Abstract
SUMMARY
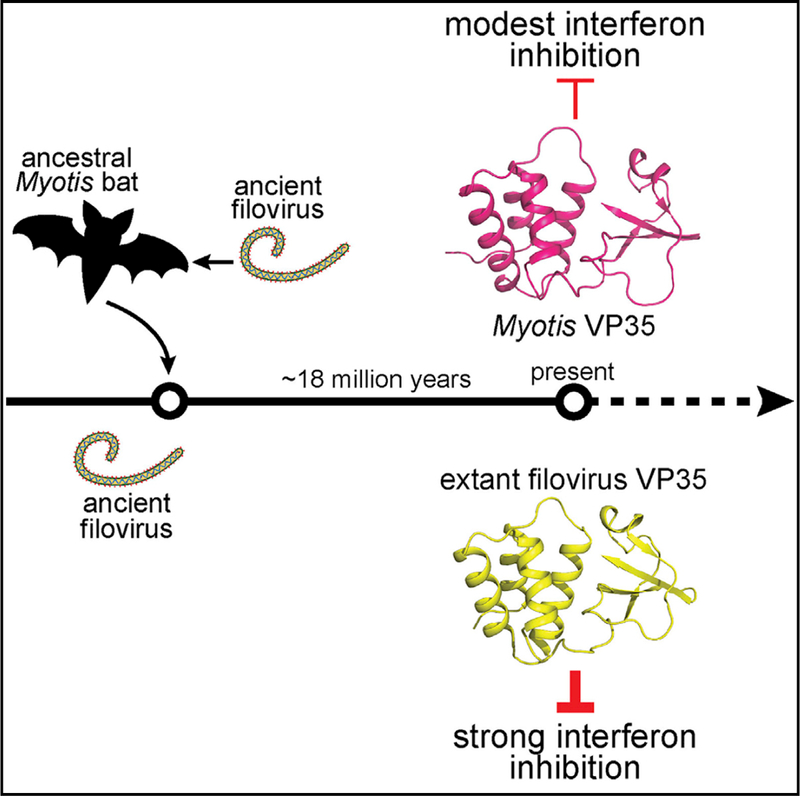
Non-retroviral integrated RNA viral sequences (NIRVs) potentially encoding ~280 amino acid homologs to filovirus VP35 proteins are present across the Myotis genus of bats. These are estimated to have been maintained for ~18 million years, indicating their co-option. To address the reasons for cooption, 16 Myotis VP35s were characterized in comparison to VP35s from the extant filoviruses Ebola virus and Marburg virus, in which VP35s play critical roles in immune evasion and RNA synthesis. The Myotis VP35s demonstrated a conserved suppression of innate immune signaling, albeit with reduced potency, in either human or Myotis cells. Their attenuation reflects a lack of dsRNA binding that in the filoviral VP35s correlates with potent suppression of interferon responses. Despite divergent function, evolution has preserved in Myotis the structure of the filoviral VP35s, indicating that this structure is critical for co-opted function, possibly as a regulator of innate immune signaling.
In Brief
Edwards et al. demonstrate that homologs to Ebola virus VP35 encoded in the genomes of bats from the Myotis genus possess striking structural homology to their viral counterparts and can inhibit interferon responses but with decreased efficiency relative to their viral homologs.
Graphical Abstract
INTRODUCTION
Non-retroviral integrated RNA viral sequences (NIRVs) are thought to reflect rare events in which RNA viruses that do not encode their own reverse transcriptase co-opt such an enzyme present in the infected cell, leading to integration of viral sequences into the germline. NIRVs are present in fungi, plants, insects, and mammals (Belyi et al., 2010; Crochu et al., 2004; Horie et al., 2010; Tanne and Sela, 2005; Taylor and Bruenn, 2009; Taylor et al., 2010, 2011). They serve as a viral fossil record, providing evidence of historical viral interactions with a host and allowing for the study of the timescale and evolution of the virus-host interaction. Beyond this, however, the biological significance of these genetic elements remains incompletely understood.
Filoviruses, which include the highly pathogenic Ebola virus (EBOV) and Marburg virus (MARV), are negative sense, singlestranded RNA viruses with cytoplasmic replication that are highly represented among mammalian NIRVs (Belyi et al., 2010; Taylor et al., 2010, 2011). Filovirus-like sequences corresponding to the nucleoprotein (NP); large (L) protein, which is the viral RNA polymerase; and viral protein of 35 kDa (VP35) have been identified in several mammals (Belyi et al., 2010; Taylor et al., 2010,2011). These include both NPand L sequences in the opossum, NP sequences in shrews and tenrecs, VP35 sequences in the tammar wallaby and Philippine tarsier, and both NP and VP35 sequences in rodents (such as the house mouse and brown rat) and mouse-eared bats (Myotis) (Belyi et al., 2010; Taylor et al., 2010, 2011). Maintenance of sequences recognizable as being of viral origin implies functional co-option. However, although NIRVs are common in eukaryotic genomes, recognizable candidates with an understood functional role are exceedingly rare.
Myotis VP35 elements, likely the result of a long interspersed nuclear element (LINE-1)-mediated insertion, retain an intact open reading frame (ORF) potentially encoding proteins of ~280 amino acids (Belyi et al., 2010; Taylor et al., 2010, 2011). This is in contrast to many other NIRVs, including the NP sequences integrated into Myotis, which consist of disrupted ORFs. Prior studies identified syntenic VP35-like ORFs in members of Old World, North American, and South American clades of Myotis bats, suggesting a single integration event that occurred an estimated 18 million years ago, before the divergence of Old World and New World species (Ruedi et al., 2013; Taylor et al., 2011). The long-term maintenance of an intact ORF, coupled with the prior identification of multiple sites within Myotis VP35 under positive selection, suggests the Myotis VP35s have been preserved to carry out a function of significance for the host (Taylor et al., 2011).
In the context of filovirus infection, VP35 proteins are innate immune suppressors and part of the viral RNA synthesis machinery (Basler etal., 2000; Mühlberger etal., 1999). Immune suppression functions include inhibition of RIG-I-like receptor (RLR) signaling to block type I interferon (IFN) production and suppress dendritic cell maturation (Basler et al., 2000; Bosio et al., 2003; Jin et al., 2010; Lubaki et al., 2016; Ramanan et al., 2012; Yen et al., 2014; Yen and Basler, 2016). These inhibitory functions correlate with VP35 capacity to bind double-stranded RNA (dsRNA) through its C-terminal IFN inhibitory domain (IID), which likely sequesters viral dsRNA from RLR recognition (Bale et al., 2012, 2013; Cárdenas et al., 2006; Dilley et al., 2017; Leung et al., 2010; Ramanan et al., 2012). Interaction of VP35 with PACT also impairs RIG-I signaling (Luthra et al., 2013). In addition, dsRNA binding-independent mechanisms of inhibition have been described (Chang et al., 2009; Prins et al., 2009). Studies on recombinant EBOV and MARV with mutated VP35s indicate that these suppressive functions are critical for efficient virus replication and for virulence in animals (Albariüo et al., 2015; Hartman et al., 2006, 2008; Prins et al., 2010b). Other VP35 functions that may contribute to innate immune evasion include inhibition of the IFN-induced antiviral protein kinase R (PKR) and counteracting microRNA (miRNA) silencing (Fabozzi et al., 2011; Feng et al., 2007; Haasnoot et al., 2007; Schümann et al., 2009; Zhu et al., 2012). For viral RNA synthesis, four viral proteins—NP, VP30, L, and VP35—are required (Mühlberger et al., 1998, 1999). In the viral RNA polymerase complex, VP35 is essential due to critical interactions with the NP and L proteins (Becker etal., 1998; Leung et al., 2015; Liu et al., 2017; Mölleret al., 2005; Prins etal., 2010a; Theriault et al., 2004; Trunschke et al., 2013; Zhu et al., 2017).
Here, we used the sequences of 16 Myotis VP35-like ORFs to directly determine, by using functional, evolutionary, and structural approaches, how features of this non-retroviral RNA virus have been maintained in the context of a multi-million-year cooption by a mammalian host.
RESULTS
Myotis VP35s Are Less Potent Suppressors of IFN-β Production than Extant Filoviral VP35s
We synthesized the originally identified Myotis VP35 ORF from Myotis lucifigus (batVP35) and amplified and sequenced an additional 15 VP35 ORFs from a variety of Myotis species, including another M. lucifigus VP35 (Figure S1) (Belyi et al., 2010). Analysis by Partitionfinder revealed that two significant partitions existed (of the three codon positions assayed), which were fit to the F81+G and K3P+G models in IQ-TREE (Trifinopoulos et al., 2016). These clades can be further divided into three groups based on the presence or absence of previously described independent deletions of 9 base pairs (bp) (blue), 39 bp (red), or no deletion (black) (Figure 1A; Figure S1) (Taylor et al., 2011). Alignment required just these two deletions, indicating that the contribution of alignment error to downstream bioinformatics analyses such as ancestral sequence reconstruction and tests of selection is low with these data. Subsequently, the ancestral Myotis VP35 sequence was reconstructed under a Jones-Tay- lor-Thornton (JTT) + gamma model of amino acid substitution using PhyloBot, a pipeline that allows for the reconstruction of extinct proteins (Hanson-Smith and Johnson, 2016).
Figure 1. Myotis VP35s Are Less Potent Suppressors of IFN-β Production than Extant Filoviral VP35s.

(A) IFN-β luciferase reporter assay in HEK293T cells in the presence of FLAG-tagged Myotis VP35 constructs (500 ng). Error bars represent the SEM for triplicate experiments. The uninfected empty vector control is indicated by the black bar labeled E; the remaining sampleswere infected with SeV. VP35 expression was assessed by western blot for the FLAG epitope tag. Western blot lanes align with the corresponding samples in the graph. Statistical significance was assessed using a oneway ANOVA and Tukey’s test, comparing columns to the SeV-infected control (white bar): ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05. E refersto empty vector. The phylogenetic tree indicates the relationship of the tested Myotis VP35s as indicated. Blue indicates a 9 base pair (bp) deletion, black indicates no deletion, and red indicates a 39 bp deletion. Numbers on the tree graph indicate branch support as estimated from approximate likelihood ratio tests and ultrafast bootstrapping. The tree scale bar represents substitutions per site for the vertical branch lengths. (B) IFN-03b2 luciferase reporter assay in HEK293T cells in the presence of the indicated FLAG-tagged VP35 constructs at decreasing concentrations (500, 50, 5, and 0.5 ng). IFN-β promoter activity and VP35 expression were assessed as in (A). (C) Western blot analysis of IRF-3 phosphorylation in HEK293T cells transfected with decreasing concentrations of the indicated FLAG-tagged VP35 and MARV NP (mNP) constructs (2, 1, and 0.5 μg) and IRF-3 (100 ng). Western blots were performed for total IRF-3 and phospho-IRF3. The phospho-IRF3 assay was repeated twice. (D) IFN-β luciferase reporter assay in M. myotis Nep cells in the presence of FLAG-tagged Myotis VP35 constructs (500 ng). IFN-β promoter activity and VP35 expression were assessed as in (A). IFN-β promoter luciferase assays were repeated at least three times. See also Figure S1.
Alignment of batVP35 to EBOV and MARV VP35s revealed amino acid conservation particularly in the IID (Figure S2). To determine whether Myotis VP35s can inhibit IFN-β production via the RIG-I signaling pathway, reporter gene assays were employed. FLAG-tagged VP35s were expressed in HEK293T cells as indicated, and Sendai virus (SeV), a known inducer of IFN-β production via the RIG-I pathway, was used to induce IFN-β promoter activity. Most Myotis VP35s, including the ancestral Myotis VP35 (Ancestral), inhibited IFN-β production to a modest extent (Figure 1A). Several Myotis VP35s had little to no inhibitory effect at the concentration tested, including M. oxyotus, whereas others, including M. davidii, exhibited increased inhibitory activity relative to the others. The variation in activity did not correlate with either the clade or the presence of base pair deletions (Figure 1A; Figure S1). To further assess potency, we selected several of the most efficient Myotis VP35s, including representatives from each sequence group (batVP35, M. nigricans, M. annectans, and M. davidii) and assessed their activity across a range of concentrations in comparison to EBOV VP35 (eVP35) in HEK293T cells (Figure 1B). As expected, eVP35 inhibited IFN-β promoter activity over a range of concentrations. In contrast, the inhibitory activity of all four Myotis VP35s was most efficient at the highest concentration tested, but inhibition titrated out quickly (Figure 1B).
Filoviral VP35 inhibition of RLR signaling prevents the virus- induced phosphorylation and activation of transcription factor IRF-3 (Basler et al., 2003; Ramanan et al., 2012). Therefore, IRF-3 phosphorylation was monitored in the absence or presence of either eVP35 or batVP35. eVP35 blocked SeV-induced IRF-3 phosphorylation over a range of concentrations, while batVP35 was effective only at the highest concentration (Figure 1C). Another control, MARV nucleoprotein (mNP), did not inhibit IRF-3 phosphorylation. These data indicate that a representative endogenous Myotis VP35 retains the ability to inhibit RLR signaling and IFN-ß production, although with decreased efficiency relative to extant filovirus VP35s.
Myotis VP35 IFN-Antagonist Function in Bat Cells
By use of an IFN-ß promoter reporter assay, we also assessed IFN-inhibitory activity of FLAG-tagged eVP35, Ancestral VP35, and the 16 Myotis VP35s in a transformed nasal epithelial (Nep) cell line from a M. myotis bat (He et al., 2014). As seen in the HEK293T cells, eVP35 potently inhibited activity of the IFN-β promoter in M. myotis cells, whereas many Myotis VP35s exhibited attenuated inhibitory activity (Figure 1D). The pattern of Myotis VP35s that inhibit SeV-induced IFN-β reporter activity in HEK293T cells does not perfectly align with inhibitory activity in M. myotis cells (Figures 1A and 1D). For example, M. oxyotus has no detectable inhibition of IFN-β promoter activity in HEK293T cells but shows modest inhibition in the M. myotis cell line. However, several Myotis VP35 ORFs show similar, attenuated, inhibitory activity in both human and Myotis cells, including batVP35, M. nigricans, M. horsfieldii, and M. riparius (Figure 1D). These data demonstrate that Myotis VP35s consistently exhibit substantially decreased IFN-inhibitory activity relative to filoviral VP35s, but many have retained some capacity to inhibit IFN-β production in both human (HEK293T) and Myotis (Nep) cell lines. batVP35 demonstrated similar, modest, IFN-β inhibitory activity in both cell lines, suggesting that batVP35 and HEK293T cells provide an appropriate model for further analysis of VP35 functions retained by Myotis VP35s.
Myotis VP35s Lack dsRNA Binding Activity
Filoviral VP35s bind dsRNA via the IID, likely sequestering dsRNA from RLRs (Bale et al., 2013; Cárdenas et al., 2006; Dilley et al., 2017; Leung et al., 2010; Ramanan et al., 2012). dsRNA binding requires amino acid residues of the central basic patch (CBP) (Leung et al., 2010; Ramanan et al., 2012). In eVP35, the CBP is composed of R305, K309, R312, K319, R322, and K339 (Leung et al., 2010). Conserved between eVP35 and MARV VP35 (mVP35) are basic residues (using eVP35 numbering) at positions R305, K309, R312, R322, and K339 (Figure S2; eVP35, asterisk; mVP35, number sign). In batVP35, positions equivalent to eVP35 residues 312, 319, 322, and 339 are basic, while the residues equivalent to 305 and 309 are not basic but instead are glutamic acid and a glycine, respectively (Figure S2). Within the 17 Myotis VP35s, the eVP35 305 equivalent is non-basic (glutamic acid [E] or glutamine [Q]) in the clade containing sequences with a 9 bp deletion (Figure S1, blue) but is a basic residue (lysine [K]) in the ancestral Myotis VP35 sequence and those in the clade containing sequences with either a 39 bp deletion (red) or no deletion (black) (Figure S1). The glycine at 309 (using eVP35 numbering) is conserved across all 16 Myotis VP35s and the ancestral reconstructed sequence (Figure S1). To determine whether batVP35 has retained dsRNA binding activity, the IIDs of eVP35 and batVP35 were expressed, purified, and used in an in vitro RNA binding assay. As expected, eVP35 bound dsRNA; however, no interaction between batVP35 and dsRNA was detected (Figure 2A). To further assess binding to dsRNA, full-length eVP35, dsRNA binding-defective eVP35 R312A, mVP35, batVP35, and the VP35s of M. davidii, M. annectans, and M. nigricans, which were the most efficient inhibitors of IFN-β promoter activity for each sequence group in HEK293T cells, and M. oxyotus, which lacks inhibition of virus-induced IFN-β reporter activity in HEK293T cells but maintains modest inhibition in the M. myotis Nep cells, were expressed in HEK293T cells. Cell lysates containing each VP35 were subjected to a polyinosinic:polycytidylic acid [poly(I:C)] pull-down (Figure 2B). As expected, eVP35 and mVP35 interacted with poly(I:C), while eVP35 R312A did not (Figure 2B). Despite expression levels greater than that of the filovirus VP35s, batVP35 again had no detectable interaction with dsRNA (Figure 2B). Furthermore, M. oxyotus, M. davidii, M. annectans, and M. nigricans did not bind the poly(I:C) beads (Figure 2B). These results indicate that Myotis VP35s lack the dsRNA binding activity characteristic of viral VP35s.
Figure 2. batVP35 Inhibits IFN-β Production Independently of dsRNA Binding.
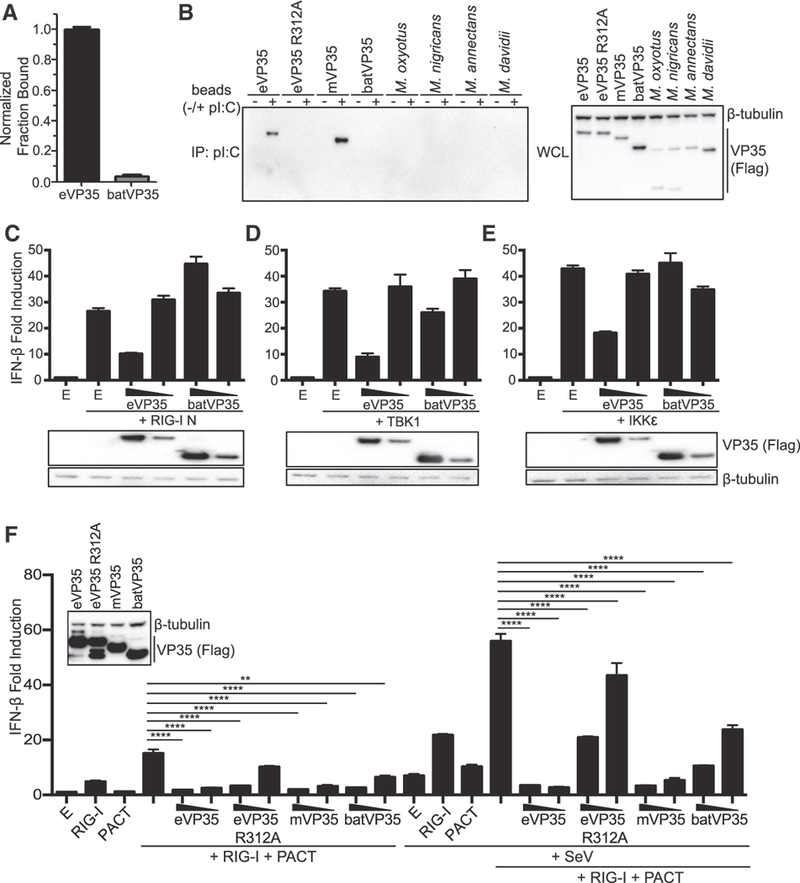
(A) In vitro dsRNA binding assay for eVP35 215–240 and batVP35 159–284. Fractional binding of batVP35 was normalized to eVP35, and error bars represent SD for the triplicate. RNA binding was assessed twice. (B) Western blot analysis of poly(I:C) pull-downs of the indicated FLAG-tagged VP35 constructs. IP, immunoprecipitation; WCL, whole-cell lysate. Poly(I:C) pull-downs were repeated twice. (C-E) IFN-β luciferase reporter assay stimulated by overexpression of (C) RIG-I N, (D) TBK1, or (E) IKKε in the presence of FLAG-tagged eVP35 or batVP35 (500 and 50 ng). Error bars represent the SEM for triplicate experiments. VP35 expression was assessed by western blot for the FLAG epitope tag, and the western blot was aligned to the corresponding samples in the graph. (F) IFN-β reporter assay in cells transfected as indicated. IFN-β promoter activity was assessed as in (C). VP35 expression was assessed for the highest concentration (500 ng) as in (C) (inset). Statistical significance was assessed using a oneway ANOVA and Tukey’s test: ****p < 0.0001, **p<0.01. E refers to empty vector. IFN-β luciferase reporter assays were repeated at least three times. See also Figures S2 and S3.
Myotis VP35 Inhibits IFN-β Production by Impairing RIG-I Activation
To determine what step or steps in the RIG-I signaling pathway Myotis VP35s target, eVP35 and batVP35 were co-expressed with a constitutively active form of RIG-I (RIG-I N), IKKe, or TBK1 in HEK293T cells (Figures 2C–2E). As previously reported, eVP35 modestly inhibited IFN-β production induced by each of these activators; however, batVP35 did not show comparable inhibitory activity, suggesting a mechanism of inhibition upstream of the kinases and activated RIG-I (Figures 2C–2E). The cellular protein PACT facilitates RIG-I activation, but eVP35 can inhibit this by binding PACT and preventing its interaction with RIG-I (Iwamura et al., 2001; Kok et al., 2011; Luthra et al., 2013). This binding and inhibition requires the CBP residues but appears to be independent of VP35 dsRNA binding activity (Luthra et al., 2013). eVP35 was therefore compared to eVP35 R312A, mVP35, and batVP35 in an IFN-β reporter gene assay to assess inhibition of PACT enhancement of RIG-I signaling (Figure 2F). In the absence of SeV, expression of RIG-I alone induced reporter activity, while expression of PACT alone did not (Figure 2F). Co-expression of PACT with RIG-I led to an enhancement of reporter activity over that of RIG-I alone, and this enhancement was efficiently inhibited by eVP35, as previously described (Figure 2F) (Luthra et al., 2013). Similarly, mVP35 blocked the effect of PACT, demonstrating that this inhibition is a conserved function of fi- loviral VP35s. Although prior work identified eVP35 dsRNA binding mutants that are unable to block PACT activation of RIG-I, eVP35 R312A blocked PACT-enhanced RIG-I activity at the highest concentration tested, demonstrating that VP35 dsRNA binding activity is not required for this relatively weak inhibition (Figure 2F). However, this effect titrates out quickly. batVP35 inhibited PACT-mediated RIG-I activation more efficiently than did eVP35 R312A, significantly inhibiting reporter activity at both concentrations tested (Figure 2F). Infection with SeV results in reporter activity that is enhanced in the presence of RIG-I. PACT expression combined with SeV infection results in a minor increase in reporter activity over infected, empty vector-transfected cells. However, co-expression of PACT with RIG-I leads to a synergistic enhancement of reporter activity (Figure 2F). Both eVP35 and mVP35 potently inhibited reporter activity in the presence of SeV infection, while eVP35 R312A showed a substantially reduced, albeit still significant, inhibition of IFN-β reporter activity. Although not as potent as the wild-type filovirus VP35s, batVP35 again significantly inhibited reporter activity (Figure 2F). eVP35 carries out its inhibitory effect at least partly through its interaction with PACT; therefore, we asked whether batVP35 can bind PACT. Similar to eVP35, mVP35 co-immunoprecipitated with PACT, suggesting mVP35 uses a mechanism of PACT inhibition comparable to eVP35 (Figure S3). As previously shown, eVP35 R312A did not detectably interact with PACT (Luthra et al., 2013). Similarly, batVP35 did not pull down PACT, despite inhibiting PACT enhancement of RIG-I (Figure S3). Therefore, batVP35 likely prevents activation of RIG-I, including activation mediated by PACT, by a dsRNA-independent mechanism, rather than targeting a downstream step to impair this pathway.
Full-Length Myotis VP35 Is Required for Inhibition of IFN-β Production
To map regions of Myotis VP35 important for inhibition of IFN-β production, truncations that include the N or C termini of eVP35 (eN VP35 or eC VP35) or batVP35 (batN VP35 or batC VP35) and chimeric constructs containing the N terminus of eVP35 and the C terminus of batVP35 (eNbatC VP35) or the N terminus of batVP35 and the C terminus of eVP35 (batNeC VP35) were constructed (Figure 3A). In the IFN-ß promoter assay, eVP35 potently inhibited IFN-β promoter activity (Figure 3B). eN VP35 exhibited decreased inhibitory activity (Figure 3B). eC VP35 retained potent inhibitory activity, demonstrating that the IID of eVP35 can function independently of its N-terminal oligomerization domain at the concentrations tested (Figure 3B). As expected, full-length batVP35 demonstrated modest inhibition of IFN-β promoter activity. However, expression of neither batN VP35 nor batC VP35 inhibited reporter activity, indicating that full-length batVP35 is required for inhibition (Figure 3B). The chimeric eNbatC VP35 and batNeC VP35 constructs ecapitulated the inhibitory patterns seen for their respective eVP35 component, preventing us from determining whether the batVP35 termini contributed to the activity detected (Figure 3B).
Figure 3. Inhibitory Activity of batVP35 Requires Full-Length Protein.
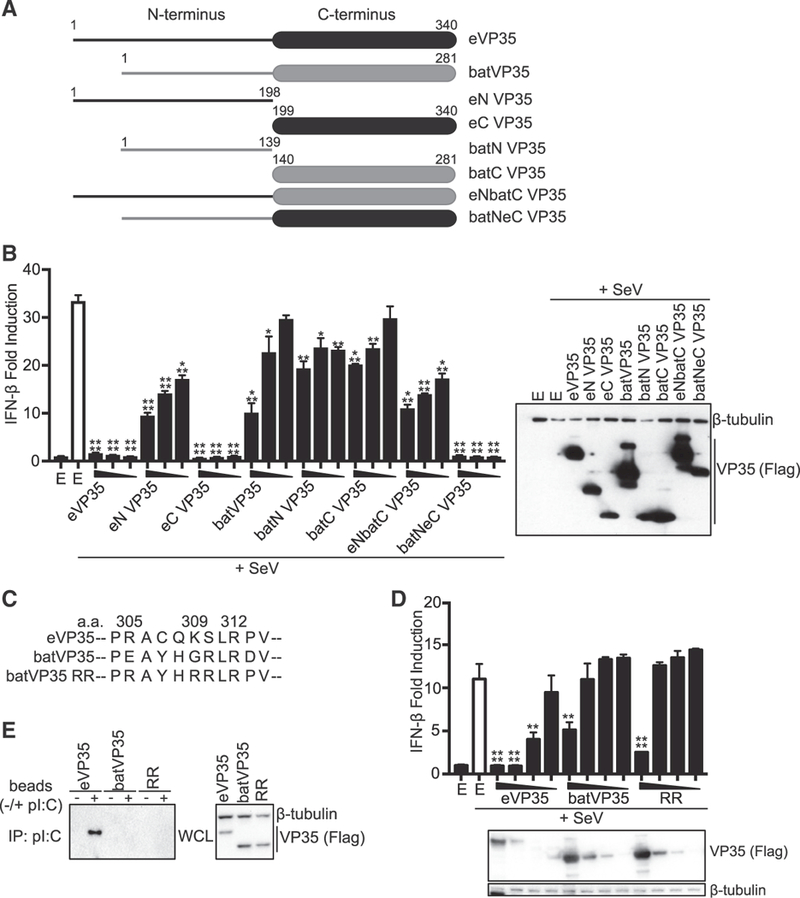
(A) Schematic diagram of eVP35 and batVP35 chimeric constructs. (B)IFN-β promoter luciferase assay in the presence of indicated FLAG-tagged VP35 constructs. The uninfected empty vector control is indicated by the black bar labeled E; the remaining samples were infected with SeV. Error bars represent the SEM for triplicate experiments. Statistical significance was assessed using an unpaired t test, comparing columns to the SeV-infected control (white bar): ****p < 0.0001, ***p < 0.001, **p < 0.01, and *p < 0.05. VP35 expression for the highest concentration was assessed by western blot for the FLAG epitope tag (right). E refers to empty vector. (C)Schematic of the generated batVP35 RR mutant. eVP35 numbering is used. (D)IFN-β reporter assay in the presence of the indicated FLAG-tagged VP35 constructs. Error bars represent the SEM for triplicate experiments. Statistical significance was assessed using a one-way ANOVA and Tukey’s test, comparing columns to the control (white bar): ****p < 0.0001, **p < 0.01. VP35 expression was assessed as in (B). E refers to empty vector. Each IFN-β lucif- erase reporter assay was repeated at least three times. (E)Western blot analysis of poly(I:C) pull-downs of the indicated FLAG-tagged VP35 constructs. IP, immunoprecipitation; WCL, whole-cell lysate. Poly(I:C) pull-downs were repeated twice.
Restoration of Basic CBP in Myotis VP35 Does Not Enhance Inhibition of IFN-β Production
Given the critical role of CBP residues in the potent IFN inhibitory activity and dsRNA binding by eVP35 and mVP35, we generated a batVP35 construct in which residues E246 and G250, positionally equivalent to eVP35 residues R305 and K309, were both mutated to arginine (batVP35 RR) (Figure 3C). As before, eVP35 potently inhibited IFN-β promoter activity while wildtype batVP35 had a low level of inhibition (Figure 3D). Restoration of the basic CBP in batVP35 RR had minimal effects on inhibitory activities (Figure 3D). Furthermore, batVP35 RR did not gain the ability to interact with dsRNA, as shown with a poly(I:C) pull-down (Figure 3E). Therefore, despite the importance of these basic residues in filovirus VP35s, mutation of these residues does not confer on batVP35 robust activity, suggesting that other residues contribute to the attenuated inhibitory activity of batVP35.
Myotis VP35 Does Not Inhibit PKR Activation or miRNA Silencing
eVP35 inhibits PKR activation, and although inhibition does not require dsRNA binding activity, loss of inhibition occurs with mutation of at least two basic amino acids in the CBP (Feng et al., 2007; Schümann et al., 2009). FLAG-tagged eVP35, eVP35 R312A, mVP35, and batVP35 were therefore assayed for inhibition of PKR phosphorylation stimulated by SeV infection. Consistent with prior studies, eVP35 and eVP35 R312A efficiently inhibited PKR phosphorylation (Figure 4A). mVP35 was similarly inhibitory, demonstrating that PKR inhibition is conserved among filoviral VP35s. However, batVP35 was unable to detectably block the SeV-induced PKR phosphorylation (Figure 4A). The same VP35 constructs were tested for inhibition of miRNA silencing using a reporter gene assay in which a luciferase reporter containing miR30 target sequences was co-transfected with plasmids expressing either the non-targeting miR21 or the targeting miR30 (Zhu et al., 2012). Whereas miR21 fails to inhibit luciferase expression, the miR30 plasmid does so. As expected, eVP35 and eVP35 R312A counteracted miRNA silencing (Figure 4B). Although this function is also conserved in mVP35, batVP35 does not block the miR30 inhibition (Figure 4B).
Figure 4. batVP35 Does Not Inhibit PKR Phosphorylation or miRNA.
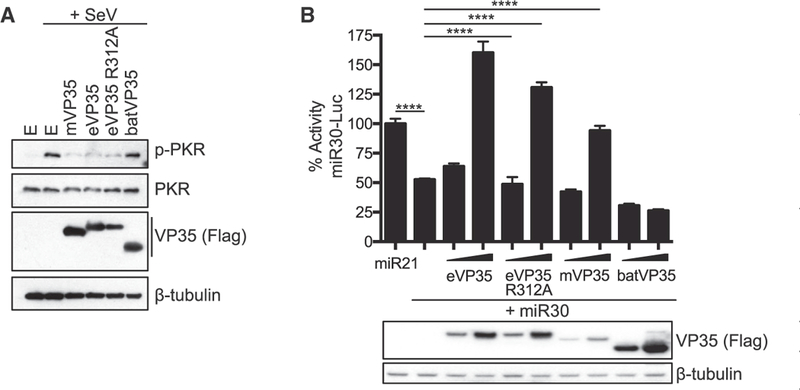
Silencing (A) Analysis of PKR phosphorylation in HEK293T cells in the presence of FLAG-tagged mVP35, eVP35, eVP35 R312A, and batVP35 (500 ng). Western blots were performed for total and phos- phorylated PKR. Three replicates were performed. E refers to empty vector control. (B) Analysis of miRNA silencing in HEK293T cells transfected with the indicated reporter, miRNA expression, and VP35 expression plasmids as indicated. Error bars represent the SEM for triplicate experiments. VP35 expression was assessed by western blot for the FLAG epitope tag. Statistical significance was assessed using a one-way ANOVA and Tukey’s test: ****p < 0.0001. The miR30-Luc luciferase reporter assay was repeated twice.
Myotis VP35s Do Not Disrupt EBOV or MARV Replication
One potential function of endogenous Myotis VP35s would be to interfere with the replication of an infecting filovirus. VP35 function in the viral RNA-dependent RNA polymerase (RDRP) complex is crucial for virus replication (Becker et al., 1998; Leung et al., 2015; Liu et al., 2017; Moller et al., 2005; Mühlberger et al., 1998, 1999; Prins et al., 2010a; Trunschke et al., 2013; Zhu et al., 2017). To determine the effect of batVP35 on replication activity, EBOV and MARV RNA synthesis was measured through the use of a minigenome system. In these assays, the RDRP complex, consisting of the NP, VP30, VP35, and L, is reconstituted in mammalian cells by transfection (Mühlberger et al., 1998, 1999). Co-transfection of a minigenome luciferase reporter containing the necessary cis-acting sequence of the EBOV or MARV genome allows for assessment of viral polymerase activity. To determine the effect of Myotis VP35s on filovirus replication, the EBOV or MARV RNA polymerase complexes were expressed in HEK293T cells in the presence of increasing concentrations of either the cognate VP35 or a subset of Myotis VP35s (Figure 5). Although both EBOV and MARV replication activity were sensitive to expression of excess cognate VP35, neither minigenome system was inhibited by overexpression of batVP35, M. nigricans, M. oxyotus, M. annectans, or M. davidii VP35s, indicating that Myotis VP35s do not impair the activity of these viral replication complexes (Figure 5). In addition, by co-immunoprecipitation assay, batVP35 did not detectably interact with EBOV NP, VP35, VP30, or a truncated form of L (L 1–505) (data not shown). Altogether, this suggests that Myotis VP35 does not interfere with the RNA synthesis of either EBOV or MARV.
Figure 5. batVP35 Does Not Interfere in EBOV or MARV.
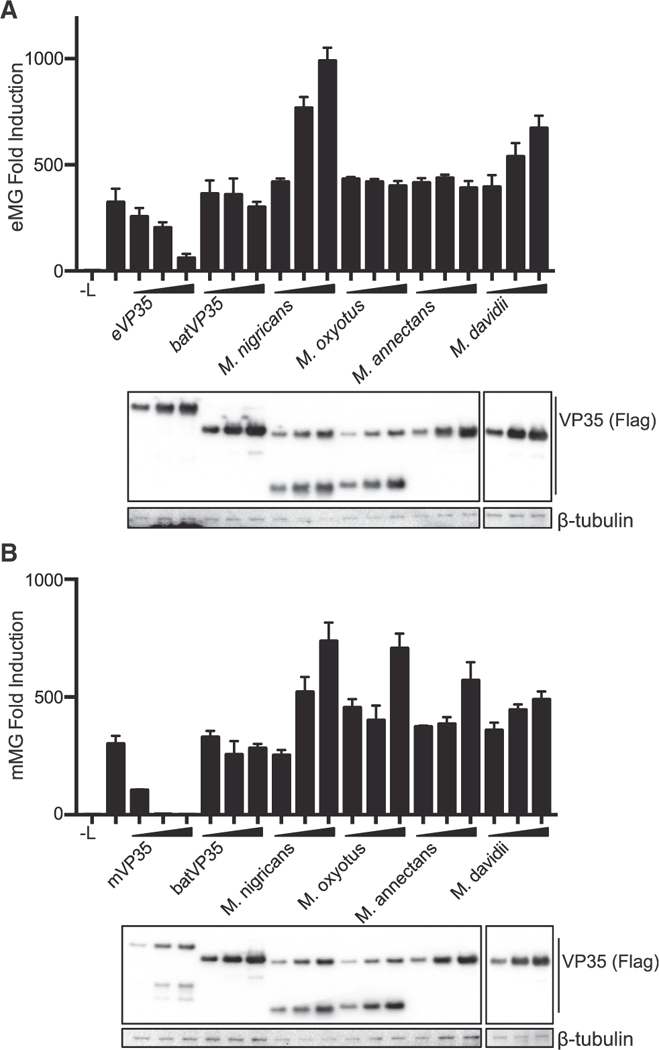
Replication HEK293T cells were transfected with the components of the (A) EBOV or (B) MARV minigenome system. -L indicates samples in which the plasmid expressing L was replaced with empty vector; in all other samples, the complete polymerase complex was transfected. Increasing concentrations of FLAG- tagged VP35 constructs were transfected as indicated. Error bars represent the SEM for representative triplicate experiments, and each minigenome assay was repeated at least three times. VP35 expression was assessed by western blot for the FLAG epitope tag.
Consistent with these functional data, the N-terminal portions of the Myotis VP35s show less homology to the filoviral VP35s compared to the C-terminal IID (Figure S2). The N-terminal domain of filoviral VP35s engages in protein-protein interactions, including the interaction of an NP binding peptide (NPBP) that engages the viral NP and a motif in eVP35 that binds the 8 kDa dynein light chain (LC8), each of which contributes to regulation of viral RNA synthesis (Kubota et al., 2009; Leung et al., 2015; Liu et al., 2017; Luthra et al., 2015; Zhu et al., 2017). The Myotis VP35s lack sequences with obvious homology to the previously described NPBP and LC8 interaction motif (Figure S2). Loss of these features, particularly the lack of the NPBP, is consistent with loss of RNA synthesis functions.
Myotis VP35 Forms Homo-oligomers
The N terminus also has a VP35 homo-oligomerization domain that is required for the maximal inhibition of IFN-β production and for filoviral VP35 function as a polymerase co-factor (Möller et al., 2005; Reid et al., 2005). A co-immunoprecipitation assay demonstrated that FLAG-tagged batVP35 can co-precipitate with hemagglutinin (HA)-tagged batVP35, consistent with oligomerization (Figure 6A). Both eVP35 and mVP35 form tetramers, as demonstrated by light scattering analyses, although work has described the crystal structure of the N terminus of mVP35 forming a trimer instead (Bruhn et al., 2017; Edwards et al., 2016). To determine the oligomeric state of batVP35, we used multi-angle light scattering coupled to size exclusion chromatography and found that it also forms a tetramer (Figure 6B). Therefore, the ability of Myotis VP35s to form homo-oligomers has been retained.
Figure 6. Conservation of Structure between batVP35 and eVP35 (A and B) batVP35 forms a tetramer.

(A) Co- immunoprecipitation assay performed with FLAG antibody on lysates of HEK293T cells expressing HA-tagged batVP35 and FLAG-tagged batVP35 as indicated. Western blots were performed for HA and FLAG. The assay was performed twice. WCL, whole-cell lysate; IP, immunoprecipitation. (B) Gel filtration elution profile of maltose binding protein (MBP)-batVP35. The theoretical monomeric molecular massforMBP-batVP35 is 75 kDa. Molecular weight determined by size exclusion chromatography with multi-angle light scattering (SEC-MALS) forthe major peak of MBP-batVP35 is 296.8 ± 6.4 kDa (three replicates). (C)Substantial structural conservation between batVP35 IID and eVP35 IID. Ribbon representation of VP35 IID structures aligned according to the four-helical bundle (root-mean-square deviation [RMSD] = 0.604 A over 331 atoms). batVP35 IID, magenta (PDB: 6DKU); eVP35 IID, yellow (PDB: 3FKE). (D and E) Electrostatic surface potential was calculated using adaptive Poisson-Boltzmann solver (APBS) from —5to+5 kBTe−1 for (D) batVP35 IID and (E) eVP35 IID (PDB: 3L25). batVP35 IID and eVP35 IID are shown in the same orientation but rotated —90° along the z axis relative to (C). (F)Purifying and diversifying selection on batVP35. batVP35 IID structure isshown as both ribbon (top) and surface (bottom) models. Residues under selection are shown as stick representations, with cyan indicating purifying selection and orange indicating diversifying selection.
Myotis VP35 C Terminus Shares Structural Homology to Filovirus VP35s
The degree of sequence homology between Myotis and filoviral VP35s is greatest in the C-terminal IID (sharing 35% and 27% amino acid identity between batVP35 IID and IIDs of eVP35 and mVP35, respectively). The structure of the batVP35 IID, containing residues 158–281, was pursued for comparison to previously reported filovirus VP35 IID structures. The X-ray crystal structure of the batVP35 IID RR mutant was solved to 2.6 A using molecular replacement with the wild-type eVP35 IID structure (PDB: 3L25, molecule A) as the search model (Figure 6C; Table S1). This demonstrated striking structural homology between batVP35 and the common filoviral VP35 protein fold (Figure 6C; batVP35, pink; eVP35, yellow). Therefore, despite limited sequence similarity to eVP35 and mVP35, this protein fold is independently conserved in batVP35 through evolution. A comparison of the surface electrostatic potentials of batVP35 and eVP35 shows that there is less basic charge along the comparable eVP35:RNA binding interface of batVP35 (Figures 6D and 6E, left). Rotation of the structure to the opposite face of the RNA binding interface reveals that batVP35 lacks much of the highly charged first basic patch, a region important in VP35 polymerase co-factor activity, consistent with the absence of conservation of residues corresponding to eVP35 R222, R225, and K248 (Figures 6D and 6E, right; Figure S2) (Prins et al., 2010a).
Myotis VP35 Residues Undergoing Diversifying and Purifying Selection Map to the IID
Analysis of the 16 Myotis VP35 sequences, excluding the ancestral reconstructed Myotis VP35, using MEME (mixed effect model of evolution) identified three residues in the IID of Myotis VP35 that underwent episodic diversifying selection (using batVP35 numbering, residues 189, 209, and 223) (Figures S1 and S2, +; Table S2). That is, these sites appeared to undergo significant episodic selection along branches leading to species of Myotis, whereas the sites have remained fixed among known genera of filoviruses (Figure S2). Those residues under diversifying selection include batVP35 residue 189, which is never a basic residue in the 16 Myotis VP35 sequences (Figure S1). In eVP35 and mVP35, the corresponding residue is a member of the first basic patch (Prins et al., 2010a). Further analysis identified nine residues under purifying selection in the Myotis VP35s (using batVP35 numbering, residues 94, 103, 132, 134, 176, 178, 181,210, and 273) (Figures S1 and S2, ~; Tables S3 and S4). The three residues under diversifying selection and five of the nine residues under purifying selection map to the batVP35 IID (batVP35 residues 176, 178, 181, 189, 209, 210, 223, and 273), with all present on the surface of the protein (Figure 6F; diversifying, orange; purifying, cyan). These analyses indicate that the prevalent evolutionary signal in Myotis VP35 is one of purifying selection, but the detection of significant diversifying selection at three sites may also indicate limited lineage-specific adaptation since their integration.
DISCUSSION
Our multidisciplinary study provides the most detailed characterization available as to how evolution has affected the structure and function of a family of mammalian NIRVs. NIRVs are rare macromutations thought to result from the integration of viral genes from RNA viruses into a host species genome through the co-option of a host reverse transcriptase (Taylor et al., 2011). Despite identification in mammalian genomes of NIRVs from RNA viruses, such as filoviruses and bornaviruses, the extent to which homology reflects conservation of structure and function is not known (Belyi et al., 2010; Taylor et al., 2010, 2011). A bornavirus nucleoprotein-like integration into the genome of the thirteen-lined ground squirrel can yield a protein capable of being incorporated into extant bornavirus ribonucleoprotein complexes and inhibit viral replication and infection (Fujino et al., 2014). However, functional roles for most described NIRVs have not yet been assessed.
Filovirus-related sequences are disproportionally represented among known mammalian NIRVs (Belyi et al., 2010; Taylor et al., 2010, 2011). Sequences related to the filovirus NP gene have been identified in at least 13 mammalian genera, Lina single genus, and VP35 in seven mammalian genera (Belyi et al., 2010; Taylor et al., 2010, 2011). The presence of such viral-like sequences in Myotis bats is intriguing given that bats serve as reservoirs for MARV and are a suspected reservoir for EBOV (Leroy et al., 2005; Towner et al., 2009). Although Myotis bats have not been specifically implicated as reservoirs for filoviruses, the presence of multiple filovirus-like elements indicates historic infections. Previous timescale analysis of the filovirus-like integrations indicated insertion of NP sequences earlier than 25 million years ago and VP35-like ORFs approximately 18 million years ago (Ruedi et al., 2013; Taylor et al., 2011). Although the NP-like ORFs are disrupted in the Myotis genome, Myotis VP35 ORFs have undergone purifying selection and have been actively maintained throughout the Myotis genus. This active maintenance suggests a functional role of the VP35-like ORF beyond that of simply a fossil record of a past virus-host interaction. Our increased genomic sampling (compared to prior efforts) bolsters evidence for the maintenance of the ORF and identifies numerous sites subject to pervasive purifying selection, many of which map to the C-terminal IID. The picture that emerges from our study is partial functional conservation of the IFN-suppressing activity of Myotis VP35s relative to filoviral VP35s.
A central feature of extant filoviral VP35s is potent suppression of RIG-I signaling and IFN production (Edwardset al., 2016; Fea- ginsand Basler, 2015; Leung etal., 2010; Ramanan etal., 2012). The most intensively studied, eVP35, inhibits IFN responses through several mechanisms (Basler et al., 2000; Leung et al., 2010; Luthra et al., 2013; Prins et al., 2009, 2010b; Yen and Basler, 2016). One crucial mechanism is thought to be the interaction of EBOV and mVP35 with dsRNA via their IID, sequestering immunostimulatory dsRNA from recognition by RLRs (Dilley et al., 2017). Of the 17 Myotis VP35 ORFs tested, many were able to inhibit RIG-I signaling in either HEK293T or M. myotis cells, although several lacked measurable inhibitory activity, with the pattern of inhibition varying between the two cell lines. Forthose that possess anti-IFN function, the degree of inhibition is substantially less than for extant filovirus VP35s. This impaired anti-IFN function correlates with a lack of dsRNA binding activity. The lack of conservation of two CBP residues in Myotis VP35 might explain the lack of dsRNA binding. However, reconstitution of the CBP in the batVP35 RR construct did not restore dsRNA binding or IFN-β inhibitory activity. The solved crystal structure of batVP35, although it contains the reconstituted CBP, is less basic than that of eVP35, potentially explaining the lack of dsRNA interaction.
Filovirus VP35s also employ dsRNA-independent mechanisms of IFN inhibition, including the inhibition of PACT enhancement of RIG-I activity through the interaction of eVP35 and PACT (Luthra et al., 2013). In addition to the previously described inhibition by eVP35, we show that mVP35 interacts with PACT and inhibits PACT activation of RIG-I, indicating a conserved filovirus VP35 anti-IFN function. eVP35 R312A retains modest inhibition of PACT-induced RIG-I activity, despite a lack of interaction with PACT. batVP35 likewise maintains the functional capacity to inhibit PACT activation of RIG-I while lacking a detectable interaction with PACT. Therefore, batVP35 appears to act upstream of RIG-I activation, potentially via inhibition of PACT, to block RIG-I signaling. Consistent with an inhibitory mechanism proximal to RIG-I activation, batVP35 lacks inhibitory activity when the kinases IKKe and TBK-1, which are upstream of IRF-3 phosphorylation but downstream of RIG-I, are overexpressed. This is in contrast to what is seen with EBOV or mVP35s in the same assay (Edwards et al., 2016; Prins et al., 2009; Ramanan et al., 2012).
A study evaluated a single Myotis VP35 ORF from M. lucifigus and concluded that the single bat-derived VP35 significantly inhibited human IFN-β promoter activity in HEK293T cells and that inhibition was comparable to that of eVP35 (Kondoh et al., 2017). However, prior studies have demonstrated that filovirus VP35s are relatively weak inhibitors of IFN responses when these are artificially induced by overexpression of signaling molecules downstream of RIG-I. In contrast, the same filovirus VP35s are potent inhibitors when the pathway is activated by canonical mechanisms (Edward et al., 2016; Ramanan et al., 2012). Therefore, under conditions in which downstream activators are overexpressed, the inhibitory activity of the Myotis VP35 may appear to be similar to that of eVP35, allowing for the erroneous conclusion that the IFN inhibitory functions of Myotis and eVP35s are comparable. In our study, we used SeV infection, a negative-sense RNA virus, to trigger an IFN response through the activation of RIG-I by its canonical mechanism. Under these conditions, one can see potent inhibition by filovirus VP35s and can more accurately assess the corresponding but lesser activity of Myotis VP35s.
Filovirus VP35s also engage in innate immune evasion tactics outside of suppressing IFN production. In contrast to filovirus VP35s, batVP35 does not inhibit PKR phosphorylation. In eVP35, it has been shown that although dsRNA binding is not required, mutation of at least two CBP residues results in a loss of inhibitory activity (Schümann et al., 2009). Therefore, the absence of inhibition of PKR phosphorylation by batVP35 could be due to the presence of non-basic residues in two positions in the CBP, at 305 and 309 (using eVP35 numbering). The lack of inhibition of miRNA gene silencing by batVP35 might also be attributable to the loss of PKR inhibition, because it has previously been suggested that eVP35 R312A counteracts miRNA silencing through its antagonism of PKR activity (Zhu et al., 2012).
The capacity of Myotis VP35 to form homo-oligomers and the substantial conservation of the C-terminal filovirus VP35 protein fold indicate purifying selection of structural elements over the evolutionary timescale. This is consistent with the hypothesis that host-virus protein-based interactions may have occurred for a substantial time following endogenization, resulting in the active maintenance of the filovirus-like VP35. The differential capacity to suppress IFN signaling by Myotis VP35s relative to extant filovirus VP35s may be attributable to the divergence of extant filovirus VP35s from the Myotis VP35s or to the divergence of extant Myotis VP35s from the ancestral bat filovirus. However, assessment of the inhibitory activity of the reconstructed, ancestral Myotis VP35 sequence suggests that the current endogenous Myotis VP35s have not evolved toward more modest IFN inhibition; instead, they have maintained for more than 18 million years the reduced, but significant, suppression of IFN induction present in the ancestral integration.
Although inhibition of EBOV or MARV RNA synthesis yielded negative data, it remains possible that Myotis VP35s could interfere with the replication of other extant viruses, including viruses that have not yet been isolated, or with the replication of ancestral filoviruses that no longer exist. It is also plausible that there was functional divergence among the differing integrated filovi- rus-like bat genes with the now-pseudogenized Myotis NP possessing an antifiloviral function while the VP35 maintained an immunoregulatory function. It seems probable that a potent suppressor of IFN responses would be selected against, because this could result in high susceptibility to viral infection. One can conceive of a scenario whereby modest inhibitory activity modulates IFN and inflammatory responses, which might otherwise be detrimental to the virus-infected host; many negative regulators of IFN and inflammatory pathways have been described (Hayden and Ghosh, 2012; Ivashkiv and Donlin, 2014). It is possible that a viral protein might be co-opted for such a purpose. Alternatively, it is possible that the Myotis VP35 was preserved to carry out an as-yet-unidentified function.
Despite evidence for functional maintenance, when and where filovirus-like VP35 ORFs are expressed in Myotis bats is not known. Limited analysis has yet to detect protein or mRNA expression of the Myotis VP35s (Taylor et al., 2010, 2011). Using qRT-PCR, we were unable to detect M. myotis VP35 mRNA in transformed cell lines of the nasal epithelium, nervus olfactorius, or brain of M. myotis (data not shown) (Heet al., 2014). We further looked for VP35 mRNA expression in the spleen of M. lucifigus and only detected a signal for the filovirus-like VP35 slightly above background (minus reverse transcriptase) levels (data not shown). However, the lack of convincing detection of Myotis VP35 mRNA in the cell lines and tissues examined does not preclude the potential for Myotis VP35 expression. The human protein syncytin, co-opted from an endogenous retroviral gene insertion and required for placental function, has tissue-specific expression, with robust presence in the human placenta, weaker expression in testis, and no detection in 21 other human tissues (Mi et al., 2000). To gain further insight into possible Myotis VP35 functions, a more exhaustive search for tissue-specific and developmentally regulated expression would be appropriate.
STAR ★ METHODS
Detailed methods are provided in the online version of this paper and include the following:
KEY RESOURCES TABLE
CONTACT FOR REAGENT AND RESOURCE SHARING
- EXPERIMENTAL MODELAND SUBJECT DETAILS
-
○Cell lines and Viruses
-
○
- METHOD DETAILS
-
○Plasmids
-
○Recombinant Protein Expression and Purification
-
○Co-immunoprecipitation Assays
-
○Poly(I:C) Pull-down Assays
-
○IRF-3 Phosphorylation Assay
-
○IFN-β Reporter Gene Assays
-
○PKR Phosphorylation Assay
-
○microRNA Silencing Reporter Assay
-
○Minigenome Assays
-
○SEC-MALS
-
○RNA filter binding assay
-
○Antibodies
-
○Western Blots
-
○Phylogenetic Analyses
-
○Analysis of Selective Pressure
-
○Crystallization, Diffraction Data Collection, Structure Determination and Refinement of Myotis VP35 IID
-
○
QUANTIFICATION AND STATISTICAL ANALYSIS
DATA AND SOFTWARE AVAILABILITY
STAR ★ METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-FLAG M2 antibody | Sigma Aldrich | Cat#F1804; RRID: AB_262044 |
| Rabbit polyclonal anti-flag antibody | Sigma Aldrich | Cat#F7425; RRID: AB_439687 |
| Mouse monoclonal anti-HA antibody | Sigma Aldrich | Cat#H3663; RRID: AB_262051 |
| Rabbit polyclonal anti-HA antibody | Sigma Aldrich | Cat#H6908; RRID: AB_260070 |
| Mouse monoclonal anti-β-tubulin antibody | Sigma Aldrich | Cat# T8328; RRID: AB_1844090 |
| Rabbit monoclonal anti-IRF3 antibody | Cell Signaling | Cat#4302S; RRID: AB_1904036 |
| Rabbit monoclonal anti-Phospho-IRF3 (Ser396) | Cell Signaling | Cat#4947S; RRID: AB_823547 |
| Rabbit polyclonal anti-PKR antibody | Cell Signaling | Cat#3072S; RRID: AB_2277600 |
| Rabbit monoclonal anti-PKR (phospho T446) antibody | Abcam | Cat#ab32036; RRID: AB_777310 |
| Mouse anti-Ebola VP35 antibody | Prins et al., 2010b | N/A |
| Bacterial and Virus Strains | ||
| E. cloni 10G chemically competent cells | Lucigen | Cat#60107 |
| E. coli BL21(DE3) | Novagen | Cat#69450–3 |
| Sendai Virus Cantell | Georgia State University | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Lipofectamine2000 | Invitrogen | Cat#11668500 |
| Protease inhibitor cocktail | Sigma Aldrich (Roche) | Cat#11836170001 |
| PhosSTOP | Sigma Aldrich (Roche) | Cat#4906845001 |
| Critical Commercial Assays | ||
| QIAGEN HiSpeed Maxi Kit | QIAGEN | Cat#12663 |
| QIAquick Gel Extraction Kit | QIAGEN | Cat#28704 |
| QIAquick PCR Purification Kit | QIAGEN | Cat#28104 |
| Dual-Luciferase Reporter Assay System | Promega | Cat#E1960 |
| Top96 Crystallization Screen | Anatrace | Cat#TOP96 |
| Deposited Data | ||
| Myotis lucifugus MN VP35 sequence | This paper | GenBank: MH431024 |
| Myotis muricola brownii VP35 sequence | This paper | GenBank: MH431025 |
| Myotis horsfieldii VP35 sequence | This paper | GenBank: MH431026 |
| Myotis blythii VP35 sequence | This paper | GenBank: MH431027 |
| Myotis oxyotus VP35 sequence | This paper | GenBank: MH431033 |
| Myotis nigricans VP35 sequence | This paper | GenBank: MH431030 |
| Myotis annectans VP35 sequence | This paper | GenBank: MH431028 |
| Myotis riparius VP35 sequence | This paper | GenBank: MH431029 |
| Myotis albescens VP35 sequence | This paper | GenBank: MH431032 |
| Myotis septentrionalis VP35 sequence | This paper | GenBank: MH431031 |
| Myotis capaccinii VP35 sequence | This paper | GenBank: MH431035 |
| Myotis myotis VP35 sequence | This paper | GenBank: MH431036 |
| Myotis velifer incautus VP35 sequence | This paper | GenBank: MH431034 |
| Crystal Structure | This paper | https://www.rcsb.org/structure/6DKU |
| Experimental Models: Cell Lines | ||
| HEK293T | ATCC | CRL-3216 |
| M. myotis nasal epithelial cells (Nep) | He et al., 2014 | N/A |
| Oligonucleotides | ||
| dsRNA used in in vitro binding: CGCAUGCG | Leung et al., 2010 | N/A |
| Recombinant DNA | ||
| pCAGGS Flag Myotis lucifugus | This paper | N/A |
| pCAGGS Flag Myotis lucifugus MN | This paper | N/A |
| pCAGGS Flag Myotis muricola brownii | This paper | N/A |
| pCAGGS Flag Myotis horsfieldii | This paper | N/A |
| pCAGGS Flag Myotis blythii | This paper | N/A |
| pCAGGS Flag Myotis oxyotus | This paper | N/A |
| pCAGGS Flag Myotis annectans | This paper | N/A |
| pCAGGS Flag Myotis riparius | This paper | N/A |
| pCAGGS Flag Myotis albescens | This paper | N/A |
| pCAGGS Flag Myotis septentrionalis | This paper | N/A |
| pCAGGS Flag Myotis capaccinii | This paper | N/A |
| pCAGGS Flag Myotis myotis | This paper | N/A |
| pCAGGS Flag Myotis velifer incautus | This paper | N/A |
| pCAGGS Flag Myotis davidii | This paper | N/A |
| pCAGGS Flag Myotis brandtii | This paper | N/A |
| pCAGGS Flag ancestral Myotis VP35 | This paper | N/A |
| pCMV-miR30 | Zeng and Cullen, 2003 | Addgene plasmid #20670 |
| pCMV-miR21 | Zeng and Cullen, 2003 | Addgene plasmid #20381 |
| pCMV-luc-miR30(P) | Zeng and Cullen, 2003 | Addgene plasmid #20875 |
| pCAGGS Flag eVP35 | Cárdenas et al., 2006 | N/A |
| pCAGGS HA eVP35 | Cárdenas et al., 2006 | N/A |
| pCAGGS Flag mVP35 | Edwards et al., 2016 | N/A |
| pCAGGS HA mVP35 | Edwards et al., 2016 | N/A |
| pCAGGS Flag IKte | Cárdenas et al., 2006 | N/A |
| pCAGGS Flag TBK1 | Cárdenas et al., 2006 | N/A |
| pCAGGS Flag RIG-I N | Edwards et al., 2016 | N/A |
| pCAGGS HA RIG-I | Edwards et al., 2016 | N/A |
| pCAGGS IRF3 | Ramanan et al., 2012 | N/A |
| pCAGGS Flag PACT | Luthra et al., 2013 | N/A |
| pM1 EBOV minigenome reporter | Edwards et al., 2015 | N/A |
| pCAGGS EBOV NP | Edwards et al., 2015 | N/A |
| pCAGGS EBOV VP30 | Edwards et al., 2015 | N/A |
| pCAGGS EBOV VP35 | Edwards et al., 2015 | N/A |
| pCAGGS EBOV L | Edwards et al., 2015 | N/A |
| pCAGGS MARV VP35 | Ramanan et al., 2012 | N/A |
| pCAGGS MARV NP | This paper | N/A |
| pCAGGS MARV VP30 | This paper | N/A |
| pM1 MARV minigenome reporter | Edwards et al., 2014 | N/A |
| pCAGGS MARV L | This paper | N/A |
| pCAGGS Flag eVP35 R312A | This paper | N/A |
| pCAGGS eVP35 R312A | Luthra et al., 2013 | N/A |
| pCAGGS Flag batVP35 RR | This paper | N/A |
| pCAGGS Flag eN VP35 | Edwards et al., 2016 | N/A |
| pCAGGS Flag eC VP35 | Edwards et al., 2016 | N/A |
| pCAGGS Flag batN VP35 | This paper | N/A |
| pCAGGS Flag batC VP35 | This paper | N/A |
| pCAGGS Flag eNbatC VP35 | This paper | N/A |
| pCAGGS Flag batNeC VP35 | This paper | N/A |
| MBP batVP35 | This paper | N/A |
| MBP batVP35 159–284 | This paper | N/A |
| Software and Algorithms | ||
| GraphPad Prism 7 | GraphPad Software | https://www.graphpad.com |
| SeaView v4.6 | Gouy et al., 2010 | http://doua.prabi.fr/software/seaview |
| Muscle | Gouy et al., 2010 | http://www.drive5.com/muscle/ |
| IQ-Tree v1.6 | Nguyen et al., 2015; Trifinopoulos et al., 2016 | http://www.iqtree.org |
| PartitionFinder v1.1 | Lanfear et al., 2012 | http://www.robertlanfear.com/partitionfinder/ |
| DataMonkey | Delport et al., 2010 | https://www.datamonkey.org |
| FigTree 1.4.3 | Rambaut, 2012 | https://bioweb.pasteur.fr/packages/pack@FigTree@1.4.3 |
| PhyloBot v10.09.2016.1 | Hanson-Smith and Johnson, 2016 | http://www.phylobot.com |
| MOLREP | Vagin and Teplyakov, 1997 | http://www.ccp4.ac.uk/dist/html/molrep.html |
| REFMAC | Murshudov et al., 1997 | https://www2.mrc-lmb.cam.ac.uk/groups/murshudov/content/refmac/SourceEtal/source.html |
| COOT | Emsley and Cowtan, 2004 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| MOLPROBITY | Chen et al., 2010 | http://molprobity.biochem.duke.edu |
| PyMOL | Schrodinger | https://www.pymol.org/2/ |
| ASTRA 6 | Wyatt Technologies | https://www.wyatt.com/products/software/astra.html |
| Other | ||
| Anti-FLAG M2 magnetic beads | Sigma-Aldrich | Cat# M8823, RRID:AB_2637089 |
| 3X FLAG peptide | Sigma-Aldrich | Cat#F4799 |
| EZview Red Anti-HA Agarose | Sigma-Aldrich | Cat#E6779 |
| Influenza Hemagglutinin (HA) peptide | Sigma-Aldrich | Cat#I2149 |
| Bolt 10% Bis-Tris Plus polyacrylamide gels | Thermo Fisher Scientific | Cat#NW00107BOX |
| Trans-Blot® Turbo Midi PVDF Transfer Packs | Bio-Rad Laboratories | Cat#170–4157 |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Christopher F. Basler (cbasler@gsu.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines and Viruses
M. myotis nasal epithelial cells (Nep) (He et al., 2014) and HEK293T cells (ATCC, CRL-3216) were maintained in DMEM, supplemented with 10% fetal bovine serum (FBS) and cultured at 37°C and 5% CO2. Sendai virus Cantell (SeV) was grown in 10 day- old embryonated chicken eggs for two days at 37°C.
METHOD DETAILS
Plasmids
Mammalian Expression
The sequence for batVP35 was synthesized (Genscript, Piscataway, NJ (codon optimized for E. coli expression)) based on a previously described Myotis lucifigus VP35 sequence and cloned with an amino-terminal Flag-tag into pCAGGS (Belyi et al., 2010). cDNA were obtained for Myotis lucifugus MN (Minnesota) (Field Museum, Chicago (FMNH)_172384), Myotis muricola brownii (FMNH_167239), Myotis horsfieldii (FMNH_177466), Myotis blythii(FMNH_140372), Myotis oxyotus (FMNH_174938), Myotis nigricans (FMNH_162544), Myotis annectans (American Museum of Natural History, Ambrose Monell Cryo Collection (AMCC_110817), Myotis riparius (AMCC_109656), Myotis albescens (AMCC_109603), Myotis septentrionalis (New York State Rabies Laboratory), Myotis capaccinii and Myotis myotis (Dr. Jordi Serra-Cobo, Barcelona University, Spain) and were used to obtain the endogenous filovirus-like VP35 open reading frames (ORFs) using the forward primer 5’GCGCGCGGCCGCATCCCTGGAG3’ and reverse primer 5’GCGCAGATCTTCAAATCTTTAAC3’. cDNA generated from a Myotis velifer incautus cell line (ATCC CRL-6012) was used to obtain the filovirus-like VP35 ORF using the forward primer 5’GCGCGCGGCCGCAATGTCCCTGGAGCAGTG C3’ and reverse primer 5’GCGCAGATCTTTAAATCTTTAACCCGAGGC3’. The resulting PCR products were cloned with N-terminal Flag-tags into pCAGGS and the sequences of the inserts were confirmed. Sequences were synthesized for Myotis davidii VP35 (GenBank: ALWT01033109.1, nucleotides 1842–2562) and Myotis brandtii VP35 (GenBank: ANKR01158691.1, nucleotides 2885–3727) and were similarly tagged and cloned (Genscript, Piscataway, NJ). Ancestral Myotis VP35 sequence reconstruction was carried out in Phylobot using the related VP35-like sequence from the tarsier as an outgroup and PROTGAMMAJTT as the substitution model (Hanson-Smith and Johnson, 2016). Phylobot uses the CODEML package of PAML to carry out empirical Bayesian ancestral sequence reconstruction. The resulting sequence was synthesized and cloned as above (Genscript, Piscataway, NJ). pCMV-miR30, pCMV-miR21 and pCMV- luc-miR30(P) were obtained from Addgene (Addgene plasmid #20670, #20381 and #20875) (Zeng and Cullen, 2003). Expression vectors for pCAGGS Flag/HA EBOV VP35 (eVP35), pCAGGS Flag/HA MARV VP35 (mVP35), pCAGGS Flag eN VP35, pCAGGS Flag eC VP35, pCAGGS Flag IKKe, pCAGGS Flag TBK1, pCAGGS Flag RIG-I N, pCAGGS HA RIG-I, pCAGGS Flag PACT, pM1 EBOV minigenome reporter (eMG), pCAGGS EBOV NP, pCAGGS EBOVVP30, pCAGGS EBOVVP35, pCAGGS EBOV L, pCAGGS MARVVP35, and pM1 MARV minigenome reporter (mMG) plasmid have previously been described (Cárdenas et al., 2006; Edwards et al., 2014, 2015, 2016; Luthra et al., 2013; Ramanan et al., 2012). MARV NP, MARV VP30 and MARV L were subcloned from Flag-tagged pCAGGS into untagged pCAGGS (Edwards et al., 2014). Overlapping PCR was used to clone eVP35 R312A, batVP35 RR and chimeric VP35s, which were cloned with N-terminal Flag-tags into pCAGGS. Bacterial Expression: eVP35 215–240, full length batVP35 and batVP35 159–284 were subcloned into a modified pET15b vector (Novagen, EMD Millipore, Billerica, MA).
Recombinant Protein Expression and Purification
Proteins were expressed as maltose binding protein (MBP) fusions in BL21(DE3) E. coli (Novagen, EMD Millipore, Billerica, MA) in LB medium. Cells were harvested and resuspended in buffer containing 25 mM sodium phosphate [pH 7.5], 50 mM NaCl, 20 mM imidazole and 5 mM 2-mercaptoethanol, and were lysed using an EmulsiFlex-C5 homogenizer (Avestin, Ottawa, Canada). Lysates were clarified by centrifugation at 42,000 × g at 10°C for 40 minutes. Proteins were purified using a series of chromatographic columns and sample purity was determined by SDS-PAGE.
Co-immunoprecipitation Assays
HEK293T cells (1 ×106) were transfected with the indicated plasmids using Lipofectamine 2000 (Thermo Fisher Scientific, MA) and at 24 hours post-transfection, cells were lysed in NP-40 lysis buffer (50 mM Tris [pH 7.5], 280 mM NaCl, 0.5% Nonidet P-40, 0.2 mM EDTA, 2 mM EGTA, 10% glycerol, protease inhibitor (cOmplete; Roche, Indianapolis, IN)). Anti-FLAG M2 magnetic beads (Sigma- Aldrich, St. Louis, MO) were incubated with lysates for one hour at 4°C, washed five times in NP-40 lysis buffer, and eluted using 3X FLAG peptide (Sigma-Aldrich, St. Louis, MO) at 4°C for 30 minutes or by boiling for five minutes in 1× sample buffer. Whole cell lysates and co-precipitation samples were analyzed by western blot.
Poly(I:C) Pull-down Assays
HEK293T cells (1 × 106) were transfected with 1 mg of the indicated VP35 plasmid using Lipofectamine 2000 (Thermo Fisher Scientific, MA). Twenty-four hours post-transfection, cells were lysed in NP-40 lysis buffer (50 mM Tris [pH 7.5], 280 mM NaCl, 0.5% Nonidet P-40, 0.2 mM EDTA, 2 mM EGTA, 10% glycerol, protease inhibitor (cOmplete; Roche, Indianapolis, IN)). Lysates were incubated with Sepharose beads (GE Healthcare, PA) either coupled or uncoupled to low molecular weight poly(I:C) (Invivogen, CA) for four hours at 4°C. Beads were washed five times in NP-40 lysis buffer and bound proteins were eluted by boiling for five minutes in 1× sample buffer. Samples were analyzed by western blot.
IRF-3 Phosphorylation Assay
HEK293T cells (5 × 105) were transfected with 2,1 or 0.5 mg of the indicated plasmids using Lipofectamine 2000 (Thermo Fisher Scientific, MA). Twenty-four hours post-transfection, cells were infected with SeV. Eight hours post-infection, cells were lysed in NP-40 lysis buffer (50 mM Tris [pH 7.5], 280 mM NaCl, 0.5% Nonidet P-40, 0.2 mM EDTA, 2 mM EGTA, 10% glycerol, protease inhibitor and phosphatase inhibitor (cOmplete and PhosSTOP; Roche, Indianapolis, IN). Lysates were analyzed by western blot.
IFN-β Reporter Gene Assays
SeV-induced reporter assay: HEK293T cells (1 × 105) were transfected with an IFN-ß firefly luciferase reporter plasmid, a constitutively expressed Renilla luciferase reporter plasmid (pRL-tk) (Promega, Madison, WI) and the indicated VP35 expression plasmids using Lipofectamine 2000 (Thermo Fisher Scientific, MA). M. myotis Nep cells (2 × 105) were transfected in the same manner. At twenty-four hours post-transfection, the cells were infected with SeV (Cantell Strain, HEK293T - 1000 HA Units, M. myotis Nep-100 HA Units) in DMEM, and 10% FBS. At eighteen hours post-treatment, cells were lysed and a dual luciferase reporter assay (Promega, Madison, WI) was performed. Firefly luciferase values were normalized to Renilla luciferase values. The assay was performed in triplicate; error bars indicated the standard error of the mean (SEM) for the triplicate. Kinase-induced reporter assay: HEK293T cells (1 × 105) were transfected with an IFN- β firefly luciferase reporter plasmid, a constitutively expressed Renilla luciferase reporter plasmid (pRL-tk) (Promega, Madison, WI), the indicated VP35 expression plasmids and the indicated activator; constitutively active form of RIG-I (Flag RIG-I N), Flag IKKe or Flag TBK1. Twenty-four hours post-transfection the cells were lysed and assayed using a dual luciferase reporter assay (Promega, Madison, WI) and analyzed as above. PACT reporter assay: HEK293T cells (1 × 105) were transfected with an IFN-ß firefly luciferase reporter plasmid, a constitutively active Renilla luciferase reporter plasmid (pRL-tk) (Promega, Madison, WI), expression plasmids encoding PACT (100 ng), RIG-I (1 ng) and indicated VP35 plasmids (500 and 5 ng) using Lipofectamine 2000 (Thermo Fisher Scientific, MA). Twenty-four hours post-transfection, cells were infected with SeV (100 HA units). At eighteen hours post-infection, cells were lysed and a dual luciferase reporter assay (Promega, Madison, WI) was performed and analyzed as above.
PKR Phosphorylation Assay
HEK293T cells (2.5 × 105) were transfected with Flag tagged eVP35, eVP35 R312A, mVP35and batVP35 (500 ng). Twenty-four hours post-transfection, cells were infected with SeV (1000 HA Units) as indicated after which virus was replaced with DMEM supplemented with 10% FBS. Eighteen hours post-infection, cells were lysed in NP-40 lysis buffer (50 mM Tris [pH 7.5], 280 mM NaCl, 0.5% Nonidet P-40, 0.2 mM EDTA, 2 mM EGTA, 10% glycerol, protease inhibitor and phosphatase inhibitor (cOmplete and PhosStop; Roche, Indianapolis, IN). Lysates were analyzed by western blot.
microRNA Silencing Reporter Assay
The assay was modified from (Zhu et al., 2012): HEK293T cells (1 × 105) were transfected using Lipofectamine 2000 (Thermo Fisher Scientific, MA) with pCMV-luc-miR30(P) firefly luciferase reporter plasmid (30 ng) containing miR30 target sites, a constitutively active Renilla luciferase reporter plasmid (5 ng), pCMV-miR30 targeting plasmid (10 ng), pCMV-miR21 non-targeting plasmid (20 ng) and the indicated pCAGGS VP35 plasmids (50 and 500 ng). Twenty-four hours post transfection the cells were lysed and assayed using a dual luciferase reporter assay. Firefly luciferase values were normalized to Renilla luciferase values and the assay was done in triplicate; error bars indicate the SEM for the triplicate.
Minigenome Assays
HEK293T cells (1 × 105) were transfected using Lipofectamine 2000 (Thermo Fisher Scientific, MA) with expression plasmidsencoding either the EBOV or MARV replication complex components; NP, L, VP30, VP35, T7, T7 promoter-driven EBOV or MARV minigenome RNA (eMG or mMG), which encode a Renilla luciferase reporter gene, and a constitutively expressed firefly luciferase plasmid (pCAGGS Firefly) that served as a transfection control. For the EBOV minigenome this consisted of 50 ng pCAGGS eNP, 100 ng pCAGGS eL, 20 ng pCAGGS eVP30, 25 ng pCAGGS VP35, 40 ng eMG, 40 ng pCAGGS T7 and 0.2 ng pCAGGS Firefly. The same concentrations of DNA were used for the MARV minigenome components, except for 25 ng of pCAGGS mVP30 was used. Additional Flag-tagged VP35 expression plasmids were transfected in increasing concentrations as indicated (20, 100 and 200 ng). Forty-eight hours post transfection the cells were lysed and assayed using a dual luciferase reporter assay (Promega, Madison, WI). The assay was performed in triplicate; error bars indicate SEM for the triplicate.
SEC-MALS
SEC-MALS experiments were performed using a DAWN-HELEOS II detector (Wyatt Technologies) coupled to a Superdex SD200 column (GE Healthcare) in (10 mM HEPES [pH 7], 150 mM NaCl, and 2 mM TCEP). 2 mg/ml sample was injected and raw data were analyzed using ASTRA 6 software (Wyatt Technologies) to determine the weight averaged molecular mass (MW). Protein concentrations were determined using the refractive index measured by an Optilab T-rEX (Wyatt Technologies) and a dn/dc = 0.185 mL x g~1.
RNA filter binding assay
Labeled RNAs (5 nM) were incubated with increasing concentrations of purified batVP35 HD protein. After 15 minutes at room temperature, samples were applied to a dot blot apparatus (Whatman) with one nitrocellulose (NC) membrane on top of one nylon (NY) membrane. Radiolabeled RNA bound to the NC and NY membranes were quantified using a Typhoon 9410 variable-mode imager, and the fraction of RNA bound to batVP35 was calculated using the following equation: fraction bound = RNA signal on NC/(RNA signal on NC + RNA signal on NY).
Antibodies
Monoclonal mouse anti-FLAG M2, polyclonal rabbit anti-Flag, monoclonal mouse anti-HA, polyclonal rabbit anti-HAand monoclonal mouse ß-tubulin antibodies were purchased from Sigma-Aldrich (St. Louis, MO). Anti-IRF-3, anti-phospho IRF-3 (Ser396) and anti-PKR were purchased from Cell Signaling (Danvers, MA). Anti-phospho PKR antibody was purchased from Abcam (Cambridge, MA). Monoclonal anti-eVP35 antibody has previously been described (Prins et al., 2010b).
Western Blots
Lysates were run on 10% Bis-Tris Plus polyacrylamide gels (Thermo Fisher Scientific, MA) and transferred to PVDF membrane (Bio-Rad, Hercules, CA). Membranes were probed with the indicated antibodies and were developed using Western Lightning Plus ECL (Perkin Elmer, Waltham, MA).
Phylogenetic Analyses
Unaligned sequences were translated in Seaview and then aligned using Muscle (Gouy et al., 2010). Seaview was then used to convert the alignment into a nucleotide-based codon alignment. Maximum likelihood analysis and model fitting was carried out using IQ-TREE using the two significant partitions identified by Partitionfinder (Lanfearet al., 2012; Nguyen et al., 2015; Trifinopouloset al., 2016). Trees, midpoint rooting, and support values (ultrafast bootstrap and approximate likelihood ratio tests) were visualized using Figtree 1.4.3 (Rambaut, 2012).
Analysis of Selective Pressure
Four tests for selection (MEME, FUBAR, FELand SLAC) were carried out using Datamonkey, a web server for HyPhy (Delport et al., 2010). We used a conservative approach to determine significance of pervasive selection which required a consensus of significant codons from at least three methods (using default levels for posterior probabilities and significance) (Table S2, S3 and S4).
Crystallization, Diffraction Data Collection, Structure Determination and Refinement of Myotis VP35 IID
Initial conditions for crystallization of Myotis lucifigus residues 158–281 containing E246R and G250R mutations were identified using commercially available screens (Anatrace). BatVP35 IID RR crystals grew in 0.1 M HEPES pH 7.5 with 25% (w/v) PEG 3350. Diffraction data was collected at the Advanced Photon Source, Argonne National Laboratory (IMCA-CAT 17-ID beamline; Argonne, IL) at 100 K. Phases were determined using molecular replacement with the native wild-type structure of eVP35 IID (PDB: 3L25 molecule A) and using MOLREP or PHASER (Read, 2001; Vagin and Teplyakov, 1997). The model was further refined using REFMAC interspaced with manual building using Coot (Emsley and Cowtan, 2004; Murshudovet al., 1997). Validation of the structure was performed using MOLPROBITY (Chen et al., 2010). Figures were prepared using PyMOL (Delano, 2002).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was performed using GraphPad Prism 7 with significance determined either by a one-way ANOVA followed by Tukey’s test or by an unpaired t test. All statistical details can be found in the figure legends and data points were considered significantly different if the p value was < 0.05.
DATA AND SOFTWARE AVAILABILITY
Generation of the ancestral Myotis VP35 sequence and analysis of Myotis VP35 phylogenetics and selective pressures used publicly available resources including: SeaView v4.6 (http://doua.prabi.fr/software/seaview), Muscle (http://www.drive5.com/muscle/), IQ-Tree v1.6 (http://www.iqtree.org), PartitionFinderv1.1 (http://www.robertlanfear.com/partitionfinder/), DataMonkey (classic version; https://www.datamonkey.org), FigTree 1.4.3 (https://bioweb.pasteur.fr/packages/pack@FigTree@1.4.3https://bioweb.pasteur.fr/packages/pack@FigTree@1.4.3) and PhyloBot v10.09.2016.1 (http://www.phylobot.com). Structural determination and refinement of batVP35 IID used publicly available resources including; MOLREP (http://www.ccp4.ac.uk/dist/html/molrep.html), REFMAC (https://www2.mrc-lmb.cam.ac.uk/groups/murshudov/content/refmac/SourceEtal/source.html), COOT (https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/) and MOLPROBITY (https://molprobity.biochem.duke.edu.)
Analysis of SEC-MALS data used ASTRA 6 (https://www.wyatt.com/products/software/astra.html). GraphPad Prism 7 v7.0c (GraphPad) is an available for purchase graphing and statistical analysis software suite. Final structure figures were prepared using the for purchase PyMOL (Schrodinger). The accession number for the crystal structure of batVP35 IID reported in this paper is PDB: 6DKU. The accession numbers for the nucleotide sequences for the Myotis VP35s reported in this paper are GenBank: MH431024, MH431025, MH431026, MH431027, MH431028, MH431029, MH431030, MH431031, MH431032, MH431033, MH431034, MH431035 and MH431036.
Supplementary Material
Highlights.
Myotis VP35s modestly inhibit interferon responses
Myotis VP35s lack dsRNA binding but target a step upstream of RIG-I activation
Myotis VP35 C terminus shares striking structural homology to filoviral VP35s
Positive and negative selection acted on the amino acid sequence of Myotis VP35s
ACKNOWLEDGMENTS
This work was supported by NIH grant AI109945 (to C.F.B., G.K.A., and D.W.L.) and by Department ofthe Defense, DefenseThreat Reduction Agency grants HDTRA1–17-1–0005 and HDTRA1–16-1–0033 (to C.F.B. and G.K.A.). C.F.B. is a Georgia Research Alliance Eminent Scholar in Microbial Pathogenesis. This research used resources at the Industrial Macromolecular Crystallography Association Collaborative Access Team (IMCA-CAT) beamline 17- ID, supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Hauptman-Woodward Medical Research Institute. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by the Argonne National Laboratory under contract DE-AC02–06CH11357. We also thank the staff at SBC/Sector 19 at theArgonne National Laboratory for beamline access. The content ofthe information does not necessarily reflect the position orthe policy ofthe federal government, and no official endorsement should be inferred. We thankAlesha Grant and Gayathri Vijayakumar for assistance with preliminary studies and Sampriti De for technical support.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes three figures and four tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.06.045.
DECLARATION OF INTERESTS
C.F.B. has an adjunct faculty appointment at the Icahn School of Medicine at Mount Sinai. The authors declare no competing interests.
REFERENCES
- Albarirño CG, Wiggleton Guerrero L, Spengler JR, Uebelhoer LS, Chak- rabarti AK, Nichol ST, and Towner JS (2015). Recombinant Marburg viruses containing mutations in the IID region of VP35 prevent inhibition of Host immune responses. Virology 476, 85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale S, Julien JP, Bornholdt ZA, Kimberlin CR, Halfmann P, Zando- natti MA, Kunert J, Kroon GJ, Kawaoka Y, MacRae IJ, et al. (2012). Marburg virus VP35 can both fully coat the backbone and cap the ends of dsRNAfor interferon antagonism. PLoS Pathog. 8, e1002916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale S, Julien JP, Bornholdt ZA, Krois AS, Wilson IA, and Saphire EO (2013). Ebolavirus VP35 coats the backbone of double-stranded RNA for interferon antagonism. J. Virol. 87, 10385–10388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler CF, Wang X, Mühlberger E, Volchkov V, Paragas J, Klenk HD, García-Sastre A, and Palese P (2000). The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. USA 97, 12289–12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler CF, Mikulasova A, Martinez-Sobrido L, Paragas J, Mühlberger E, Bray M, Klenk HD, Palese P, and García-Sastre A (2003). The Ebolavirus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77, 7945–7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker S, Rinne C, Hofsäss U, Klenk HD, and Mühlberger E (1998). Interactions of Marburg virus nucleocapsid proteins. Virology 249, 406–417. [DOI] [PubMed] [Google Scholar]
- Belyi VA, Levine AJ, and Skalka AM (2010). Unexpected inheritance: multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes. PLoS Pathog. 6, e1001030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosio CM, Aman MJ, Grogan C, Hogan R, Ruthel G, Negley D, Moha- madzadeh M, Bavari S, and Schmaljohn A (2003). Ebola and Marburg viruses replicate in monocyte-derived dendritic cells without inducing the production of cytokines and full maturation. J. Infect. Dis. 188, 1630–1638. [DOI] [PubMed] [Google Scholar]
- Bruhn JF, Kirchdoerfer RN, Urata SM, Li S, Tickle IJ, Bricogne G, and Saphire EO (2017). Crystal structure of the Marburg virus VP35 oligomerization domain. J. Virol. 91, e01085–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas WB, Loo YM, Gale M Jr., Hartman AL, Kimberlin CR, Mar- tinez-Sobrido L, Saphire EO, and Basler CF (2006). EbolavirusVP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J. Virol. 80, 5168–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TH, Kubota T, Matsuoka M, Jones S, Bradfute SB, Bray M, and Ozato K (2009). Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery. PLoS Pathog. 5, e1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crochu S, Cook S, Attoui H, Charrel RN, De Chesse R, Belhouchet M, Lemasson JJ, de Micco P, and de Lamballerie X (2004). Sequences of fla- vivirus-related RNA viruses persist in DNA form integrated in the genome of Aedes spp. mosquitoes. J. Gen. Virol. 85, 1971–1980. [DOI] [PubMed] [Google Scholar]
- Delano WL (2002). The PyMOL Molecular Graphics System (Delano Scientific). [Google Scholar]
- Delport W, Poon AF, Frost SD, and Kosakovsky Pond SL (2010). Data- monkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26, 2455–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilley KA, Voorhies AA, Luthra P, Puri V, Stockwell TB, Lorenzi H, Basler CF, and Shabman RS (2017). The Ebola virus VP35 protein binds viral immunostimulatory and host RNAs identified through deep sequencing. PLoS ONE 12, e0178717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MR, Johnson B, Mire CE, Xu W, Shabman RS, Speller LN, Leung DW, Geisbert TW, Amarasinghe GK, and Basler CF (2014). The Marburg virusVP24 protein interactswith Keap1 toactivatethecytoprotective antioxidant response pathway. Cell Rep. 6, 1017–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MR, Pietzsch C, Vausselin T, Shaw ML, Bukreyev A, and Basler CF (2015). High-throughput minigenome system for identifying small- molecule inhibitors of Ebola virus replication. ACS Infect. Dis. 1, 380–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MR, Liu G, Mire CE, Sureshchandra S, Luthra P, Yen B, Shabman RS, Leung DW, Messaoudi I, Geisbert TW, et al. (2016). Differential regulation of interferon responses by Ebola and Marburg virus VP35 proteins. Cell Rep. 14, 1632–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Fabozzi G, Nabel CS, Dolan MA, and Sullivan NJ (2011). Ebolavirus proteins suppresstheeffects ofsmall interfering RNA bydirect interaction with the mammalian RNA interference pathway. J. Virol. 85, 2512–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feagins AR, and Basler CF (2015). Lloviu virus VP24 and VP35 proteins function as innate immune antagonists in human and bat cells. Virology 485, 145–152. [DOI] [PubMed] [Google Scholar]
- Feng Z, Cerveny M, Yan Z, and He B (2007). The VP35 protein of Ebola virus inhibitstheantiviral effect mediated by double-stranded RNA-dependent protein kinase PKR. J. Virol. 81, 182–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino K, Horie M, Honda T, Merriman DK, and Tomonaga K (2014). Inhibition of Borna disease virus replication by an endogenous bornavirus-like element in the ground squirrel genome. Proc. Natl. Acad. Sci. USA 111, 13175–13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, and Gascuel O (2010). SeaViewversion 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224. [DOI] [PubMed] [Google Scholar]
- Haasnoot J, de Vries W, Geutjes EJ, Prins M, de Haan P, and Berkhout B (2007). The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 3, e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson-Smith V, and Johnson A (2016). PhyloBot: a web portal for automated phylogenetics, ancestral sequence reconstruction, and exploration of mutational trajectories. PLoS Comput. Biol. 12, e1004976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman AL, Dover JE, Towner JS, and Nichol ST (2006). Reverse genetic generation of recombinant Zaire Ebola viruses containing disrupted IRF-3 inhibitory domains results in attenuated virus growth in vitro and higher levels of IRF-3 activation without inhibiting viral transcription or replication. J. Virol. 80, 6430–6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman AL, Bird BH, Towner JS, Antoniadou ZA, Zaki SR, and Nichol ST (2008). Inhibition of IRF-3 activation by VP35 is critical for the high level of virulence of Ebola virus. J. Virol. 82, 2699–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, and Ghosh S (2012). NF-kB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 26, 203–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Korytar T, Zhu Y, Pikula J, Bandouchova H, Zukal J, and Kollner B (2014). Establishment of Myotis myotis cell lines—model for investigation of host-pathogen interaction in a natural hostforemerging viruses. PLoS ONE 9, e109795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie M, Honda T, Suzuki Y, Kobayashi Y, Daito T, Oshida T, Ikuta K, Jern P, Gojobori T, Coffin JM, and Tomonaga K (2010). Endogenous non-retroviral RNAviruselements in mammalian genomes. Nature 463,84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv LB, and Donlin LT (2014). Regulation of type I interferon responses. Nat. Rev. Immunol. 14, 36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamura T, Yoneyama M, Koizumi N, Okabe Y, Namiki H, Samuel CE, and Fujita T (2001). PACT, a double-stranded RNA binding protein acts as a positive regulator for type I interferon gene induced by Newcastle disease virus. Biochem. Biophys. Res. Commun. 282, 515–523. [DOI] [PubMed] [Google Scholar]
- Jin H, Yan Z, Prabhakar BS, Feng Z, Ma Y, Verpooten D, Ganesh B, and He B (2010). The VP35 protein of Ebolavirus impairs dendritic cell maturation induced by virus and lipopolysaccharide. J. Gen. Virol. 91, 352–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok KH, Lui PY, Ng MH, Siu KL, Au SW, and Jin DY (2011). The double-stranded RNA-binding protein PACT functions as a cellular activator of RIG-I to facilitate innate antiviral response. Cell Host Microbe 9, 299–309. [DOI] [PubMed] [Google Scholar]
- Kondoh T, Manzoor R, Nao N, Maruyama J, Furuyama W, Miyamoto H, Shigeno A, Kuroda M, Matsuno K, Fujikura D, et al. (2017). Putative endogenous filovirus VP35-like protein potentiallyfunctions as an IFN antagonist but not a polymerase cofactor. PLoS ONE 12, e0186450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T, Matsuoka M, Chang TH, Bray M, Jones S, Tashiro M, Kato A, and Ozato K (2009). Ebolavirus VP35 interacts with the cytoplasmic dynein light chain 8. J. Virol. 83, 6952–6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SY, and Guindon S (2012). Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. [DOI] [PubMed] [Google Scholar]
- Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, Delicat A, Paweska JT, Gonzalez JP, and Swanepoel R (2005). Fruit bats as reservoirs of Ebola virus. Nature 438, 575–576. [DOI] [PubMed] [Google Scholar]
- Leung DW, Shabman RS, Farahbakhsh M, Prins KC, Borek DM, Wang T, Mühlberger E, Basler CF, and Amarasinghe GK (2010). Structural and functional characterization of Reston Ebola virus VP35 interferon inhibitory domain. J. Mol. Biol. 399, 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DW, Borek D, Luthra P, Binning JM, Anantpadma M, Liu G, Harvey IB, Su Z, Endlich-Frazier A, Pan J, et al. (2015). An intrinsically disordered peptide from Ebola virus VP35 controls viral RNA synthesis by modulating nucleoprotein-RNA interactions. Cell Rep. 11, 376–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Dong S, Li G, Wang W, Liu X, Wang Y, Yang C, Rao Z, and Guo Y (2017). Structural insight into nucleoprotein conformation change chaperoned by VP35 peptide in Marburg virus. J. Virol. 91, e00825–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubaki NM, Younan P, Santos RI, Meyer M, Iampietro M, Koup RA, and Bukreyev A (2016). The Ebola interferon inhibiting domains attenuate and dysregulate cell-mediated immune responses. PLoS Pathog. 12, e1006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra P, Ramanan P, Mire CE, Weisend C, Tsuda Y, Yen B, Liu G, Leung DW, Geisbert TW, Ebihara H, et al. (2013). Mutual antagonism between the Ebola virus VP35 protein and the RIG-I activator PACT determines infection outcome. Cell Host Microbe 14, 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra P, Jordan DS, Leung DW, Amarasinghe GK, and Basler CF (2015). Ebola virus VP35 interaction with dynein LC8 regulates viral RNA synthesis. J. Virol. 89, 5148–5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, et al. (2000). Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403, 785–789. [DOI] [PubMed] [Google Scholar]
- Möller P, Pariente N, Klenk HD, and Becker S (2005). Homo-oligomerization of Marburgvirus VP35 is essential for its function in replication and transcription. J. Virol. 79, 14876–14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlberger E, Lötfering B, Klenk HD, and Becker S (1998). Three of the four nucleocapsid proteins of Marburg virus, NP, VP35, and L, are sufficient to mediate replication and transcription of Marburg virus-specific monocistronic minigenomes. J. Virol. 72, 8756–8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlberger E, Weik M, Volchkov VE, Klenk HD, and Becker S (1999). Comparison of the transcription and replication strategies of marburg virus and Ebola virus by using artificial replication systems. J. Virol. 73, 2333–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, and Dodson EJ (1997). Refinement ofmacro- molecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, and Minh BQ (2015). IQ- TREE: afast and effective stochastic algorithm forestimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins KC, Cárdenas WB, and Basler CF (2009). Ebola virus protein VP35 impairs the function of interferon regulatory factor-activating kinases IKKepsi- lon and TBK-1.J. Virol. 83, 3069–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins KC, Binning JM, Shabman RS, Leung DW, Amarasinghe GK, and Basler CF (2010a). Basic residues within the ebolavirus VP35 protein are required for its viral polymerase cofactor function. J. Virol. 84, 1058110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins KC, Delpeut S, Leung DW, Reynard O, Volchkova VA, Reid SP, Ramanan P, Cárdenas WB, Amarasinghe GK, Volchkov VE, and Basler CF (2010b). Mutations abrogating VP35 interaction with double-stranded RNA render Ebola virus avirulent in guinea pigs. J. Virol. 84, 3004–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan P, Edwards MR, Shabman RS, Leung DW, Endlich-Frazier AC, Borek DM, Otwinowski Z, Liu G, Huh J, Basler CF, and Amarasinghe GK (2012). Structural basisforMarburg virusVP35-mediated immune evasion mechanisms. Proc. Natl. Acad. Sci. USA 109, 20661–20666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A (2012). FigTree v1. 4. Molecular Evolution, Phylogenetics and Epidemiology (University of Edinburgh, Institute of Evolutionary Biology; ). [Google Scholar]
- Read RJ (2001). Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D Biol. Crystallogr. 57, 1373–1382. [DOI] [PubMed] [Google Scholar]
- Reid SP, Cárdenas WB, and Basler CF (2005). Homo-oligomerization facilitates the interferon-antagonist activity of the ebolavirus VP35 protein. Virology 341, 179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruedi M, Stadelmann B, Gager Y, Douzery EJ, Francis CM, Lin LK, Guillén-Servent A, and Cibois A (2013). Molecular phylogenetic reconstructions identify East Asia as the cradle for the evolution of the cosmopolitan genus Myotis (Mammalia, Chiroptera). Mol. Phylogenet. Evol. 69, 437–449. [DOI] [PubMed] [Google Scholar]
- Schümann M, Gantke T, and Mühlberger E (2009). EbolavirusVP35 antag- onizesPKRactivitythrough its C-terminal interferon inhibitorydomain. J. Virol. 83, 8993–8997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanne E, and Sela I (2005). Occurrence of a DNA sequence of a non-retro RNA virus in a host plant genome and its expression: evidence for recombination between viral and host RNAs. Virology 332, 614–622. [DOI] [PubMed] [Google Scholar]
- Taylor DJ, and Bruenn J (2009). The evolution of novel fungal genes from non-retroviral RNA viruses. BMC Biol. 7, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DJ, Leach RW, and Bruenn J (2010). Filoviruses are ancient and integrated into mammalian genomes. BMC Evol. Biol. 10, 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DJ, Dittmar K, Ballinger MJ, and Bruenn JA (2011). Evolutionary maintenance of filovirus-like genes in bat genomes. BMC Evol. Biol. 11, 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriault S, Groseth A, Neumann G, Kawaoka Y, and Feldmann H (2004). Rescue of Ebolavirusfrom cDNA using heterologous support proteins. Virus Res. 106, 43–50. [DOI] [PubMed] [Google Scholar]
- Towner JS, Amman BR, Sealy TK, Carroll SA, Comer JA, Kemp A, Swanepoel R, Paddock CD, Balinandi S, Khristova ML, et al. (2009). Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 5, e1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifinopoulos J, Nguyen LT,von Haeseler A, and Minh BQ (2016). W-IQ- TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44 (W1), W232–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trunschke M, Conrad D, Enterlein S, Olejnik J, Brauburger K, and Muhlberger E (2013). The L-VP35 and L-L interaction domains reside in the amino terminus ofthe Ebolavirus L protein and are potential targets for antivirals. Virology 441, 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, and Teplyakov A (1997). MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025. [Google Scholar]
- Yen BC, and Basler CF (2016). Effects of filovirus interferon antagonists on responses of human monocyte-derived dendritic cells to RNA virus infection. J. Virol. 90,5108–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen B, Mulder LC, Martinez O, and Basler CF (2014). Molecular basis for ebolavirus VP35 suppression of human dendritic cell maturation. J. Virol. 88, 12500–12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, and Cullen BR (2003). Sequence requirements for micro RNA processing and function in human cells. RNA 9, 112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Cherukuri NC, Jackel JN, Wu Z, Crary M, Buckley KJ, Bisaro DM, and Parris DS (2012). Characterization ofthe RNA silencing suppression activity of the Ebola virus VP35 protein in plants and mammalian cells. J. Virol. 86, 3038–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Song H, Peng R, Shi Y, Qi J, and Gao GF (2017). Crystal structure of the Marburg virus nucleoprotein core domain chaperoned by a VP35 peptide reveals a conserved drug target for filovirus. J. Virol. 91, e00996–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Generation of the ancestral Myotis VP35 sequence and analysis of Myotis VP35 phylogenetics and selective pressures used publicly available resources including: SeaView v4.6 (http://doua.prabi.fr/software/seaview), Muscle (http://www.drive5.com/muscle/), IQ-Tree v1.6 (http://www.iqtree.org), PartitionFinderv1.1 (http://www.robertlanfear.com/partitionfinder/), DataMonkey (classic version; https://www.datamonkey.org), FigTree 1.4.3 (https://bioweb.pasteur.fr/packages/pack@FigTree@1.4.3https://bioweb.pasteur.fr/packages/pack@FigTree@1.4.3) and PhyloBot v10.09.2016.1 (http://www.phylobot.com). Structural determination and refinement of batVP35 IID used publicly available resources including; MOLREP (http://www.ccp4.ac.uk/dist/html/molrep.html), REFMAC (https://www2.mrc-lmb.cam.ac.uk/groups/murshudov/content/refmac/SourceEtal/source.html), COOT (https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/) and MOLPROBITY (https://molprobity.biochem.duke.edu.)
Analysis of SEC-MALS data used ASTRA 6 (https://www.wyatt.com/products/software/astra.html). GraphPad Prism 7 v7.0c (GraphPad) is an available for purchase graphing and statistical analysis software suite. Final structure figures were prepared using the for purchase PyMOL (Schrodinger). The accession number for the crystal structure of batVP35 IID reported in this paper is PDB: 6DKU. The accession numbers for the nucleotide sequences for the Myotis VP35s reported in this paper are GenBank: MH431024, MH431025, MH431026, MH431027, MH431028, MH431029, MH431030, MH431031, MH431032, MH431033, MH431034, MH431035 and MH431036.


