Abstract
Heat shock factor 1 (HSF1) was discovered in 1984 as the master regulator of the heat shock response. In this classical role, HSF1 is activated following cellular stresses such as heat shock that ultimately lead to HSF1-mediated expression of heat shock proteins to protect the proteome and survive these acute stresses. However, it is now becoming clear that HSF1 also plays a significant role in several diseases, perhaps none more prominent than cancer. HSF1 appears to have a pleiotropic role in cancer by supporting multiple facets of malignancy including migration’ invasion, proliferation, and cancer cell metabolism among others. Because of these functions, and others, of HSF1, it has been investigated as a biomarker for patient outcomes in multiple cancer types. HSF1 expression alone was predictive for patient outcomes in multiple cancer types but in other instances, markers for HSF1 activity were more predictive. Clearly, further work is needed to tease out which markers are most representative of the tumor promoting effects of HSF1. Additionally, there have been several attempts at developing small molecule inhibitors to reduce HSF1 activity. All of these HSF1 inhibitors are still in preclinical models but have shown varying levels of efficacy at suppressing tumor growth. The growth of research related to HSF1 in cancer has been enormous over the last decade with many new functions of HSF1 discovered along the way. In order for these discoveries to reach clinical impact, further development of HSF1 as a biomarker or therapeutic target needs to be continued.
Keywords: HSF1, metastasis, biomarker, therapy, EMT, invasion, migration
1. INTRODUCTION
The heat shock response was initially discovered in 1962 [1] and subsequent studies identified heat shock factor 1 (HSF1) as the “master regulator” of the heat shock response [2, 3]. HSF1 is a helix-turn-helix transcription factor that binds to heat shock elements (HSEs) located in the promoters of target genes [4–7]. HSF1 is now increasingly being recognized to play a role in several diseases including cancer and neurodegenerative diseases. Complicating matters, HSF1 is overactive or overexpressed in many cancers but loses activity in neurodegenerative diseases leading to neuron death from the unfolded protein response.
HSF1 has now been recognized to play a role in many areas of tumor biology and has strong associations with patient outcomes. While the heat shock response has been associated with cancer for decades, the first mention of HSF1 having altered function in cancer was in prostate cancer [8]. Subsequently, HSF1 has been shown to have altered expression or function in colorectal [9], breast cancer [10–13], lymphomas [14, 15], hepatocellular carcinoma [16–20], oral cancers [21–23], melanoma [24, 25], pancreatic cancers [26–28], gynecologic cancers [29–32], and others [33–35].
2. HSF1 GENE AND PROTEIN STRUCTURE AND FUNCTION
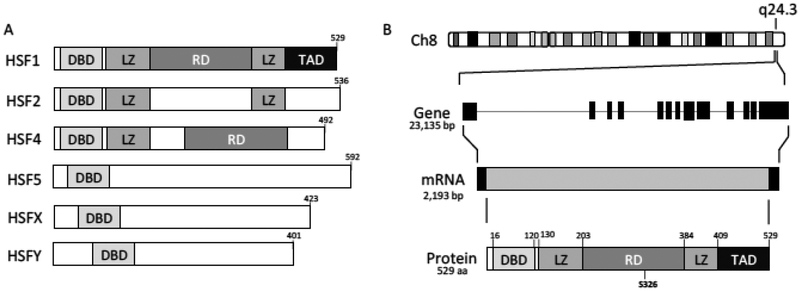
There are six human HSF genes in HSF1–2, HSF4–5, HSFX-Y (Fig. 1A). HSF1 is, by far, the most understood of these isoforms. HSF2 has been linked to early development [36] and HSF4 is involved in eye lens development [37, 38]. The function of HSF5 and HSFX is currently unknown. HSFY has been linked to male fertility [39] and HSF3 has only been identified in mice but not humans [40]. HSF1 has been most closely linked with regulating the heat shock response aside from its role in tumor development and progression. The HSF1 gene is located on the long arm of chromosome 8 (8q24.3) that results in a gene of 23,135 bp with 13 exons. The resulting 2,193 bp mRNA and 529 amino acid protein of HSF1 are constitutively expressed under physiological conditions (Fig. 1B). The mouse gene is located on chromosome 15 resulting in a protein with 553 residues [41]. The human and mouse HSF1 protein have 86% similarity. HSF1 is not frequently mutated in cancer but we have reported that the gene is amplified in 15°% of breast cancers resulting in increased mRNA expression [42]. Additionally, HSF1 mRNA is upregulated in 25% of breast cancers indicating that there are likely additional mechanisms driving increased expression of HSF 1 in breast cancers aside from gene amplification [42].
Fig. (1).
Heat Shock Factor 1 (HSF1) Gene. A) There are six human HSF isoforms all containing a homologous DNA binding domain recognizing the heat shock element (HSE). B) HSF1 is located on the long arm of chromosome 8 at q24.3. The resulting gene has 13 exons with a full gene size of 23,135 bp that transcribes to an mRNA of 2,193 bp. The resulting protein is 529 aa and has multiple functional domains. The DNA binding domain (DBD) is a winged helix-turn-helix domain. The leucine zipper (LZ) domains mediate the trimerization of HSF1 upon activation. The regulatory domain is home to many post-translational modifications that appear to regulate HSF1 transcriptional activity with Ser326 seemingly the key phosphorylation necessary for activity. The transactivation domain (TAD) of HSF1 recruits p-TEFb to release proximal promoter pausing and promote transcription elongation of target genes.
The classical heat shock response involves HSF1 activation in response to cell stress. In healthy quiescent cells, HSF1 protein is kept in a folded, inactive state and in an inhibitory protein complex with Hsp90, Hsp70, Hsp40, and the TRiC/CCt complex [43–46]. This inhibitory complex is dissolved in response to a heat stress wherein the HSPs are shuttled toward cellular proteins to protect the integrity of the proteome [46, 47]. HSF1 is subsequently unfolded via a heat-mediated mechanism [48, 49]. HSF1 then undergoes trimerization [49, 50], nuclear localization, and phosphorylation [51–53] in order to promote transcription of target genes. In this classical heat shock response mechanism, many HSF 1 targets are HSPs that further aid in protecting the proteome and aiding cell survival.
3. ROLE OF HSF1 IN TUMOR PROGRESSION
HSF1 appears to have many functions that promote tumorigenesis and tumor progression (Fig. 2). The first discovery of HSF1 playing a role in cancer was in prostate cancer cells [8]. Since that time, the number of publications related to HSF1 and cancer has steadily increased to greater than 50 publications per year since 2015. These discoveries span the breadth of tumor biology. This review will focus on the role of HSF 1 in metastasis, cell proliferation and apoptosis, and tumor metabolism as these areas are relevant to HSF1 as a biomarker and potential therapeutic target.
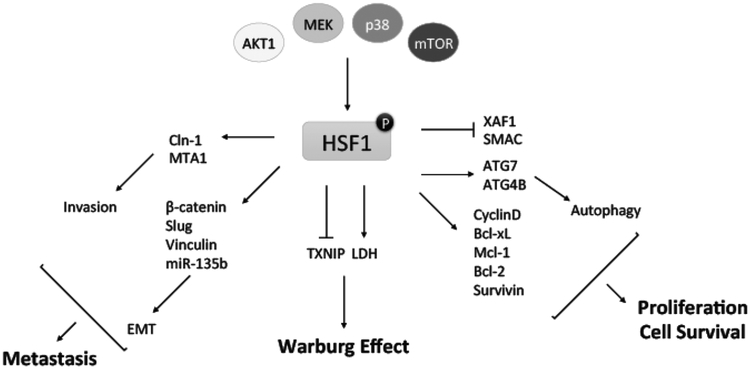
Fig. (2).
HSF1 Function in Tumorigenesis and Tumor Progression. HSF1 has been shown to be phosphorylated and activated at Ser326 by AKT1 [61], MEK [69], p38 [123], and mTOR [124]. HSF1 directly regulates multiple genes to promote migration, invasion, and EMT that ultimately translates to tumor progression and metastasis. HSF1 also appears to directly upregulate and downregulate several genes that lead to enhanced cell proliferation and survival. In addition, HSF1 has direct effects on genes that promote the Warburg effect of increasing glycolysis in cancer cells. Together these functions, and others, suggest HSF1 has multiple effects that lead to tumorigenesis and tumor progression.
3.1. Migration, Invasion, and Metastasis
The first discovery of HSF1 playing a role in cancer in prostate cancer cells showed that HSF1 had higher expression in a metastatic variant of PC-3 cells [8], suggesting that it may be altered in metastatic cancer cells. We observed a similar outcome with increased HSF1 activation in breast cancer metastatic variant cell lines compared to the parental non-metastatic cells [54]. Our data indicated that HSF1 activity is strongly associated with metastasis in a large cohort of breast cancer patients [54]. In addition, HSF1 expression or activity is increased in advanced stage tumors from multiple sites [8, 17, 24, 27] suggesting that HSF1 drives tumor progression toward metastasis. One clear piece of evidence that HSF1 plays a role in tumorigenesis and metastasis is deletion of HSF1 in the MMTV-Neu mouse model resulted in decreased breast tumor formation and lung metastasis [55]. Similarly, HSF1 overexpression increased metastasis in a hepatocellular carcinoma mouse model whereas HSF1 knockdown reduced metastasis [17]. These data indicate that HSF 1 is strongly associated with and necessary for metastasis.
Several studies indicated that HSF1 can drive migration, invasion, and anchorage-independent growth of cancer cells. For example, deletion of HSF1 from mouse embryonic fibroblasts reduces their anchorage-independent growth [56] and migration [57] suggesting that HSF 1 is critical for these functions at a basal level. Several studies have shown similar results that migration and invasion are reduced with loss of HSF1 using cancer cells from osteosarcoma [35], ovarian cancer [31], melanoma [24, 58], pancreatic cancer [27], hepatocellular carcinoma [17], breast cancer [59], and lung cancer [60]. These characteristics are key features of epithelial-to-mesenchymal transition (EMT). We and others have shown that HsF1 promotes EMT [31, 55, 60, 61]. Not only has our lab and others shown HSF1 expression promotes EMT-like changes, it also appears that HSF1 is required for TGFβ-induced EMT as well [31, 55], possibly suggesting that HSF 1 is critical to the core process of EMT rather than just one of many molecules that can initiate EMT.
There are several proposed mechanisms by which HSF1 promotes EMT-like phenotypes and metastasis. Our data indicate that HSF1 can bind and upregulate Slug, an EMT-promoting transcriptional repressor [61]. However, considering the lessened importance of Slug in EMT of breast cancer cells [62], it is likely HSF1 also utilizes additional mechanisms. For instance, HSF1 has been shown to upregulate β-catenin via HuR [63] and loss of HSF1 reduced NFκB activation and suppressed tumor growth [16]. HSF1 activation in hepatoma cells led to HSF1-driven Cln-1 expression leading to cell invasion [64]. Loss of HSF1 in a lung tumor model also showed a decrease in migration associated with a decrease in vinculin, an actin-binding protein known to promote cell migration [60]. HSF1 has been shown to regulate miRNAs such as upregulation of miR-135b, leading to upregulation of MMP2/9 and changing actin dynamics to allow for migration and invasion [65]. HSF1 has also been shown to interact with and cooperatively regulate gene expression with metastasis-associated protein 1 (MTA1) [66]. In addition, HSF1 promotes aneuploidy [67, 68], another mechanism associated with tumor progression.
A broader look at the mechanism by which HSF1 sustains tumor growth and malignancy was addressed by ChIP-Seq for HSF1 in tumors [12]. Interestingly, it was observed that HSF1 -bound genes in cancer were unique from HSF1 -bound genes during heat shock [12]. Despite a seemingly ubiquitous mechanism of HSF1 activation via trimerization and S326 phosphorylation, as well as having a similar binding sequence, the genes HSF1 regulated were unique in these two contexts. The genes involved in the cancer-specific HSF1 program involved genes from many aspects of tumor biology including energy metabolism, apoptosis, transcription, translation, and extracellular matrix [12]. Despite having a cancer-specific transcriptional program, HSF1 also has the capability to promote malignancy via its role as a stress-response protein. MEK-induced HSF1 activity has been shown to suppress apoptosis induced by the unfolded protein response in cancer cells [69]. Preventing HSF1-induced activity in response to accumulation of unfolded proteins suppressed metastasis [69]. As such, HSF1 utilizes multiple mechanisms to support tumor growth and progression toward metastasis.
Another potential mechanism for HSF1 to promote tumorigenesis and metastasis is enhancing the cancer stem cell population. We and others have shown increased HSF1 activation or expression in the cancer stem cell population [54, 63, 70]. We also showed that HSF1 expression enhanced spheroid formation in breast cancer cells and that inhibition of HSF1 reduced sphere formation and the cancer stem cell population [54]. Similarly, HSF1 knockdown also reduced spheroid formation [54, 70]. In accordance with increased HSF 1 activity in cancer stem cells, expression of Hsp27 and Hsp90 have also been linked to stemness [32, 71].
3.2. Cell Proliferation and Viability
Aside from being involved in several different areas of tumor biology, HSF1 also seems to be involved in regulating cell proliferation and turnover, a central aspect of tumor formation and growth. HSF1-null MEFs have lower proliferation rates and often showed incomplete mitosis compared to WT MEFs, suggesting that HSF1 has some impact on cell proliferation [11, 72]. Interestingly, HSF1 is directly involved in the cell cycle as it is phosphorylated at S216 leading to degradation during mitosis and mitosis is interrupted without this event [72]. HSF1 expression has also been seen to increase CyclinD levels and suppress p21 and p27 [59, 73]. While HSF1 is known to promote expression of genes that leads to enhanced cell proliferation and survival, it also seems to have a direct influence on the cell cycle.
The classical function of HSF1 is to respond to a cell stress in an effort to promote cell survival. Consistent with this function, HSF 1 induces several mechanisms to suppress apoptosis and cell death. HSF1-null MEFs showed higher rates of apoptosis compared to WT MEFs suggesting the lack of HSF1 predisposed cells to apoptosis [11]. HSF1 activation has been linked to the upregulation of several anti-apoptotic proteins including Bcl-xL, Mcl-1, Bcl-2, and survivin [59, 74–76]. The enhanced expression of these anti-apoptotic proteins has been shown to be mediated by HSF 1 -induced expression of chaperones including Hsp27 and BAG3 [59, 75, 76]. In addition to an increase in anti-apoptotic proteins, HSF1 also promotes the suppression of XAF1, an endogenous suppressor of XIAP, ultimately leading to suppression of apoptosis [77]. Furthermore, HSF1 activity suppresses expression of SMAC, an integral protein in the mitochondrial pathway of apoptosis [28].
Another interesting mechanism of cell survival promoted by HSF1 is autophagy. HSF1 was first shown to upregulate ATG7 leading to cell autophagy and desensitization to chemotherapy [78]. HSF1 was later shown to coregulate ATG7 with PSMD10 to induce autophagy and also promote resistance to sorafenib [79]. In its role as a stress response gene, HSF1 also upregulates RIPK1 in response to ER stress to promote autophagy and cell survival [80]. In addition to regulating ATG7, HSF1 was recently shown to upregulate ATG4B to promote cell-protective autophagy in response to epirubicin [81]. Thus, this is an alternative mechanism by which HSF1 seems to promote cell survival in response to cancer treatments.
3.3. Cancer Cell Metabolism
Cancer cells have a characteristic change in metabolic flux toward glycolysis known as the Warburg effect [82]. Interestingly, HSF1 seems to push cells toward the Warburg effect. Inhibition of HSF1 activity results in decreased lactate and glucose uptake suggesting that HSF1 supports these metabolic changes characteristic of tumor cells [83]. Furthermore, HSF1-null mouse embryonic fibroblasts (MEFs) has less glucose uptake and less tolerance to low glucose conditions compared to wild-type MEFs indicating that the presence of HSF1 may promote a higher dependence on glycolysis [11]. One consequence of increased flux through glycolysis is increased lactate production and HSF1 has been shown to upregulate lactate dehydrogenase (LDH) and loss of HSF1 reduces LDH levels [84, 85].
HSF 1 has interaction with several proteins that influence tumor cell metabolism. For instance, glucose-mediated activation of mTOR leads to HSF1 activation [86]. HSF1 is negatively regulated by AMPK and PGC-1α [27, 87, 88]. AMPK acts as a tumor suppressor and is a cellular metabolic stress sensor and maintains cellular energy homeostasis by detecting changes in AMP/ATP levels. AMPK can interact with HSF1 and phosphorylate S121 leading to suppression of HSF1 activity [27, 87]. Loss of AMPK resulted in enhanced HSF 1 activity and metastasis of pancreatic cancer cells [27]. PGC1-α is a metabolic transcriptional coactivator that enhances oxidative metabolism and mitochondrial biogenesis [89]. PGC1-α has been shown to directly interact with HSF1 on DNA and suppress expression of heat shock response-related HSF1 target genes as well as non-heat-related HSF1 target genes [88]. Though more work is needed to clearly understand mechanisms, HSF1 clearly has an impact on tumor cell metabolism.
4. HSF1 AS A POTENTIAL BIOMARKER AND THERAPEUTIC TARGET
4.1. HSF1 as a Cancer Biomarker
Elevated levels of HSF1 are generally associated with poor prognosis (Table 1). A genome-wide search for target sites of HSF 1 in breast cancer cell lines with different metastatic capacities was undertaken by comparing cells with high and low malignant potential alongside their non-transformed counterparts [12]. Investigators identified an HSF1-regulated transcriptional program specific to highly malignant cells and distinct from heat shock [12]. They could further document this program was active in other cancers such as colon and lung and that it contributed to the metastases and death of patients [12]. A number of clinical studies have now also documented the role of HSF1 in human cancers. These include breast cancer, colon cancer, lung cancer, endometrial cancer, hepatocellular carcinoma, oral squamous cell carcinoma and prostate cancer [90].
Table 1.
Potential HSF1 biomarkers for cancer patient outcomes.
| HSF1 Marker | Patient Outcome | Tumor Types | References |
|---|---|---|---|
| Gene Expression Signature | Overall Survival | Breast, Lung, Colon | [12] |
| Metastasis-Free Survival | Breast, Lung, Colon | [12, 54] | |
| mRNA Levels | Overall Survival | Esophageal, Breast | [23, 42] |
| Recurrence-Free Survival | Breast | [42] | |
| Incidence | Colorectal | [9] | |
| Protein Levels | Overall Survival | Esophageal, Pancreatic, Hepatocellular, Breast, Osteosarcoma, Renal | [13, 17, 23, 28, 34, 35, 90, 91] |
| Recurrence-Free Survival | Esophageal, Pancreatic, Hepatocellular, Osteosarcoma | [17, 28, 35, 91] | |
| Disease-Specific Survival | Endometrial, Renal | [30, 34] | |
| Nuclear Protein Levels | Overall Survival | Breast | [12, 13] |
| Recurrence-Free Survival | Melanoma | [24] | |
| Stromal Protein Levels | Overall Survival | Esophageal | [91] |
| Recurrence-Free Survival | Esophageal | [91] | |
| Phospho-S326 Levels | Overall Survival | Ovarian | [32] |
Breast and colon cancer illustrate well the nuances of the role of HSF1 in human cancers. Santagata et al. analyzed breast cancer samples from 1,841 participants enrolled in the Nurses’ Health Study by immunohistochemistry and correlated the expression with clinical parameters and survival outcomes [13]. Nuclear HSF1 levels were elevated in ~80% of in situ and invasive breast carcinomas. In invasive carcinomas, high HSF 1 expression was associated with high histologic grade, larger tumor size, and nodal involvement at diagnosis. Furthermore, high HSF1 levels were found to be independently associated with increased mortality, particularly in ER+ patients. Our group analyzed the TCGA cohort of approximately more than 1,000 women for the presence of mutations; these were not common (~0.2%) [42]. Copy number alterations were more common (14.8%) with almost all of these being amplification events. However, these were not associated with prognosis in all breast cancer subtypes. We further analyzed the RNA expression of HSF1 in a well annotated publically available dataset (GOBO) of breast cancer patients. In this analysis, we were able to document that high levels of HSF 1 mRNA were associated with poor overall and relapse free survival in all breast cancer patients. This was also true in a subset analysis of ER-negative (p=0.005) and in ER-positive patients (P=0.00045). There was a significant correlation with outcomes in those with or without endocrine therapy among ER+ patients [42]. Together, these studies document the importance of HSF1 in patients with breast cancer using immunohistochemistry as well as mRNA expression analysis.
Based on initial work in breast cancer, a gene expression signature consisting of 456 genes was termed an HSF1-cancer signature [12]. This signature not only identified patients with poor prognosis in breast cancer cohorts, but also identified patients with colon and lung cancer with poor outcomes. This is consistent with prior work that documented the importance of this pathway in colon cancer in a comparative analysis of normal and sporadic colon cancers (n=35) [9].
Outside of breast and colon cancer, HSF 1 also seems to serve as a prognostic biomarker in other tumor types. HSF 1 has increased expression in hepatocellular carcinoma (HCC) and high expression was associated with overall survival as well as metastasis-free survival of HCC patients [17]. This association was independent of cirrhosis status, tumor size, the number of tumor nodules, capsular formation, tumor grade, and venous invasion [17]. HSF1 expression levels were a significant predictor of endometrial cancer survival with higher levels of HSF 1 predictive of poor prognosis in both ER+ and ER- patients [30]. High expression levels of HSF1 was also a strong predictor for poor overall survival and recurrence-free survival of pancreatic cancer and melanoma [24, 28]. High HSF1 expression in esophageal carcinomas, or expression in the neighboring stromal cells of these tumors, was highly predictive of overall survival and disease-free survival [23, 91]. High HSF1 levels were also associated with poor survival in osteosarcoma patients [35]. These data were recently summarized in a meta-analysis including over 3,000 patients that concluded high HSF1 expression was significantly associated with overall survival or recurrence-free survival in all tumor types tested [90]. High HSF1 was additionally associated with many phenotypes of tumor progression such as tumor staging and tumor grade among others [90]. Another potential marker for HSF1 activity is the levels of phosphorylated HSF1 as this is required for transcriptional activity [51]. Specifically, phosphorylation at Ser326 is important for transcriptional activity and levels of HSF1 phospho-Ser326 have been shown to be predictive for overall survival in ovarian cancer [32].
The potential for HSF1 as a biomarker will require greater understanding of the basic biology of HSF1 in conditions of heat shock and cancer. Levels of HSF1 mRNA and protein have both been shown to be predictive for patient outcomes in multiple tumor types (Table 1). It is a generally held view that HSF1 is constitutively expressed in cells but requires activation for transcriptional activity. However, HSF1 has been shown to be amplified in breast cancer in 1015% of cases [10, 42]. However, the prognostic impact in breast cancer is specific to mRNA expression, but stayed insignificant by protein expression or by analyzing amplification events. Other studies suggest nuclear HSF 1 levels are important for HSF 1 activity and a predictor for patient outcome [12, 13, 24]. However, early studies on HSF1 suggested the protein is localized to the nucleus and DNA binding can occur independent of heat stress or phosphorylation [92–94]. As such, a full understanding of the steps for HSF1 activation will help considerably to choose which aspect of HSF 1 to use as a biomarker.
4.2. HSF1 as a Therapeutic Target
Therapeutic targeting of transcription factors has previously had mixed results. However, considering the importance of transcription factors in tumorigenesis and tumor progression, targeting transcription factors has become more accepted. Targeting HSF1 has remained in the pre-clinical stage of testing due to the lack of an efficacious and specific drug to inhibit HSF1 transcriptional activity.
Independent of attempts to develop an HSF 1 -specific inhibitor, several studies have shown successful repression of cancer cell growth with depletion of the HSF 1 gene. Loss of HSF1 activity by expression of a dominant negative suppressed aneuploidy and proliferation of prostate carcinoma cells as well as the proliferation of melanoma cells [25, 69, 95]. Knockdown of HSF1 also reduced the proliferation and xenograft tumor size of ovarian, glioblastoma, and pancreatic tumors [27, 28, 96]. Genetic ablation of HSF1 in p53-deficient mice suppressed lymphoma tumor incidence and enhanced tumor-free survival [15]. Similarly in breast cancer, heterozygous or homozygous deletion of HSF1 reduced tumor-free survival and incidence of metastasis in MMTV-neu mice [55]. Thus, reduction in HSF1 levels and/or activity is suppressive to cancer cell growth and tumor size.
There have been multiple attempts to develop a targeted inhibitor of HSF1 activity. KNK437 is a compound that was found to suppress expression of heat shock proteins and thermo-tolerance [97]. KNK437 has shown activity in suppressing tumorigenic properties of cancer cells, including suppressing cancer stem cell phenotypes, but these effects have primarily been observed in the role of enhancing the toxicity of other anti-tumorigenic compounds such as radiation, gemcitabine, or bortezomib [98–100]. KNK423, a metabolite of KNK437, also seemed to have inhibition of HSF1 [101]. However, both KNK437 and KNK423 have shown off-target effects indicating its specificity to HSF1 is limited [83, 102]. Another compound was discovered from a 2009 screening of a small 1300 compound library that was named “Compound 1” and was found to inhibit HSF1 activity but information on the compound itself is not currently in the public domain [103]. Another screening of >6,000 compounds from the Korea Chemical Bank identified KRIBB11 as having a suppressive effect on HSF1 activity, primarily via interaction with the transactivation domain preventing the recruitment of pTEFb and transcriptional elongation [104]. This compound has shown both in vitro suppression of cancer cell growth and in vivo suppression of tumor growth from our lab and others [54, 105, 106]. Although this compound was seemingly highly specific to HSF 1 because it directly interacts with HSF1, it appears to possibly have an off target effect on Mcl-1 [106]. A triazole nucleoside, Ly101–4b, was found to have a suppressive effect on HSF1 protein level and the subsequent expression of heat shock proteins and HSF1-regulated miR-214 [107, 108]. While the precise mechanism by which Ly101–4b suppresses HSF1 expression is unclear, the anti-tumor effect of Ly101–4b is potent in preclinical models of pancreatic cancer [107, 109]. Rohinitib (RHT), an analog of the natural compound rocaglamide A, was shown to suppress HSF1 activity [83]. This RHT compound was effective at reducing growth of several solid and hematopoietic tumor lines in vitro and in vivo with evidence of reduced HSF1 activity [83]. Subsequent modeling of RHT showed it had significant interaction with the DNA binding domain of HSF1 [110]. Another compound, named PW3405 from the Prestwick chemical library, was found to inhibit HSF1 phosphorylation and activity [111]. PW3405 was a potent inhibitor of cancer cell viability with a broad range of tumor types in vitro with the IC50 in the submicromolar range for all cell lines tested [111]. Considering the importance of HSF1 phosphorylation to its activity [51], it is possible PW3405 is inhibiting kinases upstream of HSF 1 reducing its phosphorylation and activity although this possibility has not been investigated. A chemical compound, CCT251236, was identified via an HSF1 phenotypic screen to suppress HSF1 activity measured by induction of Hsp72 [112]. This compound strongly suppressed HSF1 activity at low doses and suppressed subcutaneous xenograft tumor growth with ovarian cancer cells [112]. However, it was also found that this compound was directly interacting with pirin rather than targeting HSF1 [112]. Recently, IHSF115 was developed to inhibit HSF1 using modeling of the DNA binding domain of HSF1 and was shown to directly bind HSF1 and suppress its activity [113]. Despite predictive modeling suggesting IHSF115 would bind the DNA binding domain, it did not affect HSF 1 binding to target genes but likely suppressed HSF1-induced recruitment of transcriptional machinery [113]. This compound was able to suppress the growth of a number of cancer cell lines although the dosages needed were all in the micromolar range [113]. An alternative approach to inhibit HSF1 activity was developed through an RNA aptamer that suppresses HSF1 activity in both heat shock and cancer contexts by preventing the binding of HSF1 to target genes using a sequence specific to the DNA binding domain [114–117]. This strategy was sufficient to suppress the growth of cancer cells and suppress expression of HSF1 target genes using a plasmid-based delivery system [114]. The utility of such a system for suppressing tumor growth in vivo is unknown.
In addition to targeted small molecules directed at HSF1, there are also natural compounds known to suppress HSF1 activity. Quercetin, a plant polyphenol from the flavonoid group, was identified to inhibit the heat shock response by down regulation of HSF1 [118]. Being a natural compound, quercetin also seems to inhibit multiple targets and, therefore, is not specific to HSF1 [83]. Triptolide, a natural compound derived from tripterygium wilfordii was also discovered to suppress HSF1 activity by interfering with the HSF1 transactivation domain [119]. It has subsequently been shown to have several anticancer properties but also other cellular targets, indicating it lacks specificity to HSF1 [83, 120]. The natural compound rocaglamide A was found to have selective HSF1 inhibition by preventing HSF1 binding to target genes [83]. Analogs to this compound, in particular RHT, was found to have more specific and potent effects [83].
CONCLUSION
The role of HSF1 in cancer has become a highly discussed topic in the last decade. As recent as 2005, there were only 5 published papers indexed in MEDLINE that included HSF 1 and cancer whereas there were more than 50 published papers on HSF1 and cancer in 2,017. It has become clear that HSF 1 functions in its role as the regulator of the heat shock response in cancer cells considering that heat shock proteins themselves play a role in tumorigenesis. But it is also clear that HSF 1 has a cancer-specific transcriptional program that supports malignancy [12]. Our lab and others have shown HSF 1 to play a role in the migratory and invasive capability of cancer cells as well as metastasis. Beyond this, HSF1 appears to have a pleiotropic effect on malignancy as it has been shown to play a role in many aspects of tumor biology from DNA repair [121] to angiogenesis [122] to metabolism [83]. All of these functions of HSF1 appear to support the malignancy of cancer cells and, therefore, makes HSF1 a possible biomarker for tumors that are progressing toward metastasis. HSF1 has been shown to be a strong predictor of patient outcomes in multiple cancer types, especially breast and colon cancers. However, further development of HSF 1 as a biomarker is necessary as studies are not consistent with their evaluation of HSF1. For example, some studies have used raw HSF 1 expression as a marker, whereas others have used nuclear HSF1 or phosphorylated HSF1 at Ser326. A clear understanding of which of these markers most accurately reflect HSF1 activity will be helpful to understand mechanistically how these different detections of HSF1 contribute to tumor progression. Because HSF 1 supports tumor progression and could possibly serve as a biomarker, there have also been attempts to target HSF1 therapeutically. These agents have yet to make it to the clinic likely because many of them have obvious off-target effects and are not highly specific to HSF 1. Continued development of HSF 1 inhibitors in preclinical in vivo models with biomarkers for response to these treatments will be necessary prior to any attempt in human trials is possible.
Table 2.
Small molecular compounds that inhibit HSF1 activity.
| Selective Compounds | Effect on HSF1 | Mechanism of Action | References |
|---|---|---|---|
| KNK437 | ↓HSP expression | Unknown | [97] |
| KNK423 | ↓HSP expression | Unknown | [101] |
| “Compound 1” | ↓HSF1 phosph. and HSP expression | Unknown | [103] |
| KRIBB11 | ↓HSF1-induced transcription | Binds to transactivation domain, prevents pTEFb recruitment | [54, 104–106] |
| Ly101-4b | ↓HSF1 protein levels and HSP expression | Unknown | [108, 125] |
| Rohinitib (RHT) | ↓HSE binding and HSP expression | Binds to DNA binding domain to prevent HSE binding | [83, 110] |
| PW3405 | ↓HSF1 phosph. | Unknown | [111] |
| CCT251236 | ↓HSP expression | Unknown | [112] |
| IHSF115 | ↓HSF1 binding to HSE and HSP expression | Binds DNA binding domain to prevent HSE binding | [113] |
| HSF1 RNA Aptamer | ↓HSE binding and HSP expression | Binds DNA binding domain to prevent HSE binding | [114–117] |
| Natural Compounds | |||
| Quercetin | ↓HSF1 phosph. and HSE binding | Unknown | [83, 118, 126] |
| Triptolide | ↓HSP expression and transactivation | Inhibits HSF1-induced transcription activation | [83, 119, 120] |
| Rocaglamide A | ↓HSE binding and HSP expression | Binds to DNA binding domain to prevent HSE binding | [83] |
ACKNOWLEDGEMENTS
This work was supported by the National Cancer Institute (NCI) K22CA207575–01 (RLC) and The Catherine Peachey Fund (RLC).
Footnotes
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- [1].Ritossa F, A new puffing pattern induced by temperature shock and DNP in Drosophila. Experientia, 1962. 18: p. 571–573. [Google Scholar]
- [2].Parker CS and Topol J, A Drosophila RNA polymerase II transcription factor contains a promoter-region-specific DNA-binding activity. Cell, 1984. 36(2): p. 357–69. [DOI] [PubMed] [Google Scholar]
- [3].Topol J, Ruden DM, and Parker CS, Sequences required for in vitro transcriptional activation of a Drosophila hsp 70 gene. Cell, 1985. 42(2): p. 527–37. [DOI] [PubMed] [Google Scholar]
- [4].Amin J, Ananthan J, and Voellmy R, Key features of heat shock regulatory elements. Mol Cell Biol, 1988. 8(9): p. 3761–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Dudler R and Travers AA, Upstream elements necessary for optimal function of the hsp 70 promoter in transformed flies. Cell, 1984. 38(2): p. 391–8. [DOI] [PubMed] [Google Scholar]
- [6].Slater MR and Craig EA, Transcriptional regulation of an hsp70 heat shock gene in the yeast Saccharomyces cerevisiae. Mol Cell Biol, 1987. 7(5): p. 1906–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Xiao H and Lis JT, Germline transformation used to define key features of heat-shock response elements. Science, 1988. 239(4844): p. 1139–42. [DOI] [PubMed] [Google Scholar]
- [8].Hoang AT, et al. , A novel association between the human heat shock transcription factor 1 (HSF1) and prostate adenocarcinoma. Am J Pathol, 2000. 156(3): p. 857–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cen H, et al. , Induction of HSF1 expression is associated with sporadic colorectal cancer. World J Gastroenterol, 2004. 10(21): p. 3122–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cheng Q, et al. , Amplification and high-level expression of heat shock protein 90 marks aggressive phenotypes of human epidermal growth factor receptor 2 negative breast cancer. Breast Cancer Res, 2012. 14(2): p. R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dai C, et al. , Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell, 2007. 130(6): p. 1005–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mendillo ML, et al. , HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell, 2012. 150(3): p. 549–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Santagata S, et al. , High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc Natl Acad Sci U S A, 2011. 108(45): p. 18378–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Jego G, et al. , Dual regulation of SPI1/PU.1 transcription factor by heat shock factor 1 (HSF1) during macrophage differentiation of monocytes. Leukemia, 2014. 28(8): p. 1676–86. [DOI] [PubMed] [Google Scholar]
- [15].Min JN, et al. , Selective suppression of lymphomas by functional loss of Hsf1 in a p53-deficient mouse model for spontaneous tumors. Oncogene, 2007. 26(35): p. 5086–97. [DOI] [PubMed] [Google Scholar]
- [16].Chuma M, et al. , Heat shock factor 1 accelerates hepatocellular carcinoma development by activating nuclear factor-kappaB/mitogen-activated protein kinase. Carcinogenesis, 2014. 35(2): p. 272–81. [DOI] [PubMed] [Google Scholar]
- [17].Fang F, Chang R, and Yang L, Heat shock factor 1 promotes invasion and metastasis of hepatocellular carcinoma in vitro and in vivo. Cancer, 2012. 118(7): p. 1782–94. [DOI] [PubMed] [Google Scholar]
- [18].Jin X, Moskophidis D, and Mivechi NF, Heat shock transcription factor 1 is a key determinant of HCC development by regulating hepatic steatosis and metabolic syndrome. Cell Metab, 2011. 14(1): p. 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li S, et al. , Upregulation of heat shock factor 1 transcription ac tivity is associated with hepatocellular carcinoma progression. Mol Med Rep, 2014. 10(5): p. 2313–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhang N, et al. , HSF1 upregulates ATG4B expression and en hances epirubicin-induced protective autophagy in hepatocellular carcinoma cells. Cancer Lett, 2017. 409: p. 81–90. [DOI] [PubMed] [Google Scholar]
- [21].Ishiwata J, et al. , State of heat shock factor 1 expression as a putative diagnostic marker for oral squamous cell carcinoma. Int J Oncol, 2012. 40(1): p. 47–52. [DOI] [PubMed] [Google Scholar]
- [22].Kim SA, et al. , The antitumor effect of PLK1 and HSF1 double knockdown on human oral carcinoma cells. Int J Oncol, 2010. 36(4): p. 867–72. [DOI] [PubMed] [Google Scholar]
- [23].Tsukao Y, et al. , Overexpression of heat-shock factor 1 is associ ated with a poor prognosis in esophageal squamous cell carcinoma. Oncol Lett, 2017. 13(3): p. 1819–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kourtis N, et al. , FBXW7 modulates cellular stress response and metastatic potential through HSF1 post-translational modification. Nat Cell Biol, 2015. 17(3): p. 322–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Nakamura Y, et al. , Silencing HSF1 by short hairpin RNA decreases cell proliferation and enhances sensitivity to hyperthermia in human melanoma cell lines. J Dermatol Sci, 2010. 60(3): p. 187–92. [DOI] [PubMed] [Google Scholar]
- [26].Dudeja V, et al. , Prosurvival role of heat shock factor 1 in the pathogenesis of pancreatobiliary tumors. Am J Physiol Gastrointest Liver Physiol, 2011. 300(6): p. G948–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chen K, et al. , Loss of AMPK activation promotes the invasion and metastasis of pancreatic cancer through an HSF1-dependent pathway. Mol Oncol, 2017. 11(10): p. 1475–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liang W, et al. , Heat shock factor 1 inhibits the mitochondrial apoptosis pathway by regulating second mitochondria-derived activator of caspase to promote pancreatic tumorigenesis. J Exp Clin Cancer Res, 2017. 36(1): p. 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chen YF, et al. , Targeting HSF1 leads to an antitumor effect in human epithelial ovarian cancer. Int J Mol Med, 2017. 39(6): p. 1564–1570. [DOI] [PubMed] [Google Scholar]
- [30].Engerud H, et al. , High level of HSF1 associates with aggressive endometrial carcinoma and suggests potential for HSP90 inhibitors. Br J Cancer, 2014. 111(1): p. 78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Powell CD, et al. , The Heat Shock Transcription Factor HSF1 Induces Ovarian Cancer Epithelial-Mesenchymal Transition in a 3D Spheroid Growth Model. PLoS One, 2016. 11(12): p. e0168389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yasuda K, et al. , Phosphorylation of HSF1 at serine 326 residue is related to the maintenance of gynecologic cancer stem cells through expression of HSP27. Oncotarget, 2017. 8(19): p. 31540–31553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Cui J, Tian H, and Chen G, Upregulation of Nuclear Heat Shock Factor 1 Contributes to Tumor Angiogenesis and Poor Survival in Patients With Non-Small Cell Lung Cancer. Ann Thorac Surg, 2015. 100(2): p. 465–72. [DOI] [PubMed] [Google Scholar]
- [34].Wu PS, Chang YH, and Pan CC, High expression of heat shock proteins and heat shock factor-1 distinguishes an aggressive subset of clear cell renal cell carcinoma. Histopathology, 2017. 71(5): p. 711–718. [DOI] [PubMed] [Google Scholar]
- [35].Zhou Z, et al. , Heat shock transcription factor 1 promotes the proliferation, migration and invasion of osteosarcoma cells. Cell Prolif, 2017. 50(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Fujimoto M and Nakai A, The heat shock factor family and adaptation to proteotoxic stress. Febs j, 2010. 277(20): p. 4112–25. [DOI] [PubMed] [Google Scholar]
- [37].Fujimoto M, et al. , HSF4 is required for normal cell growth and differentiation during mouse lens development. Embo j, 2004. 23(21): p. 4297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Fujimoto M, et al. , Analysis of HSF4 binding regions reveals its necessity for gene regulation during development and heat shock response in mouse lenses. J Biol Chem, 2008. 283(44): p. 29961–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Tessari A, et al. , Characterization of HSFY, a novel AZFb gene on the Y chromosome with a possible role in human spermatogenesis. Mol Hum Reprod, 2004. 10(4): p. 253–8. [DOI] [PubMed] [Google Scholar]
- [40].Fujimoto M, et al. , A novel mouse HSF3 has the potential to activate nonclassical heat-shock genes during heat shock. Mol Biol Cell, 2010. 21(1): p. 106–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhang Y, et al. , Structural organization and promoter analysis of murine heat shock transcription factor-1 gene. J Biol Chem, 1998. 273(49): p. 32514–21. [DOI] [PubMed] [Google Scholar]
- [42].Gokmen-Polar Y and Badve S, Upregulation of HSF1 in estrogen receptor positive breast cancer. Oncotarget, 2016. 7(51): p. 84239–84245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hu Y and Mivechi NF, HSF-1 interacts with Ral-binding protein 1 in a stress-responsive, multiprotein complex with HSP90 in vivo. J Biol Chem, 2003. 278(19): p. 17299–306. [DOI] [PubMed] [Google Scholar]
- [44].Neef DW, et al. , A direct regulatory interaction between chaperonin TRiC and stress-responsive transcription factor HSF1. Cell Rep, 2014. 9(3): p. 955–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Shi Y, Mosser DD, and Morimoto RI, Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev, 1998. 12(5): p. 654–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Zou J, et al. , Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell, 1998. 94(4): p. 471–80. [DOI] [PubMed] [Google Scholar]
- [47].Morimoto RI, Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev, 1998. 12(24): p. 3788–96. [DOI] [PubMed] [Google Scholar]
- [48].Hentze N, et al. , Molecular mechanism of thermosensory function of human heat shock transcription factor Hsf1. Elife, 2016. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Rabindran SK, et al. , Regulation of heat shock factor trimer formation: role of a conserved leucine zipper. Science, 1993. 259(5092): p. 230–4. [DOI] [PubMed] [Google Scholar]
- [50].Westwood JT, Clos J, and Wu C, Stress-induced oligomerization and chromosomal relocalization of heat-shock factor. Nature, 1991. 353(6347): p. 822–7. [DOI] [PubMed] [Google Scholar]
- [51].Guettouche T, et al. , Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC Biochem, 2005. 6: p. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Larson JS, Schuetz TJ, and Kingston RE, Activation in vitro of sequence-specific DNA binding by a human regulatory factor. Nature, 1988. 335(6188): p. 372–5. [DOI] [PubMed] [Google Scholar]
- [53].Sorger PK and Pelham HR, Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell, 1988. 54(6): p. 855–64. [DOI] [PubMed] [Google Scholar]
- [54].Carpenter RL, et al. , Combined inhibition of AKT and HSF1 suppresses breast cancer stem cells and tumor growth. Oncotarget, 2017. 8(43): p. 73947–73963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Xi C, et al. , Heat shock factor Hsf1 cooperates with ErbB2 (Her2/Neu) protein to promote mammary tumorigenesis and metastasis. J Biol Chem, 2012. 287(42): p. 35646–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Khaleque MA, et al. , Induction of heat shock proteins by heregulin beta1 leads to protection from apoptosis and anchorage-independent growth. Oncogene, 2005. 24(43): p. 6564–73. [DOI] [PubMed] [Google Scholar]
- [57].O’Callaghan-Sunol C and Sherman MY, Heat shock transcription factor (HSF1) plays a critical role in cell migration via maintaining MAP kinase signaling. Cell Cycle, 2006. 5(13): p. 1431–7. [DOI] [PubMed] [Google Scholar]
- [58].Nakamura Y, et al. , Heat shock factor 1 is required for migration and invasion of human melanoma in vitro and in vivo. Cancer Lett, 2014. 354(2): p. 329–35. [DOI] [PubMed] [Google Scholar]
- [59].Meng L, Gabai VL, and Sherman MY, Heat-shock transcription factor HSF1 has a critical role in human epidermal growth factor receptor-2-induced cellular transformation and tumorigenesis. Oncogene, 2010. 29(37): p. 5204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Toma-Jonik A, et al. , Active heat shock transcription factor 1 supports migration of the melanoma cells via vinculin down-regulation. Cell Signal, 2015. 27(2): p. 394–401. [DOI] [PubMed] [Google Scholar]
- [61].Carpenter RL, et al. , Akt phosphorylates and activates HSF-1 independent of heat shock, leading to Slug overexpression and epithelial-mesenchymal transition (EMT) of HER2-overexpressing breast cancer cells. Oncogene, 2015. 34(5): p. 546–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ye X, et al. , Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature, 2015. 525(7568): p. 256–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Chou SD, et al. , HSF1 regulation of beta-catenin in mammary cancer cells through control of HuR/elavL1 expression. Oncogene, 2015. 34(17): p. 2178–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lee JH, et al. , Mitochondrial Respiratory Dysfunction Induces Claudin-1 Expression via Reactive Oxygen Species-mediated Heat Shock Factor 1 Activation, Leading to Hepatoma Cell Invasiveness. J Biol Chem, 2015. 290(35): p. 21421–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Li Y, et al. , MicroRNA-135b, a HSF1 target, promotes tumor invasion and metastasis by regulating RECK and EVI5 in hepatocellular carcinoma. Oncotarget, 2015. 6(4): p. 2421–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Khaleque MA, et al. , Heat shock factor 1 represses estrogen-dependent transcription through association with MTA1. Oncogene, 2008. 27(13): p. 1886–93. [DOI] [PubMed] [Google Scholar]
- [67].Kim EH, et al. , Heat shock factor 1-mediated aneuploidy requires a defective function of p53. Cancer Res, 2009. 69(24): p. 9404–12. [DOI] [PubMed] [Google Scholar]
- [68].Lee YJ, et al. , A novel function for HSF1-induced mitotic exit failure and genomic instability through direct interaction between HSF1 and Cdc20. Oncogene, 2008. 27(21): p. 2999–3009. [DOI] [PubMed] [Google Scholar]
- [69].Tang Z, et al. , MEK guards proteome stability and inhibits tumor-suppressive amyloidogenesis via HSF1. Cell, 2015. 160(4): p. 729–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Wang B, et al. , Heat shock factor 1 induces cancer stem cell phenotype in breast cancer cell lines. Breast Cancer Res Treat, 2015. 153(1): p. 57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Bradley E, et al. , Regulation of embryonic stem cell pluripotency by heat shock protein 90. Stem Cells, 2012. 30(8): p. 1624–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Lee YJ, et al. , HSF1 as a mitotic regulator: phosphorylation of HSF1 by Plk1 is essential for mitotic progression. Cancer Res,2008. 68(18): p. 7550–60. [DOI] [PubMed] [Google Scholar]
- [73].Yang X, et al. , HSF1 and Sp1 regulate FUT4 gene expression and cell proliferation in breast cancer cells. J Cell Biochem, 2014. 115(1): p. 168–78. [DOI] [PubMed] [Google Scholar]
- [74].Antonietti P, et al. , Interference with the HSF1/HSP70/BAG3 Pathway Primes Glioma Cells to Matrix Detachment and BH3 Mimetic-Induced Apoptosis. Mol Cancer Ther, 2017. 16(1): p. 156–168. [DOI] [PubMed] [Google Scholar]
- [75].Jacobs AT and Marnett LJ, Heat shock factor 1 attenuates 4-Hydroxynonenal-mediated apoptosis: critical role for heat shock protein 70 induction and stabilization of Bcl-XL. J Biol Chem, 2007. 282(46): p. 33412–20. [DOI] [PubMed] [Google Scholar]
- [76].Jacobs AT and Marnett LJ, HSF1-mediated BAG3 expression attenuates apoptosis in 4-hydroxynonenal-treated colon cancer cells via stabilization of anti-apoptotic Bcl-2 proteins. J Biol Chem, 2009. 284(14): p. 9176–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Wang J, et al. , HSF1 down-regulates XAF1 through transcriptional regulation. J Biol Chem, 2006. 281(5): p. 2451–9. [DOI] [PubMed] [Google Scholar]
- [78].Desai S, et al. , Heat shock factor 1 (HSF1) controls chemoresistance and autophagy through transcriptional regulation of auto-phagy-related protein 7 (ATG7). J Biol Chem, 2013. 288(13): p. 9165–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Luo T, et al. , PSMD10/gankyrin induces autophagy to promote tumor progression through cytoplasmic interaction with ATG7 and nuclear transactivation of ATG7 expression. Autophagy, 2016. 12(8): p. 1355–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Luan Q, et al. , RIPK1 regulates survival of human melanoma cells upon endoplasmic reticulum stress through autophagy. Autophagy, 2015. 11(7): p. 975–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Zhang X, Expression, correlation and prognostic significance of CD133, P57 and HSF1 in meningioma. Eur Rev Med Pharmacol Sci, 2017. 21(20): p. 4600–4605. [PubMed] [Google Scholar]
- [82].Asgari Y, et al. , Alterations in cancer cell metabolism: the Warburg effect and metabolic adaptation. Genomics, 2015. 105(5–6): p. 275–81. [DOI] [PubMed] [Google Scholar]
- [83].Santagata S, et al. , Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state. Science, 2013. 341(6143): p. 1238303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Cigliano A, et al. , Inhibition of HSF1 suppresses the growth of hepatocarcinoma cell lines in vitro and AKT-driven hepatocarcino-genesis in mice. Oncotarget, 2017. 8(33): p. 54149–54159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Zhao YH, et al. , Upregulation of lactate dehydrogenase A by ErbB2 through heat shock factor 1 promotes breast cancer cell glycolysis and growth. Oncogene, 2009. 28(42): p. 3689–701. [DOI] [PubMed] [Google Scholar]
- [86].Ma W, et al. , Glucose regulates heat shock factor 1 transcription activity via mTOR pathway in HCC cell lines. Cell Biol Int, 2015. 39(11): p. 1217–24. [DOI] [PubMed] [Google Scholar]
- [87].Dai S, et al. , Suppression of the HSF1-mediated proteotoxic stress response by the metabolic stress sensor AMPK. Embo j, 2015. 34(3): p. 275–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Minsky N and Roeder RG, Direct link between metabolic regulation and the heat-shock response through the transcriptional regulator PGC-1alpha. Proc Natl Acad Sci US A, 2015. 112(42): p. E5669–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Wu Z, et al. , Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell, 1999. 98(1): p. 115–24. [DOI] [PubMed] [Google Scholar]
- [90].Wan T, et al. , Prognostic role of HSF1 overexpression in solid tumors: a pooled analysis of 3,159 patients. Onco Targets Ther, 2018. 11: p. 383–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Liao Y, et al. , Higher heat shock factor 1 expression in tumor stroma predicts poor prognosis in esophageal squamous cell carcinoma patients. J Transl Med, 2015. 13: p. 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Baler R, Dahl G, and Voellmy R, Activation of human heat shock genes is accompanied by oligomerization, modification, and rapid translocation of heat shock transcription factor HSF1. Mol Cell Biol, 1993. 13(4): p. 2486–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Mercier PA, et al. , Xenopus heat shock factor 1 is a nuclear protein before heat stress. J Biol Chem, 1997. 272(22): p. 14147–51. [DOI] [PubMed] [Google Scholar]
- [94].Mercier PA, Winegarden NA, and Westwood JT, Human heat shock factor 1 is predominantly a nuclear protein before and after heat stress. J Cell Sci, 1999. 112 (Pt 16): p. 2765–74. [DOI] [PubMed] [Google Scholar]
- [95].Wang Y, et al. , Expression of a dominant negative heat shock factor-1 construct inhibits aneuploidy in prostate carcinoma cells. J Biol Chem, 2004. 279(31): p. 32651–9. [DOI] [PubMed] [Google Scholar]
- [96].Im CN, Yun HH, and Lee JH, Heat Shock Factor 1 Depletion Sensitizes A172 Glioblastoma Cells to Temozolomide via Suppression of Cancer Stem Cell-Like Properties. Int J Mol Sci, 2017. 18(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Yokota S, Kitahara M, and Nagata K, Benzylidene lactam compound, KNK437, a novel inhibitor of acquisition of thermotolerance and heat shock protein induction in human colon carcinoma cells. Cancer Res, 2000. 60(11): p. 2942–8. [PubMed] [Google Scholar]
- [98].Bustany S, et al. , Heat shock factor 1 is a potent therapeutic target for enhancing the efficacy of treatments for multiple myeloma with adverse prognosis. J Hematol Oncol, 2015. 8: p. 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Lee CH, et al. , Inhibition of heat shock protein (Hsp) 27 potentiates the suppressive effect of Hsp90 inhibitors in targeting breast cancer stem-like cells. Biochimie, 2012. 94(6): p. 1382–9. [DOI] [PubMed] [Google Scholar]
- [100].Taba K, et al. , KNK437 downregulates heat shock protein 27 of pancreatic cancer cells and enhances the cytotoxic effect of gemcitabine. Chemotherapy, 2011. 57(1): p. 12–6. [DOI] [PubMed] [Google Scholar]
- [101].Koishi M, et al. , The effects of KNK437, a novel inhibitor of heat shock protein synthesis, on the acquisition of thermotolerance in a murine transplantable tumor in vivo. Clin Cancer Res, 2001. 7(1): p. 215–9. [PubMed] [Google Scholar]
- [102].Oommen D and Prise KM, KNK437, abrogates hypoxia-induced radioresistance by dual targeting of the AKT and HIF-1alpha survival pathways. Biochem Biophys Res Commun, 2012. 421(3): p. 538–43. [DOI] [PubMed] [Google Scholar]
- [103].Au Q, et al. , Identification of inhibitors of HSF1 functional activity by high-content target-based screening. J Biomol Screen, 2009. 14(10): p. 1165–75. [DOI] [PubMed] [Google Scholar]
- [104].Yoon YJ, et al. , KRIBB11 inhibits HSP70 synthesis through inhibition of heat shock factor 1 function by impairing the recruitment of positive transcription elongation factor b to the hsp70 promoter. J Biol Chem, 2011. 286(3): p. 1737–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Fok JHL, et al. , HSF1 Is Essential for Myeloma Cell Survival and A Promising Therapeutic Target. Clin Cancer Res, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Kang MJ, Yun HH, and Lee JH, KRIBB11 accelerates Mcl-1 degradation through an HSF1-independent, Mule-dependent pathway in A549 non-small cell lung cancer cells. Biochem Biophys Res Commun, 2017. 492(3): p. 304–309. [DOI] [PubMed] [Google Scholar]
- [107].Chen YF, et al. , Nucleoside analog inhibits microRNA-214 through targeting heat-shock factor 1 in human epithelial ovarian cancer. Cancer Sci, 2013. 104(12): p. 1683–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Xia Y, et al. , Targeting heat shock factor 1 with a triazole nucleoside analog to elicit potent anticancer activity on drug-resistant pancreatic cancer. Cancer Lett, 2012. 318(2): p. 145–53. [DOI] [PubMed] [Google Scholar]
- [109].Cano CE, et al. , Genetic inactivation of Nupr1 acts as a dominant suppressor event in a two-hit model of pancreatic carcinogenesis. Gut, 2014. 63(6): p. 984–95. [DOI] [PubMed] [Google Scholar]
- [110].Agarwal T, et al. , Molecular docking and dynamic simulation evaluation of Rohinitib - Cantharidin based novel HSF1 inhibitors for cancer therapy. J Mol Graph Model, 2015. 61: p. 141–9. [DOI] [PubMed] [Google Scholar]
- [111].Zhang D and Zhang B, Selective killing of cancer cells by small molecules targeting heat shock stress response. Biochem Biophys Res Commun, 2016. 478(4): p. 1509–14. [DOI] [PubMed] [Google Scholar]
- [112].Cheeseman MD, et al. , Discovery of a Chemical Probe Bisamide (CCT251236): An Orally Bioavailable Efficacious Pirin Ligand from a Heat Shock Transcription Factor 1 (HSF1) Phenotypic Screen. J Med Chem, 2017. 60(1): p. 180–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Vilaboa N, et al. , New inhibitor targeting human transcription factor HSF1: effects on the heat shock response and tumor cell survival. Nucleic Acids Res, 2017. 45(10): p. 5797–5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Salamanca HH, et al. , Inhibiting heat shock factor 1 in human cancer cells with a potent RNA aptamer. PLoS One, 2014. 9(5): p. e96330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Salamanca HH, et al. , An RNA aptamer perturbs heat shock transcription factor activity in Drosophila melanogaster. Nucleic Acids Res, 2011. 39(15): p. 6729–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Wang S, et al. , Knocking down gene function with an RNA ap-tamer expressed as part of an intron. Nucleic Acids Res, 201038(15): p. e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Zhao X, et al. , An RNA aptamer that interferes with the DNA binding of the HSF transcription activator. Nucleic Acids Res, 2006. 34(13): p. 3755–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Nagai N, Nakai A, and Nagata K, Quercetin suppresses heat shock response by down regulation of HSF1. Biochem Biophys Res Commun, 1995. 208(3): p. 1099–105. [DOI] [PubMed] [Google Scholar]
- [119].Westerheide SD, et al. , Triptolide, an inhibitor of the human heat shock response that enhances stress-induced cell death. J Biol Chem, 2006. 281(14): p. 9616–22. [DOI] [PubMed] [Google Scholar]
- [120].Li XJ, Jiang ZZ, and Zhang LY, Triptolide: progress on research in pharmacodynamics and toxicology. J Ethnopharmacol, 2014. 155(1): p. 67–79. [DOI] [PubMed] [Google Scholar]
- [121].Fujimoto M, et al. , The HSF1-PARP13-PARP1 complex facilitates DNA repair and promotes mammary tumorigenesis. Nat Commun, 2017. 8(1): p. 1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Gabai VL, et al. , Heat shock transcription factor Hsf1 is involved in tumor progression via regulation of hypoxia-inducible factor 1 and RNA-binding protein HuR. Mol Cell Biol, 2012. 32(5): p. 929–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Dayalan Naidu S, et al. , Heat Shock Factor 1 Is a Substrate for p38 Mitogen-Activated Protein Kinases. Mol Cell Biol, 2016. 36(18): p. 2403–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Chou SD, et al. , mTOR is essential for the proteotoxic stress response, HSF1 activation and heat shock protein synthesis. PLoS One, 2012. 7(6): p. e39679. [DOI] [PMC free article] [PubMed] [Google Scholar]