Abstract
Vertebrate gastrointestinal tracts have co-existed with microbes over millennia. These microbial communities provide their host with numerous benefits. However, the extent to which different environmental factors contribute to the assemblage of gut microbial communities is not fully understood. The purpose of this study was to determine how the external environment influences the development of gut microbiome communities (GMCs). Faecal samples were collected from deer mice (Peromyscus maniculatus) born and raised in captivity and the wild at approximately 3–5 weeks of age. Additional samples were collected 2 weeks later, with a subset of individuals being translocated between captive and wild environments. Microbial data were analysed using 16S rRNA next-generation Illumina HiSeq sequencing methods. GMCs of deer mice were more similar between neighbours who shared the same environment, regardless of where an individual was born, demonstrating that GMCs are significantly influenced by the surrounding environment and can rapidly change over time. Mice in natural environments contained more diverse GMCs with higher relative abundances of Ruminoccocaceae, Helicobacteraceae and Lachnospiraceae spp. Future studies should examine the fitness consequences associated with the presence/absence of microbes that are characteristic of GMCs of wild populations to gain a better understanding of environment–microbe–host evolutionary and ecological relationships.
Subject terms: Microbiome, Microbial ecology, Biodiversity, Community ecology
Introduction
Microbial communities coexist in the gastrointestinal tract of vertebrates, forming tight relationships that aid in the development of host immunological, physiological, and metabolic processes during early-life stages [1–7]. Microbe–host relationships are maintained throughout the hosts’ lifetime through established exposure pathways via diet [8–10], social interactions [11, 12], biogeography [13, 14], seasonality [15, 16] and maternal sources [17–19]. Perturbations to these relationships, as demonstrated in urbanized humans [16, 20, 21], and captive animal populations [22–27], have resulted in changes in the host’s microbial diversity and community composition. Changes in gut microbiome communities (GMCs) can have important influences on host functions leading to the potential development of dysbiosis and increased susceptibility to gut-related diseases (e.g. obesity, inflammatory bowel disease, autoimmune disorders) [1, 4, 28–40]. This raises a number of concerns for captive breeding and/or reintroduction conservation initiatives regarding how changes to an animal’s natural environment may impact GMCs. However, relatively little research has been conducted on GMCs of wild animal populations [41] and their differences relative to captive populations [see refs. [22–27, 42, 43].
Understanding how changes in the environment impact GMCs may reveal important environment–host–microbe interactions and help inform the conservation of vertebrate hosts [44]. Germ-free and laboratory-based studies have limited application to animals in the wild [43, 45]; therefore, we investigated how captive and wild environments influence α-diversity and composition of deer mouse GMCs as well as the consequences of deer mice being translocated between the two environments. To the authors’ knowledge this is the first study to assess how GMCs are affected before and after individuals are translocated from captivity into the wild. In-depth classification of environment-linked microbes will also allow for an increased understanding of environment–microbe–host evolutionary and ecological interactions, a topic currently underrepresented in the literature [41]. We hypothesized that GMCs are influenced by exposure to microbes located in the surrounding environment. Gut microbiome α-diversity and community composition of deer mice were expected to homogenize with neighbouring deer mice in the same environment. Similarities between dam and offspring GMCs (i.e. maternal effects) were hypothesized to occur during the initial capture phase, with the GMCs of littermates being more similar to each other compared to offspring from other litters. However, maternal effects on the gut microbiome were expected to disappear post-translocation due to external environmental effects overwhelming maternal effects.
Methods
All methods in this study were approved by the Institutional Animal Care and Use Committee (IACUC) at Laurentian University, protocol number 2016-03-01.
Study site and sample collection
We captured deer mice (Peromyscus maniculatus) using Longworth live traps (Rogers Manufacturing Co., Canada) between May and August 2016 in Algonquin Provincial Park, Ontario, Canada (45°35′N, 78°31′W). Trapping occurred on a 220 m x 430 m grid in an area dominated by sugar maple (Acer saccharum), with traps set just prior to sunset and checked in the morning. We individually identified mice by tagging them with unique alphanumeric ear tags (National Brand and Tag co., Newport, KY). Pelage (i.e. grey) and weight (<15 g) were used to identify juvenile deer mice in the wild (between 3 and 5 weeks of age [46]). Faecal samples were collected directly from deer mice with sterilized equipment (ethanol [95%] and open flame) to limit the risk of contamination and then stored in a −20 °C freezer. Faecal samples reflect microbial communities within the lower gastrointestinal tract of hosts [47].
Experimental design
Pregnant dams were captured in May and June. Females determined to be pregnant were transferred to, and housed at, the Algonquin Wildlife Research Station. The offspring of these dams served as captive-born individuals for this experiment.
There were four main experimental groups: wild–wild (W–W), wild–captive (W–C), captive–wild (C–W), captive–captive (C–C). The first letter of the group name corresponds to where the mice were born, whereas the second letter corresponds to where the mice were during the translocation period. For example, mice in the W–C group were born in the wild and translocated to captivity, whereas C–C mice were born in captivity and remained in captivity during the translocation period. Deer mice born in captivity were first sampled at 3 weeks of age. All but two deer mice from each litter (x̅ = 4.5 juveniles/litter) in captivity were then translocated to the wild (i.e. C–W group) to the same geographic grid where wild individuals were captured, while the remaining deer mice remained in captivity (i.e. C–C group). Faecal samples were taken from all captive-born deer mice, including those that were recaptured in the wild, 2 weeks after the initial sample was acquired. Juvenile deer mice born in the wild had faecal samples collected upon capture. A subset of juvenile deer mice captured in the wild was translocated to captivity after initial faecal samples had been taken (i.e. W–C group), while the remaining individuals were left in the wild (i.e. W–W group). Two weeks after wild-born juvenile deer mice were initially sampled, they were recaptured and sampled again. We co-housed deer mice that were born in captivity at two individuals per cage, while deer mice translocated into captivity from the wild were provided with their own cage to avoid potential agonistic interactions between unrelated individuals.
Captivity conditions
We housed dams in separate cages (Plexiglas, 35 cm × 15 cm × 17 cm) with corn-cob bedding (Pestell Pet Products Easy Clean Corn-Cob Bedding), nesting materials (Anderson’s CN Crink-l’nestTM and Ancare Nestlets) and environmental enrichment (metal shower hooks). The light cycle and temperature mimicked natural conditions. Water and food (8640 Tekland Laboratories) were provided ad libitum. The captive conditions of our study were meant to reflect captive rather than sterile (laboratory) conditions.
Microbial extractions
DNA extractions from collected faecal samples were conducted using QIAamp DNA Stool Mini Kit (Qiagen, Mississauga, ON, Canada; #51504), following the manufacturer’s Isolation of DNA from Stool for Pathogen Detection instructions. A control was absent from the DNA extraction protocol because any contaminants from reagents are unlikely to overwhelm the high amount of microbial biomass in faecal samples [48]. A negative and positive control was used during PCR to ensure that there was no contamination.
Amplicon sequencing
Samples were sequenced using the Illumina MiSeq system (Illumina Biotechnology Co., San Diego, USA) by Metagenom Bio Inc. (Toronto, Canada), using their designated protocol. Broad bacterial primers were used to target the V3–V4 regions of the bacterial 16S RNA gene, Pro341F (5′-C-CTACGGGNBGCASCAG-3′) and Pro805R (5′-GACTACNVGGGTATCTAATC-C-3′). Before being pooled, samples had an index sequence of 6 bp incorporated, allowing for sequencing to be completed in a single run. A 25 µl PCR reaction was conducted using the following conditions: 5 µl of standard OneTaq buffer (5×), 0.25 µl of 25 mM dNTP, 0.5 µl of both primers, 1 µl BSA (12 mg/ml), 0.125 µl of OneTaq DNA polymerase (New England Bio, MA, USA), 1–10 ng DNA and water to reach the necessary 25 µl volume. PCR thermocycling conditions were set at: 94 °C for 5 min (initial denaturation), followed by 30 cycles of 94 °C for 30 s (denaturation), 53 °C for 45 s (annealing), 68 °C for 1 min (extension) and finished with a final extension at 68 °C for 10 min. PCRs were performed in triplicate for all samples with finished products being checked on 2% agarose gel. Bands were than excised using a MinElute gel extraction kit (Qiagen, Hilden, Germany). Purified DNA libraries were quantified via Qubit with the dsDNA HS assay kit (Life Technologies, CA, USA), with library pools spiked with 5% phix control (V3, Illumina) to improve base imbalance. Paired-end sequencing with 250 bp read lengths were conducted using MiSeq Reagent kit V2 (2 × 250 cycles) on an Illumina MiSeq platform. Raw sequences have been deposited in NCBI under the accession SRA #SRP140555.
Bioinformatics
Open-source bioinformatics tools were used to process the sequencing data. PEAR ([49]; v0.9.8) was used to pair and filter sequences to 390–590 base pairs. Primers were then trimmed from each sequence using BBduk ([50]; v37.09). USEARCH8 ([51];v10.0.240) was used to de-replicate and sort (by abundance) sequences as well as to remove singletons and doubletons. Through QIIME ([52]; v1.9.1) Operational Taxonomic Units (OTUs) were clustered using UPARSE (97% similarity), which included a chimera check step and then mapped specific OTUs. Sequences were referenced against the GreenGenes (97% confidence cut-off) database to assign taxonomic classification.
Statistical analysis
Chao1, Fisher and Shannon’s indices were calculated within QIIME and used to assess α-diversity. For Chao1 and Fisher α-diversity indices ANOVA models and post-hoc Tukey tests were used to determine differences in α-diversity between- and within-experimental groups, during initial and post-translocation sampling phases. Due to the non-normality of data for Shannon’s diversity indices, Kruskal–Wallis and post-hoc Dunn’s test were used to test for significance differences.
Turnover rate was calculated for each deer mouse GMC based on comparing differences in OTU abundances between initial capture and post-translocation samples. An ANOVA and Tukey post-hoc analysis was used to determine significant differences between GMC turnovers rates of each experimental group.
Weighted UniFrac measurements [53] were used to assess differences in GMCs between individuals as well as maternal and sex effects. Statistical differences between group GMCs and litters (for captive-born deer mice) were tested together using adonis (PERMANOVA) models, with 9999 permutations and reported F, R2 and p values, to determine if there were significant differences between captive and wild GMCs. The same approach was used to test for sex differences; however, a separate analysis was conducted for captive and wild individuals.
Variance stabilizing transformations were used to normalize GMC abundance data [54]. GMC differences between sampling periods and experimental groups were determined by evaluating changes in microbial families that made up 1% or more of GMCs. Family was the lowest classification level that provided accurate results, further classification resulted in the majority of OTUs (>75% for most samples) being unidentifiable. Mann–Whitney U tests were used to compare differences in GMCs for captive and wild deer mice during the initial capture period. Kruskal–Wallis and post-hoc Dunn tests were used to determine significant differences in microbial families between the experimental groups post-translocation. Paired Wilcoxon tests were then used to assess changes in microbial families within each experimental group. Additionally, a linear discriminant analysis of effect size (LEfSe) analysis was used to determine differences between GMCs using Kruskal–Wallis (α = 0.05) tests while incorporating biological consistency and effect size [55].
Constrained Analysis of Principal Coordinates (CAP; [56]) analysis was used to determine the top ten most enterotype-defining OTUs for captive and wild individuals (initial capture) as well as each group post-translocation. Bray–Curtis dissimilarity measurements were used in the CAP analysis, allowing abundance to be incorporated into determining which OTUs were enterotype-defining. The function ‘anova.cca’ (R package vegan [57]) was used to perform an ANOVA-like permutation test (1000 permutations) to determine significant differences between OTUs [14]. Enterotype-defining OTUs were both highly abundant as well as influential. Changes in the relative abundance of enterotype-defining OTUs in both initial and post-translocation capture periods for captive and wild GMCs were analysed through paired Wilcoxon signed-rank tests for individuals in each group.
Results
In total 3235 OTUs and 3,671,358 sequences (mean: 27,604, min: 14,916, max: 76,734, SD = ± 8764.37) were present among all deer mice, with individuals containing 444 ± 114 unique OTUs on average. In total, 69 individuals were included in the initial capture phase (n = 36 captive born; n = 33 wild born). Faecal samples from 34 deer mice were obtained during the post-translocation sampling period of the experiment: 8 captive–captive, 5 captive–wild, 13 wild–captive and 8 wild–wild. All deer mice were sampled twice, except in the W–C group where only 8 of the 13 were sampled twice. The additional five deer mice in the W–C were only used for post-translocation comparisons between groups.
The GMC composition of male and female deer mice held in captive (Weighted UniFrac: F = 1.81, R2 = 0.051, p = 0.10) and wild (Weighted UniFrac: F = 1.77, R2 = 0.054, p = 0.10) environments were only marginally different and explained only 5.1% and 5.4% of the variation, respectively. Therefore, males and females were pooled in subsequent statistical analyses.
α-diversity
During each sampling period deer mice in the wild had more diverse GMCs than captive individuals. In the initial sampling period deer mice born in the wild had significantly higher levels of α-diversity than captive-born deer mice Chao1: F1,3 = 32.23, p < 0.01 [Fig. 1a]; Fisher: F1,3 = 28.07, p < 0.01; Shannon’s: χ1,3 = 19.08, p < 0.01). Results were similar between all three α-diversity metrics, except the Shannon’s index that did not detect a significant difference between C–W and W–C groups (p = 0.10).
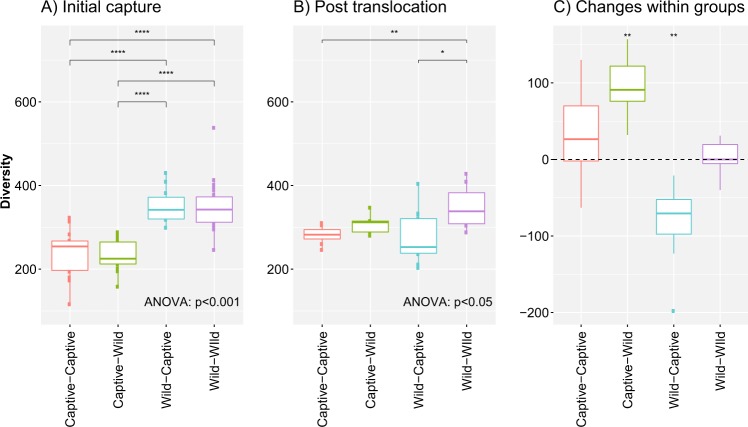
Fig. 1.
Under the Chao1 diversity index, initial capture results show that there were significant differences in α-diversity levels among the four experimental groups during the initial sampling phase (F1,3 = 32.23, p < 0.001), with deer mice born in captivity having lower α-diversity levels than deer mice born in the wild. Post-translocation significant differences were still present among the four experimental groups (F1,3 = 4.08, p < 0.05); however, only a significant difference was seen between the captive–captive and wild–wild groups (p = 0.01), and a marginal difference between wild–wild and wild–captive deer mice (p = 0.069). When looking a significant changes that occurred within groups between capture periods significant changes (p < 0.01) were seen in both translocated groups (i.e. captive–wild and wild–captive). **** represents p < 0.0001, *** represents p < 0.001, ** represents p < 0.01 and * represents p < 0.1
Significant differences were present between α-diversity levels of all four experimental groups post-translocation (Chao1: F1,3 = 4.11, p < 0.05 [Fig. 1b]; Fisher: F1,3 = 5.07, p < 0.01; Shannon: χ1,3 = 10.44, p < 0.05). Post-translocation significant differences were still present with W–W individuals having higher α-diversity scores compared to W–C counterparts (Chao1: p = 0.069; Fisher: p < 0.01; Shannon’s: p < 0.05). Similarly, differences were maintained between C–C and W–W groups (Chao1: p = 0.01; Fisher: p < 0.05; Shannon’s: p = 0.055). No significant difference in α-diversity was seen between C–C and C–W groups, or between the translocated groups. C–W deer mice showed an increase in α-diversity after being translocated to the wild; however, their α-diversity levels were only similar to W–W deer mice based on the Shannon’s diversity index.
When examining differences that occurred within groups between sampling periods, a significant increase in α-diversity was only seen for the C–W group when using Chao1 (t1,24 = 4.23, p < 0.01) α-diversity measurement. The W–C group showed a significant decrease under Chao1 (t1,24 = −4.61, p < 0.01 [Fig. 1c]) and Fisher’s (t1,24 = −4.09, p < 0.01), and a marginal decrease under Shannon’s index (Kruskal–Wallis rank sum test, p = 0.080). Individuals translocated into captivity experienced approximately the same amount of change (albeit increasing); however, it was only significant under Chao1, suggesting that a lack of significance may be due to insufficient statistical power for Fisher’s and Shannon’s indices. Control groups C–C and W–W did not show any significant changes in α-diversity between sampling periods, as predicted.
During the post-translocation sampling period W–W deer mice contained 519 (21.3%) unique OTUs, over three times more than any other group. Although 774 OTUs were shown to be shared between all the groups, only 7 OTUs were determined to compose a core microbiome (Supplemental Fig. 1), having shown evidence of membership, composition and persistence as outlined by Shade and Handelsman [58]. Significantly higher turnover rates were observed in deer mice in the C–C (p < 0.001), C–W (p < 0.001) and W–C (p < 0.001) groups compared to W–W individuals between sampling periods. Groups retained most OTUs between captures (C–C = 55.4%; W–C = 55.7%; C–W = 48.2%: W–W = 54%).
β-Diversity and community composition: initial capture
PERMANOVA analysis using Weighted UniFrac (F1,68 = 22.29, R2 = 0.22, p < 0.001) identified significant differences in the GMC composition of captive and wild individuals.
Wild and captive mice had 13 and 10 different phyla present in their GMCs, respectively. Firmicutes, Bacteroidetes and Proteobacteria were the only families to each make up ≥1% of GMCs (Fig. 2). Significant differences among dominant phyla were seen between wild and captive individuals: Firmicutes (x̅ = 68.39 ± 1.09% vs. 41.13 ± 1.29%, p < 0.001), Bacteroidetes (20.72 ± 0.53% vs. 50.22 ± 1.49%, p < 0.001) and Proteobacteria (8.53 ± 2.29% vs. 5.50 ± 4.2%, p < 0.001).
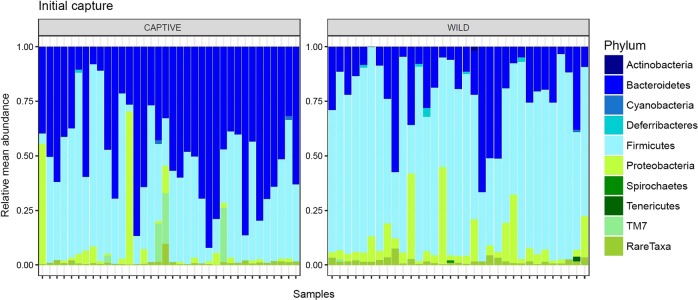
Fig. 2.
Compared differences in relative abundance between of different phyla during the initial capture period for both captive and wild deer mice. Firmicutes are shown to be dominant in wild deer mice while Bacteroidetes dominant in the gut microbiome of captive deer mice. Proteobacteria maintain a small presence in both wild and captive deer mice
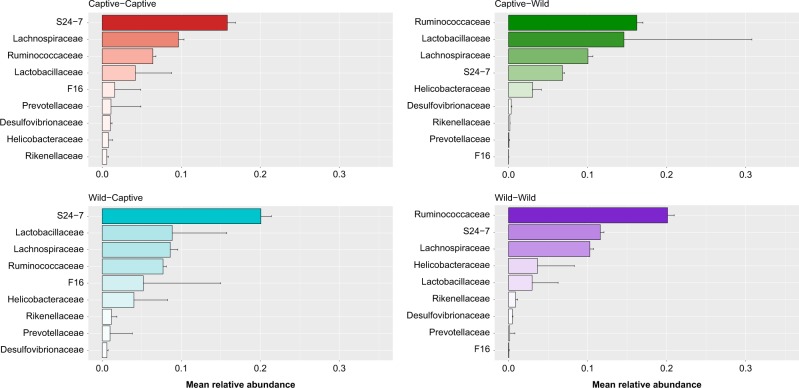
At the family level Ruminococcaceae, S24-7, Lachnospiraceae, Lactobacillaceae and Helicobacteriaceae dominated GMCs. Ruminococcaceae (p < 0.001), Lachnospiraceae (p < 0.01) and Helicobacteraceae (p < 0.01) were more abundant in wild deer mice, while S24-7 (p < 0.001) was more prevalent in captive deer mice (Fig. 3). Results from the LEfSe analysis complemented these results, indicating that when effect size was taken into account Ruminococcaceae, Lachnospiraceae and Helicobacteraceae still had higher relative abundances in wild deer mice, while S24-7 (and Enterobacteriaceae) were more abundant in captive deer mice (p < 0.05 for all, Fig. 4).
Fig. 3.
Side-by-side comparison of the mean relative abundance of families which made up at least >1% of the total gut microbiome community, within at least one individual, during the initial capture period for deer mice born in captivity and the wild. Significant differences occur between the Ruminococcocaceae (p < 0.001), Lachnospiraceae (p < 0.01), Helicobacteraceae (p < 0.01), S24-7 (p < 0.001) and Enterobacteriaceae (p < 0.05) (This relationship was driven by just two individuals.) Lactobacillus showed no significant difference between captive and wild groups during the initial sampling period
Fig. 4.
Linear discriminant analysis (LDA) effect size (LEfSe) determined that there were a number of microbial families that were more abundant in the GMCs of wild deer mice compared to GMCs of captive raised deer mice (initial sampling period), including Ruminococcaceae, Helicobacteraceae and Lachnospiraceae, which composed >1% of gut microbiome communities in deer mice. Alternatively, microbial families S24-7 and Enterobacteriaceae (composed >1% of gut microbiome communities in some captive deer mice individuals) were found to be significantly more abundant in captive deer mice
Enterotype-defining OTUs had a relative mean abundance ≥1% within at least one of the experimental groups, during either the pre- or post-translocation sampling period. The top ten enterotype-defining OTUs for GMCs of wild individuals included OTUs identified as Ruminococcaceae, Lachnospiraceae, Clostridiaceae, Helicobacteraceae [H. aurati and H. rodentium] and Lactobacillus murinus. The S24-7 microbial family contained a number of OTUs that were characteristic of GMCs in captive deer mice. OTUs identified as Lachnospiraceae were present in GMCs of both captive and wild deer mice (Fig. 5).
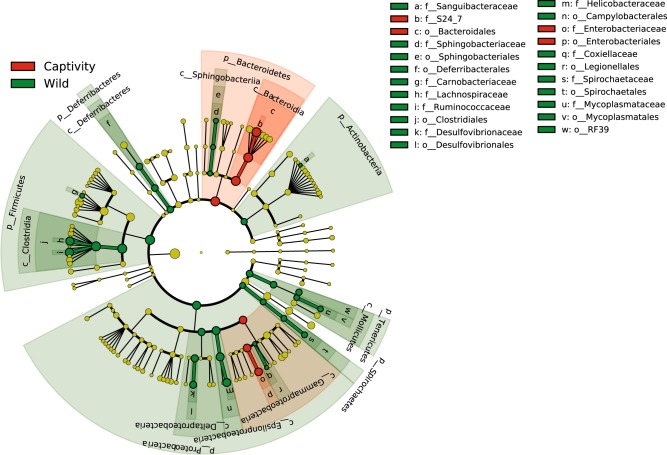
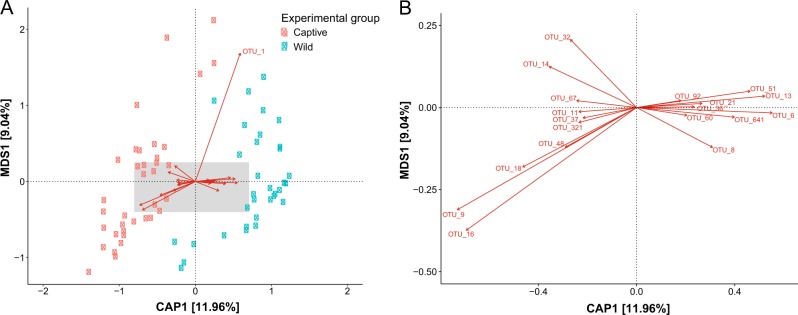
Fig. 5.
Constrained analysis of principle coordinates (CAP) ordination method was used to visualize differences between captive and wild deer mice gut microbiome communities (GMCs), during the initial sampling period. a GMCs of deer mice from the initial capture period, grouped together depending on where they were born (captivity or in the wild). The shaded section of figure a is represented in figure b, demonstrating which OTUs were the most influential in separating captive and wild enterotypes. Enterotype-defining OTUs for the GMCs of deer mice in the wild consisted for OTUs classified as Ruminococcaceae (OTU_13, OTU_51), Lachnospiraceae (OTU_21, OTU_60, OTU_92, OTU_641), Clostridiaceae (OTU_36), Helicobacter aurati (OTU_8), Helicobacter rodentium (OTU_6) and Lactobacillus murinus (OTU_1). For the captive enterotype-defining OTUs were identified as S24-7 (OTU_9, OTU_16, OTU_18, OTU_37, OTU_48, OTU_67, OTU_321), TM7 (OTU_14), Lachnospiraceae (OTU_32) and Clostridiaceae (OTU_11)
β-Diversity and community composition: post-translocation
After the translocation period GMCs of deer mice who shared the same environment clustered together (F1,33 = 3.71, R2 = 0.10, p < 0.01).
C–W deer mice post-translocation had a significantly higher abundance of Firmicutes compared to C–C deer mice (p < 0.05). Ruminococcaceae dominated GMCs of deer mice in the wild during the translocation phase and were more abundant in W–W individuals compared to the C–C (p < 0.01) and W–C groups (p < 0.01). S24-7 microbes dominated the GMC of deer mice in captivity and maintained higher relative abundances in captive deer mice compared deer mice in the wild (X2 = 6.74, p = 0.081). The relative abundance of Helicobacteraceae (X2 = 6.45, p = 0.092) was lower in C–C individuals compared to the other experiment groups (Fig. 6).
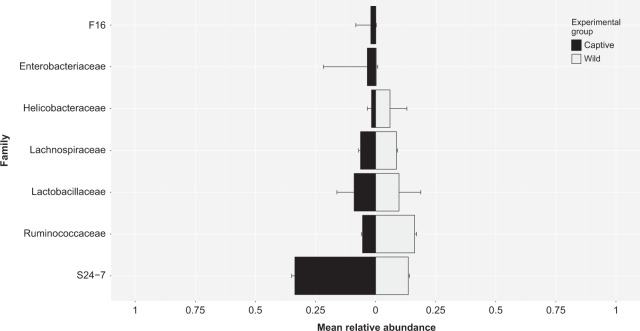
Fig. 6.
Mean relative abundance of different microbial families, which represented at least >1% or more of the total GMC within at least one individual deer mouse during the post-translocation sampling period. The microbial family S24-7 was the most dominant in deer mice that were in captivity during the 2-week translocation phase of the experiment (captive–captive and wild–captive groups). Alternatively, Ruminococcaceae was the most dominant microbial family in the GMC of deer mice who were in the wild for the translocation phase of the experiment (wild–wild and captive–wild groups)
Five OTUs identified in the wild enterotype were recognized in both initial and post-translocation stages including OTUs identified as Ruminococcaceae (OTU_13, OTU_51), Lachnospiraceae (OTU_92), Helicobacteraceae (OTU_6) and Lactobacillus murinus (OTU_1) (Supplemental Fig. 2). The remaining five OTUs belonged to Ruminoccocaceae (including Oscillospira) or within Clostridiaceae. The captive enterotype recognized three OTUs between capture periods: OTU_9 (S24-7), OTU_11 (Clostridiaceae) and OTU_14 (F16 [phylum: TM7]). Other captive enterotype OTUs during the post-translocation stage were identified as Clostridiales, Lachnospiraceae and Lactobacillus intestinalis.
Ruminococcaceae (OTU_51 and 13), Helicobacteraceae (OTU_6), Lactobacillus murinus (OTU_1) and a Lachnospiraceae (OTU_256) spp., strongly characterized the GMC of mice in the wild post-translocation, while S24-7 (OTU_9 and OTU_14), Lactobacillus intestinalis (OTU_2), Clostridiaceae (OTU_11) and Lachnospiraceae (OTU_7) spp. were heavily characteristic of deer mice in captivity post-translocation (Fig. 7).
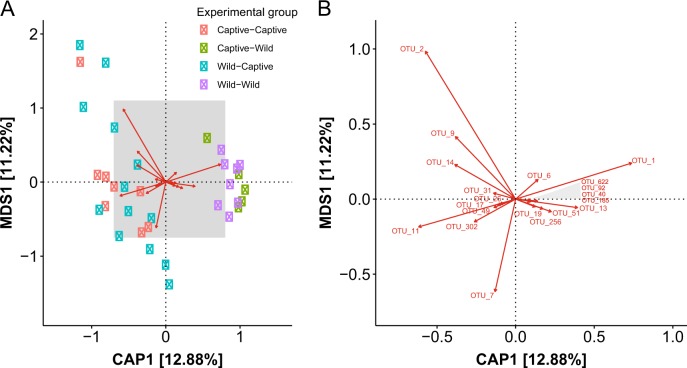
Fig. 7.
Constrained analysis of principle coordinates (CAP) ordination method was used to visualize differences between captive and wild deer mice gut microbiome communities (GMCs). a During the post-translocation sampling period. GMCs of deer mice from the post-translocation sampling period grouped together depending on the environment they were in during the translocation phase of the experiment. The shaded section of figure a is represented in figure b demonstrating which operational taxonomic units (OTUs) were the most influential in separating captive and wild enterotypes post-translocation. Enterotype-defining OTUs for the GMCs of deer mice in the wild post-translocation consisted for OTUs classified as: Ruminococcaceae (OTU_13*, OTU_51*, OTU_40, OTU_185, OTU_19 [Oscillopsira]), Lachnospiraceae (OTU_92*, OTU_622), Clostridiaceae (OTU_256), Helicobacter rodentium (OTU_6*) and Lactobacillus murinus (OTU_1*). For the captive enterotype-defining OTUs were identified as S24-7 (OTU_9*, OTU_49), TM7 (OTU_14*), Lachnospiraceae (OTU_7, OTU_17), Clostridiaceae (OTU_11*, OTU_302), Eubacterium (OTU_26, [Eubacterium tortuosum] OTU_31) and Lactobacillus intestinalis (OTU_2). *indicates that an OTU was determined to be enterotype defining for both initial and post-translocation sampling periods
β-Diversity and community composition: within groups comparisons
Captive–captive
Individuals in the C–C group had a marginal increase in Firmicutes between sampling periods (p = 0.08). At the family level an increase were seen in Lachnospiraceae (p = 0.059) as well as decreases in Lactobacillaceae (p = 0.059).
Between capture periods C–C individuals experienced increases among OTUs identified as Clostridiaceae (OTU_11, p < 0.05; OTU_39, p = 0.059; OTU_36, p = 0.08) and Lachnospiraceae (OTU_21, p < 0.05; OTU_7, p = 0.08). Significant decreases between capture periods were seen in Lactobacillus murinus (OTU_1, p < 0.05) and a S24-7 spp. (OTU_37, p < 0.05).
Captive–wild
C–W individuals experienced consistent changes among Firimicutes (increase, p = 0.059), Bacteroidetes (decrease, p = 0.059) and Proteobacteria (increase, p = 0.059) between capture periods. Although the sample size is small (n = 5), consistent changes in the relative abundance of phyla suggests that observed changes provide evidence for biologically meaningful relationships. Strong relationships were seen among Helicobacteraceae (increase, p = 0.059) and S24-7 (decrease, p = 0.059) families upon translocation to the wild. Ruminoccocaceae (p = 0.11) experienced an increasing trend (Supplemental Fig. 3), but was only found to be significant when accounting for effect size.
C–W individuals experienced increases among OTUs that were identified as Helicobacteraceae, Ruminococcaceae and some Lachnospiraceae. Decreases were seen among Lactobacillus and S24-7 groups. All relationships observed for C–W individuals reported a p = 0.059, suggesting that the small sample size for this group is limiting statistical power.
Wild–captive
At the family level significant decreases were seen in Helicobacteraceae (p = 0.014) and Ruminococcaceae (p = 0.042) (Supplemental Fig. 4). Helicobacteraceae and Ruminococcaceae in W–C displayed opposite results compared to individuals from C–W, suggesting they might be tightly linked to external environmental factors.
Opposite to C–W, W–C individuals exhibited a decrease in OTU 6 (Helicobacter rodentium, p = 0.052) as well as significant decreases (p < 0.05) in Ruminoccocaceae (OTU 13, 185, 40), Lachnospiraceae (OTU 60, 622, 641, 92) and Clostridiaceae (OTU 256) OTUs.
Wild–wild
Rikenellaceae (p = 0.052) and Ruminococcaceae (p = 0.076) marginally increased between sampling periods. No significant changes were seen at the OTU level.
Maternal influences
During the initial capture phase GMCs were more similar between littermates than offspring from other litters in captive deer mice (F1.68 = 2.32, R2 = 0.18, p < 0.001). Post-translocation the GMCs of captive-born littermates translocated to the wild did not cluster with their siblings that remained in captivity (F1,33 = 0.46, R2 = 0.05, p = 0.96; Supplemental Fig. 5). Due to low samples sizes there was not enough statistical power to separately test for significant clustering between siblings within each environment; however, siblings did not appear to cluster together in captivity or the wild post-translocation.
Discussion
We assessed GMC α-diversity and composition of captive and wild deer mice as well as changes that occurred within GMCs following reciprocal translocation events between captive and wild environments. The gut microbiome of both captive and wild deer mice were dominated by Firmicutes, Bacteroidetes and Proteobacteria; however, clear differences in α-diversity and community composition were present between wild and captive populations.
Wild vs. captive
Deer mice in the natural environment had higher levels of gut microbiome α-diversity than captive deer mice. This pattern mirrors results demonstrated in humans [16, 20, 21, 59] and animals [22–26, 45]. Compared to captive animals, wild conspecifics are exposed to more diverse microbial meta-communities via environmental sources (e.g. larger ranges, seasonality, social interactions, expansive diets) that can serve to increase the diversity of GMCs [11–15, 45, 60–62]. However, evidence suggests that gut microbiome composition is constrained by neutral dispersion limitations [13, 14, 60] (i.e. microbes have equal opportunities at successfully colonizing an environment; the more exposure a host has to a microbial species the more likely that species is to persist in the host’s GMC [63, 64]); therefore, continued exposure to diverse microbial meta-communities is necessary to maintain gut microbiome diversity. Nonetheless, in some cases conspecifics have not displayed differences in GMC α-diversity between captive and wild environments including lizards [42], even-toed ungulates (i.e. Cetartiodactyla) and two myrmecophagus species [26]. The maintenance of high α-diversity levels in captivity likely varies among taxa, with results between studies needing to be interpreted cautiously. Understanding the reasons for these conflicting results justifies further study of GMCs in wild and captive populations.
Firmicutes dominated the gut microbiome of wild deer mice, whereas Bacteroidetes were the dominant phyla in captive deer mice. Proteobacteria was less abundant than Firmicutes and Bacteroidetes in both wild and captive deer mice; however, the relative abundance of Proteobacteria was higher in wild mice compared to mice in captivity. Firmicutes have been shown to dominate GMCs in previous studies examining wild populations of mammals [8, 10, 22, 23, 26, 45, 65], including wild wood mice (Apodemus sylvaticus) [15] and house mice (Mus musculus domesticus) [14, 66]. This suggests that the Bacteroidetes dominated captive enterotype is divergent from the Firmicutes dominated enterotype seen in natural populations. Firmicutes have been associated with digestion efficiency [67] and may be crucial for wild animals that need to maximize energy extraction from their diet due to threats that are not present in captive environments (e.g. food scarcity, predation).
Differences between the GMCs of wild and captive deer mice were driven by changes in the relative abundance of particular microbial families. Ruminococcaceae, Lachnospiraceae and Helicobacteraceae were indicative of GMCs in wild deer mice, while S24-7 was associated with microbial communities in captive mice. Ruminococcaceae and Lachnospiraceae have previously been associated with healthy human microbiomes [68] and are known to perform functional roles as degraders of complex plant material [69–71]. Helicobacteraceae (ε-Proteobacteria) contains microbes that are opportunistic pathogens [72]; however, this family appears to be consistently present in the gut microbiome of wild murine populations [15, 66, 73]. Mice may potentially be acting as reservoir hosts to pathogenic and potentially zoonotic Helicobacteraceae strains [74]. S24-7 microbes are associated with maternal-based diets (i.e. milk from mothers) and the breakdown of proteins and carbohydrates [75, 76]; however, it is difficult speculate on the functional influence of S24-7 because the role of these microbes within GMCs is not yet fully understood.
Post-translocation: between-group comparisons
GMC α-diversity and composition from deer mice varied based on an individual’s surrounding environment. The gut microbiome of deer mice translocated to the wild increased levels of α-diversity and developed communities that were similar to native wild mice. The opposite was displayed in mice translocated to captivity. Neighbouring individuals likely acquire microbes from the same microbial pools [13, 14] resulting in rapid changes and reduced variation in GMCs between individuals in the same environment [10]. Upon being translocated to the wild the GMCs of C–W mice increased in diversity, presumably because they were exposed to diverse microbial meta-communities that were likely absent from captive environments. It is important to note that animals brought into captivity rapidly lost GMC diversity, suggesting that gut microbiome studies on wild populations should not be conducted in captive settings.
Deer mice in the wild retained fewer microbial species (i.e. OTUs) between capture periods than deer mice in captivity. Microbial communities continually experience turnover resulting in different microbial compositions across multiple time points [62, 77], due to microorganisms being continually lost or gained as species compete for limited resources. Wild deer mice would have been exposed to more diverse microbial meta-communities causing increased resource competition and encounter rates among a higher number of rare species that were likely replaced between capture periods.
Ruminococcaceae was more abundant in wild deer mice compared to mice in captivity, while Helicobacteriaceae maintained the lowest abundance in deer mice that were never exposed to the wild. These results suggest that Ruminococcaceae and Helicobacteraceae are more prominent in wild environments and rely on continued exposure to the natural environment to maintain high relative abundances within GMCs.
Post-translocation: within-group comparisons
Translocated groups showed nearly identical differences in terms of how much α-diversity was either lost or gained. The α-diversity of C–C individuals’ increased slightly, perhaps due to a transition from a maternal to lab-chow-based diet, resulting in similar α-diversity levels between animals in captivity post-translocation. C–W mice regained some natural GMC diversity upon being translocated to the wild, however, failed to achieve the same level of α-diversity as W–W mice. This may have been caused by translocated mice having reduced range sizes compared to native mice [78]. Similarly, free range (i.e. outdoor enclosures) Tasmanian devils were shown to contain intermediate gut microbiome α-diversity levels compared to captive and wild populations [22], suggesting that limited ranges may inhibit the ability for individuals to achieve the α-diversity levels as high as wild conspecifics. These results complement the idea of GMC being controlled by neutral dispersal limitations; however, further research should be conducted to determine the influence of range size on GMC diversity.
The relative abundance of Ruminococcaceae and Helicobacteraceae was dependent on exposure to the wild. Translocated groups demonstrated significant reciprocal changes in the relative abundance of these families (including OTUs further identified as Oscillospira and Helicobacter rodentium [72]) that were characteristic of wild deer mice GMCs. The associated between Oscillospira and the wild environment may be a result of wild mice continually experiencing periods of fasting, leaness and glycan degradation [79]. The impact of Ruminococcaceae (particularly Oscillospira) and Helicobacteraceae on host fitness should be further studied since these families are prominent in GMC of wild deer mice, yet only maintain lower relative abundances in captivity. Conflicting findings concerning the relationship between Ruminococcaceae captivity have been demonstrated in desert woodrats (Neotoma lepida), which increased in Ruminococcus and Coprococcus (Lachnospiraceae genus) upon entering captivity [43]. Such contradictions within the literature suggest that differences between taxa (e.g. diet, physiology) need to be considered before attempting to interpret results between studies.
OTUs belonging to Lachnospiraceae and Clostridiaceae also increased in relative abundance in deer mice translocated to the wild while showing the opposite in reciprocally transplanted mice. However, captive deer mice also contain a number of Lachnospiraceae and Clostridiaceae OTUs that were characteristic of their GMC. In a similar scenario two different Lactobillaceae species were identified as being characteristic in the GMC of captive and wild mice (wild—Lactobicillus murinus; captivity—L. intestinalis). These families likely contain a number of microbes that are functionally redundant, suggesting that animals in captivity may be able to maintain certain GMC functions despite a loss of diversity [80]. It is important to note that this study did not examine the GMC functional diversity between captive and wild deer mice, however, does provide speculative evidence that functional diversity can be preserved in captivity.
Maternal and sex influences
Maternal influences were present during the initial sampling of captive-born deer mice; however, post-translocation C–W deer mice contained GMCs that were more similar to wild deer mice than their siblings that remained in captivity. Although maternal effects have been demonstrated in lab-based studies there is growing evidence that in natural populations maternal influences on microbiome communities only occur in early-life stages and are short lasting [18, 81, 82]. Sex differences explained relatively little variation within GMC of wild and captive mice, likely as a result of individuals being reproductively immature and captive mice co-habituating. A number of previous studies on murine species have revealed similar results where sex differences in GMCs were absent or explained little variation [15, 66, 81, 82].
Conclusion
GMC α-diversity and community composition of deer mice were shown to be dependent on the surrounding environment. Upon entering a new environment GMCs rapidly homogenized with neighbouring individuals in terms of α-diversity and community composition. The natural environment promoted increased diversity and relative abundance of Ruminoccocaceae, Helicobacteraceae and Lachnospiraceae. Ruminoccocaceae and Lachnospiraceae have previously associated with maintaining healthy microbiomes and digestion, suggesting that animals in captivity may be deprived of microbes that have important functional roles in wild environments. Future studies are needed to determine how later-in-life changes in GMC diversity and community composition may impact host immunological, physiological and metabolic processes that are typically established during early-life stages. Researchers should also be cautious when attempting to analyse GMCs of wild animals in captive conditions.
Supplementary information
Acknowledgements
We thank A. Sawyer and S. Wilkes for their assistance in the field collecting data. This work has been funded by the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant and the Canadian Foundation of Innovation (CFI)—Leaders Opportunity Grant to A.I.S.-H.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-019-0345-8) contains supplementary material, which is available to authorized users.
References
- 1.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 2.Manson JM, Rauch M, Gilmore MS. The commensal microbiology of the gastrointestinal tract. Adv Exp Med Biol. 2008;7:15–28. doi: 10.1007/978-0-387-09550-9_2. [DOI] [PubMed] [Google Scholar]
- 3.O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–93. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–23. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu J, Gordon JI. Honor thy symbionts. Proc Natl Acad Sci USA. 2003;100:10452–9. doi: 10.1073/pnas.1734063100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hooper LV. Bacterial contributions to mammalian gut development. Trends Microbiol. 2004;12:129–34. doi: 10.1016/j.tim.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Hooper LV, Littman DR, Macpherson AJ, Program MP. Interactions between the microbiota and the immune system. Science. 2015;336:1268–73. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–51. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–4. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Linnenbrink M, Künzel S, Fernandes R, Nadeau M, Rosenstiel P, et al. Dietary history contributes to enterotype-like clustering and functional metagenomic content in the intestinal microbiome of wild mice. Proc Natl Acad Sci USA. 2014;111:e2703–10. doi: 10.1073/pnas.1402342111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gavish Y, Kedem H, Messika I, Cohen C, Toh E, Munro D, et al. Association of host and microbial species diversity across spatial scales in desert rodent communities. PLoS ONE. 2014;9:1–9. doi: 10.1371/journal.pone.0109677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koch H, Schmid-hempel P. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc Natl Acad Sci USA. 2011;108:19288–92. doi: 10.1073/pnas.1110474108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lankau EW, Hong P, Mackie RI. Ecological drift and local exposures drive enteric bacterial community differences within species of Galápagos iguanas. Mol Ecol. 2012;21:1779–88. doi: 10.1111/j.1365-294X.2012.05502.x. [DOI] [PubMed] [Google Scholar]
- 14.Linnenbrink M, Wang J, Hardouin Ea, Künzel S, Metzler D, Baines JF. The role of biogeography in shaping diversity of the intestinal microbiota in house mice. Mol Ecol. 2013;22:1904–16. doi: 10.1111/mec.12206. [DOI] [PubMed] [Google Scholar]
- 15.Maurice CF, Knowles SC, Ladau J, Pollard KS, Fenton A, Pedersen AB, et al. Marked seasonal variation in the wild mouse gut microbiota. ISME J. 2015;9:2423–34. doi: 10.1038/ismej.2015.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smits SA, Leach J, Sonnenburg ED, Gonzalez CG, Lichtman JS, Reid G, et al. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science. 2017;806:1–18. doi: 10.1126/science.aan4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friswell MK, Gika H, Stratford IJ, Theodoridis G, Telfer B, Wilson ID, et al. Site and strain-specific variation in gut microbiota profiles and metabolism in experimental mice. PLoS ONE. 2010;5:e8584. doi: 10.1371/journal.pone.0008584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lucas FS, Heeb P. Environmental factors shape cloacal bacterial assemblages in great tit Parus major and blue tit P. caeruleus nestlings. J Avian Biol. 2005;36:510–6. doi: 10.1111/j.0908-8857.2005.03479.x. [DOI] [Google Scholar]
- 19.Neu J. Perinatal and neonatal manipulation of the intestinal microbiome: a note of caution. Nutr Rev. 2007;65:282–5. doi: 10.1111/j.1753-4887.2007.tb00305.x. [DOI] [PubMed] [Google Scholar]
- 20.Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B, et al. The microbiome of uncontacted Amerindians. Sci Adv. 2015;1:e1500183. doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martínez I, Stegen JC, Maldonado-Gómez MX, Eren MA, Siba PM, Greenhill AR, et al. The gut microbiota of rural Papua New Guineans: composition, diversity patterns, and ecological processes. Cell Rep. 2015;11:527–38. doi: 10.1016/j.celrep.2015.03.049. [DOI] [PubMed] [Google Scholar]
- 22.Cheng Y, Fox S, Pemberton D, Hogg C, Papenfuss AT, Belov K. The Tasmanian devil microbiome—implications for conservation and management. Microbiome. 2015;3:76. doi: 10.1186/s40168-015-0143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villers LM, Jang SS, Lent CL, Lewin-Koh SC, Norosoarinaivo JA. Survey and comparison of major intestinal flora in captive and wild ring-tailed lemur (Lemur catta) populations. Am J Primatol. 2008;184:175–84. doi: 10.1002/ajp.20482. [DOI] [PubMed] [Google Scholar]
- 24.Xenoulis PG, Gray PL, Brightsmith D, Palculict B, Hoppes S, Steiner JM, et al. Molecular characterization of the cloacal microbiota of wild and captive parrots. Vet Microbiol. 2010;146:320–5. doi: 10.1016/j.vetmic.2010.05.024. [DOI] [PubMed] [Google Scholar]
- 25.Nakamura N, Amato KR, Garber P, Estrada A, Mackie RI, Gaskins HR. Analysis of the hydrogenotrophic microbiota of wild and captive black howler monkeys (Alouatta pigra) in Palenque national park, Mexico. Am J Primatol. 2011;73:909–19. doi: 10.1002/ajp.20961. [DOI] [PubMed] [Google Scholar]
- 26.McKenzie Valerie J., Song Se Jin, Delsuc Frédéric, Prest Tiffany L., Oliverio Angela M., Korpita Timothy M., Alexiev Alexandra, Amato Katherine R., Metcalf Jessica L., Kowalewski Martin, Avenant Nico L., Link Andres, Di Fiore Anthony, Seguin-Orlando Andaine, Feh Claudia, Orlando Ludovic, Mendelson Joseph R., Sanders Jon, Knight Rob. The Effects of Captivity on the Mammalian Gut Microbiome. Integrative and Comparative Biology. 2017;57(4):690–704. doi: 10.1093/icb/icx090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson TM, Rogers TL, Carlini AR, Brown MV. Diet and phylogeny shape the gut microbiota of Antarctic seals: a comparison of wild and captive animals. Environ Microbiol. 2013;15:1132–45. doi: 10.1111/1462-2920.12022. [DOI] [PubMed] [Google Scholar]
- 28.Moloney RD, Desbonnet L, Clarke G, Dinan TG, Cryan JF. The microbiome: stress, health and disease. Mamm Genome. 2014;25:49–74. doi: 10.1007/s00335-013-9488-5. [DOI] [PubMed] [Google Scholar]
- 29.Dillion JR, Vennard CT, Buckling A, Charnley AK. Diversity of locust gut bacteria protects against pathogen invasion. Ecol Lett. 2005;8:1291–8. doi: 10.1111/j.1461-0248.2005.00828.x. [DOI] [Google Scholar]
- 30.Sonnenburg JL, Angenent LT, Gordon JI. Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nat Immunol. 2004;5:569–73. doi: 10.1038/ni1079. [DOI] [PubMed] [Google Scholar]
- 31.Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. 2016;535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 32.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:1626–32. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 33.Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–93. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–90. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 35.Mulder IE, Schmidt B, Lewis M, Delday M, Stokes CR, Bailey M, et al. Restricting microbial exposure in early life negates the immune benefits associated with gut colonization in environments of high microbial diversity. PLoS ONE. 2011;6:e28279. doi: 10.1371/journal.pone.0028279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ley RE, Bäckhed F, Turnbaugh P, Lozupone Ca, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–5. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cahenzli J, Balmer ML, Mccoy KD. Microbial-immune cross-talk and regulation of the immune system. Immunology. 2013;138:12–22. doi: 10.1111/j.1365-2567.2012.03624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. 2016;352:539–44. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cash HL, Hooper LV. Commensal bacteria shape intestinal immune system development. ASM News. 2005;71:77–83. [Google Scholar]
- 40.Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci USA. 2002;99:15451–5. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hird SM. Evolutionary biology needs wild microbiomes. Front Microbiol. 2017;8:1–10. doi: 10.3389/fmicb.2017.00725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kohl KD, Brun A, Magallanes M, Brinkerhoff J, Laspiur A, Acosta JC, et al. Gut microbial ecology of lizards: insights into diversity in the wild, effects of captivity, variation across gut regions and transmission. Mol Ecol. 2017;26:1175–89. doi: 10.1111/mec.13921. [DOI] [PubMed] [Google Scholar]
- 43.Kohl KD, Dearing MD. Wild-caught rodents retain a majority of their natural gut microbiota upon entrance into captivity. Environ Microbiol Rep. 2014;6:191–5. doi: 10.1111/1758-2229.12118. [DOI] [PubMed] [Google Scholar]
- 44.Redford KH, Segre JA, Salafsky N, Del Rio CM, Mcaloose D. Conservation and the microbiome. Conserv Biol. 2012;26:195–7. doi: 10.1111/j.1523-1739.2012.01829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kreisinger J, Cížková D, Vohánka J, Piálek J. Gastrointestinal microbiota of wild and inbred individuals of two house mouse subspecies assessed using high throughput parallel pyrosequencing. Mol Ecol. 2014;23:5048–60. doi: 10.1111/mec.12909. [DOI] [PubMed] [Google Scholar]
- 46.Banfield AW. The mammals of Canada. 1st edn. Toronto and Buffalo: University of Toronto Press; 1974. [Google Scholar]
- 47.Suzuki TA, Nachman MW. Spatial heterogeneity of gut microbial composition along the gastrointestinal tract in natural populations of house mice. PLoS ONE. 2016;11:1–15. doi: 10.1371/journal.pone.0163720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiss S, Amir A, Hyde ER, Metcalf JL, Song SJ, Knight R. Tracking down the sources of experimental contamination in microbiome studies. Genome Biol. 2014;15:1–3. doi: 10.1186/s13059-014-0564-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–20. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brian B. BBMap. Web. https://jgi.doe.gov/data-and-tools/bbtools/bb-tools-user-guide/bbduk-guide/
- 51.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 52.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. Correspondence QIIME allows analysis of high-throughput community sequencing data Intensity normalization improves color calling in SOLiD sequencing. Nat Publ Gr. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLOS Comput Biol. 2014;10:1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anderson MJ, Willis TJ. Canonical analysis of principal coordinates: a useful method of constrained ordination for ecology. Ecol Soc Am. 2003;84:511–25. [Google Scholar]
- 57.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. vegan: Community Ecology Package. 2018. R package version 2.5-1. https://CRAN.R-project.org/package=vegan.
- 58.Shade A, Handelsman J. Beyond the Venn diagram: the hunt for a core microbiome. Environ Microbiol. 2012;14:4–12. doi: 10.1111/j.1462-2920.2011.02585.x. [DOI] [PubMed] [Google Scholar]
- 59.Turroni S, Rampelli S, Centanni M, Schnorr SL, Consolandi C, Severgnini M, et al. Enterocyte-associated microbiome of the Hadza hunter-gatherers. Front Microbiol. 2016;7:1–12. doi: 10.3389/fmicb.2016.00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burns AR, Stephens WZ, Stagaman K, Wong S, Rawls JF, Guillemin K, et al. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J. 2016;10:655–64. doi: 10.1038/ismej.2015.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adair KL, Douglas AE. Making a microbiome: the many determinants of host-associated microbial community composition. Curr Opin Microbiol. 2017;35:23–29. doi: 10.1016/j.mib.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 62.Bobbie CB, Mykytczuk NCS, Schulte-hostedde AI. Temporal variation of the microbiome is dependent on body region in a wild mammal (Tamiasciurus hudsonicus). FEMS Microbiol Ecol. 2017;93:fix081. [DOI] [PubMed]
- 63.Chave J. Neutral theory and community ecology. Ecol Lett. 2004;7:241–53. doi: 10.1111/j.1461-0248.2003.00566.x. [DOI] [Google Scholar]
- 64.Hubbell SP. The unified neutral theory of biodviersity and biogeography. Princeton, New Jersey, USA: Princeton University Press; 2001. [Google Scholar]
- 65.Uenishi G, Fujita S, Ohashi G, Kato A, Yamauchi S, Matsuzawa T, et al. Molecular analyses of the intestinal microbiota of chimpanzees in the wild and in captivity. Am J Primatol. 2007;69:367–76. doi: 10.1002/ajp.20351. [DOI] [PubMed] [Google Scholar]
- 66.Weldon L, Abolins S, Lenzi L, Bourne C, Riley EM, Viney M. The gut microbiota of wild mice. PLoS ONE. 2015;10:e0134643. doi: 10.1371/journal.pone.0134643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Turnbaugh PJ, Ley RE, Mahowald Ma, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 68.Schubert AM, Rogers MaM, Ring C, Mogle J, Petrosino JP, Young VB, et al. Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy. mBio. 2014;5:1–9. doi: 10.1128/mBio.01021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mackie RI, Aminov RI, Hu W, Klieve AV, Ouwerkerk D, Sundset MA, et al. Ecology of uncultivated Oscillospira species in the rumen of cattle, sheep, and reindeer as assessed by microscopy and molecular approaches. Appl Environ Microbiol. 2003;69:6808–15. doi: 10.1128/AEM.69.11.6808-6815.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Biddle A, Stewart L, Blanchard J, Leschine S. Untangling the genetic basis of fibrolytic specialization by lachnospiraceae and ruminococcaceae in diverse gut communities. Diversity. 2013;5:627–40. doi: 10.3390/d5030627. [DOI] [Google Scholar]
- 71.Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol. 2008;6:121–31. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 72.Myles MH, Livingston RS, Franklin CL. Pathogenicity of Helicobacter rodentium in A/JCr and SCID mice. Comp Med. 2004;54:549–57. [PubMed] [Google Scholar]
- 73.Baxter NT, Wan JJ, Schubert AM, Jenior ML, Myers P, Schloss PD. Intra- and interindividual variations mask interspecies variation in the microbiota of sympatric Peromyscus populations. Appl Environ Microbiol. 2015;81:396–404. doi: 10.1128/AEM.02303-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wasimuddin, Čížková Dagmar, Bryja Josef, Albrechtová Jana, Hauffe Heidi C., Piálek Jaroslav. High Prevalence and Species Diversity of Helicobacter spp. Detected in Wild House Mice. Applied and Environmental Microbiology. 2012;78(22):8158–8160. doi: 10.1128/AEM.01989-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Serino M, Luche E, Gres S, Baylac A, Bergé M, Cenac C, et al. Metabolic adaptation to a high-fat diet is associated with a change in the gut microbiota. Gut. 2012;61:543–53. doi: 10.1136/gutjnl-2011-301012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Evans CC, LePard KJ, Kwak JW, Stancukas MC, Laskowski S, Dougherty J, et al. Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat diet-induced obesity. PLoS ONE. 2014;9:e92193. doi: 10.1371/journal.pone.0092193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Banks PB, Norrdahl K, Korpimäki E. Mobility decisions and the predation risks of reintroduction. Biol Conserv. 2002;103:133–8. doi: 10.1016/S0006-3207(01)00110-0. [DOI] [Google Scholar]
- 79.Konikoff T, Gophna U. Oscillospira: a central, enigmatic component of the human gut microbiota. Trends Microbiol. 2016;24:523–4. doi: 10.1016/j.tim.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 80.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kovacs A, Ben-Jacob N, Tayem H, Halperin E, Iraqi FA, Gophna U. Genotype is a stronger determinant than sex of the mouse gut microbiota. Microb Ecol. 2011;61:423–8. doi: 10.1007/s00248-010-9787-2. [DOI] [PubMed] [Google Scholar]
- 82.Campbell JH, Foster CM, Vishnivetskaya T, Campbell AG, Yang ZK, Wymore A, et al. Host genetic and environmental effects on mouse intestinal microbiota. ISME J. 2012;6:2033–44. doi: 10.1038/ismej.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.