 4963 for 50 μM OWRWR, 4877 for 50 μM RWRWR, and 2617 for 50 μM TAT (Fig. 4b). Using the population mean alone would imply that the ARG peptide yields the best uptake for all cells; however, the lowest cluster for ARG is comparable in size and normalized fluorescent as those for OWRWR and TAT suggesting that most of the population of cells will experience similar degrees of peptide uptake for all three peptides. This variation between population mean and population distribution was not as extreme when cells were incubated with 10 μM peptide solutions suggesting that the heterogeneous uptake of peptide is concentration dependent. In fact, the population distribution for cells incubated with 10 μM RWRWR, TAT, and ARG all follow a similar trend of three subpopulations with the largest cluster having a normalized fluorescence of ~1200 (Fig. 4a). Interestingly, a similar distribution of intracellular fluorescence was observed for HeLa cells that were stably transfected with green fluorescent protein, GFP-HeLa (Fig. 4b). This confirms prior findings about transfection efficiencies and how some cells exhibit a high degree of transfection while others experience a lesser degree of transfection depending on cell cycle among other factors[30]. These results also suggest that CPP uptake may follow a similar trend as cellular transfection with small subpopulations exhibiting very high transfection efficiencies.
Fig 4.
Distribution and quantification of distinct subpopulation of cells based on CPP uptake. (a) The normalized fluorescence signals of intact HeLa cells incubated with 10 μM peptide solutions quantified by fluorescent microscopy and clustered into 3–5 subpopulations. The number inside each bubble corresponds to the relative size of the cluster for each subpopulation. The mean (red ‘X’) and median (red vertical line) of the entire population is shown for comparison. (b) A similar quantification and clustering as shown in (A) but for HeLa cells incubated with 50 μM peptide solution.
Overall CPP uptake efficiency correlates to intracellular peptide distribution in a concentration-dependent manner.
An analysis was performed to examine how peptide concentration affected the intracellular peptide distribution in cells. To accomplish this, the coefficient of variance (COV) of intracellular fluorescence was measured for each individual cell analyzed, and the correlation between intracellular peptide distribution and overall uptake signal was studied and compared for different cohorts. Here, the COV was defined as the standard deviation of the intracellular fluorescent signals measured for the pixels within the ROI defined around the cell by their mean value. As a result, the COV is a dimensionless metric for the intracellular heterogeneity of peptide distribution or the punctateness level of the peptide taken up by the cell. A poor intracellular distribution is often associated with peptide isolation in membrane-bound vesicles which limits the ability of the CPP to uniformly deliver a cargo to a cell.[31] Population histograms of peptide uptake signals and intracellular heterogeneities (COV) for HeLa cells incubated with 10 μM OWRWR peptide are presented in Figs. 5a and 5b. Similar analyses were performed for all datasets for all four peptides (OWRWR, RWRWR, TAT, and ARG) at both concentrations (10 μM and 50 μM) which are presented in Figs. S1–S5 (see ESM). Non-linear curve fitting performed on these plots revealed that the cells demonstrate a logarithmic normal distribution in terms of overall CPP uptake (Fig. 5a), while they demonstrate a Gaussian (normal) distribution in terms of intracellular heterogeneity (Fig. 5b). Comparing the results in Figs. 5a–b to results obtained for the other peptides (see ESM Figs. S1–S4) identified a common trend among all cohorts (e.g., in all cases). It was found that cells demonstrated a logarithmic normal distribution in terms of CPP uptake and a normal distribution in terms of intracellular heterogeneity. Comparing the histograms for the two concentrations of each peptide presented in Fig. 5 and ESM Figs. S1–S4 showed that increasing the incubation concentration shifts the peak of the intracellular heterogeneity histogram towards lower values. Similarly, increasing the concentrations for OWRWR, RWRWR, and ARG shifts the peak of uptake signals toward higher values. The only exception to this trend was for the TAT peptide for which this effect was insignificant. Similarly, the population histogram for the GFP-expressing HeLa cells (see ESM Fig. S5) follows the same logarithmic trend as CPP treated cells. The correlation between overall uptake efficiency (Fig. 5a) and intracellular heterogeneity (Fig. 5b) for the model 10 μM OWRWR experiment is illustrated at the single-cell level in Fig. S6A (see ESM In this plot, data demonstrating an exponential decay as numerically simulated by non-linear curve fitting illustrates the reverse relationship between intracellular heterogeneity and uptake efficiency meaning that overall uptake efficiency is higher in cells with more homogeneous intracellular peptide distribution.
To illustrate and compare this correlation among all cohorts, the simulated curves for all datasets are provided in Fig. 5c with their median values presented in Fig. 5d. Raw single-cell data used to generate the simulated curves in Fig. 5c are provided in Fig. S6B (see ESM). It was observed in Figs. 5c and ESM Fig. S6B that cells treated with 10 μM peptide solutions clustered within a small range in areas with lower intensities and higher intracellular heterogeneities, although this occurred to variant extents for different peptides. By increasing the concentration of each peptide, fluorescence intensities expanded to a wider range to include areas with significantly higher intensities (with the exception of TAT for which the effect was not significant). Moreover, in most cases (with the exception of ARG for which the effect was not significant) the intracellular heterogeneities decreased. Similar trends were previously described qualitatively in off-chip efforts characterizing CPP uptake and internalization mechanisms[32]. Additionally, a single cell study performed by Cao et al[33]. recently identified a direct quantitative correlation between fluorescence intensity of cells and the cytosolic/nuclear localization of native proteins.
As far as the relative results among different conditions are concerned, comparing the median population values of intracellular heterogeneity and fluorescent intensity (Fig. 5d) identified three groupings by where the median values appeared on the plot. The upper left quadrant, which contains median values for 10 μM OWRWR, 10 μM RWRWR, and 10 μM TAT, is representative of highly punctate intracellular distribution of peptides coupled with a relatively low median values for overall peptide uptake. When comparing the median population values of intracellular heterogeneity and fluorescent intensity (Fig. 5d), three groupings could be identified by where the median values appeared on the plot. The upper left quadrant, which contains median values for 10 μM OWRWR, 10 μM RWRWR, and 10 μM TAT, is representative of highly punctate intracellular distribution of peptides coupled with a relatively low values for overall peptide uptake. This implies that these three peptides at this incubation concentration will have a poorer intracellular distribution coupled with a lower degree of internalization. The lower right quadrant is representative of cells with a lower level of intracellular heterogeneity (or punctateness) coupled with a high or very high overall peptide uptake efficiency. These three conditions include cells incubated with 50 μM peptide solutions of TAT, RWRWR, and OWRWR. This implies that increasing peptide concentration results in lower intracellular heterogeneity which correlates to higher intracellular homogeneity and higher overall peptide uptake efficiency. Intriguingly, the one exception to this finding was the TAT peptide which found that increasing the concentration decreased the intracellular heterogeneity yet did not significantly increase the overall uptake efficiency. This may be attributed to the high cytotoxicity of this peptide at high concentrations[34]. Intracellular peptide distribution is an important characteristic of peptide-cell interaction that is often used to develop hypotheses and make conclusions about peptide uptake mechanism[32, 35]. This important parameter is usually investigated through live cell microscopy and described qualitatively based on analyzing a small number of cells versus being quantified for a large number of cells[32]. The microfluidic droplet trapping array presented here facilitated an efficient analysis to accomplish this goal.
Cell size does not affect peptide uptake efficiency but does affect the net amount of internalized peptide.
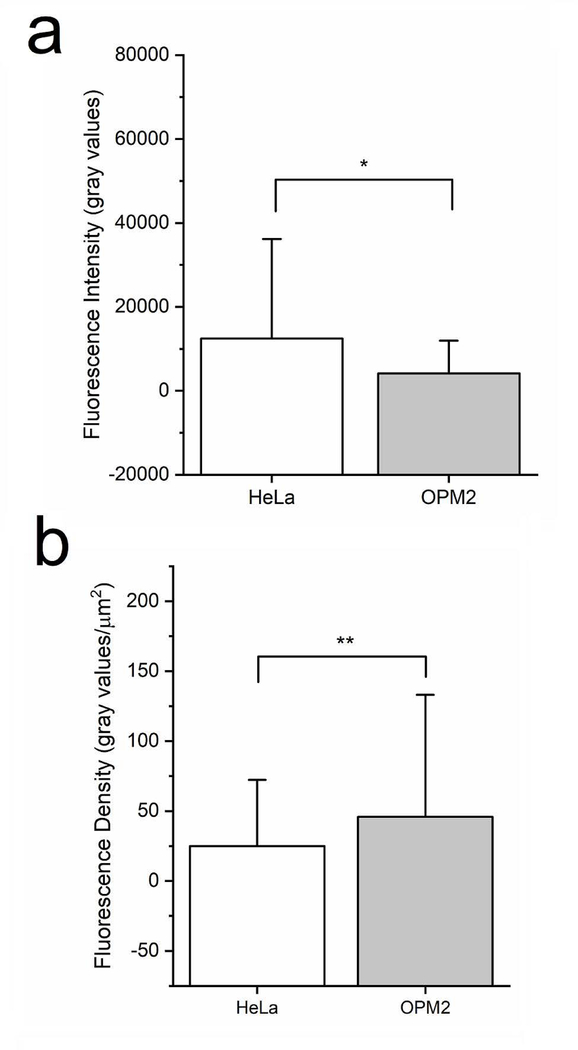
The microfluidic platform was finally used to identify the relationship between peptide uptake and cell type by comparing the uptake of one of the CPPs across an adherent (HeLa) and a suspension (OPM-2) that are significantly different in size. Current bulk approaches like fluorometry oftentimes use fluorescent intensity as a metric of peptide uptake; however, this value can be biased when comparing uptake efficiencies between two different types of cells, especially those that differ in size. To explore this, peptide uptake was studies in the OPM-2 cell line. OPM-2 cells are a suspension, myeloma-derived cell line with an average measured cross-sectional area of 90 μm2 compared to HeLa cells which exhibit an average measured cross-sectional area of 500 μm2. Both cell lines were incubated with 50 μM ARG solutions and single cell analysis of intracellular fluorescent signal was performed (Fig. 6). As expected, the mean fluorescent signal was significantly lower for OPM-2 cells compared to HeLa cells under similar experimental conditions (p<0.00001). Since HeLa and OPM-2 cells are significantly different in size, it was hypothesized that the observed difference was due to the smaller size of the OPM-2 cells. To account for size, the mean fluorescent density was calculated which divided the median fluorescent signal by the average cross-sectional area for the population of cells. This new metric, which removed the bias of cell size, confirmed this hypothesis and illustrated that unlike overall fluorescent output, the fluorescent density was comparable between the two cells lines exposed to the same peptide at the same concentration (p>0.05)
Fig 6.
Effect of cell size on peptide uptake efficiency. HeLa cells (average cross-sectional area of 500 μm2) and OPM-2 cells (average cross-sectional area 90 μm2) were incubated with 50 μM ARG for 60 minutes at 37 °C. (a) The mean fluorescence intensity for the entire population of cells was compared for HeLa (n=97 cells) and OPM2 (n=26 cells). *denotes p<0.00001 (b) The mean fluorescent intensity values for the entire population of cells was divided by the average cellular cross-sectional area for both HeLa and OPM2 cells to calculate the fluorescence density. **denotes p>0.05.
Conclusions
In this paper, a microfluidic droplet generator and overhead trapping array was used to perform single cell analysis on the concentration dependent uptake of four different cell penetrating peptides. Cells treated with 10 μM and 50 μM peptide solutions were analyzed in terms of peptide uptake efficiency, intracellular distribution, and the correlation between them. Single cell analysis results identified that increasing the initial peptide concentration results in a more homogeneous intracellular peptide distribution and a stronger uptake efficiency. All histograms followed a logarithmic normal trend. Kernel density functions numerically simulated the distribution histograms predicted cell behavior in terms of uptake efficiency and intracellular heterogeneity. The correlation between peptide concentration and intracellular heterogeneity was illustrated in a scatter plot for a model experiment (10 μM OWRWR) and numerically simulated using an exponential decay function to predict the behavior. Finally, repeating a select experiment (50 μM ARG) with a suspension cell line (OPM-2) identified a significant difference in fluorescent signals, but normalizing the results by cell area resulted in similar fluorescent densities which revealed that the difference observed could mainly be attributed the smaller size of the OPM-2 cells. This work demonstrates the application of a microfluidic droplet trapping array for quantifying the uptake of fluorescent cell penetrating peptides in single intact cells and presents a statistical approach for performing single cell analysis of the samples. The microfluidic device and analytical approach can be further utilized to quantify enzyme activity using intracellular fluorescence tied to fluorescent based reporters to provide new insight on population heterogeneity associated with clinical samples.
Supplementary Material
Acknowledgements
This work was supported by grants from the National Institute of Biomedical Imaging and Bioengineering (R03EB02935) and the National Science Foundation (CBET1509713) awarded to ATM. The authors would like to thank Dr. Nancy Allbritton (University of North Carolina) for providing the GFP-HeLa and OPM-2 cells. The authors would also like to thank Dr. Ted Gauthier (LSU AgCenter Biotechnology Lab) for assistance in the synthesis and purification of the peptides used in this study. The authors would also like to thank Joseph B. Balhoff for assistance in producing the graphical abstract and Riad Elkhanoufi and Wayne Wortmann III for some assistance with device fabrication.
Footnotes
Compliance with ethical standards
Conflict of interest: The authors have no conflicts to declare.
References
- 1.Hamley I Small Bioactive Peptides for Biomaterials Design and Therapeutics. Chemical Reviews. 2017;117(24):14015–41. doi: 10.1021/acs.chemrev.7b00522. [DOI] [PubMed] [Google Scholar]
- 2.Craik D, Fairlie D, Liras S, Price D. The Future of Peptide-based Drugs. Chemical Biology & Drug Design. 2013;81(1):136–47. doi: 10.1111/cbdd.12055. [DOI] [PubMed] [Google Scholar]
- 3.Juan G-VC. MM. Fluorescent Reporters and Biosensors for Probing the Dynamic Behavior of Protein Kinases. proteomes. 2015;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kovarik M, Allbritton N. Measuring enzyme activity in single cells. Trends in Biotechnology. 2011;29(5):222–30. doi: 10.1016/j.tibtech.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Carlo D, Lee L. Dynamic single-cell analysis for quantitative biology. Anal Chem. 2006;78(23):7918–25. [DOI] [PubMed] [Google Scholar]
- 6.Cai L, Friedman N, Xie X. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440(7082):358–62. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Zhan Y, Bao N, Lu C. Quantitative measurement of quantum dot uptake at the cell population level using microfluidic evanescent-wave-based flow cytometry. Lab on a Chip. 2012;12(8):1441–5. doi: 10.1039/c2lc21298f. [DOI] [PubMed] [Google Scholar]
- 8.Murphy T, Zhang Q, Naler L, Ma S, Lu C. Recent advances in the use of microfluidic technologies for single cell analysis. Analyst. 2018;143(1):60–80. doi: 10.1039/c7an01346a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reece A, Xia B, Jiang Z, Noren B, McBride R, Oakey J. Microfluidic techniques for high throughput single cell analysis. Current Opinion in Biotechnology. 2016;40:90–6. doi: 10.1016/j.copbio.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Bao N, Paris L, Wang H, Geahlen R, Lu C. Detection of kinase translocation using microfluidic electroporative flow cytometry. Anal Chem. 2008;80(4):1087–93. doi: 10.1021/ac702065e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shang L, Cheng Y, Zhao Y. Emerging Droplet Microfluidics. Chemical Reviews. 2017;117(12):7964–8040. doi: 10.1021/acs.chemrev.6b00848. [DOI] [PubMed] [Google Scholar]
- 12.Du G, Fang Q, den Toonder J. Microfluidics for cell-based high throughput screening platformsd-A review. Analytica Chimica Acta. 2016;903:36–50. doi: 10.1016/j.aca.2015.11.023. [DOI] [PubMed] [Google Scholar]
- 13.Pekin D, Skhiri Y, Baret J, Le Corre D, Mazutis L, Ben Salem C et al. Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Lab on a Chip. 2011;11(13):2156–66. doi: 10.1039/c1lc20128j. [DOI] [PubMed] [Google Scholar]
- 14.Sjostrom S, Joensson H, Svahn H. Multiplex analysis of enzyme kinetics and inhibition by droplet microfluidics using picoinjectors. Lab on a Chip. 2013;13(9):1754–61. doi: 10.1039/c3lc41398e. [DOI] [PubMed] [Google Scholar]
- 15.Jin S, Lee S, Lee B, Jeong S, Peter M, Lee C. Programmable Static Droplet Array for the Analysis of Cell-Cell Communication in a Confined Microenvironment. Analytical Chemistry. 2017;89(18):9722–9. doi: 10.1021/acs.analchem.7b01462. [DOI] [PubMed] [Google Scholar]
- 16.Toseland CP. Fluorescent labeling and modification of proteins. J Chem Biol. 2013;6(3):85–95. doi: 10.1007/s12154-0130094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brouzes E, Medkova M, Savenelli N, Marran D, Twardowski M, Hutchison J et al. Droplet microfluidic technology for single-cell high-throughput screening. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(34):14195–200. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khorshidi M, Rajeswari P, Wahlby C, Joensson H, Svahn H. Automated analysis of dynamic behavior of single cells in picoliter droplets. Lab on a Chip. 2014;14(5):931–7. doi: 10.1039/c3lc51136g. [DOI] [PubMed] [Google Scholar]
- 19.Jackson-Holmes E, McDevitt T, Lu H. A microfluidic trap array for longitudinal monitoring and multi-modal phenotypic analysis of individual stem cell aggregates. Lab on a Chip. 2017;17(21):3634–42. doi: 10.1039/c7lc00763a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Safa N, Anderson JC, Vaithiyanathan M, Pettigrew JH, Pappas GA, Liu D et al. CPProtectides: Rapid uptake of well‐folded β-hairpin peptides with enhanced resistance to intracellular degradation. Peptide Science. 2018:e24092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian Z, LaRochelle J, Jiang B, Lian W, Hard R, Selner N et al. Early Endosomal Escape of a Cyclic Cell-Penetrating Peptide Allows Effective Cytosolic Cargo Delivery. Biochemistry. 2014;53(24):4034–46. doi: 10.1021/bi5004102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazutis L, Gilbert J, Ung W, Weitz D, Griffiths A, Heyman J. Single-cell analysis and sorting using droplet-based microfluidics. Nature Protocols. 2013;8(5):870–91. doi: 10.1038/nprot.2013.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian Z, Liu T, Liu Y, Briesewitz R, Barrios A, Jhiang S et al. Efficient Delivery of Cyclic Peptides into Mammalian Cells with Short Sequence Motifs. Acs Chemical Biology. 2013;8(2):423–31. doi: 10.1021/cb3005275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brock G, Pihur V, Datta S, Datta S. clValid: An R Package for Cluster Validation. Journal of Statistical Software. 2008;25(4):122. [Google Scholar]
- 25.Charrad M, Ghazzali N, Bioteau V, Niknafs A. NbClust: An R package for Determining the Relevant Number of Clusters in a Data Set. Journal of statistical software. 2014;61(6):1–36. [Google Scholar]
- 26.Collins DJ, Neild A, deMello A, Liu A-Q, Ai Y. The Poisson distribution and beyond: methods for microfluidic droplet production and single cell encapsulation. Lab on a Chip. 2015;15(17):3439–59. doi: 10.1039/C5LC00614G. [DOI] [PubMed] [Google Scholar]
- 27.Handl J, Knowles J, Kell D. Computational cluster validation in post-genomic data analysis. Bioinformatics. 2005;21(15):3201–12. doi: 10.1093/bioinformatics/bti517. [DOI] [PubMed] [Google Scholar]
- 28.Dunn C Well-separated clusters and optimal fuzzy partitions. Journal of Cybernetics. 1974;4:95–104. [Google Scholar]
- 29.ROUSSEEUW P SILHOUETTES - A GRAPHICAL AID TO THE INTERPRETATION AND VALIDATION OF CLUSTER-ANALYSIS. J Comput Appl Math. 1987;20:53–65. doi: 10.1016/0377-0427(87)90125-7. [DOI] [Google Scholar]
- 30.Brunner S, Sauer T, Carotta S, Cotten M, Saltik M, Wagner E. Cell cycle dependence of gene transfer by lipoplex polyplex and recombinant adenovirus. Gene Therapy. 2000;7(5):401–7. doi: 10.1038/sj.gt.3301102. [DOI] [PubMed] [Google Scholar]
- 31.El-Sayed A, Futaki S, Harashima H. Delivery of Macromolecules Using Arginine-Rich Cell-Penetrating Peptides: Ways to Overcome Endosomal Entrapment. Aaps Journal. 2009;11(1):13–22. doi: 10.1208/s12248-008-9071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brock R The Uptake of Arginine-Rich Cell-Penetrating Peptides: Putting the Puzzle Together. Bioconjugate Chemistry. 2014;25(5):863–8. doi: 10.1021/bc500017t. [DOI] [PubMed] [Google Scholar]
- 33.Cao Z, Geng S, Li L, Lu C. Detecting intracellular translocation of native proteins quantitatively at the single cell level. Chemical Science. 2014;5(6):2530–5. doi: 10.1039/c4sc00578c. [DOI] [Google Scholar]
- 34.Gomez JA, Chen J, Ngo J, Hajkova D, Yeh I-J, Gama V et al. Cell-Penetrating Penta-Peptides (CPP5s): Measurement of Cell Entry and Protein-Transduction Activity. pharmaceuticals. 2010;3:3594–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duchardt F, Fotin-Mleczek M, Schwarz H, Fischer R, Brock R. A comprehensive model for the cellular uptake of cationic cell-penetrating peptides. Traffic. 2007;8(7):848–66. doi: 10.1111/j.1600-0854.2007.00572.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.