Abstract
As a fascinating and complicated nanomotor, chloroplast ATP synthase comprises nine subunits encoded by both the nuclear and plastid genomes. Because of its uneven subunit stoichiometry, biogenesis of ATP synthase and expression of plastid-encoded ATP synthase genes requires assistance by nucleus-encoded factors involved in transcriptional, post-transcriptional, and translational steps. In this study, we report a P-class pentatricopeptide repeat (PPR) protein BFA2 (Biogenesis Factor required for ATP synthase 2) that is essential for accumulation of the dicistronic atpH/F transcript in Arabidopsis chloroplasts. A loss-of-function mutation in BFA2 results in a specific reduction of more than 3/4 of chloroplast ATP synthase, which is likely due to the absence of dicistronic atpH/F transcript. BFA2 protein contains 22 putative PPR motifs and exclusively localizes in the chloroplast. Bioinformatics and Electrophoretic Mobility Shift Assays (EMSA) analysis showed that BFA2 binds to the consensus sequence of the atpF-atpA intergenic region in a sequence-specific manner. However, translation initiation of the atpA was not affected in the bfa2 mutant. Thus, we propose that the chloroplast PPR protein BFA2 mainly acts as barrier to prevent the atpH/F transcript degradation by exoribonucleases by binding to the consensus sequence of the atpF-atpA intergenic region.
Keywords: chloroplast ATP synthase, PPR protein, gene expression, photosynthesis, stability
Introduction
Chloroplasts in photosynthetic eukaryotes are thought to have originated from cyanobacteria through endosymbiosis. During evolution, most of the genes from the cyanobacterial ancestor were transferred to the nucleus of the host cell and chloroplasts have only retained about 100 genes (Martin et al., 2002). These plastid genes encode proteins required for transcription and translation as well as the essential components of photosynthetic complexes. To ensure efficient gene expression, chloroplasts require a vast number of nuclear-encoded protein factors facilitating transcription, RNA stabilization, splicing, editing, and translation (Stern et al., 2010; Barkan, 2011). Among these factors, pentatricopeptide repeat (PPR) proteins are highly prominent and involved in various steps of RNA metabolism and protein translation (Schmitz-Linneweber and Small, 2008). There are hundreds of PPR proteins in land plants most of which function in chloroplast and mitochondrial gene expression (Barkan and Small, 2014). PPR proteins comprise a large class of proteins with tandem arrays of a 35-amino-acid degenerate motif (Small and Peeters, 2000). According to the PPR motif type, PPR proteins can be divided into two major subfamilies, P and PLS. While P-type PPR proteins contain only P (35 amino acids) motifs with one or more tandem arrays, PLS-class PPR proteins have tandem triplet arrays of P, L (35–36 amino acids), and S (31 amino acids) motifs. Extensive studies showed that the P-class PPR proteins are involved in RNA stabilization, cleavage, and splicing as well as in the activation of translation (Barkan and Small, 2014). A few P-class PPR proteins also contain a small-MutS-related (SMR) motif at their C-terminus, which was recently shown to have RNA endonuclease activity in vitro (Zhou et al., 2017). The PLS-class PPR proteins usually contain C-terminal E and DYW motifs which are required for RNA editing (Shikanai, 2015).
Chloroplast ATP synthase is a multi-subunit complex located in the thylakoid membranes. It produces ATP from ADP by utilizing the proton motive force generated by photosynthetic electron transport. Chloroplast ATP synthase is composed of the two CFo and CF1 modules, and they contain five and four subunits with the stoichiometry α3β3γ1ε1δ1 and I1II1III14IV1, respectively (Hahn et al., 2018), encoded by both the nuclear and chloroplast genomes. Chloroplast-encoded ATP synthase subunits arise from two polycistronic chloroplast transcription units, the large (atpI/H/F/A) and the small (atpB/E) atp operons. Both operons are transcribed by the plastid-encoded RNA polymerase (PEP) and several sigma factors are required (Malik Ghulam et al., 2012).
During the past decade, several nucleus-encoded factors have been shown to be involved in the expression of atp genes. For the large atp operon, P-class PPR protein PPR10 binds to the intergenic regions of atpI-atpH and psaJ-rpl33 (Pfalz et al., 2009). The binding of PPR10 to the 5′ end of atpH not only stabilizes atpH transcripts by blocking 5′→3′ exoribonucleases but also alters the structure of the 5′ end of atpH to promote activation of translation initiation (Prikryl et al., 2011). The atpF gene contains a single intron which belongs to the group-II intron family. Splicing of the atpF intron requires several protein factors such as CRS1, RNC1, WHY1, WTF1, MatK, and AEF1/MPR25 (Till et al., 2001; Watkins et al., 2007; Prikryl et al., 2008; Kroeger et al., 2009; Zoschke et al., 2010; Yap et al., 2015). Besides splicing, PPR protein AEF1/MPR25 is also required for editing atpF RNA in Arabidopsis (Yap et al., 2015). In the chloroplast of Chlamydomonas reinhardtii, the TDA1 protein is involved in the trapping and translation activation of atpA transcripts (Eberhard et al., 2011). In the case of the small atp operon, the PPR-SMR protein ATP4/SVR7 as well as the ATP1 protein have been proposed to be involved in the translation of the atpB/E mRNA in maize and Arabidopsis (McCormac and Barkan, 1999; Zoschke et al., 2012, 2013).
In this study, we report the characterization of a chloroplast PPR protein called BFA2 (Biogenesis Factors required for ATP synthase 2) that binds to the atpF-atpA intergenic region in a sequence-specific manner. Our results demonstrated that binding of BFA2 to the 3′-UTR of atpH/F is essential for stabilization of atpH/F RNA.
Materials and Methods
Plant Material and Growth Conditions
Arabidopsis plants were grown on soil in the greenhouse (80 μmol photons m-2 s-1, 16 h photoperiod, 23oC) for 3–4 weeks. The bfa2-1 mutant was isolated from a collection of pSKI015 insertion Arabidopsis lines using the FluorCam imaging fluorometer (FC 800-C, PSI, Czech Republic) (Zhang et al., 2016). The bfa2-2 mutant (SAIL_571_H02) was obtained from NASC and its T-DNA insertion site was confirmed by genomic PCR and subsequent sequencing of the PCR products. For complementation analysis, a genomic DNA fragment of the BFA2 gene (3753 bp) was cloned into the pBI121 binary vector. The resulting construct was transformed into Agrobacterium tumefaciens C58C and then introduced into bfa2-1 and bfa2-2 plants by floral dip transformation.
RNA Extraction, RNA Blotting, and cRT-PCR Assay
Total RNA was isolated from rosette leaves using TRIzol Reagent (Invitrogen Life Technologies). For RNA blot analyses, a total of 5 μg (for atpB and Actin 7) or 2.5 μg (for atpI, atpH, atpE, atpF, atpF intron, and atpA) RNA was fractioned by electrophoresis on 1.5% formaldehyde-containing agarose gels and blotted onto nylon membranes (Hybond-N+, GE Healthcare). The RNA was fixed by UV crosslinking (HL-2000 HybriLinker). Pre-hybridization and hybridization were carried out at 50oC with the DIG Easy Hyb (Roche) buffer. The probes were amplified from DNA and labeled with digoxigenin-11-dUTP according to the manufacturer’s instructions. Signals were visualized with chemiluminescence analyzer or X-film.
For circular RT-PCR (cRT-PCR) analysis, total RNA was treated with RNase-free DNase I (Takara) to remove the residual DNA before further analysis. 10 μg of total RNA was self-ligated for 2 h at 25oC with 10 U of T4 RNA ligase (New England Biolabs). After ligation, RNA was extracted and resuspended in 10 μl of DEPC-treated water. Reverse transcription was performed using 20 pmol of primer and 5 μg of self-ligated RNA for 1 h at 42oC with 200 U of M-MLV reverse transcriptase (Thermo). After transcription, 1/20th of cDNA was used in a single PCR amplification reaction and the DNA products were then cloned in the pMD-18T vector for sequencing. The primers used this experiment are listed in Supplementary Table S2.
Subcellular Localization of GFP Protein
For subcellular localization of GFP protein, the first 200 amino acids (to ensure the complete targeting information of BFA2, the N-terminal 200 amino acids including the first PPR motif were used) of BFA2 were fused in-frame with GFP in the pBI221 vector. The chloroplast and mitochondrial markers were constructed according to Zhang et al. (2016). The resulting constructs were transformed into Arabidopsis protoplasts by PEG-mediated transformation and the protoplasts were placed in darkness for 16 h at 23oC. Transient GFP expression was observed using a confocal laser scanning microscope (LSM 510 Meta; Zeiss).
Electrophoretic Mobility Shift Assays
To express the recombinant BFA2-MBP protein, the cDNA sequence encoding amino acids 62–904 of BFA2 was subcloned into the plasmid pMAL-c5x (New England Biolabs). Expression was induced in E. coli BL21 strain with 0.3 mM isopropyl β-D-1-thiogalacopyranoside for 20 h at 16oC. Purification of the recombinant protein was performed according to the New England Biolabs protocol. The RNA probe (5′-UAUAGGCAUUAUUUUUUUUUCU-3′, atpF sRNA) was chemically synthesized, and its 5′-end was labeled by biotin (Takara Co., Ltd.). For competition assays, a specific probe (nonlabeled atpF sRNA) and a nonspecific probe (5′-UUAUGACGAUACUCGGUAGCAUAGAUAUAA-3′; 5′-end of the ndhA mRNA) were chemically synthesized.
Recombinant BFA2-MBP was incubated with biotinylated atpF sRNA for 30 min at 20oC in the binding buffer (10 mM HEPES, pH 7.5, 20 mM KCl, 2 mM MgCl2, 1 mM DTT, 5% glycerol, 1 μg tRNA). Subsequently, the reactions were resolved on 6% native polyacrylamide gels containing 2.5% glycerol. The signal was detected with the chemiluminescent detection kit (Thermo, 89880). For competition assays, specific probe and nonspecific probes were added in the reaction buffer.
Other Methods
Polyclonal antibody against BFA2 was raised in rabbits using the recombinant BFA2 protein (amino acids 62–300 of BFA2). Chlorophyll fluorescence analysis, thylakoid membrane and stromal protein isolation, BN-PAGE, 2D/SDS-PAGE, and immunoblot analysis were performed as previously described (Li et al., 2019). The gH+ was monitored with the Dual PAM-100 according to previously described methods (Rott et al., 2011; Zhang et al., 2018). Polysome association analyses were performed as previously described (Zhang et al., 2018). The rRNAs were stained by Super GelRed (US Everbright Inc., Suzhou, China) and used as fractionation and loading controls. Chloroplast protein labeling and chase was performed as previously described (Zhang et al., 2016, 2018). Immunoblot signals were detected with a Pro-light HRP Chemiluminescent Kit (TIANGEN) and visualized with a LuminoGraph chemiluminescence analyzer (ATTO). Antibodies against CF1α (PHY0311), CF1β (PHY0312), CF1γ (PHY0313), CF1ε (PHY0314), CF1δ (PHY0315), CFoI (PHY0316), CFoII (PHY0170S), PsaA (PHY0342), PsaD (PHY0343), D1 (PHY0057), D2 (PHY0060), Cyt f (PHY0321), and NdhN (PHY0335) were obtained from PhytoAB (United States).
Accession Numbers
Sequence data from this article can be found in GenBank/EMBL/DDBJ databases under accession number AtBFA2 (AT4G30825, Arabidopsis thaliana), GmBFA2 (Glyma.04G155800, Glycine max), OsBFA2 (Os09g25550, Oryza sativa), ZmBFA2 (XP_008662784, Zea mays), NsBFA2 (XP_009792607.1, Nicotiana sylvestris), SbBFA2 (SORBIDRAFT_07g007540, Sorghum bicolor), PpBFA2-A (Pp3c16_4140, Physcomitrella patens), PpBFA2-B (Pp3c5_2530, Physcomitrella patens). The aligned sequences of atpF-atpA can be found in the chloroplast genomes of Arabidopsis thaliana (At; NC_000932), Glycine max (Gm; NC_021650), Nicotiana sylvestris (Ns; NC_007500.1), Oryza sativa (Os; NC_001320), Zea mays (Zm; NC_001666). Physcomitrella patens (Pp, NC_005087), and Selaginella moellendorffii (Sm, nc_013086).
Results
The bfa2 Mutants Are Defective in Normal Accumulation of the Chloroplast ATP Synthase
While the bfa2-1 mutant was isolated by screening T-DNA mutant pools (Zhang et al., 2016), bfa2-2 was obtained from the European Arabidopsis Stock Centre (NASC). Both mutants show high levels of nonphotochemical quenching (NPQ) upon illumination with actinic light (80 μmol photons m-2 s-1) (Figure 1A,B). During illumination, photosynthetic electron transport induces accumulation of protons in the thylakoid lumen, which persists after illumination for 40 s in the wild-type (WT) plants and triggers the induction of NPQ (Figure 1C). Because of the activation of chloroplast ATP synthase in the light, protons accumulated in the thylakoid lumen move out rapidly through the ATP synthase to produce ATP, resulting in the relaxation of NPQ within 2 min of illumination (Figure 1C). In contrast, the relaxation of NPQ is less efficient in the bfa2 mutants and NPQ is maintained at high levels compared with WT (Figure 1C). Conductivity of the thylakoids to protons, gH+ (thylakoid conductivity), is usually used to monitor the activity of chloroplast ATP synthase in vivo (Cruz et al., 2001). The level of gH+ in bfa2 is indeed reduced to ∼2/3 of the WT level with an irradiance of 628 μmol photons m-2 s-1 as actinic light (Figure 1D), implying that the high-NPQ phenotype can be ascribed to the low activity of the chloroplast ATP synthase in bfa2.
FIGURE 1.
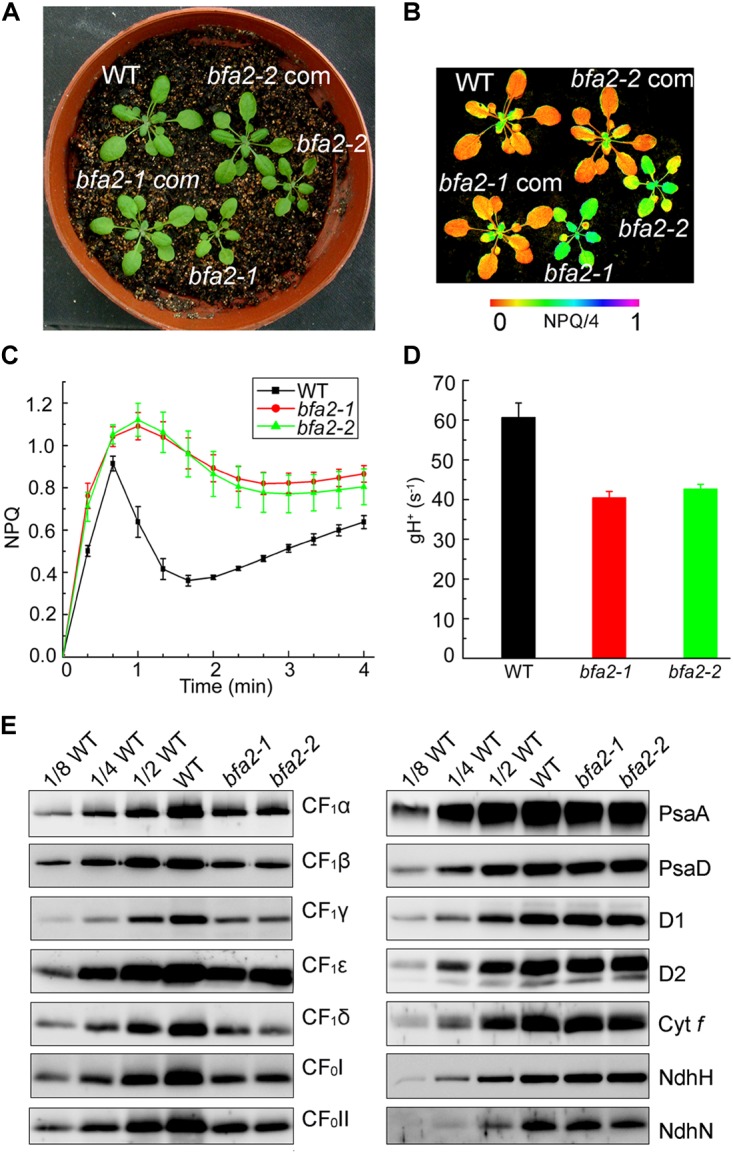
Identification and characterization of the bfa2 mutants. (A) Phenotype of the bfa2 mutants. bfa2-1 com and bfa2-2 com, complemented lines with overexpression of BFA2 in bfa2-1 and bfa2-2, respectively. (B) Image of NPQ. The image was captured upon illumination (80 μmol photons m-2 s-1) for 2 min. Values for NPQ/4 are indicated at the bottom on a false color scale. (C) Time course of NPQ induction. NPQ induction kinetics was measured upon illumination with AL (Actinic light) light (80 μmol photons m-2 s-1) for 4 min. Means ± SD (n = 4). (D) H+ conductivity through ATP synthase (gH+) under an irradiance of 628 μmol photons m-2 s-1. Means ± SD (n = 4). (E) Immunoblot analysis of the thylakoid membrane proteins in bfa2 and WT plants. Proteins were loaded on an equal chlorophyll basis and blots were probed with the indicated antibodies.
The seedling size of bfa2 is smaller than that of WT after germination for 25 days on soil (Figure 1A). To further characterize the phenotype of bfa2, several photosynthetic parameters were measured. Fv/Fm, the ratio between variable and maximum fluorescence, that represents the maximum quantum yield of photosystem II (PSII) was found to be comparable between WT and bfa2 plants (0.79 ± 0.01 for both genotypes), indicating that the function of PSII is not affected. We also investigated the dependence of ETR (electron transport rate through PSII) and NPQ on irradiance. While the ETR is significantly reduced in bfa2 at an irradiance above 200 μmol photons m-2 s-1, the level of NPQ is higher in bfa2 than in WT at all light intensities investigated (Supplementary Figures S1A,B), implying that protons over-accumulate in the thylakoid lumen of bfa2 and that photosynthetic linear electron transport is inhibited. Analysis of the dependence of 1-qL and the oxidation of the donor side of PSI on irradiance showed that photosynthetic electron transport is significantly restricted between PSII and PSI in bfa2 compared to WT plants (Supplementary Figures S1C,D). All of these photosynthetic properties in bfa2 are similar to those of mutants that accumulate low amounts of chloroplast ATP synthase (Zoschke et al., 2012, 2013; Rühle et al., 2014; Fristedt et al., 2015; Grahl et al., 2016; Zhang et al., 2016, 2018).
Immunoblot analysis showed that the levels of the chloroplast ATP synthase subunits in bfa2 are reduced to ∼25–50% of those of wild-type plants (Figure 1E). In contrast, accumulation of PSI (PsaA and PsaD), PSII (D1 and D2), Cytochrome b6f (Cyt f), and NADH dehydrogenase-like (NDH) complex (NdhH and NdhN) in bfa2 was as in WT (Figure 1E). Consistent with these results, blue native-PAGE (BN-PAGE) and subsequent two dimensional (2D) SDS-PAGE analysis showed that formation of the NDH-PSI supercomplex, PSII supercomplexes, PSII dimer, PSI monomer and other chlorophyll-containing complexes was not affected in bfa2 (Supplementary Figure S2A). Although the levels of CF1α/β/γ were reduced to about one quarter in bfa2, the remaining subunits were assembled into the intact ATP synthase and CF1 subcomplex (Supplementary Figure S2B), which accounts for the ∼2/3 activity of ATP synthase in bfa2 and for its photoautotrophic growth (Figure 1A,D). Taken together, we conclude that accumulation of chloroplast ATP synthase is specifically impaired whereas other thylakoid protein complexes are not affected in bfa2. Similar to the bfa1 and bfa3 mutants we characterized previously (Zhang et al., 2016, 2018), bfa2 is also a mutant that accumulates lower amounts of chloroplast ATP synthase.
BFA2 Is a PPR Protein Present in the Chloroplast Stroma
Map-based cloning detected a 17-nucleotide deletion (2130–2146 bp) in the coding region of AT4G30825 in bfa2-1, resulting in a premature stop codon (Figure 2A). A T-DNA was inserted in the same gene in the bfa2-2 mutant. Furthermore, overexpression of AT4G30825 under the control of the 35S promoter of cauliflower mosaic virus in the bfa2-1 and bfa2-2 mutants fully rescued their phenotype (Figure 1A,B). From these results, we conclude that the AT4G30825 gene corresponds to BFA2 that is required for full chloroplast ATP synthase activity in vivo.
FIGURE 2.
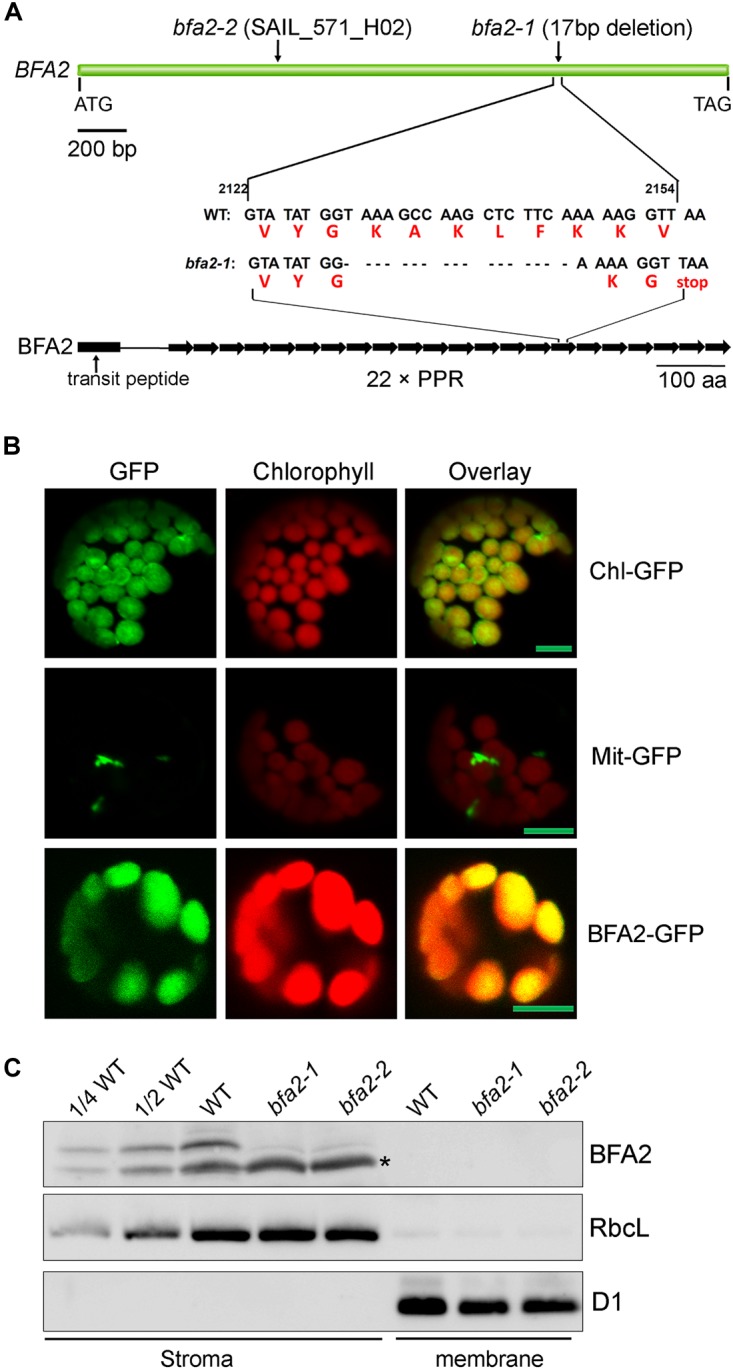
Characterization of the BFA2 protein. (A) Schematic representation of BFA2 gene (Top panel) and BFA2 protein (bottom panel). Positions for nucleotide deletion in baf2-1 and T-DNA insertion in bfa2-2 are indicated. Each right arrow represents one PPR domain. The 17-nucleotide deletion results in a premature stop codon at the position of the 16th PPR motif in BFA2. (B) Subcellular localization of BFA2 by GFP assay. The first 200 amino acids of BFA2 were fused with GFP (BFA2-GFP) and expressed in Arabidopsis protoplasts. The signal of GFP was visualized using a confocal laser scanning microscope. Chl-GFP and Mit-GFP represent chloroplast and mitochondrial controls, respectively. Bars = 5 μm. (C) Immunolocalization of BFA2. Intact chloroplasts isolated from WT and bfa2 mutants were fractionated into stromal and membrane fractions. Proteins were separated by SDS-PAGE and immunodetected with antibodies against BFA2, RbcL, and D1. The series of WT dilutions is indicated. A major nonspecific band detected in the stromal fractions with BFA2 antibody is indicated by an asterisk. A weak band above the major nonspecific band detected in bfa2 stroma also appears to be nonspecific.
The BFA2 gene encodes a PPR protein of 904 amino acid residues with unknown function (Figure 2A). Sequence analysis revealed that the BFA2 protein belongs to the P subfamily and comprises 22 PPR motifs spanning amino acid residues 139–904 (Figure 2A and Supplementary Figure S3). The last PPR motif only contains 32 residues and may represent an incomplete PPR motif (Supplementary Figure S3). Genes with significant sequence identity (more than 50%) to BFA2 are found in eudicotyledons and monocotyledons (Supplementary Figure S4). A blast search also revealed two proteins (PpBFA2-A and PpBFA2-B) in Physcomitrella patens (P. patens) with low sequence identity to BFA2 (35–38%, Supplementary Figure S4). No genes significantly related to BFA2 were found in Selaginella moellendorffii and Chlamydomonas. This fact implies that BFA2 may have evolved when land plants including bryophytes originated and was probably lost in the lycophytes during evolution.
BFA2 is predicted to have a putative chloroplast transit peptide of 61 amino acids at its N-terminus. To confirm its chloroplast localization, the DNA region coding for the first 200 amino acids of BFA2 was fused in-frame with GFP in the pBI221 vector and the resulting vector was introduced into Arabidopsis protoplasts by transient transformation. Analysis by confocal laser scanning microscopy showed that the BFA2-GFP signal co-localizes with the chloroplast fluorescence, indicating that BFA2 is targeted to the chloroplast (Figure 2B). To further determine the precise location of BFA2 within chloroplasts, a polyclonal antibody against recombinant BFA2 protein was raised. A signal with a molecular mass of ∼100 kDa (the predicted molecular mass of mature BFA2 is 94 kDa) was detected in the stromal fractions isolated from WT plants, but absent in the stromal fraction from bfa2 mutants as well as in the thylakoid membranes from WT and bfa2 plants (Figure 2C). These results indicate that BFA2 is localized to the chloroplast stroma.
BFA2 Is Required for Accumulation of the atpH/F Transcript
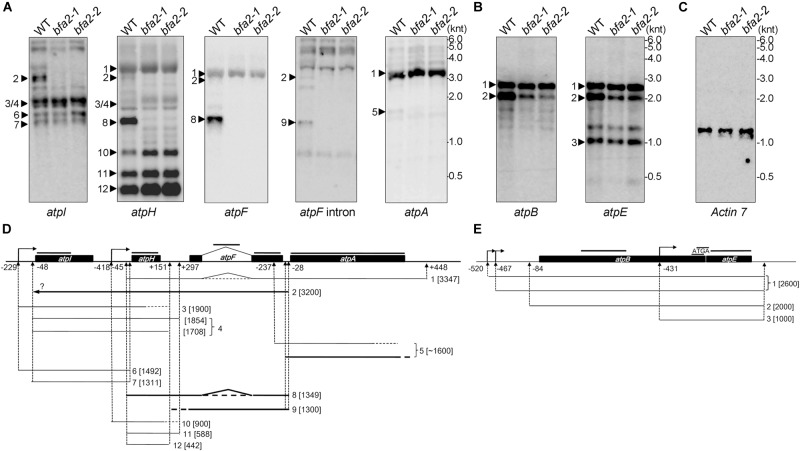
Since the PPR proteins are well known to be involved in organelle gene expression, it is very likely that the expression of one or more chloroplast genes encoding ATP synthase subunits is affected in the bfa2 mutants. To investigate this possibility, we performed RNA gel blot analysis with probes for the large (atpI/H/F/A) and the small (atpB/E) atp operons (Figure 3). For the large atp operon, the most striking difference is that the dicistronic atpH/F transcript is barely detected in the bfa2 mutants (Transcript 8; Figure 3A,D), indicating that BFA2 is essential for accumulation of this transcript. However, the level of the monocistronic atpH transcripts (transcripts 10, 11, and 12) in the bfa2 mutants is higher than that in WT (Figure 3A), excluding the possibility that absence of the atpH/F transcripts in bfa2 is due to deficient cleavage between atpI and atpH. RNA blot analysis using atpI, atpH, atpF exon, and atpF intron probes also detected a ∼3 kb transcript in WT plants that was absent in the bfa2 mutants (transcript 2, Figure 3A,D). Given the detection of this transcript with these four probes and its size, it is likely that this transcript corresponds to the unspliced atpI/H/F transcript (transcript 2, Figure 3D). The monocistronic unspliced atpF transcript was detected with the atpF intron probe in the WT plants but was absent in the bfa2 mutants (transcript 9, Figure 3A).
FIGURE 3.
RNA blot analysis of the plastid-encoded ATP synthase genes. (A–C) Total RNA isolated from WT and bfa2 mutants was probed with digoxigenin-labeled probes corresponding to the genes in the large (A) and small (B) atp operons. A blot with the Actin 7 probe was used as a loading control (C). The positions of RNA markers are shown in kilonucleotides (knt). (D,E) Partial transcript map of the atpI/H/F/A (D) and atpB/E (E) operons in Arabidopsis. The map was generated based on the transcript size in (A,B) as well as on previous reports (Malik Ghulam et al., 2012, 2013). Transcripts absent in bfa2 are shown as bold black lines. The positions of the probes are indicated above the operons. The numbers refer to the distance upstream of the initiation codon (-) and downstream of the termination codon (+).
The atpA RNA was mainly detected in the polycistronic atpH/F/A transcript (transcript 1, Figure 3A), which is inconsistent with previous reports (Malik Ghulam et al., 2013). In addition to this main polycistronic mRNA, the atpA probe also detected a fuzzy weak band around 1600 nucleotides in WT, but the level of this band was significantly reduced in the bfa2 mutants (transcript 5, Figure 3A,D). As discussed by Malik Ghulam et al. (2013), the monocistronic atpA transcript is present in very low amounts in vivo and usually possesses truncated 3′ ends whereas most of the 5′ ends of this RNA map at positions -237 (inside the atpF mRNA) and -50 (just overlapping with the 3′ end of atpF) relative to the atpA start codon (Figure 3D; Malik Ghulam et al., 2013). Thus, the weak bands detected in our RNA blot (transcript 5 and several bands below transcript 5) correspond most likely to the monocistronic atpA transcript with different 5′ ends, overlapping the atpF 3′ end, and truncated 3′ ends (Figure 3D). Reduction of transcript 5 in the bfa2 mutants indicates that some type of monocistronic atpA transcript is unstable in the absence of BFA2.
In the case of the small atp operon atpB/E, two major bands can be detected by the atpB probe (Figure 3B). The monocistronic atpE transcript can also be detected by the atpE probe (Figure 3B). While the upmost band represents the primary dicistronic atpB/E transcript with two isoforms (-520 and -467), the second band corresponds to the processed dicistronic atpB/E transcript ending at -84 (Figure 3E, Malik Ghulam et al., 2012). Our results show that the level of the -84 processed atpB/E dicistronic mRNA is significantly reduced in the bfa2 mutants compared with WT plants (Figure 3B,E). The level of primary dicistronic atpB/E and the monocistronic atpE transcripts are identical in the bfa2 mutants compared with WT plants (Figure 3E). Reduction of the processed dicistronic atpB/E transcript was also observed in the bfa1-1 and cgl160 mutants, in which assembly of the chloroplast ATP synthase CF1 and CFo subcomplexes, respectively, is less efficient (Rühle et al., 2014; Zhang et al., 2018). Thus, reduction of processed dicistronic atpB/E likely represents a secondary effect due to impairment in the assembly of chloroplast ATP synthase.
Translation Initiation of atpA Is Not Affected in the Absence of BFA2
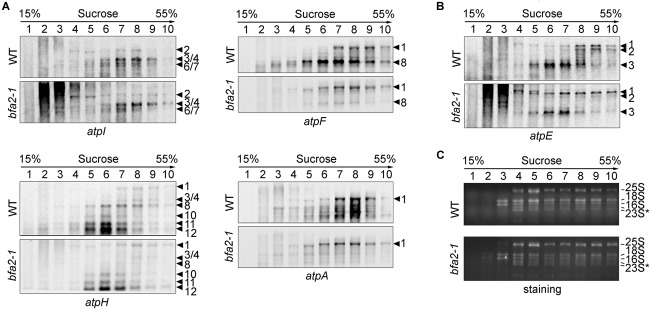
To rule out the possibility that reduction of the chloroplast ATP synthase in bfa2 is due to defects in the translation of atp mRNAs, we performed a polysome association analysis to investigate translation initiation (Figure 4). Our results show that the distribution of the atpH/F/A mRNAs in bfa2 was slightly shifted toward lower molecular weight fractions compared with WT (transcript 1, Figure 4A). The distribution of other transcripts in the atpI/H/F/A operon, such as monomeric atpH, was almost identical between bfa2-1 and WT plants. For the atpB/E operon, a clear shift of primary dicistronic atpB/E transcript toward lower molecular weight fractions in the bfa2-1 mutant compared with the wild type was observed (transcript 1, Figure 4B). The shift of the primary dicistronic atpB/E transcript is also observed in the bfa1 and cgl160 mutants and is unlikely to be the cause for the low accumulation of chloroplast ATP synthase in bfa2 (Zhang et al., 2018).
FIGURE 4.
Analysis of polysome association of the plastid transcripts encoding ATP synthase subunits. (A,B) Total leaf extracts from wild-type and bfa2-1 plants were fractionated by centrifugation on 15–55% sucrose density gradients. After centrifugation, the sucrose gradients were divided into 10 fractions of equal volume for RNA isolation. The isolated RNAs were blotted with DIG-labeled DNA probes corresponding to the plastid atpA, atpF, atpH, and atpI (A) as well as atpE (B) transcripts. The numbers to the right of the panels correspond to the corresponding transcripts illustrated in Figure 3D,E. (C) Staining of the rRNA was used as fractionation and loading control. 23S∗, two breakdown products of the chloroplast 23S rRNA. The numbers on the right indicate sedimentation coefficients of the major rRNAs.
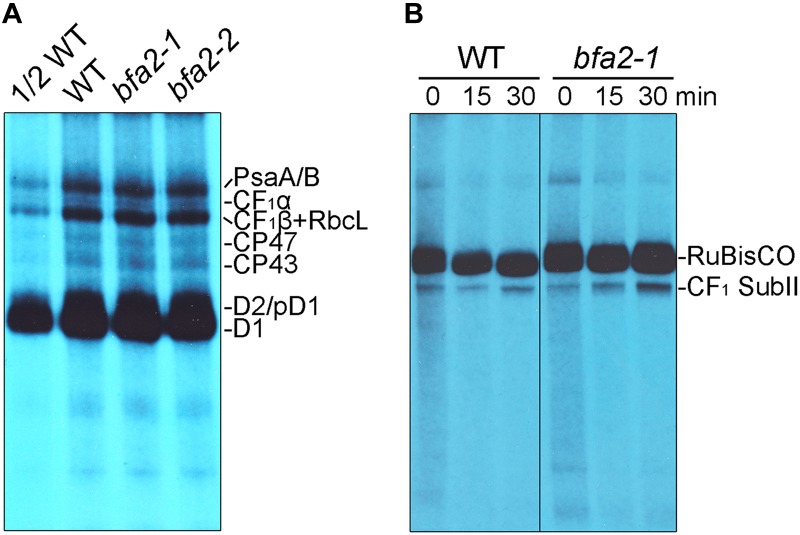
To investigate whether the alteration of the polysome association with atpH/F/A and primary dicistronic atpB/E transcripts in bfa2-1 is responsible for the defect in chloroplast ATP synthase accumulation, in vivo protein labeling of the chloroplast proteins with [35S]-Met was performed (Figure 5). Cycloheximide, an inhibitor of cytosolic translation, was added to avoid interference with the synthesis of nucleus-encoded proteins. After labeling, thylakoid membranes were isolated and the newly synthesized thylakoid proteins were separated by SDS-PAGE. Radiolabeled thylakoid proteins were detected by autoradiography. The results showed that, as expected, the levels of the newly synthesized PsaA/B, CP47, CP43, D2/pD1, and D1 protein were comparable between WT and bfa2 mutants (Figure 5A), which is consistent with the fact that bfa2 is specifically defective in accumulation of chloroplast ATP synthase. For the chloroplast ATP synthase CF1α subunit, a very weak signal was detected below the PsaA/B subunits and its level is identical in both WT and two bfa2 mutant genotypes (Figure 5A). The levels of newly synthesized CF1β subunits of as well as RbcL contamination in thylakoids were also identical in bfa2 and WT plants after labeling for 20 min (Figure 5A).
FIGURE 5.
In vivo protein labeling analysis of plastid-encoded proteins. (A) Pulse labeling of chloroplast thylakoid proteins of WT and bfa2 mutants. Chloroplast thylakoid proteins were extracted from primary leaves of 12 days old seedlings after labeling with [35S]-Met for 20 min, and separated by SDS-urea-PAGE. The labeled proteins were visualized using autoradiography. (B) Pulse-chase labeling of chloroplast stromal protein complexes of WT and bfa2-1 plants. After pulse labeling for 20 min (indicated as 0 min), the leaves were chased with cold Met for 15 and 30 min. Total soluble protein complexes extracted from the leaves were separated by CN-PAGE and the newly assembled complexes were detected by autoradiography.
CF1α and CF1β are components of the ATP synthase CF1 subcomplex. To further prove that protein synthesis of CF1α and CF1β and their subsequent incorporation into functional CF1 is not affected in bfa2, we analyzed the assembly of the CF1 subcomplex in the chloroplast stroma by pulse-chase labeling. We designated this subcomplex CF1 SubII in our previous report (Figure 5B, Zhang et al., 2018), and it contains subunits of CF1α, CF1β, CF1γ, CF1ε, and CF1δ, but not CFoI, which is the product of atpF (Zhang et al., 2016). Our results show that formation of CF1 SubII is as efficient in bfa2-1 as in WT plants after pulse-labeling for 20 min and a subsequent chase for 15 and 30 min (Figure 5B). These results are different from those obtained with bfa1 and bfa3 (Zhang et al., 2016, 2018), further confirming that synthesis of CF1α and CF1β is not affected in bfa2, although the level of processed dicistronic atpB/E was reduced and polysome association with atpH/F/A and primary dicistronic atpB/E was slightly altered in the bfa2 mutants (Figure 3, 4).
Taken together, we conclude that absence of the dicistronic atpH/F is the primary cause for the decreased accumulation of chloroplast ATP synthase in bfa2, and that BFA2 is likely directly required for the accumulation of the RNAs with a 3′-end or 5′-end mapping between atpF and atpA (Figure 3A,D). The BFA2 protein belongs to the P class PPR proteins and this class of proteins can act as barrier to prevent the RNA degradation by exoribonucleases (Barkan and Small, 2014). Since PPR10 binds to the 5′ termini of atpH/F, we hypothesize that BFA2 binds to the 3′ termini of atpH/F as well as to other transcripts overlapping the intergenic region of atpF-atpA (Figure 3D).
BFA2 Binds to the Consensus Sequence in the atpF 3′-UTR and atpA 5′-UTR
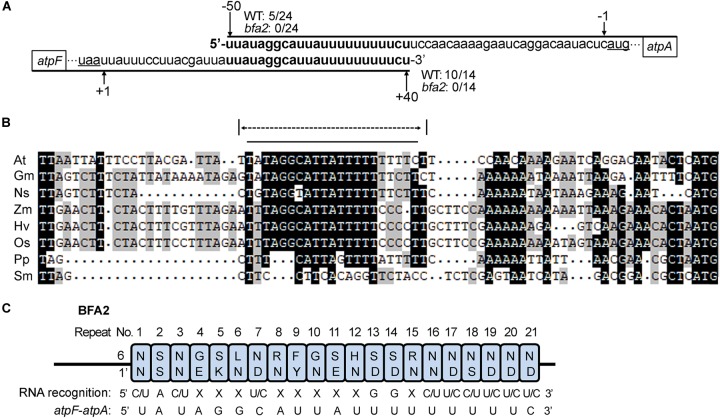
A small RNA (sRNA) corresponding to the atpF 3′ region was detected in barley and rice and this sRNA is predicted to be the footprint of uncharacterized PPR-like proteins (Ruwe and Schmitz-Linneweber, 2012; Zhelyazkova et al., 2012). These facts led us to propose that the sRNA from the atpF-atpA intergenic region is the footprint of BFA2 in Arabidopsis. To confirm this hypothesis, we first determined the transcript termini in the atpF-atpA intergenic region by circularization RT-PCR (cRT-PCR) (Supplementary Figure S5). Consistent with the results reported by Malik Ghulam et al. (2013), our results show that most clones (10 out of 14) had their 3′ end at position +40 from the atpF stop codon and 5 out of 24 clones had their 5′ end at position -50 from the atpA start codon in WT (Figure 6A and Supplementary Table S1). However, neither 3′ ends of atpF nor 5′ ends of atpA were mapped to these two positions in bfa2 (Figure 6A and Supplementary Table S1). These results suggest that BFA2 binds to the atpF-atpA intergenic region to stabilize the corresponding mRNA in vivo. Moreover, the overlapping region comprises 23 residues and is basically consistent with the number of PPR motifs (22 PPR motifs) of the BFA2 protein (Figure 6A).
FIGURE 6.
Position of the BFA2-dependent RNA termini. (A) Overlapping transcript termini in the atpF/A intergenic region. Nucleotide positions relative to the start codon of atpA (-50 and -1) and stop codon of atpF (+40 and +1) are shown above and below the sequences, respectively. The numbers of 5/24, 0/24, 10/14, and 0/14 represent the ratio of the clones with a 5′ or 3′ end positions from the WT or bfa2 samples in the cRT-PCR analyses (Supplementary Figure S5 and Supplementary Table S1). (B) Alignment of the atpF/A intergenic regions (from the stop codon of atpF to the start codon of atpA). The overlapping transcript termini in the atpF/A intergenic regions and the sequence predicted for BFA2 binding are indicated by the dotted and black lines, respectively. Nucleotides U are indicated by T. At, Arabidopsis thaliana; Gm, Glycine max; Os, Oryza sativa; Zm, Zea mays; Ns, Nicotiana sylvestris; Hv, Hordeum vulgare; Pp, Physcomitrella patens; Sm, Selaginella moellendorffii. (C) Prediction of the nucleotide-binding site for BFA2. The residues that determine nucleotide-binding specificity (residues 6 and 1’ in Supplementary Figure S3) in each PPR motif were extracted according to Barkan et al. (2012). The overlapping sequence in the atpF/A intergenic RNA (from the second nucleotide) is shown.
Alignment analysis showed that the overlapping region (except for the first residue) in the atpF 3′-UTR and atpA 5′-UTR in Arabidopsis is highly conserved in Angiosperms, but not in Physcomitrella patens and Selaginella moellendorffii (Figure 6B), which is in line with the fact that two proteins (PpBFA2-A and PpBFA2-B) in Physcomitrella patens (P. patens) show low sequence identity with BFA2 (35–38%) and that no BFA2-like protein was found in S. moellendorffii (Supplementary Figure S4). Although the last 6 residues vary among different plant species, most residues in this region are U and C (Figure 6B). Since the first residue is not conserved (Figure 6B), BFA2 may bind to the 22 conserved residues from the second residue in the sRNA. To confirm our hypothesis, the potential binding sequence of BFA2 was predicted according to the PPR code established previously (Barkan et al., 2012). As shown in Figure 6C, the 21 nucleotides predicted to bind by the 21 PPR motifs of BFA2 are (C/U)A(C/U)XXX(U/C)XXXXXGGX(C/U)(U/C)(C/U)(U/C)(U/C)(U/C). While X represents any nucleotide that cannot be precisely predicted, the nucleotides in parentheses are optional. Among the 21 nucleotides, 10 of them match with the corresponding residues in the overlapping transcript termini of the atpF-atpA intergenic region (Figure 6C). For the 5th and 11th PPR motifs, serine (S) was identified at position 6 (Figure 6C). It has been suggested that S6 shows a strong correlation with purines (Barkan et al., 2012), which is consistent with fact that G and A were found in the corresponding position of the atpF-atpA intergenic region (Figure 6C). These results support our view that BFA2 binds to the overlapping transcript termini in the atpH-atpA intergenic region starting from the second residue.
In vitro electrophoretic mobility shift assays (EMSA) were performed. Recombinant mature BFA2 protein fused with the MBP (maltose-binding protein) tag was expressed in Escherichia coli (E. coli) and purified (Figure 7A). The molecular mass of the purified fusion protein is about 130 kDa and is consistent with the predicted molecular mass of BFA2-MBP (136 kDa). The biotinylated RNA corresponding to the overlapping transcript termini in atpF-atpA was chemically synthesized and incubated with the BFA2-MBP fusion protein. The BFA2-RNA complex can be detected when the protein molar concentration is three times higher than that of the RNA (Figure 7B). There was no shift when the biotinylated RNA was incubated with MBP protein (Figure 7B). A set of competition assays were performed to confirm the binding specificity of BFA2. The 5′ end of ndhA mRNA has been shown to be the binding site of PGR3 (PROTON GRADIENT REGULATION 3) (Cai et al., 2011). Even addition of 1000-fold amount of cold ndhA mRNA did not affect the formation of the BFA2-RNA complex (Figure 7C). However, the addition of >30-fold amount of unlabeled atpF-atpA RNA significantly inhibited the binding of BFA2 with the labeled RNA probe (Figure 7D). These results clearly demonstrate that BFA2 protein binds to the atpF-atpA intergenic region in a sequence-specific manner.
FIGURE 7.
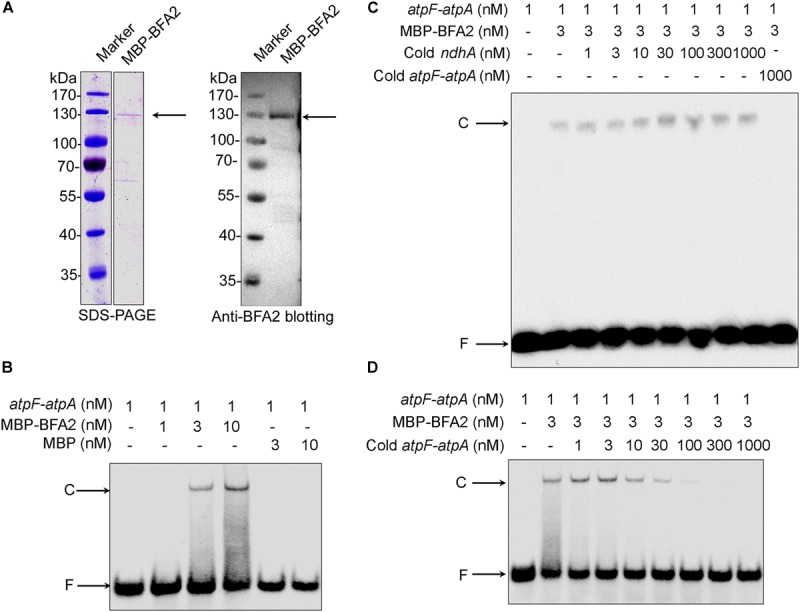
Gel mobility shift assays of the RNA binding activity of BFA2. (A) Purified MBP-BFA2 used in the RNA binding assays. The purified recombinant BFA2 was separated by SDS-PAGE and visualized by Coomassie Brilliant Blue (CBB) staining (left) and immunoblotting with anti-BFA2 antibody (right). (B) Gel mobility shift assays demonstrating that MBP-BFA2 binds with high affinity to the atpF sRNA. The concentrations of labeled sRNA probe, recombinant BFA2, and recombinant MBP are indicated above each lane. C and F indicate the BFA2-RNA complex and free labeled RNA, respectively. (C,D) Competition experiments using nonlabeled ndhA sRNA (C) and nonlabeled atpF-atpA sRNA (D). The concentrations of probes and protein are indicated above the gel. In each reaction, 1 μg yeast tRNA was added to lower the background of nonspecific RNA binding.
Discussion
The plastid-encoded atpF gene encodes the CFoI subunit of the chloroplast ATP synthase. CFoI interacts with the atpG product CFoII to form the peripheral stalk holding CFo and CF1 together (Rühle and Leister, 2015). In chloroplasts, the atpF RNA is solely detected in the polycistronic atpH/F/A and dicistronic atpH/F transcripts (Figure 3A; Malik Ghulam et al., 2013). Analysis of chloroplast small RNAs (sRNAs) in rice and barely reveals two sRNAs mapping at the two ends of dicistronic atpH/F mRNA, respectively (Ruwe and Schmitz-Linneweber, 2012; Zhelyazkova et al., 2012). Both of them are predicted to represent footprints of PPR proteins (Zhelyazkova et al., 2012; Malik Ghulam et al., 2013). While the sRNA at the 5′-end of atpH/F includes the binding site for PPR10 (Pfalz et al., 2009; Prikryl et al., 2011), the putative PPR protein binding to the 3′-end of the dicistronic atpH/F transcript was not yet known. In this study, we provide evidence that P-class PPR protein BFA2 binds to this site.
Our conclusion is mainly supported by the following evidence. (1) The level of the chloroplast ATP synthase is specifically reduced in the absence of BFA2, while accumulation of other thylakoid complexes is not affected (Figure 1E and Supplementary Figure S2). This is also consistent with the photosynthetic properties detected in bfa2 (Figure 1 and Supplementary Figure S1). (2) Dicistronic atpH/F transcript is absent in bfa2 and other transcripts with termini in the intergenic region of atpF-atpA also appear to be unstable in the absence of BFA2 (Figure 3). (3) The BFA2 binding site was predicted to cover the overlapping region between the 3′ end of atpF and the 5′ end of atpA (Figure 6). (4) EMSA analyses showed that BFA2 protein binds to the atpF-atpA intergenic region in a sequence-specific manner (Figure 7). Sequence alignment analysis showed that BFA2 belongs to the P-class PPR proteins with 22 PPR motifs (Supplementary Figure S3). Our results suggest that BFA2 acts analogously to other typical PPR proteins such as PPR10, PGR3, and HCF152, by directly binding to the atpF-atpA intergenic region to prevent degradation of BFA2-dependent transcripts by blocking exoribonucleases acting either from the 5′ or 3′ ends (Barkan and Small, 2014). However, because several nucleotides that bind to the PPR motifs in BFA2 can not be precisely predicted (Figure 6C), BFA2 may have another binding site(s) in the chloroplast-encoded genes, which need to be investigated in the further analyses.
For some P-class PPR proteins like PPR10, they not only act as site-specific barriers to protect target RNA segments by blocking exoribonuclease intruding from either direction, but also remodel the structure of ribosome-binding sites in the target RNA to enhance translation (Prikryl et al., 2011). Since BFA2 binds to the intergenic regions of atpF-atpA, which is just upstream of the start codon of atpA, is it possible that binding of BFA2 in this region releases the ribosome binding site of atpA? In Arabidopsis, monomeric atpA transcript was barely detectable in chloroplasts (Malik Ghulam et al., 2013; Figure 3). Thus, atpA translation should arise from the polycistronic atpH/F/A transcript. Although polysome association with atpH/F/A transcript was slightly reduced in the bfa2 mutant (Figure 4A), CF1α synthesis and subsequent assembly into CF1 were not affected (Figure 5). These facts suggest that binding of BFA2 in the intergenic region of atpF-atpA is not required for the translation of atpA. However, we cannot fully rule out the possibility that BFA2 is involved in the activation of atpA translation since no solid evidence was obtained by more direct approaches like polysome profiling.
Our results demonstrate that absence of dicistronic atpH/F transcript is the main cause of the low ATP synthase accumulation phenotype of bfa2 (Figure 3–5). The dicistronic atpH/F transcript is barely detectable in bfa2 (Figure 3). This raises the question of how the atpF product CFoI can accumulate to about one-quarter in bfa2 as compared to WT (Figure 1)? One possibility is that atpF translation proceeds to a small extent from the polycistronic atpH/F/A transcript which accumulates normally in the bfa2 mutants (Figure 3).
Homologs of BFA2 are found in angiosperms, consistent with the highly conserved intergenic regions of atpF-atpA among angiosperms (Figure 6B and Supplementary Figure S4, Zhelyazkova et al., 2012). Moreover, two putative BFA2 homologs were found in P. patens although they display low sequence identity with BFA2 from higher plants (Supplementary Figure S4). However, although a ∼20 nt sequence in the atpF-atpA regions from P. patens chloroplasts shows high similarity to the BFA2-binding sequence of higher plants, a 3 nt deletion was found in this sequence (Figure 6B). Moreover, this sequence is located just downstream of the stop codon of atpF (Figure 6B). It is reasonable to assume that translation termination may be affected if the BFA2-like proteins in P. patens bind to this region. Thus, detailed analyses are necessary to clarify the function of these two proteins in P. patens.
In summary, our genetic approaches have identified a P-class PPR protein BFA2, which is specifically required for the normal accumulation of chloroplast ATP synthase. We have demonstrated that BFA2 binds to the intergenic region of atpF-atpA and mainly acts as a site-specific barrier to protect atpH/F mRNA by blocking exoribonuclease degradation from the 3′-direction. Thus, stabilization of the atpH/F transcript requires two independent PPR proteins, PPR10 and BFA2, to protect the mRNA against exoribonucleases.
Significance Statement
In this study, we discovered a chloroplast PPR protein BFA2, which protects target mRNAs from degradation by exoribonucleases by binding to the consensus sequence of the atpF-atpA intergenic region.
Author Contributions
LZ, WL, and LP conceived the study and designed the experiments. LZ, WL, WZ, and LC performed the experiments. LZ and WL produced the figures. LZ, WL, J-DR, and LP wrote the manuscript. LP supervised the whole study. All authors analyzed the data.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank NASC for providing the mutant seeds.
Footnotes
Funding. This work was supported by the National Science Foundation for Young Scientists of China (31500196 and 31700202), the China Postdoctoral Science Foundation (2017M621511), and the funds from the Shanghai Engineering Research Center of Plant Germplasm Resources (17DZ2252700) and the Science and Technology Commission of Shanghai Municipality (18DZ2260500).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00446/full#supplementary-material
References
- Barkan A. (2011). Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 155 1520–1532. 10.1104/pp.110.171231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., Rojas M., Fujii S., Yap A., Chong Y. S., Bond C. S., et al. (2012). A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 8:e1002910. 10.1371/journal.pgen.1002910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., Small I. (2014). Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant. Biol. 65 415–442. 10.1146/annurev-arplant-050213-040159 [DOI] [PubMed] [Google Scholar]
- Cai W., Okuda K., Peng L., Shikanai T. (2011). PROTON GRADIENT REGULATION 3 recognizes multiple targets with limited similarity and mediates translation and RNA stabilization in plastids. Plant J. 67 318–327. 10.1111/j.1365-313X.2011.04593.x [DOI] [PubMed] [Google Scholar]
- Cruz J. A., Sacksteder C. A., Kanazawa A., Kramer D. M. (2001). Contribution of electric field (ΔΨ) to steady-state transthylakoid proton motive force (pmf) in vitro and in vivo. Control of pmf Parsing into Δψ and ΔpH by ionic strength. Biochemistry 40 1226–1237. 10.1021/bi0018741 [DOI] [PubMed] [Google Scholar]
- Eberhard S., Loiselay C., Drapier D., Bujaldon S., Girard-Bascou J., Kuras R., et al. (2011). Dual functions of the nucleus-encoded factor TDA1in trapping and translation activation of atpA transcripts in Chlamydomonas reinhardtii chloroplasts. Plant J. 67 1055–1066. 10.1111/j.1365-313X.2011.04657.x [DOI] [PubMed] [Google Scholar]
- Fristedt R., Martins N. F., Strenkert D., Clarke C. A., Suchoszek M., Thiele W., et al. (2015). The thylakoid membrane protein CGL160 supports CF1CFo ATP synthase accumulation in Arabidopsis thaliana. PLoS One 10:e0121658. 10.1371/journal.pone.0121658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahl S., Reiter B., Gügel I. L., Vamvaka E., Gandini C., Jahns P., et al. (2016). The Arabidopsis protein CGLD11 is required for chloroplast ATP synthase accumulation. Mol. Plant 9 885–899. 10.1016/j.molp.2016.03.002 [DOI] [PubMed] [Google Scholar]
- Hahn A., Vonck J., Mills D. J., Meier T., Kühlbrandt W. (2018). Structure, mechanism, and regulation of the chloroplast ATP synthase. Science 360:eaat4318. 10.1126/science.aat4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger T. S., Watkins K. P., Friso G., van Wijk K. J., Barkan A. (2009). A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc. Natl. Acad. Sci. U.S.A. 106 4537–4542. 10.1073/pnas.0812503106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Liu B., Zhang J., Kong F., Zhang L., Meng H., et al. (2019). OHP1, OHP2, and HCF244 form a transient functional complex with the photosystem II reaction center. Plant Physiol. 179 195–208. 10.1104/pp.18.01231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik Ghulam M., Courtois F., Lerbs-Mache S., Merendino L. (2013). Complex processing patterns of mRNAs of the large ATP synthase operon in Arabidopsis chloroplasts. PLoS One 8:e78265. 10.1371/journal.pone.0078265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik Ghulam M., Zghidi-Abouzid O., Lambert E., Lerbs-Mache S., Merendino L. (2012). Transcriptional organization of the large and the small ATP synthase operons, atpI/H/F/A and atpB/E, in Arabidopsis thaliana chloroplasts. Plant Mol. Biol. 79 259–272. 10.1007/s11103-012-9910-5 [DOI] [PubMed] [Google Scholar]
- Martin M., Rujan T., Richly E., Hansen A., Cornelsen S., Lins T., et al. (2002). Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. U.S.A. 99 12246–12251. 10.1073/pnas.182432999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormac D. J., Barkan A. (1999). A nuclear gene in maize required for the translation of the chloroplast atpB/E mRNA. Plant Cell 11 1709–1716. 10.1105/tpc.11.9.1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J., Bayraktar O. A., Prikryl J., Barkan A. (2009). Site-specific binding of a PPR protein defines and stabilizes 5′and 3′mRNA termini in chloroplasts. EMBO J. 28 2042–2052. 10.1038/emboj.2009.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prikryl J., Rojas M., Schuster G., Barkan A. (2011). Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl. Acad. Sci. U.S.A. 108 415–420. 10.1073/pnas.1012076108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prikryl J., Watkins K. P., Friso G., van Wijk K. J., Barkan A. (2008). A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res. 36 5152–5165. 10.1093/nar/gkn492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rott M., Martins N. F., Thiele W., Lein W., Bock R., Kramer D. M., et al. (2011). ATP synthase repression in tobacco restricts photosynthetic electron transport, CO2 assimilation, and plant growth by over acidification of the thylakoid lumen. Plant Cell 23 304–321. 10.1105/tpc.110.079111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rühle T., Leister D. (2015). Assembly of F1F0-ATP synthases. Biochim. Biophys. Acta 1847 849–860. 10.1016/j.bbabio.2015.02.005 [DOI] [PubMed] [Google Scholar]
- Rühle T., Razeghi J. A., Vamvaka E., Viola S., Gandini C., Kleine T., et al. (2014). The Arabidopsis protein CONSERVED ONLY IN THE GREEN LINEAGE160 promotes the assembly of the membranous part of the chloroplast ATP synthase. Plant Physiol. 165 207–226. 10.1104/pp.114.237883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruwe H., Schmitz-Linneweber C. (2012). Short non-coding RNA fragments accumulating in chloroplasts: footprints of RNA binding proteins. Nucleic Acids Res. 40 3106–3116. 10.1093/nar/gkr1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Small I. (2008). Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 13 663–670. 10.1016/j.tplants.2008.10.001 [DOI] [PubMed] [Google Scholar]
- Shikanai T. (2015). RNA editing in plants: machinery and flexibility of site recognition. Biochim. Biophys. Acta 1847 779–785. 10.1016/j.bbabio.2014.12.010 [DOI] [PubMed] [Google Scholar]
- Small I. D., Peeters N. (2000). The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25 46–47. 10.1016/S0968-0004(99)01520-0 [DOI] [PubMed] [Google Scholar]
- Stern D. B., Goldschmidt-Clermont M., Hanson M. R. (2010). Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 61 125–155. 10.1146/annurev-arplant-042809-112242 [DOI] [PubMed] [Google Scholar]
- Till B., Schmitz-Linneweber C., Williams-Carrier R., Barkan A. (2001). CRS1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA 7 1227–1238. 10.1017/S1355838201010445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins K. P., Kroeger T. S., Cooke A. M., Williams-Carrier R. E., Friso G., Belcher S. E., et al. (2007). A ribonuclease III domain protein functions in group II intron splicing in maize chloroplasts. Plant Cell 19 2606–2623. 10.1105/tpc.107.053736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap A., Kindgren P., Colas des Francs-Small C., Kazama T., Tanz S. K., Toriyama K., et al. (2015). AEF1/MPR25 is implicated in RNA editing of plastid atpF and mitochondrial nad5, and also promotes atpF splicing in Arabidopsis and rice. Plant J. 81 661–669. 10.1111/tpj.12756 [DOI] [PubMed] [Google Scholar]
- Zhang L., Duan Z., Zhang J., Peng L. (2016). BIOGENESIS FACTOR REQUIRED FOR ATP SYNTHASE 3 facilitates assembly of the chloroplast ATP synthase complex. Plant Physiol. 171 1291–1306. 10.1104/pp.16.00248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Pu H., Duan Z., Li Y., Liu B., Zhang Q., et al. (2018). Nucleus-encoded protein BFA1 promotes efficient assembly of the chloroplast ATP synthase coupling factor 1. Plant Cell 30 1770–1788. 10.1105/tpc.18.00075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelyazkova P., Hammani K., Rojas M., Voelker R., Vargas-Suarez M., Börner T., et al. (2012). Protein-mediated protection as the predominant mechanism for defining processed mRNA termini in land plant chloroplasts. Nucleic Acids Res. 40 3092–3105. 10.1093/nar/gkr1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Lu Q., Li Q., Wang L., Ding S., Zhang A., et al. (2017). PPR-SMR protein SOT1 has RNA endonuclease activity. Proc. Natl. Acad. Sci. U.S.A. 114 E1554–E1563. 10.1073/pnas.1612460114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R., Kroeger T., Belcher S., Schottler M. A., Barkan A., Schmitz-Linneweber C. (2012). The pentatricopeptide repeat-SMR protein ATP4 promotes translation of the chloroplast atpB/E mRNA. Plant J. 72 547–558. 10.1111/j.1365-313X.2012.05081.x [DOI] [PubMed] [Google Scholar]
- Zoschke R., Nakamura M., Liere K., Sugiura M., Börner T., Schmitz-Linneweber C. (2010). An organellar maturase associates with multiple group II introns. Proc. Natl. Acad. Sci. U.S.A. 107 3245–3250. 10.1073/pnas.0909400107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R., Qu Y., Zubo Y. O., Börner T., Schmitz-Linneweber C. (2013). Mutation of the pentatricopeptide repeat-SMR protein SVR7 impairs accumulation and translation of chloroplast ATP synthase subunits in Arabidopsis thaliana. J. Plant Res. 126 403–414. 10.1007/s10265-012-0527-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.