Abstract
Recently, the modification of the epigenetic status of somatic cell nuclear transfer (SCNT) embryos by treatment with histone deacetylase inhibitors (HDACis) has made it possible to alter epigenetic traits and improve the developmental competence of these embryos. In the current study, we examined the effects of an HDACi, quisinostat (JNJ), on the in vitro development of porcine cloned embryos and their epigenetic nuclear reprogramming status. SCNT embryos were cultured under various conditions, and we found that treatment with 100 nM JNJ for 24 h post activation could improve blastocyst formation rates compared to the control (P < 0.05). Therefore, this was chosen as the optimal condition and used for further investigations. To explore the effects of JNJ on the nuclear reprogramming of early stage embryos and how it improved cloning efficiency, immunofluorescence staining and quantitative real-time PCR were performed. From the pseudo-pronuclear to 2-cell stages, the levels of acetylation of histone 3 at lysine 9 (AcH3K9) and acetylation of histone 4 at lysine 12 (AcH4K12) increased, and global DNA methylation levels revealed by anti-5-methylcytosine (5-mC) antibody staining were decreased in the JNJ-treated group compared to the control (P < 0.05). However, JNJ treatment failed to alter AcH3K9, AcH4K12, or 5-mC levels at the 4-cell embryo stage. Moreover, JNJ treatment significantly upregulated the expression of the development-related genes OCT4, SOX2, and NANOG, and reduced the expression of genes related to DNA methylation (DNMT1, DNMT3a, and DNMT3b) and histone acetylation (HDAC1, HDAC2, and HDAC3). Together, these results suggest that treatment of SCNT embryos with JNJ could promote their developmental competence by altering epigenetic nuclear reprogramming events.
Keywords: Histone deacetylase inhibitor, Porcine in vitro culture, Quisinostat, Somatic cell nuclear transfer
Cloning mammals by somatic cell nuclear transfer (SCNT) has become an established procedure that has been successfully achieved in many species [1,2,3,4]. This technique has shown great potential for application in both animal science and biomedical research. In particular, cloned pigs have been used as a model for human genetic disease and for pig-to-human xenotransplantation because of the similarities to humans in their anatomy and physiology [5, 6]. However, the efficiency of porcine SCNT remains extremely low, and one of the reasons for this is incomplete epigenetic nuclear reprogramming [7]. To date, the mechanism of epigenetic reprogramming during SCNT remains largely unknown. However, reprogramming seems to have an important role in establishing nuclear totipotency in cloned embryos [8]. This process starts when the donor nuclear cell is present in the recipient ooplasm, and its configuration is quite different from that of a germ cell or an embryonic nucleus [9], which is greatly modified coincident with zygotic genome activation (ZGA) [10].
Many studies have tried to improve the efficiency of SCNT by altering the nuclear reprogramming status of early stage embryos [11, 12]. A widely studied epigenetic modification is histone acetylation, which is controlled by two major enzyme systems: histone acetyltransferases (HATs) and histone deacetylases (HDACs) [12]. These enzymes are important factors in the regulation of chromatin dynamics and gene expression [13]. An increase in the acetylation level of histones causes the loose binding of DNA to nucleosomes, chromatin relaxation, and the activation of gene transcription [14]. To study the role of histone acetylation in nuclear reprogramming, various classes of histone deacetylase inhibitors (HDACis) have been investigated, and the results indicated that HDACis, such as A (TSA) [2, 4, 15,16,17,18,19], valproic acid (VPA), scriptaid [3], oxamflatin [20,21,22], sodium butyrate (NaBu) [23], and m-carboxycinnamic acid bishydroxamide (CBHA) [24, 25] could improve the efficiency of SCNT embryo development in various species. However, it is not yet known if other novel compounds could also improve the development of porcine SCNT embryos and their mechanisms of action need to be investigated.
The current study investigated the potency of quisinostat (JNJ; JNJ-26481585), a potent, second generation pyrimidyl-hydroxamic acid HDACi, that strongly inhibits both Class I and II HDACs [26, 27]. JNJ has been studied mostly in cancer research for its antitumor and pharmacodynamic activities in various tumors [28]. In preclinical tumor models, it has been reported to show superior efficacy compared to other HDACis, including vorinostat, R306465, panobinostat, CRA-24781, and mocetinostat [26]. However, only a recent study has tested its efficacy for improving the in vitro development of porcine SCNT embryos [29].
Therefore, the purpose of this study was to investigate the effects of JNJ treatment on the in vitro development of porcine embryos derived by SCNT, and to optimize the conditions for JNJ treatment. To further elucidate the effects of JNJ treatment, the global acetylation levels of histone 3 at lysine 9 (AcH3K9) and histone 4 at lysine 12 (AcH4K12), and the global levels of 5-methylcytocine (5-mC) DNA methylation were assessed by specific immunofluorescence staining. Moreover, we analyzed the effects of JNJ on the expression levels of genes related to embryo development and reprogramming in blastocysts using quantitative real-time PCR (RT-qPCR).
Materials and Methods
Chemicals
All chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless specifically stated otherwise.
Oocyte collection and in vitro maturation
The collection of porcine cumulus-oocyte-complexes (COCs) and in vitro maturation (IVM) were performed as described previously [30]. Briefly, porcine ovaries were obtained from a local slaughter house and transported to the laboratory in a thermos bottle containing sterile saline at 30–35°C within 3 h. The follicular fluid from antral follicles 3–6 mm in diameter was aspirated using an 18 G needle attached to a 10 ml syringe. COCs were recovered and washed in tissue culture medium-199 (TCM-199, Invitrogen, Carlsbad, CA, USA) containing 5 mM sodium hydroxide, 2 mM sodium bicarbonate, 10 mM N-[2-Hydroxyethyl] piperazine-N'-[2-ethanesulfonic acid] (HEPES), 0.3% polyvinyl alcohol (PVA), and 1% Pen-Strep (Invitrogen). Only oocytes surrounded by a minimum of three cumulus cell layers and with an evenly granulated cytoplasm were selected. On average, 50 COCs were placed into IVM medium containing TCM-199 supplemented with 0.57 mM cysteine, 0.91 mM sodium pyruvate, 5 µl/ml insulin transferrin selenium solution (ITS-A) 100 × (Invitrogen), 10 ng/ml epidermal growth factor (EGF), 10% porcine follicular fluid (vol/vol), 10 IU/ml equine chorionic gonadotropin (eCG), and 10 IU/ml human chorionic gonadotropin (hCG). Subsequently, the COCs were cultured at 38.5°C under 5% CO2 in 95% humidified air for 21–22 h. They were then washed and transferred into fresh hormone-free IVM medium and cultured under the same conditions for an additional 21–22 h to complete their maturation.
Nuclear donor cell preparation
Nuclear donor cell cultures were established from ear skin tissue of an adult pig as described previously [30]. Briefly, the ear punch sample was rinsed four times with phosphate-buffered saline (PBS, Gibco, Grand Island, NY, USA) and the hair was removed. The ear punch was minced into small pieces using sterile scissors and cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) containing 10% fetal bovine serum (FBS; Gibco) (v/v), 1 mM sodium pyruvate, and 100 IU/ml each of penicillin and streptomycin at 38.5°C in an atmosphere of 5% CO2 for a week. Fibroblasts showing over 90% confluence were trypsinized, rinsed, and recultivated for further passaging. Nuclear donor cells derived from passages seven and eight were used for SCNT. A single-cell suspension was prepared by standard trypsinization and resuspension in Tyrode’s albumin lactate pyruvate (TALP)-HEPES medium prior to nuclear transfer.
SCNT and embryo culture
SCNT was performed as previously described [31]. After 42–44 h of IVM, COCs were completely denuded by gentle pipetting in TALP-HEPES medium containing 0.1% hyaluronidase. Denuded oocytes with a first polar body and uniform ooplasm were selected and stained with 5 µg/ml Hoechst 33342 in TALP-HEPES for 10 min. Enucleation was performed using an 18 µm inner diameter glass pipette by removing the first polar body and a small amount of surrounding cytoplasm in 4 µl TALP-HEPES microdrops containing 5 µg/ml cytochalasin B. To confirm that the nuclear material had been removed completely, the expelled cytoplasm was examined under ultraviolet radiation. After this, a single intact donor cell with good morphology and size was placed into the perivitelline space of each enucleated oocyte. Fusion was performed by equilibrating oocyte-cell couplets with fusion medium (0.28 M mannitol solution containing 0.5 mM HEPES and 0.1 mM MgSO4), placing them between a pair of platinum electrodes connected to an electric pulsing machine (LF101; Nepa Gene, Chiba, Japan) in 20 µl microdrops of fusion medium, and applying a single direct current (DC) pulse of 1.2 kV/cm for 30 µsec. Couplets were then washed and cultured in porcine zygote medium-5 (PZM-5; Funakoshi Corporation, Tokyo, Japan) for 1 h. For activation, reconstructed SCNT embryos were equilibrated with activation medium (0.28 M mannitol solution containing 0.5 mM HEPES, 0.1 mM CaCl2, and 0.1 mM MgSO4), transferred into an activation chamber filled with activation medium, and activated with a single DC pulse of 1.5 kV/cm for 30 µsec using a BTX ElectroCell Manipulator 2001 (BTX, San Diego, CA, USA). Approximately 10–12 SCNT embryos were cultured in 15 µl PZM-5 droplets covered with mineral oil at 38.5°C in a humidified atmosphere of 5% CO2, 5% O2, and 90% N2.
Preparation of JNJ and treatment protocol
JNJ was dissolved in dimethyl sulfoxide (DMSO) to achieve stock solutions of 10 µM, 100 µM, and 500 µM, and stored at –20°C. Working solutions were prepared immediately before use by diluting 1 µl of each stock solution in 1 ml PZM-5 medium to obtain 10, 100, and 500 nM JNJ, respectively. For the control, 1 µl of DMSO was diluted in 1 ml PZM-5 medium.
To determine the optimal concentration of JNJ, PZM-5 medium containing various concentrations of JNJ (0, 10, 100, and 500 nM) was used as the in vitro culture (IVC) medium immediately following reconstructed embryo activation for the first 24 h of IVC, then the SCNT embryos were transferred into JNJ-free PZM-5 medium for further culture. To determine the optimal treatment duration, 100 nM JNJ was included in the culture medium for 0, 6, 24, and 48 h post-activation.
Embryo evaluation and total blastocyst cell counts
In each experimental group, the cleavage and blastocyst formation rates were examined at 48 h and 168 h, respectively. To count the total cell numbers of the blastocysts, 15–20 blastocysts from each group were collected, washed in TALP-HEPES medium, and stained with 5 µg/ml Hoechst 33342 in TALP-HEPES for 10 min. After rinsing with TALP-HEPES medium, they were placed into 100% glycerol droplets, mounted with a cover slip, and evaluated under a fluorescence microscope (Nikon, Tokyo, Japan).
Evaluation of AcH3K9, AcH4K12, and 5-mC levels by immunofluorescence staining
Indirect immunofluorescence staining was performed to evaluate and compare the levels of histone acetylation and DNA methylation after SCNT. In each group, a total of 15 to 20 embryos at various stages (pseudo-pronuclear; PNC, 2-cell, and 4-cell) were collected, washed three times in distilled water (DW) containing 1% PVA (w/v) solution, fixed for at least 1 h in 4% paraformaldehyde (w/v) in PVA, permeabilized with 1% Triton X-100 (v/v) in DW for 30 min at 25°C, washed five times in 1% PVA in DW, and transferred into DW containing 2% bovine serum albumin (BSA; w/v) solution for 4 h to block the non-specific sites. After blocking, the embryos were incubated with the primary antibody rabbit polyclonal antibody against histone AcH3K9 and AcH4K12 (Upstate Biotechnology, Lake Placid, NY, USA; diluted 1:200) in DW containing 2% BSA solution at 4°C overnight. After washing three times, the embryos were incubated with goat anti-rabbit fluorescein isothiocyanate-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA, USA, diluted 1:200) in 2% BSA solution for 3 h at 25°C.
To stain methylated DNA, fixed embryos were washed extensively with DW containing 1% PVA and permeabilized with 1% Triton X-100 (v/v) in DW for 30 min. After this, they were treated with 50 µg/ml RNase A for 1 h at 37°C in the dark, treated with 3 M HCl for 30 min at 25°C, and then neutralized in 100 mmol/l Tris-HCl (pH 8.5) for 20 min before being blocked with DW containing 2% BSA. The embryos were incubated with 5-mC mouse monoclonal antibody (NA81 mAb; Calbiochem, San Diego, CA, USA; diluted 1:200) at 4°C overnight, incubated at 25°C with the secondary antibody Alexa Fluor 488-labeled goat anti-mouse IgG (1:200) in 2% BSA solution for 3 h, and washed three times in 2% PVA in DW, and DNA was counterstained with 25 µg/ml Hoechst 33342 for 3 min. The samples were mounted on glass slides and examined under an epifluorescence microscope (TE2000-S; Nikon) with similar exposure time. The mean fluorescence intensity levels were measured using ImageJ software (Version 1.49q; National Institutes of Health, Bethesda, MD, USA) and normalized to control embryos.
Quantitative real-time PCR
All samples were washed with diethylpyrocarbonate-treated water (DEPC) and stored at –80°C until RNA was extracted. The primer sequences for all the genes were synthesized according to our previous studies [32] and are presented in Table 1. Total mRNA was extracted from over 50 blastocysts from the control and treatment (100 nM for 24 h) groups using TRIzol reagent (Invitrogen) in accordance with the manufacturer’s protocol, and the total RNA concentration was quantified on a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). The total mRNA from these samples was used to generate complementary DNA (cDNA) using amfiRivert cDNA Synthesis Platinum Master Mix (GenDEPOT, Houston, TX, USA) according to the manufacturer’s protocol. Each reaction mixture contained 1 µl cDNA, 0.4 µl (10 pmol/µl) forward primer, 0.4 µl (10 pmol/µl) reverse primer, 10 µl SYBR Premix Ex Taq (TaKaRa, Otsu, Japan), and 8.2 µl of DEPC in a PCR plate (Micro-Amp Optical 96-Well Reaction Plate, Applied Biosystems, Singapore) and was amplified with the Applied Biosystems StepOneTM Real-Time PCR Systems (Applied Biosystems, Waltham, MA, USA). Thermal cycling conditions began with an initial denaturation at 95°C for 10 min, followed by 40 PCR cycles of 95°C for 15 sec, 60°C for 1 min, and 72°C for 1 min. Each transcript sample was quantified in three replicates using the 2–∆∆CT method [32], and the mRNA levels of all genes were normalized to the housekeeping gene Glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The average expression level of each gene in the control group was set as 1 for ease of comparison.
Table 1. Primer sequences used for real-time PCR.
| Genes | Primer sequences (5’- 3’) | Product size (bp) | Accession No. |
|---|---|---|---|
| GAPDH | F: GTCGGTTGTGGATCTGACCT | 207 | NM_001206359 |
| R: TTGACGAAGTGGTCGTTGAG | |||
| OCT4 | F: TTTGGGAAGGTGTTCAGCCAAACG | 198 | NM_001113060 |
| R: TCGGTTCTCGATACTTGTCCGCTT | |||
| SOX2 | F: ATGCACAACTCGGAGATCAG | 130 | NM_001123197 |
| R: TATAATCCGGGTGCTCCTTC | |||
| NANOG | F: GGTTTATGGGCCTGAAGAAA | 98 | NM_001129971 |
| R: GATCCATGGAGGAAGGAAGA | |||
| DNMT1 | F: TCGAACCAAAACGGCAGTAC | 215 | NM_001032355 |
| R: CGGTCAGTTTGTGTTGGACA | |||
| DNMT3a | F: CTGAGAAGCCCAAGGTCAAG | 200 | NM_001097437 |
| R: GTACTGATACGCGCACTCCA | |||
| DNMT3b | F: AGTGTGTGAGGAGTCCATTG | 133 | XM_013985277 |
| R: GCTTCCGCCAATCACCAGT | |||
| HDAC1 | F: CGCATGACTCACAATTTGCT | 211 | XM_013999116 |
| R: AGCCATCAAATACCGGACAG | |||
| HDAC2 | F: ACAGGAGACTTGAGGGAT | 232 | XM_001925318 |
| R: CACATTTAGCGTGACCTT | |||
| HDAC3 | F: GCTGCTGGACGGATGAGA | 108 | NM_001243827 |
| R: CTGGATGGAGCGTGAAGT | |||
F: Forward primer; R: Reverse primer.
Experimental design
To find the optimal conditions for JNJ treatment during IVC, SCNT embryos were treated with various concentrations of JNJ (0, 10, 100, and 500 nM) for 24 h after activation. The concentration that caused the highest percentage of embryos to develop (100 nM) was used for various durations of treatment (0, 6, 24, and 48 h) during IVC. Cleavage and blastocyst formation rates were observed at 48 and 168 h, respectively. To further assess the development of SCNT embryos among the experimental groups, blastocysts from each experimental group were stained with Hoechst 33342 and observed by fluorescence microscopy to enable the number of blastocyst cells to be counted. Moreover, to investigate the effects of JNJ on histone acetylation, the levels of AcH3K9 and AcH4K12 were determined in PNC, 2-cell, and 4-cell stage embryos. The relative expression of genes involved in development, DNA methylation, and histone acetylation were also determined.
Statistical analysis
Data are expressed as the mean values ± standard error of the mean (SEM). The gene expression and histone acetylation levels were compared between the two groups using a Student’s t-test, and the development data from the four groups were analyzed using univariate analysis of variance (ANOVA) followed by a Duncan’s multiple range test using SPSS 23.0 (SPSS, Chicago, IL, USA) statistical software. Differences were considered significant at P < 0.05, and all experiments were repeated at least three times.
Results
Effects of different JNJ concentrations on porcine embryo development
To determine the optimal concentration for JNJ treatment, SCNT embryos were cultured in various concentrations of JNJ (0, 10, 100, and 500 nM) for 24 h during IVC. A total of 561 embryos were cultured in different concentrations over six replicates. As shown in Table 2, no significant differences were observed in cleavage rates or the total number of blastocyst cells among the treatment groups. However, within the same incubation time, SCNT embryos cultured with 100 nM JNJ had a higher rate of blastocyst formation than embryos cultured in the other concentrations (P < 0.05).
Table 2. Effects of different concentrations of quisinostat treatment during IVC on embryonic development after somatic cell nuclear transfer.
| JNJ Concentration (nM) | No. of embryos cultured |
No. of embryos developed to (mean ± SEM, %) |
Total blastocyst cell number (mean ± SEM) |
|
|---|---|---|---|---|
| ≥ 2 cells | Blastocyst | |||
| 0 | 142 | 103 (72.83 ± 1.28) | 19 (13.97 ± 1.37) a | 50.08 ± 2.87 |
| 10 | 141 | 105 (75.36 ± 2.03) | 22 (15.61 ± 1.03) a | 48.92 ± 2.90 |
| 100 | 139 | 106 (76.99 ± 1.65) | 32 (23.50 ± 1.30) b | 53.52 ± 2.28 |
| 500 | 139 | 100 (72.11 ± 1.78) | 20 (15.21 ± 2.29) a | 50.27 ± 1.85 |
Replication number = six. a, b Values with different superscripts in the same column are significantly different (P < 0.05).
Effects of different JNJ treatment durations on porcine embryo development
Based on our previous results, treatment with 100 nM of JNJ was used for further investigations. To optimize the JNJ treatment, we determined the embryo developmental rates in groups treated with JNJ for various durations (Table 3). A total of 380 embryos were cultured with JNJ for different durations in five replicates. The highest percentage of blastocysts formed was observed for SCNT embryos supplemented with 100 nM JNJ for 24 h post activation, which was significantly higher than that for the control group (P < 0.05). However, no beneficial effect was observed on the cleavage rate or total blastocyst cell numbers.
Table 3. Effects of different durations of quisinostat treatment during IVC on embryonic development after somatic cell nuclear transfer.
| Duration (h) | No. of embryos cultured |
No. of embryos developed to (mean ± SEM, %) |
Total blastocyst cell number (mean ± SEM) |
|
|---|---|---|---|---|
| ≥ 2 cells | Blastocyst | |||
| 0 | 95 | 68 (72.07 ± 5.80) | 12 (12.59 ± 1.06) a | 49.15 ± 3.14 |
| 6 | 95 | 70 (73.57 ± 3.63) | 17 (17.71 ± 3.21) ab | 46.05 ± 2.37 |
| 24 | 96 | 75 (78.10 ± 3.23) | 21 (21.94 ± 3.04) b | 47.70 ± 1.76 |
| 48 | 94 | 66 (70.83 ± 6.35) | 13 (14.17 ± 2.80) ab | 49.66 ± 2.85 |
Replication number = five. a, b Values with different superscripts in the same column are significantly different (P < 0.05).
Assessment of global histone acetylation (AcH3K9 and AcH4K12) and global DNA methylation (5-mC) levels in PNC, 2-cell, and 4-cell embryo stages
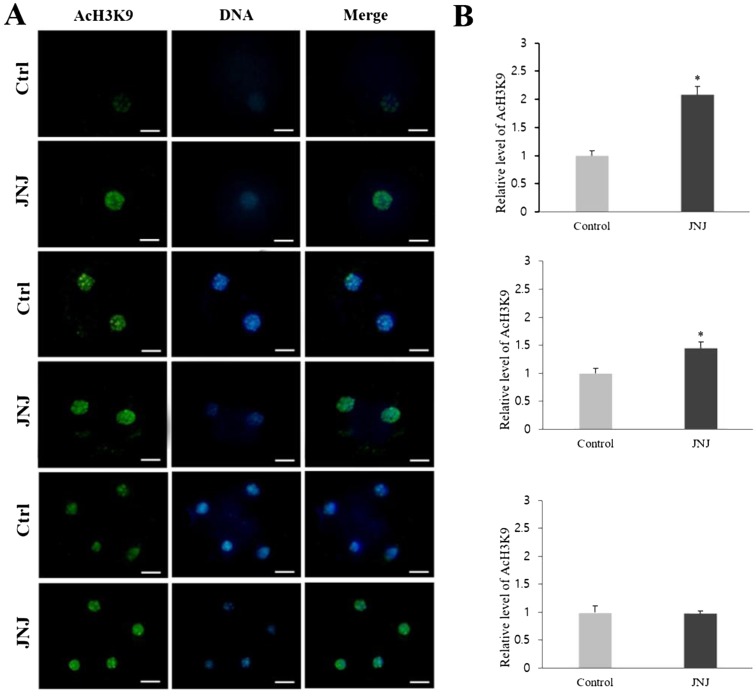
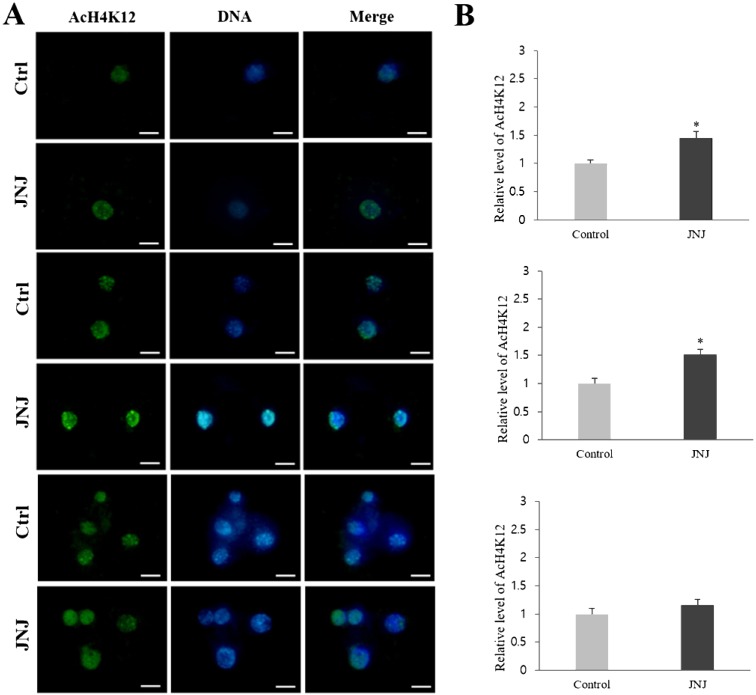
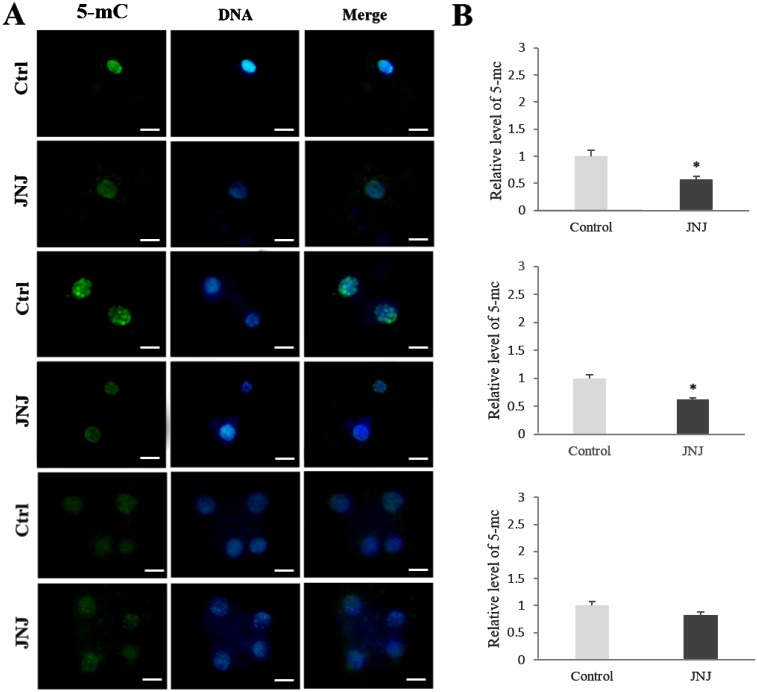
To elucidate how JNJ affects the preimplantation development of porcine SCNT embryos, the levels of AcH3K9, AcH4K12, and 5-mC in PNC, 2-cell, and 4-cell stages of embryos were investigated. The AcH3K9 and AcH4K12 signals were detected by immunofluorescence staining and all experiments were replicated three times. At the PNC and 2-cell stages, the intensity of the AcH3K9 and AcH4K12 signals in JNJ-treated embryos was significantly higher than that in control embryos. However, the relative intensity of 5-mC staining in the JNJ treated group was significantly lower than that of the control (P < 0.05) at the PNC and 2-cell stages. However, at the 4-cell stage, there were no significant differences in the intensities between the groups (Figs. 1, 2 and 3, P < 0.05).
Fig. 1.
Global acetylation levels of H3K9 in pseudo-pronuclear (PNC), 2-cell, and 4-cell stage SCNT embryos. (A) Immunofluorescence staining (green) in the control (Ctrl) and quisinostat treatment (JNJ, 100 nM for 24 h) groups. DNA was stained with Hoechst 33342 (blue), and merged images are shown. Scale bars indicate 50 µm. (B) Quantifications are shown as intensities relative to control embryos (set as 100%). Replication number = three. Asterisk above columns of the same development stage indicates statistically significant differences (P < 0.05).
Fig. 2.
Global acetylation levels of H4K12 in pseudo-pronuclear (PNC), 2-cell, and 4-cell stage SCNT embryos. All details are described in the legend for Fig. 1.
Fig. 3.
Levels of 5-mC DNA methylation in pseudo-pronuclear (PNC), 2-cell, and 4-cell stage SCNT embryos. All details are described in the legend for Fig. 1.
Relative expression levels of genes related to development, DNA methylation, and histone acetylation, determined by RT-qPCR
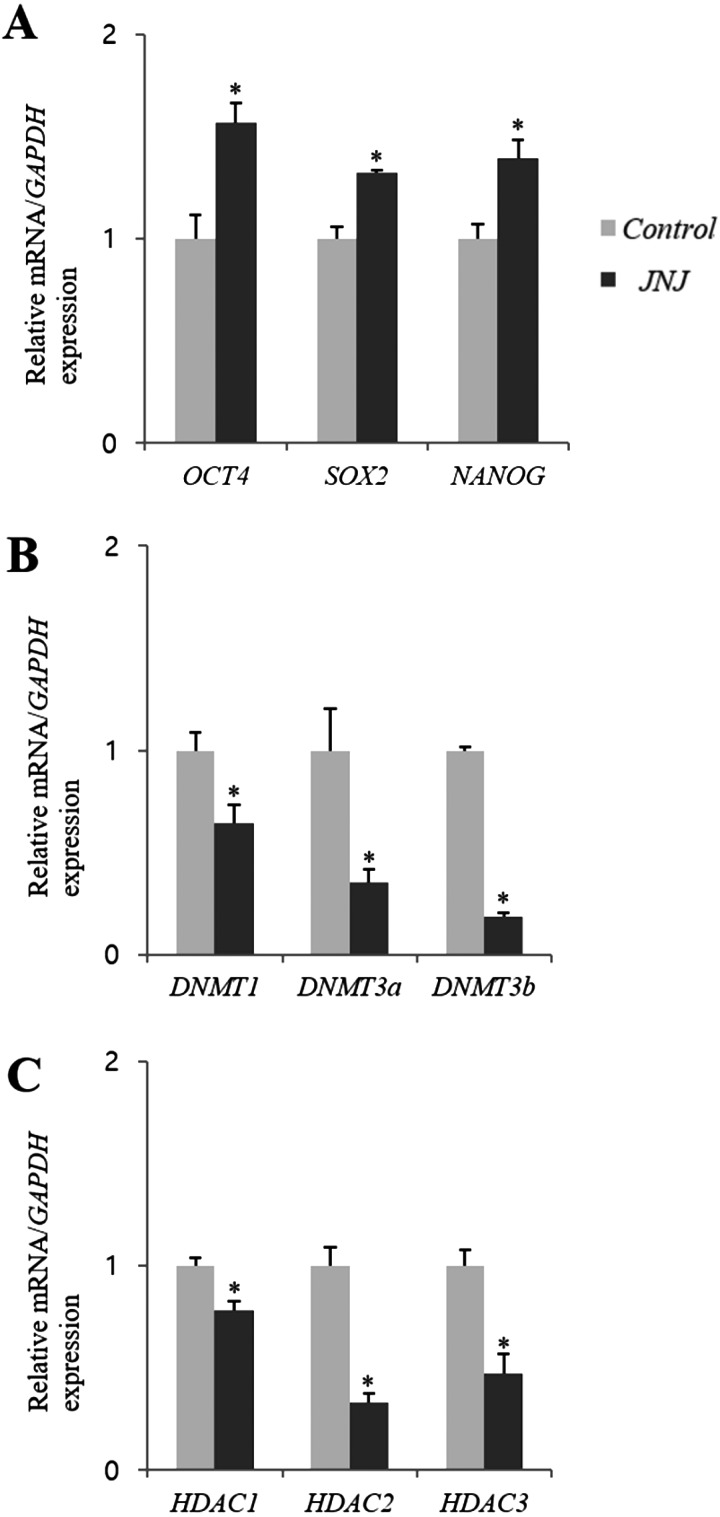
The relative abundance of nine selected genes in control and JNJ treated blastocysts is presented in Fig. 4. The relative transcript abundance of the embryonic development-related genes OCT4, Sox2, and Nanog in the JNJ treated group was significantly higher than that in the control group (P < 0.05). The expression levels of the DNA methylation-related genes DNMT1, DNMT3a, and DNMT3b in JNJ-treated blastocysts were significantly lower than those in the control group (P < 0.05). In addition, the expression levels of the histone acetylation-related genes HDAC1, HDAC2, and HDAC3 in the JNJ treatment group were lower than those in the control group (P < 0.05).
Fig. 4.
Comparison of mRNA expression levels (mean ± SEM) of genes related to developmental competence (A), global DNA methylation (B), and global histone acetylation (C) between the control and quisinostat treatment (JNJ, 100 nM for 24 h) groups. For the same mRNA, bars with asterisks are significantly different (P < 0.05). The experiment was replicated three times.
Discussion
Epigenetic reprogramming is an essential process for establishing totipotency after fertilization, and occurs abruptly during the early stages of embryonic development [8]. Histone acetylation, one of the major types of epigenetic marks, has a significant effect on the reprogramming process and affects the in vitro development of SCNT embryos [12, 33, 34]. Increasing histone acetylation could promote the accessibility of transcription factors to nucleosomal DNA, whereas the deacetylation of histones may cause chromatin condensation and thus decrease essential transcriptional activities during the early development of embryos [34, 35]. Many efforts have been made to modify the level of histone acetylation using HDACis, and it is well known that the treatment of donor cells and/or early stage SCNT embryos under optimized conditions with various HDACis could improve their in vitro developmental competence [17, 22, 25]. However, to date, the precise mechanism underlying histone acetylation in mammalian species remains unclear.
The application of epigenetic modification agents, such as HDACis and DNA methyltransferase inhibitors (DNMTis), might be a promising way to improve the developmental competence of SCNT embryos. In pigs, a high number of high quality cloned embryos need to be transferred into a surrogate mother to produce offspring [36]. It is well known that porcine organs are considered a valuable resource for xenotransplantation research due to the many similarities between pigs and humans in both anatomy and physiology [37]. Hence, we need to address the low efficiency of porcine SCNT, which has been mainly attributed to their aberrant epigenetic status.
In the present study, with the aim of improving SCNT embryonic development, we cultured porcine SCNT embryos with various concentrations of JNJ and for different durations to determine the optimal conditions for this treatment. The treatment of SCNT embryos with 100 nM JNJ resulted in a significant increase in blastocyst formation rates compared to the control. However, no differences were observed in the cleavage formation rates or total blastocyst cell numbers among the groups (Supplementary Fig. 1: online only). Interestingly, a recent study reported that treatment with 10 nM JNJ notably improved the blastocyst formation rate of porcine SCNT embryos [29]. The ten times higher optimal concentration in our study might be due to differences in the embryo activation protocol, IVC medium, and/or the nuclear donor cells used in the two studies [19, 38]. Similarly, several studies have also reported that the optimal concentration of trichostatin A, one of the most well-known HDACis, ranged from 5 to 50 nM under different conditions [18, 39]. Moreover, although there was no dose-dependent detrimental effect on the percentage of cleaved embryos, blastocyst development was significantly decreased at the highest concentration (500 nM) of JNJ compared to the 100 nM treated group. This indicates a negative effect of the HDACi at a high concentration, as reported in previous studies [16, 20]. Furthermore, the temporal effects of JNJ exposure were examined. We found that culturing SCNT embryos in 100 nM JNJ for 24 h resulted in the highest percentage of blastocysts forming, and this was significantly higher than the control. Altogether, in our study, the optimal conditions for treating SCNT embryos with JNJ during IVC were 100 nM JNJ for 24 h.
Among the various histone types, acetylation of histone H3 has a crucial role facilitating gene expression in SCNT embryos, which may affect their developmental competence [34]. Furthermore, it has been suggested that acetylation of AcH4K12 is essential for the initiation of embryonic gene activation, which is necessary for the further development of early stage embryos [40]. In the present study, to determine the effect of JNJ on nuclear reprogramming using histone acetylation as a readout, we examined the global acetylation levels of AcH3K9 and AcH4K12 in PNC, 2-cell, and 4-cell stage embryos using immunofluorescence staining. Similar to Jin et al., our results showed that JNJ treatment increased AcH3K9 levels in PNC embryos [29]. We also found that the level of AcH4K12 was increased in PNC embryos. Additionally, treatment with JNJ increased both AcH3K9 and AcH4K12 levels in 2-cell SCNT embryos. These results support the hypothesis that treatment with a HDACi affects the activities of HATs and HDACs, leading to decreased deacetylation and increased acetylation levels, resulting in enhanced transcriptional activities due to changes in chromatin configuration [41]. In addition, decreases in the global methylation levels of 5-mC in PNC and 2-cell stage embryos were observed after treatment with JNJ. The effects of HDACis are not limited to promoting the acetylation of histones, and may indirectly affect another epigenetic event, DNA methylation, through converging pathways, and disruption of one of these two epigenetic marks may inevitably affect the other [42]. A recent study suggested that HDACis could regulate DNMT1 levels by inhibiting the activating phosphorylation of MAP kinase I (ERK), and ultimately, DNA methylation [43]. In contrast, no notable changes were observed in either AcH3K9 or AcH4K12 levels in 4-cell stage SCNT embryos. We hypothesize that JNJ could have degraded in the IVC medium by this time, resulting in a sub-inhibitory level. Additionally, it is possible that the 4-cell stage embryos were less sensitive to JNJ treatment or simply had higher acetylation/lower methylation levels than the remainder of the cohort. This pattern of acetylation and methylation has also been observed in several previous porcine and bovine studies [15, 44].
To further investigate the mechanism behind the improved porcine SCNT efficiency, the relative expression of nine genes related to development (OCT4, SOX2, and NANOG); histone deacetylation (HDAC1, HDAC2, and HDAC3); and DNA methylation (DNMT1, DMNT3a, and DNMT3b) were analyzed in porcine SCNT blastocysts using RT-qPCR. Our results showed that JNJ treatment noticeably upregulated the expression of OCT4, SOX2, and NANOG and decreased the expression levels of genes related to histone acetylation and DNA methylation in JNJ-treated blastocysts compared to untreated controls.
OCT4/POU5F1 is a key regulator of pluripotency and cell differentiation, and is widely used to identify pluripotent cells in various species. A lack of OCT4 expression in embryos may cause an inhibition of development and a failure to form the inner cell mass [21, 45]. SOX2 is another important regulator that cooperates with OCT4 to form an OCT-SOX enhancer that functions to stably maintain the pluripotency of embryonic stem cells [46, 47]. Furthermore, the OCT-SOX enhancer has a role in regulating the expression of NANOG, which is a critical transcription factor involved in self-renewal and the differentiation of pluripotent stem cells [48].
HDAC1–3, which are members of the class I HDACs that are mainly expressed in oocytes and early stage embryos [49], have been marked as candidate genes involved in ZGA. Decreased expression of these genes could affect chromatin modulation and reprogramming, and promote transcriptional activities. On the other hand, hyperacetylation causes relaxation of the binding between nucleosomes and DNA, and can affect DNA methylation, another crucial epigenetic mechanism, which is regulated through the activities of DNA methyltransferases, including DNMT1, 3a, and 3b, that are important for embryonic development [50, 51]. Moreover, it has also been suggested that inhibition of HDACs could promote DNMT1 degradation through the ubiquitin-dependent proteosomal pathway [52]. The downregulation of DNA methylation-related genes may contribute to the reduction of 5-mC levels, which could encourage early embryonic development [53]. Taken together, these observations suggest that JNJ treatment improves porcine SCNT developmental competence through the positive regulation of genes related to development, histone acetylation, and DNA methylation.
In this study, we focused on the application and optimization of JNJ treatment to improve porcine SCNT efficiency. Our results demonstrated that JNJ treatment under optimal conditions (100 nM for 24 h) could improve the developmental competence of porcine SCNT embryos. This improvement seems to be associated with the positive regulation of global histone acetylation levels and the enhanced expression of genes related to development, histone acetylation, and DNA methylation by JNJ. However, further studies are required to elucidate the mechanism of action of JNJ.
Supplementary
Acknowledgments
This study was supported by the National Research Foundation (#2015R1C1A2A01054373; 2016M3A9B6903410; 2018R1D1A1B 07048765), Hansung Science High School R&E program funded by the Seoul Metropolitan of eduaction, the BK21 PLUS Program, and the Research Institute for Veterinary Science.
References
- 1.Jin JX, Lee S, Taweechaipaisankul A, Kim GA, Lee BC. The HDAC inhibitor LAQ824 enhances epigenetic reprogramming and in vitro development of porcine SCNT embryos. Cell Physiol Biochem 2017; 41: 1255–1266. [DOI] [PubMed] [Google Scholar]
- 2.Kohda T, Kishigami S, Kaneko-Ishino T, Wakayama T, Ishino F. Gene expression profile normalization in cloned mice by trichostatin A treatment. Cell Reprogram 2012; 14: 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Panda SK, George A, Saha A, Sharma R, Singh AK, Manik RS, Chauhan MS, Palta P, Singla SK. Effect of scriptaid, a histone deacetylase inhibitor, on the developmental competence of Handmade cloned buffalo (Bubalus bubalis) embryos. Theriogenology 2012; 77: 195–200. [DOI] [PubMed] [Google Scholar]
- 4.Shi LH, Ai JS, Ouyang YC, Huang JC, Lei ZL, Wang Q, Yin S, Han ZM, Sun QY, Chen DY. Trichostatin A and nuclear reprogramming of cloned rabbit embryos. J Anim Sci 2008; 86: 1106–1113. [DOI] [PubMed] [Google Scholar]
- 5.Ekser B, Cooper DKC, Tector AJ. The need for xenotransplantation as a source of organs and cells for clinical transplantation. Int J Surg 2015; 23(Pt B): 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim GA, Lee EM, Jin JX, Lee S, Taweechaipaisankul A, Hwang JI, Alam Z, Ahn C, Lee BC. Generation of CMAHKO/GTKO/shTNFRI-Fc/HO-1 quadruple gene modified pigs. Transgenic Res 2017; 26: 435–445. [DOI] [PubMed] [Google Scholar]
- 7.Niemann H. Epigenetic reprogramming in mammalian species after SCNT-based cloning. Theriogenology 2016; 86: 80–90. [DOI] [PubMed] [Google Scholar]
- 8.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science 2001; 293: 1089–1093. [DOI] [PubMed] [Google Scholar]
- 9.Rybouchkin A, Kato Y, Tsunoda Y. Role of histone acetylation in reprogramming of somatic nuclei following nuclear transfer. Biol Reprod 2006; 74: 1083–1089. [DOI] [PubMed] [Google Scholar]
- 10.Østrup O, Andersen IS, Collas P. Chromatin-linked determinants of zygotic genome activation. Cell Mol Life Sci 2013; 70: 1425–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fulka J, Fulka H, Slavik T, Okada K, Fulka J., Jr. DNA methylation pattern in pig in vivo produced embryos. Histochem Cell Biol 2006; 126: 213–217. [DOI] [PubMed] [Google Scholar]
- 12.Ma P, Schultz RM. Histone deacetylase 1 (HDAC1) regulates histone acetylation, development, and gene expression in preimplantation mouse embryos. Dev Biol 2008; 319: 110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho B, Kim HJ, Kim H, Sun W. Changes in the Histone Acetylation Patterns during the Development of the Nervous System. Exp Neurobiol 2011; 20: 81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao J, Hao Y, Ross JW, Spate LD, Walters EM, Samuel MS, Rieke A, Murphy CN, Prather RS. Histone deacetylase inhibitors improve in vitro and in vivo developmental competence of somatic cell nuclear transfer porcine embryos. Cell Reprogram 2010; 12: 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iager AE, Ragina NP, Ross PJ, Beyhan Z, Cunniff K, Rodriguez RM, Cibelli JB. Trichostatin A improves histone acetylation in bovine somatic cell nuclear transfer early embryos. Cloning Stem Cells 2008; 10: 371–379. [DOI] [PubMed] [Google Scholar]
- 16.Tsuji Y, Kato Y, Tsunoda Y. The developmental potential of mouse somatic cell nuclear-transferred oocytes treated with trichostatin A and 5-aza-2′-deoxycytidine. Zygote 2009; 17: 109–115. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Su J, Wang L, Xu W, Quan F, Liu J, Zhang Y. The effects of 5-aza-2′- deoxycytidine and trichostatin A on gene expression and DNA methylation status in cloned bovine blastocysts. Cell Reprogram 2011; 13: 297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cong P, Zhu K, Ji Q, Zhao H, Chen Y. Effects of trichostatin A on histone acetylation and methylation characteristics in early porcine embryos after somatic cell nuclear transfer. Anim Sci J 2013; 84: 639–649. [DOI] [PubMed] [Google Scholar]
- 19.Guo Z, Lv L, Liu D, Fu B. Effects of trichostatin A on pig SCNT blastocyst formation rate and cell number: A meta-analysis. Res Vet Sci 2018; 117: 161–166. [DOI] [PubMed] [Google Scholar]
- 20.Su J, Wang Y, Li Y, Li R, Li Q, Wu Y, Quan F, Liu J, Guo Z, Zhang Y. Oxamflatin significantly improves nuclear reprogramming, blastocyst quality, and in vitro development of bovine SCNT embryos. PLoS One 2011; 6: e23805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park SJ, Park HJ, Koo OJ, Choi WJ, Moon JH, Kwon DK, Kang JT, Kim S, Choi JY, Jang G, Lee BC. Oxamflatin improves developmental competence of porcine somatic cell nuclear transfer embryos. Cell Reprogram 2012; 14: 398–406. [DOI] [PubMed] [Google Scholar]
- 22.Hou L, Ma F, Yang J, Riaz H, Wang Y, Wu W, Xia X, Ma Z, Zhou Y, Zhang L, Ying W, Xu D, Zuo B, Ren Z, Xiong Y. Effects of histone deacetylase inhibitor oxamflatin on in vitro porcine somatic cell nuclear transfer embryos. Cell Reprogram 2014; 16: 253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar BM, Maeng GH, Lee YM, Lee JH, Jeon BG, Ock SA, Kang T, Rho GJ. Epigenetic modification of fetal fibroblasts improves developmental competency and gene expression in porcine cloned embryos. Vet Res Commun 2013; 37: 19–28. [DOI] [PubMed] [Google Scholar]
- 24.Song Y, Hai T, Wang Y, Guo R, Li W, Wang L, Zhou Q. Epigenetic reprogramming, gene expression and in vitro development of porcine SCNT embryos are significantly improved by a histone deacetylase inhibitor--m-carboxycinnamic acid bishydroxamide (CBHA). Protein Cell 2014; 5: 382–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agrawal H, Selokar NL, Saini M, Singh MK, Chauhan MS, Palta P, Singla SK, Manik RS. Epigenetic alteration of donor cells with histone deacetylase inhibitor m-carboxycinnamic acid bishydroxymide improves the in vitro developmental competence of buffalo (Bubalus bubalis) cloned embryos. Cell Reprogram 2018; 20: 76–88. [DOI] [PubMed] [Google Scholar]
- 26.Arts J, King P, Mariën A, Floren W, Beliën A, Janssen L, Pilatte I, Roux B, Decrane L, Gilissen R, Hickson I, Vreys V, Cox E, Bol K, Talloen W, Goris I, Andries L, Du Jardin M, Janicot M, Page M, van Emelen K, Angibaud P. JNJ-26481585, a novel “second-generation” oral histone deacetylase inhibitor, shows broad-spectrum preclinical antitumoral activity. Clin Cancer Res 2009; 15: 6841–6851. [DOI] [PubMed] [Google Scholar]
- 27.Carol H, Gorlick R, Kolb EA, Morton CL, Manesh DM, Keir ST, Reynolds CP, Kang MH, Maris JM, Wozniak A, Hickson I, Lyalin D, Kurmasheva RT, Houghton PJ, Smith MA, Lock R. Initial testing (stage 1) of the histone deacetylase inhibitor, quisinostat (JNJ-26481585), by the Pediatric Preclinical Testing Program. Pediatr Blood Cancer 2014; 61: 245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venugopal B, Baird R, Kristeleit RS, Plummer R, Cowan R, Stewart A, Fourneau N, Hellemans P, Elsayed Y, McClue S, Smit JW, Forslund A, Phelps C, Camm J, Evans TR, de Bono JS, Banerji U. A phase I study of quisinostat (JNJ-26481585), an oral hydroxamate histone deacetylase inhibitor with evidence of target modulation and antitumor activity, in patients with advanced solid tumors. Clin Cancer Res 2013; 19: 4262–4272. [DOI] [PubMed] [Google Scholar]
- 29.Jin L, Guo Q, Zhu HY, Xing XX, Zhang GL, Xuan MF, Luo QR, Luo ZB, Wang JX, Yin XJ, Kang JD. Quisinostat treatment improves histone acetylation and developmental competence of porcine somatic cell nuclear transfer embryos. Mol Reprod Dev 2017; 84: 340–346. [DOI] [PubMed] [Google Scholar]
- 30.Taweechaipaisankul A, Jin JX, Lee S, Kim GA, Lee BC. The effects of canthaxanthin on porcine oocyte maturation and embryo development in vitro after parthenogenetic activation and somatic cell nuclear transfer. Reprod Domest Anim 2016; 51: 870–876. [DOI] [PubMed] [Google Scholar]
- 31.Jin JX, Lee S, Taweechaipaisankul A, Kim GA, Lee BC. Melatonin regulates lipid metabolism in porcine oocytes. J Pineal Res 2017; 62: e12388. [DOI] [PubMed] [Google Scholar]
- 32.Lee S, Jin JX, Taweechaipaisankul A, Kim GA, Ahn C, Lee BC. Sonic hedgehog signaling mediates resveratrol to improve maturation of pig oocytes in vitro and subsequent preimplantation embryo development. J Cell Physiol 2018; 233: 5023–5033. [DOI] [PubMed] [Google Scholar]
- 33.Wee G, Shim JJ, Koo DB, Chae JI, Lee KK, Han YM. Epigenetic alteration of the donor cells does not recapitulate the reprogramming of DNA methylation in cloned embryos. Reproduction 2007; 134: 781–787. [DOI] [PubMed] [Google Scholar]
- 34.Yamanaka K, Sugimura S, Wakai T, Kawahara M, Sato E. Acetylation level of histone H3 in early embryonic stages affects subsequent development of miniature pig somatic cell nuclear transfer embryos. J Reprod Dev 2009; 55: 638–644. [DOI] [PubMed] [Google Scholar]
- 35.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet 2002; 3: 662–673. [DOI] [PubMed] [Google Scholar]
- 36.Song K, Hyun SH, Shin T, Lee E. Post-activation treatment with demecolcine improves development of somatic cell nuclear transfer embryos in pigs by modifying the remodeling of donor nuclei. Mol Reprod Dev 2009; 76: 611–619. [DOI] [PubMed] [Google Scholar]
- 37.Niu D, Wei HJ, Lin L, George H, Wang T, Lee IH, Zhao HY, Wang Y, Kan Y, Shrock E, Lesha E, Wang G, Luo Y, Qing Y, Jiao D, Zhao H, Zhou X, Wang S, Wei H, Güell M, Church GM, Yang L. Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9. Science 2017; 357: 1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez-Diaz MA, Che L, Albornoz M, Seneda MM, Collis D, Coutinho AR, El-Beirouthi N, Laurin D, Zhao X, Bordignon V. Pre- and postimplantation development of swine-cloned embryos derived from fibroblasts and bone marrow cells after inhibition of histone deacetylases. Cell Reprogram 2010; 12: 85–94. [DOI] [PubMed] [Google Scholar]
- 39.Kishigami S, Mizutani E, Ohta H, Hikichi T, Thuan NV, Wakayama S, Bui HT, Wakayama T. Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem Biophys Res Commun 2006; 340: 183–189. [DOI] [PubMed] [Google Scholar]
- 40.Wang F, Kou Z, Zhang Y, Gao S. Dynamic reprogramming of histone acetylation and methylation in the first cell cycle of cloned mouse embryos. Biol Reprod 2007; 77: 1007–1016. [DOI] [PubMed] [Google Scholar]
- 41.Kretsovali A, Hadjimichael C, Charmpilas N. Histone deacetylase inhibitors in cell pluripotency, differentiation, and reprogramming. Stem Cells Int 2012; 2012: 184154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaissière T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res 2008; 659: 40–48. [DOI] [PubMed] [Google Scholar]
- 43.Sarkar S, Abujamra AL, Loew JE, Forman LW, Perrine SP, Faller DV. Histone deacetylase inhibitors reverse CpG methylation by regulating DNMT1 through ERK signaling. Anticancer Res 2011; 31: 2723–2732. [PubMed] [Google Scholar]
- 44.Jin JX, Li S, Hong Y, Jin L, Zhu HY, Guo Q, Gao QS, Yan CG, Kang JD, Yin XJ. CUDC-101, a histone deacetylase inhibitor, improves the in vitro and in vivo developmental competence of somatic cell nuclear transfer pig embryos. Theriogenology 2014; 81: 572–578. [DOI] [PubMed] [Google Scholar]
- 45.Kirchhof N, Carnwath JW, Lemme E, Anastassiadis K, Schöler H, Niemann H. Expression pattern of Oct-4 in preimplantation embryos of different species. Biol Reprod 2000; 63: 1698–1705. [DOI] [PubMed] [Google Scholar]
- 46.Keramari M, Razavi J, Ingman KA, Patsch C, Edenhofer F, Ward CM, Kimber SJ. Sox2 is essential for formation of trophectoderm in the preimplantation embryo. PLoS One 2010; 5: e13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, Ko MS, Niwa H. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol 2007; 9: 625–635. [DOI] [PubMed] [Google Scholar]
- 48.Herrmann D, Dahl JA, Lucas-Hahn A, Collas P, Niemann H. Histone modifications and mRNA expression in the inner cell mass and trophectoderm of bovine blastocysts. Epigenetics 2013; 8: 281–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan H, O’brien MJ, Wigglesworth K, Eppig JJ, Schultz RM. Transcript profiling during mouse oocyte development and the effect of gonadotropin priming and development in vitro. Dev Biol 2005; 286: 493–506. [DOI] [PubMed] [Google Scholar]
- 50.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S, Mitsuya K, Okano M, Koseki H. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 2007; 450: 908–912. [DOI] [PubMed] [Google Scholar]
- 51.Zlatanova J, Caiafa P, Van Holde K. Linker histone binding and displacement: versatile mechanism for transcriptional regulation. FASEB J 2000; 14: 1697–1704. [DOI] [PubMed] [Google Scholar]
- 52.Zhou Q, Agoston AT, Atadja P, Nelson WG, Davidson NE. Inhibition of histone deacetylases promotes ubiquitin-dependent proteasomal degradation of DNA methyltransferase 1 in human breast cancer cells. Mol Cancer Res 2008; 6: 873–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang S, Tang B, Fan C, Shi L, Zhang X, Sun L, Li Z. Effect of DNMT inhibitor on bovine parthenogenetic embryo development. Biochem Biophys Res Commun 2015; 466: 505–511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.