Abstract
Cochlear implants (CIs) use electrode arrays that are surgically inserted into the cochlea to stimulate nerve endings to replace the natural electro-mechanical transduction mechanism and restore hearing for patients with profound hearing loss. Post-operatively, the CI needs to be programmed. Traditionally, this is done by an audiologist who is blind to the positions of the electrodes relative to the cochlea and relies on the patient’s subjective response to stimuli. This is a trial-and-error process that can be frustratingly long (dozens of programming sessions are not unusual). To assist audiologists, we have proposed what we call IGCIP for image-guided cochlear implant programming. In IGCIP, we use image processing algorithms to segment the intra-cochlear anatomy in pre-operative CT images and to localize the electrode arrays in post-operative CTs. We have shown that programming strategies informed by image-derived information significantly improve hearing outcomes for both adults and pediatric populations. We are now aiming at deploying these techniques clinically, which requires full automation. One challenge we face is the lack of standard image acquisition protocols. The content of the image volumes we need to process thus varies greatly and visual inspection and labelling is currently required to initialize processing pipelines. In this work we propose a deep learning-based approach to automatically detect if a head CT volume contains two ears, one ear, or no ear. Our approach has been tested on a data set that contains over 2,000 CT volumes from 153 patients and we achieve an overall 95.97% classification accuracy.
Keywords: Cochlear implant, image-guided cochlear implant programming, image classification, Convolutional Neural Networks
PURPOSE:
Cochlear implants (CIs) have been one of the most successful prosthetics in the past decades [1]. With a CI, an array of electrodes that is surgically inserted into the cochlea is used to stimulate auditory nerve endings, thus replacing the natural electro-mechanical transduction mechanism and restoring hearing for patients with profound hearing loss. Postoperatively, CIs need to be programmed to tune the implant for each recipient, e.g., to assign a frequency range to individual contacts such that they are activated when a frequency within that range is detected in the input signal and to adjust activation levels. In clinical practice, this is done by an audiologist who is blind to the positions of the electrodes relative to the cochlea and relies on the subjective patients’ response to stimuli, e.g., whether they can hear a signal or rank pitches. This is a trial-and-error process that has remained essentially unchanged since the mid-80s and can be frustratingly long (dozens of programming sessions are not unusual). In recent years, we have introduced what we call IGCIP for image-guided cochlear implant programming [2]. In IGCIP, we use image processing algorithms to segment the intra-cochlear anatomy in pre-operative CT images and to localize the electrode array in post-operative CTs [3–8]. Using this information, we have designed techniques to recommend CI processor settings to assist audiologists in programming the implants. We have shown that this leads to improvement in patient outcomes [9–11].
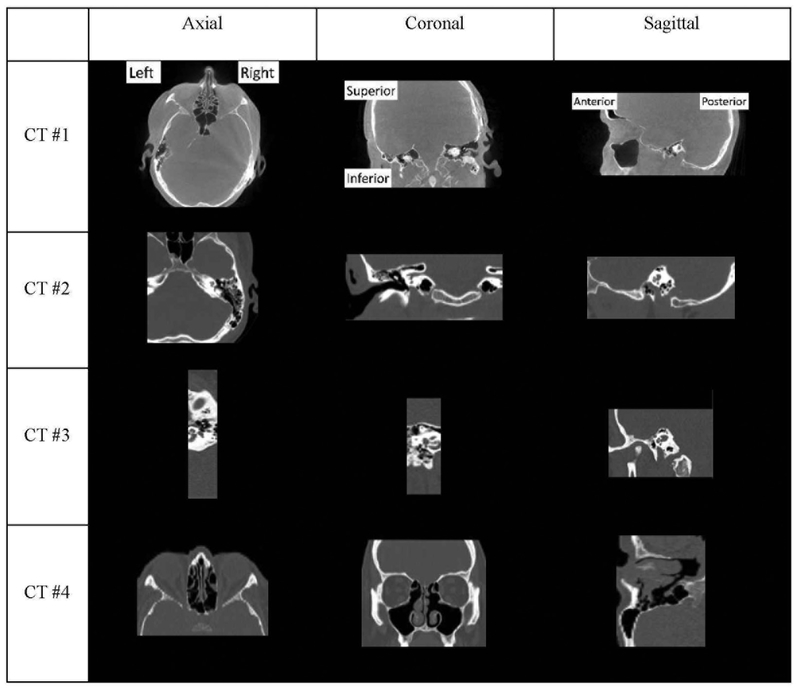
Our long term objective is to automate the series of image processing steps that support IGCIP to permit its clinical deployment. One barrier to full automation is the lack of standardized image acquisition protocols. This results in datasets which contain images that include very different portions of the head. They can include both inner ears, one inner ear (left or right), or sometimes neither. Figure 1 shows 4 examples to illustrate the range of images we need to be able to process. CT #1 includes the whole head so both ears are visible. In CT #2, although both the full right half and a fairly large portion of the left half of the head are included, only the right inner ear is visible. CT #3 includes only a very narrow portion of the head, but the whole right inner ear is visible. CT #4 includes only an anterior portion of the head and neither inner ear is visible.
Figure 1.
4 examples from our dataset. Orientations of the slices are labeled in CT #1 and apply to other examples as well.
In our current IGCIP process, when a new volume is received, it is visually inspected and assigned a label to document its content for proper processing in subsequent steps. In this work, we aim to replace this visual inspection step.
METHODS:
In recent years, convolutional neural networks (CNN or ConvNets) have been proposed as a solution for a wide range of problems such as image classification, object detection, semantic segmentation and other high-level computer vision tasks. The impressive performance they have achieved makes them the preferred solution for an increasing number of applications (see [12–15] for representative examples). CNN designed specifically for detection tasks include R-CNN [16], fast R-CNN [17], SPP-net [18], faster R-CNN [19] and YOLO [20]. These networks permit detecting the presence of a set of pre-defined objects and localizing each of them with a bounding box in 2D images. Extending these algorithms to 3D data sets requires substantially more resources in terms of hardware, model complexity, training data and training time but solutions to this problem have been proposed. Work that is particularly relevant to or own work is presented in [21, 22] in which authors use a 2D CNN to detect whether the anatomical structure of interest is present using slices extracted from axial, coronal and sagittal views of the 3D CT volumes. The algorithm is validated in three different CT datasets [22]. Similarly, Mamani et al. [23] used 2D multi-label convolutional neural networks for each orthogonal view of the thorax-abdomen CT scans for the detection of four human organs. In the work described herein, we propose to use axial slices of the head CTs to train a CNN to determine whether a new head CT volume includes one or both inner ears, to facilitate automating our IGCIP pipeline.
The dataset that we use in this study consists of 1,593 CT volumes obtained from 322 patients. We have more volumes than patients because it is common that multiple acquisitions are performed and that multiple reconstructions, and thus volumes, are produced from the same acquisition. For each patient, both the pre-operative CT and post-operative CT are included. The CTs are acquired on several scanners. The volumes in our dataset also include regions of very different sizes, ranging from 10 mm to 256 mm in the left-right and anterior-posterior dimensions, and from 52 mm to 195 mm in the superior-inferior dimension. The voxel dimensions range from 0.14 mm to 2 mm in left-right and anterior-posterior directions and from 0.14 mm to 2.5 mm in superior-inferior direction. We use CTs of half of the patients to train the CNN and to select parameters for 3D volume classification and CTs of the other half of the patients to do testing. Specifically the first half is split into a training set and a validation set, for training the CNN model and for optimizing parameters of volume-wise classification, respectively.
We first resample all CT volumes to isotropic 0.8 × 0.8 × 0.8 mm3 voxels using trilinear interpolation. All CT volumes are visually checked and are assigned to one of four categories: category 1, no ear; category 2, both ears; category 3, only the right ear; and category 4, only the left ear. As we have mentioned, we split the image volumes into (1) a training set, (2) a validation set and (3) a test set. The number of patients and number of CT volumes in each set are shown in the second and third rows of Table 1. Unfortunately, the data set we currently have at our disposal is very unbalanced in terms of the content. About 80% of the image volumes include both ears and about 20% include a single left or right ear. Image volumes which do not include any ear do exist but are very rare. If we build a machine learning system and search for the best parameters to maximize the overall accuracy using unbalanced training set and validation sets, the optimal setting will tend to classify all images into the majority class. To tackle this problem, we need to balance the number of samples in the four categories. To do so, we cropped the original CT volumes in the validation set to make more image volumes that include a single left ear, or right ear, or no ear and add them back to make the validation set have roughly equal numbers of volumes from each category. The same balancing operation is done for the test set. Since we use 2D slices to train the network, in the training set, we only need to make sure the number of slices that we sampled, instead of the number of CT volumes is the same for each of the four categories. No artificial data thus needs to be added to balance the training set. After adding the artificial CT volumes, the total numbers of CT volumes in each set are shown in the fourth row of Table 1. For each image volume in the training set, we manually localize the inner ears. This is done by selecting one point around the cochlea, as shown in Figure 2. As we have mentioned, the images are obtained with different protocols. This results in different intensity ranges. We normalize each image’s intensity to a uniform range, i.e., [0, 1].
Table 1.
Numbers of patients and CT volumes in each group
| Group | Training | Validation | Testing |
|---|---|---|---|
| # of patients | 120 | 49 | 153 |
| # of original CTs | 563 | 235 | 795 |
| # of CTs after adding artificial CTs | 563 | 732 | 2484 |
Figure 2.
The yellow marker is the landmark we use as the position of the left inner ear. From top to bottom, they are the axial, coronal and sagittal views, respectively.
At the current stage of the work, we assume that we know the orientation of the volume and we base our approach on axial images. To train the network, we use slices in the training volumes and we assign each slice to one of the four previously mentioned categories, i.e., category 1, no ear; category 2, both ears; category 3, only the right ear; and category 4, only the left ear. A slice is assigned to category 1 if either the CT volume it belongs to is in category 1, or if the CT volume belongs to categories 2–4 but the distances between the ears and the slice are larger than a threshold dt. Here, we empirically choose dt = 10 mm. A slice is assigned to category 2 if it comes from a CT volume in category 2 and the distances between its ears and the slice are less than dt. A slice is assigned to category 3 if it comes from a CT volume in category 3 and the distance between its ear’s distance and the slice is less than dt. Finally, a slice is assigned to category 4 if it comes from a volume in category 4 and the distance between its ear and the slice is less than dt. We augment the training volumes by applying reasonable translations, scaling and rotations to existing CT volumes and extract additional slices from them. By doing data augmentation, we have generated 100,000 slices from the training CT volumes to train the network. Because the size of the regions covered by the images varies from volume to volume, resampling to isotropic pixels leads to slices with different number of pixels, which cannot be accommodated by the network we use. To address this issue we symmetrically crop or pad the slices to make them 224 × 224 pixels which is the size of the network’s input layer.
In this work, we use the AlexNet [12] architecture that is pre-trained on the ImageNet data set. Figure 3 shows the architecture of this network (more details can be found in [12]). It has five convolutional layers and three fully-connected layers. At each convolutional layer, multiple filters are used for convolution with the input raw images or feature maps. The output feature maps are shown in the figure as stacked squares. The number of feature maps obtained after each layer is shown on the left of the feature maps. The size of the feature maps is shown on the right. Following convolution, max pooling is applied to reduce the dimensionality of the feature space. Finally, a non-linear activation function, here a rectified linear unit (ReLU) is applied to the feature maps. The following fully-connected layers are the same as layers used in traditional artificial neural networks. A Softmax function is applied to the output of the third fully-connected layer to generate probabilities which sum to 1. In the AlexNet architecture, the size of the output layer is 1000.
Figure 3.
Architecture of AlexNet
To adapt the architecture to our needs, we change the size of the output layer from 1000 units to 4 and we reinitialize the weights of the last fully-connected layer. Since the first layers of the pre-trained CNN are generic feature extractors, they do not need substantial update. We thus fine-tune the CNN by keeping the learning rate of the first 7 layers 1/10 of that of the last layer. We use the categorical cross entropy between ground truth labels and the output as the loss function and minimize it. The network is trained using stochastic gradient descent using a batch size of 256. We adopted the simplified learning rate adjustment strategy of the original AlexNet paper. The initial learning rate of the last layer is 0.01 and gets 10 times smaller after each 10,000 iterations. AlexNet is designed for RGB images. Here we tested two strategies for generating 3-channel inputs: (1) For each position, we simply use three copies of the same axial slice at this position, one per channel. (2) For each position, besides the slice at this position, we also extracted the slice that is above it and the slice below it to constitute the 3-channel input. By using neighboring slices, we are able to capture extra spatial information.
When using the trained network to label a new volume, we preprocess it the way we do for image volumes in the training set, i.e., we resample it, and crop or zero-pad it as required. Slices at each position are then input to the CNN to obtain the probabilities that it belongs to each of the four categories. Suppose the number of slices in the test volume is q, the output we produce is a q × 4 matrix [pn, pb, pr, pl]. Here, the four column vectors of dimensionality q, pn, pb, pr, and pl, represent the probabilities that the slices belong to the “no ear”, “both ears”, “right ear” and “left ear” categories, respectively. Each row in the matrix represents the probabilities of the corresponding slice in the volume. Figure 4 shows four representative examples. The images show coronal views of four CT volumes. The two four-curve groups on the right show pn, pb, pr, and pl, respectively. The group on the left is produced by the model using input generating strategy (1) and the group on the right is produced by the model using input generating strategy (2). The x−axis is the probability. The y−axis is the slice number. The images and the plots have been aligned to help relating the content of the image and the curves. In the example shown in (a), the image volume covers the right ear. The probability curves show that for those slices close to the inner ear, the “right ear” probability is nearly 1. For other slices, the “no ear” probability is nearly 1. The CT volume in (b) covers the whole head. The probability curves show that for those slices close to the inner ear, the “both ears” probability is nearly 1. For other slices, the “no ear” probability is nearly 1. (c) & (d) show two examples in which the probability curves are not as neat as those in (a) & (b). The CT volume in (c) contains only the left ear. The probabilities of the slices being “left ear” are higher but the values are not close to 1 and the number of consecutive slices having high “left ear” probabilities are fewer compared to that in (a) & (b). We can see a similar phenomenon in (d), in which the CT contains only the right ear. This could be due to the visually noticeable image noise in (c) and imaging artifact in (d). However, in both (c) and (d), the overall responses at the ground-truth channel produced by the model using input generating strategy (2) are stronger than those produced by the model using input generating strategy (1). This could be attributed to the incorporation of the extra spatial information.
Figure 4.
Four examples, (a)-(d). For each example, the image on the left is a coronal slice of the CT. The two plots represent the probabilities of the slice series containing no ear, both ears, right ear and left ear, generated by models using strategy (1) and (2).
The last step in our approach is to assign each volume to a category based on the probability curves. A straightforward criterion, which we currently use, is to find the class c (c = l, r or b) such that there exist a threshold probability pt and k consecutive indices i, i+1,…, i+k−1, such that, . If there is no such c, we predict that the volume does not include any ear. If there are multiple cs, we choose the category copt for which the probability curve has the maximal average value. The performance of our algorithm depends on the value of k and pt and, to find the optimal values of them, we do a grid search in the validation set. The optimal values for k and pt are: k = 3 and pt = 0.56 for the model trained using input generating strategy (1) and k = 4 and pt = 0.63 for the model trained using input generating strategy (2).
RESULTS:
In the validation set, the classification error rates for models trained using strategy (1) and (2) are 4.64% and 3.83%, respectively. Table 2 & 3 are the results we have obtained with our validation set under the optimal k & pt settings, when the input generating strategy (1) and (2) are used, respectively. Similarly, Table 4 & 5 show the results we have obtained with our test set when input generating strategy (1) and (2) are used, respectively. In the test set, using input generating strategy (1), we have achieved an overall labelling accuracy of 94.28%. Using input generating strategy (2), we have achieved an overall labelling accuracy of 95.97%. The detection accuracy when using strategy (2) is thus slightly higher than that of using strategy (1).
Table 2.
Detection results in the validation set produced by the model using input generating strategy (1)
| Predicted: no ear | Predicted: both ears | Predicted: right ear | Predicted: left ear | |
|---|---|---|---|---|
| Actual: no ear | 171 | 1 | 4 | 7 |
| Actual: both ears | 0 | 183 | 0 | 0 |
| Actual: right ear | 6 | 2 | 167 | 8 |
| Actual: left ear | 2 | 2 | 2 | 177 |
Table 3.
Detection results in the validation set produced by the model using input generating strategy (2)
| Predicted: no ear | Predicted: both ears | Predicted: right ear | Predicted: left ear | |
|---|---|---|---|---|
| Actual: no ear | 177 | 1 | 2 | 3 |
| Actual: both ears | 0 | 183 | 0 | 0 |
| Actual: right ear | 6 | 1 | 176 | 0 |
| Actual: left ear | 3 | 11 | 1 | 168 |
Table 4.
Detection results in the test set produced by the model using input generating strategy (1)
| Predicted: no ear | Predicted: both ears | Predicted: right ear | Predicted: left ear | |
|---|---|---|---|---|
| Actual: no ear | 579 | 10 | 14 | 18 |
| Actual: both ears | 1 | 619 | 0 | 1 |
| Actual: right ear | 27 | 14 | 554 | 26 |
| Actual: left ear | 17 | 11 | 3 | 590 |
Table 5.
Detection results in the test set produced by the model using input generating strategy (2)
| Predicted: no ear | Predicted: both ears | Predicted: right ear | Predicted: left ear | |
|---|---|---|---|---|
| Actual: no ear | 596 | 9 | 5 | 11 |
| Actual: both ears | 2 | 617 | 0 | 2 |
| Actual: right ear | 12 | 13 | 595 | 1 |
| Actual: left ear | 14 | 28 | 3 | 576 |
CONCLUSIONS:
Automatic labelling of head CT images with CNNs appears achievable. So far we have tested our approach on 2484 image volumes and we have reached a very encouraging success rate. We have achieved higher accuracy when using three neighboring slices as input to the CNN, compared to that when we replicate a single slice twice. This improvement could be attributed to the consideration of extra spatial information in the additional dimension other than the two dimensions in a single 2D slice.
Since in this work, we only used 2D slices or a limited number of neighboring slices to train the CNN, the unique 3D nature of CTs is not exploited thoroughly. As our next step, to leverage such information, we plan to develop efficient 3D algorithms which take the whole volume as an input unit. Also, besides determining the presence or absence of inner ears, we plan to enable our algorithm to accurately localize them, which will further facilitate the following image processing steps in our IGCIP pipeline.
ACKNOWLEDGMENT:
This research has been supported by grants NIH R01 DC014037, R01 DC014462 from the National Institute of Deafness and Other Communications Disorders. The experiments are conducted under the support of Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University. The content is solely the responsibility of the authors and does not necessarily represent the official views of this institute.
REFERENCES
- [1].NIDCD Fact Sheet: Cochlear Implants, “National institute on deafness and other communication disorders,” NIH Publication No. 11–4798, https://www.nidcd.nih.gov/sites/default/files/Documents/health/hearing/FactSheetCochlearImplant.pdf (2011).
- [2].Noble JH, et al. “Image-guidance enables new methods for customizing cochlear implant stimulation strategies.” IEEE Transactions on Neural Systems and Rehabilitation Engineering 21 5 (2013): 820–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Noble JH and Dawant BM, “Automatic graph-based localization of cochlear implant electrodes in CT,” Med Image Comput Comput Assist Interv, vol. 9350, pp. 152–159, October 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Reda FA, McRackan TR, Labadie RF, Dawant BM, and Noble JH, “Automatic segmentation of intra-cochlear anatomy in post-implantation CT of unilateral cochlear implant recipients,” Med Image Anal, vol. 18, pp. 605–615, April 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Reda Fitsum A., et al. “An artifact-robust, shape library-based algorithm for automatic segmentation of inner ear anatomy in post-cochlear-implantation CT” Medical Imaging 2014: Image Processing. Vol. 9034 International Society for Optics and Photonics, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Noble JH, Labadie RF, Majdani O, and Dawant BM, “Automatic segmentation of intracochlear anatomy in conventional CT,” IEEE Trans Biomed Eng, 58(9), pp. 2625–2632, September 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhao Y, Dawant BM, Labadie RF, and Noble JH, “Automatic localization of cochlear implant electrodes in CT,” Med Image Comput Comput Assist Interv, vol. 17, pp. 331–338, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhao Y, Dawant BM, and Noble JH, “Automatic localization of cochlear implant electrodes in CTs with a limited intensity range”, Proc. SPIE 10133, Medical Imaging 2017: Image Processing, 101330T (February 24, 2017) [Google Scholar]
- [9].Noble JH, et al. “Clinical evaluation of an image-guided cochlear implant programming strategy.” Audiology and Neurotology 196 (2014): 400–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Noble JH, et al. “Initial results with image-guided cochlear implant programming in children.” Otology & neurotology: official publication of the American Otological Society, American Neurotology Society [and] European Academy of Otology and Neurotology 372 (2016): e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Labadie RF, Noble JH, Hedley-Williams AJ, Sunderhaus LW, Dawant BM, and Gifford RH, “Results of Postoperative, CT-based, Electrode Deactivation on Hearing in Prelingually Deafened Adult Cochlear Implant Recipients,” Otol Neurotol, vol. 37, pp. 137–45, February 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Krizhevsky A, Sutskever I, and Hinton GE. “Imagenet classification with deep convolutional neural networks.” Advances in neural information processing systems 2012. (pp. 1097–1105). [Google Scholar]
- [13].Simonyan K, and Zisserman A. “Very deep convolutional networks for large-scale image recognition.” arXiv preprint arXiv:1409.1556 (2014). [Google Scholar]
- [14].He K, et al. “Deep residual learning for image recognition.” Proceedings of the IEEE conference on computer vision and pattern recognition 2016. (pp. 770–778). [Google Scholar]
- [15].Gao H, et al. “Densely connected convolutional networks.” arXiv preprint arXiv:1608.06993 (2016). [Google Scholar]
- [16].Girshick R, et al. “Rich feature hierarchies for accurate object detection and semantic segmentation.” Proceedings of the IEEE conference on computer vision and pattern recognition 2014. (pp. 580–587) [Google Scholar]
- [17].<He j/>K., et al. “Spatial pyramid pooling in deep convolutional networks for visual recognition” European Conference on Computer Vision Springer, Cham, 2014. (pp. 346–361) [Google Scholar]
- [18].Girshick R. “Fast r-cnn.” arXiv preprint arXiv:1504.08083 (2015). [Google Scholar]
- [19].Ren S, et al. “Faster R-CNN: Towards real-time object detection with region proposal networks.” Advances in neural information processing systems 2015. (pp. 91–99). [Google Scholar]
- [20].Redmon J, et al. “You only look once: Unified, real-time object detection.” Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. (pp. 779–788). [Google Scholar]
- [21].Vos B. de, et al. “2D image classification for 3D anatomy localization: employing deep convolutional neural networks” Medical Imaging: Image Processing. Vol. 9784, International Society for Optics and Photonics, 2016. [Google Scholar]
- [22].de Vos B, et al. “ConvNet-Based Localization of Anatomical Structures in 3D Medical Images.” IEEE Transactions on Medical Imaging 36 7 (2017): 1470–1481. [DOI] [PubMed] [Google Scholar]
- [23].Mamani GEH, et al. “Organ detection in thorax abdomen CT using multi-label convolutional neural networks” SPIE Medical Imaging Vol 10134 International Society for Optics and Photonics, 2017. [Google Scholar]