Abstract
Chitinase catalyzes the hydrolysis of chitin β-1,4 linkages. However, plants cannot produce chitin, suggesting that plant chitinases do not have the same function as animals. This study investigated the chitinase gene family in tomato and divided into eight groups via phylogenetic analyses with Arabidopsis and rice members. Conserved gene structures and motif arrangements indicated their functional relevance with each group. These genes were nonrandomly distributed across the tomato chromosomes, and tandem duplication contributed to the expansion of this gene family. Synteny analysis also established orthology relationships and functional linkages between Arabidopsis and tomato chitinase genes. Several positive selection sites were identified, which may contribute to the functional divergence of the protein family in evolution. In addition, differential expression profiles of the tomato chitinase genes were also investigated at some developmental stages, or under different biotic and abiotic stresses. Finally, functional network analysis found 124 physical or functional interactions, implying the diversity of physiological functions of the family proteins. These results provide a foundation for the exploration of the chitinase genes in plants and will offer some insights for further functional studies.
Keywords: chitinase, phylogenetic analyses, evolution, selective pressure, expression, functional network
1. Introduction
Chitin is a type of glycopolymer of N-acetyl-d-glucosamine in nature, which is widely distributed among the insect exoskeleton, the crustacean shell, and the fungal cell wall [1]. As a molecular model for identifying potential pathogens, chitin participates in the innate immune system of animals and plants. Chitin oligosaccharides have been shown to induce a variety of defensive responses through some chitin elicitor-binding proteins in plants [2,3,4]. Hydrolysis of the chitin was catalyzed by chitinase. Plants do not produce chitin, therefore, plant chitinases have been indicated to hydrolyze the cell walls of pathogens and to release elicitors for defense reactions [5]. Structurally, the chitinase protein adopts a (β/α)8 fold with an α-helix and a five stranded β-sheet insertion [6]. Based on their amino acid sequence similarity, plant chitinases are generally divided into six groups [7]. Among these groups, Classes III and V belong to the GH (glycosyl hydrolase) 18 family, and the remains belong to the GH19 family. Class I chitinases have the chitin binding domain (CBD) at the N-terminal region, while Classes II and III do not contain it [8]. Class V possesses two CBDs and a C-terminal extension. Only one half of the CBD was found in Class VI chitinases [9]. The divergent sequences and structures among different groups implied that they may originate from different ancestral genes [10,11].
Chitinase activity can be induced by infection of the tissue with fungal or pathogens in many plants [12,13,14,15,16]. Through blocking mycelium cells and inducting downstream defense pathways, the induced plant chitinases are involved in the defense response against pathogen and fungal infection [17]. Overexpression of the chitinase and the glucanase genes can enhance the protection against fungal attack in transgenic tobacco [18]. Moreover, chitinases are also involved in the growth and development of plants. Mutation of the Arabidopsis Class II chitinase causes cell morphological abnormalities and length changes of the internode and root [19]. Similar study on chitinase affecting root growth has also been reported in rice [20]. Mutation of the OsCTL1 gene reduces cellulose content and mechanical strength, suggesting a possible mechanism for cellulose biosynthesis and cell wall remodeling [21]. Other biological processes, such as cell separation or loosening, embryonal development, and programmed cell death, are associated with plant chitinase activity [22,23,24,25,26]. In addition, some plant hormones and abiotic stress also regulate the expression and activity of chitinases [27,28,29]. For instances, jasmonates and wounding can induce the expression of BjCHI and SafChia genes [30,31]. Some chitinases are upregulated by heavy metals including cadmium, and arsenic [32]. Moreover, salt, cold and drought stresses promote the transcription of some chitinase genes [28,31]. All these suggest that chitinases may be involved in a variety of growth processes and stress responses.
Plants are easily attacked by various pathogens and other environmental factors, and it seems reasonable to expect that selection will favor functional divergence in the evolution process. A previous study indicated that positive selection can drive the divergence of ChiA and ChiB duplicated genes in the GH19 family [33]. This variation has spurred plants to obtain some ability to recognize a wide diversity of pathogens [34]. The rapid evolution of plant chitinases implies the coevolutionary interactions between plants and pathogens. Furthermore, the changes of these proteins can explain the diversity of disease resistance [35]. In the evolutionary process, several chitinases have retained the chitin-binding ability while lost hydrolyzing ability, which may affect a number of biological processes as described above [11].
As one of the most economically important vegetables in the world, tomato (Solanum lycopersicum) has been chosen as one model species for studies of fruit development [36], domestication [37], and stress responses [38]. A total of 25 and 49 chitinase family genes have been identified in Arabidopsis and rice, respectively [29]. Only few tomato chitinases have been functionally identified to date [23,39] and no systematic investigation of chitinase protein family has been reported in the tomato. The completed sequencing of the tomato provides an opportunity to deduce the chitinase gene family and infer its evolutionary history [40]. This study identified 43 tomato chitinases via database analysis. Their phylogeny, structure, chromosomal location and duplication, and synteny were then investigated, followed by selective pressure analysis and expression profiles. Finally, a functional network analysis of the tomato chitinase proteins was also conducted. This study provides several useful insights for further evolutional and functional investigation of this gene family.
2. Materials and Methods
2.1. Identification of the Chitinase Genes in Tomato
To identify potential chitinase genes in the tomato plant genome (S. lycopersicum), first, all chitinase sequences of Arabidopsis and rice [29] were used as queries to perform BLAST searches against the tomato database [40] in the phytozome database (http://www.phytozome.net) [41] with a threshold of -1 E. Furthermore, all candidate proteins were further used to perform new queries in this database. SMART [42] and Pfam [43] were used to further examine the obtained sequences, and those without common domains (PF00182, PF00187, or PF00704) were excluded. The physicochemical parameters and subcellular localization of the chitinase proteins were predicted using the ProtParam tool [44] and the TargetP 1.1 server [45], respectively.
2.2. Phylogeny, Gene Organization, and Conserved Motif Analysis of the Chitinase Gene Family
Multiple sequence alignment of all predicted chitinase proteins was performed with the MUSCLE method [46]. Next, MEGA6 [47] was used to construct a neighbor joining (NJ) tree with 1000 bootstrap replications, a p-distance substitution model, and pairwise deletion gaps parameters. Gene organization was inferred from the phytozome database (http://www.phytozome.net) by comparing the genomic sequence and the CDS sequence of chitinase members. Additionally, the MEME program [48] was used to identify the conserved motifs with the following parameters: zero or one motif in each sequence, six and 50 width of motifs, and a maximum of 12 motifs.
2.3. Chromosomal Location and Duplication Time Inference
Annotation information of the chitinase genes in the tomato database was used to determine their chromosomal locations. Chitinase paralogous gene pairs were determined via their phylogeny and were used as reference for pairwise DNA coding sequence alignments using the MUSCLE method [46]. K-Estimator 6.0 [49] was used to estimate Ka (the number of nonsynonymous substitutions per nonsynonymous site) and Ks (the number of synonymous substitutions per synonymous site) values for these pairwise alignments. Finally, the Ks value was used to calculate the approximate date of the duplication event (T = Ks/2λ), assuming clock-like rates (λ) of 1.5 × 10−8 synonymous/substitution site/year for tomato [50].
2.4. Analyses of Synteny
The SynMap server of the CoGe [51] was used to identify syntenic gene blocks between Arabidopsis and tomato with the following parameters: DAGchainer algorithm, a maximum distance of 20 genes between two matches, and a minimum of five aligned gene pairs.
2.5. Site-Specific Selection Assessment and Testing
The selective pressure was estimated by calculating the synonymous rate (Ks) and the nonsynonymous rate (Ka) at each codon. Three evolutionary models [M8 (beta + w ≥ 1), M7 (beta), and M5 (gamma)] were used to describe how the characteristics evolve in probabilistic terms using a Bayesian inference approach [52]. Each model used different biological assumptions to assume a statistical distribution that accounts for heterogeneous Ka/Ks values among sites. Next, eight discrete categories were used to approximate the distribution. Finally, the Ka/Ks values were computed by calculating the expectation of a posterior distribution [52]. The Phyre2 Server [53] was used to predict the three-dimensional structure of the chitinase protein (Solyc10g055820.1) in the GH19-A group. I-Mutant2.0 [54] was then used to estimate the effects of point mutations on protein folding stability. The free energy change value [∆∆G (kcal/mol)] was used to infer the protein stability. When ∆∆G exceeds zero, the stability of the mutant protein increases; in contrast, when ∆∆G is below zero, the stability decreases.
2.6. Transcriptome-Based Expression Analysis
Transcriptome data (GSE33507) was used to investigate the expression analysis of the tomato chitinase genes in different development stages. Two to four biological replicates were performed. The average fragments per kilobase of transcripts per million fragments mapped (FRKM) was used as the unit of measurement to estimate the expression levels of each transcript. The Log2 ratio of the sample signal to control signal was used to indicate its relative expression level. The HeatMapper Plus tool of BAR (http://bar.utoronto.ca) was used to access their relative expression levels.
2.7. Plant Materials, Stress Treatments, RNA Isolation, Quantitative Real-Time PCR (qRT-PCR) Analysis, and Promoter Sequence Analysis
One-week-old tomato (S. lycopersicum L. cv Hezuo 908) seedlings were used to examine the expression patterns of chitinase genes under different stress treatments. Plants were grown in liquid MS culture in a plant growth chamber at 23 ± 1 °C with a 14-h light/10-h dark photoperiod. Tomato seedlings were kept at 4 ± 1 and 42 ± 1 °C for 3 h for cold and heat treatments, respectively. Tomato seedlings were put in 150mM NaCl for 24 h for salt stress. For the drought treatment, the seedlings were dried between folds of tissue paper at 23 ± 1 °C for 3 h. Control (CK) seedlings were grown at 23 ± 1 °C with normal irrigation. For the biotic treatment, we first cultured the mycelia of Sclerotinia sclerotiorum on PDA medium for about four days. The mycelia of ~3 mm agar plugs in diameter were excised from the edges of growing colonies and were upended onto the tomato leaves for 24 h and 48 h for pathogen infection treatment, respectively. We used the normally grown tomato leaves as controls. Three replicates were performed per sample. After different abiotic and biotic stress treatments, total RNA of each sample was extracted with the Trizol total RNA extraction kit (Sangon, Shanghai, China, SK1321) and treated with DNase I (TakaRa, Dalian, China). Reverse transcription was performed using M-MLV (TakaRa) with random primers. Quantitative assays were performed on each cDNA dilution using the SYBR Green Master Mix (TakaRa) with an ABI 7500 sequence detection system, according to the manufacturer’s protocol. Eleven tomato chitinase genes were selected for qRT-PCR analysis with the expression level of the Actin gene (Solyc01g104770.2) as endogenous control. The specific primers are shown in Table S1. The 2−∆∆CT method [55] was used to calculate the relative expression level of each sample. Through comparing the expressed sequence tag with the genomic sequence, we first define the TSS (transcription start site) of each chitinase gene in tomato, Arabidopsis, and rice. Here, 1000 bp upstream promoter sequences were used to identify their cis-elements using PLACE [56].
2.8. Network Assembly
The STRING database (http://string-db.org) [57] was used to assemble the protein–protein interaction networks. This database includes several interaction sources, such as, textmining, experiments, coexpression, neighborhood, fusion, co-occurrence databases, and so on [57]. All predicted tomato chitinase proteins were submitted to this database. The minimum required interaction score was set to medium confidence (0.400). Max number of interactors showed no more than 20 on the first shell, and no more than 10 on the second shell.
3. Results
3.1. Identification and Phylogenetic Analysis of the Chitinase Gene Family in Tomato
To identify the putative chitinase family genes in the tomato genome, a BLAST search of the phytozome database (http://www.phytozome.net) was first performed using the with methods described above. As a result, 43 putative chitinase genes were identified in tomato. Most of these tomato chitinase genes encode highly hydrophilous residues of 60–386 amino acids in length, with a predicted isoelectric point (pI) range of 4.41 to 9.6 (Table 1). Further subcellular localization prediction indicated that more than 79% of the candidate chitinase proteins are likely localized in the secretory pathway, which is similar to the results of Arabidopsis and rice chitinases [29].
Table 1.
Chitinase genes identified in tomato. Mw/Kda means molecular weight (kilodalton). GRAVY stands for the grand average of hydropathy. S, C, and M mean secretory pathway, chloroplast, and mitochondrion, respectively, in TargetP localization.
| Gene ID | Chrome Location | Length | Mw/Kda | pI | GRAVY | Domains | TargetP Localization |
|---|---|---|---|---|---|---|---|
| Solyc01g095250.1 | ch01:86587643..86588539 (+) | 298 | 31.91 | 4.51 | −0.022 | GH18 | S (0.995) |
| Solyc01g095260.1 | ch01:86590501..86591397 (+) | 298 | 31.95 | 4.41 | −0.019 | GH18 | S (0.993) |
| Solyc01g095310.1 | ch01:86626834..86627742 (+) | 302 | 32.35 | 5.34 | −0.05 | GH18 | S (0.994) |
| Solyc01g097270.2 | ch01:88179851..88181214 (−) | 212 | 22.69 | 8.36 | −0.295 | GH19 | S (0.910) |
| Solyc01g097280.2 | ch01:88183566..88184361 (−) | 199 | 21.49 | 8.78 | −0.37 | GH19 | S (0.959) |
| Solyc02g061770.2 | ch02:33290113..33291907 (+) | 263 | 28.52 | 8.69 | −0.17 | GH19 | S (0.953) |
| Solyc02g082920.2 | ch02:46550982..46552140 (+) | 253 | 27.63 | 5.93 | −0.265 | GH19 | S (0.988) |
| Solyc02g082930.2 | ch02:46554240..46555813 (+) | 247 | 26.58 | 4.68 | −0.228 | GH19 | S (0.841) |
| Solyc02g082960.2 | ch02:46564405..46566106 (+) | 273 | 30.14 | 9.56 | −0.194 | GH19 | S (0.887) |
| Solyc03g116190.1 | ch03:65691845..65692213 (−) | 122 | 13.98 | 8.09 | −0.202 | GH19 | S (0.896) |
| Solyc03g116230.2 | ch03:65723988..65725192 (−) | 295 | 30.59 | 5.38 | −0.196 | GH19 | S (0.982) |
| Solyc03g116240.2 | ch03:65733982..65736687 (−) | 87 | 9.26 | 4.94 | 0.526 | GH19 | S (0.985) |
| Solyc04g072000.2 | ch04:59066373..59068261 (−) | 276 | 30.01 | 4.58 | −0.294 | GH19 | S (0.994) |
| Solyc05g050130.2 | ch05:60108919..60110761 (−) | 292 | 31.26 | 8.46 | 0.087 | GH18 | S (0.972) |
| Solyc06g053380.2 | ch06:36128793..36130489 (−) | 289 | 31.81 | 5.37 | −0.284 | GH19 | S (0.977) |
| Solyc07g005080.1 | ch07:112239..113536 (+) | 386 | 41.83 | 4.82 | −0.125 | GH18 | Other (0.554); C (0.337) |
| Solyc07g005090.2 | ch07:113980..115328 (−) | 371 | 41.51 | 8.66 | −0.111 | GH18 | S (0.899) |
| Solyc07g005100.2 | ch07:123941..126335 (+) | 376 | 41.85 | 9.34 | −0.271 | GH18 | S (0.951) |
| Solyc07g009500.1 | ch07:4618503..4618766 (+) | 87 | 9.49 | 8.36 | −0.298 | GH19 | S (0.704) |
| Solyc07g009510.1 | ch07:4622506..4622769 (+) | 87 | 9.49 | 8.36 | −0.298 | GH19 | S (0.704) |
| Solyc07g009530.1 | ch07:4681799..4681990 (+) | 63 | 6.49 | 7.48 | 0.733 | GH19 | S (0.995) |
| Solyc07g026990.1 | ch07:32915139..32915324 (−) | 61 | 7.43 | 7.77 | −0.387 | GH19 | Other (0.784) |
| Solyc09g098540.2 | ch09:72338607..72340235 (−) | 319 | 35.56 | 7.95 | −0.205 | GH19 | S (0.964) |
| Solyc10g017970.1 | ch10:6248916..6249098 (−) | 60 | 6.17 | 5.57 | 0.497 | GH19 | S (0.992) |
| Solyc10g017980.1 | ch10:6273805..6273996 (+) | 63 | 6.56 | 7.48 | 0.471 | GH19 | S (0.995) |
| Solyc10g055790.1 | ch10:57408394..57408612 (−) | 72 | 7.72 | 7.51 | 0.408 | GH19 | S (0.976) |
| Solyc10g055800.1 | ch10:57410647..57411787 (−) | 329 | 35.41 | 8.64 | −0.423 | GH19 | S (0.838) |
| Solyc10g055810.1 | ch10:57417284..57418434 (−) | 322 | 34.35 | 6.19 | −0.248 | GH19 | S (0.989) |
| Solyc10g055820.1 | ch10:57435941..57437097 (−) | 322 | 34.89 | 6.36 | −0.348 | GH19 | S (0.975) |
| Solyc10g068350.1 | ch10:65525076..65525336 (−) | 86 | 9.23 | 7.65 | −0.494 | GH19 | Other (0.600); C (0.419) |
| Solyc10g074360.1 | ch10:57700740..57701259 (−) | 154 | 17.16 | 8.51 | −0.161 | GH19 | Other (0.677); M (0.523) |
| Solyc10g074380.1 | ch10:57722610..57722870 (−) | 64 | 7.19 | 9.6 | −1.008 | GH19 | Other (0.504); C (0.363) |
| Solyc10g074390.1 | ch10:57732393..57733097 (−) | 216 | 22.81 | 8.47 | −0.315 | GH19 | S (0.964) |
| Solyc10g074400.1 | ch10:57738774..57739374 (−) | 173 | 18.43 | 9.11 | −0.381 | GH19 | S (0.436); M (0.435) |
| Solyc10g074460.1 | ch10:57954087..57956593 (−) | 194 | 20.89 | 8.36 | −0.132 | GH19 | S (0.966) |
| Solyc11g005890.1 | ch11:710226..711943 (−) | 295 | 33.33 | 5.7 | −0.052 | GH18 | S (0.876) |
| Solyc11g020130.1 | ch11:10197594..10198487 (+) | 163 | 17.39 | 7.75 | −0.111 | GH19 | S (0.995) |
| Solyc11g020450.1 | ch11:11116611..11117328 (−) | 206 | 22.18 | 8.17 | −0.675 | GH19 | M (0.368) |
| Solyc11g020530.1 | ch11:11364445..11364994 (−) | 167 | 18.29 | 8.47 | −0.499 | GH19 | M (0.470) |
| Solyc11g072750.1 | ch11:55960411..55961439 (−) | 342 | 38.89 | 6.89 | −0.139 | GH18 | M (0.597) |
| Solyc11g072760.1 | ch11:55962509..55963429 (−) | 306 | 34.63 | 6.29 | −0.037 | GH18 | S (0.909) |
| Solyc11g072830.1 | ch11:56024533..56025453 (+) | 306 | 33.74 | 4.79 | −0.15 | GH18 | S (0.974) |
| Solyc12g098810.1 | ch12:66187159..66189951 (+) | 328 | 36.44 | 6.32 | −0.242 | GH19 | S (0.962) |
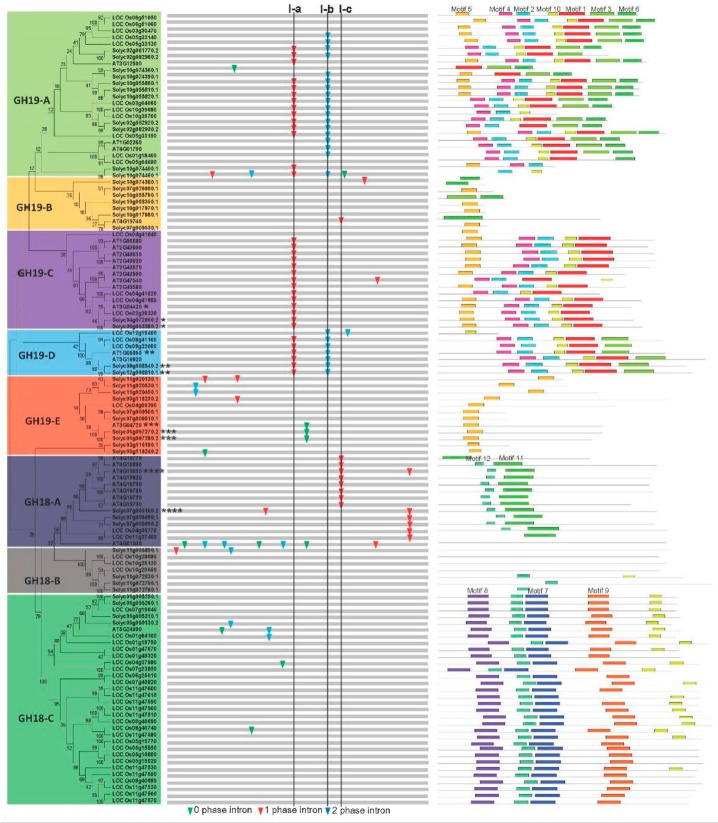
To assess the evolutionary relationship of the chitinase genes in the tomato, Arabidopsis, and rice, phylogenetic analyses were performed based on a NJ method using MEGA6 [47]. The 120 predicted chitinase proteins were classified into two major evolutionary branches: GH19 and GH18, which were further divided into the following eight groups: GH19-A, GH19-B, GH19-C, GH19-D, GH19-E, GH18-A, GH18-B, and GH18-C (Figure 1). Further evidences, such as gene organization and conserved motif distribution (described below), also support this classification. The GH18-C group is the largest with 32 members, representing 26.7% of the total number of chitinase genes, while GH19-D and GH18-B groups are the smallest, with only seven genes. In addition, one eudicot-specific chitinase clade formed the GH19-B group (Figure 1). The distribution of the chitinase genes in different species and groups was also assessed (Table S2). Similar to Arabidopsis and rice, tomato chitinases are not distributed evenly in these eight phylogeny groups. Nearly half of the tomato chitinase genes are distributed in GH19-A and GH19-E groups. With regard to rice, ~53% of the chitinase members are distributed in GH18-C.
Figure 1.
Phylogenetic relationship, gene structure, and motif composition of the chitinase genes. The phylogenetic tree is constructed and classified into eight groups. Classification of GH18 and GH19 is based on the SMART and Pfam databases [42,43]. Four syntenic paralogs were marked with * for pairs 1, ** for pairs 2, *** for pairs 3, and **** for pairs 4 as indicated in Figure 3. The insertion positions of 0, 1, and 2 phase introns are marked with green, red, and blue inverted triangles, respectively. Different motifs predicted by MEME [48] of the chitinase proteins are displayed by different colored boxes.
3.2. Gene Organization and Conserved Motif Distribution of the Chitinases
To further examine the organizational diversity of these chitinase genes, their exon–intron structures were inferred by comparing both their coding sequences and their genome sequences. A detailed illustration of the chitinase gene organization is shown in Figure 1. In general, the chitinase gene organization was well conserved, especially in CH19-A and CH19-D groups, which supports a common origin. Furthermore, three introns (I-a, I-b, and I-c) were designated to demonstrate the feature of exon–intron evolution of chitinases. Intron I-a is conserved among most members of the putative chitinase genes in the groups GH19-A, GH19-B, and GH19-D. Intron I-b was only found in the groups GH19-A and GH19-D. Intron I-c is specific for Group CH18-A. In addition, some intron loss or gain has occurred within several evolutionary branches (Figure 1).
To further examine the diversification of chitinase proteins, several conserved motifs were investigated using MEME [48]. As a result, 12 conserved motifs were found in the predicted chitinase proteins (Figure 1; Table S3). Similar motif compositions of each group not only provided additional evidence in support of the results of the phylogenetic analyses, but also implied functional relevance. Furthermore, several distinct motifs were found in specific groups. For instance, motifs 7, 8, and 9 are restricted to the CH18-C group, and motif 11 is unique to the GH18-A group. Interestingly, we did not find any of these 12 motifs in most members of GH18-B group, suggesting a high degree of divergence between them.
3.3. Chromosomal Localization, Gene Duplication and Synteny Analyses
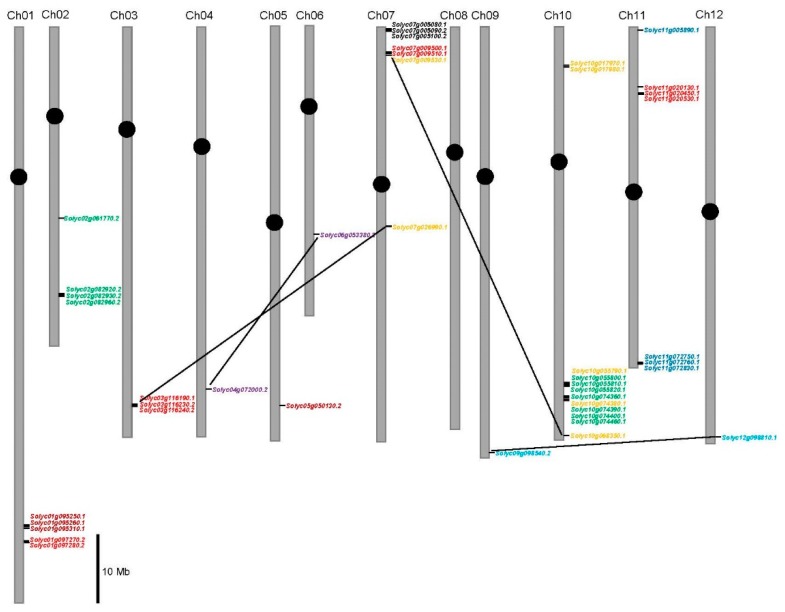
To further investigate the relationship between the genetic divergence and gene duplication of the tomato chitinase gene family, their chromosomal locations were determined. The results indicated that 43 tomato chitinase genes are located on 11 different chromosomes, and chromosome 8 does not contain any chitinase gene (Figure 2). Among the identified chitinase genes, twelve are present on chromosome 10; seven on chromosomes 7 and 11; five on chromosome 1; four on chromosome 2; three on chromosome 3; and one on each of chromosomes 4, 5, 6, 9, and 12 (Figure 2). Furthermore, several chitinase gene clusters containing 34 tandem members were found on chromosomes 1, 2, 3, 7, 10, and 11. Consequently, over 79% of tomato chitinase genes originate from tandem duplication. Four pairs of paralogous chitinase genes (Solyc03g116190.1-Solyc07g026990.1, Solyc04g072000.2-Solyc06g053380.2, Solyc07g009530.1-Solyc10g068350.1, and Solyc09g098540.2-Solyc12g098810.1) were putative segmental duplication events (Figure 2).
Figure 2.
Positions of the chitinase family genes on the tomato chromosomes. Scale stands for a 10 Mb chromosomal distance. Segmental duplicate genes are linked by a black line. Chitinase members of different groups are marked with different colors: GH19-A members in green; GH19-B in saffron yellow; GH19-C in purple; GH19-D in light blue; GH19-E in red; GH18-A in black; GH18-B in dark blue; and GH18-C in deep red, respectively.
To better understand the evolutionary history of the chitinase family, the Ks values were used to estimate the duplication timing of 16 pairs of paralogous genes (Table 2). The Ks values of the tomato chitinase paralogues ranged from 0.01677 to 1.45233, suggesting that these duplication events occurred approximately 0.56–48.41 million years ago (Mya). Furthermore, duplication events were found in eight of 16 pairs of paralogous chitinase genes to have occurred at approximately 11.91–27.94 Mya. This period coincided with the recent large-scale duplication event of the tomato genome [50], implying that the expansion of the tomato chitinase gene family mainly occurred during the recent large-scale duplication event period. The initial (Solyc07g005080.1–Solyc07g005090.2) and the final (Solyc10g017970.1–Solyc07g017980.1) tandem duplication occurred at about 48.41 and 0.56 Mya on the chitinase cluster region of the tomato chromosomes 7 and 10, respectively.
Table 2.
Inference of duplication time of chitinase paralogous pairs in tomato. Ka and Ks were calculated with K-Estimator 6.0 [49]. The approximate date of the duplication event was inferred from the formula (T = Ks/2λ) as described in methods.
| Paralogous Pairs | Ka | Ks | Ka/Ks | Data (Mya) | Duplication Style | |
|---|---|---|---|---|---|---|
| Solyc10g055810.1 | Solyc10g055820.1 | 0.07390 | 0.35721 | 0.20688 | 11.91 | Tandem duplication |
| Solyc10g074400.1 | Solyc10g074460.1 | 0.07391 | 0.17341 | 0.42622 | 5.78 | Tandem duplication |
| Solyc02g061770.2 | Solyc02g082960.2 | 0.18678 | 0.83819 | 0.22284 | 27.94 | Tandem duplication |
| Solyc02g082920.2 | Solyc02g082930.2 | 0.17407 | 0.63165 | 0.27579 | 21.06 | Tandem duplication |
| Solyc10g055790.1 | Solyc10g074380.1 | 1.35815 | 0.70977 | 1.91351 | 23.66 | Tandem duplication |
| Solyc10g017970.1 | Solyc10g017980.1 | 0 | 0.01667 | 0 | 0.56 | Tandem duplication |
| Solyc01g097270.2 | Solyc01g097280.2 | 0.09717 | 0.57283 | 0.16963 | 19.09 | Tandem duplication |
| Solyc07g009500.1 | Solyc07g009510.1 | 0 | 0.14145 | 0 | 4.72 | Tandem duplication |
| Solyc11g020450.1 | Solyc11g020530.1 | 0.05676 | 0.06623 | 0.85701 | 2.21 | Tandem duplication |
| Solyc11g072750.1 | Solyc11g072760.1 | 0.07842 | 0.2463 | 0.31839 | 8.21 | Tandem duplication |
| Solyc07g005080.1 | Solyc07g005090.2 | 0.46577 | 1.45233 | 0.32071 | 48.41 | Tandem duplication |
| Solyc01g095250.1 | Solyc01g095260.1 | 0.01671 | 0.14434 | 0.11577 | 4.81 | Tandem duplication |
| Solyc03g116190.1 | Solyc07g026990.1 | 1.30202 | 1.25040 | 1.04128 | 41.68 | Transposition |
| Solyc07g009530.1 | Solyc10g068350.1 | 1.10203 | 0.77691 | 1.41848 | 25.89 | Transposition |
| Solyc04g072000.2 | Solyc06g053380.2 | 0.18915 | 0.72624 | 0.26045 | 24.21 | Transposition |
| Solyc09g098540.2 | Solyc12g098810.1 | 0.14047 | 0.67931 | 0.20678 | 22.64 | Transposition |
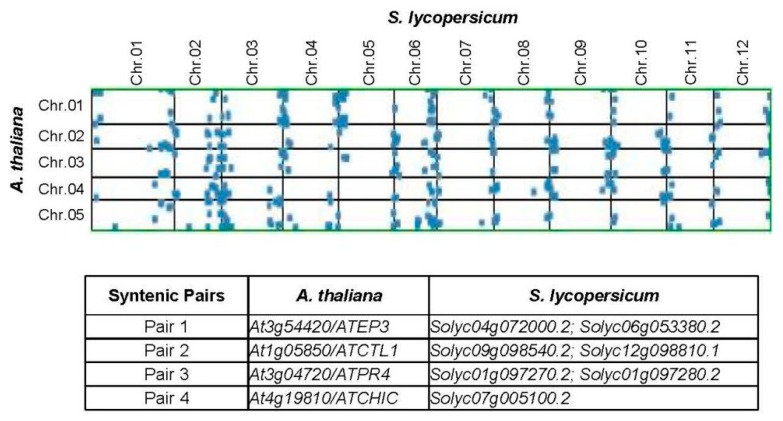
Identification of potential orthologue genes contributes to the preliminary understanding of the chitinases in other species. Next, synteny analyses were performed between Arabidopsis and tomato genomes using the SynMap server [51]. Four ortholog pairs were identified in syntenic blocks (Figure 3). Solyc04g072000.2 is microsyntenic to Solyc06g053380.2 and syntenic to At3g54420 (ATEP3) (ortholog pair 1). At1g05850 (ATCTL1) is orthologous to Solyc09g098540.2 and Solyc12g098810.1 (ortholog pair 2). ATCTL1 is involved in both lignin accumulation and root system architecture [58,59]. Solyc09g098540.2 and Solyc12g098810.1 belong to GH19-D and are likely connected to the development of plant root cell wall, since At3g16920 (ATCTL2) is clustered in this clade [60]. In addition, both Solyc01g097270.2 and Solyc097280.2 are syntenic to At3g04720 (ATPR4), which controls the resistance to pathogen infection [61].
Figure 3.
Synteny analyses of the chitinase genes in Arabidopsis and tomato genomes. Four syntenic paralogs were found and also marked with * for pairs 1, ** for pairs 2, *** for pairs 3, and **** for pairs 4 in Figure 1.
3.4. Selective Pressure Analysis among Different Amino Acid Sites
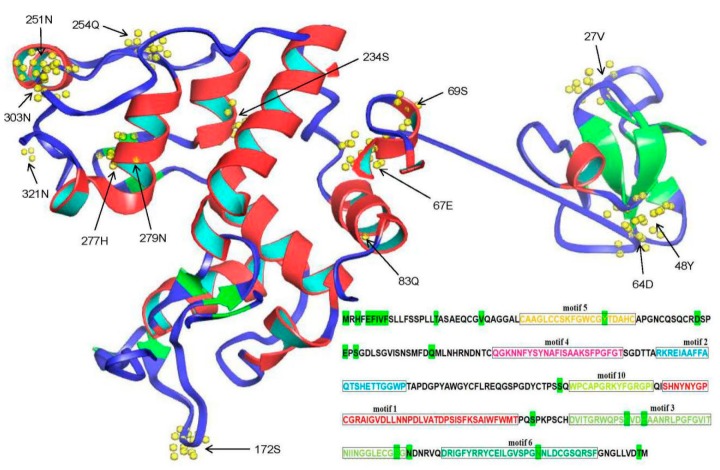
During the process of evolution, positive selection sites often affect the protein structure and their function [62]. Phylogenetic results indicated that eight groups were generated after chitinase duplication in the three investigated plant species. To further explore which amino acid change was subjected to positive selection after duplication, variable Ka/Ks values were investigated among different chitinase sites in each group. The results indicated that the Ka/Ks values differed for each group (Table 3). Compared to the values for the others, the Ka/Ks values are comparatively higher in GH19-B, GH19-E, and GH18-A, indicating a faster evolutionary rate within the three groups (Table 3). Despite these differences, all Ka/Ks values are below 1, suggesting that the chitinase proteins are under purifying selection. However, some positive selection sites, such as chitinase members in GH19-A and GH19-B (as predicted by the M5 model) and GH19-E, GH18-A, and GH18-C (as predicted by the M5 and M8 models), were also found in this analysis (Table 3). However, no positively selected sites were found with the M7 selection model. As an example, a detailed distribution position of the predicted positive selection sites in GH19-A via the M5 model is shown in Figure 4. Twenty-two sites were found to be under positive selection. Among them, eight sites were located in the signal peptide region of the chitinase protein (Solyc10g055820.1). Since the signal peptides will be removed in the mature protein, the tertiary structure of this chitinase does not contain this segment. Therefore, only 14 predicted amino acid sites are shown here (Figure 4). Further analyses indicated that six sites were located in α-helixes and five in β sheets. As described above, seven conserved motifs (motifs 5, 4, 2, 10, 1, 3, and 6) were found in most members of the GH19-A group (Figure 1). Of the 22 predicted positive selection sites, only five sites were located in the conserved motifs, containing three sites (251N, 254Q, and 277H) in motif 3 and one site (48Y and 303N) in motifs 5 and 6, respectively. In summary, more than 77% of the positive selection sites are located in the less conserved region of the chitinase protein. To further investigate whether these positive selection sites can cause changes in protein stability, I-Mutant2.0 [54] was used to estimate the effects of point mutations on the protein folding stability on the chitinase protein (Solyc10g055820.1). As shown in Table S4, most of the detected sites under positive selection can decrease the stability of the chitinase protein.
Table 3.
Likelihood values and parameter estimates of the chitinase genes. Three evolutionary modes—M8 (beta + w ≥ 1), M7 (beta), and M5 (gamma)—were used in this analysis. PSS: positive selection site.
| Branches | Models | Ka/Ks | Log-Likelihood | Number of PSS |
|---|---|---|---|---|
| GH19-A | M8 | 0.3927 | −15690.8 | 0 |
| M7 | 0.3748 | −15691.3 | 0 | |
| M5 | 0.4004 | −15694.2 | 22 | |
| GH19-B | M8 | 0.4587 | −2023.77 | 0 |
| M7 | 0.5158 | −2024.59 | 0 | |
| M5 | 0.6367 | −2029.54 | 6 | |
| GH19-C | M8 | 0.3059 | −9827.03 | 0 |
| M7 | 0.2989 | −9829.36 | 0 | |
| M5 | 0.3199 | −9831.15 | 0 | |
| GH19-D | M8 | 0.2091 | −5411.06 | 0 |
| M7 | 0.2046 | −5414.26 | 0 | |
| M5 | 0.2182 | −5409.51 | 0 | |
| GH19-E | M8 | 0.6195 | −6088.14 | 10 |
| M7 | 0.4399 | −6099.04 | 0 | |
| M5 | 0.5484 | −6091.2 | 13 | |
| GH18-A | M8 | 0.5305 | −15342.2 | 47 |
| M7 | 0.4669 | −15380.5 | 0 | |
| M5 | 0.5576 | −15367.2 | 57 | |
| GH18-B | M8 | 0.3478 | −5749.57 | 0 |
| M7 | 0.3589 | −5751.15 | 0 | |
| M5 | 0.3503 | −5749.08 | 0 | |
| GH18-C | M8 | 0.3696 | −20704.8 | 16 |
| M7 | 0.3409 | −20745.7 | 0 | |
| M5 | 0.3923 | −20736.3 | 33 |
Figure 4.
Distribution of positive selection sites of chitinase members in CH19-A group predicted by M5 model. Solyc10g055820.1 was used to predict the tertiary structure of the chitinase protein with the Phyre2 Server [53]. Predicted positive selection amino acid sites are displayed and marked with arrowheads or bright green. In addition, distribution of the seven conserved motifs are also marked different colors here.
3.5. Divergent Expression Profile of Tomato Chitinase Genes in Different Tissues
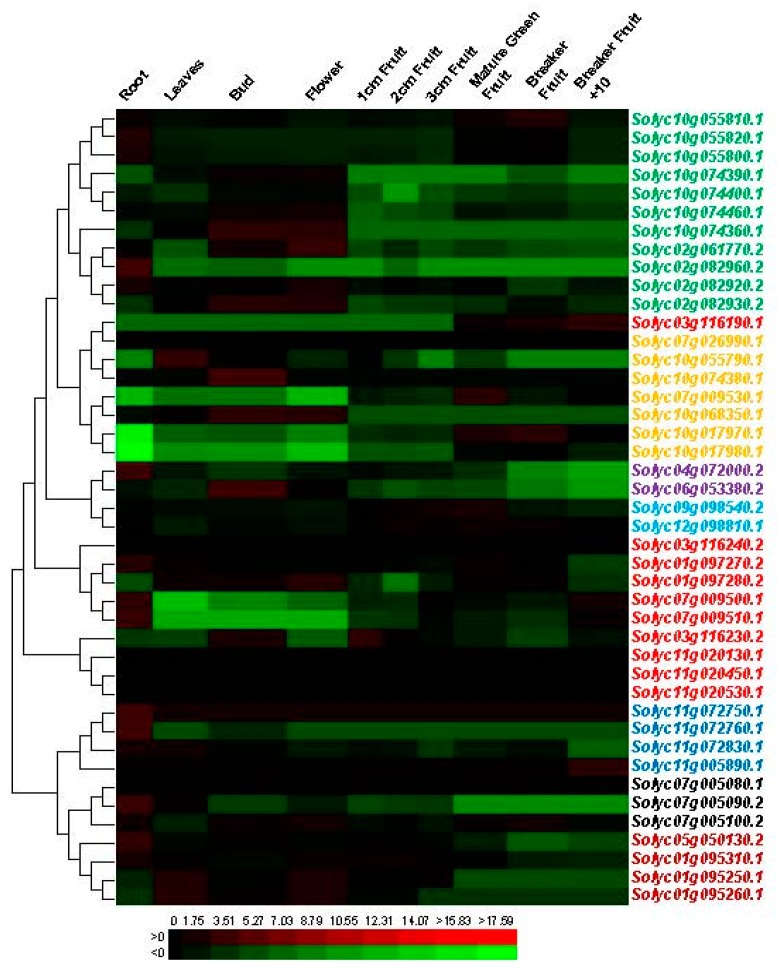
To further understand the roles of specific chitinase genes in different tomato tissues, we investigated the expression profiles of these genes using available transcriptome data (GSE33507). Transcription profiles of the chitinase genes were collected and analyzed in 10 different tissues (Figure 5; Table S5). The results showed that the chitinase genes showed diverse expression profiles among the different tissues, suggesting distinct roles for a variety of developmental stages. Several chitinase genes were significantly abundant in some tissues. For instance, Solyc10g074360.1, Solyc02g082930.2, Solyc10g068350.1, and Solyc06g053380.2 transcripts accumulated more during the bud and flower stages than in the other tissues. The highest level of Solyc02g082960.2, Solyc04g072000.2, Solyc11g072760.1 and Solyc07g005090.2 gene transcripts are found in the root, whereas expression of the Solyc02g082930.2, Solyc11g005890.1, and Solyc07g009530.1 was highest at the fruit development stage. As described above, 16 pairs of paralogous chitinase genes in tomato were identified to originate from duplication (Table 2). We also examined the expression profiles of these paralogous genes and found that none of them showed similar expression patterns (Figure 5).
Figure 5.
Expression patterns of the chitinase genes in different developmental stages of tomato. The Log2 ratio of the sample signal to control signal was used to indicate its relative expression level. Heat maps reflect the strength of relative expression. Members of different groups are marked with different colors (please refer to Figure 2).
3.6. Divergent Expression Profile of Tomato Chitinase Genes under Various Abiotic and Biotic Stresses
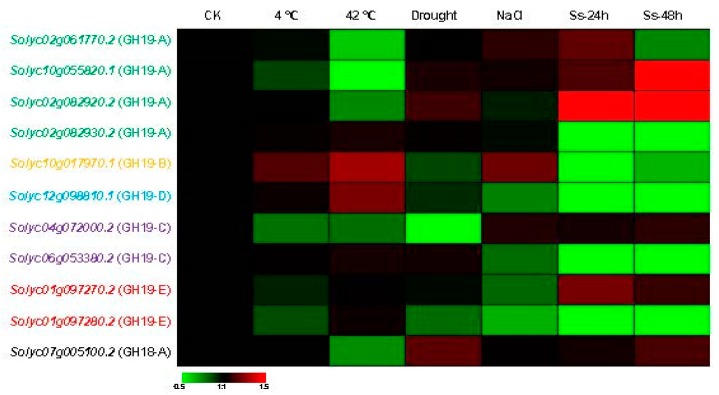
To further determine the involvement of tomato chitinase genes in the response to various stresses, we further analyzed the expression patterns of 11 genes under low temperature (4 °C), high temperature (42 °C), drought, salt, and S. sclerotiorum stresses via qRT-PCR (Figure 6; Table S6). The expression levels of seven and five tested chitinase genes were enhanced under low and high temperature stress, respectively. More than half of the analyzed tomato chitinase genes showed a higher expression level under drought stress. Under salt treatment, 10 of the 11 selected tomato chitinase genes displayed elevated expression, while Solyc01g097280.2 showed to be downregulation. For S. sclerotiorum infection, the expression levels of more than 64 percent detected chitinase genes increased during this pathogen stress, and this trend was particularly evident after 48 h of treatment (Figure 6). Interestingly, the Solyc10g017970.1 gene was upregulated under low temperature, high temperature, salt, and S. sclerotiorum stresses, suggesting that this chitinase gene may be involved in these biotic and abiotic responses. The Solyc04g072000.2 gene showed an enhancement of the transcript level under salt and S. sclerotiorum stresses and the expression level of this gene decreased in response to the other three stresses, implying that the Solyc04g072000.2 gene may be involved in the salt and pathogen tolerance of the tomato. In addition, the functional divergence of duplicated genes was also investigated. Two pairs of duplicated chitinase genes (Solyc02g082920.2-Solyc02g082930.2 and Solyc01g097270.2-Solyc01g097280.2) did not show similar expression patterns in response to these stresses. For examples, high temperature and pathogen infection can induce the expression of Solyc02g082930.2 gene but decrease the transcript level of the Solyc02g082920.2 gene.
Figure 6.
Expression profiles of the eleven tomato chitinase genes under different biotic and abiotic stresses by qRT-PCR. The fold change between the treated sample and the control sample as the relative expression level. Heat maps reflect the strength of relative expression. Ss stands for S. sclerotiorum. Chitinase genes of different groups are marked with different colors (please refer to Figure 2).
To further explore the potential regulatory mechanism of the chitinase genes responsive abiotic stress in tomato, Arabidopsis and rice, we also identified their cis-elements using the PLACE [56]. A few abiotic response regulatory elements were found here. They are cold responsive element (S000407), low temperature-responsive element (S000157), heat shock element (S0000030), cold/drought-responsive element (S000153), drought-responsive element (S000414 and S000174), salt- and pathogen-responsive element (S000453) and pathogen-responsive element (S000390). The results indicated that all chitinase genes contained multiple cis-elements in their promoters (Table S7), suggesting these abiotic stresses regulated their expression. And we cannot find any same or highly similar distribution of these regulatory elements among paralogs, implying divergent expression patterns in the duplicated genes.
3.7. Protein–Protein Interaction Network Analysis of the Chitinases in Tomato
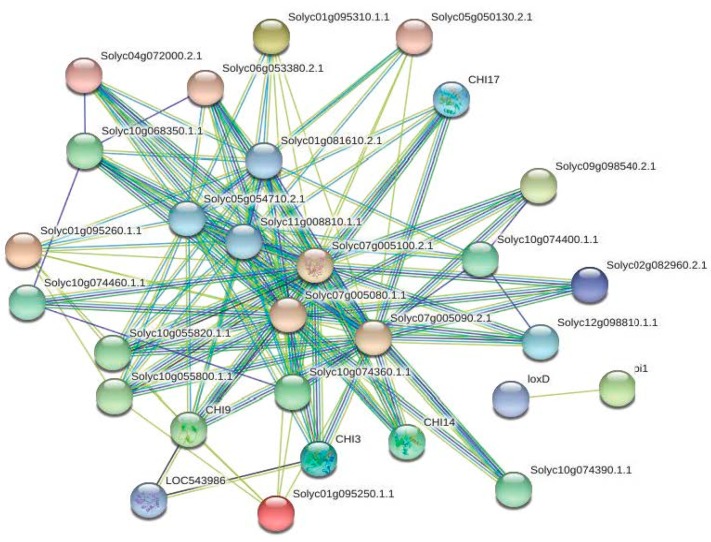
To further investigate which proteins can potentially interact with members of the chitinase family, a protein interaction network was assembled with the STRING database [57]. The network was based on either experimental or predicted interactions. As a result, 24 of the 43 tomato chitinases appeared in the network, exhibiting 124 interactions by a total of 29 unique genes (Figure 7; Table S8). Among these, 95 interactions occurred between the tomato chitinase proteins and Solyc07g005080.1 and Solyc07g005100.2 interacted with other 20 chitinases (Figure 7; Table S8). Beta-hexosaminidase, lipoxygenase and beta-1,3-glucanase were predicted as the main interaction partners of tomato chitinases. For instances, three beta-hexosaminidases (Solyc01g081610.2, Solyc05g054710.2, and Solyc11g008810.1.1) were predicted to interact with chitinases (CHI3: Solyc02g082920.2; CHI9: Solyc10g055810.1; CHI14: Solyc02g061770.2; CHI17: Solyc02g082930.2). Solyc01g097270.2 (pi1) was predicted to interact with loxD (Solyc03g122340.2) in tomato. Furthermore, one tomato beta-1,3-glucanase protein (LOC543986: Solyc01g008620.2) was observed to interact with both CHI3 (Solyc02g082920.2) and CHI9 (Solyc10g055810.1). These results implied functional diversity of tomato chitinases.
Figure 7.
Functional network assembly of the tomato chitinase genes. 124 interactions were exhibited among a total of 29 genes.
4. Discussion
In this study, tomato chitinase genes were identified and compared with data in Arabidopsis and rice. Phylogenetic analyses divided them into eight groups. This is different from previous classification of chitinase proteins that were divided six groups (Classes I–VI) according to their sequence similarity [7]. We also aligned the tomato chitinase protein sequences, and found some signatures of GH18 and GH19 family and catalytic or binding residues (Figure S1). By comparing our data with previous classifications [7,63], we found that GH18-A and GH18-C members in this study belong to the Class V and Class III, respectively. Chitinase members in GH19-A and GH19-C have the catalytic residues, they should be classified as the Class I. CH19-D members should belong to the Class II because they do not contain chitin-binding domain. Gene organization and conserved motif distribution provide additional evidences for the evolutionary relationships of multigene families [64,65]. Intron gain and loss can increase the complexity of gene organization throughout evolution [66]. The diversification of the exon–intron organization of chitinase family genes among different groups may contribute to the structural diversity of this family during plant evolution. The structural diversity of chitinase proteins (Figure 1; Table S3) can also mediate a number of biological processes such as stress responses, growth, and developmental operations, which has led to neofunctionalization [11]. Moreover, a similar motif composition was found in each group, suggesting functional relevance. A previous study has shown that the DxDxE sequence of motif 11 is important for the catalysis in family GH-18 enzymes [67]. At the same time, this diverse motif composition implies functional diversification among different groups.
Chromosomal localization and gene duplication studies showed that 79% of tomato chitinase genes originated from tandem duplication (Figure 2). In other words, tandem duplication predominated in tomato chitinase gene expansion during evolution. This is similar to the results of Arabidopsis and rice, where most of the chitinase genes were also reported to originate from tandem duplication events [29]. Duplication of chromosomal segments can amplify the number of family genes, while the duplication of family genes (such as tandem duplication) can also affect the genetic composition of the chromosomal segments. Over the course of evolution, the duplicated genes will change their functions, i.e., genetic divergence of the duplication genes will occur throughout evolution [68]. Next, synteny analyses between the Arabidopsis and tomato genomes were also found several potential orthologue genes: ATEP3, ATCTL1, ATCTL2, and ATPR4 (Figure 3). The transcript of the ATEP3 gene was accumulated rapidly after inoculation with Xanthomonas campestris, suggesting that ATEP3 protein is involved in the initial events of the hypersensitive reaction [17]. However, specific abiotic stresses did not significantly affect ATEP3 gene expression [28]. Based on this, both of these chitinases in tomato might be responsible for host defense against pathogens, but not for abiotic stress tolerance. ATCTL1 is involved in both lignin accumulation and root system architecture [58,59]. In addition, ATCTL2 and ATPR4 are connected to the development of the plant root cell wall and the resistance to pathogen infection, respectively [60,61]. In this study, both Solyc09g098540.2 and Solyc12g098810.1 were found to cluster with ATCTL2, and Solyc01g097270.2 and Solyc097280.2 are syntenic to ATPR4. Therefore, these identified tomato chitinases might have functions in cell wall architecture and disease resistance, which requires further experimental verification.
Positively selected amino acid sites can alter protein structure and function in evolution [50]. Some studies have indicated that purifying selection promotes subfunctionalization of the duplicated genes, and positive selection accelerates neofunctionalization of the duplicated genes [66,68]. In this study, we found that the selective pressure from different chitinase groups are different (Table 3), and several positive selection sites located at different protein positions were also identified (Figure 4). It suggests that selection spurs the amino acid diversity at some residues. Interestingly, we also found that most positive selection sites were located to the variable regions of chitinase proteins, implying that these sites might change the protein structures and thus increase their function divergence.
Furthermore, the divergent expression profile of tomato chitinase genes was investigated in different tissues and under some biotic and abiotic stresses (Figure 5 and Figure 6), suggesting this gene family may be involved in a variety of physiological processes [69,70]. For duplicated genes, a change of expression pattern is one of the characteristics of the functional divergence. Several studies have reported that different transcription patterns occurred in duplicated genes [66,71]. In our study, the diverse expression profile of the chitinase genes might be the result of subfunctionalization or neofunctionalization processes in tomato. Over the process of biological evolution, one copy of these duplicated genes may acquire new functions, while the other may maintain the original function. Alternatively, both copies may undergo subfunctionalization and are uniquely expressed in different tissues or under different stresses [72,73]. Over the process of evolution, the loss or increase of specific domains (such as CBD) may cause functional changes in the family protein. Consequently, the chitinase family protein can not only hydrolyze chitin, but also be involved in the growth and development of plants as described above. The number increase and functional divergence of chitinase family genes are an adaptation of organisms to external environmental changes. All of these findings suggest that functional divergence has occurred among these duplicated genes, and that the products of these genes might play different roles in the tomato development or in a specific stress response. In addition, these 11 genes were distributed in six of the eight identified groups with different motif distributions (Figure 1). The structural difference may affect their functions.
Network analysis of the tomato chitinases indicated that most interactions occurred between the tomato chitinase proteins (Figure 7; Table S8), indicating that some chitinase genes may be neighborhood, co-occurrence, or coexpressed. Such a joint participation characteristic of the tomato chitinase genes is of great significance for the defense response against pathogens and for the regulation of growth and development. Additionally, several beta-hexosaminidases, lipoxygenases, and beta-1,3-glucanases were predicted as the main interaction partners of tomato chitinases (Figure 7; Table S8). Beta-hexosaminidase is an important glycosidase, which is involved in important signal transduction, cell division, and cell integrity events [74,75]. Arabidopsis beta-hexosaminidase (AtHEXO1) can generate paucimannosidic N-glycans on vacuolar glycoproteins, while AtHEXO3 functions on secreted glycoproteins [76,77]. They are involved in hydrolysis processes of fruit ripening [78,79]. In this study, three beta-hexosaminidases were predicted to interact with CHI3, CHI9, CHI14, and CHI17, suggesting that the chitinase family proteins may be involved in the molecular function of beta-hexosaminidases in tomato. A similar result has indicated that tomato beta-hexosaminidase can hydrolyze chitin under acidic conditions [79]. Lipoxygenases catalyze the dioxygenation of polyunsaturated fatty acids to form hydroperoxides participating in leaf senescence, synthesis of jasmonic acid, and defense responses to both abiotic and biotic stress [80,81,82]. Solyc01g097270.2 (pi1) was predicted to interact with loxD (Solyc03g122340.2) in this study, implying the coordination or relevance of their functions as described above. As members of the pathogenesis-related protein 2 (PR-2) family, beta-1,3-glucanase catalyzes the hydrolytic cleavage of the beta-1,3-D glucosidic linkages in beta-1,3-glucans. It is well known that plant beta-1,3-glucanase plays important roles in various stress responses as well as developmental processes [83,84]. Interestingly, in our network analysis, one beta-1,3-glucanase protein (LOC543986: Solyc01g008620.2) was observed to interact with both CHI3 and CHI9 (Figure 7). These results indicated the diversity of chitinase binding proteins, which was helpful to understand their functional roles.
5. Conclusions
In this study, a comparative analysis of the chitinase gene family was performed. Eight groups were divided via phylogenetic analyses. Exon–intron structure and motif distribution were highly conserved in each group, implying their functional conservation. Chromosomal localization indicated that tandem duplication is the main way for plant chitinases expansion in evolution. Synteny analyses suggest functional relevance. Selection analyses identified several significant site-specific selective constraints acting on most chitinase paralogs after gene duplication, leading to functional divergence. Differential expression patterns of the chitinase genes suggested functional divergence, especially for the duplicated genes. Moreover, functional network analyses also identified several potential correlated genes. This study will provide useful insights for further functional investigation of this family gene.
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/8/3/52/s1, Table S1: Primers using in this study, Table S2: Distribution of the chitinase genes in different species and groups, Table S3: Sequence consensus of 12 motif identified in this study, Table S4: The effects of point mutations on folding stability on the chitinase protein (Solyc10g055820.1), Table S5: The raw transcriptome data (GSE33507) used in this study to investigate the expression profiles of the chitinase genes in different tomato tissues, Table S6: Raw data for the qRT-PCR in this study, Table S7: cis-elements analysis of the promoters in the chitinase genes. Table S8: Interaction data generated by STRING in this study, Figure S1: Alignment of the tomato chitinase proteins. Chitinase names of different groups in our classification are marked with different colors: GH19-A members in green; GH19-B in saffron yellow; GH19-C in purple; GH19-D in light blue; GH19-E in red; GH18-A in black; GH18-B in dark blue; and GH18-C in deep red, respectively (refer to Figure 2). Chitinase 18 signature is marked with bright green background. Two chitinase 19 signatures are marked with grey and red background. Catalytic residues of chitinase 18 are written in red. Crystallyn signature is marked with yellow background. C-terminal extension for vacuolar targeting is marked with blue background. Pink stands for the catalytic activity of Class I chitinases.
Author Contributions
J.C. designed and supervised the experiments, J.C. and X.T. carried out the experiments. J.C. wrote the manuscript. All authors read and approved the manuscript.
Funding
This project is supported by grants from the Jiangsu University “Youth Backbone Teacher Training Project”.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Debono M., Gordee R.S. Antibiotics that inhibit fungal cell wall development. Annu. Rev. Microbiol. 1994;48:471–497. doi: 10.1146/annurev.mi.48.100194.002351. [DOI] [PubMed] [Google Scholar]
- 2.Day R.B., Okada M., Ito Y., Tsukada K., Zaghouani H., Shibuya N., Stacey G. Binding site for chitin oligosaccharides in the soybean plasma membrane. Plant Physiol. 2001;126:1162–1173. doi: 10.1104/pp.126.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaku H., Nishizawa Y., Ishii-Minami N., Akimoto-Tomiyama C., Dohmae N., Takio K., Minami E., Shibuya N. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA. 2006;103:11086–11091. doi: 10.1073/pnas.0508882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu T., Liu Z., Song C., Hu Y., Han Z., She J., Fan F., Wang J., Jin C., Chang J., et al. Chitin-induced dimerization activates a plant immune receptor. Science. 2012;336:1160–1164. doi: 10.1126/science.1218867. [DOI] [PubMed] [Google Scholar]
- 5.Hadwiger L.A., Beckman J.M. Chitosan as a Component of Pea-Fusarium solani Interactions. Plant Physiol. 1980;66:205–211. doi: 10.1104/pp.66.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohnuma T., Numata T., Osawa T., Mizuhara M., Lampela O., Juffer A.H., Skriver K., Fukamizo T. A class V chitinase from Arabidopsis thaliana: Gene responses, enzymatic properties, and crystallographic analysis. Planta. 2011;234:123–137. doi: 10.1007/s00425-011-1390-3. [DOI] [PubMed] [Google Scholar]
- 7.Neuhaus J.M., Fritig B., Linthorst H.J.M., Meins F., Mikkelsen J.D., Ryals J. A revised nomenclature for chitinase genes. Plant Mol. Biol. Rep. 1996;14:102–104. doi: 10.1007/BF02684897. [DOI] [Google Scholar]
- 8.Liu G., Wei X., Qin Y., Qu Y. Characterization of the endoglucanase and glucomannanase activities of a glycoside hydrolase 45 protein from Penicillium decumbens 114-2. J. Gen. Appl. Microbiol. 2010;56:223–229. doi: 10.2323/jgam.56.223. [DOI] [PubMed] [Google Scholar]
- 9.Berglund L., Brunstedt J., Nielsen K.K., Chen Z., Mikkelsen J.D., Marcker K.A. A proline-rich chitianse from Beta vulgaris. Plant Mol. Biol. 1995;27:211–216. doi: 10.1007/BF00019193. [DOI] [PubMed] [Google Scholar]
- 10.Tyler L., Bragg J.N., Wu J., Yang X., Tuskan G.A., Vogel J.P. Annotation and comparative analysis of the glycoside hydrolase genes in Brachypodium distachyon. BMC Genom. 2010;11:600. doi: 10.1186/1471-2164-11-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kesari P., Patil D.N., Kumar P., Tomar S., Sharma A.K., Kumar P. Structural and functional evolution of chitinase-like proteins from plants. Proteomics. 2015;15:1693–1705. doi: 10.1002/pmic.201400421. [DOI] [PubMed] [Google Scholar]
- 12.Herget T., Schell J., Schreier P.H. Elicitor-specific induction of one member of the chitinase gene family in Arachis hypogaea. Mol. Gen. Genet. 1990;224:469–476. doi: 10.1007/BF00262442. [DOI] [PubMed] [Google Scholar]
- 13.Bowles D.J. Defense-related proteins in higher plants. Annu. Rev. Biochem. 1990;59:873–907. doi: 10.1146/annurev.bi.59.070190.004301. [DOI] [PubMed] [Google Scholar]
- 14.Zhao J., Wang J., An L., Doerge R.W., Chen Z.J., Grau C.R., Meng J., Osborn T.C. Analysis of gene expression profiles in response to Sclerotinia sclerotiorum in Brassica napus. Planta. 2007;227:13–24. doi: 10.1007/s00425-007-0586-z. [DOI] [PubMed] [Google Scholar]
- 15.Shrestha C.L., Ona I., Muthukrishnan S., Mew T.W. Chitinase levels in rice cultivars correlate with resistance to the sheath blight pathogen Rhizoctonia solani. Eur. J. Plant Pathol. 2007;120:69–77. doi: 10.1007/s10658-007-9199-4. [DOI] [Google Scholar]
- 16.Wang J., Tian N., Huang X., Chen L.Y., Schlappi M., Xu Z.Q. Tall fescue turf grass class I chitinase is activated by fungal elicitors, dehydration ethylene and mechanical wounding. Plant Mol. Biol. 2009;27:305–314. doi: 10.1007/s11105-008-0086-8. [DOI] [Google Scholar]
- 17.Gerhardt L.B., Sachetto-Martins G., Contarini M.G., Sandroni M., de Lima V.M., Cordeiro M.C., de Oliveira D.E., Margis-Pinheiro M. Arabidopsis thaliana class IV chitinase is early induced during the interaction with Xanthomonas campestris. FEBS Lett. 1997;419:69–75. doi: 10.1016/S0014-5793(97)01332-X. [DOI] [PubMed] [Google Scholar]
- 18.Zhu Y., Maher E.A., Masoud S., Dixon R.A., Lamb C.J. Enhanced protection against fungal attack by constitutive co-expression of chitinase and glucanase genes in transgenic tobacco. Biotechnology. 1994;12:807–812. doi: 10.1038/nbt0894-807. [DOI] [Google Scholar]
- 19.Zhong R., Kays S.J., Schroeder B.P., Ye Z.H. Mutation of a chitinase-like gene causes ectopic deposition of lignin, aberrant cell shapes, and overproduction of ethylene. Plant Cell. 2002;14:165–179. doi: 10.1105/tpc.010278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu J., Wang Y., Kim S.G., Jung K.H., Gupta R., Kim J., Park Y., Kang K.Y., Kim S.T. A secreted chitinase-like protein (OsCLP) supports root growth through calcium signaling in Oryza sativa. Physiol. Plant. 2017;161:273–284. doi: 10.1111/ppl.12579. [DOI] [PubMed] [Google Scholar]
- 21.Wu B., Zhang B., Dai Y., Zhang L., Shang-Guan K., Peng Y., Zhou Y., Zhu Z. Brittle Culm15 encodes a membrane-associated chitinase-like protein required for cellulose biosynthesis in rice. Plant Physiol. 2012;159:1440–1452. doi: 10.1104/pp.112.195529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts J.A., Whitelaw C.A., Gonzalez-Carranza Z.H., McManus M.T. Cell separation processes in plants—Models, mechanisms and manipulation. Ann. Bot. 2000;86:223–235. doi: 10.1006/anbo.2000.1203. [DOI] [Google Scholar]
- 23.Eriksson E.M., Bovy A., Manning K., Harrison L., Andrews J., De Silva J., Tucker G.A., Seymour G.B. Effect of the colorless non-ripening mutation on cell wall biochemistry and gene expression during tomato fruit development and ripening. Plant Physiol. 2004;136:4184–4197. doi: 10.1104/pp.104.045765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agusti J., Merelo P., Cercos M., Tadeo F.R., Talon M. Ethylene-induced differential gene expression during abscission of citrus leaves. J. Exp. Bot. 2008;59:2717–2733. doi: 10.1093/jxb/ern138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerhardt L.B., Magioli C., Perez A.B.U.C.M., Margis R., Martins G.S., Pinheiro M.M. Atchit IV gene expression is stimulated under abiotic stresses and is spatially and temporally regulated during embryo development. Genet. Mol. Biol. 2004;27:118–123. doi: 10.1590/S1415-47572004000100020. [DOI] [Google Scholar]
- 26.Passarinho P.A., Van Hengel A.J., Fransz P.F., de Vries S.C. Expression pattern of the Arabidopsis thaliana AtEP3/AtchitIV endochitinase gene. Planta. 2001;212:556–567. doi: 10.1007/s004250000464. [DOI] [PubMed] [Google Scholar]
- 27.Van Loon L.C., Rep M., Pieterse C.M. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006;44:135–162. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- 28.Takenaka Y., Nakano S., Tamoi M., Sakuda S., Fukamizo T. Chitinase gene expression in response to environmental stresses in Arabidopsis thaliana: Chitinase inhibitor allosamidin enhances stress tolerance. Biosci. Biotechnol. Biochem. 2009;73:1066–1071. doi: 10.1271/bbb.80837. [DOI] [PubMed] [Google Scholar]
- 29.Grover A. Plant chitinases: Genetic diversity and physiological roles. Crit. Rev. Plant Sci. 2012;31:57–73. doi: 10.1080/07352689.2011.616043. [DOI] [Google Scholar]
- 30.Wu X.F., Wang C.L., Xie E.B., Gao Y., Fan Y.L., Liu P.Q., Zhao K.J. Molecular cloning and characterization of the promoter for the multiple stress-inducible gene BjCHI1 from Brassica juncea. Planta. 2009;229:1231–1242. doi: 10.1007/s00425-009-0911-9. [DOI] [PubMed] [Google Scholar]
- 31.Lopez R.C., Gomez-Gomez L. Isolation of a new fungi and woundinduced chitinase class in corms of Crocus sativus. Plant Physiol. Biochem. 2009;47:426–434. doi: 10.1016/j.plaphy.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 32.Bekesiova B., Hraska S., Libantova J., Moravcikova J., Matusikova I. Heavy-metal stress induced accumulation of chitinase isoforms in plants. Mol. Biol. Rep. 2008;35:579–588. doi: 10.1007/s11033-007-9127-x. [DOI] [PubMed] [Google Scholar]
- 33.Tiffin P. Comparative evolutionary histories of chitinase genes in the Genus zea and Family poaceae. Genetics. 2004;167:1331–1340. doi: 10.1534/genetics.104.026856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mondragón-Palomino M., Meyers B.C., Michelmore R.W., Gaut B.S. Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res. 2002;12:1305–1315. doi: 10.1101/gr.159402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bishop J.G., Dean A.M., Mitchell-Olds T. Rapid evolution in plant chitinases: Molecular targets of selection in plant-pathogen coevolution. Proc. Natl. Acad. Sci. USA. 2000;97:5322–5327. doi: 10.1073/pnas.97.10.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klee H.J., Giovannoni J.J. Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 2011;45:41–59. doi: 10.1146/annurev-genet-110410-132507. [DOI] [PubMed] [Google Scholar]
- 37.Schauer N., Semel Y., Balbo I., Steinfath M., Repsilber D., Selbig J., Pleban T., Zamir D., Fernie A.R. Mode of inheritance of primary metabolic traits in tomato. Plant Cell. 2008;20:509–523. doi: 10.1105/tpc.107.056523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cuartero J., Bolarin M.C., Asins M.J., Moreno V. Increasing salt tolerance in the tomato. J. Exp. Bot. 2006;57:1045–1058. doi: 10.1093/jxb/erj102. [DOI] [PubMed] [Google Scholar]
- 39.Danhash N., Wagemakers C.A., van Kan J.A., de Wit P.J. Molecular characterization of four chitinase cDNAs obtained from Cladosporium fulvum-infected tomato. Plant Mol. Biol. 1993;22:1017–1029. doi: 10.1007/BF00028974. [DOI] [PubMed] [Google Scholar]
- 40.Tomato Genome Consortium The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Letunic I., Doerks T., Bork P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43:D257–D260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., et al. Pfam: The protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker J.M. The Proteomics Protocols Handbook. Humana Press; New York, NY, USA: 2007. pp. 571–607. [Google Scholar]
- 45.Emanuelsson O., Brunak S., von Heijne G., Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 46.Edgar R.C. MSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bailey T.L., Williams N., Misleh C., Li W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Comeron J.M. K-Estimator: Calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics. 1999;15:763–764. doi: 10.1093/bioinformatics/15.9.763. [DOI] [PubMed] [Google Scholar]
- 50.Blanc G., Wolfe K.H. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004;16:1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haug-Baltzell A., Stephens S.A., Davey S., Scheidegger C.E., Lyons E. SynMap2 and SynMap3D: Web-based whole-genome synteny browsers. Bioinformatics. 2017;33:2197–2198. doi: 10.1093/bioinformatics/btx144. [DOI] [PubMed] [Google Scholar]
- 52.Stern A., Doron-Faigenboim A., Erez E., Martz E., Bacharach E., Pupko T. Selecton 2007: Advanced models for detecting positive and purifying selection using a Bayesian inference approach. Nucleic Acids Res. 2007;35:W506–W511. doi: 10.1093/nar/gkm382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Capriotti E., Fariselli P., Casadio R. I-Mutant2.0: Predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33:W306–W310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 56.Higo K., Ugawa Y., Iwamoto M., Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hermans C., Porco S., Verbruggen N., Bush D.R. Chitinase-like protein CTL1 plays a role in altering root system architecture in response to multiple environmental conditions. Plant Physiol. 2010;152:904–917. doi: 10.1104/pp.109.149849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hossain M.A., Noh H.N., Kim K.I., Koh E.J., Wi S.G., Bae H.J., Lee H., Hong S.W. Mutation of the chitinase-like protein-encoding AtCTL2 gene enhances lignin accumulation in dark-grown Arabidopsis seedlings. J. Plant Physiol. 2010;167:650–658. doi: 10.1016/j.jplph.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 60.Sánchez-Rodríguez C., Bauer S., Hématy K., Saxe F., Ibáñez A.B., Vodermaier V., Konlechner C., Sampathkumar A., Rüggeberg M., Aichinger E., et al. Chitinase-like1/pom-pom1 and its homolog CTL2 are glucan-interacting proteins important for cellulose biosynthesis in Arabidopsis. Plant Cell. 2012;24:589–607. doi: 10.1105/tpc.111.094672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qi L., Yan J., Li Y., Jiang H., Sun J., Chen Q., Li H., Chu J., Yan C., Sun X., et al. Arabidopsis thaliana plants differentially modulate auxin biosynthesis and transport during defense responses to the necrotrophic pathogen Alternaria brassicicola. New Phytol. 2012;195:872–882. doi: 10.1111/j.1469-8137.2012.04208.x. [DOI] [PubMed] [Google Scholar]
- 62.Morgan C.C., Shakya K., Webb A., Walsh T.A., Lynch M., Loscher C.E., Ruskin H.J., O’Connell M.J. Colon cancer associated genes exhibit signatures of positive selection at functionally significant positions. BMC Evol. Biol. 2012;12:114. doi: 10.1186/1471-2148-12-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tobias P.A., Christie N., Naidoo S., Guest D.I., Külheim C. Identification of the Eucalyptus grandis chitinase gene family and expression characterization under different biotic stress challenges. Tree Physiol. 2017;37:565–582. doi: 10.1093/treephys/tpx010. [DOI] [PubMed] [Google Scholar]
- 64.Hou Z., Cao J. Comparative study of the P2X gene family in animals and plants. Purinergic Signal. 2016;12:269–281. doi: 10.1007/s11302-016-9501-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cao J., Li X., Lv Y. Dynein light chain family genes in 15 plant species: Identification, evolution and expression profiles. Plant Sci. 2017;254:70–81. doi: 10.1016/j.plantsci.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 66.Cao J., Li X. Identification and phylogenetic analysis of late embryogenesis abundant proteins family in tomato (Solanum lycopersicum) Planta. 2015;241:757–772. doi: 10.1007/s00425-014-2215-y. [DOI] [PubMed] [Google Scholar]
- 67.Vaaje-Kolstad G., Houston D.R., Rao F.V., Peter M.G., Synstad B., van Aalten D.M., Eijsink V.G. Structure of the D142N mutant of the family 18 chitinase ChiB from Serratia marcescens and its complex with allosamidin. Biochim. Biophys. Acta. 2004;1696:103–111. doi: 10.1016/j.bbapap.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 68.Moore R.C., Purugganan M.D. The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 2005;8:122–128. doi: 10.1016/j.pbi.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 69.Kashyap P., Deswal R. A novel class I Chitinase from Hippophae rhamnoides: Indications for participating in ICE-CBF cold stress signaling pathway. Plant Sci. 2017;259:62–70. doi: 10.1016/j.plantsci.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 70.Malolepszy A., Kelly S., Sørensen K.K., James E.K., Kalisch C., Bozsoki Z., Panting M., Andersen S.U., Sato S., Tao K., et al. A plant chitinase controls cortical infection thread progression and nitrogen-fixing symbiosis. Elife. 2018;7:e38874. doi: 10.7554/eLife.38874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li X., Cao J. Late embryogenesis abundant (LEA) gene family in maize: Identification, evolution, and expression profiles. Plant Mol. Biol. Rep. 2016;34:15–28. doi: 10.1007/s11105-015-0901-y. [DOI] [Google Scholar]
- 72.Lynch M., Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cao J. Molecular Evolution of the Vacuolar Iron Transporter (VIT) Family Genes in 14 Plant Species. Genes. 2019;10:144. doi: 10.3390/genes10020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Altmann F., Schwihla H., Staudacher E., Glössl J., März L. Insect cells contain an unusual, membrane-bound β-N-acetylglucosaminidase probably involved in the processing of protein N-glycans. J. Biol. Chem. 1995;270:17344–17349. doi: 10.1074/jbc.270.29.17344. [DOI] [PubMed] [Google Scholar]
- 75.Wells L., Whelan S.A., Hart G.W. O-GlcNAc: A regulatory post-translational modification. Biochem. Biophys. Res. Commun. 2003;302:435–441. doi: 10.1016/S0006-291X(03)00175-X. [DOI] [PubMed] [Google Scholar]
- 76.Liebminger E., Veit C., Pabst M., Batoux M., Zipfel C., Altmann F., Mach L., Strasser R. β-N-acetylhexosaminidases HEXO1 and HEXO3 are responsible for the formation of paucimannosidic N-glycans in Arabidopsis thaliana. J. Biol. Chem. 2011;286:10793–10802. doi: 10.1074/jbc.M110.178020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Castilho A., Windwarder M., Gattinger P., Mach L., Strasser R., Altmann F., Steinkellner H. Proteolytic and N-glycan processing of human α1-antitrypsin expressed in Nicotiana benthamiana. Plant Physiol. 2014;166:1839–1851. doi: 10.1104/pp.114.250720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jagadeesh B.H., Prabha T.N. β-hexosaminidase, an enzyme from ripening bell capsicum (Capsicum annuum var variata) Phytochemistry. 2002;61:295–300. doi: 10.1016/S0031-9422(02)00181-4. [DOI] [PubMed] [Google Scholar]
- 79.Jagadeesh B.H., Prabha T.N., Srinivasan K. Multiple forms of β-hexosaminidase in ripening tomato (Lycopersicum esculantum L.) Indian J. Plant Physiol. 2006;11:13–19. [Google Scholar]
- 80.Andreou A., Feussner I. Lipoxygenases—Structure and reaction mechanism. Phytochemistry. 2009;70:1504–1510. doi: 10.1016/j.phytochem.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 81.Shen J., Tieman D., Jones J.B., Taylor M.G., Schmelz E., Huffaker A., Bies D., Chen K., Klee H.J. A 13-lipoxygenase, TomloxC, is essential for synthesis of C5 flavour volatiles in tomato. J. Exp. Bot. 2014;65:419–428. doi: 10.1093/jxb/ert382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Springer A., Kang C., Rustgi S., von Wettstein D., Reinbothe C., Pollmann S., Reinbothe S. Programmed chloroplast destruction during leaf senescence involves 13-lipoxygenase (13-LOX) Proc. Natl. Acad. Sci. USA. 2016;113:3383–3388. doi: 10.1073/pnas.1525747113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vaddepalli P., Fulton L., Wieland J., Wassmer K., Schaeffer M., Ranf S., Schneitz K. The cell wall-localized atypical β-1,3 glucanase ZERZAUST controls tissue morphogenesis in Arabidopsis thaliana. Development. 2017;144:2259–2269. doi: 10.1242/dev.152231. [DOI] [PubMed] [Google Scholar]
- 84.Mestre P., Arista G., Piron M.C., Rustenholz C., Ritzenthaler C., Merdinoglu D., Chich J.F. Identification of a Vitis vinifera endo-β-1,3-glucanase with antimicrobial activity against Plasmopara viticola. Mol. Plant Pathol. 2017;18:708–719. doi: 10.1111/mpp.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.