Abstract
About 10% of male infertile patients show abnormalities in spermatogenesis. The microdeletion of azoospermia factor a (AZFa) region of the Y chromosome is thought to be a cause of spermatogenic failure. However, candidate gene responsible for the spermatogenic failure in AZFa deleted patients has not been elucidated yet. Using mice, we explored the function of Ddx3y, a strong candidate gene in the Azfa region, and Ddx3x, a Ddx3y paralog on the X chromosome, in spermatogenesis. We first generated Ddx3y KO male mice using CRISPR/Cas9 and found that the Ddx3y KO male mice show normal spermatogenesis, produce morphologically normal spermatozoa, and sire healthy offspring. Because Ddx3x KO males were embryonic lethal, we next generated chimeric mice, which contain Ddx3x and Ddx3y double KO (dKO) germ cells, and found that the dKO germ cells can differentiate into spermatozoa and transmit their mutant alleles to offspring by normal mating. We conclude that Ddx3x and Ddx3y are dispensable for spermatogenesis at least in mice. Unlike human, mice have an additional Ddx3y paralog D1pas1, that has been reported to be essential for spermatogenesis. These findings suggest that human and mouse DDX3 related proteins have distinct differences in their functions.
Keywords: Azoospermia factor region, Chimeric analysis, CRISPR/Cas9, Male infertility, Y chromosome
Recent findings have observed infertility occurring among in one in six pairs of couples in developed countries, with half of the infertility due to male factors [1]. About 10% of male infertile patients are categorized as non-obstructive azoospermia (NOA), which is defined as no sperm in the ejaculate due to failure of spermatogenesis [2]. Artificial reproductive technologies, such as ICSI and ROSI, can be applicable even if a few spermatozoa or haploid spermatids are observed in the testis [3, 4]. When spermatogenesis is arrested at the meiotic stage, or when only the somatic Sertoli cells (but not germ cells) are present in the seminiferous tubules of testes (Sertoli cell-only syndrome: SCOS), an effective treatment is limited. Less than 3% of infertile patients who suffer from meiotic arrest or SCOS can be explained by abnormal hormone secretion (hypogonadotropic hypogonadism), such as low FSH and/or LH, and can be treated by hormonal therapy to promote sperm production [2, 5]. The other molecular mechanisms behind NOA are still unclear.
A microdeletion of the Y chromosome is one of the major genetic factors of NOA. In 1976, Tiepolo and Zuffardi examined chromosomes of six NOA patients and found a novel deletion in the long arm of the Y chromosome (Yq11) [6]. The mutation was believed to be de novo as it was not found in their farther or brothers. In 1996, Vogt et al. analyzed Yq11 of 370 male infertile patients and found that 12 of them had different microdeletions [7]. These microdeletions are classified into three subgroups, named Azoospermia Factor (AZF) regions (AZFa, AZFb, and AZFc). The deletion of the AZFa region is associated with SCOS, which is the severest symptom [7, 8]. However, a candidate gene responsible for spermatogenic failure in AZFa deleted patients has not been elucidated yet.
The human AZFa region includes three genes, USP9Y, DDX3Y, and UTY [9,10,11,12]. Like in human, the mouse orthologous genes (Usp9y, Ddx3y, and Uty) locate close to each other, in the short arm of the Y chromosome [13]. In 2006 and 2009, deletion of USP9Y gene has been reported in men whose fathers also contained the deletion, suggesting that USP9Y is not the causative gene [14, 15]. In addition, the Uty knockout (KO) mice generated by TALEN-based genome editing [16, 17] did not show any problems with fertility [18]. Therefore, DDX3Y (Ddx3y in mice) is thought to be the candidate factor in AZFa deletion induced spermatogenic failure [19]. Indeed, DDX3Y has a higher mutation rate in SCOS patients than the other two genes in the AZFa region [20], and DDX3Y protein localization is restricted only in testes, specifically in spermatogonia and early spermatocytes, albeit with low mRNA expression of DDX3Y in various organs [21].
DDX3Y has a paralog on the X chromosome, DDX3X, and their homology at the amino acid and nucleotide sequence are 91% and 88%, respectively. In mice, homologies between Ddx3y and Ddx3x in amino acid sequences and nucleotides are 90% and 84%, respectively. DDX3X protein is expressed in the brain, kidney, ovary, and testis [21]. Interestingly, it is suggested that mouse DDX3X protein is localized in germ cells of the testes, including spermatogonia [22]. At present, functions of DDX3Y and DDX3X in spermatogenesis remains unclear.
In this study, we analyzed the functions of Ddx3y and Ddx3x in spermatogenesis, using mice. We first generated Ddx3y KO male mice by CRISPR/Cas9 [23, 24] and analyzed spermatogenesis. Because Ddx3x KO males show embryonic lethality, we next generated chimeric mice, which contain spermatogenic cells derived from ES cells mutated for both Ddx3x and Ddx3y, and analyzed spermatogenesis in the double KO (dKO) ES chimeric mice.
Materials and Methods
Animals
All animal experiments were conducted in accordance with the guidelines of “Animal experiment rules” established by the Research Institute for Microbial Diseases, Osaka University, and were approved by the Animal Care and Use Committee of the Research Institute for Microbial Diseases, Osaka University (#Biken-AP-H25-02). B6D2F1, ICR, and C57BL/6NCr mice were purchased from CLEA (Tokyo, Japan) or SLC (Shizuoka, Japan).
Plasmid construction and genotyping
Construction of sgRNA/CAS9 expressing plasmids, pX330 (#42230, Addgene, Cambridge, MA, USA), were performed by ligating oligos into the BbsI site of each plasmid as described previously [25, 26]. Potential off-target sites were searched using Bowtie software (http://bowtie-bio.sourceforge.net/index.shtml) as described previously [25]. We chose gRNAs whose 14 bases at the 3’ end, plus the NGG site, only matched the target site. The sgRNA target sequences for generating Ddx3y KO mice were 5’- GTCTGTGATAAGGACAGTTC -3’ (sgRNA #1) and 5’- TATTTCAGTGATCGTGGAAG -3’ (sgRNA#2). The sgRNA target sequence for generating Ddx3x KO mice was 5’- GTGGCAGTGGAAAATGCGCT -3’. The following primer sets were used for genotyping PCR of Ddx3y: Forward primer 5’- CATGCCCTCATCTCAATATCCCATAAGGT -3’ and Reverse primer 5’- GGATAGCCATTGTTGGACTAGTTGGACA -3’, The following primer sets were used for genotyping PCR of Ddx3x: Forward primer 5’- CCAAGGCTTCTTTATGAGCCGACG -3’ and Reverse primer 5’- CCACCTGCGGGTCACTAATCAAC -3’.
Pronuclear injection of mouse fertilized eggs
B6D2F1 superovulated females were mated with B6D2F1 males, then fertilized eggs were collected 20 h after hCG. Circular pX330 plasmids were injected into one of the pronuclei at 5 ng/μl [25, 26]. The eggs were cultured in KSOM overnight [27], and the two-cell stage embryos were transferred into the oviducts of pseudopregnant ICR females.
Genome editing in mouse ES cells and generation of chimeric mice
The EGR-G01 ES cells [28] (1 × 103–4) were seeded on mouse embryonic fibroblasts (MEF) in a 6-well plate and transfected with pX330 (total 1.0 µg) using Lipofectamine LTX & PLUS technology (Thermo Fisher Scientific, MA, USA). A pPGK-puro plasmid (0.1 µg) was co-transfected. After 14–18 h, the cells were selected with puromycin (0.1 µg/ml) for 48 h, then grown for 5 to 6 more days, picked, and transferred onto MEF cells in 96-well plates. After 48–72 h of culture, each ES cell clone was split in duplicate, for freezing, and DNA harvesting. After PCR amplification and direct sequencing, the positive clones were thawed and expanded to analyze their karyotypes. The mutant ES cell clones with normal karyotypes were injected into 8-cell ICR embryos, and the chimeric blastocysts were transplanted into the uteri of 2.5 dpc pseudopregnant females [29].
cDNA synthesis and RT-PCR
Testes were collected from C57BL/6NCr and Ddx3y KO male mice. These samples were homogenized in TRIzol (Thermo Fisher Scientific). The total RNA was reverse-transcribed to cDNA using SuperScript III First Strand Synthesis System for RT-PCR (Thermo Fisher Scientific). Five ng of cDNA was used for PCR with primer sets and KOD DNA Polymerase (KOD-Fx Neo, TOYOBO, Osaka, Japan). The following primer sets were used for detecting Ddx3y: Forward primer 5’- ATGAGTCAAGTGGCAGCGG -3’ and Reverse primer 5’- TCAATTGCCCCACCAGTCAACTGCC -3’.
Prediction of domain structure
The predicted amino acid sequences of wild-type DDX3Y and DDX3X, or mutant DDX3Y and DDX3X were submitted to SMART web tool (http://smart.embl-heidelberg.de/).
Morphology analysis of spermatozoa
The spermatozoa collected from cauda epididymis of male mice were incubated in TYH medium [30] for 10 min and dispersed in PBS.
Morphology analysis of embryos
Ddx3x heterozygous females were mated with Ddx3y KO males. Mouse embryos were collected at embryonic day 10.5 (E10.5), with the visualization of the copulatory plug considered to be E0.5. Genotype of Ddx3x was verified by PCR and DNA sequencing. To determine the sex of embryos, primers amplifying Uba1 (ubiquitin-like modifier activating enzyme 1) (X chromosome) and Uba1y (ubiquitin-like modifier activating enzyme 1) (Y chromosome) were used. The size of the PCR product for Uba1 or Uba1y is 211-bp or 183-bp, respectively. The primers used were 5’-TGGTCTGGACCCAAACGCTGTCCACA -3’and 5’- GGCAGCAGCCATCACATAATCCAGATG -3’.
Mating test
Male mice were caged with 2 B6D2F1 females for 3 months. The number of delivered pups was counted. Frozen spermatozoa from Ddx3y disrupted males (X/YDdx3y-em2) will be available through RIKEN BRC (http://en.brc.riken.jp/index.shtml) and CARD R-BASE (http://cardb.cc.kumamoto-u.ac.jp/transgenic/). The stock ID number of Ddx3y KO mouse strain is 09785 (Riken BRC) or 2432 (CARD), respectively.
Histological analysis of testis
After mating test, males were sacrificed by cervical dislocation following anesthesia. Testes were fixed in 4% paraformaldehyde in PBS and were processed for plastic sectioning using Technovit® 8100 (Mitsui chemicals, Tokyo, Japan) according to the manufacturer's instruction. Briefly, fixed testes were washed in PBS at 4ºC for an hour, dehydrated in acetone at 4ºC for an hour, infiltrated with in mixed solution of Technovit 8100 basic solution and hardener 1 (1.5 ml of basic solution plus 9 mg of hardener 1 per sample) at 4ºC for 2–6 h, and then embedded after adding 50 µl of hardener 2. For analysis of Ddx3y KO mouse testes, 5 µm sections were treated with 1% periodic acid for 10 min, followed by treatment with Schiff's reagent (Wako, Osaka, Japan) for 20 min. The sections were stained with Mayer's hematoxylin solution prior to imaging, and observed under a microscope. For analysis of EGFP-labeled spermatozoa in Ddx3x and Ddx3y dKO ES chimeric mouse testes, 5 µm sections were stained with 65 μM Hoechst 33342 (Life Technologies) in PBS for 5 min, and observed under a fluorescence microscope.
Statistical analysis
All values are shown as the mean ± SD of at least three independent experiments. Statistical analyses were performed using Student’s t-test for comparing the number of pups (litter size) derived from wild-type females mated with wild-type or Ddx3y KO males, and using Tukey HSD test for comparing litter size derived from Ddx3x heterozygous females mated with Ddx3y KO males.
Results
Generation of Ddx3y deficient mice
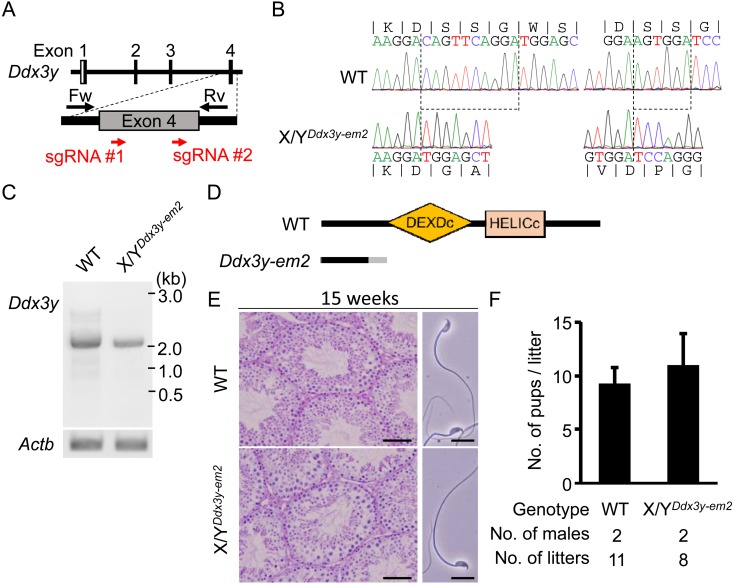
In order to generate Ddx3y KO mice, we performed genome editing by pronuclear injection of fertilized eggs using the CRISPR/Cas9 system. Mouse Ddx3y consists of 17 exons. We designed 2 single guide RNAs (sgRNAs) targeting the fourth exon of Ddx3y (#1 and #2) (Fig. 1A). We perfomed pronuclear injection of a combination of pX330s expressing human codon-optimized Cas9 and sgRNA #1 or #2. Mutations of Ddx3y in founder mice (F0) were confirmed by PCR and sequencing. From 94 injected eggs, 2 out of 5 male offspring (40%) had mutations in Ddx3y. One male had two small indels, 10-bp and 6-bp at #1 and #2 target sites, respectively, that resulted in the frame shift (X/YDdx3y- em2) (Fig. 1B). The 8-week-old X/YDdx3y-em2 founder male was mated with 2 of wild-type BDF1 females, and produced male offspring (F1) to establish the Ddx3y mutant line for subsequent analysis.
Fig. 1.
Ddx3y deficient male mice have normal spermatozoa and sire offspring. (A) Design of sgRNAs for generating Ddx3y KO mice. Red arrows indicate the location of sgRNAs. Black arrows indicate primers used for genotyping. White and gray boxes show untranslated region and coding sequences, respectively. (B) Genomic sequence of wild-type and Ddx3y KO (YDdx3y-em2) alleles; 10-bp and 6-bp are deleted (dashed lines). (C) RT-PCR of full length transcripts of Ddx3y from wild-type and Ddx3y KO testis cDNA. Actb (actin beta) was used as control. (D) Predicted protein product of the wild-type and Ddx3y KO allele. Orange and pink boxes represent the DEXDc and HELICc RNA-helicase domains respectively. The gray region in Ddx3y KO indicates amino acid sequence differing from wild-type. (E) Testicular sections (Left) and sperm morphology (Right) of 15-week-old wild-type and Ddx3y KO mice. Testicular sections were stained with hematoxylin and PAS. Scale bar: 50 µm (Left). Sperm morphology of wild-type and Ddx3y KO. Scale bars: 20 µm (Right). (F) Average litter size derived from wild-type and Ddx3y KO males. Error bars represent standard deviation (SD).
To predict whether any potential DDX3Y protein generated in the mutant is functional, we next amplified the entire ORF region of the mutated Ddx3y by RT-PCR, sequenced the cDNA, and examined potential reading frames. As expected, the Ddx3y band from cDNA of X/YDdx3y-em2 mice showed a similar size to that in wild-type mice (Fig. 1C), without any other detectable bands. We confirmed the cDNA from the mutant sequence showed a frameshift mutation with a stop codon before the RNA helicase domain (DEXDc) (Fig. 1D and Supplementary Fig. 1: online only), suggesting that only non-functional protein without an RNA helicase domain can be translated. Therefore, X/YDdx3y- em2 males were considered KO mice.
Ddx3y deficient male mice have normal spermatozoa and sire offspring
There is a possibility that the founder Ddx3y KO mice (F0) generated by pronuclear injection were genotypically mosaic [31]. Thus, we used the male offspring (F1) for phenotypic analysis. To analyze spermatogenesis in Ddx3y KO mice (F1), the testis histology was observed by Hematoxylin-PAS staining. Ddx3y KO males showed no abnormality in spermatogenesis at 15 weeks of age (Fig. 1E left), suggesting that Ddx3y is not required for spermatogenesis or the maintenance of spermatogonial stem cells. We next collected spermatozoa from cauda epididymis of Ddx3y KO mice and found that the KO spermatozoa showed normal morphology (Fig. 1E right). Indeed, the litter size derived from Ddx3y KO males was comparable to that from WT males (Fig. 1F).
Ddx3x is required for development
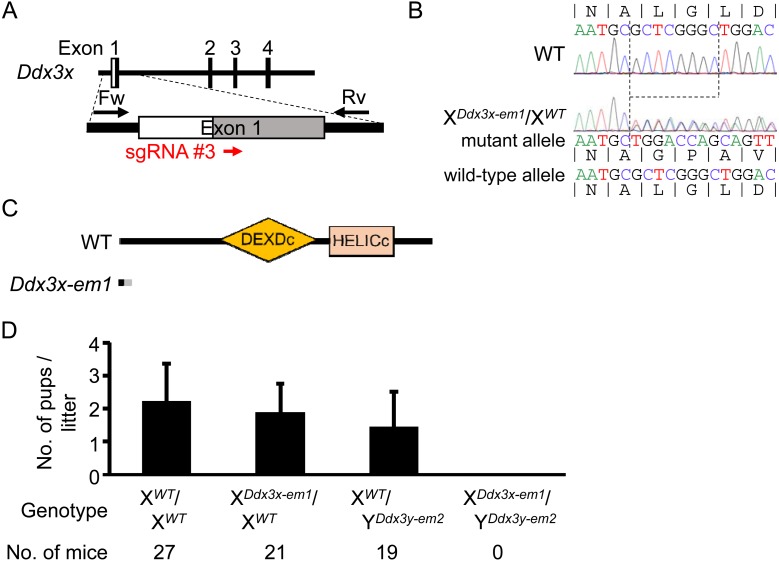
Mouse Ddx3x also consists of 17 exons. We designed sgRNA #3 targeting the first exon of Ddx3x (Fig. 2A) and performed pronuclear injection of pX330s targeting Ddx3y (#2) and Ddx3x (#3) simultaneously. From 119 injected eggs, 2 out of 24 offspring (8.3%), a male and a female, had a mutation in Ddx3x. The male had an in-frame mutation (12-bp deletion) in Ddx3x and no mutation in Ddx3y (XDdx3x-em1/YWT). The female had a frame shift mutation (8-bp deletion) in one allele of Ddx3x (XDdx3x- em1/XWT), and produced female offspring (F1) with the same mutant allele (Fig. 2B). Sequencing of the allele identified a premature stop codon before the DEXDc sequences (Fig. 2C and Supplementary Fig. 2A: online only), suggesting that only a non-functional protein without an RNA helicase domain can be translated. The F1 XDdx3x- em1/XWT females were mated with XWT/YDdx3y- em2 males, but dKO males (XDdx3x-em1/YDdx3y- em2) were never obtained (Fig. 2D), suggesting the importance of Ddx3x in mouse embryonic development. To determine whether Ddx3x is important for male embryonic development, we analyzed embryos at E10.5 and found only one dKO male out of 34 embryos (2.9%), which was lower than the Mendelian ratio (25.0%), and the dKO male was also smaller than wild-type embryos (Supplementary Fig. 2B and 2C). These results are consistent with a recent report showing that Ddx3x KO males exhibit embryonic lethality because of abnormal embryogenesis and placental dysfunction [32].
Fig. 2.
Ddx3x and Ddx3y double KO mice are embryonic lethal. (A) Design of the sgRNA for generating Ddx3x KO mice. Red arrows indicate the location of the sgRNA. Black arrows indicate primers for genotyping. White and gray boxes show untranslated region and coding sequences, respectively. (B) Genomic sequence of wild-type (XWT) and Ddx3x KO (XDdx3x-em1) alleles; 8-bp is deleted (dashed lines). (C) Predicted protein product of the wild-type and Ddx3x KO allele. Orange and pink boxes represent the DEXDc and HELICc RNA-helicase domains respectively. The gray region in Ddx3x KO indicates amino acid sequence differing from wild-type. (D) Average litter size delivered from Ddx3x heterozygous mutant females mated with Ddx3y hemizygous mutant males. Error bars represent SD.
Establishing Ddx3x and Ddx3y double KO ES cells using CRISPR/Cas9
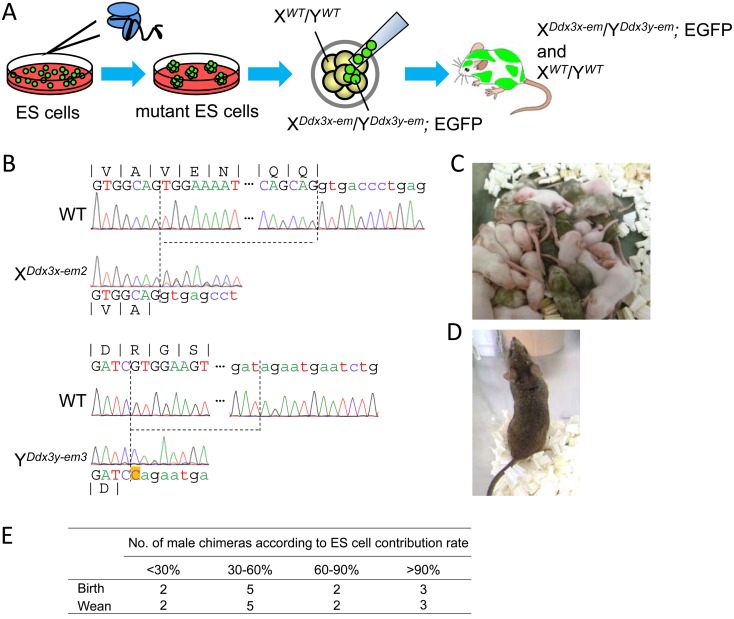
In order to bypass the embryonic lethality of Ddx3x KO males, we utilized chimeric analysis (Fig. 3A). In 2016, we reported that chimeric mice derived from ES cells carrying a biallelic mutation in a lethal gene can overcome embryonic lethality due to the presence of wild-type cells [29]. This chimeric approach is a useful tool to analyze the function of lethal genes in spermatogenesis [29, 33]. We first transfected ES cells with pX330s, targeting Ddx3x (#3) and Ddx3y (#2) (Fig. 1A and 2A), into EGR-G01 ES cells [28]. The EGR-G01 ES cells were used as they ubiquitously express EGFP in the cytoplasm of all cell types and the acrosome of spermatozoa [28]. All ES cell clones (8/8) had mutations in both Ddx3x and Ddx3y. To generate chimeric mice, we used one ES cell line, which had frame shift mutations in both Ddx3x (29-bp deletion) and Ddx3y (125-bp deletion and 1-bp insertion) (XDdx3x-em2/YDdx3y- em3) (Fig. 3B). The obtained chimeric male mice were all viable (13/13) even though ES cell contribution rate, judged by coat color, was 90% or higher (Fig. 3C–E). We decided to use the mice showing high contribution rate (> 90%) for the analysis of spermatogenesis.
Fig. 3.
Establishment of the Ddx3x and Ddx3y double KO ES cell clones using CRISPR/Cas9 system. (A) Schematic of chimeric male mice generation containing Ddx3x and Ddx3y double KO cells (EGFP positive). The image is adapted from ref. [29]. (B) (Upper region) Genomic sequence of wild-type and Ddx3x KO (XDdx3x-em2) alleles; 29-bp is deleted (dashed lines). (Lower region) Genomic sequences of wild-type and Ddx3y KO (YDdx3y-em3) alleles; 125-bp is deleted (dashed lines). Orange box in Ddx3y-em3 allele indicates 1-bp (cytosine) insertion. Exons are in upper case letters and introns in lower case. (C) Male and female chimeric pups and ICR pups delivered from foster mothers. ES cell contribution is judged by coat color of mice: agouti (ES cells) and white (ICR). (D) Adult male chimera with more than 90% of coat color derived from ES cells. (E) Viability of male chimeras from various ES cell contribution rates. All 13 chimeras are viable after 4 weeks of age (wean) with a high contribution of ES cells.
Ddx3x and Ddx3y double KO germ cells in chimeric males differentiate into spermatozoa and transmit their mutant alleles to offspring
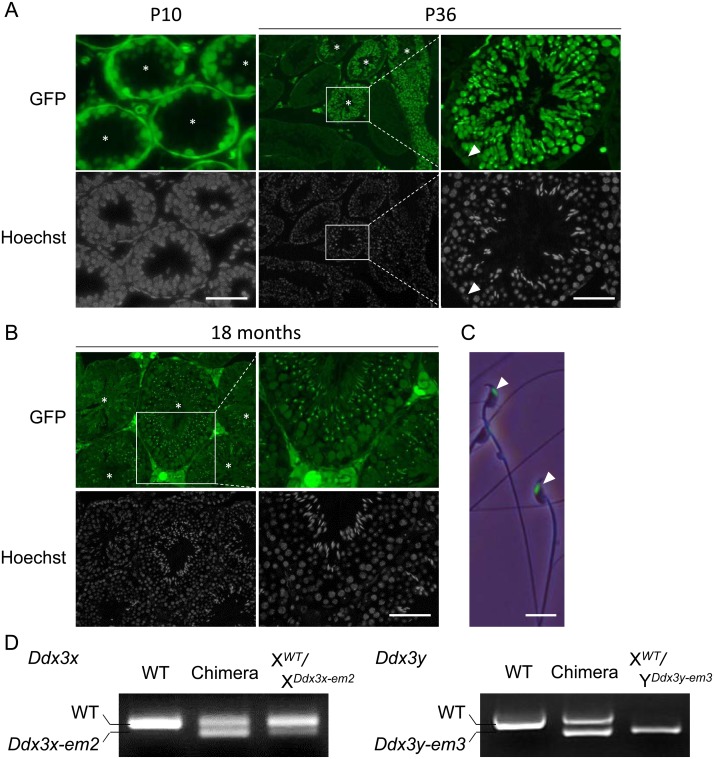
To confirm the contribution of Ddx3x and Ddx3y dKO ES cells to the germ lineage, we generated chimeic mice with the dKO ES clone and examined testicular sections at postnatal day 10 (P10). Germ cells were labeled with EGFP, indicating the contribution of dKO ES cells to germ cells (Fig. 4A). At P36, spermatids, spermatozoa, and Sertoli cells were labeled with EGFP in some seminiferous tubules (Fig. 4A). Spermatozoa labeled with EGFP were also present in 18-month-old chimeric mouse testes (Fig. 4B), indicating that Ddx3x and Ddx3y is not cell autonomously required for spermatogenesis or maintenance of spermatogonial stem cells. No morphological abnormalities were observed in EGFP-labeled spermatozoa collected from cauda epididymis of 8-week-old chimeric mice (Fig. 4C). Finally, mating these chimeric male mice with wild-type female mice showed successful transmission of Ddx3x or Ddx3y mutant allele to the next generation of female or male mice, respectively (Fig. 4D).
Fig. 4.
Ddx3x and Ddx3y double KO germ cells in chimeric males produce functional spermatozoa. (A) (Left) Testicular sections of Ddx3x and Ddx3y double KO ES chimeric males at postnatal day 10 (P10). Asterisks (*) indicate seminiferous tubules which contain ES cell contributed germ cells (EGFP positive). Scale bar: 50 µm. (Middle) Testicular sections of Ddx3x and Ddx3y double KO ES chimeric males at P36. Asterisks (*) indicate seminiferous tubules which contain ES cell contributed germ cells (EGFP positive). (Right) Magnified images of middle panel. White arrowhead indicates a Sertoli cell labeled with EGFP. Scale bars: 50 µm. (B) Testicular sections of double KO ES chimeric males at 18 months of age. Asterisks (*) indicate seminiferous tubules which contain ES cell contributed germ cells (EGFP positive). Scale bar: 50 µm. (C) Sperm morphology of double KO ES chimeric males. White arrowheads indicate spermatozoa derived from ES cells (EGFP positive heads of spermatozoa). Scale bar: 10 µm. (D) Genotyping of F1 females (Left; XWT/XDdx3x-em2) and males (Right; XWT/YDdx3y-em3) derived from chimeras. Ddx3x KO (29-bp deletion) band is slightly smaller than the wild-type. Ddx3y KO (125-bp deletion and 1-bp insertion) band is slightly smaller than the wild-type.
Discussion
We investigated the function of mouse Ddx3y, one of three genes present in the Azfa region, and mouse Ddx3x, an X chromosomal gene homologous to Ddx3y, in spermatogenesis. It has been difficult to knockout Y chromosome related genes by conventional gene targeting with homologous recombination in ES cells because the Y chromosome is composed of a variety of repetitive DNA sequences [34, 35]. Recently developed genome editing techniques, such as TALEN and CRISPR/Cas9 systems enable us to target genes with specific 20–30 bp short sequences. Here, we succeeded in generating Ddx3y KO male mice by using CRISPR/Cas9 system. We found that Ddx3y KO male mice show no abnormality in spermatogenesis, produce normal morphology of spermatozoa, and sire offspring. We conclude that Ddx3y is dispensable for spermatogenesis at least in mice. In addition, because Ddx3x KO males were found to be embryonic lethal, we generated chimeric mice, which contain Ddx3x and Ddx3y dKO germ cells, and found that these dKO germ cells can differentiate into spermatozoa and transmit their mutant alleles to offspring. Because both Ddx3x and Ddx3y mRNAs are expressed in germ cells [36] and are predicted as non-secretory proteins lacking signal peptides, it is unlikely that wild-type germ or somatic cells present in chimeric mice compensate for the function of Ddx3x and Ddx3y in dKO germ cells. Thus, we conclude that the both Ddx3x and Ddx3y are not required for spermatogenesis at least in mice.
Recently it was reported that when human iPS cell lines with AZFa deletions were xenotransplanted to mouse testes, spermatogenic failure of the derived germ cell-like cells were partially rescued by introduction of DDX3Y [37]. This suggests that DDX3Y is functional during spermatogenesis in human. Interestingly, in mice, Ddx3y has an autosomal paralog on chromosome 1, D1pas1, which is thought to be pseudogene in human [38, 39]. D1pas1 has 87% and 80% homology with Ddx3y at the amino acid and nucleotide sequence level respectively, respectively, and the mRNA expression is testis specific [36]. Based on our finding, we hypothesize that D1pas1 has similar functions to Ddx3y, and thus can mask the phenotype of Ddx3y KO mice. Consistent with our hypothesis, it has been recently reported that spermatogenic failure is observed in D1pas1 KO mice, indicating the requirement of D1pas1 for spermatogenesis [40]. Because we showed no spermatogenic failure in Ddx3y KO mice, D1pas1 might overcome Ddx3y deficiency. However, D1pas1 mRNA is expressed in spermatocytes, but not in spermatogonia [36], and D1pas1 KO germ cells arrest in late pachytene spermatocytes [40], indicating that the spermatogenic failure in D1pas1 KO mice is milder than that in AZFa-deleted SCOS patients. These reports suggest that mouse D1pas1 alone is not sufficient to function as a substitute for human AZFa genes. Thus, it will be useful to conduct studies to determine whether Ddx3y and D1pas1 compensate for each other in mice by generating Ddx3y and D1pas1 dKO mice.
Supplementary
Acknowledgments
We thank Dr Julio M. Castaneda for critical reading of the manuscript; the member of both Department of Experimental Genome Research and NPO for Biotechnology Research and Development for experimental assistance. This research was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT)/ Japan Society for the Promotion of Science (JSPS) KAKENHI Grants (JP17J09669 to TM, JP25112007 and JP17H01394 to MI), AMED under Grant Number JP18fk0210006h0003 and JP18gm5010001h0002 to MI, Takeda Science Foundation Grants to MI, NIH grant P01HD087157 and R01HD088412 to MI, and The Bill & Melinda Gates Foundation (Grand Challenges Explorations grant OPP1160866) to MI.
References
- 1.Kumar N, Singh AK. Trends of male factor infertility, an important cause of infertility: A review of literature. J Hum Reprod Sci 2015; 8: 191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar R. Medical management of non-obstructive azoospermia. Clinics (Sao Paulo) 2013; 68(Suppl 1): 75–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palermo G, Joris H, Devroey P, Van Steirteghem AC. Pregnancies after intracytoplasmic injection of single spermatozoon into an oocyte. Lancet 1992; 340: 17–18. [DOI] [PubMed] [Google Scholar]
- 4.Tesarik J, Mendoza C. Spermatid injection into human oocytes. I. Laboratory techniques and special features of zygote development. Hum Reprod 1996; 11: 772–779. [DOI] [PubMed] [Google Scholar]
- 5.Fraietta R, Zylberstejn DS, Esteves SC. Hypogonadotropic hypogonadism revisited. Clinics (Sao Paulo) 2013; 68(Suppl 1): 81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tiepolo L, Zuffardi O. Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human Y chromosome long arm. Hum Genet 1976; 34: 119–124. [DOI] [PubMed] [Google Scholar]
- 7.Vogt PH, Edelmann A, Kirsch S, Henegariu O, Hirschmann P, Kiesewetter F, Köhn FM, Schill WB, Farah S, Ramos C, Hartmann M, Hartschuh W, Meschede D, Behre HM, Castel A, Nieschlag E, Weidner W, Gröne HJ, Jung A, Engel W, Haidl G. Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet 1996; 5: 933–943. [DOI] [PubMed] [Google Scholar]
- 8.Kamp C, Huellen K, Fernandes S, Sousa M, Schlegel PN, Mielnik A, Kleiman S, Yavetz H, Krause W, Küpker W, Johannisson R, Schulze W, Weidner W, Barros A, Vogt PH. High deletion frequency of the complete AZFa sequence in men with Sertoli-cell-only syndrome. MHR: Basic science of reproductive medicine 2001; 7: 987–994. [DOI] [PubMed] [Google Scholar]
- 9.Lahn BT, Page DC. Functional coherence of the human Y chromosome. Science 1997; 278: 675–680. [DOI] [PubMed] [Google Scholar]
- 10.Jones MH, Furlong RA, Burkin H, Chalmers IJ, Brown GM, Khwaja O, Affara NA. The Drosophila developmental gene fat facets has a human homologue in Xp11.4 which escapes X-inactivation and has related sequences on Yq11.2. Hum Mol Genet 1996; 5: 1695–1701. [DOI] [PubMed] [Google Scholar]
- 11.Brown GM, Furlong RA, Sargent CA, Erickson RP, Longepied G, Mitchell M, Jones MH, Hargreave TB, Cooke HJ, Affara NA. Characterisation of the coding sequence and fine mapping of the human DFFRY gene and comparative expression analysis and mapping to the Sxrb interval of the mouse Y chromosome of the Dffry gene. Hum Mol Genet 1998; 7: 97–107. [DOI] [PubMed] [Google Scholar]
- 12.Sargent CA, Boucher CA, Kirsch S, Brown G, Weiss B, Trundley A, Burgoyne P, Saut N, Durand C, Levy N, Terriou P, Hargreave T, Cooke H, Mitchell M, Rappold GA, Affara NA. The critical region of overlap defining the AZFa male infertility interval of proximal Yq contains three transcribed sequences. J Med Genet 1999; 36: 670–677. [PMC free article] [PubMed] [Google Scholar]
- 13.Mazeyrat S, Saut N, Sargent CA, Grimmond S, Longepied G, Ehrmann IE, Ellis PS, Greenfield A, Affara NA, Mitchell MJ. The mouse Y chromosome interval necessary for spermatogonial proliferation is gene dense with syntenic homology to the human AZFa region. Hum Mol Genet 1998; 7: 1713–1724. [DOI] [PubMed] [Google Scholar]
- 14.Krausz C, Degl’Innocenti S, Nuti F, Morelli A, Felici F, Sansone M, Varriale G, Forti G. Natural transmission of USP9Y gene mutations: a new perspective on the role of AZFa genes in male fertility. Hum Mol Genet 2006; 15: 2673–2681. [DOI] [PubMed] [Google Scholar]
- 15.Luddi A, Margollicci M, Gambera L, Serafini F, Cioni M, De Leo V, Balestri P, Piomboni P. Spermatogenesis in a man with complete deletion of USP9Y. N Engl J Med 2009; 360: 881–885. [DOI] [PubMed] [Google Scholar]
- 16.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 2009; 326: 1509–1512. [DOI] [PubMed] [Google Scholar]
- 17.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science 2009; 326: 1501. [DOI] [PubMed] [Google Scholar]
- 18.Shpargel KB, Sengoku T, Yokoyama S, Magnuson T. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet 2012; 8: e1002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Flynn O’Brien KL, Varghese AC, Agarwal A. The genetic causes of male factor infertility: a review. Fertil Steril 2010; 93: 1–12. [DOI] [PubMed] [Google Scholar]
- 20.Foresta C, Ferlin A, Moro E. Deletion and expression analysis of AZFa genes on the human Y chromosome revealed a major role for DBY in male infertility. Hum Mol Genet 2000; 9: 1161–1169. [DOI] [PubMed] [Google Scholar]
- 21.Ditton HJ, Zimmer J, Kamp C, Rajpert-De Meyts E, Vogt PH. The AZFa gene DBY (DDX3Y) is widely transcribed but the protein is limited to the male germ cells by translation control. Hum Mol Genet 2004; 13: 2333–2341. [DOI] [PubMed] [Google Scholar]
- 22.Sekiguchi T, Iida H, Fukumura J, Nishimoto T. Human DDX3Y, the Y-encoded isoform of RNA helicase DDX3, rescues a hamster temperature-sensitive ET24 mutant cell line with a DDX3X mutation. Exp Cell Res 2004; 300: 213–222. [DOI] [PubMed] [Google Scholar]
- 23.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science 2013; 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013; 153: 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mashiko D, Fujihara Y, Satouh Y, Miyata H, Isotani A, Ikawa M. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci Rep 2013; 3: 3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abbasi F, Miyata H, Ikawa M. Revolutionizing male fertility factor research in mice by using the genome editing tool CRISPR/Cas9. Reprod Med Biol 2017; 17: 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho Y, Wigglesworth K, Eppig JJ, Schultz RM. Preimplantation development of mouse embryos in KSOM: augmentation by amino acids and analysis of gene expression. Mol Reprod Dev 1995; 41: 232–238. [DOI] [PubMed] [Google Scholar]
- 28.Fujihara Y, Kaseda K, Inoue N, Ikawa M, Okabe M. Production of mouse pups from germline transmission-failed knockout chimeras. Transgenic Res 2013; 22: 195–200. [DOI] [PubMed] [Google Scholar]
- 29.Oji A, Noda T, Fujihara Y, Miyata H, Kim YJ, Muto M, Nozawa K, Matsumura T, Isotani A, Ikawa M. CRISPR/Cas9 mediated genome editing in ES cells and its application for chimeric analysis in mice. Sci Rep 2016; 6: 31666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toyoda Y, Yokoyama M, Hoshi T. Studies on fertilization of mouse eggs in vitro. Jpn J Anim Reprod 1971; 16: 152–157. [Google Scholar]
- 31.Yen ST, Zhang M, Deng JM, Usman SJ, Smith CN, Parker-Thornburg J, Swinton PG, Martin JF, Behringer RR. Somatic mosaicism and allele complexity induced by CRISPR/Cas9 RNA injections in mouse zygotes. Dev Biol 2014; 393: 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen CY, Chan CH, Chen CM, Tsai YS, Tsai TY, Wu Lee YH, You LR. Targeted inactivation of murine Ddx3x: essential roles of Ddx3x in placentation and embryogenesis. Hum Mol Genet 2016; 25: 2905–2922. [DOI] [PubMed] [Google Scholar]
- 33.Oura S, Miyata H, Noda T, Shimada K, Matsumura T, Morohoshi A, Isotani A, Ikawa M. Chimeric analysis with newly established EGFP/DsRed2-tagged ES cells identify HYDIN as essential for spermiogenesis in mice. Exp Anim 2018. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reijo R, Lee TY, Salo P, Alagappan R, Brown LG, Rosenberg M, Rozen S, Jaffe T, Straus D, Hovatta O, et al. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat Genet 1995; 10: 383–393. [DOI] [PubMed] [Google Scholar]
- 35.Quintana-Murci L, Fellous M. The human Y chromosome: the biological role of a “functional wasteland”. J Biomed Biotechnol 2001; 1: 18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vong QP, Li Y, Lau YF, Dym M, Rennert OM, Chan WY. Structural characterization and expression studies of Dby and its homologs in the mouse. J Androl 2006; 27: 653–661. [DOI] [PubMed] [Google Scholar]
- 37.Ramathal C, Angulo B, Sukhwani M, Cui J, Durruthy-Durruthy J, Fang F, Schanes P, Turek PJ, Orwig KE, Reijo Pera R. DDX3Y gene rescue of a Y chromosome AZFa deletion restores germ cell formation and transcriptional programs. Sci Rep 2015; 5: 15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Session DR, Lee GS, Wolgemuth DJ. Characterization of D1Pas1, a mouse autosomal homologue of the human AZFa region DBY, as a nuclear protein in spermatogenic cells. Fertil Steril 2001; 76: 804–811. [DOI] [PubMed] [Google Scholar]
- 39.Kim YS, Lee SG, Park SH, Song K. Gene structure of the human DDX3 and chromosome mapping of its related sequences. Mol Cells 2001; 12: 209–214. [PubMed] [Google Scholar]
- 40.Inoue H, Ogonuki N, Hirose M, Hatanaka Y, Matoba S, Chuma S, Kobayashi K, Wakana S, Noguchi J, Inoue K, Tanemura K, Ogura A. Mouse D1Pas1, a DEAD-box RNA helicase, is required for the completion of first meiotic prophase in male germ cells. Biochem Biophys Res Commun 2016; 478: 592–598. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.