Abstract
The active layer of soil overlaying permafrost in the Arctic is subjected to annual changes in temperature and soil chemistry, which we hypothesize to affect the overall soil microbial community. We investigated changes in soil microorganisms at different temperatures during warming and freezing of the active layer soil from Svalbard, Norway. Soil community data were obtained by direct shotgun sequencing of total extracted RNA. No changes in soil microbial communities were detected when warming from −10 to −2 °C or when freezing from −2 to −10 °C. In contrast, within a few days we observed changes when warming from −2 to +2 °C with a decrease in fungal rRNA and an increase in several OTUs belonging to Gemmatimonadetes, Bacteroidetes and Betaproteobacteria. Even more substantial changes occurred when incubating at 2 °C for 16 days, with declines in total fungal potential activity and decreases in oligotrophic members from Actinobacteria and Acidobacteria. Additionally, we detected an increase in transcriptome sequences of bacterial phyla Bacteriodetes, Firmicutes, Betaproteobacteria and Gammaproteobacteria—collectively presumed to be copiotrophic. Furthermore, we detected an increase in putative bacterivorous heterotrophic flagellates, likely due to predation upon the bacterial community via grazing. Although this grazing activity may explain relatively large changes in the bacterial community composition, no changes in total 16S rRNA gene copy number were observed and the total RNA level remained stable during the incubation. Together, these results are showing the first comprehensive ecological evaluation across prokaryotic and eukaryotic microbial communities on thawing and freezing of soil by application of the TotalRNA technique.
Subject terms: Biogeochemistry, Environmental sciences
Introduction
Permafrost-affected soil systems cover up to 20% of the total land area in the northern hemisphere [1] and are estimated to contain up to 472 Pg of organic carbon in the first upper meter [2], which is equivalent to more than half of the atmospheric carbon content [3]. The Arctic is currently experiencing a temperature increase at a higher rate than the global average [4, 5], with documented impacts on permafrost-affected soils via thawing and increasing depth of the above active layers [6, 7]. Active layer soil is defined as the overlaying permafrost soils being exposed to seasonal freezing and thawing. Permafrost thaw will likely lead to an enhanced degradation of the large carbon pool in the newly established active layer by a wide diversity of microorganisms, including bacteria, archaea, fungi and protozoa [8–10]. Thus, there is a growing interest in incorporating microbial parameters into modelling of the changing Arctic environments [11, 12]. Even though active layer permafrost soils are exposed to harsh conditions like freeze/thaw cycles, sub-zero temperatures and low nutrient availability most of the year, these soils harbour a high diversity of bacteria [13, 14], fungi [15, 16] and protozoa [8]. Microbial communities in active layer soils are dynamic and show relatively large annual changes in biomass [17–20], with a peak before thawing of the soil followed by a fast decline shortly after [17, 18]. Accordingly, bacterial community composition in active layer soils is also dynamic [21, 22], with distinct winter and summer bacterial communities [22], and altered soil fungal community structures between seasons in the Arctic and at lower latitudes [20, 23].
Active layer soil may be exposed to several freeze/thaw cycles during spring and autumn, and it has been shown that these cycles change the microbial community composition [24, 25]. Soil freezing may result in lysis of microbial cells, resulting in available and easily degradable organic substrates from dead microorganisms for survivors upon thawing [26].
Several authors have proposed the concept of copiotrophic/oligotrophic lifestyles to classify bacterial groups as a response to nutrient concentration in soils ecosystems, and that large phylogenetic groups fall into these lifestyle groups [27–31]. Copiotrophic bacteria respond relatively quickly to an increase in labile nutrient availability, show a high growth rate and have a high (>5) copy number of the 16S rRNA gene, whereas oligotrophic bacteria typically have low-growth rates, respond slowly to an increase in labile nutrient availability and have few (</=<2) copies of the 16S rRNA gene [27]. These trophic lifestyles may indicate which bacterial groups will respond as the soil thaws and nutrient availability increases.
Cultivation biases, providing access to only a minority of soil microorganisms [32], has spurred the use of molecular techniques to study microbial communities, especially in soil where biodiversity levels are relatively high [33]. The majority of these studies are based on DNA, such as direct shotgun metagenomics and analysis of PCR amplicons from amplified 16S rRNA gene fragments or fungal ITS2, but interpretation in terms of activity and functions is hampered by a large fraction of extracellular DNA [34]. In combination with the low turnover of microbial DNA, the extracellular DNA pool may obscure short-term changes in microbial communities caused by thawing. In contrast, RNA has a faster turnover than DNA and is presumed to be produced only by metabolically active microorganisms [35]. Studies of the active soil microbial community using RNA-based techniques were pioneered by Urich et al. [36], who isolated total soil RNA and used shotgun cDNA sequencing and bioinformatics tools to annotate putative rRNA reads against databases using BLAST. This approach has subsequently been used to study microbial communities of Arctic soils [37, 38]. These studies mainly focused on microbial community changes following thawing and did not reveal the microbial responses to (i) increasing temperatures before transition from frozen to thawed conditions (an important aspect because the main future temperature increase in the Arctic is expected to happen during winter [39]), and (ii) re-freezing. In addition, it has been demonstrated that predation of certain bacteria by protists may change the composition of the microbial community [40], but little is known about interactions among different trophic levels of soil microorganisms following short-term (days) perturbations, like thawing.
In order to understand the microbial response to not only thawing but also to increasing sub-zero temperatures and to freezing, we investigated the changes in rRNA-based active layer soil microbial communities to gradually increasing and decreasing temperature in the range from −10 to 2 °C. We hypothesized that only a subset of bacterial and fungi taxa would respond to an increase in temperature from −10 to −2 °C, and that bacterial–protozoan interactions will influence the microbial response to thawing events.
Materials and methods
Soil sampling and preparation
An active layer soil core was collected on 18th of April 2014 in Sassendalen, Svalbard, Norway (Latitude 78.270961, Longitude 17.228315, 5 m above sea level). The area has a mean annual air temperature of −6 °C and a mean annual precipitation of ~200 mm water equivalent [41]. After removal of the snow cover, the soil core (0–14-cm depth) was sampled using a core catcher (8-cm diameter) with a motorized hand drill. The core was transported in a cooling box to a −18 °C freezer at the University Centre in Svalbard and then to Denmark by airplane in a Styrofoam box with cooling elements to avoid thawing. Measurements by a temperature logger and visual inspection of the core indicated that it did not thaw during transport. At the University of Copenhagen, the core was kept at −18 °C until sample preparation, which was conducted in a −15 °C freezelab. Contamination during sample preparation was avoided by sterilizing all surfaces by UV exposure and washing all equipment in ethanol before autoclaving. In addition, we avoided soil potentially contaminated during drilling by only sampling the unexposed interior of the core as described by Bang-Andreasen et al. [42]. In brief, plant cover was scraped off the surface. Then the soil core was broken and the inner core soil was aseptically sampled with a sterilized bore head (16 mm) from the newly exposed surface and homogenized by smashing the soil in a sterile plastic bag with a hammer, obtaining an appropriate grain size range of 1–10 mm.
Soil incubation
Two-gram subsamples were placed in 5-ml sterile conical plastic tubes (Eppendorf AG, Hamburg, Germany) to a total of 40 replicates. The samples were incubated in a custom-made temperature chamber based on a recirculating cooling liquid (Bath fluid Thermal G, JULABO GmbH, Seelbach, Germany) with a recirculating Cooler (FL300, JULABO GmbH, Seelbach, Germany). The base of the device was an aluminium plate (28 × 15.5 × 4 cm, L × W × H) with drilled-in canals for the cooling liquid. On top of this, a steel box (21.5 × 14.5 × 9.5 cm, L × W × H) was placed and filled with Ottawa sand (Fisher Scientific, Leicestershire, UK) to evenly distribute the temperature. The entire device was then covered with Styrofoam to stabilize the temperature, which was monitored by five temperature sensors distributed in the sand. During incubation, the standard error of the mean of the temperature measurements by the five sensors was ≤0.1 °C. Samples were incubated for 26 days, starting with a pre-incubation step at −10 °C for 48 h, then a gradual increase to +2 °C over 6 days, followed by +2 °C for 16 days and a freezing phase with decreasing temperature over 6 days to −10 °C. Samples were obtained at two-day intervals at −10, −6, −2 and +2 °C during the thawing phase; these samples will be referred to as T−10°C, T−6°C, T−2°C and T2°C, respectively. Following the 16 days at +2 °C, samples were obtained at two-day intervals +2, −2, −6 and −10 °C in the freezing phase; these samples will be referred to as F2°C, F−2°C, F−6°C and F−10°C, respectively (see Fig. 1). At each of the eight sampling points, five replicates were snap-frozen in liquid nitrogen and subsequently stored at −80 °C until RNA extraction.
Fig. 1.
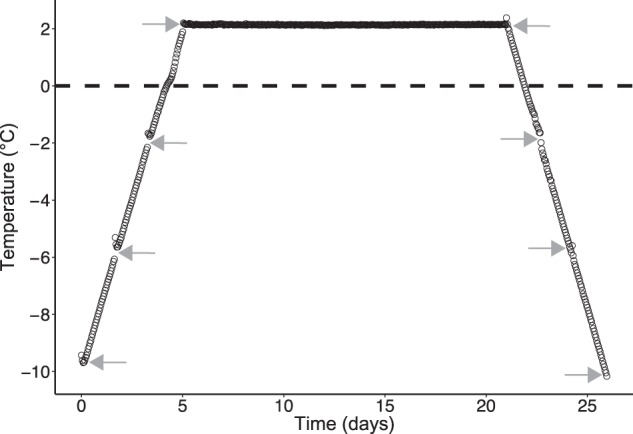
Measured incubation temperature of active layer permafrost samples shown as average of five temperature loggers (black circles); Standard error of mean ≤ 0.1 °C. Grey arrows show time of sampling for RNA extraction
Soil analyses
Grain-size distribution was analysed by dissolving soil samples in 0.01 M tetrasodium pyrophosphate and demineralized H2O with 2 min of ultrasound and then analysed by laser diffraction on a Mastersizer 2000 (Malvern Instruments, Malvern, UK). Soil pH was measured in a second incubation run, using the same incubation chamber. This was done because the whole sample needed to be snap-frozen to preserve the RNA in the first incubation run. The second run had the same temperature setup as the first run. A total of 36 samples of 2 g soil was incubated in same type of 5-ml plastic tubes, as described above. We selected four time points for the pH measurements: T−6°C, T2°C, F2°C and F−6°C. A total of 16 g was collected at each time point: four replicates with 4 g (two tubes pooled per replicate) of soil were each extracted in 20 mL ddH2O for 10 min at 5 °C on a shaker. All extracts were filtered through Whatman GF-D filters and measured on pH meter. The total carbon and nitrogen contents were measured in solid samples by Dumas combustion (1020 °C) on an elemental analyser (CE 1110, Thermo Electron, Milan, Italy). Subsamples of 10 mg of dried and finely homogenized (ball mill) material were weighed out into tin combustion cups for elemental analysis. Soil standards (Elemental Microanalysis, Okehampton, UK) were used for elemental analyser mass calibration. For pH measurement, the temperature incubation was repeated with the sample experimental setup using a subset of the homogenized soil.
RNA and DNA isolation and sequencing library preparation
Total RNA isolation was done from 2 g of soil using RNA PowerSoil total RNA isolation kit (MO BIO Laboratories, Carlsbad, CA, USA) with phenol:chloroform:isoamyl alcohol in the ratio 25:24:1 (Sigma-Aldrich, Copenhagen, Denmark) following the instructions of the manufacturer, resulting in 100-μl RNA extracts. DNA was co-isolated with RNA PowerSoil DNA Elution Accessory Kit (MO BIO Laboratories, Carlsbad, CA, USA) following the instructions of the manufacturer. DNase treatment on a 10-μl subsample was performed using DNase max (MO BIO Laboratories) by following the manufacturer's instructions. The quality of DNase-treated RNA was checked with a Bioanalyser 2100 (Agilent Technologies, Glostrup, Denmark). This step was used to exclude one replicate from each time point, by selecting the replicate with the lowest RNA integrity number (RIN) number (28S/23S to18S/16S ratio). RNA quantity was estimated using Qubit 2.0 (Thermo Fisher Scientific, Hvidovre, Denmark) with the Qubit RNA HS Assay Kit. For sequencing library preparation, the NebNext Ultra directional RNA Library prep kit for Illumina (New England Biolabs, Herlev, Denmark) was used with 100 ng of RNA from each sample. Library preparation was carried out according to the manufacturer's instructions. Samples were sequenced using a HiSeq 2500 (Illumina) in rapid mode, 150 base-paired end reads, at the National High-throughput DNA Sequencing Centre, University of Copenhagen.
Quantitative PCR
Quantitative PCR was performed to estimate the numbers of 16S rRNA gene and transcript copies in the soil samples. A 17-μl aliquot of each RNA extract was DNase treated as described above and reverse transcribed using random hexamer primers (Thermo Scientific, Schwerte, Germany) and RevertAid Reverse Transcriptase (Thermo Scientific). Reverse transcription was performed by incubation at 25 °C for 10 min, 50 °C for 30 min and 85 °C for 5 min. cDNA samples were stored at −80 °C until further analysis. Three replicate PCR amplifications were performed on each DNA and cDNA sample using the primers 341 F (5′-CCTACGGGAGGCAGCAG-3′) and 518 R (5′-ATTACCGCGGCTGG-3′) [43]. For producing a standard, E. coli K27 was grown in Luria–Bertani medium for 2 days at room temperature to a desired density, after which DNA was isolated and the number of cells were counted in a microscope.
Bioinformatics framework
Quality filtering, merging and ribosomal filtering of reads were done using BioDSL (http://maasha.github.io/BioDSL/). In brief, raw sequences were filtered into two parts, one for potential mRNA and one for ribosomal RNA separated by SortMeRNA v. 2.0 [44] (http://bioinfo.lifl.fr/sortmerna/sortmerna.php). Reads were then cleaned by removing Illumina adapter sequences and low-quality reads (>20 Phred score). Finally, reads were paired using the following parameters: minimum overlap = 20 bp, and 20% mismatch allowed. Reads that could not be paired were discarded. From each sample, 2,000,000 quality checked and cleaned reads were randomly picked in order to reduce the dataset for computational efficiency. Potential ribosomal reads were used to find the closest matching microorganism based on ribosomal small subunit (SSU) in the SILVA database release 111, using Metaxa2 version 2.1.3 [45] with default settings (Reliability score cutoff = 80). For this study, we only used the results matching SSU. The complete OTU table can be seen in a supplementary datasheet.
Data and statistical analyses
Statistical analyses of the Physiochemical parameters were done with the multcomp package in R using ANOVA with a post-hoc Tukey HSD correction test (p < 0.05). The alpha diversity analysis was carried out as described in previous work [46]. R software version 3.0.2 (R Development Core [47]) and PAST software [48] were used to calculate the alpha diversity indices, which was done on rarefied data at 750,000 reads per sample in order to avoid biases that may come from uneven read numbers. Alpha diversity was assessed by measuring observed richness and the Shannon index (H). Statistical analysis was carried out as described for the soil Physiochemical parameters.
The beta diversity and multivariate analyses were done with the raw and non-rarefied contingency tables using the R with the package vegan [49] and ade4 [50]. Because the contingency tables have features with abundance variation larger than 1000-fold, a log10 transformation was applied to account for this dispersion. Principal component analysis (PCA) was performed after centre-scaling normalization. A pattern search was applied to the original PCAs by grouping replicates together in order to perform a between-group analysis (BGA). The statistical significance of the selected grouping factor was tested with a Monte–Carlo simulation involving 10,000 permutations. Complementing PERMANOVA tests were performed on the Bray–Curtis dissimilarity profiles using 10,000 permutations in order to assess the significance of the tested factors. The major relative abundance-driven changes in the phylogenetic composition of profiles were identified by means of ANOVA with post-hoc Tukey HSD correction test and false discovery rate (FDR) multiple correction test (p < 0.05). Significant changes in large taxonomic groups were also investigated separately for each thaw and freezing treatments to identify potential temperature-related effects. Finally, fine changes in the soil microbiome at the operational taxonomic unit (OTU) (97% identity) level were extracted using generalized linear model and negative binomial regression (nbGLM) using the R package edgeR [51]. This was conducted in order to determine OTUs responding in a similar manner to thawing and freezing. The significance of changes in OTU abundance was inferred with a likelihood ratio test (LRT) under correction by a post-hoc false discovery rate test with the package edgeR in R (p < 0.05 following FDR correction). Groups of OTUs that changed their abundance upon thawing and freezing were plotted in a heatmap of centred and scaled counts using a combination of the following R packages: gplots[52], vegan [49], rioja [53] and RcolorBrewer [54], as previously described [33]. OTUs were clustered and classified in functional response groups (FRGs, i.e., groups of organisms responding similarly as a function of environmental conditions) as previously proposed [46], using complete hierarchical cluster dendrograms based on Euclidean distance. Functional response groups were manually determined with the lowest number of groups possible. The selected groups were validated via a Monte–Carlo simulation involving 10,000 permutations between groups and the remaining non-significant OTUs, and visualized in a BGA.
Average 16S rRNA gene copy numbers in the microbial community were calculated to test the hypothesis that bacteria with a high number of 16S rRNA gene copies respond quickly to thawing. From Metaxa2 output, the bacterial fasta file (average of 711,123 reads per sample) was used to characterize the bacterial composition using QIIME v. 1.9.1 [55] to find the closest matching bacteria using closed-reference OTU-picking with default settings. 16S rRNA copy number for Greengenes v13.5 OTUs was obtained from the PICRUSt pipeline [56]. For each sample, we calculated the mean 16S rRNA gene copy number weighted by the relative abundances of each OTU; an R script for this analysis is available at: https://github.com/Russel88/MicEco. Significant differences between the thawing and freezing event was tested using Welch’s t test (p <0.05). The estimate of phylum level copy numbers was obtained by finding the median and 90% quantile copy number of the OTUs in our dataset. The phylum-level fold change was estimated with the log2 ratio of the mean relative abundance for all freeze samples divided by the mean relative abundance for all thaw samples.
Results
Soil Physiochemical parameters
The average temperature during incubation and time of sampling can be seen in Fig. 1. The temperature measurements during incubation had a standard error of the mean of ≤0.1 °C. The slight increase in temperature after each sampling point is due to the opening of the chamber. Physiochemical properties of the soil measured prior to incubation were as follows. Water content was 39.6% (±1.4 SE), total carbon and nitrogen were 7.19% (±0.32 SE) and 0.48% (±0.02 SE) of soil dry weight, respectively. Soil texture distribution was clay (<2 µm) 3.1% (±0.4 SE), silt (2–63 µm) 55.4% (±5.7 SE) and sand (>63 µm) 41.5% (±6.2 SE). The pH of the soil was measured to be 7.5 (±0.02 SE), 7.6 (±0.04 SE), 7.4 (±0.03 SE) and 7.4 (±0.0 SE) for T−6°C, T2°C, F2°C and F−6°C, respectively.
RNA concentration and 16S rRNA gene transcripts
RNA concentration after DNAse treatment showed no difference between samples (ANOVA, p = 0.06), though with a slightly higher concentration in the freezing samples. The mean ± standard error of the mean (SE) RNA concentration of all samples was 1.19 ± 0.04 µg g−1 of soil (fresh weight). The 16S rRNA copy number did not differ between sampling points (ANOVA, p = 0.06), and the mean ± SE 16S rRNA transcript was 2.92 ± 0.5 × 108 g−1 of soil (fresh weight). The same observation for 16S RNA gene copies was seen with no difference between samples (ANOVA, p = 0.23). The mean ± SE 16S rRNA gene was 1.60 ± 0.5 × 108 g-1 soil.
Overall microbial community composition
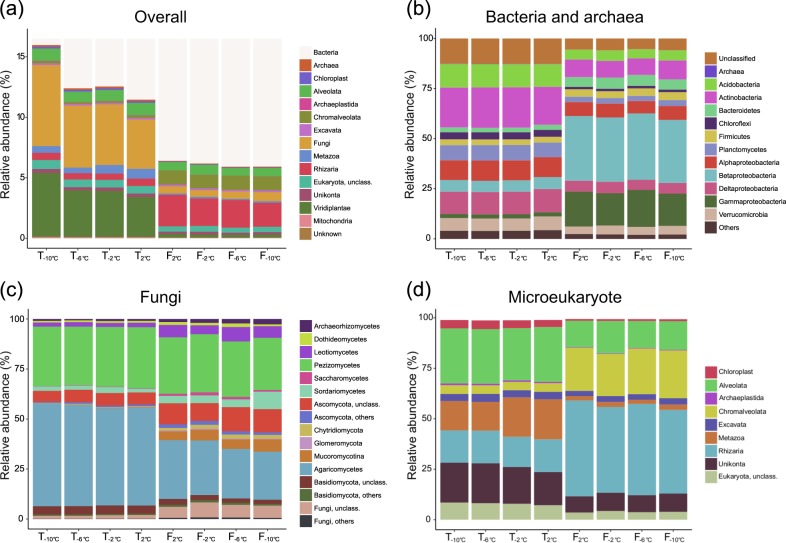
To study the microbial community, we screened two million randomly selected reads from each sample for rRNA transcripts, which resulted in 785,989 ± 2972 reads (average ± standard error of the mean, n = 32) per sample matching 16S or 18S rRNA transcripts. A total of 9763 distinct OTUs were detected of which 7154 belonged to Bacteria and Archaea, 651 to Fungi and 1958 to other Eukaryotes. The taxonomic composition of the community, including prokaryotes, Fungi and microeukaryotes (aka non-fungal/non-plant sequences) can be seen in Fig. 2. Bacteria constituted 84.0−94.1% of the total number of SSU reads, Archaea 0.05−0.15%, Fungi 0.6−6.7%, Rhizaria 0.5−2.6%, Viridiplantae 0.2–5.2% and other Eukaryotes 2.7–3.3%. At the highest taxonomic level (see Fig. 2a), only the relative abundance of Fungi significantly decreased in relative abundance from 6.7 ± 0.5 to 4.1 ± 0.1% when the temperature increased (ANOVA, p = 0.037 following the FDR correction, see Supplementary Figure S2). In contrast, the relative abundance of all these major taxonomic groups changed significantly when kept at 2 °C for 16 days. Three groups increased in relative abundance over the two sampling points, including Bacteria (88.5 ± 0.4 to 93.6 ± 0.3%), Chromalveolata (0.16 ± 0.01 to 1.16 ± 0.06%) and Rhizaria (0.62 ± 0.04 to 2.61 ± 0.23%).
Fig. 2.
Changes in three-domain community profile of the microbiota in active layer permafrost during a thawing and a freezing event. The taxonomic profile are based on small subunit ribosomal RNA, extracted from total RNA sequences. The bar charts display the relative abundance in each taxonomic group. a Overall soil community, b bacterial and archaeal community, c fungal community and d microeukaryotic community
The three groups that decreased most drastically during the period at 2 °C for 16 days were Fungi (4.08 ± 0.10 to 0.58 ± 0.10%), Metazoa (0.81 ± 0.28 to 0.12 ± 0.01%) and Viridiplantae (3.23 ± 0.34 to 0.30 ± 0.06%). The ratio between reads assigned to bacteria and fungi as calculated on real counts, increased from 17.4 ± 1.0 to 156.8 ± 9.4 (n = 16) between T2°C and F2°C (ANOVA, p < 0.0001), but did not change within the thaw and freeze events, see Fig. 3.
Fig. 3.
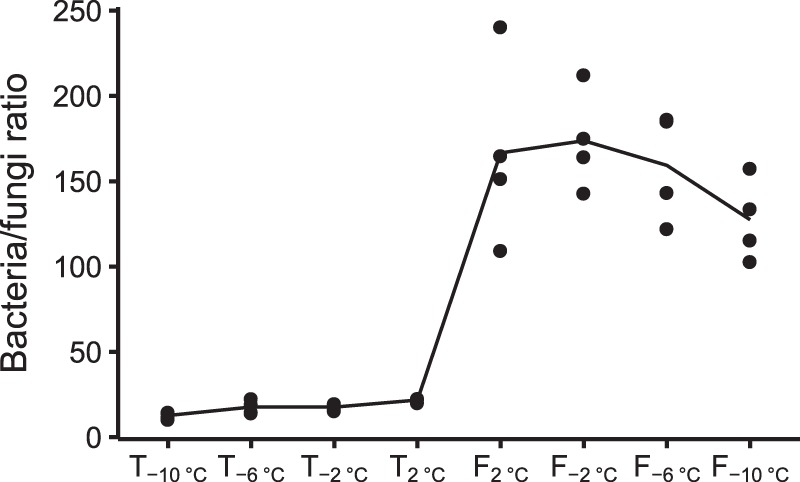
Ratio between bacterial and fungal small subunit (SSU) rRNA reads calculated on raw sequence counts. Black dots are the ratio between bacteria- and fungi-assigned reads calculated from four replicates. Black line is mean of four replicates
Prokaryote community composition
Major changes in the relative abundance of prokaryotic phyla and proteobacterial classes were found between the two sampling points at 2 °C (Fig. 2b). While most of these groups significantly decreased in abundance during this period, the majority (58.3%) of the 16S rRNA sequences belonged to a few phylogenetic groups enriched after 16 days at 2 °C. These were Beta- and Gammaproteobacteria, Firmicutes, Bacteroidetes and candidate division TM7 (Figure S1). The Firmicutes were dominated by the genus Paenibacillus in the freezing samples, representing 37.3 ± 1.20% of the reads. The relative abundance of Betaproteobacteria increased from 5.95 ± 0.09 to 32.26 ± 0.44% and Gammaproteobacteria increased from 2.15 ± 0.05 to 17.48 ± 2.29%. In contrast, Acidobacteria and Actinobacteria decreased from 11.20 ± 0.09 to 4.91 ± 0.34% and from 18.98 ± 0.14 to 8.75 ± 0.71%, respectively, between the two time points at 2 °C. Only Gemmatimonadetes, Bacteroidetes and Betaproteobacteria responded to the increase in temperature from −10 to +2 °C (Supplementary Figure S2), and Aquificae was the only bacterial phyla that significantly responded to the freezing event as its relative abundance decreased between −2 and −6 °C (p = 0.03). Significant changes among prokaryotes at OTU level are described later.
Fungal community composition
The fungal community was dominated by two classes, Pezizomycetes (relative abundance of 29.8 ± 0.8 and 27.8 ± 2.3% in thawing and freezing samples, respectively) and Agaricomycetes (49.8 ± 1.3 and 26.3 ± 2.3% in thawing and freezing samples, respectively), see Fig. 2c. The overall trend of fungi was a decrease in 18S rRNA copies (Fig. 2a), which was also confirmed by an EdgeR test showing that fungal OTUs totalling 97.2% of the relative abundance at T2°C significantly decreased after 16 days at 2 °C. In contrast, only six fungal OTUs corresponding to a total of 0.2% of the relative abundance, significantly increased between the two sampling times. These OTUs were assigned to Unclassified Mucoromycotina (0.0224 ± 0.002% at F2°C), zygomycete sp. AM-2008a (0.002 ± 0.0002% at F2°C), Tetracladium marchalianum (0.001 ± 0.0001% at F2°C), T. setigerum (0.0008 ± 0.0002% at F2°C), Mortierella parvispora (0.0003 ± 0.0001% at F2°C) and Nephridiophaga blattellae (0.0002 ± 0.000 % at F2°C), see supplementary datasheet.
Microeukaryote community composition
Microeukaryote community is the eukaryote community, excluding fungi and plants. None of the major groups significantly changed within the thawing or the freezing event (Fig. 2d). However, the relative abundance of all these groups, except Excavata, changed between the two events (p < 0.05, ANOVA with a FDR correction). These changes included a major increase in the relative abundance of Rhizaria from 16.0 ± 0.9 to 45.3 ± 2.73% and of Chromalveolata from 4.3 ± 0.2 to 22.4 ± 1.3% following 16 days at 2 °C. During this period, the relative abundance of Alveolata and Metazoa decreased from 27.1 ± 1.3 and 17.3 ± 2.9% to 13.7 ± 2.0 and 2.3 ± 0.4%, respectively.
Alpha and beta diversity
A clear effect in favour of higher diversity in thaw profiles compared with the freeze profiles was observed. No clear effect of the temperature changes on alpha diversity was found (Supplementary Figure S3), but observed richness and Shannon index decreased significantly (p < 0.05) between thawing and freezing samples from 4075 ± 31 to 3761 ± 49 and from 5.1 ± 0.0 to 4.5 ± 0.1, respectively.
Beta diversity analysis based on all OTUs was applied in order to study the clustering of samples in the experimental setup. The constrained ordination analysis by grouping replicates under Monte–Carlo simulation revealed a significant, non-random distribution of the samples according to each temperature condition (p < 0.0001; Fig. 4). The first component explained most of the variance (55.67%) and clearly segregated the RNA profiles based on the treatment (freeze vs. thaw). The second component explained only 8.96% of the variance and was mainly separating the T2°C samples from the T−10°C, T−6°C and T−2°C samples.
Fig. 4.
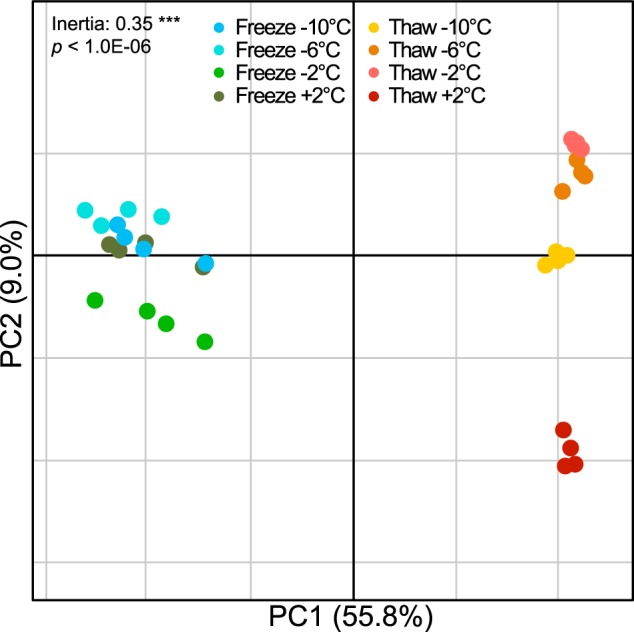
Between-group analysis (BGA) of the soil microbial communities. The figure shows constrained principal component analysis (PCA) of the microbiome profiles after applying sample grouping, according to replicates for frozen and thaw samples along recorded temperature. Non-random distribution of the BGA grouping was tested using a Monte–Carlo simulation with 10.000 permutations (p<1.0E-04***)
These observations were confirmed by the PERMANOVA on Bray–Curtis dissimilarity, where 62% of the variance was attributed to thaw versus freeze (p < 0.0001, Supplementary Table S1), although the changing temperature did not show any significant trend (4–8% of the variance). Similar analyses were conducted on the fungal and microeukaroyte communities, and the same pattern was observed with clustering of the thawing samples and freezing samples and no clear trend within each event (Supplementary Figure S4).
Prokaryotic functional response groups (FRGs)
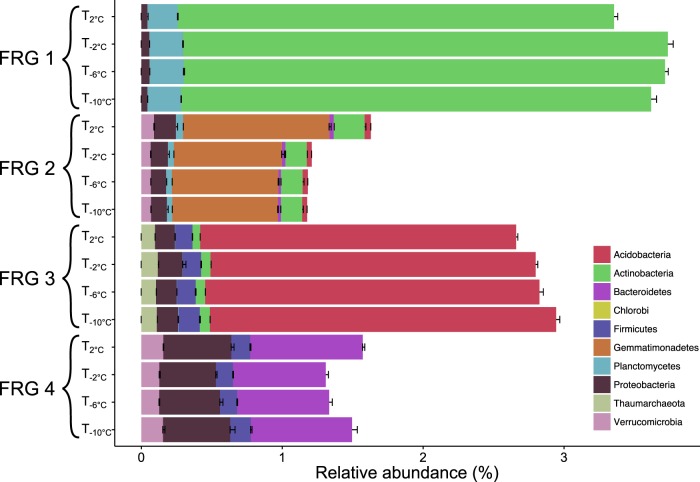
We investigated the changes in relative abundance at OTU level for prokaryotes within the thawing samples (T−10°C, T−6°C, T−2°C and T2°C) and freezing samples (F−10°C, F−6°C, F−2°C and F2°C). The relative abundance of 62 OTUs changed significantly within the thawing event (negative binomial regression, p < 0.05, see Supplementary Figure S5). Four different groups were identified representing four different responses upon thawing of the soil. Each of the prokaryotic functional response groups (FRGs) contained unique phylum-level specific OTUs (Fig. 5).
Fig. 5.
Composition and abundance of the four functional response groups (FRG) representing a group of organisms that respond similarly to environmental changes. The figure represents the weighted abundance of phylogenetic groups in each FRGs for each temperature during both thawing and freezing events. Statistical coherency of the four FRGs was verified under a Monte–Carlo simulation results, where random groups of the same size were sampled from the whole operational taxonomic unit (OTU) pool to test the robustness of defined FRGs (p < 1.0E-5)
Both FRG 1 and FRG 3 responded negatively to increasing temperature when above -2 °C. FRG 1 had an increase until T−2°C and then decreased at T2°C, whereas the OTUs in FRG 3 had a steady decrease during the entire thawing event. FRG 1 included 16 OTUs and was dominated by Actinobacteria, representing 92.1% of all OTUs within the group. FRG 3 consisted of 10 OTUs and was dominated by Acidobacteria, which represented 83.5% of the OTUs in the group. FRG 2 and FRG 4 represented OTUs, which increased in relative abundance when temperature increased from −2 to +2 °C. FRG 2, with 19 OTUs, increased throughout the thawing event and was dominated by Gemmatimonadetes. The relative abundance of OTUs in FRG 4 decreased between T−10°C and T−2°C and then increased at T2°C. This group was represented by 16 OTUs within Bacteroidetes and Proteobacteria representing 49.5 and 31.4% of OTUs, respectively.
In the freezing event, the relative abundance of 39 OTUs changed significantly and four FRGs were established, but no phylum-level patterns within these groups were revealed (Supplementary Figure S6).
16S rRNA gene copy number and response to thawing
In order to test the hypothesis that the number of 16S rRNA gene copies within a bacterium correlates with its response to thawing, the average 16S rRNA gene copy number weighted by the relative abundance of each OTU was calculated. The average 16S rRNA gene copy number significantly increased from 2.19 ± 0.00 at the thawing to 3.35 ± 0.05 at the freezing event (Welch’s test, DF = 15.02, p = 1.14E-12, Fig. 6).
Fig. 6.
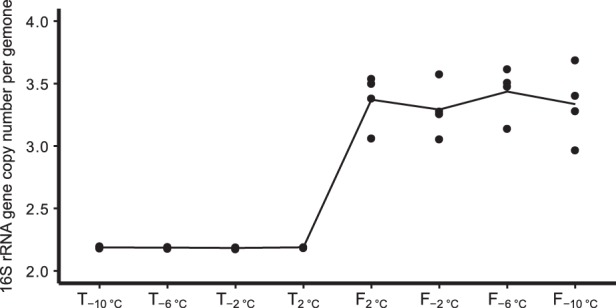
Mean 16S rRNA gene copy number per genome of the four replicates from each time point. Calculated as mean 16S rRNA gene copy number weighted by the relative abundances of each operational taxomomic unit (OTU) based on the PICRUSt pipeline
A linear regression of the log2-fold change in relative abundance of each OTU between thawing and freezing samples against the 16S rRNA gene copy number revealed that the gene copy number explained 10.2% of the variance in the log2-fold change (p < 0.0001, Fig. 7).
Fig. 7.
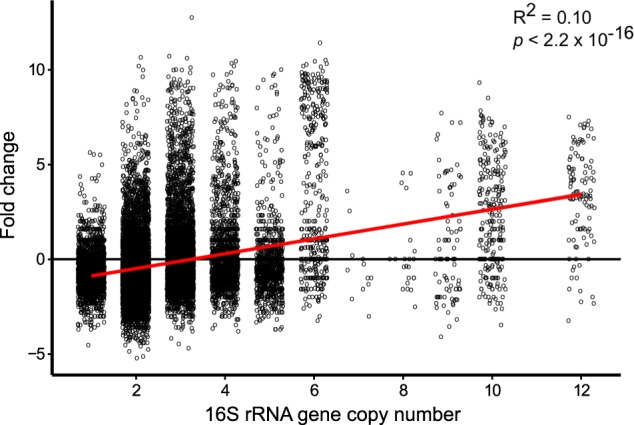
Dot plot of the log2-fold change between T2°C and F2°C and the 16S copy number for each operational taxonomic unit (OTU) based on the PICRUSt pipeline. Red line are linear regression
Discussion
Here, we present the first detailed analysis of changes in microbial and microeukaryotic communities during thawing and freezing of an active layer permafrost soil using a direct shotgun RNAseq procedure, which is not biased by PCR amplification. The rRNA level cannot directly be linked to cell activity; instead it should be seen as an indicator for potential cell activity [35]. Further, we are aware that the rRNA content in cells differs considerably and that the number of rRNA transcript copies cannot be directly related to the number of cells. However, rRNA transcript copy numbers can be used for relative comparison of the different experimental conditions on each microbial taxa. We show that major changes in the soil microbiome occur after thaw with only small alterations at sub-zero temperatures, indicating that fluctuation in water availability is a main driver for changes in the community composition. Despite the knowledge that microbes are active and able to grow under sub-zero temperature [57, 58], we observed no changes in the microbial community at changing temperatures below the freezing point. In a previous study, looking at season changes in a potentially active microbial community over a course of one season, we observed very little variation during the winter months, November till April [22]. During this period, the temperature ranged from −14.3 to −3 °C. We believe that the lack of changes in the microbial composition could be due to time of incubation at sub-zero degrees and additionally, the turnover time of rRNA transcripts is decreased at low temperatures.
For all temperatures, the majority of rRNA was produced by bacteria. A large shift in the bacterial community was observed between T2°C and F2°C, which may be linked to the environmental selection for different life strategies conserved at phylum level, and potentially linked to trophic strategies as previously shown [27]. During the experiment, we did not detect any changes in RNA concentration and 16S rRNA copy number. This was likely due to an increase in abundance of heterotrophic flagellates predating on bacteria and thereby limiting potential increases in total RNA concentration and 16S rRNA copy number due to enhanced bacterial activity. The present data enabled us to reveal the dynamics of the soil microbial community upon thawing, which may have implications for our understanding of the degradation of soil organic matter during the short Arctic summer.
Temperature response of the bacterial community
The clear microbiome structure differences observed between thawing and freezing events were linked to the incubation time at 2 °C. Indeed, the molecular pictures obtained via RNA from the same communities ended up looking completely different based on this decisive 2 °C threshold. Even though this separation was mainly driven by large abundance changes in bacterial community composition, fungi and microeukaroytes also showed the same trend, indicating that all the microbial components of this system were impacted. These changes in bacterial composition upon a thawing event are consistent with other studies involving thaw of Arctic soils [25, 59–61]. Furthermore, the overall composition of the bacterial community is consistent with other studies of Arctic soil bacterial communities [13, 22, 37]. The relative abundance of most sequences from potentially active bacterial phyla decreased between T2°C and F2°C, whereas only sequences from Bacteriodetes, Firmicutes and the Beta-Gammaproteobacterial classes increased. The shifts in the potential active bacterial composition at phylum level and proteobacterial class level may be explained by the copiotrophic and oligotropic lifestyle proposed by several authors [27, 28, 30, 31, 62]. Members of the phyla Bacteriodetes and Beta- and Gammaproteobacteria have previously been associated with a copiotrophic lifestyle [27, 63]. We found that their increase between samples T2°C and F2°C when thawing, likely corresponds to enhanced nutrient availability. There is little agreement on whether members of the Firmicutes have a copiotrophic or oligotrophic lifestyle. Another explanation for the increase in Firmicutes rRNA copy number is the capacity of several members within this phylum to produce endospores, including Paenibacillus [64, 65]. In endospore-forming bacteria, the rRNA level is as high or even higher in dormant cells compared with vegetative cells [66]. Thus, the increase in rRNA affiliated to Firmicutes may be a conserved response characteristic from this phylum to an initial increase in nutrient availability and/or due to preparation for dormancy as key nutrients are depleted.
The relative abundance of Actinobacteria and Acidobacteria decreased between the sampling points T2°C and F2°C. Several Acidobacteria taxa have generally been reported as oligotrophic [27, 67, 68], and may be outcompeted by fast-growing bacteria when thawing enhances nutrient availability. In contrast, Actinobacteria have been reported as partly copiotrophic [69–71], which could explain the decrease in potential activity of this phyla. Actinobacteria have been shown to be sensitive to freeze–thaw cycles in active layer soil [25], and to seasonal changes on glacial forefields [72] and in active layer soil [22].
In addition to the change in the prokaryote community during incubation at 2 °C for 16 days, we observed an increase in the relative abundance of Gemmatimonadetes, Bacteroidetes and Betaproteobacteria when temperature increased from −2 to 2 °C. Members of these three taxa responded quickly, within 1.5 days, to soil thawing. The increase in Bacteroidetes and Betaproteobacteria can again be explained by their rapid response to increased nutrient availability [27]. The life strategy of the phylum Gemmatimonadetes have not been studied in soil as far as we know; nevertheless, a study by Aanderud and Lennon [73] showed that Gemmatimonadetes responded rapidly to re-wetting of a soil by incorporating H218O into DNA. A more refined analysis at the OTU level using the Functional Response Group (FRG) analysis proposed by Nunes et al. [74] also showed a rapid response in the relative abundance of rRNA for some bacterial taxa. The analysis resulted in four functional response groups (FRGs) of which two were related to thawing. OTUs belonging to FRG 1 and FRG 3, decreased as the soil thawed, i.e., when temperature increased from −2 to 2 °C. Most OTUs in FRG 1 belonged to Actinobacteria, while FRG 3 was dominated by OTUs affiliated with Acidobacteria. Actinobacteria have been shown to respond negatively to an increase in soil moisture content [75–77], which could explain the decrease in relative abundance of the actinobacterial OTUs when water availability increases upon thawing. In contrast, members of FRG 2 and FRG 4 increased as the temperature increased from −2 to 2 °C. FRG 2 was dominated by the genus Gemmatimonas (Gemmatimonadetes). The ecological role of Gemmatimonas in soil systems is not well-understood due to the lack of strains isolated from soil: the only representative of Gemmatimonas, G. aurantica was isolated from a wastewater treatment plant [78]. Further, even though OTUs assigned to Gemmatimonas have been found in Arctic soils [22, 78, 79], little is known about the response of Gemmatimonas spp. to thawing of soil and changes in nutrient availability. FRG 4 was dominated by unclassified Bacteriodetes and an unclassified betaproteobacterium. The response of these OTUs may be explained by their lifestyle as explained above.
In contrast to our initial hypothesis, very few members of the bacterial community responded to the temperature increase from −10 to −2 °C. We also observed negligible changes in the relative abundance of rRNA-based OTUs when the temperature decreased from −2 to −10 °C. Freezing is likely resulting in drastic decrease in nutrient availability and hence induce starvation. Bacteria decrease the number of 16S rRNA upon starvation [80], but the low temperature may also lower the turnover of rRNA.
Average 16S rRNA copy number in bacterial community
The number of rRNA operons in bacterial genomes is correlated with growth rates [81, 82], and during early succession, soils are dominated by bacteria with a high number of rRNA operons, whereas bacteria with low numbers dominate during late succession phases [83, 84]. In a glacier forefield, the average rRNA gene copy number per cell was highest close to the glacier and decreased with distance from the glacier, which indicates that copiotrophic bacteria have an advantage in the initial phases of soil development [83]. To further investigate our hypothesis that the relative abundance of copiotrophic r-selected bacteria increase upon thawing of the soil, we estimated the number of 16S rRNA gene copies for each OTU, and then found the average 16S rRNA gene copy number for each sample. We observed an increase in the average 16S rRNA gene copy number per cell in the bacterial community after thawing, indicating a dominance of copiotrophic bacteria within 16 days of thawing. We hypothesize that a decrease in the 16S rRNA gene copy number per cell would occur if the soil was incubated at 2 °C for longer than 16 days as oligotrophic bacteria are expected to dominate later.
A linear regression between the log2-fold change in relative abundance of each OTU due to thawing and the 16S rRNA gene copy numbers revealed that 10.2% of the variation in the change could be explained by the copy number. Also, the average 16S rRNA gene copy number per cell of the 10 most abundant phyla and their log2-fold changes revealed that phyla assumed to be dominated by copiotrophic taxa increased the most upon thawing (Supplementary Figure S7). This further contributes to the hypothesis that copiotrophic bacteria dominated the soil within the first 16 days of thawing.
Temperature response of microeukaryotes
As seen for bacteria, thawing caused a marked shift in the microeukaryote community. Freeze–thaw cycles disturb the physical structure of the soil and hereby liberate organic substrate for the primary decomposers, for example bacteria, which will subsequently be grazed by protozoa that, along with the metazoan nematodes, are the primary grazing predators [85]. The thawing and the changed soil structure will create a larger water filled environment likely leading to an increased predation. Hence, to understand changes in abundance and diversity in the bacterial community, we must focus on the dynamics of their grazing predators. The microeukaryotes that increased during the thaw period can all be included in the large paraphyletic group called heterotrophic flagellates. This group includes various stramenopiles, for example bicocoecids, a group containing many unknown species in soil [86], and the common heterotrophic chrysophyte Spumella. Kinetoplastids, e.g., Neobodo, and in particular cercozoa, a very abundant group in soil Cercozoa [87], also strongly increased; in particular glissomonads—mainly Neoheteromita (formerly Heteromita) [88]. Heterotrophic flagellates are smaller than other protozoa, i.e., amoebae and ciliates [89] and therefore on average they have much shorter doubling times—down to a few hours, which is comparable with those of bacteria [90]. These small flagellates are faster colonizers and flourish at an earlier successional stage than larger protozoa [91]. Therefore, they are able to take advantage of the short period of thaw. In contrast, the other bacterivorous protozoa as ciliates and amoebae, and also metazoan nematodes, annelids and arthropods declined during the thaw period. Soil protozoa builds up populations during periods of growth and concomitantly form cysts; cyst formation is an intricate process and can hardly be done in response to the onset of unfavourable conditions but must rather be accomplished before initiation of a new frost period. Thus, a certain percentage of the actively growing cells are transferred to a cyst-bank, a percentage that increases drastically when cells are crowded [92]. Not all cysts will survive a frost period; they will be degraded by bacteria after onset of thaw and time is required to build up new populations. The thaw period in this study was only long enough to build up populations of heterotrophic flagellates. Longer thaw periods are most likely needed to keep up the population of the larger microeukaryotes.
We observed no change in 16S rRNA copy number using quantitative PCR or total RNA concentration after extraction and DNase treatment, which indicate that the number of potentially active microorganisms remained stable during the experiment. Furthermore, we detected a decline in dissolved organic carbon and an increase in CO2 production between the two events indicating microbial activity and potential growth. We suggest that the stable total RNA and 16S rRNA concentrations are a consequence of grazing of bacteria by heterotrophic flagellates [85]. As discussed above, the relative abundance of potentially active copiotrophic bacteria increased upon thawing, which likely leads to growth of these bacteria and initiates grazing by the fast-growing heterotrophic flagellates. Even though protozoa are known to prey selectively on specific phylogenetic bacterial groups [93, 94], their grazing may not be limited to fast-growing bacteria but may also involve slow- or non-growing bacteria. This could explain the decrease in the relative abundance of oligotrophic bacteria following thaw and the stable RNA level.
Temperature response of the fungal community
The fungal community structure showed high similarity to communities in other Arctic soils [15, 95, 96], with a dominance of Agaricomycetes (Basidiomycota) and Ascomycota. However, in contrast to other studies, we found that the dominant class of Ascomycota was Pezizomycetes and not Leotiomycetes.
The fungal proportion of the microbial community declined dramatically when the soil started to thaw. In alpine soil systems, the ratio between fungi and bacteria shows strong seasonal variation with fungal dominance during winter, which indicates that fungi respond more dramatically to changes in soil temperature than bacteria [19, 20]. In accordance, Lipson et al. [19] showed that fungal hyphal length decreased during summer. This may explain the decline in the relative abundance of fungi following thaw. However, destruction of hyphae during our soil homogenization and disruption of the connection between plant-associated fungi and their plant partner may also explain the decline. Physical disturbance and soil handling can lead to breakage of fungal hyphae [97–99] and may expose fungal rRNA to degradation by other soil microorganisms. Of the 58 fungal OTUs that decreased between T2°C and F2°C, 17 were assigned as ectomycorrhizal fungi (according to Tedersoo et al. [15]). These 17 OTUs correspond to a total relative abundance of 3.2%, but this percentage is a low estimate, as 20 of the 58 OTUs could not be assigned to an ecological function. This may be explained by the homogenization of the samples and removal of plants from the soil core prior to homogenization, since ectomycorrhizal fungi form symbiotic relationship with plants. Furthermore, we were unable to detect any significant changes in single celled fungal group Saccharomycotina using nbGLM between the T2°C and F2°C, indicating that this fungal group were unaffected by the changes. Despite the general decline in the relative abundance of fungi, five fungal OTUs increased between T2°C and F2°C. The two most dominating of these were an OTU assigned to an unclassified Mucoromycotina and an OTU assigned to zygomycete sp. AM-2008a. Zygomycetes and members of Mucoromycotina have been described as fast growing and are often among the primary fungal colonizers in systems with high concentrations of simple carbohydrates [100–103]. They increase their abundance if mycorrhizal fungi are disconnected from their plant partner [104]. These groups also harbour members that proliferate in low-temperature environments. Thus, fungi belonging to the Zygomycete orders Mucorales and Mortierellales that were isolated from snow in a subalpine forest in Colorado, USA, had fast growth rates at −2 °C [102, 105], and fungi isolated close to a Svalbard glacier all belonged to Zygomycetes [106]. The combination of activity at low temperature and fast growth rates enables these fungi to increase their potential activity soon after soil thawing. The dramatic decline in fungal rRNA reads may have implications for the functionality of newly thawed soil because fungi play important roles in the degradation of recalcitrant soil organic matter, such as lignin, hemicellulose and cellulose [107]. Concomitant to the fungal decline, two bacterial phyla with members known to degrade complex organic matter, Acidobacteria [68, 108, 109] and Actinobacteria [107, 110, 111], declined. This decline could be caused by the increased flagellate grazing on these slow growing taxa following the thawing induced increase in fast-growing copiotrophic bacteria. The increase in potentially copiotrophic bacteria and fungi degrading mainly labile soil organic matter [27], and the concomitant decrease in oligotrophic bacteria, may delay degradation of recalcitrant soil organic matter when active layer soil thaws.
Conclusions
The RNAseq-based method allowed us to investigate the dynamics of all major members of the soil microbial community during a thawing and freezing event. Pronounced changes were observed in the microbial communities after the soil was thawed and kept at 2 °C for 16 days, for example a drastic decline in fungal potential activity. For bacteria, these changes could be related to the potential copiotrophic and oligotrophic lifestyle of the different bacterial taxa, with the relative abundance of potentially active copiotrophic bacteria increasing even within 1.5 days of thawing, showing a fast response of the these bacteria groups to thaw. As copiotrophic bacteria increased, heterotrophic flagellates grazing on bacteria also increased. These observations led us to hypothesize that protozoan grazing was not limited to the surge in copiotrophic bacteria, but also affected presumed oligotrophic bacterial taxa that are likely to possess important roles in the degradation of plant litter and soil organic matter. Thus, the combination of a decrease in potential fungal activity and likely grazing of bacteria by protozoa could potentially delay the degradation of more complex organic matter upon thawing of active layer soil. This may have implications for our understanding of plant nutrient availability and CO2 emission from Arctic soils.
Supplementary information
Acknowledgements
This work was supported by the Danish National Research Foundation (CENPERM DNRF100), Danish Geocenter for sequencing funding, Danish Council for Independent Research (DFF-4002-00274) and the EU MSC Action 675546: MicroArctic. The authors thank Pia Bach Jacobsen for help and technical support in the laboratory, Samuel Faucherre for sampling the soil core, and Annelise Kjøller for discussions on the fungal community changes. The authors would also like to thank Martin Asser Hansen for bioinformatics support.
Data availability
Complete OTU tables are available in the Supplementary datasheet. Sequence data generated in this study was deposited in the NCBI Sequence Read Archive and are accessible through accession number SRP124869.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-019-0351-x) contains supplementary material, which is available to authorized users.
References
- 1.Zhang T, Barry RG, Knowles K, Heginbottom J, Brown J. Statistics and characteristics of permafrost and ground‐ice distribution in the Northern Hemisphere. Polar Geogr. 1999;23:132–54. [Google Scholar]
- 2.Hugelius G, Strauss J, Zubrzycki S, Harden JW, Schuur EAG, Ping CL, et al. Estimated stocks of circumpolar permafrost carbon with quantified uncertainty ranges and identified data gaps. Biogeosciences. 2014;11:6573–93. [Google Scholar]
- 3.Siegenthaler U, Sarmiento J. Atmospheric carbon dioxide and the ocean. Nature. 1993;365:119–25. [Google Scholar]
- 4.Screen JA, Simmonds I. The central role of diminishing sea ice in recent Arctic temperature amplification. Nature. 2010;464:1334–7. doi: 10.1038/nature09051. [DOI] [PubMed] [Google Scholar]
- 5.Serreze M, Barrett A, Stroeve J, Kindig D, Holland M. The emergence of surface-based Arctic amplification. Cryosphere. 2009;3:11. [Google Scholar]
- 6.Zhang T. Spatial and temporal variability in active layer thickness over the Russian Arctic drainage basin. J Geophys Res. 2005; 110, D16101.
- 7.Hayes DJ, Kicklighter DW, McGuire AD, Chen M, Zhuang Q, Yuan F, et al. The impacts of recent permafrost thaw on land–atmosphere greenhouse gas exchange. Environ Res Lett. 2014;9:045005. [Google Scholar]
- 8.Geisen S, Tveit AT, Clark IM, Richter A, Svenning MM, Bonkowski M, et al. Metatranscriptomic census of active protists in soils. ISME J. 2015;9:2178–90. doi: 10.1038/ismej.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pautler BG, Simpson AJ, Mcnally DJ, Lamoureux SF, Simpson MJ. Arctic permafrost active layer detachments stimulate microbial activity and degradation of soil organic matter. Environ Sci Technol. 2010;44:4076–82. doi: 10.1021/es903685j. [DOI] [PubMed] [Google Scholar]
- 10.Schuur EAG, Bockheim J, Canadell JG, Euskirchen E, Field CB, Goryachkin SV, et al. Vulnerability of permafrost carbon to climate change: implications for the global carbon cycle. BioScience. 2008;58:701. [Google Scholar]
- 11.Graham DE, Wallenstein MD, Vishnivetskaya TA, Waldrop MP, Phelps TJ, Pfiffner SM, et al. Microbes in thawing permafrost: the unknown variable in the climate change equation. ISME J. 2012;6:709–12. doi: 10.1038/ismej.2011.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradley JA, Arndt S, Šabacká M, Benning LG, Barker GL, Blacker JJ, et al. Microbial dynamics in a high Arctic glacier forefield: a combined field, laboratory, and modelling approach. Biogeosciences. 2016;13:5677–96. [Google Scholar]
- 13.Chu H, Fierer N, Lauber CL, Caporaso J, Knight R, Grogan P. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ Microbiol. 2010;12:2998–3006. doi: 10.1111/j.1462-2920.2010.02277.x. [DOI] [PubMed] [Google Scholar]
- 14.Neufeld JD, Mohn WW. Unexpectedly high bacterial diversity in arctic tundra relative to boreal forest soils, revealed by serial analysis of ribosomal sequence tags. Appl Environ Microbiol. 2005;71:5710–8. doi: 10.1128/AEM.71.10.5710-5718.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tedersoo L, Bahram M, Põlme S, Kõljalg U, Yorou NS, Wijesundera R, et al. Global diversity and geography of soil fungi. science. 2014;346:1256688. doi: 10.1126/science.1256688. [DOI] [PubMed] [Google Scholar]
- 16.Timling I, Walker DA, Nusbaum C, Lennon NJ, Taylor DL. Rich and cold: diversity, distribution and drivers of fungal communities in patterned-ground ecosystems of the North American Arctic. Microb Ecol. 2014;23:3258–72. doi: 10.1111/mec.12743. [DOI] [PubMed] [Google Scholar]
- 17.Jefferies RL, Walker NA, Edwards KA, Dainty J. Is the decline of soil microbial biomass in late winter coupled to changes in the physical state of cold soils? Soil Biol Biochem. 2010;42:129–35. [Google Scholar]
- 18.Larsen KS, Grogan P, Jonasson S, Michelsen A. Respiration and microbial dynamics in two subarctic ecosystems during winter and spring thaw: effects of increased snow depth. Arct Antarct Alp Res. 2007;39:268–76. [Google Scholar]
- 19.Lipson D, Schadt C, Schmidt S. Changes in soil microbial community structure and function in an alpine dry meadow following spring snow melt. Microb Ecol. 2002;43:307–14. doi: 10.1007/s00248-001-1057-x. [DOI] [PubMed] [Google Scholar]
- 20.Schadt CW, Martin AP, Lipson DA, Schmidt SK. Seasonal dynamics of previously unknown fungal lineages in tundra soils. science. 2003;301:1359–61. doi: 10.1126/science.1086940. [DOI] [PubMed] [Google Scholar]
- 21.McMahon SK, Wallenstein MD, Schimel JP. A cross-seasonal comparison of active and total bacterial community composition in Arctic tundra soil using bromodeoxyuridine labeling. Soil Biol Biochem. 2011;43:287–95. [Google Scholar]
- 22.Schostag M, Stibal M, Jacobsen CS, Bælum J, Taş N, Elberling B, et al. Distinct summer and winter bacterial communities in the active layer of Svalbard permafrost revealed by DNA-and RNA-based analyses. Front Microbiol. 2015;6:399. doi: 10.3389/fmicb.2015.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zifcakova L, Vetrovsky T, Howe A, Baldrian P. Microbial activity in forest soil reflects the changes in ecosystem properties between summer and winter. Environ Microbiol. 2016;18:288–301. doi: 10.1111/1462-2920.13026. [DOI] [PubMed] [Google Scholar]
- 24.Kumar N, Grogan P, Chu H, Christiansen CT, Walker VK. The effect of freeze-thaw conditions on arctic soil bacterial communities. Biol (Basel) 2013;2:356–77. doi: 10.3390/biology2010356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Männistö MK, Tiirola M, Häggblom MM. Effect of freeze-thaw cycles on bacterial communities of Arctic tundra soil. Microb Ecol. 2009;58:621–31. doi: 10.1007/s00248-009-9516-x. [DOI] [PubMed] [Google Scholar]
- 26.Schimel JP, Clein JS. Microbial response to freeze-thaw cycles in tundra and taiga soils. Soil Biol Biochem. 1996;28:1061–6. [Google Scholar]
- 27.Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology. 2007;88:1354–64. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- 28.Fierer N, Lauber CL, Ramirez KS, Zaneveld J, Bradford MA, Knight R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012;6:1007–17. doi: 10.1038/ismej.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nemergut DR, Cleveland CC, Wieder WR, Washenberger CL, Townsend AR. Plot-scale manipulations of organic matter inputs to soils correlate with shifts in microbial community composition in a lowland tropical rain forest. Soil Biol Biochem. 2010;42:2153–60. [Google Scholar]
- 30.Philippot L, Andersson SG, Battin TJ, Prosser JI, Schimel JP, Whitman WB, et al. The ecological coherence of high bacterial taxonomic ranks. Nat Rev Microbiol. 2010;8:523–9. doi: 10.1038/nrmicro2367. [DOI] [PubMed] [Google Scholar]
- 31.Ramirez KS, Craine JM, Fierer N. Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob Chang Biol. 2012;18:1918–27. [Google Scholar]
- 32.Stewart EJ. Growing unculturable bacteria. J Bacteriol. 2012;194:4151–60. doi: 10.1128/JB.00345-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacquiod S, Stenbæk J, Santos SS, Winding A, Sørensen SJ, Priemé A. Metagenomes provide valuable comparative information on soil microeukaryotes. Res Microbiol. 2016;167:436–50. doi: 10.1016/j.resmic.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 34.Carini P, Marsden PJ, Leff JW, Morgan EE, Strickland MS, Fierer N. Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat Microbiol. 2016;2:16242. doi: 10.1038/nmicrobiol.2016.242. [DOI] [PubMed] [Google Scholar]
- 35.Blazewicz SJ, Barnard RL, Daly RA, Firestone MK. Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J. 2013;7:2061–8. doi: 10.1038/ismej.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Urich T, Lanzén A, Qi J, Huson DH, Schleper C, Schuster SC. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One. 2008;3:e2527. doi: 10.1371/journal.pone.0002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tveit A, Schwacke R, Svenning MM, Urich T. Organic carbon transformations in high-Arctic peat soils: key functions and microorganisms. ISME J. 2013;7:299–311. doi: 10.1038/ismej.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hultman J, Waldrop MP, Mackelprang R, David MM, McFarland J, Blazewicz SJ, et al. Multi-omics of permafrost, active layer and thermokarst bog soil microbiomes. Nature. 2015;521:208–12. doi: 10.1038/nature14238. [DOI] [PubMed] [Google Scholar]
- 39.Kirtman B, Power S, Adedoyin A, Boer G, Bojariu R, Camilloni I et al. Near-term climate change: projections and predictability. In: Stocker TF, Qin D, Plattner GK, Tignor M, Allen SK, Boschung J, et al.(eds.) Climate Change: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge, United Kingdom and New York, NY, USA 2013.
- 40.Rønn R, McCaig AE, Griffiths BS, Prosser JI. Impact of protozoan grazing on bacterial community structure in soil microcosms. Appl Environ Microbiol. 2002;68:6094–105. doi: 10.1128/AEM.68.12.6094-6105.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ingólfsson Oacute. Fingerprints of Quaternary glaciations on Svalbard. Geol Soc Spec Publ. 2011;354:15–31. [Google Scholar]
- 42.Bang-Andreasen T, Schostag M, Priemé A, Elberling B, Jacobsen CS. Potential microbial contamination during sampling of permafrost soil assessed by tracers. Sci Rep. 2017;7:43338. doi: 10.1038/srep43338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lane D. 16S/23S rRNA sequencing. Nucleic acid techniques in bacterial systematics: Edited by E. Stackebrandt & M. Goodfellow. Chichester: Wiley. 125–75, 1991.
- 44.Kopylova E, Noé L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28:3211–7. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 45.Bengtsson-Palme J, Hartmann M, Eriksson KM, Pal C, Thorell K, Larsson DG, et al. METAXA2: improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Mol Ecol Resour. 2015;15:1403–14. doi: 10.1111/1755-0998.12399. [DOI] [PubMed] [Google Scholar]
- 46.Jacquiod S, Brejnrod A, Morberg SM, Abu Al‐Soud W, Sørensen SJ, Riber L. Deciphering conjugative plasmid permissiveness dynamics in wastewater microbiomes. Microb Ecol 2017;26:3556–3571. [DOI] [PubMed]
- 47.Team Rc. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. p. 2013. [Google Scholar]
- 48.Hammer Ø, Harper D, Ryan P. PAST-palaeontological statistics, ver. 1.89. Palaeontol Electronica. 2001;4:1–9.
- 49.Oksanen J, Kindt R, Legendre P, O’Hara B, Stevens MHH, Oksanen MJ, et al. The vegan package. Community Ecol Package. 2007;10:631–7. [Google Scholar]
- 50.Dray S, Dufour AB. The ade4 package: implementing the duality diagram for ecologists. J Stat Softw. 2007;22:1–20. [Google Scholar]
- 51.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warnes GR, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A et al. gplots: Various R programming tools for plotting data. R package version 2, 2009. https://CRAN.R-project.org/package=gplots.
- 53.Juggins S (2009). rioja: analysis of Quaternary science data. R package version 0.5-6. http://cranr-projectorg/package=rioja.
- 54.Neuwirth E. RColorBrewer: ColorBrewer palettes. R package version 1, 2011. https://cran.rproject.org/web/packages/RColorBrewer/.
- 55.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–21. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tuorto SJ, Darias P, McGuinness LR, Panikov N, Zhang T, Haggblom MM, et al. Bacterial genome replication at subzero temperatures in permafrost. ISME J. 2014;8:139–49. doi: 10.1038/ismej.2013.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nikrad MP, Kerkhof LJ, Häggblom MM. The subzero microbiome: microbial activity in frozen and thawing soils. FEMS Microbiol Ecol. 2016;92:fiw081. doi: 10.1093/femsec/fiw081. [DOI] [PubMed] [Google Scholar]
- 59.Coolen MJ, van de Giessen J, Zhu EY, Wuchter C. Bioavailability of soil organic matter and microbial community dynamics upon permafrost thaw. Environ Microbiol. 2011;13:2299–314. doi: 10.1111/j.1462-2920.2011.02489.x. [DOI] [PubMed] [Google Scholar]
- 60.Deng J, Gu Y, Zhang J, Xue K, Qin Y, Yuan M, et al. Shifts of tundra bacterial and archaeal communities along a permafrost thaw gradient in Alaska. Microb Ecol. 2015;24:222–34. doi: 10.1111/mec.13015. [DOI] [PubMed] [Google Scholar]
- 61.Mackelprang R, Waldrop MP, DeAngelis KM, David MM, Chavarria KL, Blazewicz SJ, et al. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature. 2011;480:368–71. doi: 10.1038/nature10576. [DOI] [PubMed] [Google Scholar]
- 62.Placella SA, Brodie EL, Firestone MK. Rainfall-induced carbon dioxide pulses result from sequential resuscitation of phylogenetically clustered microbial groups. Proc Natl Acad Sci USA. 2012;109:10931–6. doi: 10.1073/pnas.1204306109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cleveland CC, Nemergut DR, Schmidt SK, Townsend AR. Increases in soil respiration following labile carbon additions linked to rapid shifts in soil microbial community composition. Biogeochemistry. 2007;82:229–40. [Google Scholar]
- 64.Onyenwoke RU, Brill JA, Farahi K, Wiegel J. Sporulation genes in members of the low G+C Gram-type-positive phylogenetic branch (Firmicutes) Arch Microbiol. 2004;182:182–92. doi: 10.1007/s00203-004-0696-y. [DOI] [PubMed] [Google Scholar]
- 65.Paredes CJ, Alsaker KV, Papoutsakis ET. A comparative genomic view of clostridial sporulation and physiology. Nat Rev Microbiol. 2005;3:969–78. doi: 10.1038/nrmicro1288. [DOI] [PubMed] [Google Scholar]
- 66.Chambon P, Deutscher. MP, Kornberg. A. Biochemical studies of bacterial sporulation and germination X. Ribosomes and nucleic acids of vegetative cells and spores of bacillus megaterium. J Biol Chem. 1968;243:5110–6. [PubMed]
- 67.Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 2009;3:442–53. doi: 10.1038/ismej.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kielak AM, Barreto CC, Kowalchuk GA, van Veen JA, Kuramae EE. The ecology of Acidobacteria: moving beyond genes and genomes. Front Microbiol. 2016;7:744 [DOI] [PMC free article] [PubMed]
- 69.Eilers KG, Lauber CL, Knight R, Fierer N. Shifts in bacterial community structure associated with inputs of low molecular weight carbon compounds to soil. Soil Biol Biochem. 2010;42:896–903. [Google Scholar]
- 70.Pan Y, Cassman N, de Hollander M, Mendes LW, Korevaar H, Geerts RH, et al. Impact of long-term N, P, K, and NPK fertilization on the composition and potential functions of the bacterial community in grassland soil. FEMS Microbiol Ecol. 2014;90:195–205. doi: 10.1111/1574-6941.12384. [DOI] [PubMed] [Google Scholar]
- 71.Buelow HN, Winter AS, Van Horn DJ, Barrett JE, Gooseff MN, Schwartz E, et al. Microbial Community Responses to Increased Water and Organic Matter in the Arid Soils of the McMurdo Dry Valleys, Antarctica. Front Microbiol. 2016;7:1040. doi: 10.3389/fmicb.2016.01040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lazzaro A, Brankatschk R, Zeyer J. Seasonal dynamics of nutrients and bacterial communities in unvegetated alpine glacier forefields. Appl Soil Ecol. 2012;53:10–22. [Google Scholar]
- 73.Aanderud ZT, Lennon JT. Validation of heavy-water stable isotope probing for the characterization of rapidly responding soil bacteria. Appl Environ Microbiol. 2011;77:4589–96. doi: 10.1128/AEM.02735-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nunes Inês, Jacquiod Samuel, Brejnrod Asker, Holm Peter E., Johansen Anders, Brandt Kristian K., Priemé Anders, Sørensen Søren J. Coping with copper: legacy effect of copper on potential activity of soil bacteria following a century of exposure. FEMS Microbiology Ecology. 2016;92(11):fiw175. doi: 10.1093/femsec/fiw175. [DOI] [PubMed] [Google Scholar]
- 75.Barnard RL, Osborne CA, Firestone MK. Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 2013;7:2229–41. doi: 10.1038/ismej.2013.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.LeBlanc JC, Gonçalves ER, Mohn WW. Global response to desiccation stress in the soil actinomycete Rhodococcus jostii RHA1. Appl Environ Microbiol. 2008;74:2627–36. doi: 10.1128/AEM.02711-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McHugh TA, Koch GW, Schwartz E. Minor changes in soil bacterial and fungal community composition occur in response to monsoon precipitation in a semiarid grassland. Microb Ecol. 2014;68:370–8. doi: 10.1007/s00248-014-0416-3. [DOI] [PubMed] [Google Scholar]
- 78.Zhang H, Sekiguchi Y, Hanada S, Hugenholtz P, Kim H, Kamagata Y, et al. Gemmatimonas aurantiaca gen. nov., sp. nov., a Gram-negative, aerobic, polyphosphate-accumulating micro-organism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov. Int J Syst Evol Microbiol. 2003;53:1155–63. doi: 10.1099/ijs.0.02520-0. [DOI] [PubMed] [Google Scholar]
- 79.Wilhelm RC, Niederberger TD, Greer C, Whyte LG. Microbial diversity of active layer and permafrost in an acidic wetland from the Canadian High Arctic. Can J Microbiol. 2011;57:303–15. doi: 10.1139/w11-004. [DOI] [PubMed] [Google Scholar]
- 80.Deutscher MP. Degradation of stable RNA in bacteria. J Biol Chem. 2003;278:45041–4. doi: 10.1074/jbc.R300031200. [DOI] [PubMed] [Google Scholar]
- 81.Klappenbach JA, Dunbar JM, Schmidt TM. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol. 2000;66:1328–33. doi: 10.1128/aem.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roller BR, Stoddard SF, Schmidt TM. Exploiting rRNA operon copy number to investigate bacterial reproductive strategies. Nat Microbiol. 2016;1:16160. doi: 10.1038/nmicrobiol.2016.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nemergut DR, Knelman JE, Ferrenberg S, Bilinski T, Melbourne B, Jiang L, et al. Decreases in average bacterial community rRNA operon copy number during succession. ISME J. 2016;10:1147–56. doi: 10.1038/ismej.2015.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shrestha PM, Noll M, Liesack W. Phylogenetic identity, growth-response time and rRNA operon copy number of soil bacteria indicate different stages of community succession. Environ Microbiol. 2007;9:2464–74. doi: 10.1111/j.1462-2920.2007.01364.x. [DOI] [PubMed] [Google Scholar]
- 85.Rønn R, Vestergård M, Ekelund F. Interactions between bacteria, protozoa and nematodes in soil. Acta Protozool. 2015;51:223–35. [Google Scholar]
- 86.Harder CB, Ekelund F, Karpov SA. Ultrastructure and phylogenetic position of Regin rotiferus and Otto terricolus genera et species novae (Bicosoecida, Heterokonta/Stramenopiles) Protist. 2014;165:144–60. doi: 10.1016/j.protis.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 87.Harder Christoffer Bugge, Rønn Regin, Brejnrod Asker, Bass David, Al-Soud Waleed Abu, Ekelund Flemming. Local diversity of heathland Cercozoa explored by in-depth sequencing. The ISME Journal. 2016;10(10):2488–2497. doi: 10.1038/ismej.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Howe AT, Bass D, Vickerman K, Chao EE, Cavalier-Smith T. Phylogeny, taxonomy, and astounding genetic diversity of Glissomonadida ord. nov., the dominant gliding zooflagellates in soil (Protozoa: Cercozoa) Protist. 2009;160:159–89. doi: 10.1016/j.protis.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 89.Ekelund F, Rønn R. Notes on protozoa in agricultural soil with emphasis on heterotrophic flagellates and naked amoebae and their ecology. FEMS Microbiol Rev. 1994;15:321–53. doi: 10.1111/j.1574-6976.1994.tb00144.x. [DOI] [PubMed] [Google Scholar]
- 90.Fenchel T (2013). Ecology of Protozoa: The biology of free-living phagotropic protists. Springer-Verlag. Berlin.
- 91.Altenburger A, Ekelund F, Jacobsen CS. Protozoa and their bacterial prey colonize sterile soil fast. Soil Biol Biochem. 2010;42:1636–9. [Google Scholar]
- 92.Ekelund F, Frederiksen HB, Ronn R. Population dynamics of active and total ciliate populations in arable soil amended with wheat. Appl Environ Microbiol. 2002;68:1096–101. doi: 10.1128/AEM.68.3.1096-1101.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ekelund F, Saj S, Vestergård M, Bertaux J, Mikola J. The “soil microbial loop” is not always needed to explain protozoan stimulation of plants. Soil Biol Biochem. 2009;41:2336–42. [Google Scholar]
- 94.Pedersen AL, Winding A, Altenburger A, Ekelund F. Protozoan growth rates on secondary-metabolite-producing Pseudomonas spp. correlate with high-level protozoan taxonomy. FEMS Microbiol Lett. 2011;316:16–22. doi: 10.1111/j.1574-6968.2010.02182.x. [DOI] [PubMed] [Google Scholar]
- 95.Choma M, Barta J, Santruckova H, Urich T. Low abundance of Archaeorhizomycetes among fungi in soil metatranscriptomes. Sci Rep. 2016;6:38455. doi: 10.1038/srep38455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gittel A, Barta J, Kohoutova I, Mikutta R, Owens S, Gilbert J, et al. Distinct microbial communities associated with buried soils in the Siberian tundra. ISME J. 2014;8:841–53. doi: 10.1038/ismej.2013.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Frey S, Elliott E, Paustian K. Bacterial and fungal abundance and biomass in conventional and no-tillage agroecosystems along two climatic gradients. Soil Biol Biochem. 1999;31:573–85. [Google Scholar]
- 98.Beare M, Hu S, Coleman D, Hendrix P. Influences of mycelial fungi on soil aggregation and organic matter storage in conventional and no-tillage soils. Appl Soil Ecol. 1997;5:211–9. [Google Scholar]
- 99.Shamlou PA, Makagiansar H, Ison A, Lilly M, Thomas C. Turbulent breakage of filamentous microorganisms in submerged culture in mechanically stirred bioreactors. Chem Eng Sci. 1994;49:2621–31. [Google Scholar]
- 100.Hanson CA, Allison SD, Bradford MA, Wallenstein MD, Treseder KK. Fungal taxa target different carbon sources in forest soil. Ecosystems. 2008;11:1157–67. [Google Scholar]
- 101.Richardson M. The ecology of the Zygomycetes and its impact on environmental exposure. Clin Microbiol Infect. 2009;15:2–9. doi: 10.1111/j.1469-0691.2009.02972.x. [DOI] [PubMed] [Google Scholar]
- 102.Schmidt SK, Wilson KL, Meyer AF, Gebauer MM, King AJ. Phylogeny and ecophysiology of opportunistic “snow molds” from a subalpine forest ecosystem. Microb Ecol. 2008;56:681–7. doi: 10.1007/s00248-008-9387-6. [DOI] [PubMed] [Google Scholar]
- 103.Shanthi S, Vittal BPR. Fungi associated with decomposing leaf litter of cashew (Anacardium occidentale) Mycology. 2010;1:121–9. [Google Scholar]
- 104.Lindahl BD, de Boer W, Finlay RD. Disruption of root carbon transport into forest humus stimulates fungal opportunists at the expense of mycorrhizal fungi. ISME J. 2010;4:872–81. doi: 10.1038/ismej.2010.19. [DOI] [PubMed] [Google Scholar]
- 105.Schmidt SK, Wilson KL, Monson RK, Lipson DA. Exponential growth of “snow molds” at sub-zero temperatures: an explanation for high beneath-snow respiration rates and Q 10 values. Biogeochemistry. 2008;95:13–21. [Google Scholar]
- 106.Tsuji M, Uetake J, Tanabe Y. Changes in the fungal community of Austre Brøggerbreen deglaciation area, Ny-Ålesund, Svalbard, High Arctic. Mycoscience. 2016;57:448–51. [Google Scholar]
- 107.Perez J, Munoz-Dorado J, de la Rubia T, Martinez J. Biodegradation and biological treatments of cellulose, hemicellulose and lignin: an overview. Int Micro. 2002;5:53–63. doi: 10.1007/s10123-002-0062-3. [DOI] [PubMed] [Google Scholar]
- 108.Eichorst SA, Kuske CR, Schmidt TM. Influence of plant polymers on the distribution and cultivation of bacteria in the phylum Acidobacteria. Appl Environ Microbiol. 2011;77:586–96. doi: 10.1128/AEM.01080-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rawat SR, Mannisto MK, Bromberg Y, Haggblom MM. Comparative genomic and physiological analysis provides insights into the role of Acidobacteria in organic carbon utilization in Arctic tundra soils. FEMS Microbiol Ecol. 2012;82:341–55. doi: 10.1111/j.1574-6941.2012.01381.x. [DOI] [PubMed] [Google Scholar]
- 110.Vetrovsky T, Steffen KT, Baldrian P. Potential of cometabolic transformation of polysaccharides and lignin in lignocellulose by soil Actinobacteria. PLoS One. 2014;9:e89108. doi: 10.1371/journal.pone.0089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Warren R. Microbial hydrolysis of polysaccharides. Annu Rev Microbiol. 1996;50:183–212. doi: 10.1146/annurev.micro.50.1.183. [DOI] [PubMed] [Google Scholar]
- 112.Padmanabhan P, Padmanabhan S, DeRito C, Gray A, Gannon D, Snape J, et al. Respiration of 13C-labeled substrates added to soil in the field and subsequent 16S rRNA gene analysis of 13C-labeled soil DNA. Appl Environ Microbiol. 2003;69:1614–622. doi: 10.1128/AEM.69.3.1614-1622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Complete OTU tables are available in the Supplementary datasheet. Sequence data generated in this study was deposited in the NCBI Sequence Read Archive and are accessible through accession number SRP124869.