Abstract
Glial cell line-derived neurotrophic factor (GDNF) promotes the survival of postnatal—but not embryonic—mouse dorsal root ganglion cells in vitro, despite the fact that its receptors are expressed at both ages. To understand this difference, we have performed an oligonucleotide microarray experiment. We found that several hundred genes were regulated between embryonic and postnatal stages, and that several important classes of genes were differentially regulated by GDNF treatment, including genes related to translation and to phenotypic specification and maturation. Interestingly, a set of genes related to cell adhesion, cytoskeleton and cellular morphology were consistently down-regulated by GDNF, suggesting a previously uncharacterized role for GDNF in repressing neurite growth and/or branching. This nuclear program initiated by GDNF was functionally confirmed in cultures of embryonic wild-type neurons sustained with nerve growth factor and in bax−/− neurons that survive in the absence of trophic support.
Keywords: neurotrophic‖microarray
Glial cell line-derived neurotrophic factor (GDNF) is the prototypic member of a transforming growth factor-β subfamily that includes neurturin, persephin, and artemin. Originally isolated as a trophic factor for midbrain dopaminergic neurons (1), it has recently been implicated in processes as diverse as kidney formation, neuronal survival, enteric nervous system (ENS) precursor proliferation, and neuromuscular junction plasticity. The GDNF-family receptors are multicomponent complexes formed by the tyrosine kinase receptor Ret and one of the GDNF family receptor (GFR)-α glycosylphosphatidylinositol (GPI)-linked receptors, which determines ligand specificity (2–7).
GFRα1 and to a lesser extent GFRα2 have been shown to be the major determinants of GDNF signaling through Ret (8). GDNF and GFRα1 are expressed throughout the nervous system and peripheral organs during development, with prominent expression in the basal ganglia, thalamic nucleus, cerebellum, and spinal cord, as well as in skin, whisker pad, kidney, stomach, skeletal muscle, and testis (9–11).
GDNF exerts a variety of effects at different developmental stages and on many different cell types. In enteric as well as motor neurons, GDNF stimulates neurite growth (12, 13), causes hyperinnervation (14), leads to a rapid enhancement of transmitter release at neuromuscular junctions (15), and its receptors GFRα1 and GFRα2 are rapidly regulated by depolarization in sympathetic, parasympathetic, and sensory neurons (16), all suggesting the importance of activity-dependent plasticity of the GDNF response during development. In the developing ENS, GDNF also has a strong mitogenic effect on ENS precursor cells, and GDNF-null mutant mice lack ENS neurons. GDNF plays an inductive role in the epithelial mesenchymal interactions necessary for the metanephric kidney development by regulating proliferation and branching (17, 18). Gene deletions show that GDNF, Ret, and GFRα1 are all required for kidney formation and ENS development (19–23). In dorsal root ganglia (DRG), a subset of small cells expressing all of the receptor components for GDNF signaling (Ret and GFRα1 as well as GFRα2) are rescued by GDNF treatment after sciatic nerve injury (24), and GDNF also enhances the survival of cultured postnatal DRG neurons (25).
We examined the differential trophic effects of GDNF on DRGs at embryonic and postnatal stages and analyzed the transcriptional program in response to GDNF stimulation by using oligonucleotide microarrays and real-time PCR with a data mining tool integrating diverse sources of information.
Materials and Methods
Nonradioactive in Situ Hybridization.
In situ hybridization was performed as described (25), with probes as described in (26).
GDNF Stimulation of Intact Ganglia.
DRG were dissected from embryonic day (E)14 and postnatal day (P)14 mice and incubated in Ham's F14 (Imperial Chemical Industries) supplemented with 2 mM glutamine/0.35% bovine serum albumin/60 ng/ml progesterone/16 μg/ml putrescine/400 ng/ml l-tyroxine/38 ng/ml sodium selenite/340 ng/ml triiodo-thyronine/60 μg/ml penicillin/100 μg/ml streptomycin. After 6 h, 20 ng/μl recombinant GDNF protein was added to one group, and all samples were incubated 2 h more. The DRGs were then snap-frozen on dry ice.
Microarray Hybridization.
Total RNA, isolated by using RNeasy (Qiagen, Chatsworth, CA), was converted to cDNA and biotinylated cRNA produced according to the manufacturer's instructions (Affymetrix, Santa Clara, CA). Each sample gave 20–30 μg of total RNA which resulted in 3–5 μg cDNA. One microgram was used to produce about 20–30 μg labeled cRNA.
The biotinylated cRNA was hybridized to Mu19K and Mu11K gene chips at 45°C for 16 h, automatically washed and stained on a fluidics station, and then scanned on a GeneArray scanner (all from Affymetrix). The reported average difference between match and mismatch probes was used as an indicator of mRNA abundance. To compensate for differences in overall hybridization signal, each chip was normalized to have a mean of 100. After normalization, the mRNA levels of actin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were 7,600 ± 1,400 and 3,300 ± 470, respectively (mean ± SD). Normalization reduced the SDs of these probes from around 90% of the mean to about 15%, showing that the normalization procedure worked.
Real-Time PCR.
Primer design.
Primer pairs were designed for 85 UniGene clusters. The full-length mRNA sequence was used wherever possible. Each pair was aimed at the 3′-most 500 bps of the transcript; each was designed to yield a short amplicon (50–150 bp) and to have a melting temperature of about 59°C.
Amplification.
cDNA (1–2 μg) was diluted to 500 μl. For each reaction, 1 μl of diluted cDNA was mixed with 300 pmol of each primer in 25 μl (final volume) SYBR Green Master Mix (Applied Biosystems). Real-time PCR was performed in an ABI 5700 (Applied Biosystems) with the following thermal profile: 5 min at 50°C, 10 min at 94°C, followed by 40 cycles of 30 s at 94°C and 30 s at 60°C. All amplifications were run in triplicate.
Controls.
Intron-specific primers or mock cDNA synthesis (omitting reverse transcriptase) were used to control for genomic contamination. A no-template control was performed for each primer pair. After each amplification, a melting curve was obtained, and any primer pair that showed multiple peaks was removed from further analysis. Actin-β was selected as endogenous control based on a comparison of six different potential controls GAPDH, acidic ribosomal phosphoprotein PO (ARPB), hypoxanthine phosphoribosyltransferase (HPRT), TATA box-binding protein (TBP), and Trf receptor (TRFR) across the four samples. In addition, PCR products were gel-separated to confirm a band of the expected size.
Calculations.
ΔCT was computed by subtracting CT (the number of cycles to reach threshold) for actin from each primer pair CT. The expression level for each gene expressed in units of actin-β was then taken as 2−ΔCT. For convenience, these units were normalized so that the actin mRNA level was 10,000. Standard deviations were generally smaller than 20% of the mean, and 2.5-fold differences were generally significant (P < 0.05 by Student's t test).
Survival and Neurite Growth Assays.
C57/Bl6 mice were used for the survival and neurite-length assays, and bax−/− animals were used for the neurite-polarity assay. Approximately 100 DRGs were dissected and collected in PBS/glucose for the survival assay; for the neurite-length and -polarity assays, individual DRGs were used. Otherwise the procedures were similar. To dissociate the neurons, the DRGs were incubated at 37°C with trypsin/DNase 0.05% (GIBCO/BRL and Sigma). After removal of the trypsin solution, the ganglia were washed once with DMEM/10% (vol/vol) heat-inactivated horse serum and washed twice with defined medium (see below). The ganglia then were gently triturated with a fire-polished Pasteur pipette to give a single-cell suspension. Nonneuronal cells were eliminated by preplating. The neurons were plated on 24-well plates (Nunc) precoated with polyornithine (0.5 mg/ml for 30 min; Sigma) and laminin (20 μg/ml, overnight; GIBCO/BRL) in a defined medium (Ham's F14 supplemented with 2 mM glutamine/0.35% BSA/60 ng/ml progesterone/16 μg/ml putrescine/400 ng/ml l-tyroxine/38 ng/ml sodium selenite/340 ng/ml triiodo-thyronine/60 μg/ml penicillin/100 μg/ml streptomycin). For the neurite-length and -polarity assays, neurons were plated at low density. Nerve growth factor (NGF; neurite length assay) was added at 10 ng/ml, and GDNF was added at 10 ng/ml. The primary cultures were maintained for 48 h (survival and neurite-length assay) or 72 h (neurite-polarity assay) at 37°C in a humidified incubator under 5% CO2. After 4 h of incubation, the neurons were recognized clearly by their morphology under phase-contrast optics, and the number of plated neurons was counted. Neurons were stained with antineurofilament (200 kDa) polyclonal antibody (Affiniti Research Products, Exeter, U.K.) at 1:500 by using the Vectastain ABC kit (Vector Laboratories) according to the manufacturer's instructions. For measurements of GFRα1 and Ret positive neurons, cultures were stained with a Rabbit anti-GFRα1 antibody (1:300; ref. 27) or rabbit anti-Ret antibody (2 μg/ml, Immunobiologicals, Lisle, IL). Bax+/− mice were bred to produce bax+/+, bax+/−, and bax−/− offspring. The plug date was considered E0. Animals were genotyped by PCR (primer sequences have been published; ref. 28). In the neurite-length assay, the length of the longest neurite was measured with a grid and plotted in a frequency histogram by unit length.
Results
We selected developing DRG as a model of neurotrophic action. To determine at what ages GDNF could act on these ganglia, we first performed in situ hybridization for the two receptors (Ret and GFRα1) required for GDNF activation of downstream tyrosine kinase signaling. Second, we examined the trophic actions of recombinant GDNF on cultured ganglia at two selected ages. Third, we analyzed the transcriptional program in response to GDNF stimulation at the two ages with oligonucleotide microarrays and real-time PCR. And fourth, prompted by the results of the microarray hybridization, we analyzed the effects of GDNF on neurite growth and branching in vitro.
Differential Actions of GDNF at Embryonic and Adult Stages.
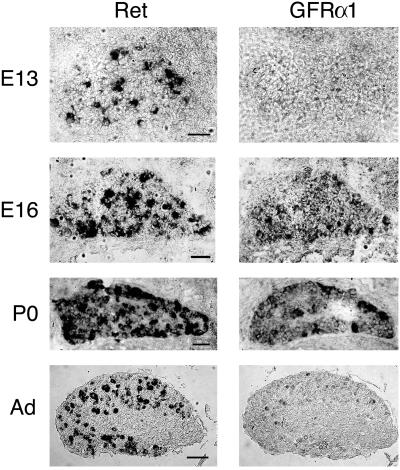
By using in situ hybridization, we found that the receptors for GDNF were expressed in DRGs from early embryonic stages through adulthood. As shown in Fig. 1, Ret tyrosine kinase receptor mRNA was widely expressed in DRG from E13 to P0 (as shown, ref. 25), extending to adult stages. mRNA for the coreceptor GFRα1 was expressed similarly, although in adult ganglia the expression was limited to a restricted subset of cells, as has been shown (24). Thus, the two receptors required for GDNF action were present in the ganglia throughout the ages studied. We selected E14 and P14 ganglia for further study because they seemed to be at opposite extremes of a changing pattern of expression.
Figure 1.
Nonradioactive in situ hybridization for the detection of Ret and GFRα1 mRNAs in developing and postnatal murine DRGs at the indicated stages. Ret was highly expressed during development, at birth, and in the adult (Ad). GFRα1 was expressed in a diffuse pattern at E13, but was up-regulated at E16 and remained relatively high at birth. Only a subpopulation of neurons expressed ret and/or GFRα1 in the adult. Thus, both GDNF receptors were present throughout the ages studied in this work, albeit in varying patterns and levels. (Bars = 100 μm.)
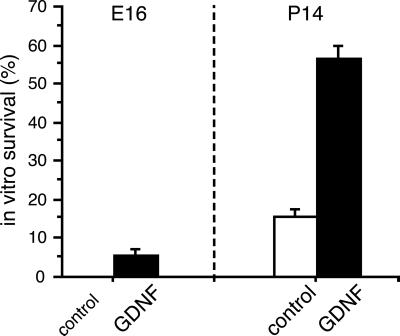
Dissociated DRG neurons from C57/Bl6 mice were cultured in serum-free defined medium. Under these conditions, most cells from both embryonic and postnatal ganglia died within 48 h in vitro in the absence of trophic support (25). A total of 60% of the postnatal cells, but only 5% of the embryonic cells, were rescued by the addition of recombinant GDNF, as shown in Fig. 2. Thus, although receptors for GDNF were present at both ages, embryonic cells did not respond to GDNF treatment by any significant survival.
Figure 2.
Sensory neuron survival after GDNF treatment. Dissociated DRG neurons were cultured for 48 h in the presence or absence of GDNF. At E16, only 5% of the cells were rescued by the growth factor (Left), despite the fact that receptors for GDNF were present. In contrast, almost 60% of the neurons were rescued by GDNF at P14 (Right), showing that, at this age, GDNF is a true neurotrophic factor for these cells and indicating that their intrinsic phenotype or identity must have changed during maturation. Error bars show standard deviations (n = 3).
We then designed an oligonucleotide microarray and real-time PCR experiment to explore patterns of gene expression underlying the different effects of GDNF on embryonic and postnatal DRGs.
Oligonucleotide Microarray Experiment.
We used a strategy of gene discovery based on the pairwise comparison of samples after stimulation with GDNF. Intact DRGs were dissected from embryonic and postnatal animals and cultured for 6 h in serum-free defined medium. Recombinant GDNF then was added to half of the cultures, followed by an additional 2 h of incubation. RNA extracted from these cultures was labeled and hybridized to oligonucleotide microarrays containing ≈34,000 probe sets.
Probes showing more than 5-fold difference between unstimulated and stimulated cells at either age and with an absolute difference at least two times the mean probe intensity were selected for further analysis (n = 197, 0.9%). The selected genes were manually classified according to cellular role, and 85 genes were selected for real-time PCR confirmation. Of the 85 primer pairs, 9 were discarded for various reasons (amplification in no-template controls, imperfect melting curve, etc.).
In most cases, the magnitude of the regulation was significantly smaller by real-time PCR (the median-fold change was 1.75 for real-time PCR but 7.5 for the microarrays), perhaps because of a highly nonlinear response of the microarrays close to their detection limit. Twenty genes were consistently regulated more than 2-fold in both real-time PCR and microarray experiments, although several more seemed to be regulated by real-time PCR but with smaller magnitude compared with the microarray data. Table 1 summarizes the results. Down-regulation is indicated by negative numbers; confirmed absolute changes greater than 2-fold are in bold. Accession numbers to Unigene are indicated by a leading “Mm” and to The Institute for Genomic Research (TIGR) by “TC”. Fold changes apparently greater than ±20, which arose because of a transition from or to undetectable levels, are in parentheses to indicate this fact.
Table 1.
All GDNF-regulated genes successfully confirmed by real-time PCR
| E14 | P14 | Accession | Gene |
|---|---|---|---|
| −5.2 | 3.2 | Mm.297 | A-X actin |
| 1.0 | −19.1 | Mm.4968 | Translation elongation factor 2 |
| 2.1 | −5.1 | Mm.16323 | Translation initiation factor 4A2 |
| 1.2 | −12.0 | Mm.1391 | GATA-binding protein 2 |
| −5.8 | 6.9 | Mm.2344 | Guanine nucleotide binding protein β-1 |
| −8.4 | −1.7 | Mm.5079 | Hydroxysteroid 11-β dehydrogenase 2 |
| −1.5 | −7.9 | Mm.20354 | Kinesin light chain 1 |
| −9.9 | −17.8 | Mm.7362 | Lamin B2 |
| −9.3 | 3.1 | Mm.4071 | Laminin receptor 1 (LBP67) |
| −1.6 | −6.6 | TC35356 | Neuritin |
| 3.4 | (−38) | Mm.7414 | Neuron specific gene family member 1 |
| 1.6 | (−165) | Mm.3210 | Neuronatin |
| −4.1 | 5.6 | Mm.66 | Ribosomal protein S4, X-linked |
| −6.3 | −1.4 | Mm.1493 | SRY-box containing gene 10 (Sox10) |
| 1.1 | −13.6 | TC38163 | Tubulin β-4 |
| −4.4 | (403) | Mm.18538 | ESTs |
| (2362) | 15.0 | Mm.20417 | ESTs |
| −5.0 | 6.6 | Mm.2498 | ESTs |
| −5.8 | 2.8 | Mm.5063 | ESTs |
| 7.8 | 2.5 | Mm.30134 | 60S Ribosomal protein L44 |
Comparing Age-Regulated and GDNF-Regulated Transcripts.
By our criteria (more than 5-fold regulated and more than two times the mean), 310 genes (0.8%) changed between E14 and P14, showing that major transcriptional changes had taken place between the two ages.
Partly because of our selection criteria, these transcripts were more abundant than average genes, with a mean level of 422 ± 67 (mean ± 95% confidence interval; the mean of all genes was 100 by design). In contrast, the 195 genes (0.6%) changed by GDNF treatment at either age were less abundant, with a mean level of 261 ± 36. One explanation may be that developmentally regulated genes code for many structural and metabolic proteins that are normally expressed at high levels, whereas growth factor stimulation would preferentially act on less abundant cellular components such as transcription factors or signaling molecules.
Despite that only 0.8% and 0.6% of the genes were regulated by age or GDNF stimulation, respectively, a full 39% of the age-regulated genes also were regulated by GDNF. These genes had a mean abundance (291 ± 42) not significantly different from that of GDNF-regulated genes in general but much lower than that of age-regulated genes in general, suggesting that a low-abundance subset of the developmentally active genes were responsive to growth factor stimulation.
These figures were probably far short of the true extent of gene regulation by either age or trophic factor stimulation; in particular, the requirement that the absolute change must be two times the mean excluded a large majority of genes whose expression levels were too low to be assayed confidently by the microarrays. This conjecture was corroborated by the fact that when using real-time PCR, we found two genes (Δ-like homolog 1 and Ferritin light chain 2) apparently regulated in an opposite direction from that indicated by the microarrays but which would have otherwise passed our criteria. If 2.6% (2/76) is taken as an independent measure of the extent of GDNF gene regulation, a further 890 genes must have gone undetected by the microarrays.
Regulation by GDNF at E14 was not predictive of regulation by GDNF at P14 (Pearson's correlation coefficient = −0.12), confirming the differential effects of GDNF at the two ages. This result was expected both because the cells were different morphologically and in terms of gene expression profiles, but also because the subpopulation expressing GDNF receptors may have changed during DRG development.
Age-Regulated Genes.
The differential effects of GDNF at E14 and P14 could be caused by the changing phenotype of DRG cells during development. Indeed, over 300 genes changed between ages according to our stringent criteria. Among those confirmed by real-time PCR as down-regulated by age were Ncam—a neuron-specific cell-adhesion molecule—and cerebroglycan, a cell-surface heparan sulfate proteoglycan known to be transiently expressed by immature neurons and that typically disappears after axon outgrowth and after cell migration has been completed (29). This finding would suggest that E14 DRGs were dominated by young neurons still in the process of differentiation, whereas P14 DRGs had settled already in an adult phenotype.
Major Classes of GDNF-Activated Genes.
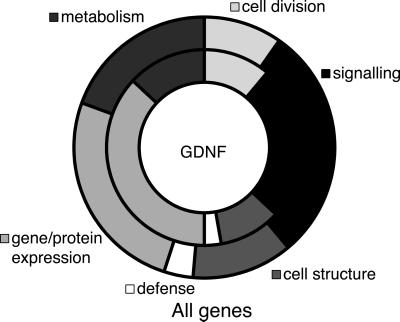
As expected for a growth factor, most of the genes activated by GDNF were either signaling genes or gene and protein expression-related genes, as shown in Fig. 3 (inner circle, GDNF-regulated genes). In the latter category, a disproportionate number of genes were involved in translation (ribosomal proteins, tRNA synthesis, and translation factors), suggesting a role for GDNF in a direct or indirect regulation of the translation machinery. For example, the ribosomal proteins S4 and L44 were both induced at P14, whereas elongation factor 2 and initiation factor 4a2 were down-regulated (confirmed by real-time PCR). Compared with a random sample of the probes present on the chip (Fig. 3, outer circle), gene/protein expression-related genes were much more common among the GDNF-regulated probes (+44%). In contrast, genes related to metabolism (−33%), cell structure (−19%) and cell defense (−20%) were all less common among the regulated genes.
Figure 3.
Classes of GDNF-regulated genes. We manually classified the 195 GDNF-regulated genes as well as 303 genes selected at random from the microarrays. The graph shows all of the major classes of genes in two aligned rings (inner ring, GDNF; outer ring, all genes). About half of the genes in either group were unknown and showed no useful homologies to previously described genes (omitted from graph). Among the rest, there was a clear bias toward gene/protein expression among the GDNF-regulated genes (+44% compared with all genes) at the expense of genes related to metabolism (−33%). There was also a smaller than average percentage of genes related to defense (−20%) and cell structure (−19%), whereas the remaining categories showed only small differences (<15%).
A particularly interesting target of Ret signaling discovered in this experiment was SRY-box containing gene 10 (Sox10), a transcription factor expressed in developing and maturing neural crest, glial cell precursors, and mature oligodendrocytes, which was down-regulated about 2-fold at both E14 and P14 (confirmed by real-time PCR). Sox10 has previously been shown to induce c-ret expression by binding with Pax3 to adjacent sites in the c-ret promoter (30), suggesting the existence of an autocrine or paracrine feedback loop.
A variety of cytoskeletal, structural, and cell adhesion-related genes, including lamin B2, kinesin light chain and neuritin (confirmed by real-time PCR), were down-regulated at both ages, suggesting a role for GDNF in regulating neuritogenesis and cellular morphology, in addition to its trophic action. Although tubulin β-4 and neuronatin were found down-regulated only at P14 in the microarray experiment (Table 1), real-time PCR indicated down regulation at both E14 and P14. In addition, A-X actin (31), an actin variant known to be expressed in melanoma cell lines, and a laminin-binding protein (LBP67) were down-regulated at E14. The fact that a range of genes involved in neurite growth were down-regulated enforces a concept of GDNF preventing neurite initiation and growth. It should be noted, however, that our results show that proportionally, fewer neurons express c-Ret postnatally than embryonically. Thus, the postnatal GDNF-dependent signal is expected to be diluted, compared with the early stage.
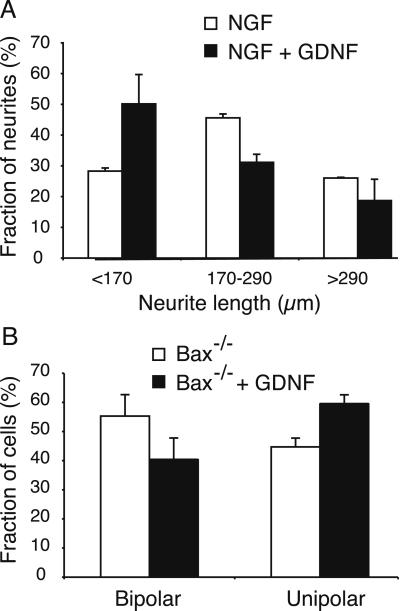
To find out whether this general down-regulation of cytoskeletal, structural, and cell adhesion-related genes had any functional consequences for these cells, we performed the following experiments. Dissociated embryonic sensory neurons from E12 bax−/− mice, which have been shown previously to survive indefinitely in culture in the absence of trophic support (28, 32, 33), were cultured in serum-free defined medium with or without the addition of recombinant GDNF. Immunohistological staining for GFRα1 or Ret showed that a majority of the neurons expressed detectable levels (61% and 84%, respectively), indicating that more than half of the cultured neurons would be expected to respond to GDNF. We then counted the number of neurons showing unipolar (single axon extended) or bipolar (bidirectional axons extended) morphologies. In contrast to the effects of other neurotrophic factors on sensory neurons (33), GDNF stimulation caused a marked decrease in the number of bipolar neurons, with a corresponding increase in the number of unipolar neurons. As shown in Fig. 4B, GDNF stimulation caused a significant reduction in the percentage of bipolar cells from 55% to 40% (P ≈ 0.01, by Student's t test). In an independent assay with wild-type mice, sensory neurons were cultured in the presence of NGF (10 ng/ml), which has both survival and neurite growth-promoting activities. The neurite length was scored and classified into three groups; those shorter than 170 μm, those between 170 and 290 μm, and those longer than 290 μm. GDNF caused a marked reduction of the length of the neurites (Fig. 4A). Both these results support a role for GDNF in suppressing neurite initiation and growth in these sensory neurons.
Figure 4.
Neurite growth assays. (A) Dissociated wild-type DRG neurons were maintained with NGF in the presence or absence of GDNF. The longest neurite was measured, and the percentages of neurites shorter than 170 μm, between 170–290 μm, and longer than 290 μm were plotted. With GDNF treatment, the percentage of cells containing shorter neurites increases markedly. Error bars show standard deviations (n = 2). (B) Dissociated bax−/− DRG neurons were cultured in the presence or absence of GDNF, and the number of neurons showing bipolar and unipolar morphologies was counted. With GDNF treatment, the percentage of cells with bipolar morphology was significantly reduced (P < 0.02 by Student's t test), as shown (Left). At the same time, the percentage of cells showing unipolar morphology increased (by P < 0.02). Together, these results indicate that GDNF has a suppressing effect on neurite branching or growth. Error bars show standard deviations (n = 3).
Discussion
We have found a differential response to GDNF stimulation of DRG neurons at embryonic and postnatal stages, despite the fact that its receptors are present at both ages—a difference that can be explained by the different gene-expression profiles of the two cell populations. Postnatal, but not embryonic, DRG neurons can be rescued from cell death in culture by the addition of GDNF. We have shown that during development, these cells undergo major changes in gene expression, including 5-fold changes of at least several hundred transcripts. We found differential responses to GDNF stimulation in that translation-related genes were affected mainly at P14, as was the transcription factor Sox10. In contrast, several cytoskeletal, structural, and cell adhesion-related genes were consistently down-regulated at both ages, suggesting a common mechanism acting to suppress neurite growth or branching and similar processes at the two ages. In cell culture, we found, by examining the morphology of DRG neurons after GDNF stimulation, that GDNF indeed has a suppressive effect on neurite growth and/or branching.
We have previously found that a dynamic spatial and temporal regulation of GDNF and its ligand-binding receptor within the follicle–sinus complex correlate with development of the distinct subclasses of sensory nerve endings (27). In that study, we also found that ligand and receptor association seem to be intricately linked to a local Schwann cell–axon interaction essential for sensory terminal formation. Based on these results, we hypothesized that GDNF plays a different role other than survival during embryogenesis, and that it could participate in terminal innervation. Our expression-profiling experiment confirms that GDNF plays distinct embryonic and postnatal roles. Interestingly, one of the major consistent findings was that a number of cytoskeletal, structural, and cell adhesion-related genes involved in neurite outgrowth were down-regulated. This down-regulation corresponded to a suppression of neurite growth and/or branching in cultures of bax−/− mice. GDNF has been shown to stimulate neuronal survival (25), elicit proliferation in the ENS and kidney (17, 18), and modulate synaptic plasticity (15). Our results reveal a previously uncharacterized activity of GDNF and identifies the nuclear program executing this response during nervous system development.
The factors implicated by our microarray hybridization support a wide variety of cytochemical responses and morphological changes. Neuronatin is a transmembrane transformation-suppression protein suggested to be a cell adhesion molecule or receptor that is down-regulated in PC12 cells upon NGF-induced differentiation (34). Lamin B2 is a member of the lamin family of nuclear envelope intermediate filaments that are involved in the reorganization of the nucleus during the cell cycle, and which have been shown to be regulated during cellular differentiation (35, 36). Tubulin β-4 is one isoform of the β-subunit of microtubules required for axonal growth and consequently is up-regulated during neurite growth. Kinesin light chain 1 forms two subunits of the kinesin heterotetramer, which is one of the motor proteins of the microtubular transport system in neurons, and has been shown to be necessary for axonal transport in Drosophila (37). Neuritin, finally, is a GPI-linked protein originally isolated from hippocampus, which is induced by activity and by the activity-regulated neurotrophins brain-derived neurotrophic factor (BDNF) and NT-3, and which causes neurite outgrowth and arborization in primary hippocampal and cortical cultures (38). Together, the consistent down-regulation of all of these factors embryonically and postnatally by GDNF support a suppressing effect of GDNF on neurite growth or branching, an effect that we were able to confirm in dissociated cultures of DRG neurons.
In addition to the above, both A-X actin and LBP67 (34/67 kDa laminin receptor) were down-regulated by GDNF exclusively in the embryonic DRG (confirmed by real-time PCR). Little is known of A-X actin except that it reduces invasiveness of melanoma cell lines when highly expressed by increased organization of stress fibers (31, 39). Laminin is a major component of basement membranes that strongly induces neurite initiation and outgrowth (40). The non-integrin LBP67 is involved in cell adhesion to laminin (41). Another laminin-binding protein (LBP110) has been shown to be expressed in neural crest cells as they colonize the bowel, and these neurons are known to be restricted in fate to either ENS or sympathetic neurons (42). The finding that LBP67 is expressed in the sensory lineage of the neural crest indicates a potential role for this protein in differentiation and neurite outgrowth of DRG neurons. Thus, the down-regulation of LBP67 by GDNF could have immediate effects on neurite outgrowth. It should be noted that trophic factor-independent neurite outgrowth was induced by culturing the neurons on laminin-coated dishes, thus supplying ligand for LBP67.
The loss of Ret function in humans leads to inherited disorders that arise because of neural crest defects. Congenital megacolon (Hirschsprung's disease) is characterized by the absence of enteric neurons in the colon (intestinal agangliosis) and is often caused by mutations in the Ret-receptor gene (43, 44). The symptoms of Waardenburg-Hirschsprung's disease (Waardenburg syndrome 4; WS4) are very similar to Hirschsprung's disease but also include deafness and pigmentary disorders. This disease has been genetically linked to mutations in the high mobility group (HMG) box transcription factor Sox10; Sox10 mouse mutants also show intestinal agangliosis and melanoblast deficits (45). A failure of Sox10 regulation of the basic helix-loop-helix transcription factor micropthalmia (MITF) gene has been linked to the melanocyte deficits of WS4 (46–48). In intestinal agangliosis, c-ret is a putative Sox10 target gene. Pax3 and Sox10 have adjacent binding sites in the c-ret enhancer and activate c-ret expression (30); a failure of c-ret expression leads to intestinal agangliosis (21). Our results show that Ret activation markedly down-regulates Sox10 both at early embryonic and postnatal stages (microarray data confirmed by real-time PCR) and, therefore, show the existence of a previously unknown regulatory feedback–inhibition mechanism. During normal development, both Sox10 and c-ret are expressed in the migrating neural crest (49, 50). Later in DRG development, Sox10 is down-regulated in neurons while the expression persists in Schwann cell precursors, where (it has been postulated) it participates in Schwann cell specification (49). In contrast, c-ret expression persists in neurons but not Schwann cells (26). Thus, c-ret expression and activation might participate in Sox10 down-regulation during development, in which case it could be postulated to play an essential role in determining neuron vs. glial phenotypes.
Acknowledgments
We thank Lennart Hansson, Anders Thelin, and the staff at the molecular biology unit, AstraZeneca, Mölndal, Sweden for continuous support and interest in this work. We thank Lotta Johansson for secretarial assistance. This research was supported by the Swedish Medical Research Council, the Swedish Cancer Society, AstraZeneca, and the European Union Biotechnology Program QLG3–1999-00602.
Abbreviations
- GDNF
glial cell line-derived neurotrophic factor
- ENS
enteric nervous system
- GFR
GDNF family receptor
- DRG
dorsal root ganglia
- En
embryonic day n
- Pn
postnatal day n
References
- 1.Lin L F, Doherty D, Lile J, Bektesh S, Collins F. Science. 1993;260:1130–1132. doi: 10.1126/science.8493557. [DOI] [PubMed] [Google Scholar]
- 2.Klein R D, Sherman D, Ho W H, Stone D, Bennett G L, Moffat B, Vandlen R, Simmons L, Gu Q M, Hongo J A, et al. Nature (London) 1997;387:717–721. doi: 10.1038/42722. [DOI] [PubMed] [Google Scholar]
- 3.Buj-Bello A, Adu J, Pinon L, Horton A, Thompson J, Rosenthal A, Chinchetru M, Buchman V L, Davies A M. Nature (London) 1997;387:721–724. doi: 10.1038/42729. [DOI] [PubMed] [Google Scholar]
- 4.Baloh R H, Tansey M G, Golden J P, Creedon D J, Heuckeroth R O, Keck C L, Zimonjic D B, Popescu N C, Johnson E M, Milbrandt J. Neuron. 1997;18:793–802. doi: 10.1016/s0896-6273(00)80318-9. [DOI] [PubMed] [Google Scholar]
- 5.Worby C A, Vega Q C, Zhao Y, Chao H, Seasholtz A F, Dixon J E. J Biol Chem. 1996;271:23619–23622. doi: 10.1074/jbc.271.39.23619. [DOI] [PubMed] [Google Scholar]
- 6.Trupp M, Arenas E, Fainzilber M, Nilsson A-S, Sieber B-A, Grigoriou M, Kilkenny C, Salazar-Grueso E, Pachnis V, Arumäe U, et al. Nature (London) 1996;381:785–789. doi: 10.1038/381785a0. [DOI] [PubMed] [Google Scholar]
- 7.Treanor J J S, Goodman L, de Sauvage F, Stone D M, Poulsen K T, Beck C D, Gray C, Armanini M P, Pollock R A, Hefti F, et al. Nature (London) 1996;382:80–83. doi: 10.1038/382080a0. [DOI] [PubMed] [Google Scholar]
- 8.Trupp M, Raynoschek C, Belluardo N, Ibanez C F. Mol Cell Neurosci. 1998;11:47–63. doi: 10.1006/mcne.1998.0667. [DOI] [PubMed] [Google Scholar]
- 9.Golden J P, DeMaro J A, Osborne P A, Milbrandt J, Johnson E M., Jr Exp Neurol. 1999;158:504–528. doi: 10.1006/exnr.1999.7127. [DOI] [PubMed] [Google Scholar]
- 10.Trupp M, Ryden M, Jornvall H, Funakoshi H, Timmusk T, Arenas E, Ibanez C F. J Cell Biol. 1995;130:137–148. doi: 10.1083/jcb.130.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trupp M, Belluardo N, Funakoshi H, Ibanez C F. J Neurosci. 1997;17:3554–3567. doi: 10.1523/JNEUROSCI.17-10-03554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schafer K H, Mestres P. Exp Brain Res. 1999;125:447–452. doi: 10.1007/s002210050702. [DOI] [PubMed] [Google Scholar]
- 13.Zurn A D, Winkel L, Menoud A, Djabali K, Aebischer P. J Neurosci Res. 1996;44:133–141. doi: 10.1002/(SICI)1097-4547(19960415)44:2<133::AID-JNR5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen Q T, Parsadanian A S, Snider W D, Lichtman J W. Science. 1998;279:1725–1729. doi: 10.1126/science.279.5357.1725. [DOI] [PubMed] [Google Scholar]
- 15.Ribchester R R, Thomson D, Haddow L J, Ushkaryov Y A. J Physiol (London) 1998;512:635–641. doi: 10.1111/j.1469-7793.1998.635bd.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doxakis E, Wyatt S, Davies A M. Development (Cambridge) 2000;127:1477–1487. doi: 10.1242/dev.127.7.1477. [DOI] [PubMed] [Google Scholar]
- 17.Heuckeroth R O, Lampe P A, Johnson E M, Milbrandt J. Dev Biol. 1998;200:116–129. doi: 10.1006/dbio.1998.8955. [DOI] [PubMed] [Google Scholar]
- 18.Pepicelli C V, Kispert A, Rowitch D H, McMahon A P. Dev Biol. 1997;192:193–198. doi: 10.1006/dbio.1997.8745. [DOI] [PubMed] [Google Scholar]
- 19.Pichel J G, Shen L, Sheng H Z, Granholm A C, Drago J, Grinberg A, Lee E J, Huang S P, Saarma M, Hoffer B J, et al. Nature (London) 1996;382:73–76. doi: 10.1038/382073a0. [DOI] [PubMed] [Google Scholar]
- 20.Sanchez M P, Silos-Santiago I, Frisen J, He B, Lira S A, Barbacid M. Nature (London) 1996;382:70–73. doi: 10.1038/382070a0. [DOI] [PubMed] [Google Scholar]
- 21.Schuchardt A, D'Agati V, Larsson-Blomberg L, Costantini F, Pachnis V. Nature (London) 1994;367:380–383. doi: 10.1038/367380a0. [DOI] [PubMed] [Google Scholar]
- 22.Enomoto H, Araki T, Jackman A, Heuckeroth R O, Snider W D, Johnson E M, Jr, Milbrandt J. Neuron. 1998;21:317–324. doi: 10.1016/s0896-6273(00)80541-3. [DOI] [PubMed] [Google Scholar]
- 23.Moore M W, Klein R D, Farinas I, Sauer H, Armanini M, Phillips H, Reichardt L F, Ryan A M, Carver-Moore K, Rosenthal A. Nature (London) 1996;382:76–79. doi: 10.1038/382076a0. [DOI] [PubMed] [Google Scholar]
- 24.Bennett D L, Michael G J, Ramachandran N, Munson J B, Averill S, Yan Q, McMahon S B, Priestley J V. J Neurosci. 1998;18:3059–3072. doi: 10.1523/JNEUROSCI.18-08-03059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baudet C, Mikaels A, Westphal H, Johansen J, Johansen T E, Ernfors P. Development (Cambridge) 2000;127:4335–4344. doi: 10.1242/dev.127.20.4335. [DOI] [PubMed] [Google Scholar]
- 26.Naveilhan P, Elshamy V M, Emfors P. Eur J Neurosci. 1997;9:1450–1460. doi: 10.1111/j.1460-9568.1997.tb01499.x. [DOI] [PubMed] [Google Scholar]
- 27.Fundin B T, Mikaels A, Westphal H, Ernfors P. Development (Cambridge) 1999;126:2597–2610. doi: 10.1242/dev.126.12.2597. [DOI] [PubMed] [Google Scholar]
- 28.White F A, Keller-Peck C R, Knudson C M, Korsmeyer S J, Snider W D. J Neurosci. 1998;18:1428–1439. doi: 10.1523/JNEUROSCI.18-04-01428.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stipp C S, Litwack E D, Lander A D. J Cell Biol. 1994;124:149–160. doi: 10.1083/jcb.124.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lang D, Chen F, Milewski R, Li J, Lu M M, Epstein J A. J Clin Invest. 2000;106:963–971. doi: 10.1172/JCI10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sadano H, Taniguchi S, Kakunaga T, Baba T. J Biol Chem. 1988;263:15868–15871. [PubMed] [Google Scholar]
- 32.Lentz S I, Knudson C M, Korsmeyer S J, Snider W D. J Neurosci. 1999;19:1038–1048. doi: 10.1523/JNEUROSCI.19-03-01038.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deckwerth T L, Elliott J L, Knudson C M, Johnson E M, Jr, Snider W D, Korsmeyer S J. Neuron. 1996;17:401–411. doi: 10.1016/s0896-6273(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 34.Joseph R, Tsang W, Dou D, Nelson K, Edvardsen K. Brain Res. 1996;738:32–38. doi: 10.1016/0006-8993(96)00768-8. [DOI] [PubMed] [Google Scholar]
- 35.Moir R D, Spann T P, Goldman R D. Int. Rev. Cytol. 1995. 141–82. [DOI] [PubMed] [Google Scholar]
- 36.Moir R D, Spann T P, Lopez-Soler R I, Yoon M, Goldman A E, Khuon S, Goldman R D. J Struct Biol. 2000;129:324–334. doi: 10.1006/jsbi.2000.4251. [DOI] [PubMed] [Google Scholar]
- 37.Gindhart J G, Jr, Desai C J, Beushausen S, Zinn K, Goldstein L S. J Cell Biol. 1998;141:443–454. doi: 10.1083/jcb.141.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naeve G S, Ramakrishnan M, Kramer R, Hevroni D, Citri Y, Theill L E. Proc Natl Acad Sci USA. 1997;94:2648–2653. doi: 10.1073/pnas.94.6.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taniguchi S, Kawano T, Kakunaga T, Baba T. J Biol Chem. 1986;261:6100–6106. [PubMed] [Google Scholar]
- 40.Cho S I, Ko J, Patton B L, Sanes J R, Chiu A Y. J Neurobiol. 1998;37:339–358. [PubMed] [Google Scholar]
- 41.Kazmin D A, Hoyt T R, Taubner L, Teintze M, Starkey J R. J Mol Biol. 2000;298:431–445. doi: 10.1006/jmbi.2000.3680. [DOI] [PubMed] [Google Scholar]
- 42.Howard M J, Gershon M D. J Neurobiol. 1998;35:341–354. doi: 10.1002/(sici)1097-4695(19980615)35:4<341::aid-neu1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 43.Edery P, Lyonnet S, Mulligan L M, Pelet A, Dow E, Abel L, Holder S, Nihoul F C, Ponder B A, Munnich A. Nature (London) 1994;367:378–380. doi: 10.1038/367378a0. [DOI] [PubMed] [Google Scholar]
- 44.Romeo G, Ronchetto P, Luo Y, Barone V, Seri M, Ceccherini I, Pasini B, Bocciardi R, Lerone M, Kaariainen H, et al. Nature (London) 1994;367:377–378. doi: 10.1038/367377a0. [DOI] [PubMed] [Google Scholar]
- 45.Lane P W, Liu H M. J Hered. 1984;75:435–439. doi: 10.1093/oxfordjournals.jhered.a109980. [DOI] [PubMed] [Google Scholar]
- 46.Lee M, Goodall J, Verastegui C, Ballotti R, Goding C R. J Biol Chem. 2000;275:37978–37983. doi: 10.1074/jbc.M003816200. [DOI] [PubMed] [Google Scholar]
- 47.Potterf S B, Furumura M, Dunn K J, Arnheiter H, Pavan W J. Hum Genet. 2000;107:1–6. doi: 10.1007/s004390000328. [DOI] [PubMed] [Google Scholar]
- 48.Bondurand N, Pingault V, Goerich D E, Lemort N, Sock E, Caignec C L, Wegner M, Goossens M. Hum Mol Genet. 2000;9:1907–1917. doi: 10.1093/hmg/9.13.1907. [DOI] [PubMed] [Google Scholar]
- 49.Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. J Neurosci. 1998;18:237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pusch C, Hustert E, Pfeifer D, Sudbeck P, Kist R, Roe B, Wang Z, Balling R, Blin N, Scherer G. Hum Genet. 1998;103:115–123. doi: 10.1007/s004390050793. [DOI] [PubMed] [Google Scholar]