The authors evaluated tumor cell density using a transfer learning method that generates individualized patient models, grounded in the wealth of population data, while also detecting and adjusting for interpatient variabilities based on each patient's own histologic data. They collected 82 image-recorded biopsy samples, from 18 patients with primary GBM. With multivariate modeling, transfer learning improved performance (r = 0.88) compared with one-model-fits-all (r = 0.39). They conclude that transfer learning significantly improves predictive modeling performance for quantifying tumor cell density in glioblastoma.
Abstract
BACKGROUND AND PURPOSE:
MR imaging–based modeling of tumor cell density can substantially improve targeted treatment of glioblastoma. Unfortunately, interpatient variability limits the predictive ability of many modeling approaches. We present a transfer learning method that generates individualized patient models, grounded in the wealth of population data, while also detecting and adjusting for interpatient variabilities based on each patient's own histologic data.
MATERIALS AND METHODS:
We recruited patients with primary glioblastoma undergoing image-guided biopsies and preoperative imaging, including contrast-enhanced MR imaging, dynamic susceptibility contrast MR imaging, and diffusion tensor imaging. We calculated relative cerebral blood volume from DSC-MR imaging and mean diffusivity and fractional anisotropy from DTI. Following image coregistration, we assessed tumor cell density for each biopsy and identified corresponding localized MR imaging measurements. We then explored a range of univariate and multivariate predictive models of tumor cell density based on MR imaging measurements in a generalized one-model-fits-all approach. We then implemented both univariate and multivariate individualized transfer learning predictive models, which harness the available population-level data but allow individual variability in their predictions. Finally, we compared Pearson correlation coefficients and mean absolute error between the individualized transfer learning and generalized one-model-fits-all models.
RESULTS:
Tumor cell density significantly correlated with relative CBV (r = 0.33, P < .001), and T1-weighted postcontrast (r = 0.36, P < .001) on univariate analysis after correcting for multiple comparisons. With single-variable modeling (using relative CBV), transfer learning increased predictive performance (r = 0.53, mean absolute error = 15.19%) compared with one-model-fits-all (r = 0.27, mean absolute error = 17.79%). With multivariate modeling, transfer learning further improved performance (r = 0.88, mean absolute error = 5.66%) compared with one-model-fits-all (r = 0.39, mean absolute error = 16.55%).
CONCLUSIONS:
Transfer learning significantly improves predictive modeling performance for quantifying tumor cell density in glioblastoma.
Surgical debulking and radiation therapy represent first-line treatments for glioblastoma (GBM), which rely heavily on image guidance to delineate tumor from adjacent nontumoral brain. Contrast-enhanced MR imaging currently serves as the clinical standard for image guidance, but its diagnostic accuracy remains limited. Specifically, contrast-enhanced MR imaging localizes contrast-enhancing tumor for surgical resection and/or biopsy but poorly identifies nonenhancing invasive tumor in the surrounding T2-weighted/FLAIR abnormality.1–3 This invasive tumor segment can represent a substantial proportion of overall burden for many GBM tumors and contributes to recurrent disease and poor survival if left unresected.4–6 In addition to surgical guidance, contrast-enhanced MR imaging also fails to localize nonenhancing tumor during dosimetric radiation treatment planning. To compensate, most radiation oncologists must apply submaximal doses across the entire nonenhancing T2/FLAIR volume, which risks toxicity to normal brain and undertreatment of nondetected bulk tumor.7,8 These issues underscore the need to improve the image-based detection and targeted treatment of the nonenhancing tumor segment in GBM.
Advanced MR imaging techniques can help characterize nonenhancing tumor by measuring an array of biophysical features that complement contrast-enhanced MR imaging. These include tissue cell density (TCD) on diffusion-weighted imaging,2,9 white matter infiltration on diffusion tensor imaging,2,10,11 and microvessel morphology on dynamic susceptibility contrast perfusion MR imaging.2,12–14 Multiple studies have used image-guided biopsies to compare these advanced MR imaging features with TCD in a spatially accurate manner.3,5,9,11,15–22 These studies have revealed promising trends between MR imaging signal and tumor abundance but also surprising discrepancies in correlations among studies. For instance, some groups have reported that higher TCD correlates negatively with lower fractional anisotropy (FA) on DTI, presumably from greater white matter tract disruption.5,11 Yet, opposite (positive) correlations between FA and tumor content have also been reported.21 Similarly, published studies have differed on whether TCD correlates negatively or positively with mean diffusivity (MD) measures on DWI.9,11,16,19 These discrepancies present obvious challenges for developing generalized MR imaging–based models to prospectively quantify TCD and extent of invasion.
We hypothesized that the relationship between MR imaging signal (for any given contrast) and regional TCD demonstrates patient-to-patient variability within a given cohort. This may underlie the aforementioned discrepancies among published reports in the literature. In this study, we set out to quantify interpatient variability and determine whether it can strengthen the predictive accuracy of MR imaging–based models for quantifying TCD and invasion. Specifically, we have developed a transfer learning (TL) approach that quantifies concordance and variability of MR imaging–histologic relationships across patients. TL builds 1 model for each patient to account for potential interpatient variabilities in MR imaging–histologic relationships, while coupling the estimation processes of each patient-specific model to allow knowledge transfer between models. As proof of concept, we trained and cross-validated this TL approach in a cohort of patients with primary GBM using multiparametric MR imaging and spatially matched image-guided biopsies. Our overarching goal was to optimize predictive models that guide targeted treatment for the problematic nonenhancing tumor segment of GBM.
Materials and Methods
Acquisition and Processing of Clinical MR Imaging and Histologic Data
Patient Recruitment.
We recruited patients with clinically suspected primary GBM undergoing preoperative stereotactic MR imaging for first-line surgical resection before any treatment, as per our institutional review board protocol at Barrow Neurological Institute. All patients provided written and informed consent before enrollment. The patient cohort presented here has also been described in previous studies.3,6
Preoperative MR Imaging Acquisition Protocol.
We acquired preoperative 3T MR imaging (Sigma HDx; GE Healthcare, Milwaukee, Wisconsin) within 1 day of stereotactic surgery, including T2-weighted, T1-weighted precontrast, and T1-weighted postcontrast (T1 + C) sequences.3,6,14 T1 + C images were acquired after completion of dynamic susceptibility contrast perfusion MR imaging following a total Gd-DTPA dosage of 0.15 mmol/kg.3,6,14 In brief, for the DSC protocol, we administered an IV preload dose (0.1 mmol/kg of Gd-DTPA) to minimize T1-weighted leakage errors, after which we administered a second IV bolus injection (0.05 mmol/kg of Gd-DTPA) during the 3-minute DSC acquisition (gradient-echo echo-planar imaging: TR/TE/flip angle = 1500/20 ms/60°, matrix = 128 × 128, thickness = 5 mm).3,6,14 We derived postcontrast T2*WI (EPI+C) from the initial-source DSC volume.3,23 DTI acquisition consisted of spin-echo EPI (TR/TE = 10,000/85.2 ms, matrix = 256 × 256, FOV = 30 cm, thickness = 3 mm, 30 directions, array spatial sensitivity encoding technique, b=0, 1000).3,6
Surgical Biopsy.
Neurosurgeons used T1 + C and T2-weighted imaging to guide stereotactic biopsies following the smallest possible diameter craniotomies to minimize brain shift, as previously described.3,6 On average, we collected 5–6 tissue specimens from each tumor, selecting targets ≥1 cm apart from both T1 + C enhancing regions and nonenhancing T2-weighted hyperintense regions (so called brain-around-tumor) in pseudorandom fashion from different poles of the enhancing lesion periphery while avoiding centrally necrotic regions on the basis of the clinical feasibility as per clinical protocol. Enhancing and nonenhancing regions were distinguished on the basis of visual assessment by the neurosurgeon at the time of surgical biopsy and/or resection, as per clinical protocol. The target volume of each biopsy sample was approximately 125 mg. The neurosurgeons recorded biopsy locations via screen capture to allow subsequent coregistration with multiparametric MR imaging datasets. The neurosurgeon visually validated stereotactic imaging locations with corresponding intracranial anatomic landmarks, such as vascular structures and ventricle margins, before recording specimen locations.
Histologic Analysis and TCD Measurements from Image-Localized Biopsies.
Tissue specimens (volume = 125 mg) were flash frozen in liquid nitrogen in the operating suite and stored in a −80°C freezer until subsequent retrieval for embedding in an optimal cutting temperature compound and sectioning (thickness = 4 mm) in a −20C° cryostat (Microm-HM-550; Richard-Allan Scientific Company, Canton, Michigan) using a Microtome Blade (Thermo Scientific, Waltham, Massachusetts).3,6 Hematoxylin-eosin–stained slides were reviewed blinded to diagnosis by our neuropathologist (J.M.E.) to assess tumor content. Taking into account all visible cells (neurons, inflammatory cells, reactive glia, tumor cells, and so forth), we estimated the percentage of tumor nuclei (ie, tumor cells relative to all visible cells) (0%–100%), rounded to the nearest fifth percentile, and recorded it for each tissue sample as a ±5% range of TCD (eg, 30%–40%, 75%–85%, and so forth).3,6 This method served as a compromise between the resolution of TCD measurements and the precision of the neuropathologist's estimates.
Image Signal Normalization, DSC and DTI Analysis, Image Coregistration, and ROI analysis.
We normalized the signal for T1 + C, T2-weighted, and EPI+C image datasets using the Simple Insight Segmentation and Registration Toolkit (SimpleITK, Version 1.0.1; http://www.simpleitk.org/)24 in Python (Version 3.6.2; https://www.python.org/). The CurvatureFlow ITK algorithm was applied to remove image noise,25 and the N4ITK algorithm to correct for image-intensity nonuniformity bias that could be due to factors such as local magnetic field heterogeneity.26 Following these corrections, the CSF of the lateral ventricles was used as a reference tissue to normalize the intensity distributions of each dataset using a previously described linear scaling process.27 For DTI, we generated mean diffusivity and fractional anisotropy maps.5 For DSC, we generated relative cerebral blood volume (rCBV) maps using IB Neuro (Imaging Biometrics, Elm Grove, Wisconsin), using leakage correction28,29 and white matter normalization.3,6,14 We coregistered all images using ITK tools (https://itk.org/) and IB Suite (Imaging Biometrics), using the DTI B0 anatomic volume as the coregistration target.3,6,14 A board-certified neuroradiologist (L.S.H.) placed ROIs (8 × 8 voxels) for all coregistered multiparametric MR imaging datasets at the stereotactic biopsy locations.3,6,14 We selected this ROI size (0.93 × 0.93 cm) to encompass a broader area relative to the expected biopsy dimensions (125 mg). This selection helped to offset any potential minor shifts in biopsy localization (at the time of tissue sampling) that might otherwise result in misregistration. We measured mean signal values within ROIs for all MR imaging contrasts to correlate with spatially matched estimates of TCD from corresponding biopsies.
Statistical Analysis and Predictive Modeling
Univariate Statistical Analysis and Predictive Modeling.
To broadly survey potential associations between MR imaging signal and TCD, we performed univariate analysis using linear regression and Pearson correlations for each contrast (eg, T1 + C, rCBV, and so forth) against spatially matched biopsies across the entire patient cohort. We used the false discovery rate to adjust for multiple testing.30 To quantify intersubject variability, we also plotted MR imaging signal-versus-TCD in each patient separately.
Multivariate Statistical Analysis and Predictive Modeling Using the Conventional One-Model-Fits-All Approach.
To evaluate the potential complementary function of multiple combined MR imaging contrasts to characterize TCD, we performed multivariable linear regression to correlate MR imaging contrasts with histologic quantification of TCD across all biopsies and patients with GBM.16,22 This entailed the conventional approach in which a single static multivariable model was applied uniformly to all biopsy samples within the cohort (ie, one-model-fits-all [OMFA]). We evaluated a range of combined MR imaging contrasts to create various linear regression OMFA models. For instance, a model using signal intensities on rCBV, MD, and FA would be defined by the following: F(x) = ax1 + bx2 + cx3 + d, where the predicted TCD, F(x), was a linear function of signal intensity on rCBV, MD, and FA maps (x1, x2, and x3, respectively).
Transfer Learning Predictive Modeling.
Interpatient variability is a known limiting factor for many types of predictive models. For instance, a single model applied uniformly across all patients within a cohort (ie, one-model-fits-all) is unable to adjust for likely interpatient variabilities in MR imaging–histologic relationships. In contrast, individual models developed for each patient using only that patient's data (ie, one-model-each-patient) are unable to benefit from general population patterns and thus inherently have small sample sizes.
TL represents a compromise between the 2 aforementioned approaches. TL is a subfield of Machine Learning (ML), with various algorithms having been developed to allow knowledge transfer in jointly building different-but-inherently related models.31,32 We have previously published a TL algorithm under the Bayesian framework.33 With our TL algorithm, 1 model is built for each patient, but a Bayesian framework is used to bias the model parameter-estimation process toward the population pattern in the case in which the individual data are not sufficient to precisely determine the individual's variation. In essence, the population pattern represents a generalized model, and TL uses each patient's own MR imaging and biopsy data to modify that generalized model to more appropriately fit that particular individual patient. Thus, TL models can account for potential variabilities in MR imaging–histologic relationships across different patients, while also coupling the estimation processes of each patient-specific model to allow knowledge transfer between models. We detailed TL theory and methodology in the On-line Appendix and in our previous publication.33
Leave One-Out Cross-Validation.
To reduce overfitting, we used leave-one-out cross-validation (LOOCV) for all OMFA and TL model training. Briefly, all samples except 1 (randomly selected in each patient) were used to train the predictive model, while the excluded sample served as the test case to generate predicted TCD. We repeated this iteratively so that all 82 samples served as the test case.3,34 We then plotted model-predicted TCD against actual TCD for all biopsies to determine cross-validated Pearson correlation coefficients and mean absolute error rates (the difference between predicted versus actual TCD) for each model.
Results
Patients and Biopsy Samples
We collected 82 image-recorded biopsy samples (median = 4/patient, range = 2–14) from 18 patients with primary GBM (9:9 = female/male, median age = 60 years, range = 18–81 years). We collected ≥3 biopsies from 14/18 patients. In total, 33 biopsy samples originated from regions of nonenhancement (ie, brain-around-tumor), with 14 of 18 patients contributing at least 1 biopsy from nonenhancing regions. The remaining 49 biopsy samples originated from enhancing tumor regions.
OMFA Univariate and Multivariate Correlations between TCD and MR Imaging
We performed univariate analysis (Table 1) across all 82 samples comparing TCD with the 6 MR imaging features individually. We found significant correlations for T1 + C (r = 0.36, P < .001), rCBV (r = 0.33, P < .001), and FA (r = −0.24, P < .03), though only T1 + C and rCBV remained significant after correcting for multiple comparisons (P = .01). Our observed T1 + C correlation with TCD supports the long-held assumption that regions of MR imaging enhancement (and higher T1 + C signal) generally correspond with higher tumor content compared with peripherally nonenhancing regions (with lower T1 + C signal).35 This assumption underlies the clinical rationale for guiding surgical cytoreduction based on MR imaging enhancement. However, because nonenhancing regions, by definition, lack MR imaging enhancement, then T1 + C would presumably have much lower correlation with TCD within nonenhancing regions containing invasive tumor content. In fact, separate subgroup analysis (On-line Table) showed much lower correlation values between T1 + C and TCD when restricted to only nonenhancing biopsy samples. Meanwhile, the same subgroup analysis showed more consistent rCBV correlations across enhancing and nonenhancing biopsy subgroups, suggesting the potential utility of rCBV as a biomarker for both tumor segments. We also address any potential risks of overfitting by performing LOOCV to estimate the predictive performance of rCBV for quantifying TCD. Using the generalized one-model-fits-all approach, we generated a single variable linear regression model, with rCBV as the sole predictor input, and plotted predicted-versus-actual TCD (n = 82 samples). This generalized OMFA approach (based on rCBV alone) demonstrated poor performance, with a low correlation coefficient (r = 0.27) and high error (mean absolute error = 17.79%) (Table 2). We then evaluated other MR imaging features (combined with rCBV) using multivariable linear regression analysis. Regardless of the combination of MR imaging features (with rCBV), the generalized multivariate OMFA models failed to significantly improve performance (Table 2).
Table 1:
Univariate correlations for spatially matched MRI features vs TCD (n = 82 biopsies, 18 patients)
| MRI Feature | Pearson Correlation Coefficient (r) | P Value | FDR-Corrected P Value |
|---|---|---|---|
| T1 + Ca | +0.36 | <.001 | .01 |
| T2WI | +0.13 | .25 | .38 |
| rCBVa | +0.33 | <.001 | .01 |
| EPI + C | −0.02 | .85 | .85 |
| FA | −0.24 | .03 | .06 |
| MD | +0.03 | .78 | .85 |
Note:—FDR indicates false discovery rate; EPI + C, post-contrast T2*WI.
Statistical significance after correcting for multiple comparisons.
Table 2:
OMFA vs transfer learning for various MRI model predictorsa
| Predictors for Model | OMFA, LOOCV (r), and MAE | TL, LOOCV (r), and MAE | ||
|---|---|---|---|---|
| rCBV | 0.27 | 17.79 | 0.53 | 15.19 |
| rCBV, EPI + C | 0.25 | 18.03 | 0.63 | 11.65 |
| rCBV, FA | 0.34 | 17.24 | 0.58 | 11.31 |
| rCBV, MD | 0.28 | 17.74 | 0.60 | 11.93 |
| rCBV, T1 + C | 0.33 | 16.69 | 0.69 | 11.15 |
| rCBV, T2WI | 0.26 | 17.96 | 0.59 | 12.30 |
| rCBV, FA, MD | 0.32 | 17.47 | 0.66 | 11.93 |
| rCBV, T1 + C, T2WI | 0.35 | 16.61 | 0.75 | 9.03 |
| rCBV, T1 + C, FA | 0.39 | 16.55 | 0.73 | 9.07 |
| rCBV, T1 + C, MD | 0.35 | 16.77 | 0.74 | 9.41 |
| rCBV, T2WI, FA | 0.32 | 17.47 | 0.64 | 10.94 |
| rCBV, T2WI, MD | 0.26 | 18.02 | 0.64 | 11.15 |
| rCBV, T1 + C, FA, MD | 0.37 | 16.78 | 0.85 | 6.73 |
| rCBV, T2WI, FA, MD | 0.30 | 17.68 | 0.69 | 10.95 |
| rCBV, T1 + C, T2WI, FA | 0.37 | 16.79 | 0.73 | 9.41 |
| rCBV, T1 + C, T2WI, MD | 0.34 | 16.88 | 0.78 | 7.01 |
| rCBV, T1 + C, T2WI, FA, MD | 0.35 | 17.05 | 0.88 | 5.66 |
| rCBV, T1 + C, T2WI, FA, MD, EPI + C | 0.34 | 17.17 | 0.86 | 6.09 |
Note:—MAE indicates mean absolute error.
OMFA models were generated on the basis of linear regression analysis. Both (r) and MAE were determined using LOOCV to plot model-predicted TCD against actual TCD from spatially matched biopsies (n = 82).
Individualized Patient Plots and Interpatient Variability
To investigate interpatient variability, we separately plotted MR imaging signal versus TCD for each of the 14 (of 18) patients who contributed ≥3 separate biopsies (which allowed for LOOCV, as described in the Materials and Methods section above). The other 4 contributed <3 biopsies, which was insufficient for LOOCV analysis. Patient-by-patient plots are also shown in On-line Figs 1–6. Of the MR imaging correlations with TCD, rCBV demonstrated the greatest consistency across patients, with 13/14 patients (92.5%) having a positive correlation (r > 0.00), though these varied in strength from patient to patient (range of r = 0.07 to 0.95) (On-line Fig 1). Conversely, FA correlations showed greater variability (r = −0.75 to +0.78), with only 57.1% (8/14) of patients having negative correlations with TCD (versus 42.9% with positive correlations) (On-line Fig 2). MD correlations were also highly variable (range, r = −0.96 to +0.78), with 50% of patients split between negative and positive correlations (On-line Fig 3). These data suggest that while certain MR imaging features may be highly correlated with tumor content in a particular patient (or subset of patients), other patients may exhibit weaker or even opposite correlations that mask the overall effect in group analysis. We recognize that some of these individual plots may have small sample sizes that limit the statistical confidence of the correlation coefficients. Therefore, the individual plot coefficient values themselves should be viewed with this limitation in mind. At the same time, the directionality of the individual correlation plots (ie, positive-versus-negative) appears to show distinct population trends, and we intend to use the results here to illustrate some of the interpatient variabilities that may exist between TCD and imaging measurements. These interpatient variabilities motivate the use of the transfer learning approach detailed in the next section.
Transfer Learning
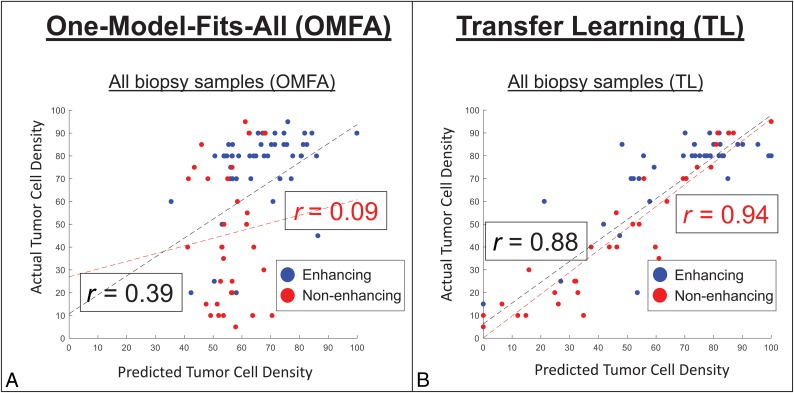
On the basis of the results from univariate analysis and individual scatterplots, we prioritized rCBV for training of TL and constrained knowledge transfer from patients with an arbitrary threshold for correlation between rCBV and TCD (r ≥ 0.10). By means of LOOCV, the TL model with rCBV as the sole model predictor improved the correlation between predicted-versus-actual TCD (r = 0.53, P < .001, n = 82), compared with the generalized OMFA model, which also used rCBV as the sole predictor (r = 0.27 using LOOCV) (Table 2). We then quantified incremental gains from adding other MR imaging contrasts to the rCBV-based TL model. As shown in Table 2, individualized transfer learning models consistently improved correlation coefficients and mean error rates compared with corresponding generalized OMFA models. Figure 1 shows the scatterplots for actual-versus-predicted TCD using the highest performing TL model, which incorporated rCBV, T1 + C, FA, and MD as model predictors. After LOOCV, this model achieved a Pearson correlation of r = 0.88 (P < .001) across all samples (n = 82), which further increased among nonenhancing T2/FLAIR samples alone (r = 0.94, P < .001, n = 33). By comparison, the generalized OMFA approach, using the same model predictors, achieved much lower correlation coefficients for all samples (r = 0.39, n = 82) and nonenhancing samples alone (r = 0.09, n = 33). As shown in Fig 2, the TL model can be used to generate color overlay maps of predicted TCD that correspond with actual TCD from spatially matched biopsies throughout different nonenhancing regions in the same GBM tumor.
Fig 1.
Transfer learning improves predictive performance for quantifying TCD. Scatterplots show the correlations between actual (y-axis) versus-model-predicted (x-axis) tumor cell densities using multivariable modeling (rCBV, T1 + C, MD, and FA) and the generalized (A) OMFA versus the individualized (B) TL approaches. Blue and red dots denote biopsy samples from enhancing (n = 49) and nonenhancing (n = 33) regions, respectively. Regression lines and Pearson correlations (r) are shown for all 82 samples (black lines and text) versus only the 33 nonenhancing samples (red lines and text). Correlations using TL are significantly higher compared with the generalized OMFA approach.
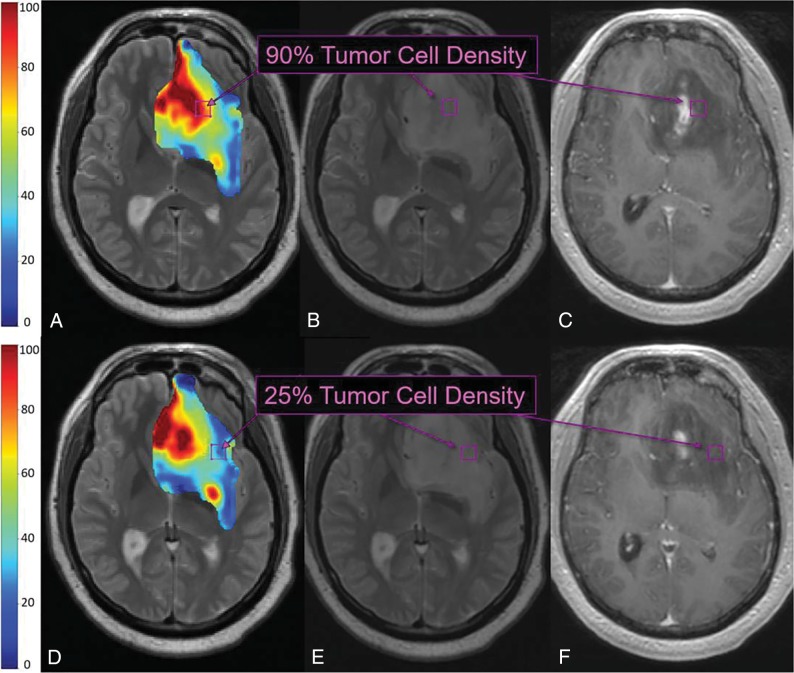
Fig 2.
TL maps and multiple biopsies in a 71-year-old patient with primary GBM. TL-based color map overlay on a T2-weighted image (A) shows predicted regional TCD ranging from 0% to 100% (blue to red) throughout the segmented tumor region (based on the T2-weighted signal abnormality). Histologic analysis of the top biopsy (purple boxes B and C) yielded 90% TCD, corresponding to red regions of high TL-predicted TCD (purple box, A). TL-based color map (D) from a more caudal T2-weighted slice shows blue regions of low TL-predicted TCD (blue/green, purple box), corresponding to 25% TCD on histologic analysis of the bottom biopsy (purple boxes, E and F).
Discussion
In this study, we correlated a panel of MR imaging features with regional tumor cell density using image-guided biopsies and spatially matched MR imaging measurements. In particular, the advanced MR imaging features (ie, rCBV, MD, FA) have been widely published for their potential to characterize TCD within the nonenhancing tumor segment of GBM. On univariate analysis, we found that rCBV was the only advanced MR imaging feature to significantly correlate with TCD after correcting for multiple comparisons (P = .01) (besides the conventional MR imaging feature T1 + C). This low-moderate positive correlation (r = 0.33, P < .001) remains concordant with other published studies on rCBV,18–20 which have all consistently reported positive correlations with TCD. In contrast, our analysis showed no significant correlation for MD. The correlation for FA (r = −0.24) showed a P value of .06 after correcting for multiple comparisons, which trends toward significance. These results are not entirely discordant with the literature because past studies have lacked general consensus on how MD and FA correlate with TCD. For instance, Stadlbauer et al11 and Price et al5 reported negative correlations between FA and TCD, while Beppu et al21 reported positive correlations. Similarly, Ellingson et al9 and Chang et al16 reported negative correlations between MD and TCD, while Stadlbauer et al11 reported positive correlations.
We hypothesized that the correlations observed in our study (as well as in the literature) may be impacted by interpatient variability in how MR imaging signal relates to TCD. To gain insight to this variability, we analyzed individualized plots of MR imaging signal versus tumor content within our cohort. In keeping with trends in the literature, rCBV correlations were highly consistent with regard to direction, with 92.5% of patients showing positive correlations. We did, however, observe a wide range of correlation strengths (range, r = +0.07 to +0.95), which explains the low-moderate group correlation. In contrast to rCBV, we observed highly variable correlations for both FA (range, r = −0.75 to +0.78) and MD (range, r = −0.96 to +0.78), with conflicting directions across patients. These discrepancies echo the discordance among studies in the literature and underscore the challenge of using a one-model-fits-all approach to generalize heterogeneity across patients.
TL addresses this challenge by estimating models for each patient by iteratively identifying patient similarity and leveraging it for robust model building using a Bayesian framework.33 Our results strongly suggest that TL should prioritize rCBV, given its high interpatient consistency and statistically significant correlations in univariate analysis. This assertion is supported further by the consistency of reported rCBV correlations in the literature.18–20 The application of TL to an rCBV-based model predictor significantly improved correlation (r = 0.53) compared with the one-model-fits-all approach (r = 0.27). TL further improved performance when combining rCBV with other MR imaging features in a multivariate fashion (r = 0.88), particularly for the nonenhancing T2/FLAIR samples (r = 0.94). This represents a substantial increase in performance compared with the conventional one-model-fits-all approach in our study (r = 0.30) (Table 2) and those previously published.16,22
Of note, the TL method presented here requires MR imaging and histologic input from at least 2 image-localized biopsies (per patient) to make predictive inferences for the remaining “unknown” (ie, unbiopsied) regions throughout each patient's tumor. Our analysis shows that these patient-specific inputs dramatically increase predictive performance for quantifying regional TCD on an individualized basis. While retrospective in nature, these results offer proof of concept that TL can help prospectively guide surgical biopsy and/or resection in a patient-specific manner. As part of our intraoperative workflow, we are currently integrating real-time neuropathologists' estimates of TCD (adding ∼2–3 minutes of analysis per sample) from frozen surgical specimens to support prospective TL-based neurosurgical guidance. These real-time estimates allow active updating of the TL model intraoperatively, accomplished in <5 minutes given standard computing hardware, to guide surgical targeting for the remaining unresected tumor regions. Future prospective studies in a larger cohort of patients will also allow us to evaluate how the tissue-sampling methodology (eg, number and distribution of biopsy locations) in individual patients might be optimized to improve predictive accuracy and model uncertainty. Additionally, individualized TL-based models can also guide dosimetric radiation planning in the postoperative setting. By delineating regional populations of residual nonenhancing tumor in the otherwise nonspecific T2/FLAIR segment, individualized TL-based maps will facilitate more nuanced radiation-planning strategies to prescribe higher doses with increasing TCD while sparing the dose to normal nontumoral brain.
We recognize several limitations to this study. First, like all other previously published reports, our study lacks a dedicated validation set. In fact, the conventional approach in the literature has been to report correlation coefficients from training data alone, which are prone to overfitting.3,5,9,11,15–22 By comparison, our study is the first to use more rigorous cross-validation testing (through LOOCV) to offset potential risks of overfitting, whereby each biopsy sample in our cohort is treated as an “unseen” or unknown case to test predictive performance.3 While cross-validation strengthens our confidence in these initial findings, prospective validation in future studies will ultimately be needed. Second, we have used neuropathologists' estimates of TCD as the benchmark measure of tumor content. We believe that this represents a more clinically relevant metric compared with total cell density, which can comprise both tumoral and nontumoral components (eg, astrocytes, microglia).9,16
Nonetheless, we are currently pursuing automated quantification of cellular density in our histologic samples.16 This type of standardization will help facilitate TL implementation across different institutions in the future. Finally, we recognize that image distortions and brain shift following craniotomy could lead to misregistration errors. To compensate, neurosurgeons used small craniotomy sizes to minimize brain shift and also visually validated stereotactic image location with intracranial neuroanatomic landmarks to help correct for random brain shifts. Rigid-body coregistration of multiparametric imaging also helped reduce possible geometric distortions.2,3,12,18 Overall, our experience suggests that combined misregistration is approximately 1–2 mm from both brain shift and registration techniques, which is similar to that from previous studies using stereotactic needle biopsy.11 To help minimize the effects of these potential misregistration errors, we selected larger ROI sizes relative to the expected biopsy tissue volumes. Thus, any minor shifts in the recorded biopsy locations (at the time of tissue sampling) would, in all likelihood, remain colocalized to the specified ROI.
Conclusions
We present an MR imaging–based transfer learning approach that optimizes individualized models of TCD and extent for patients with GBM. These models show particularly high predictive performance for the nonenhancing infiltrative tumor segment that is problematic to the diagnosis and treatment of GBM. Particularly, relevant clinical applications include surgical guidance for the extent of resection and dosimetric radiation targeting of nonenhancing residual tumor during postoperative adjuvant care.
ABBREVIATIONS:
- FA
fractional anisotropy
- GBM
glioblastoma
- LOOCV
leave-one-out cross-validation
- MD
mean diffusivity
- OMFA
one-model-fits-all
- rCBV
relative CBV
- T1 + C
T1-weighted postcontrast
- TCD
tumor cell density
- TL
transfer learning
- EPI+C
post-contrast T2*WI
Footnotes
Disclosures: Leland S. Hu—RELATED: Grant: National Institutes of Health, Comments: National Institutes of Health/National Institute of Neurological Disorders and Stroke: R21-NS082609, National Institutes of Health/National Cancer Institute: U01-CA220378, R01-CA221938, P50-CA108961*; UNRELATED: Grants/Grants Pending: National Institutes of Health, Comments: National Institutes of Health/National Institute of Neurological Disorders and Stroke: R21NS082609, National Institutes of Health/National Cancer Institute: U01-CA220378*; Patents (Planned, Pending or Issued): patent application title: Methods for Using Machine Learning and Mechanistic Models for Cell Density Mapping of Glioblastoma with Multiparametric MRI, Patent Application No. 62/684,096, application type: Provisional, Country: USA, filing date: June 12, 2018, Mayo Clinic Case No. 2017–498. Hyunsoo Yoon—RELATED: Grant: National Institutes of Health U01.* Leslie C. Baxter—RELATED: Grant: several grants from the National Cancer Institute (National Institutes of Health).* Amylou C. Dueck—RELATED: Grant: National Institutes of Health, Comments: R21-NS082609 and U01-CA220378.* Peter Nakaji—UNRELATED: Consultancy: Carl Zeiss Meditec, Comments: microscope company that does tumor imaging and fluorescence work; Payment for Lectures Including Service on Speakers Bureaus: Carl Zeiss Meditec, Comments: microscope company that does tumor imaging and fluorescence work, for which I sometimes lecture; Patents (Planned, Pending or Issued): GT Medical Technologies, Comments: creates brachytherapy solutions for recurrent brain tumors, not related to current work. I was a founder and inventor; Stock/Stock Options: GT Medical Technologies, Comments: creates brachytherapy solutions for recurrent brain tumors, not related to current work; *Money paid to the individual (P.N.). Other: Stryker, SpiWay, Thieme. Yanzhe Xu—RELATED: Grant: National Institutes of Health U01.* Lujia Wang—RELATED: Grant: National Institutes of Health U01.* Andrea J. Hawkins-Daarud—RELATED: Grant: National Institutes of Health.* Pamela R. Jackson—RELATED: Grant: National Institutes of Health.* Jing Li—RELATED: Grant: R21-NS082609, U01-CA220378.* Teresa Wu—RELATED: Grant: R21-NS082609, U01-CA220378.* Chad Quarles—RELATED: Grant: National Institutes of Health.* Kristin R. Swanson—RELATED: Grant: National Institutes of Health, James S. McDonnell Foundation, Ivy Foundation, Arabidopsis Biological Resource Center.* Mithun G. Sattur—UNRELATED: Stock/Stock Options: MRI interventions. *Money paid to the institution.
This work was supported by R21-NS082609, R01-CA221938, U01-CA220378, P50-CA108961, R01-CA158079 of the National Cancer Institute; the Mayo Clinic Foundation; the James S. McDonnell Foundation; the Ivy Foundation; and the Arizona Biomedical Research Commission.
References
- 1. Sarkaria JN, Hu LS, Parney IF, et al. Is the blood-brain barrier really disrupted in all glioblastomas? A critical assessment of existing clinical data. Neuro Oncol 2018;20:184–91 10.1093/neuonc/nox175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barajas RF Jr, Hodgson JG, Chang JS, et al. Glioblastoma multiforme regional genetic and cellular expression patterns: influence on anatomic and physiologic MR imaging. Radiology 2010;254:564–76 10.1148/radiol.09090663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hu LS, Ning S, Eschbacher JM, et al. Multi-parametric MRI and texture analysis to visualize spatial histologic heterogeneity and tumor extent in glioblastoma. PLoS One 2015;10:e0141506 10.1371/journal.pone.0141506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baldock AL, Ahn S, Rockne R1, et al. Patient-specific metrics of invasiveness reveal significant prognostic benefit of resection in a predictable subset of gliomas. PLoS One 2014;9:e99057 10.1371/journal.pone.0099057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Price SJ, Jena R, Burnet NG, et al. Improved delineation of glioma margins and regions of infiltration with the use of diffusion tensor imaging: an image-guided biopsy study. AJNR Am J Neuroradiol 2006;27:1969–74 [PMC free article] [PubMed] [Google Scholar]
- 6. Hu LS, Ning S, Eschbacher JM, et al. Radiogenomics to characterize regional genetic heterogeneity in glioblastoma. Neuro Oncol 2017;19:128–37 10.1093/neuonc/now135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Corwin D, Holdsworth C, Rockne RC, et al. Toward patient-specific, biologically optimized radiation therapy plans for the treatment of glioblastoma. PLoS One 2013;8:e79115 10.1371/journal.pone.0079115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pafundi DH, Laack NN, Youland RS, et al. Biopsy validation of 18F-DOPA PET and biodistribution in gliomas for neurosurgical planning and radiotherapy target delineation: results of a prospective pilot study. Neuro Oncol 2013;15:1058–67 10.1093/neuonc/not002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ellingson BM, Malkin MG, Rand SD, et al. Validation of functional diffusion maps (fDMs) as a biomarker for human glioma cellularity. J Magn Reson Imaging 2010;31:538–48 10.1002/jmri.22068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mohsen LA, Shi V, Jena R., et al. Diffusion tensor invasive phenotypes can predict progression-free survival in glioblastomas. Br J Neurosurg 2013;27:436–41 10.3109/02688697.2013.771136 [DOI] [PubMed] [Google Scholar]
- 11. Stadlbauer A, Ganslandt O, Buslei R., et al. Gliomas: histopathologic evaluation of changes in directionality and magnitude of water diffusion at diffusion-tensor MR imaging. Radiology 2006;240:803–10 10.1148/radiol.2403050937 [DOI] [PubMed] [Google Scholar]
- 12. Hu LS, Eschbacher JM, Dueck AC, et al. Correlations between perfusion MR imaging cerebral blood volume, microvessel quantification, and clinical outcome using stereotactic analysis in recurrent high-grade glioma. AJNR Am J Neuroradiol 2012;33:69–76 10.3174/ajnr.A2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hu LS, Baxter LC, Smith KA, et al. Relative cerebral blood volume values to differentiate high-grade glioma recurrence from posttreatment radiation effect: direct correlation between image-guided tissue histopathology and localized dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging measurements. AJNR Am J Neuroradiol 2009;30:552–58 10.3174/ajnr.A1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hu LS, Eschbacher JM, Heiserman JE, et al. Reevaluating the imaging definition of tumor progression: perfusion MRI quantifies recurrent glioblastoma tumor fraction, pseudoprogression, and radiation necrosis to predict survival. Neuro Oncol 2012;14:919–30 10.1093/neuonc/nos112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. LaViolette PS, Mickevicius NJ, Cochran EJ, et al. Precise ex vivo histological validation of heightened cellularity and diffusion-restricted necrosis in regions of dark apparent diffusion coefficient in 7 cases of high-grade glioma. Neuro Oncol 2014;16:1599–1606 10.1093/neuonc/nou142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chang PD, Malone HR, Bowden SG, et al. A multiparametric model for mapping cellularity in glioblastoma using radiographically localized biopsies. AJNR Am J Neuroradiol 2017;38:890–98 10.3174/ajnr.A5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eidel O, Neumann JO, Burth S, et al. Automatic analysis of cellularity in glioblastoma and correlation with ADC using trajectory analysis and automatic nuclei counting. PLoS One 2016;11:e0160250 10.1371/journal.pone.0160250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barajas RF Jr, Phillips JJ, Parvataneni R, et al. Regional variation in histopathologic features of tumor specimens from treatment-naive glioblastoma correlates with anatomic and physiologic MR imaging. Neuro Oncol 2012;14:942–54 10.1093/neuonc/nos128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sadeghi N, D'Haene N, Decaestecker C, et al. Apparent diffusion coefficient and cerebral blood volume in brain gliomas: relation to tumor cell density and tumor microvessel density based on stereotactic biopsies. AJNR Am J Neuroradiol 2008;29:476–82 10.3174/ajnr.A0851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Price SJ, Green HA, Dean AF, et al. Correlation of MR relative cerebral blood volume measurements with cellular density and proliferation in high-grade gliomas: an image-guided biopsy study. AJNR Am J Neuroradiol 2011;32:501–06 10.3174/ajnr.A2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Beppu T, Inoue T, Shibata Y, et al. Fractional anisotropy value by diffusion tensor magnetic resonance imaging as a predictor of cell density and proliferation activity of glioblastomas. Surg Neurol 2005;63:56–61; discussion 61 10.1016/j.surneu.2004.02.034 [DOI] [PubMed] [Google Scholar]
- 22. Durst CR, Raghavan P, Shaffrey ME, et al. Multimodal MR imaging model to predict tumor infiltration in patients with gliomas. Neuroradiology 2014;56:107–15 10.1007/s00234-013-1308-9 [DOI] [PubMed] [Google Scholar]
- 23. Semmineh NB, Xu J, Skinner JT, et al. Assessing tumor cytoarchitecture using multiecho DSC-MRI derived measures of the transverse relaxivity at tracer equilibrium (TRATE). Magn Reson Med 2015;74:772–84 10.1002/mrm.25435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lowekamp BC, Chen DT, Ibáñez L, et al. The design of simple eITK. Front Neuroinform 2013;7:45 10.3389/fninf.2013.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sethian JA. Level Set Methods and Fast Marching Methods: Evolving Interfaces in Computational Geometry, Fluid Mechanics, Computer Vision, and Materials Science. New York: Cambridge University Press, 1999 [Google Scholar]
- 26. Tustison NJ, Avants BB, Cook PA, et al. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 2010;29:1310–20 10.1109/TMI.2010.2046908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mitchell JR, Jones C, Karlik SJ, et al. MR multispectral analysis of multiple sclerosis lesions. J Magn Reson Imaging 1997; 7:499–511 10.1002/jmri.1880070309 [DOI] [PubMed] [Google Scholar]
- 28. Hu LS, Kelm Z, Korfiatis P, et al. Impact of software modeling on the accuracy of perfusion MRI in glioma. AJNR Am J Neuroradiol 2015;36:2242–49 10.3174/ajnr.A4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boxerman JL, Schmainda KM, Weisskoff RM. Relative cerebral blood volume maps corrected for contrast agent extravasation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. AJNR Am J Neuroradiol 2006;27:859–67 [PMC free article] [PubMed] [Google Scholar]
- 30. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 31. Weiss K, Khoshgoftaar TM, Wang D. A survey of transfer learning. J Big Data 2016;3:9 [Google Scholar]
- 32. Pan SJ, Yang Q. A survey on transfer learning. IEEE Trans Knowl Data Eng 2009;22:1345–59 [Google Scholar]
- 33. Zou N, Baydogan M, Zhu Y, et al. A transfer learning approach for predictive modeling of degenerate biological systems. Technometrics 2015;57:362–73 10.1080/00401706.2015.1044117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brown R, Zlatescu M, Sijben A, et al. The use of magnetic resonance imaging to noninvasively detect genetic signatures in oligodendroglioma. Clin Cancer Res 2008;14:2357–62 10.1158/1078-0432.CCR-07-1964 [DOI] [PubMed] [Google Scholar]
- 35. Dalrymple SJ, Parisi JE, Roche PC, et al. Changes in proliferating cell nuclear antigen expression in glioblastoma multiforme cells along a stereotactic biopsy trajectory. Neurosurgery 1994;35:1036–44; discussion 1044–55 10.1227/00006123-199412000-00004 [DOI] [PubMed] [Google Scholar]