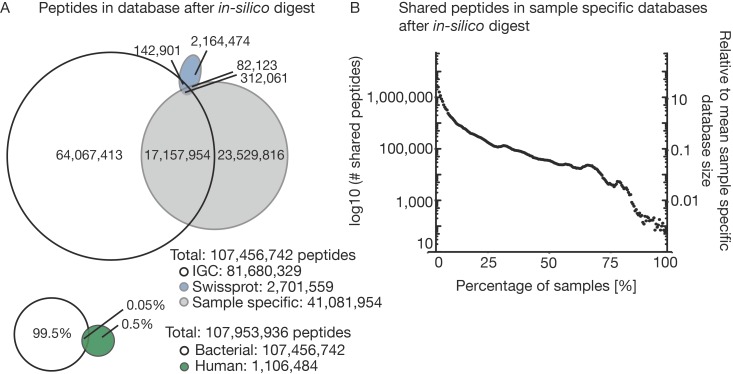
Figure 2.
In silico comparison of four different databases. Four different databases (Integrated Genome Reference Catalog (IGC), SWISS-PROT bacteria, SWISS-PROT human and sample specific metagenome-based databases) were digested in silico, and the possible search space was compared. (A) Venn diagram of the resulting peptides after in silico digestion comparing the three bacterial databases and all bacterial databases combined versus the peptides from the in silico digested human database. (B) Number of shared peptides in the 212 sample specific databases against the percentage of samples. The right axis indicates to which the percentage of the average sample specific database the number of shared peptides corresponds.