Abstract
The human opportunistic pathogen Pseudomonas aeruginosa strain PA14 infects both plants and animals. Previously, using plants to screen directly for P. aeruginosa virulence-attenuated mutants, we identified a locus, pho34B12, relevant in mammalian pathogenesis. Here, nonsense point mutations in the two opposing ORFs identified in the pho34B12 locus revealed that one of them, mvfR (multiple virulence factor Regulator), is able to control all of the phenotypes that mutant phoA34B12 displays. Both genetic and biochemical evidence demonstrate that the mvfR gene encodes a LysR-like transcriptional factor that positively regulates the production of elastase, phospholipase, and of the autoinducers, 3oxo-dodecanoyl homoserine lactone (PAI I) and 2-heptyl-3-hydroxy-4-quinolone (PQS), as well as the expression of the phnAB operon, involved in phenazine biosynthesis. We demonstrate that the MvfR protein is membrane-associated and acts as a transcriptional activator until cells reach stationary phase, when a unique negative feedback mechanism is activated to signal the down-regulation of the MvfR protein. This work reveals an unprecedented virulence mechanism of P. aeruginosa and identifies a unique indispensable player in the P. aeruginosa quorum-sensing cascade.
Pseudomonas aeruginosa is a well known opportunistic pathogen that primarily causes infections in immunocompromised individuals and patients with cystic fibrosis (1, 2). Its rather impermeable membrane (1), the presence of β-lactamase (2, 3), and its various efflux systems (4–6) all make P. aeruginosa the leading cause of nosocomial infections and hospital-acquired pneumonia (7).
Despite detailed knowledge about some of the extracellular proteins and several surface-associated components identified in P. aeruginosa, many virulence factors essential for pathogenicity and the mechanisms by which they coordinately function during pathogenesis remain to be elucidated. By using a multihost system consisting of plants, nematodes, and animals, and a systematic genetic approach, we have identified a large number of virulence-associated genes encoding either unidentified or previously identified virulence factors (reviewed in refs. 8 and 9). Uncovering loci involved in pathogenesis is facilitated by our high-throughput approach; thus, the challenge lies in the identification of functions.
The pho34B12 locus is one of the novel virulence genes of P. aeruginosa identified by using the strategy outlined previously. Previous studies have shown that mutation at this locus reduced the ability of P. aeruginosa strain PA14 to cause disease in plants and animals. Partial characterization of the pho34B12 locus indicated that one of the potential genes contained in this locus might encode a novel quorum-sensing (QS) transcriptional regulator controlling the production of the virulence-associated factors (elastase, phospholipase, and pyocyanin) relevant to mammalian pathogenesis (10). Pyocyanin seems to contribute to the persistence of P. aeruginosa in the lungs of patients with cystic fibrosis (11). Although little is known about the nature of the enzymes that catalyze the formation of pyocyanin in P. aeruginosa, the conversion of chorismate to anthranilate is thought to be a key step in the pyocyanin biosynthesis pathway, catalyzed by the anthranilate synthase encoded by the phnA and phnB genes (12). Genetic studies also identified the phzABCDEFG operon as containing genes required for the synthesis of pyocyanin (13–15). Various in vitro studies using purified pyocyanin have shown that pyocyanin inhibits mammalian cell respiration, disrupts the beating of human cilia, inhibits the growth of epidermal cells, and inhibits the release of IL-2, which leads to the inhibition of T lymphocyte proliferation and Ig secretion by B-lymphocytes (16, 17).
In this article, we further dissect the complex cell density-dependent regulatory circuit of P. aeruginosa by determining the role of a previously unknown gene, mvfR, in the QS system. Furthermore, the biosynthetic pathway of pyocyanin as well as its effects on the pathogenesis of diseases produced by Pseudomonads is further characterized. Here, both genetic and biochemical evidence show that the mvfR encodes a novel LysR-like transcriptional regulator. MvfR positively regulates the expression of the phnAB operon and controls the production of elastase, phospholipase, autoinducer I [3-oxo-dodecanoyl homoserine lactone (PAI I); ref. 18] and the autoinducer-like molecule 2-heptyl-3-hydroxy-4-quinolone (PQS; ref. 19), as well as the levels of various P. aeruginosa-secreted proteins. We demonstrate that a unique and unprecedented negative feedback mechanism is used to regulate the function of the MvfR protein, a member of the QS-associated regulatory cascade.
Materials and Methods
Bacterial Strains, Media, and Growth Conditions.
We used Escherichia coli strains TOP 10 and BL21 (DE3); P. aeruginosa strain UCBPP-PA14; TnphoA-mutagenized UCBPP-PA14 mutant strain (20); pho34B12 (10); PAO1 (21); and PAO1 isogenic mutant strains lasR−, rhlR−, and lasR−rhlR− (22). Luria–Bertani medium was used for growth of both E. coli and P. aeruginosa strains at 37°C.
Site-Directed Mutagenesis.
The plasmid pLGR34B12 (10), with a 3.7-kb EcoRI fragment containing the entire pho34B12 locus, was used for mutagenesis. In the pho34B12 region, ORFs 1 and 2 are arranged in a way that the first nucleotide of the ORF1 codon is the third nucleotide of the ORF2 codon and vice versa. Therefore, the point mutations were introduced by means of PCR into the first nucleotide of a codon in both ORFs. In ORF1, a glutamine (no. 262 of 427 amino acids) was converted to a stop codon by changing CAG to TAG, and in ORF2, a glutamic acid (no. 151 of 344 amino acids) was switched to a stop codon by changing GAG to TAG. The single-nucleotide change in each ORF was confirmed by sequencing. The mutagenized 3.7-kb EcoRI fragments were then subcloned into the SmaI site of pCVD (23) to generate two plasmids, each containing mutated ORF1 (pHCORF1*) and mutated MvfR (pHCORF2*). These two plasmids were then used to replace the pho34B12 gene with TnphoA insertion via homologous recombination as described in ref. 23. The resulting mutants are designated as ORF1* and ORF2*, respectively.
Virulence Studies in Plant and Animal Models.
Assessment of P. aeruginosa growth in Arabidopsis and determination of mortalities in the thermal injury mouse model were performed as described (10).
Analysis of Pyocyanin and Autoinducers.
Quantitation of pyocyanin from cell-free P. aeruginosa cultures was performed as described (12). Autoinducers were extracted and analyzed by a standard procedure with HPLC (24).
Construction of mvfR-lacZ and phnAB-lacZ Transcriptional Fusions.
To construct the mvfR-lacZ fusion, a 473-bp fragment (447-bp upstream of the start codon and 26-bp downstream) was PCR-amplified and cloned into the EcoRI/BamHI site of pPCS1002 (25) to obtain pHCmvfR-lacZ. To construct the phnAB-lacZ fusion, a 522-bp fragment (478-bp upstream of the start codon of phnA and 44-bp downstream) was PCR-amplified and cloned into the EcoRI/BamHI site of pPCS1002 to obtain pHCphnAB-lacZ, which was introduced by means of electroporation into P. aeruginosa strains PA14 and ORF2*.
β-Galactosidase Assay.
P. aeruginosa strains containing the lacZ transcriptional fusions were grown overnight to OD600 = 2.5–3.0 or as indicated. The β-galactosidase assays were carried out as described (26).
Overexpression and Purification of MvfR in E. coli.
A 1035-bp fragment containing the entire coding region of mvfR was PCR-amplified and cloned into the XhoI/HindIII site of pBAD/HisA (Invitrogen) to obtain pBAD/His-mvfR. Overexpression and purification of the MvfR protein was conducted according to the instruction manual from Xpress System Protein Purification kit (Invitrogen). The purified recombinant protein was treated with enterokinase to remove the N-terminal His-tag and used in the gel electrophoresis mobility-shift assay.
Gel Electrophoresis Mobility-Shift Assay.
Two pairs of complementary oligonucleotides from the promoter region of the phnAB operon were synthesized. One pair is 210–160 bp (P1) upstream of the methionine start codon, and the other is 484–434 bp (P2) upstream. About 100 ng of each oligonucleotide pair was annealed, and radio-labeled DNA was obtained by end-labeling with 32P by using T4 polynucleotide kinase and [γ-32P]ATP. The DNA-binding assays were carried out at room temperature with 4,000 cpm of the radio-labeled probes and 0.5 μg of the purified MvfR protein. Then the reaction mix was subject to 6% PAGE and autoradiography.
Construction of the mvfR-Glutathione S-Transferase (GST) Translational Fusion.
A ≈1.5-kb fragment containing both the promoter and the entire coding region (excluding the stop codon) of mvfR, a ≈0.7-kb fragment containing the entire coding region of the GST gene from Schistosoma japonicum, and a ≈0.6-kb fragment of the mvfR 3′ region downstream of the stop codon were cloned into HindIII/XbaI, XbaI/KpnI, and KpnI/EcoRI sites of pUCP19 vector (27), respectively, to obtain pUCP19/mvfR-GST. The pUCP19/mvfR-GST was then introduced into Pseudomonas strains by means of electroporation as described (28).
Cell Fractionation.
Bacterial strains were grown to the optical density as indicated, and cells were pelleted and fractionated as described (29). The additional fractionation of the membrane into inner and outer membranes was carried out by using sucrose gradient centrifugation at 100,000 × g (30). The separation of outer membrane from inner membrane was monitored by measuring the activities of NADH oxidase (inner membrane marker) in each membrane fraction (31).
Immunoblot Analysis.
Proteins of cell fractions were separated on 10% SDS-polyacrylamide gels and transferred to Immobilon-P membrane (Millipore) following the manufacturer's instructions. The proteins from different preps were prepared from the same number of cells. The immunoblot analyses were performed with anti-GST mAb following the manufacturer's instructions (Upstate Biotechnology, Lake Placid, NY).
Results
The Mutation in ORF2 Is Responsible for the Mutant Phenotype of phoA34B12.
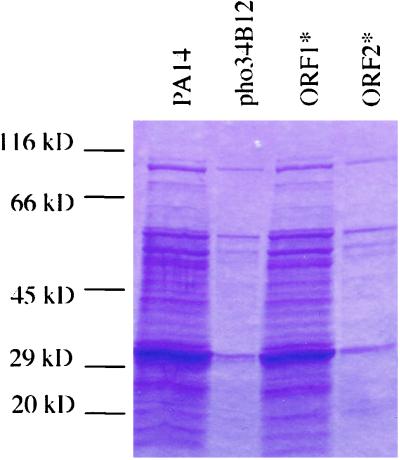
Previous studies identified two overlapping ORFs in opposite orientations in the pho34B12 locus (ref. 10; Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org). Northern blot analyses showed that both ORFs are transcribed (data not shown). Both ORF1 and ORF2 loci are also present in the genome of PAO1 strain, and ORF2 was identified with an ID number of PA1003. However, although ORF1 is transcribed, it is not predicted as an ORF in the annotated genome. To determine the role each ORF plays, a nonsense point mutation was introduced into each of them by PCR and marker-exchanged into the chromosome of the wild-type strain PA14 by homologous recombination. As described in Materials and Methods, a glutamine at position 262 of ORF1 and a glutamic acid at position 151 of ORF2 were replaced by a stop codon, respectively. Both mutants were tested for virulence in plants and animals, using an Arabidopsis leaf-infiltration assay (20) and a mouse thermal injury model (32). The phenotypes of both point mutants were then compared with those of the pho34B12 mutant and are summarized in Table 1. Mutation in ORF1 (ORF1*) does not affect the wild-type phenotype. In contrast, and as in mutant phoA34B12, the mutant ORF2* results in an attenuated virulence phenotype in both plants and animals, as demonstrated by about 320-fold less growth in Arabidopsis and 35% mortality in mice. In addition, as exhibited by pho34B12, ORF2* exhibits lack of pyocyanin production and decreased levels of elastase, phospholipase, and of various exoproteins secreted (Fig. 1). The importance of the ORF2 locus in the QS is further demonstrated in that the ORF2* mutation results in decreased levels of the P. aeruginosa QS signal molecules PAI I and PQS (Table 1). The mutant phenotypes of ORF2* can be complemented by a plasmid containing a wild-type copy of ORF2 (data not shown).
Table 1.
Phenotypic analysis of wild-type PA14, pho34B12 mutant, and two point mutants, ORF1* and ORF2*
| Strain | Growth in Arabidopsis, cfu per leaf disc† | Mortality in burn mouse model, 5 × 105 cfu‡ | Pyocyanin | Secreted proteins | Auto-inducers (PAI I and PQS) |
|---|---|---|---|---|---|
| PA14 | 1.6 × 107 | 100% | 100% | Normal | 100% |
| 34B12 | 6.8 × 105 | 60% | <10% | Reduced | ∼10% |
| ORF1* | 2.8 × 107 | 100% | 170% | Normal | 100% |
| ORF2* | 5.0 × 104 | 35% | <10% | Reduced | ∼10% |
cfu, colony-forming unit.
The numbers shown represent the bacterial growth four days post infection. Four different samples were taken by using two-leave discs per sample.
Eight to ten mice were used per experiment.
Figure 1.
Coomassie blue stain of exoproteins isolated from wild-type (lane 1) and mutant strains, pho34B12 (lane 2), ORF1* (lane 3), and ORF2* (lane 4). Extracellular proteins were isolated from equal amounts of wild-type and mutant bacterial cells grown in Luria–Bertani media (OD600 = 2.5–3.0), separated on a 4–20% gradient polyacrylamide gel, and stained with Coomassie brilliant blue R-250.
MvfR (ORF2) Functions Independently of QS Regulators lasR and rhlR.
Because the site-directed mutagenesis studies demonstrate the importance of the ORF2 locus (here after called mvfR, for multiple virulence factors regulator) in the pathogenesis of P. aeruginosa-induced infection, subsequent efforts were focused on it.
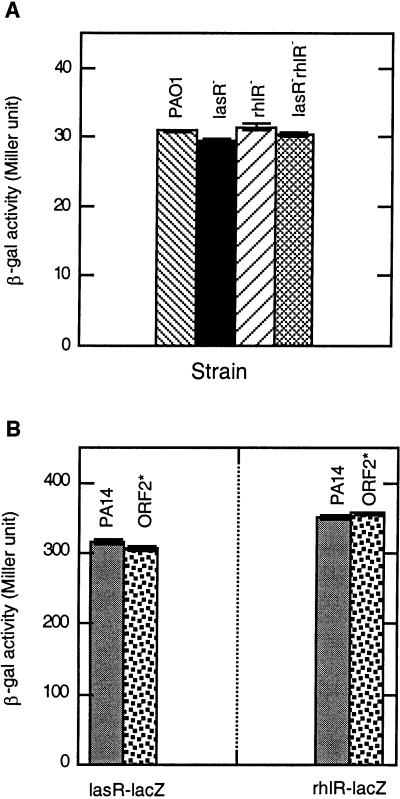
The phenotypic analysis of the ORF2* mutant indicates that the mvfR gene regulates the production of autoinducers PAI I and PQS, components of the QS cascade. Therefore, we attempted to determine the relationship between mvfR and the two known QS regulators, lasR and rhlR, of P. aeruginosa. By using β-galactosidase transcriptional fusions of the genes mvfR, lasR, and rhlR, we tested whether mvfR and the lasR and rhlR regulators control the expression of each other. First, the expression of the transcriptional fusion mvfR-lacZ in the wild-type P. aeruginosa strain PAO1, as well as in the PAO1 isogenic mutants lasR−, rhlR−, and lasR−rhlR− (22), was studied by measuring β-galactosidase activity. The β-galactosidase activity in the three mutant strains was not significantly different from that of the wild-type strain (Fig. 2A), indicating that the expression of mvfR is not controlled by either of the two QS regulators. Second, the expression of lasR-lacZ and rhlR-lacZ transcriptional fusions in PA14 and in the ORF2* mutant strain were essentially the same, indicating that mvfR does not regulate the expression of either of the two QS regulators (Fig. 2B). Both experiments indicate that although mvfR regulates QS-dependent components, at transcription level, it functions independently of the known regulators of the QS circuit.
Figure 2.
(A) The expression of the mvfR-lacZ transcriptional fusion in wild-type PAO1 and mutant strains lasR−, rhlR−, and lasR−rhlR−. Plasmid containing mvfR-lacZ transcriptional fusion was introduced into strains PAO1, lasR−, rhlR−, and lasR−rhlR−. β-galactosidase activity was measured in these strains at OD600 = 2.5–3.0. (B) The expression of lasR-lacZ and rhlR-lacZ in wild-type PA14 and mutant strain ORF2*. Plasmids containing either lasR-lacZ or rhlR-lacZ were introduced into wild-type PA14 and mutant ORF2*. β-galactosidase activity was measured in these strains at OD600 = 2.5–3.0.
mvfR Encodes a Transcriptional Regulator of the LysR Family That Positively Regulates the Expression of the phnAB Operon.
Because mvfR controls diverse pathogenicity functions, it is likely that mvfR may encode a regulator. Indeed, the predicted protein sequence of mvfR contains a helix–turn–helix (HTH) motif at the N terminus that bears strong similarity to the HTH signature motif belonging to the LysR family of transcription regulators (33). In addition, the blastp search shows that the first 280 amino acids of MvfR share significant amount of similarity (62–71%) to a few LysR-like transcriptional regulators (data not shown). Because mutation in mvfR leads to a deficiency in pyocyanin and mvfR is located next to phnAB (Fig. 6), it is likely that MvfR regulates the expression of the phnAB operon. Therefore, a plasmid containing phnAB-lacZ transcriptional fusion was constructed and introduced into both wild-type PA14 and the mutant strain ORF2*. The expression of lacZ was determined in the two strains by measuring their β-galactosidase activity. MvfR positively regulates the expression of the phnAB operon because the β-galactosidase activity of the phnAB-lacZ in the PA14 and ORF2* was 500 and 100 Miller units, respectively. Thus the β-galactosidase activity in the ORF2* strain is about 5-fold lower than in the wild type (Fig. 7, which is published as supporting information on the PNAS web site). RNA blot analysis of these two strains using the phnAB coding sequence as the nucleotide probe confirmed these results (data not shown).
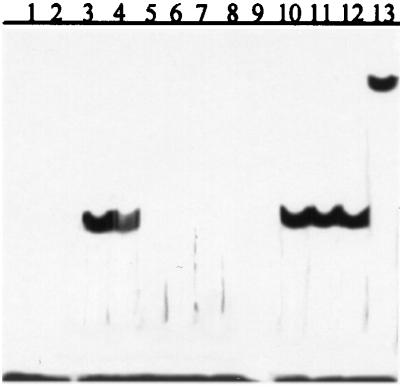
To determine whether the MvfR protein physically interacts with the promoter of the phnAB operon, a gel electrophoresis mobility-shift assay was performed in which a 51-bp fragment (P1) about 185-bp upstream of the start codon of phnA was used as a DNA probe. P1 contains a consensus sequence, found in the target genes' promoters with which the LysR-like transcription factors interact. Although the exact transcription start point in phnAB operon has not been identified, a putative −10-bp sequence was found about 120-bp upstream of the start codon of phnA. Another 51-bp fragment (P2), located at about 460-bp upstream of the start codon, was used for the competition-binding assay. As illustrated in Fig. 3, radio-labeled P1 was incubated in the absence (lane 1) and presence of MvfR protein purified from E. coli (lane 3). The appearance of a shifted band in lane 3 indicates that MvfR does bind to P1, and this binding is specific because it can be competed away by excess non-radio-labeled P1 (Fig. 3, lanes 4–8) but not by an excess amount of P2 (Fig. 3, lanes 10–12). That the shifted band (complex) contains MvfR protein is confirmed by the formation of a supershifted band after the addition of a polyclonal Ab raised against the C terminus of MvfR to the binding reaction (Fig. 3, lane 13).
Figure 3.
The MvfR protein binds specifically to the promoter of the phnAB operon. Lane 1, radio-labeled P1 (a 51-bp sequence 185-bp upstream of the start codon of phnAB operon) only; lane 2, blank; lanes 3–8, MvfR + radio-labeled P1 + x-fold cold P1. Lane 3, x = 0; lane 4, x = 10; lane 5, x = 20; lane 6, x = 40; lane 7, x = 80; lane 8, x = 160; lane 9, blank; lanes 10–12, MvfR + radio-labeled P1 + y-fold cold P2 (a 51-bp sequence 460-bp upstream of the start codon of phnA/B operon). Lane 10, y = 20; lane 11, y = 40; lane 12, y = 80; and lane 13, MvfR + radio-labeled P1 + anti-MvfR polyclonal Ab.
Cytoplasmic Membrane-Bound MvfR Protein Is Cleaved When Cells Reach Stationary Phase, and Modification of MvfR Protein Is Regulated by a Secreted Signal That Requires Functional MvfR.
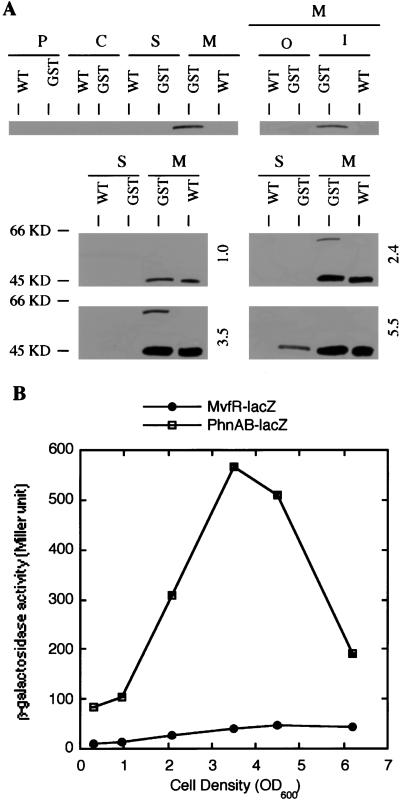
Previous studies showed that the phoA34B12 mutant has a positive alkaline phosphatase activity, indicating that the product of the pho34B12 gene is either membrane-spanning or secreted and that the phoA gene is in-frame with mvfR. A motif search on mvfR also identified a putative signal peptide at the extreme N terminus of MvfR, suggesting that MvfR is a secreted protein. However, this finding is inconsistent with a role for MvfR as a transcription factor that binds to DNA. Hence, localization of the MvfR protein was necessary. Immunoblot studies using polyclonal Ab against MvfR resulted in ambiguous signals. Therefore, a GST-MvfR translational fusion protein was generated in which the GST gene from S. japonicum was linked in-frame to the C terminus of MvfR. The ability of this fusion protein to represent the endogenous MvfR was confirmed in that the plasmid containing the fusion protein was able to complement the pyocyanin-deficient phenotype of the ORF2* strain. The mvfR-GST fusion was then introduced into the wild-type strain, and the transformant was designated PA14/GST. A mAb against the GST protein detected the fusion protein in the immunoblot assays. The control in all of the assays was the wild-type strain lacking the fusion protein. The expected size of the fusion protein band is about 64 kDa, the sum of MvfR (38 kDa as predicted) and GST (26 kDa). Immunoblot assays were performed on fractionated cell extracts from different bacterial growth stages (Fig. 4A). No fusion protein was detected when cells were in the early exponential growth phase (i.e., OD600 = 1.0); however, a protein band of about 64 kDa, unique to the PA14/GST strain, was detected in the membrane fraction, when cells reached the late exponential phase (i.e., OD600 = 2.4 and 3.5; Fig. 4A). The 64-kDa band was detected in the cytoplasmic membrane when the membrane fraction was further separated into cytoplasmic membrane and outer membrane. Interestingly, when cells grew to the stationary phase (i.e., OD600 = 5.5), the band of 64 kDa in the membrane fraction diminished and an additional band of about 48 kDa, unique to strain PA14/GST, was then detected in the supernatant (Fig. 4A). These data suggest that the expression of mvfR is cell density-dependent and peaks at the late exponential phase. The results also show that MvfR protein is cytoplasmic membrane-associated and that it is cleaved when cells reach the stationary phase.
Figure 4.
(A) Cellular localization of the MvfR protein at different growth phases. Plasmid containing the MvfR-GST translational fusion was introduced into PA14 strain. Protein extracts from cell fractionations of both PA14 and transformant strains grown to cell density as indicated were prepared, separated on a 10% polyacrylamide gel, and blotted onto Immobilon-P [poly(vinylidene difluoride) (PVDF)] membranes. A mAb against GST was used to detect the MvfR-GST fusion. The numbers to the right indicate the cell density (OD600). WT, wild-type PA14 strain; GST, PA14 strain containing MvfR-GST translational fusion; P, periplasmic; C, cytoplasmic; S, secreted; M, membrane; O, outer; I, inner. (B) The expression of mvfR and phnAB at different growth phases. β-galactosidase activities in PA14 strain containing either mvfR-lacZ or phnAB-lacZ transcriptional fusion were measured at the growth phases as indicated on the graph.
It was important to ascertain that the cleavage of MvfR represents its own activity regulation and is not a consequence of cell lysis at stationary phase or protein turnover. The lacZ expression of phnAB operon, which is regulated by MvfR, was determined in PA14 and compared with the expression and modification of the MvfR protein at different growth phases. The β-galactosidase activities of phnAB-lacZ transcriptional fusion were low when cells were at early exponential phase, they increased during cell growth, peaked at the late exponential phase, and started to decrease when cells grew into stationary phase (Fig. 4B). This pattern is consistent with the profile of the mvfR expression and its modification.
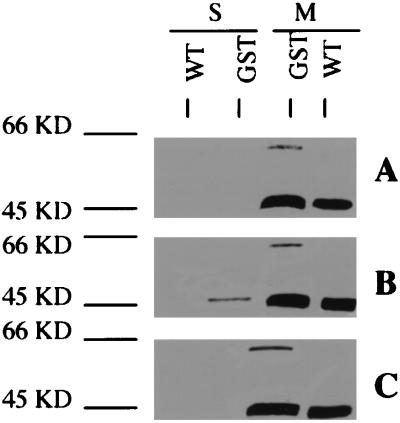
However, the correlation of the expression pattern of the phnAB operon with that of mvfR alone is not sufficient to distinguish between the options of activity regulation vs. cell lysis or protein turnover, because the former is a coordinated and regulated procedure and the latter two are usually not. To determine whether the modification of MvfR protein is regulated, we tested whether the modification of the MvfR protein can be triggered before cells reach the stationary phase. In this experiment, both PA14 and PA14/GST strains were grown to late exponential phase (OD600 = 2.5–3.0, when the expression of mvfR is maximal), and the cells were pelleted and resuspended in the cell-free supernatant from the PA14 strain grown to stationary phase (OD600 > 5.0). The cells were incubated at 37°C for 1 h before the fractionated cell extracts were prepared. Although the cells are at the late exponential phase, the band of 48 kDa was already detected in the supernatant of PA14/GST strain treated with the cell-free supernatant of PA14 from the stationary culture (Fig. 5B). In contrast, the 48-kDa band was absent from the supernatant of PA14/GST strain grown to the same stage in untreated Luria–Bertani media (Fig. 5A). The 48-kDa band was not also detected in extracellular fraction of PA14/GST cells treated with the mutant ORF2* supernatant, instead of the wild type (Fig. 5C).
Figure 5.
Translocation and cleavage of MvfR in response to extracellular signals from wild-type PA14 (B) and mutant strain ORF2* (C). Both PA14 strain (WT) and PA14 strain containing MvfR-GST translational fusion (GST) were grown in Luria–Bertani media until OD600 reached 2.5–3.0. Then the cells were harvested and treated for 1 h with cell-free cultures of PA14 and ORF2* strains grown to stationary phase (OD600 > 5.0). Protein preps from supernatant (s) and membrane (m) fractions of those treated (B and C) and untreated (A) cells were separated on a 10% polyacrylamide gel and blotted onto Immobilon-P [poly(vinylidene difluoride) (PVDF)] membranes. A mAb against GST was used to detect the MvfR-GST fusion.
The authenticity of the 48-kDa fragment visualized was confirmed further by sequencing of the first five N-terminal amino acid residues. These residues exactly matched the MvfR protein sequence amino acids from position 147 to 151. The predicted molecular weight of cleaved MvfR (147–344; 22 kDa) plus GST (26 kDa) is 48 kDa, matching the size of fragment we observed in the extracellular fraction. We also probed the cell fractionations of PA14/GST grown to late exponential (OD600 = 2.5–3.0) and stationary phases (OD600 > 5.0) with anti-secB (a cytoplasmic protein marker; A. Economou, personal communication) Ab in immunoblot assays. SecB was present only in the cytoplasmic fractions in both growth stages and was absent in the supernatant fractions (data not shown). We therefore conclude that the cleavage of MvfR protein is a regulated process rather than a consequence of cell lysis or protein turnover. Moreover, we show that the signal for the modification of MvfR protein is secreted and is controlled by the mvfR gene itself.
Discussion
In this article, we molecularly and biochemically characterized a P. aeruginosa virulence regulator, MvfR, that seems to be associated with QS and use a unique negative “autoregulation” mechanism.
We demonstrated that mvfR regulates the expression of the phnAB operon. Therefore, does the mvfR gene have other target genes and what are they? The mutation in mvfR affects the levels of a number of secreted proteins indicating that mvfR does have other target genes in addition to those involved in pyocyanin biosynthesis. It is possible that mvfR either directly regulates the expression of these secreted proteins or indirectly controls them by regulating some other transcription factors or proteins involved in secretion. The latter possibility is supported by the findings that mutation of mvfR causes reduction in production of one of the two major autoinducers in P. aeruginosa, PAI I and the autoinducer-like molecule, PQS (19). In P. aeruginosa, the lasI/lasR system has been shown to positively regulate the xcp operons, which encode for the apparatus proteins required for the second step of type II secretion (general secretion pathway)—the delivery of secreted proteins from the periplasmic space across the outer membrane (34). The reduction in PAI I as displayed by the mvfR mutant, therefore, would impair the function of the lasI/lasR system, result in a deficiency in Xcp proteins, and cause a pleiotropic defect in type II secretion. In addition to being a LysR-like transcription regulator, MvfR has been shown to be membrane-associated. Another example of a membrane-associated regulator is the nodD gene of Rhizobium leguminosarum, a member of the LysR family of transcription regulators (35). The MvfR protein does not contain any typical membrane-spanning sequence. Proteinase K treatment of the spheroplasts of PA14/GST and immunoblot analysis indicated that MvfR protein was resistant to proteinase K treatment (data not shown), implying that MvfR does not have any apparent periplasmic domains. Moreover, cell fractionation in the presence of 1 M NaCl did not remove MvfR from the membrane fraction (data not shown), which suggests that the association of MvfR protein with membrane is not an experimental artifact. Thus, the MvfR protein seems to be an amphipathic protein similar to the LuxR protein from Vibrio fischeri (36). The membrane-associated position of MvfR as a transcription factor may reflect its crucial role in translating environmental signals into basic cellular activities (i.e., gene expression).
Interestingly, MvfR negatively autoregulates its function through translocation and modification. In prokaryotes, the examples of regulation of transcription factors by both the changes in cellular localization and modification of the molecules have been reported for the sporulation-specific transcription factors σE and σK of Bacillus subtilis (37, 38). However, the regulatory mechanism for MvfR is unprecedented because σE and σK require the cleavage of their signal peptides followed by the release of the proteins from the membrane into cytoplasm. We show that the MvfR protein is delivered from cytoplasmic membrane to the extracellular space (rather than into cytoplasm), accompanied by the cleavage of the molecule. The cleavage results in inactivation of the MvfR protein because the transcription level of phnAB significantly decreases when MvfR is secreted and cleaved, whereas cleavage and translocation of σE and σK leads to activation of the molecules. Interestingly, the sequence analysis of mvfR has identified a putative signal peptide about 24-aa long at the N terminus of the predicted amino acid sequence. Although the phoA+ phenotype of pho34B12 mutant suggests that this signal peptide is bona fide, our results demonstrate that the cleavage of MvfR at the stationary phase occurs in the middle of the molecule rather than at the predicted cleavage site of the signal peptide. Whether the cleavage of the protein occurs before or after the secretion of MvfR and whether this signal peptide plays a role during the function and down-regulation of MvfR remains to be elucidated.
In addition to the expression of phnAB, we also examined the effect of MvfR modification on pyocyanin production by adding concentrated cell-free supernatant from both wild-type PA14 and ORF2* mutants, grown to stationary phase, to freshly inoculated culture of strain PA14/GST. We found that the supernatants from both PA14 and ORF2* were able to inhibit the production of pyocyanin by PA14/GST (data not shown). Because mvfR positively regulates pyocyanin production, this result is also in support of a modification of MvfR protein leading to inactivation or down-regulation of MvfR. However, the inhibitory effect of ORF2* supernatant is somewhat surprising, because mvfR controls the signal(s) for its own modification. One possibility is that ORF2* mutant strain may still produce some signals for the modification of MvfR, although at a much lower level than wild type but sufficient to modify MvfR. Alternatively, the presence of other factor(s) in the supernatant, independent of mvfR, which also could trigger the inhibition of pyocyanin production is possible. More experiments are necessary to distinguish between these possibilities.
MvfR protein belongs to the LysR family of transcription regulators, in which most members negatively regulate themselves (33). MvfR protein, however, does not regulate its own transcription because the expression of mvfR is not affected in the ORF2* mutant (data not shown). Because the translocation and modification of MvfR lead to inactivation of the protein as a transcription activator and depend on signal(s) controlled by mvfR, it could be considered as a novel means of autoregulation for LysR-like transcription regulators that involves negative feedback.
It seems that the regulation of mvfR not only includes a posttranslational negative autoregulation but is also cell-density dependent, because its expression increases as cell density rises. The expression studies with QS regulators lasR, rhlR, and gacA indicate that at the transcription level, mvfR operates independently of these regulators and vice versa, even though they all play a role in the regulation of pyocyanin production and other QS-dependent components. However, it is possible that mvfR may interact at posttranscriptional level with these QS regulators. In addition, because mvfR positively regulates the production of the PAI I autoinducer, it is possible that mvfR activates the expression of the lasI gene that catalyzes biosynthesis of PAI I.
On the basis of our evidence, we propose a working model of how MvfR may function as a transcription regulator at different growth phases. The level of MvfR protein is very low at early exponential growth phase and increases to reach a maximum at late exponential phase. At these growth phases, MvfR is associated with the cytoplasmic membrane and functions as a transcription factor that positively regulates the expression of QS-related virulence factors and a number of secreted proteins. At the stationary phase, secreted product(s) regulated by mvfR itself reach(es) a critical level and signal(s) the down-regulation of mvfR. Consequently, the MvfR protein is secreted into the supernatant with cleavage. The complicated regulation of mvfR and its role in regulation of diverse cellular activities indicate that mvfR is not only an important regulator of pathogenesis alone, but is also a key component in the crosstalk of different signal transduction pathways in P. aeruginosa in response to various developmental and environmental cues. The importance of mvfR as a regulator of pathogenesis is emphasized further by the fact that proper functioning of mvfR is required for the virulence of P. aeruginosa in multiple host systems, including plants (10), nematodes (15), the fruit fly Drosophila (G. Lau and L.G.R., unpublished data), and mouse (10). Therefore, mvfR should be considered as a fundamentally essential virulence gene that plays an indispensable role in the cell density-dependent regulatory circuit and pathogenesis of P. aeruginosa.
Supplementary Material
Acknowledgments
We thank Barbara Iglewski (Univ. of Rochester, Rochester, NY) for lasR-lacZ and rhlR-lacZ transcriptional fusions as well as lasR−, rhlR−, and lasR−rhlR− mutant strains and Anastasios Economou (Univ. of Crete, Crete, Greece) for anti-SecB Ab. This work was supported by Cystic Fibrosis Foundation Research Award 99G0 (to L.G.R.). H.C. and B.G. were supported by the Damon Runyon–Walter Winchell Cancer Research Foundation and by a Shriners Research Fellowship, respectively.
Abbreviations
- QS
quorum-sensing
- GST
glutathione S-transferase
- PAI I
3oxo-dodecanoyl homoserine lactone
- PQS
2-heptyl-3-hydroxy-4-quinolone
References
- 1.Hancock R E W. Clin Infect Dis. 1998;1:S93–S99. doi: 10.1086/514909. [DOI] [PubMed] [Google Scholar]
- 2.Hancock R E W, Woodruff W A. Rev Infect Dis. 1988;10:770–775. doi: 10.1093/clinids/10.4.770. [DOI] [PubMed] [Google Scholar]
- 3.Philippon L N, Naas T, Bouthers A T, Barakett V, Nordmann P. Antimicrob Agents Chemother. 1997;41:2188–2195. doi: 10.1128/aac.41.10.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechere J C. C Antimicrob Agents Chemother. 1997;41:2540–2543. doi: 10.1128/aac.41.11.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poole K, Krebes K, McNally C, Neshat S. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poole K, Gotoh N, Tsujimoto H, Zaho Q, Wada A, Yamasaki T, Neshat S, Yamagishi J, Nishino T. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 7.Jarvis W R, Martone W J. J Antimicrob Chemother. 1992;29:S19–S24. doi: 10.1093/jac/29.suppl_a.19. [DOI] [PubMed] [Google Scholar]
- 8.Rahme L G, Ausubel F M, Cao H, Drenkard E, Goumnerov B C, Lau G W, Mahajan-Miklos S, Plotnikova J, Tan M-W, Tsongalis J, et al. Proc Natl Acad Sci USA. 2000;97:8815–8821. doi: 10.1073/pnas.97.16.8815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao H, Baldini R, Rahme L G. Annu Rev Phytopathol. 2001;39:259–284. doi: 10.1146/annurev.phyto.39.1.259. [DOI] [PubMed] [Google Scholar]
- 10.Rahme L G, Tan M-W, Le L, Wong S M, Tompkins R G, Calderwood S B, Ausubel F M. Proc Natl Acad Sci USA. 1997;94:13245–13250. doi: 10.1073/pnas.94.24.13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson R, Sykes D A, Watson D, Rutman A, Taylor G W, Cole P J. Infect Immun. 1988;56:2515–2517. doi: 10.1128/iai.56.9.2515-2517.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Essar D W, Eberly L, Hadero A, Crawford I P. J Bacteriol. 1990;172:884–900. doi: 10.1128/jb.172.2.884-900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mavrodi D V, Ksenzenko V N, Bonsall R F, Cook R J, Boronin A M, Thomashow L S. J Bacteriol. 1998;180:2541–2548. doi: 10.1128/jb.180.9.2541-2548.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chugani S, Whiteley M, Lee K, D'Argenio D, Manoil C, Greenberg E. Proc Natl Acad Sci USA. 2001;98:2752–2757. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahajan-Miklos S, Tan M-W, Rahme L G, Ausubel F M. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 16.Wilson R, Pitt T, Taylor G, Watson D, MacDermot J, Sykes D, Roberts D, Cole P. J Clin Invest. 1987;79:221–229. doi: 10.1172/JCI112787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulmer A, Pryjma J, Tarnok Z, Ernst M, Flad H. Infect Immun. 1990;58:808–815. doi: 10.1128/iai.58.3.808-815.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pearson J P, Gray K M, Passador L, Tucker K D, Eberhard A, Iglewski B H, Greenberg E P. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pesci E C, Milbank J B J, Pearson J P, McKnight S, Kende A S, Greenberg E P, Iglewski B H. Proc Natl Acad Sci USA. 1999;96:11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rahme L G, Stevens E J, Wolfort S F, Shao J, Tompkins R G, Ausubel F M. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 21.Holloway B W. J Gen Microbiol. 1955;13:572–581. doi: 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
- 22.Pearson J P, Pesci E C, Iglewski B H. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donnenberg M S, Kaper J B. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eberhard A, Burlingame A, Eberhard C, Kenyon G, Nealson K, Oppenheimer N. Biochemistry. 1981;20:2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- 25.Albus A M, Pesci E C, Runyen-Janecky L J, West S E, Iglewski B H. J Bacteriol. 1997;179:3928–3935. doi: 10.1128/jb.179.12.3928-3935.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 27.West S E, Schweizer H P, Dall C, Sample A K, Runyen-Janecky L J. Gene. 1994;148:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 28.Bloemberg G V, O'Toole G A, Lugtenberg B J, Kolter R. Microbiology. 1997;63:4543–4551. doi: 10.1128/aem.63.11.4543-4551.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas A W, Topping A W, Slater J H, Weightman A J. J Bacteriol. 1992;174:1941–1947. doi: 10.1128/jb.174.6.1941-1947.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hancock R E W, Nikaido H. J Bacteriol. 1978;136:381–390. doi: 10.1128/jb.136.1.381-390.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osborn M J, Gander J E, Parisi E, Carson J. J Biol Chem. 1972;247:3962–3972. [PubMed] [Google Scholar]
- 32.Stevens E J, Ryan C M, Friedberg J S, Barnhill R L, Yarmush M L, Tompkins R G. J Burn Care Rehabil. 1994;15:232–235. doi: 10.1097/00004630-199405000-00005. [DOI] [PubMed] [Google Scholar]
- 33.Schell M A. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 34.Lazdunski A, Filloux A, Michel G, Foglino M, Murgier M, Latifi A, Chapon V, Bleves S, Bally M. In: Molecular Biology of Pseudomonads. Nakazawa T, Furukawa K, Haas D, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 427–437. [Google Scholar]
- 35.Schlaman H R M, Spaink H P, Okker R J H, Lugtenberg B J J. J Bacteriol. 1989;171:4686–4693. doi: 10.1128/jb.171.9.4686-4693.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolibachuk D, Greenberg E P. J Bacteriol. 1993;175:7307–7312. doi: 10.1128/jb.175.22.7307-7312.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hofmeister A E M, Londono-Vallejo A, Harry E, Stragier P, Losick R. Cell. 1995;83:219–226. doi: 10.1016/0092-8674(95)90163-9. [DOI] [PubMed] [Google Scholar]
- 38.Hofmeister A E M. J Bacteriol. 1998;180:2426–2433. doi: 10.1128/jb.180.9.2426-2433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.