Abstract
Background:
Occupational exposure to the most widely used diisocyanate, 4,4’-methylene diphenyl diisocyanate (MDI), is a cause of occupational asthma (OA). Early recognition of MDI exposure and sensitization is essential for the prevention of MDI-OA.
Objective:
Identify circulating microRNAs (miRs) as novel biomarkers for early detection of MDI exposure and prevention of MDI-OA.
Materials and methods:
Female BALB/c mice were exposed to one of three exposure regimens: dermal exposure to 1% MDI in acetone; nose-only exposure to 4580 ± 1497 μg/m3 MDI-aerosol for 60 minutes; or MDI dermal exposure/sensitization followed by MDI-aerosol inhalation challenge. Blood was collected and miRCURY™ miRs qPCR Profiling Service was used to profile circulating miRs from dermally exposed mice. Candidate miRs were identified and verified from mice exposed to three MDI-exposure regimens by TaqMan® miR assays.
Results:
Up/down-regulation patterns of circulating mmu-miRs-183-5p, -206-3p and -381-3p were identified and verified. Circulating mmu-miR-183-5p was upregulated whereas mmu-miRs-206-3p and -381-3p were downregulated in mice exposed via all three MDI exposure regimens.
Discussion and conclusion:
Upregulation of circulating miR-183-5p along with downregulation of circulating miRs-206-3p and -381-3p may serve as putative biomarkers of MDI exposure and may be considered as potential candidates for validation in exposed human worker populations.
Keywords: Occupational Asthma (OA); Diisocyanates (dNCOs); 4,4’-methylene diphenyl diisocyanate (MDI); Circulating microRNAs (miRs)
Introduction
Diisocyanates (dNCOs) are low molecular weight cross-linkers used in polyurethane production. Methylene diphenyl diisocyanate (MDI) is the most widely used dNCO globally (Allport et al., 2003), where it is utilized in spray foam insulation, truck bed liners, wood products and adhesives. MDI is a potent respiratory system and skin irritant/sensitizer and cause of allergic contact dermatitis (ACD) and occupational asthma (OA) (Bernstein et al., 1993, NIOSH, 1994a, NIOSH, 1994b, Redlich and Karol, 2002, Lofgren et al., 2003, NIOSH, 2004, Jan et al., 2008, Engfeldt et al., 2013). In an occupational setting, workers can be exposed and sensitized to MDI in liquid, vapor, or aerosol form or to MDI-coated particles such as wood dust (Woellner et al., 1997), which may potentially lead to the development of MDI-associated OA (MDI-OA). To limit MDI exposure, the Occupational Safety and Health Administration (OSHA) currently sets the permissible exposure limit (PEL) for MDI at 0.2 mg/m3 (0.02 ppm). In addition, the American Conference of Governmental Industrial Hygienists (ACGIH) has established a threshold limit value (TLV) of MDI at 0.005 ppm (0.05 mg/m3) (ACGIH, 1999). Urine test methods using hydrolyzed 4,4’-Diaminodiphenylmethane (MDA) as a surveillance marker for MDI exposure have been suggested (Schutze et al., 1995, Skarping and Dalene, 1995, Skarping et al., 1995). However, the half-life of MDI metabolites in urine has been determined to be between 59–82 hours (Skarping et al., 1995, Dalene et al., 1997); therefore, these assays may only provide information on a worker with short term MDI exposure, rather than a longer-term cumulative exposure which may be more relevant in terms of the development of MDI sensitization. Detection of immune sensitization to MDI and/or early recognition of MDI-OA may be effective in slowing the progression of the disease (Wang and Petsonk, 2004); however, it can be difficult to recognize MDI-OA due to the lack of sensitive and reliable diagnostic tests for MDI exposure and sensitization.
In the clinical setting, physicians diagnose MDI-OA using patient histories of MDI exposure, though this is predictive in only 30–46% of cases (Malo et al., 1991), or through an MDI specific inhalation challenge (SIC) which is expensive and poses possible health risks to the patient. The discovery of novel biomarkers for the early detection of MDI exposure and sensitization in workers is needed for early intervention to prevent subsequent MDI-OA. Significant effort has been focused on developing MDI-specific antibody-based tests for determination of MDI exposure and sensitization; however, these biomarkers are not sensitive enough to be used in a clinical setting (Keskinen et al., 1988, Cartier et al., 1989, Wass and Belin, 1989, Tee et al., 1998, Ott et al., 2007, Budnik et al., 2013). By using proteomic methods, Hur et. al. first identified that ferritin and transferrin levels in serum may serve as biomarkers for MDI-OA (Hur et al., 2008); however, the serum ferritin and transferrin levels failed to serve as biomarker for another closely-related diisocyanate, toluene 2,4-diisocyanate (TDI) (Sastre et al., 2010). In addition, Haenen S et. al. identified several candidate biomarkers such as hemopexin for TDI-OA using a mouse model; however, these biomarkers for TDI-OA were not validated in humans (Haenen et al., 2010, Haenen et al., 2012, Haenen et al., 2014). Other serological biomarkers such as matrix metalloproteinases-9 (MMP-9), interleukin-8 (IL-8), and vascular endothelial growth factor (VEGF) have also been suggested as biomarkers for TDI-OA (Kim et al., 2011); however, usefulness of these serum cytokines as markers for MDI-OA has yet to be demonstrated. Given the limited success of previous attempts to determine immunological, serological, and protein biomarkers for diagnosing MDI-OA, development of state-of-the art novel techniques and classes of biomarkers to identify early MDI exposure and sensitization is needed.
MicroRNAs (miRs) are single-stranded RNA molecules ranging from 19 to 24 nucleotides in length with the ability to regulate diverse cellular processes through post-transcriptional regulation of target gene expression. The functional role of miRs in disease pathogenesis is an emerging field of study, and miRs are being utilized for disease diagnosis, prognosis, and evaluation of treatment response (Mendell and Olson, 2012, Srinivasan et al., 2013). miRs may function by suppressing translation of target genes or by causing target messenger RNA (mRNA) degradation through imperfect binding to the 3’ untranslated region (UTR) (Bartel, 2009). In addition to binding to the 3’UTR, miRs may be capable of binding to 5’UTRs, exons of mRNA and even DNA elements (Lytle et al., 2007, Orom et al., 2008, Place et al., 2008, Fang and Rajewsky, 2011, Zhou and Rigoutsos, 2014). In some reports, miRs have been shown to upregulate target gene expression (Vasudevan et al., 2007, Orom et al., 2008, Cordes et al., 2009, Lin et al., 2011, Truesdell et al., 2012). The majority of miRs are found in the intracellular region; however, many miRs may be found in the extracellular environment, including serum, plasma and other biological fluids (Valadi et al., 2007, Chen et al., 2008, Weber et al., 2010, Arroyo et al., 2011). Extracellular miRs commonly bind with RNA-binding proteins (RBPs), high-density lipoprotein particles (HDL) or are enclosed within lipid vesicles (exosomes, microvesicles, etc.) in the extracellular environment (Arroyo et al., 2011, Turchinovich et al., 2011, Vickers et al., 2011). These extracellular miRs are reported to be relatively stable compared to other RNA species (Chen et al., 2008, Mitchell et al., 2008, Turchinovich et al., 2011). Given their relatively high stability, and the fact that their expression levels have been associated with multiple disease development processes, extracellular miRs are considered to be good molecular candidates for biomarkers of disease. Circulating miRs, a subset of these extracellular miRs, are emerging as a novel class of minimally invasive biomarkers in different diseases including cancers (Mitchell et al., 2008, Kosaka et al., 2010), cardiovascular diseases (Gupta et al., 2010, Tijsen et al., 2012, Xu et al., 2012), diabetes (Guay and Regazzi, 2013), and other diseases. Recent research on circulating miRs has mainly focused on their association with cancer and other diseases. Until recently, few reports focused on asthma-associated circulating miRs have been published (Wang et al., 2015, Kho et al., 2016, Panganiban et al., 2016, Davis et al., 2017, Milger et al., 2017). Currently, there is no published research on MDI exposure associated circulating miRs, and we hypothesize that circulating miRs can be identified and used for detection of MDI exposure.
This report is focused on characterizing the response of circulating miRs to MDI exposure using a murine model. MDI exposure was performed dermally, via nose-only inhalation, and via nose-only challenge following dermal exposure/sensitization. Up/down-regulation patterns of mmu-miRs-183-5p, 206-3p and -381-3p were found consistently across three experimental murine models suggesting their putative roles as novel biomarkers for MDI exposure.
Clinical significance:
MDI exposure is a cause of occupational asthma. Early detection of MDI exposure/sensitization and timely removal of workers from exposure to MDI are essential for prevention of MDI-OA disease progression. The upregulation of circulating miR-183-5p, and downregulation of circulating miRs-206-3p and -381-3p can potentially be used to detect MDI exposure and/or sensitization.
Materials and methods
Chemicals and Reagents
HPLC grade acetone, 3Å molecular sieve (4–8 mesh), and 98% 4,4’-methylene diphenyl diisocyanate were acquired from Sigma-Aldrich (St. Louis, MO). Fatal-Plus® (sodium pentobarbital) euthanasia solution was acquired from Vortech Pharmaceuticals, Ltd. (Dearborn, MI). Dry acetone was prepared by incubating 10 ml HPLC grade acetone on 3 Å molecular sieve for a minimum of 24 hours to adsorb water.
Animals
Female BALB/c mice, 6–8 week old, were purchased from Taconic (Germantown, New York). Mice were acclimated at least for 5 days before being randomly assigned into different treatment groups. Mice were housed in ventilated plastic cages with hardwood chip bedding at 5 animals per cage. Each animal cage was enriched with a section of polyvinyl chloride (PVC) pipe (1.5” O.D. × 6”) to acclimate animals to the nose-only restraint device. Further acclimation to the restraint device was performed by three stays of increasing duration (15 minutes, 30 minutes, 60 minutes) on three consecutive days in the restraint device. Device acclimation was well-tolerated. An NIH-31 modified 6% irradiated rodent diet (Harlan Teklad) and tap water were administered ad libitum. Housing facilities were maintained at 68–72 °F and 36–57% relative humidity with a 12-hour light-dark cycle. All animal experiments were performed in the AAALAC, International-accredited National Institute for Occupational Safety and Health animal facility in accordance with an institutionally-approved animal care and use protocol (Protocol # 16-JH-m-009).
MDI exposure
MDI dermal exposures were performed on groups of 5 mice by applying a single dose of 1% (w/v) MDI in dry acetone or dry acetone only (vehicle control; Ctl) on the dorsal surface of each ear (25 μl per ear) daily on 3 consecutive days. Mice were euthanized, ear thickness was measured using a caliper, ears and blood were collected 24 hours after final MDI dermal exposure. To verify candidate circulating miRs identified from miRCURY™ miRs qPCR Profiling Service, additional groups of 5 mice were dermally exposed to 1% MDI or vehicle Ctl. MDI aerosol exposures were performed on groups of 5 mice by exposing the animals, via a nose-only inhalation exposure system (NOIES) to MDI aerosol or pure house air (Control) for 1 hour as previously described (Hettick et al., 2018). For each 1 hour aerosol exposure, the total of 4580 ± 1497 μg/m3 of MDI concentrations were achieved and maintained. About half of the total MDI aerosol (2243 ± 903.8 μg/m3) generated during the 1 hour exposure were particles less than 3.0 μm in diameter. MDI dermal exposure/aerosol challenge was performed on groups of five mice by applying 25 μl 1% MDI/acetone (w/v) dose on the dorsal surface of each ear on days 1, 2, 3, 14, 15, and 16. The second dermal exposure on days 14, 15, and 16 served as a booster exposure to achieve a potential better sensitization response as previously described (Vanoirbeek et al., 2004, Selgrade et al., 2006). On day 21, the animals were nose-only exposed to MDI aerosol using the NOIES for 1 hour, similar to the aerosol-only group. For both MDI aerosol exposures and MDI dermal exposure/aerosol challenge groups, mice were euthanized at 4 hours and 24 hours after MDI aerosol exposure.
Euthanasia, serum, tissue collection, and processing
Animals were euthanized via intraperitoneal injection of sodium pentobarbital. Following a non-reflexive response to a toe pinch test, exsanguination was performed via cardiac puncture and blood was placed into serum collection tubes (Blood Collection Microtainer Tube with Serum Separator, Becton Dickinson, San Jose, CA), centrifuged, and serum was collected and stored at −80 °C for subsequent RNA analysis. Ears from MDI dermal exposure experiments were collected and stored at −80 °C for RNA isolation and subsequent gene expression analysis.
Tissue RNA isolation, reverse transcription, and Real-Time PCR
MDI or dry-acetone exposed ears were processed for total RNA isolation using a Tissue Lyser II (Qiagen, Hilden, Germany) in mirVana™ miR lysis/binding buffer (Thermo Fisher Scientific). Tissue total RNA was isolated using mirVana™ miR Isolation Kit (Thermo Fisher Scientific). The concentration and purity of the RNA was determined using a ND-1000 spectrophotometer (Thermo Fisher Scientific). 200 ng of total RNA was subjected to first strand cDNA synthesis using a High-Capacity cDNA Synthesis Kit (Thermo Fisher Scientific) according to manufacturer’s protocol on a Mastercycler Pro thermocycler (Eppendorf, Hauppauge, NY). For analysis of mRNA expression, TaqMan® Universal PCR Master Mix (Thermo Fisher Scientific), cDNA, and mouse-specific mRNA TaqMan® assays were combined and PCR was performed according to manufacturer’s protocol. Assays used in this study include: Il-1β (Mm00434228_m1), Il-2 (Mm00434256_m1), Il-4 (Mm00445259_m1), Il-5 (Mm00439646_m1), Il-6 (Mm00446190_m1), Il-13 (Mm00434204_m1), Tnf-α (Mm00443258_m1), Inf-γ (Mm01168134_m1), Fcer1a (FcεRI) (Mm00438867_m1) and B2m (Mm00437762_m1). Real-time PCR was performed on an ABI PRISM 7500 (Thermo Fisher Scientific) with the following cycling conditions: 95 °C for 10 minutes followed by 40 cycles of 95 °C for 15 seconds and 60 °C for 1 minute. B2m served as the endogenous reference control. The relative expression levels of mRNAs were calculated using the comparative ΔΔCt method as previously described (Lin et al., 2011, Sharma et al., 2014, Lin et al., 2015).
Circulating miRs qPCR array profiling assay
The MDI dermal exposure mouse serum was used to characterize candidate circulating miRs for MDI exposure biomarker identification. Serum samples were sent to Exiqon (Vedbaek, Denmark) for the miRCURY™ miRs qPCR Profiling Service to profile circulating miRs. A total of 310 miRs were detected in both control and MDI exposed samples by miRCURY™ miRs qPCR Profiling Service. The heat map for all 310 miRs were generated by the online analysis tool, CIMminer (https://discover.nci.nih.gov/cimminer/).
Verification of candidate circulating miRs
200 μL sera from independent, separate groups of MDI-dermal exposure, MDI-aerosol exposure, and MDI dermal exposure/MDI-aerosol challenge mice were used for total RNA isolation using mirVana™ PARIS™ Kit (Thermo Fisher Scientific, Waltham, MA) following the manufacturer’s protocol for liquid samples. The concentration and purity of the RNA were determined using an ND-1000 spectrophotometer (Thermo Fisher Scientific). To synthesize cDNA for miRs assays, 10 ng of serum total RNA from each treatment was subjected to reverse transcription (RT) using a TaqMan® miR Reverse Transcription Kit (Thermo Fisher Scientific) and specific TaqMan RT primers included in the miR assays following the manufacturer’s protocol. Specific TaqMan miR assays (Thermo Fisher Scientific) were acquired: mmu-miR-16-5p (Assay ID #000391), mmu-miR-30d-3p (#002305), mmu-miR-127-3p (#000452), mmu-miR-153-3p (#001191), mmu-miR-181a-1-3p (#000516), mmu-miR-183-5p (#002269), mmu-miR-192-3p (#002272), mmu-miR-206-3p (#000510), mmu-miR-381-3p (#000571), mmu-miR-433-3p (#001028), and mmu-miR-744-3p (#002325). RT was performed on a Mastercycler Pro thermocycler (Eppendorf, Hauppauge, NY) with the following cycling conditions: 16 °C for 30 minutes, 42 °C for 30 minutes, followed by a final step of 85 °C for 5 minutes to inactivate the reverse transcriptase. To generate enough miR cDNA template for following real-time PCR reaction, the cDNA was pre-amplified using specific miR TaqMan assays and a PreAmp Master Mix (Thermo Fisher Scientific) following the manufacturer’s instructions. The PreAmp primer pool consisted of 0.2× TaqMan primers specific for each of the candidate miRs. The pre-amplification cycling conditions were as follows: 95 °C for 10 min followed by 14 cycles of 95 °C for 15 seconds and 60 °C for 4 minutes.
After the pre-amplification step, the products were diluted 200-fold with RNase-free water and served as template for real-time PCR reaction. Real-time PCR was performed as described above. Circulating mmu-miR-16-5p was previously reported as being constantly expressed in serum (Ng et al., 2009, Tanaka et al., 2009, Wang et al., 2009, Zhu et al., 2009) and was used as an endogenous control in this study. The relative expression levels of miRs were calculated using the comparative ΔΔCt method as described previously (Lin et al., 2011, Sharma et al., 2014, Lin et al., 2015).
Candidate circulating miRs in silico pathway analysis
Predicted targets of candidate circulating miRs were obtained from 7 in silico algorithms, including DIANA-microT (Maragkakis et al., 2009a, Maragkakis et al., 2009b), miRanda (Enright et al., 2003, Betel et al., 2008), mirBridge (Tsang et al., 2010), PicTar (Krek et al., 2005), PITA (Kertesz et al., 2007), rna22 (Miranda et al., 2006), and TargetScan (Lewis et al., 2005, Grimson et al., 2007, Friedman et al., 2009). Furthermore, two experimentally-validated databases were queried, TarBase (Papadopoulos et al., 2009) and miRecords (Xiao et al., 2009) using the web-based tool miRsystem (http://mirsystem.cgm.ntu.edu.tw/) (Lu et al., 2012). Pathway enrichment analysis of candidate miRs target genes was conducted using the miRsystem online tool with three pathway databases: KEGG (Kanehisa et al., 2008), BioCarta (Nishimura, 2001), and Reactome (Matthews et al., 2009).
Statistical analysis
Data were analyzed using either the unpaired t-test (two-tailed) in MDI-dermal exposure experiments (Figures 1&3) or one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison ad hoc post-test in MDI-aerosol exposure and MDI-dermal exposure/MDI-aerosol challenge experiments (Figures 4&5). Statistical analyses were performed in GraphPad Prism 7.0 (GraphPad Software, La Jolla, CA). The empirical P-values of each identified enriched pathway were determined by using the default settings of miRsystem (Lu et al., 2012). Differences were considered significant when the analysis yielded P<0.05.
Results
Examination of immunological responses, skin irritancy and sensitization potential of MDI dermal exposure
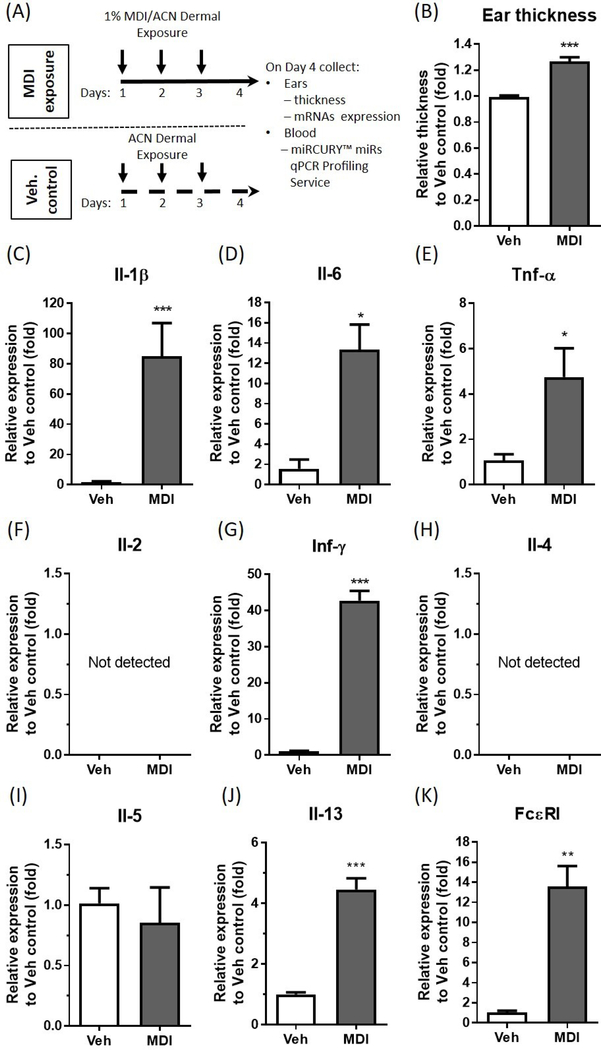
To confirm that exposure to MDI would result in possible inflammation, irritation and sensitization of the animals, 1% MDI (in dry acetone, w/v) was painted on each ear of randomly grouped BALB/c mice for 3 consecutive days followed by collection of ears and blood on day 4 (Figure 1A). Ears were assayed for swelling and were used for RNA isolation. Expressions of inflammatory cytokines, Th1 and 2 cytokines, and the high affinity IgE receptor were measured by qRT-PCR, (Figure 1B-K). This 1% MDI dosing regimen was selected because 1% TDI dermal exposure was proven sufficient to induce sensitization (Anderson et al., 2014). Furthermore, Selgrade et. al. reported that 1% MDI dermal exposure on the shaved back of BALB/c mice was sufficiently to induce serum IgE and Th2 cytokines (Selgrade et al., 2006). Average ear swelling was significantly increased four days following 1% MDI exposure (Figure 1B). Inflammatory cytokines (Il-1β, Il-6, and Tnf-α,) mRNA levels were significantly increased in the ear four days following 1% MDI exposure (Figure 1 C-E), indicating that dermal MDI exposure may cause an inflammation response. To examine whether this dermal exposure regimen may result in possible sensitization responses, we measured Th1 (Il-2 and Inf-γ), Th2 cytokines (Il-4, -5, and -13) and high affinity IgE receptor mRNA levels in the ear. Inf-γ mRNA level was significantly upregulated 42.7-fold (Figure 1G) whereas Il-13 mRNA level was significantly increased 4.46-fold (Figure 1J) in the ear four days following 1% MDI exposure compared to vehicle control. The expression of Th1 Il-2 mRNA (Figure 1F) and Th2 Il-4 mRNA (Figure 1H) were not detectable in either MDI or vehicle control exposed ears whereas the Th2 Il-5 mRNA was not changed (Figure 1I). Given that the expression level of the α subunit of the high-affinity IgE receptor (Fc epsilon RI; FcεRI) in human mast cells and basophil has been found to be directly correlated with serum IgE levels (Gomez et al., 2007), and the presence of MDI-specific IgE or increase of total IgE level indicates that there is systemic sensitization to MDI (Wisnewski et al., 2011, Pollaris et al., 2016), we used FcεR1 mRNA expression in the ear as a measure of possible sensitization. FcεR1 mRNA was significantly increased 13.56-fold four days following 3 consecutive days of 1% MDI exposure (Figure 1K). These data suggest that dermal exposure to 1% MDI induces significant immunological responses in the ear, and it may cause sensitization to MDI.
Figure 1. Examination of immunological responses, skin irritancy and sensitization potential of MDI dermal exposure.
Experimental timelines showing MDI exposed time points, sample collection and analyses (A). Relative ear thickness change as determined 4 days following 1% MDI/ACN exposure (B) (N=30; bars, s.e.m.). Ear mRNA expression of the inflammatory cytokines Il-1β (C), Il-6 (D), Tnf-α (E), Th1 cytokines Il-2 (F) and Inf-γ (G), Th2 cytokines Il-4 (H), Il-5 (I) and Il-13 (J) as well as high affinity IgE receptor, FcεRI (K) were determined 4 days following MDI exposure via RT-qPCR (N=3; bars, s.e.m.). MDI: 4,4’-methylene diphenyl diisocyanate; Veh: vehicle; ACN: acetone. (*P<0.05; **P<0.01; ***P<0.001).
Circulating miR profiling and verification in murine model
To identify potential circulating miRs as biomarkers for MDI dermal exposure, we used miRCURY™ miRs qPCR Profiling Service (Exiqon) to profile serum miR changes between dermal MDI-exposed and control mice. Of the total 752 miRs profiled, 310 miRs were detected in both MDI-exposed and control mice (Figure 2). To refine the list of candidate miRs, we identified the top 20 differentially expressed miRs between the two groups (Table 1). Of these 20 miRs, we constrained our analysis to the 10 miRs that have human homologs. We then independently verified the expression of these 10 miRs in-house using TaqMan® miR stem-loop qRT-PCR assays on another separate groups of mice dermally exposed to MDI and vehicle control (Figure 3A). Compared with serum RNA isolated from control mice, mmu-miR-183-5p was upregulated 82.1-fold (Figure 3B), whereas mmu-miR-206-3p and mmu-miR-381-3p were downregulated 1.59-fold (Figure 3C) and 6.31-fold (Figure 3D), respectively. Of the putative differentially expressed miRs identified in the miRCURY™ miRs qPCR Profiling Service, mmu-miRs-127, -192-3p, -181a-1-3p, -433-3p and -744-3p (Figure 3E-I) failed verification (by RT-PCR) between MD-Iexposed and control mice. Furthermore, mmu-miRs-30d and -153-3p were not detected in either MDI-exposed or control mice (Figure 3J and K). Moving forward, we considered only circulating mmu-miRs-183-5p, -206-3p and -381-3p as potential biomarkers for dermal MDI exposure.
Figure 2. Heat map shows circulating miRs detected in both dermal MDI-exposed and vehicle control exposed mice.
Serum were collected from mice dermally exposed to either 1% MDI in dry acetone (MDI) or vehicle control (CTL), and sent to Exiqon in Denmark for miRCURY™ miRs qPCR Profiling Service to profile miR expressions. Total 310 miRs were detected in both MDI and control mice. The color scale shown at the bottom illustrates the relative expression level of a miR across all samples: red color represents an expression level above mean, green color represents expression lower than the mean. (Please see Figure 2 online at (https://doi.org/10.1080/1354750X.2018.1508308) for the color version).
Table 1. Top 20 candidate circulating miRs identified by miRCURY™ miRs qPCR Profiling Service.
Microarray data shows differently expressed circulating miRs from dermal MDI exposure compare to vehicle control. The 20 most differentially expressed serum miR fold-changes are shown from MDI-dermal exposed mice compared to acetone (ACN) control (CTL). Microarray analysis for miR was performed with RNA extracted after 4 days following 1% MDI/ACN exposure to MDI or ACN skin painted mice.
| microRNAs | Mature microRNA sequences | Human homologues | Fold changes compare to CTL |
|---|---|---|---|
| mmu-miR-1983 | CUCACCUGGAGCAUGUUUUCU | N/A | −12 |
| mmu-miR-30d-3p | CUUUCAGUCAGAUGUUUGCUGC | hsa-miR-30d-3p | −11 |
| mmu-miR-381-3p | UAUACAAGGGCAAGCUCUCUGU | hsa-miR-381-3p | −8.5 |
| mmu-miR-667-3p | UGACACCUGCCACCCAGCCCAAG | N/A | −8 |
| mmu-miR-376b-3p | AUCAUAGAGGAACAUCCACUU | N/A | −7.8 |
| mmu-miR-744-3p | CUGUUGCCACUAACCUCAACCU | hsa-miR-744-3p | −7.6 |
| mmu-miR-206-3p | UGGAAUGUAAGGAAGUGUGUGG | hsa-miR-206 | −7.1 |
| mmu-miR-92b-3p | UAUUGCACUCGUCCCGGCCUCC | N/A | −6.3 |
| mmu-miR-672-5p | UGAGGUUGGUGUACUGUGUGUGA | N/A | −5.8 |
| mmu-miR-127-3p | UCGGAUCCGUCUGAGCUUGGCU | hsa-miR-127-3p | −5.4 |
| mmu-miR-181a-1-3p | ACCAUCGACCGUUGAUUGUACC | hsa-miR-213 | −4.6 |
| mmu-miR-300-3p | UAUGCAAGGGCAAGCUCUCUUC | N/A | −4.5 |
| mmu-miR-192-3p | CUGCCAAUUCCAUAGGUCACAG | hsa-miR-192-3p | 4.5 |
| mmu-miR-202-5p | UUCCUAUGCAUAUACUUCUUU | N/A | 4.5 |
| mmu-miR-201-5p | UACUCAGUAAGGCAUUGUUCUU | N/A | 4.6 |
| mmu-miR-153-3p | UUGCAUAGUCACAAAAGUGAUC | hsa-miR-153-3p | 4.8 |
| mmu-miR-351-5p | UCCCUGAGGAGCCCUUUGAGCCUG | N/A | 5.5 |
| mmu-miR-433-3p | AUCAUGAUGGGCUCCUCGGUGU | hsa-miR-433-3p | 9.3 |
| mmu-miR-183-5p | UAUGGCACUGGUAGAAUUCACU | hsa-miR-183-5p | 11 |
| mmu-miR-1195 | UGAGUUCGAGGCCAGCCUGCUCA | N/A | 13 |
Figure 3. Verification of candidate circulating miRs by stem-loop qRT-PCR on MDI dermal exposed mice.
Experimental timelines showing MDI exposed time points, sample collection and analysis in additional separate groups of mice (A). Serum total RNA were isolated by miRVana™ PARIS™ isolation kit, reverse transcribed, preamplified and subjected to TaqMan miR stem-loop qRT-PCR. Circulating miR expressions of candidate (B) mmu-miR-183-5p, (C) mmu-miR-206-3p, (D) mmu-miR-381-3p, (E) mmu-miR-127, (F) mmu-miR-192-3p, (G) mmu-miR-181a-1-3p, (H) mmu-miR-433-3p, (I) mmu-miR-744-3p, (J) mmu-miR-30d and (K) mmu-miR-153-3p changes were determined 4 days following 1% MDI/ACN exposure (N=3; bars, s.e.m). MDI: 4,4’-methylene diphenyl diisocyanate; Veh: Vehicle; ACN: acetone. (*P<0.05; **P<0.01).
Circulating miR validation in nose-only aerosol exposure murine model
Occupational exposure routes to MDI are thought to be mostly through inhalation and dermal contact (NIOSH, 2004). At room temperature, MDI is not volatile (vapor pressure is approximately at 10−5 mm Hg). However, during application, MDI is frequently heated and/or aerosolized via a spray gun, thus generating respirable vapor and/or aerosols. To mimic MDI occupational airway aerosol exposure, we have developed a nose-only aerosol exposure mouse model (Hettick et al., 2018). We further determined whether the circulating mmu-miRs-183-5p, -206-3p, and -381-3p expression changes in response to aerosol exposure, as it does for dermal exposure. Compared to mice exposed to house air only, mmu-miR-183-5p was upregulated 16.47-fold in serum collected at 4 hours (4h) post MDI aerosol exposure and 36.12-fold in serum collected at 24 hours (24h) after MDI aerosol exposure (Figure 4 A), whereas mmu-miR-206-3p was downregulated 10.17-fold at 4h and 3.29-fold at 24h post MDI exposure (Figure 4 B). In addition, serum mmu-miR-381-3p levels were downregulated 7.94-fold at 4 h and 3.65-fold at 24h after MDI-aerosol exposure compared to air only control (Figure 4C). These results suggest that circulating mmu-miRs-183-5p, -206-3p and -381-3p can also be used as putative biomarkers for detection of short term airway exposure to MDI.
Figure 4. Verification of candidate serum miRs by stem-loop qRT-PCR on nose-only MDI-aerosol exposed mice.
Serum total RNA was isolated using the miRVana™ PARIS™ isolation kit, reverse transcribed, preamplified and subjected to miR stem-loop qRT-PCR. Circulating miR expressions of candidate (A) mmu-miR-183-5p, (B) mmu-miR-206-3p, and (C) mmu-miR-381-3p were determined at 4 hours or 24 hours after 1 hour of nose-only MDI-aerosol exposure (N=3; bars, s.e.m). MDI: 4,4’-methylene diphenyl diisocyanate. (**P<0.01; ***P<0.001).
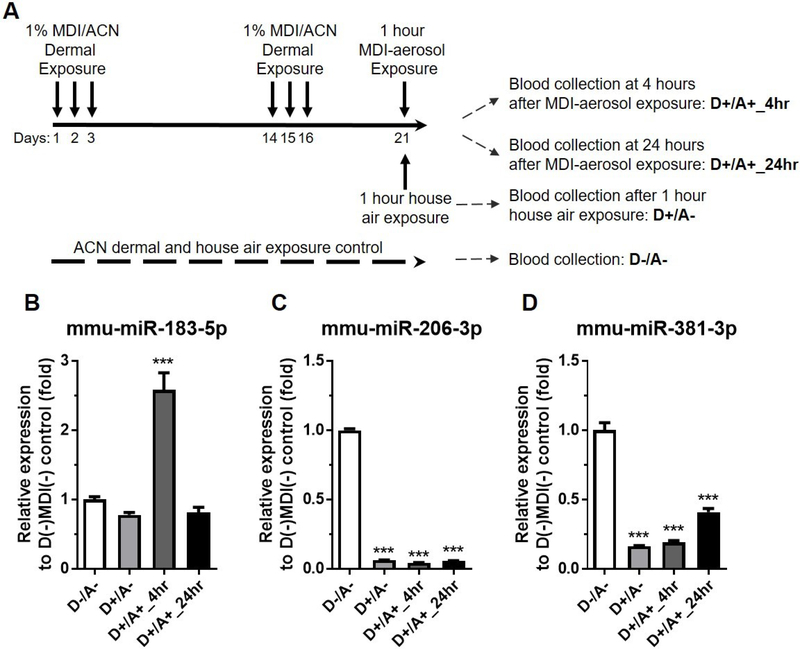
Dermal exposed MDI aerosol challenge murine model
MDI dermal exposure followed by MDI inhalation has been associated with MDI-OA attack (Rattray et al., 1994, Petsonk et al., 2000). To investigate whether candidate miRs can serve as MDI exposure biomarkers over a longer term (21 days), as well as in an industrially-relevant exposure model, we developed a MDI-dermal exposure followed by MDI-aerosol challenge murine model suggestive of OA exposure (Figure 5A). Examination of candidate miRs in the house-air exposed control mice (D−/A− vs. D+/A−) failed to demonstrate a change in mmu-miR-183-5p level (Figure 5B) but caused 15.6-fold and 6.15-fold downregulation of serum mmu-miRs-206-3p and -381-3p (Figure 5C&D), indicating that only downregulation of both circulating miRs-206-3p and -381-3p may serve as biomarker for long term repeated dermal MDI exposure. Circulating mmu-miR-183-5p was upregulated 2.58-fold at 4hr following aerosol challenge (D+/A+) but returned to control levels 24hr after compared to D−/A− control (Figure 5B). Circulating mmu-miR-206-3p level was downregulated 23.33-fold at 4hr and 18.47-fold 24hr after MDI aerosol exposure (D+/A+) compared to control (D−/A−) (Figure 5C). In addition, circulating mmu-miR381-3p levels were downregulated 5.22-fold at 4hr and 2.45-fold at 24hr after MDI exposure (D+/A+) compared to control (D−/A−) (Figure 5D). The non-responsiveness of circulating mmu-miR-183-5p from repeated MDI-dermal exposure (D−/A− vs. D+/A−, Figure 5B) may be due to desensitization of the upregulation of circulating mmu-miR-183-5p or the result of rapid clearance from circulation through an unknown mechanism(s). This putative mechanism(s) of non-responsiveness of circulating mmu-miR-183-5p from repeated dermal MDI exposure may be the cause of the comparatively small 2.58-fold upregulation at 4hr following MDI aerosol challenge (D+/A+_4hr, Figure 5B) versus a higher 16.47-fold upregulation at 4hr following MDI aerosol only exposure (Figure 4A). Similarly, this putative mechanism(s) may also be the cause for no observed upregulation of circulating mmu-miR-183-5p at 24hr following MDI aerosol challenge (D+/A+_24hr, Figure 5B). The rapid upregulation of circulating mmu-miR-183-5p after MDI dermal exposure only (Figure 3B) and MDI aerosol exposure only (Figure 4A) represents the early response for acute MDI exposure. Both circulating mmu-miRs-206-3p and -381-3p were downregulated in MDI dermal exposure (Figure 3C&D) and were rapidly downregulated in MDI aerosol exposure (see MDI-4h vs. Air; Figure 4B&C); therefore, both rapid upregulation of circulating mmu-miR-183-5p and rapid downregulation of circulating miRs-206-3p and -381-3p may serve as biomarkers for acute (~4hr post-exposure) MDI exposure. In addition, downregulation of circulating miRs-206-3p and -381-3p may serve as good biomarkers for detection of MDI aerosol exposure on longer time scales (24 hours +).
Figure 5. Determination of candidate circulating miRs in dermal exposed/sensitized followed by nose-only MDI-aerosol challenged murine model.
(A) Experimental timelines showing MDI exposed time points, routes. Expression levels of candidate circulating (B) mmu-miR-183-5p, (C) mmu-miR-206-3p, and (D) mmu-miR-381-3p were compared to control mice without dermal and airway MDI-aerosol exposure control (N=3; bars, s.e.m.). MDI: 4,4’-methylene diphenyl diisocyanate; ACN: acetone. (***P<0.001).
Candidate circulating miR targets and pathway analysis
To explore the biological mechanisms underlying the roles of identified circulating miRs in relation to MDI exposure and MDI-OA, we performed target identification of human homolog miRs, hsa-miR-183-5p, hsa-miR-206-3p, and hsa-miR-381-3p on miRsystem (Lu et al., 2012). In silico analysis of target prediction returned 446 target genes for hsa-miR-183-5p (Supplemental Table S1), 699 target genes for hsa-miR-206 (Supplemental Table S2), 622 target genes for hsa-miR-381-3p (Supplemental Table S3). 89 genes were co-targeted by both hsa-miRs-206 and 381-3p (Supplemental Table S4), and 1142 genes can be targeted by either hsa-miR-206 or hsa-miR-381-3p (Supplemental Table S5).
Pathway analysis against KEGG, Reactome, and BIOCARTA databases showed a total of 79 pathways are potentially impacted by hsa-miR-183-5p (Table 2). Twelve enriched pathways had ≥8 potential genes that potentially can be targeted by hsa-miR-183-5p and the majority of the pathways were related to human diseases, such as HIV-1 relative pathways, prion disease, Type I diabetes mellitus, Huntington’s disease, and long-term depression. Pathway analysis for potential co-targets of hsa-miR-206 and -381-3p revealed 38 pathways. Among those pathways, five of the pathways were related to the immune system, such as Fc gamma R-mediated phagocytosis, NFAT pathway, chemokine signaling pathway, FMLP pathway, and leukocyte transendothelial migration (Table 3). Fourteen of these pathways had ≥9 potential genes that could potentially be co-targetted by both hsa-miR-206, and -381-3p. The predicted co-targets also were involved in the signaling transduction pathways, such as signaling by PDGF, downstream signal transduction, G alpha (Q) signaling events and signaling to Erks.
Table 2.
Pathways enriched in potential hsa-miR-183-5p targets.
| Databases | Pathwaysa | Pathway ID | Genesb | Targetc | Empirical P-value |
|---|---|---|---|---|---|
| REACTOME | Neuronal system | REACT_13685 | 289 | 20 | 0.0010 |
| REACTOME | Opioid signaling | REACT_15295 | 80 | 9 | 0.0012 |
| KEGG | Gastric acid secretion | 4971 | 74 | 8 | 0.0012 |
| REACTOME | Platelet homeostasis | REACT_23876 | 81 | 8 | 0.0012 |
| REACTOME | Potassium channels | REACT_75908 | 99 | 9 | 0.0013 |
| REACTOME | Activation of kainate receptors upon glutamate binding | REACT_21312 | 30 | 5 | 0.0019 |
| REACTOME | G-protein beta gamma signaling | REACT_19388 | 28 | 4 | 0.0020 |
| REACTOME | Glucagon-type ligand receptors | REACT_18377 | 33 | 4 | 0.0025 |
| REACTOME | Thrombin signaling through proteinase activated receptors (PARS) | REACT_21384 | 32 | 5 | 0.0027 |
| REACTOME | ADP signaling through P2Y purinoceptor 1 | REACT_19140 | 25 | 5 | 0.0030 |
| REACTOME | Formation of HIV-1 elongation complex containing HIV-1 Tat | REACT_6346 | 42 | 4 | 0.0034 |
| REACTOME | HIV-1 transcription elongation | REACT_6274 | 42 | 4 | 0.0034 |
| REACTOME | Tat-mediated elongation of the HIV-1 transcript | REACT_6162 | 42 | 4 | 0.0034 |
| REACTOME | Transmission across chemical synapses | REACT_13477 | 190 | 15 | 0.0052 |
| REACTOME | Glucagon signaling in metabolic regulation | REACT_1665 | 33 | 5 | 0.0053 |
| REACTOME | Inwardly rectifying K+ channels | REACT_75918 | 31 | 5 | 0.0059 |
| REACTOME | Activation of G protein gated potassium channels | REACT_75831 | 25 | 4 | 0.0064 |
| REACTOME | G protein gated potassium channels | REACT_75780 | 25 | 4 | 0.0064 |
| REACTOME | Inhibition of voltage gated Ca2+ channels via G beta gamma γ subunits | REACT_25004 | 25 | 4 | 0.0064 |
| REACTOME | Signal amplification | REACT_20524 | 31 | 5 | 0.0066 |
| REACTOME | G beta gamma signaling through PI3Kgamma | REACT_19290 | 25 | 4 | 0.0074 |
| REACTOME | G alpha (z) signaling events | REACT_19333 | 45 | 6 | 0.0075 |
| REACTOME | Formation of HIV-1 elongation complex in the absence of HIV-1 Tat | REACT_22201 | 42 | 4 | 0.0076 |
| REACTOME | Formation of RNA Pol II elongation complex | REACT_1845 | 42 | 4 | 0.0076 |
| REACTOME | RNA polymerase II transcription elongation | REACT_833 | 42 | 4 | 0.0076 |
| REACTOME | Neurotransmitter receptor binding and downstream transmission in the postsynaptic cell | REACT_15370 | 136 | 10 | 0.0077 |
| REACTOME | Integration of energy metabolism | REACT_1505 | 125 | 10 | 0.0101 |
| REACTOME | G-protein activation | REACT_15457 | 28 | 4 | 0.0115 |
| REACTOME | Transcription-coupled NER (TC-NER) | REACT_1628 | 44 | 3 | 0.0117 |
| REACTOME | mRNA capping | REACT_1470 | 28 | 3 | 0.0118 |
| REACTOME | RNA Pol II CTD phosphorylation and interaction with CE | REACT_6237 | 26 | 3 | 0.0118 |
| REACTOME | RNA Pol II CTD phosphorylation and interaction with CE | REACT_975 | 26 | 3 | 0.0118 |
| REACTOME | Regulation of insulin secretion | REACT_18325 | 98 | 8 | 0.0126 |
| REACTOME | G alpha (q) signaling events | REACT_18283 | 186 | 8 | 0.0134 |
| REACTOME | RNA polymerase II pre-transcription events | REACT_22107 | 58 | 4 | 0.0137 |
| REACTOME | Pausing and recovery of TAT-mediated HIV-1 elongation | REACT_6143 | 31 | 3 | 0.0146 |
| REACTOME | TAT-mediated HIV-1 elongation arrest and recovery | REACT_6344 | 31 | 3 | 0.0146 |
| REACTOME | Aquaporin-mediated transport | REACT_23887 | 47 | 5 | 0.0148 |
| REACTOME | Regulation of water balance by renal aquaporins | REACT_24023 | 40 | 5 | 0.0151 |
| REACTOME | Dual incision reaction in TC-NER | REACT_2222 | 28 | 3 | 0.0153 |
| REACTOME | Formation of transcription-coupled NER (TC-NER) repair complex | REACT_1941 | 28 | 3 | 0.0153 |
| REACTOME | Regulation of insulin secretion by glucagon-like peptide-1 | REACT_18274 | 43 | 5 | 0.0169 |
| REACTOME | DNA repair | REACT_216 | 108 | 5 | 0.0177 |
| REACTOME | Inhibition of insulin secretion by adrenaline noradrenaline | REACT_18339 | 29 | 4 | 0.0177 |
| REACTOME | Nucleotide excision repair | REACT_1826 | 49 | 3 | 0.0189 |
| KEGG | Leishmaniasis | 5140 | 72 | 5 | 0.0191 |
| KEGG | Prion diseases | 5020 | 36 | 4 | 0.0209 |
| REACTOME | HIV-1 elongation arrest and recovery | REACT_6259 | 31 | 3 | 0.0212 |
| REACTOME | Pausing and recovery of HIV-1 elongation | REACT_6244 | 31 | 3 | 0.0212 |
| REACTOME | Class B 2 (Secretin family receptors) | REACT_18372 | 90 | 5 | 0.0212 |
| REACTOME | Formation of the early elongation complex | REACT_846 | 32 | 3 | 0.0231 |
| REACTOME | Formation of the HIV-1 early elongation complex | REACT_6319 | 32 | 3 | 0.0231 |
| REACTOME | Transcription of the HIV genome | REACT_6233 | 61 | 4 | 0.0243 |
| REACTOME | Interferon signaling | REACT_25229 | 110 | 5 | 0.0281 |
| REACTOME | HIV-1 transcription initiation | REACT_6332 | 39 | 3 | 0.0297 |
| REACTOME | RNA polymerase II HIV-1 promoter escape | REACT_6253 | 39 | 3 | 0.0297 |
| REACTOME | RNA polymerase II promoter escape | REACT_2089 | 39 | 3 | 0.0297 |
| REACTOME | RNA Polymerase II transcription initiation | REACT_1851 | 39 | 3 | 0.0297 |
| REACTOME | RNA polymerase II transcription initiation and promoter clearance | REACT_834 | 39 | 3 | 0.0297 |
| REACTOME | RNA polymerase II transcription pre-initiation and promoter opening | REACT_1655 | 39 | 3 | 0.0297 |
| KEGG | Huntington’s disease | 5016 | 183 | 8 | 0.0309 |
| KEGG | Long-term depression | 4730 | 70 | 6 | 0.0311 |
| KEGG | Type I diabetes mellitus | 4940 | 43 | 4 | 0.0316 |
| REACTOME | RNA polymerase II transcription | REACT_1366 | 101 | 5 | 0.0330 |
| KEGG | Tight junction | 4530 | 132 | 9 | 0.0351 |
| KEGG | Chemokine signaling pathway | 4062 | 189 | 10 | 0.0359 |
| REACTOME | Degradation of beta-catenin by the destruction complex | REACT_11063 | 67 | 4 | 0.0360 |
| REACTOME | Signaling by wnt | REACT_11045 | 67 | 4 | 0.0360 |
| REACTOME | Metabolism of carbohydrates | REACT_474 | 126 | 5 | 0.0370 |
| REACTOME | GABA receptor activation | REACT_25199 | 53 | 5 | 0.0388 |
| REACTOME | G alpha (S) signaling events | REACT_19327 | 125 | 6 | 0.0407 |
| REACTOME | Interferon gamma signaling | REACT_25078 | 73 | 4 | 0.0444 |
| REACTOME | Costimulation by the CD28 family | REACT_19344 | 77 | 6 | 0.0464 |
| REACTOME | NFκB and MAP KINASES activation mediated by TLR4 signaling repertoire | REACT_25281 | 71 | 6 | 0.0473 |
| REACTOME | Mitotic prometaphase | REACT_682 | 92 | 5 | 0.0477 |
| REACTOME | Activation of GABA B receptors | REACT_25330 | 38 | 4 | 0.0488 |
| REACTOME | GABA B receptor activation | REACT_25031 | 38 | 4 | 0.0488 |
| REACTOME | M phase | REACT_910 | 96 | 5 | 0.0499 |
| REACTOME | TRAF6 Mediated induction of proinflammatory cytokines | REACT_6782 | 68 | 6 | 0.0499 |
Human diseases relative pathways are shown in bold font
indicates the total number of genes involve in the given pathway
indicates the numbers of potential hsa-miR-183-5p targets in the pathways.
Table 3.
Pathways enriched in both hsa-miRs-206 and -381-3p cotargets.
| Databases | Pathwaysa | Pathway ID | Genesb | Targetc | Empirical P-value |
|---|---|---|---|---|---|
| REACTOME | Hemostasis | REACT_604 | 467 | 36 | 0.0003 |
| KEGG | Adherens junction | 4520 | 73 | 11 | 0.0004 |
| REACTOME | Platelet activation signaling and aggregation | REACT_798 | 205 | 19 | 0.0007 |
| BIOCARTA | Biocarta EDG1 pathway | 27 | 5 | 0.0032 | |
| KEGG | Bacterial invasion of epithelial cells | 5100 | 70 | 9 | 0.0041 |
| KEGG | Fc gamma R-mediated phagocytosis | 4666 | 94 | 9 | 0.0084 |
| BIOCARTA | Biocarta NFAT pathway | 54 | 6 | 0.0114 | |
| KEGG | Chemokine signaling pathway | 4062 | 189 | 13 | 0.0130 |
| KEGG | Regulation of actin cytoskeleton | 4810 | 213 | 16 | 0.0131 |
| KEGG | Shigellosis | 5131 | 61 | 7 | 0.0149 |
| REACTOME | Neuronal system | REACT_13685 | 289 | 17 | 0.0160 |
| REACTOME | Platelet degranulation | REACT_318 | 78 | 7 | 0.0160 |
| BIOCARTA | Biocarta FMLP pathway | 37 | 5 | 0.0177 | |
| REACTOME | Response to elevated platelet cytosolic Ca2+ | REACT_1280 | 83 | 7 | 0.0184 |
| KEGG | Vibrio cholera infection | 5110 | 54 | 5 | 0.0192 |
| KEGG | Pentose phosphate pathway | 30 | 26 | 4 | 0.0196 |
| REACTOME | Signaling by Robo receptor | REACT_19351 | 32 | 5 | 0.0197 |
| REACTOME | Platelet homeostasis | REACT_23876 | 81 | 8 | 0.0201 |
| BIOCARTA | Biocarta MPR pathway | 34 | 5 | 0.0211 | |
| REACTOME | Sphingolipid metabolism | REACT_19323 | 32 | 4 | 0.0211 |
| REACTOME | Metabolism of carbohydrates | REACT_474 | 126 | 9 | 0.0223 |
| KEGG | Epithelial cells signaling in helicobacter pylori infection | 5120 | 68 | 7 | 0.0327 |
| BIOCARTA | Biocarta MET pathway | 37 | 6 | 0.0328 | |
| BIOCARTA | Biocarta keratinocyte pathway | 46 | 6 | 0.0337 | |
| KEGG | Spliceosome | 3040 | 127 | 9 | 0.0347 |
| KEGG | Leukocyte transendothelial migration | 4670 | 116 | 8 | 0.0353 |
| REACTOME | Formation and maturation of mRNA transcript | REACT_2039 | 185 | 12 | 0.0354 |
| BIOCARTA | Biocarta PPARA pathway | 58 | 6 | 0.0367 | |
| REACTOME | Signaling by PDGF | REACT_16888 | 122 | 11 | 0.0369 |
| REACTOME | Downstream signal transduction | REACT_17025 | 93 | 8 | 0.0379 |
| KEGG | Vascular smooth muscle contraction | 4270 | 126 | 8 | 0.0381 |
| REACTOME | G alpha (q) signaling events | REACT_18283 | 186 | 9 | 0.0393 |
| REACTOME | Transmission across chemical synapses | REACT_13477 | 190 | 11 | 0.0399 |
| KEGG | Focal adhesion | 4510 | 199 | 16 | 0.0420 |
| BIOCARTA | Biocarta VEGF pathway | 29 | 5 | 0.0457 | |
| KEGG | RNA degradation | 3018 | 57 | 4 | 0.0457 |
| BIOCARTA | Biocarta CREB pathway | 27 | 4 | 0.0482 | |
| REACTOME | Signaling to erks | REACT_12058 | 35 | 4 | 0.0495 |
immune system relative pathways are shown in bold font
indicates the total number of genes involve in the given pathway
indicates the numbers of hsa-miRs-206 and -381-3p cotargets in the pathways.
Discussion
Circulating miRs have been used as novel minimally invasive biomarkers for a variety of diseases since Mitchell et al. first reported that circulating miRs had potential for detection of different cancers in 2008 (Mitchell et al., 2008). Since then, few studies have focused on identifying circulating miRs in asthma until recently (Wang et al., 2015, Kho et al., 2016, Panganiban et al., 2016, Davis et al., 2017, Milger et al., 2017). The use of circulating miRs for detection of dNCO exposure and dNCO-associated OA have not been previously reported. To our knowledge, this study is the first report to determine circulating miR biomarkers for detection of MDI exposure in murine models. We identified that circulating mmu-miRs-183-5p, -206-3p and -381-3p levels were either upregulated or downregulated after MDI exposure and determined that the up/down-regulation patterns of these three circulating miRs may potentially serve as biomarkers for detection of MDI exposure.
Recent studies identified circulating hsa-miRs-15a, -16, -21, -27a, -29c, -30d-5p, -125b, -126, -133b, -206, -223, -299-5p, -342, -425, -1260a, and -3162-3p may serve as novel biomarkers for allergic rhinitis or asthma diagnosis (Wang et al., 2015, Kho et al., 2016, Panganiban et al., 2016, Davis et al., 2017, Milger et al., 2017). In those reports, there is not a single circulating miR identified that was repeatedly identified between studies. In our study, the murine homolog to circulating hsa-miR-206 was identified as a component of a putative biomarker pattern for MDI exposure. Expression of hsa-miR-206 has been shown to be elevated in patients with allergic rhinitis, but similar in patients with asthma and heathy subjects (Panganiban et al., 2016). This study shows that the level of miRs-206 and -381-3p are decreased in mouse after MDI exposure, indicating that the MDI-triggered downregulation of both circulating miRs-206 and -381-3p is specific, and may be used to distinguish MDI exposure from other conditions.
The physiological roles that miRs-183-5p, -206-3p, and -381-3p play in association with asthma and MDI exposure are currently unknown and are worthy of subsequent functional studies. For this study, we used pathway enrichment assays to predict the functional roles in association with MDI exposure. Many pathways enriched in potential genes regulated by hsa-miR-183-5p were associated with HIV-related diseases; however, the single most significant pathway identified is the neuronal system. One of the protein targets of hsa-miR-183-5p (Table 2, and Supplemental Table S1) identified in the neuronal system is the potassium calcium-activated channel subfamily M regulatory beta subunit 1 (KCNMB1), which is associated with lung diseases such as COPD and asthma (Seibold et al., 2008, Cao et al., 2014). In the airway smooth muscle (ASM), the increase of intracellular calcium concentration triggered by muscarinic acetylcholine receptors activation cause ASM contraction, and consequently, an asthma attack. The large conductance, Ca2+ and voltage-dependent K+ (BK) channels decrease intracellular calcium concentration leading to ASM relaxation; therefore, the BK channel proteins become potential treatment targets for COPD and asthma (Pelaia et al., 2002). The BK channels are composed of a pore forming α-subunit which is encoded by KCNMA1 and a regulatory β-subunit which is encoded by KCNMB1. Interestingly, hsa-miR183-5p was elevated and has been shown to downregulate KCNMB1 expression in COPD lung tissues (Cao et al., 2014). In our study, we identified that hsa-miR-183-5p was elevated after MDI exposure, and subsequent studies should examine if MDI exposure-related elevation of hsa-miR-183-5p will downregulate KCNMB1 in the lung ASM, leading to accumulation of intracellular calcium, smooth muscle contraction, and subsequent asthmatic symptoms.
Among the several different pathways that are enriched in genes potentially regulated by both hsa-miRs-206-3p and -381-3p, we found that the immune related pathways were highly enriched. Interestingly, all enriched immune related pathways involve calcium related signaling. One of the important calcium related signaling pathways is the NFAT (Nuclear Factors of Activated T-Cells) transcription factors activation signaling pathways. NFAT transcription factors were originally described as important transcription regulators in naive T cells and differentiated effector T cells (Macian, 2005), but have been found to be expressed in many diverse cell types in the immune system including dendritic cells (Goodridge et al., 2007, Zanoni et al., 2009), mast cells (Monticelli et al., 2004, Klein et al., 2006), B-Cells (Berland and Wortis, 2003, Winslow et al., 2006), NK (Natural Killer) T cells (Lazarevic et al., 2009), and other cell types. The activation of NFATs is induced by receptor-coupled calcium signaling, which includes activation of calcium binding calmodulin and activation of the calmodulin-dependent phosphatase, calcineurin. Activated calcineurin dephosphorylates and activates cytosolic NFATs, which translocate into the nucleus and mediate gene expression (Feske et al., 2003, Hogan et al., 2003). Associated with other transcription factors such as AP-1 (Macian et al., 2001), NFATs can mediate expression of a number of immunologically important genes, including Th1 type cytokines: IL-2 and INFγ (Peng et al., 2001), Th2 type cytokines: IL-4, IL-5, and IL-13 (Zhang et al., 1999, Burke et al., 2000, Macian et al., 2000), and inflammatory relative genes: IL-3, GM-CSF, and TNFα (Cockerill et al., 1995, Bert et al., 2000, Macian et al., 2000). Cell-surface proteins such as CD40L, FasL and CTLA4 on immune related cells are also regulated by NFATs (Im and Rao, 2004, Macian, 2005). Dysregulation of NFAT signaling has been associated with asthma (Hodge et al., 1996, Keen et al., 2001, van Rietschoten et al., 2001, Diehl et al., 2002, Rengarajan et al., 2002a, Rengarajan et al., 2002b, Koch et al., 2015). Our observation that the murine homologs of circulating hsa-miRs-206-3p and -381-3p were decreased after MDI exposure suggests the interesting hypothesis that calcium dependent NFAT signaling is activated in MDI exposed cells.
Anderson et. al. first reported upregulation of miRs-21, -22, -27b, -31, -126, -155, -210, and -301a from draining lymph node (dLN) of TDI dermally sensitized mice (Anderson et al., 2014). In a follow-up mechanistic study, it was determined that miR-210 upregulation may inhibit regulatory T cell (Treg) function during TDI sensitization (Long et al., 2016). Given that TDI and MDI are both aromatic diisocyanates containing the same reactive moiety (N=C=O) and that both are potent sensitizers, we would expect that MDI and TDI dermal exposure should cause similar miR responses. However, significant changes to expression were not observed for circulating miRs-21, -22, -27b, -31, -126, -155, -210, and -301a between MDI dermal exposed and control mice (Table 1 and data not shown). It is as yet unclear whether the differences observed between these results and those of Anderson et al. are due to differences in isocyanate (TDI vs. MDI), time point (4 days vs. 4/24 hr), or tissue type (draining lymph node vs. serum). Furthermore, detailed mechanistic understanding of how specific miRs are selected and released into circulation from the intracellular environment remains unclear. Evidence has shown that intracellular miRs can be released into extracellular environment through packaging into exosomes (Valadi et al., 2007), microvesicles (Hunter et al., 2008), apoptotic bodies (Zernecke et al., 2009), binding to high density lipoprotein (HDL) (Vickers et al., 2011) and AGO protein complex (Arroyo et al., 2011, Turchinovich et al., 2011). Certainly, circulating miRs reflect a systemic response to the chemical exposure rather than a tissue- or cell-specific response. Future studies are planned to investigate whether the up/down-regulation of miRs-183-5p, -206-3p, and -381-3p identified in current study are observed in the dLN cell population.
Conclusion
We identified that circulating mmu-miRs-183-5p, -206-3p and -381-3p are up/down-regulated after MDI exposure in murine models. Because these miRs are conserved in humans, the findings support the hypothesis that circulating miRs-183-5p, -206-3p and -381-3p may serve as easily obtainable, measurable, circulating biomarkers of MDI-exposure in workers. Given that miRs have been shown to have prognostic value in many diverse diseases, including cancers (Mitchell et al., 2008, Ng et al., 2009, Zhu et al., 2009, Kosaka et al., 2010, Freres et al., 2016, Heishima et al., 2017) and are involved in other inflammatory processes (Zampetaki et al., 2010, Hromadnikova et al., 2014, Xu et al., 2014), future studies on the predictive value of these miRs, and their pathophysiological mechanisms in relation to MDI exposure/sensitization and MDI-OA is needed. Furthermore, these circulating miRs should be evaluated in exposed worker populations for validation as biomarkers of MDI exposure. Finally, pathway analysis of two downregulated miRs (miRs-206-3p and -381-3p) suggests that calcium-dependent NFAT signaling is a candidate pathway for functional studies on diisocyanate induced disease pathogenesis.
Supplementary Material
Acknowledgments
Disclosure Statement
The authors declare that they have no competing financial interests. This work was supported by the National Institute for Occupational Safety and Health (NIOSH) intramural funds. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the National Institute for Occupational Safety and Health, Centers for Disease Control and Prevention.
References
- Acgih, 1999. TLVs and BEIs: Threshold Limit Values for Chemical Substances and Physical Agents; Biological Exposure Indices. American Conference of Governmental Industrial Hygenists Cincinnati, Ohio. [Google Scholar]
- Allport DC, Gilbert DS & Outterside SM, 2003. MDI and TDI : a safety, health and the environment : a source book and practical guide New York: J. Wiley. [Google Scholar]
- Anderson SE, Beezhold K, Lukomska E, Richardson J, Long C, Anderson K, Franko J, Meade BJ & Beezhold DH, 2014. Expression kinetics of miRNA involved in dermal toluene 2,4-diisocyanate sensitization. J Immunotoxicol, 11, 250–9. [DOI] [PubMed] [Google Scholar]
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF & Tewari M, 2011. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A, 108, 5003–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP, 2009. MicroRNAs: target recognition and regulatory functions. Cell, 136, 215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berland R & Wortis HH, 2003. Normal B-1a cell development requires B cell-intrinsic NFATc1 activity. Proc Natl Acad Sci U S A, 100, 13459–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein DI, Korbee L, Stauder T, Bernstein JA, Scinto J, Herd ZL & Bernstein IL, 1993. The low prevalence of occupational asthma and antibody-dependent sensitization to diphenylmethane diisocyanate in a plant engineered for minimal exposure to diisocyanates. J Allergy Clin Immunol, 92, 387–96. [DOI] [PubMed] [Google Scholar]
- Bert AG, Burrows J, Hawwari A, Vadas MA & Cockerill PN, 2000. Reconstitution of T cell-specific transcription directed by composite NFAT/Oct elements. J Immunol, 165, 5646–55. [DOI] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS & Sander C, 2008. The microRNA.org resource: targets and expression. Nucleic Acids Res, 36, D149–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnik LT, Preisser AM, Permentier H & Baur X, 2013. Is specific IgE antibody analysis feasible for the diagnosis of methylenediphenyl diisocyanate-induced occupational asthma? Int Arch Occup Environ Health, 86, 417–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke TF, Casolaro V & Georas SN, 2000. Characterization of P5, a novel NFAT/AP-1 site in the human IL-4 promoter. Biochem Biophys Res Commun, 270, 1016–23. [DOI] [PubMed] [Google Scholar]
- Cao Z, Zhang N, Lou T, Jin Y, Wu Y, Ye Z & Pan J, 2014. microRNA-183 down-regulates the expression of BKCabeta1 protein that is related to the severity of chronic obstructive pulmonary disease. Hippokratia, 18, 328–32. [PMC free article] [PubMed] [Google Scholar]
- Cartier A, Grammer L, Malo JL, Lagier F, Ghezzo H, Harris K & Patterson R, 1989. Specific serum antibodies against isocyanates: association with occupational asthma. J Allergy Clin Immunol, 84, 507–14. [DOI] [PubMed] [Google Scholar]
- Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J & Zhang CY, 2008. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res, 18, 997–1006. [DOI] [PubMed] [Google Scholar]
- Cockerill PN, Bert AG, Jenkins F, Ryan GR, Shannon MF & Vadas MA, 1995. Human granulocyte-macrophage colony-stimulating factor enhancer function is associated with cooperative interactions between AP-1 and NFATp/c. Mol Cell Biol, 15, 2071–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN & Srivastava D, 2009. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature, 460, 705–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalene M, Skarping G & Lind P, 1997. Workers exposed to thermal degradation products of TDI- and MDI-based polyurethane: biomonitoring of 2,4-TDA, 2,6-TDA, and 4,4’-MDA in hydrolyzed urine and plasma. Am Ind Hyg Assoc J, 58, 587–91. [DOI] [PubMed] [Google Scholar]
- Davis JS, Sun M, Kho AT, Moore KG, Sylvia JM, Weiss ST, Lu Q & Tantisira KG, 2017. Circulating microRNAs and association with methacholine PC20 in the Childhood Asthma Management Program (CAMP) cohort. PLoS One, 12, e0180329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl S, Chow CW, Weiss L, Palmetshofer A, Twardzik T, Rounds L, Serfling E, Davis RJ, Anguita J & Rincon M, 2002. Induction of NFATc2 expression by interleukin 6 promotes T helper type 2 differentiation. J Exp Med, 196, 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engfeldt M, Isaksson M, Zimerson E & Bruze M, 2013. Several cases of work-related allergic contact dermatitis caused by isocyanates at a company manufacturing heat exchangers. Contact Dermatitis, 68, 175–80. [DOI] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C & Marks DS, 2003. MicroRNA targets in Drosophila. Genome Biol, 5, R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z & Rajewsky N, 2011. The impact of miRNA target sites in coding sequences and in 3’UTRs. PLoS One, 6, e18067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feske S, Okamura H, Hogan PG & Rao A, 2003. Ca2+/calcineurin signalling in cells of the immune system. Biochem Biophys Res Commun, 311, 1117–32. [DOI] [PubMed] [Google Scholar]
- Freres P, Wenric S, Boukerroucha M, Fasquelle C, Thiry J, Bovy N, Struman I, Geurts P, Collignon J, Schroeder H, Kridelka F, Lifrange E, Jossa V, Bours V, Josse C & Jerusalem G, 2016. Circulating microRNA-based screening tool for breast cancer. Oncotarget, 7, 5416–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB & Bartel DP, 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res, 19, 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez G, Jogie-Brahim S, Shima M & Schwartz LB, 2007. Omalizumab reverses the phenotypic and functional effects of IgE-enhanced Fc epsilonRI on human skin mast cells. J Immunol, 179, 1353–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodridge HS, Simmons RM & Underhill DM, 2007. Dectin-1 stimulation by Candida albicans yeast or zymosan triggers NFAT activation in macrophages and dendritic cells. J Immunol, 178, 3107–15. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP & Bartel DP, 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell, 27, 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guay C & Regazzi R, 2013. Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat Rev Endocrinol, 9, 513–21. [DOI] [PubMed] [Google Scholar]
- Gupta SK, Bang C & Thum T, 2010. Circulating microRNAs as biomarkers and potential paracrine mediators of cardiovascular disease. Circ Cardiovasc Genet, 3, 484–8. [DOI] [PubMed] [Google Scholar]
- Haenen S, Clynen E, De Vooght V, Schoofs L, Nemery B, Hoet PH & Vanoirbeek JA, 2012. Proteome changes in auricular lymph nodes and serum after dermal sensitization to toluene diisocyanate in mice. Proteomics, 12, 3548–58. [DOI] [PubMed] [Google Scholar]
- Haenen S, Clynen E, Nemery B, Hoet PHM & Vanoirbeek J.a.J., 2014. Biomarker discovery in asthma and COPD: Application of proteomics techniques in human and mice. EuPA Open Proteomics, 4, 101–12. [Google Scholar]
- Haenen S, Vanoirbeek JA, De Vooght V, Maes E, Schoofs L, Nemery B, Hoet PH & Clynen E, 2010. Proteome analysis of multiple compartments in a mouse model of chemical-induced asthma. J Proteome Res, 9, 5868–76. [DOI] [PubMed] [Google Scholar]
- Heishima K, Ichikawa Y, Yoshida K, Iwasaki R, Sakai H, Nakagawa T, Tanaka Y, Hoshino Y, Okamura Y, Murakami M, Maruo K, Akao Y & Mori T, 2017. Circulating microRNA-214 and -126 as potential biomarkers for canine neoplastic disease. Sci Rep, 7, 2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettick JM, Law BF, Lin CC, Wisnewski AV & Siegel PD, 2018. Mass spectrometry-based analysis of murine bronchoalveolar lavage fluid following respiratory exposure to 4,4’-methylene diphenyl diisocyanate aerosol. Xenobiotica, 48, 626–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge MR, Ranger AM, Charles De La Brousse F, Hoey T, Grusby MJ & Glimcher LH, 1996. Hyperproliferation and dysregulation of IL-4 expression in NF-ATp-deficient mice. Immunity, 4, 397–405. [DOI] [PubMed] [Google Scholar]
- Hogan PG, Chen L, Nardone J & Rao A, 2003. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev, 17, 2205–32. [DOI] [PubMed] [Google Scholar]
- Hromadnikova I, Kotlabova K, Hympanova L, Doucha J & Krofta L, 2014. First trimester screening of circulating C19MC microRNAs can predict subsequent onset of gestational hypertension. PLoS One, 9, e113735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, Nana-Sinkam SP, Jarjoura D & Marsh CB, 2008. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One, 3, e3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur GY, Choi GS, Sheen SS, Lee HY, Park HJ, Choi SJ, Ye YM & Park HS, 2008. Serum ferritin and transferrin levels as serologic markers of methylene diphenyl diisocyanate-induced occupational asthma. J Allergy Clin Immunol, 122, 774–80. [DOI] [PubMed] [Google Scholar]
- Im SH & Rao A, 2004. Activation and deactivation of gene expression by Ca2+/calcineurin-NFAT-mediated signaling. Mol Cells, 18, 1–9. [PubMed] [Google Scholar]
- Jan RL, Chen SH, Chang HY, Yeh HJ, Shieh CC & Wang JY, 2008. Asthma-like syndrome in school children after accidental exposure to xylene and methylene diphenyl diisocyanate. J Microbiol Immunol Infect, 41, 337–41. [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T & Yamanishi Y, 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res, 36, D480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen JC, Sholl L, Wills-Karp M & Georas SN, 2001. Preferential activation of nuclear factor of activated T cells c correlates with mouse strain susceptibility to allergic responses and interleukin-4 gene expression. Am J Respir Cell Mol Biol, 24, 58–65. [DOI] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U & Segal E, 2007. The role of site accessibility in microRNA target recognition. Nat Genet, 39, 1278–84. [DOI] [PubMed] [Google Scholar]
- Keskinen H, Tupasela O, Tiikkainen U & Nordman H, 1988. Experiences of specific IgE in asthma due to diisocyanates. Clin Allergy, 18, 597–604. [DOI] [PubMed] [Google Scholar]
- Kho AT, Sharma S, Davis JS, Spina J, Howard D, Mcenroy K, Moore K, Sylvia J, Qiu W, Weiss ST & Tantisira KG, 2016. Circulating MicroRNAs: Association with Lung Function in Asthma. PLoS One, 11, e0157998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kim JE, Choi GS, Kim HY, Ye YM & Park HS, 2011. Serum cytokines markers in toluene diisocyanate-induced asthma. Respir Med, 105, 1091–4. [DOI] [PubMed] [Google Scholar]
- Klein M, Klein-Hessling S, Palmetshofer A, Serfling E, Tertilt C, Bopp T, Heib V, Becker M, Taube C, Schild H, Schmitt E & Stassen M, 2006. Specific and redundant roles for NFAT transcription factors in the expression of mast cell-derived cytokines. J Immunol, 177, 6667–74. [DOI] [PubMed] [Google Scholar]
- Koch S, Reppert S & Finotto S, 2015. NFATc1 deletion in T lymphocytes inhibits the allergic trait in a murine model of asthma. Clin Exp Allergy, 45, 1356–66. [DOI] [PubMed] [Google Scholar]
- Kosaka N, Iguchi H & Ochiya T, 2010. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci, 101, 2087–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, Macmenamin P, Da Piedade I, Gunsalus KC, Stoffel M & Rajewsky N, 2005. Combinatorial microRNA target predictions. Nat Genet, 37, 495–500. [DOI] [PubMed] [Google Scholar]
- Lazarevic V, Zullo AJ, Schweitzer MN, Staton TL, Gallo EM, Crabtree GR & Glimcher LH, 2009. The gene encoding early growth response 2, a target of the transcription factor NFAT, is required for the development and maturation of natural killer T cells. Nat Immunol, 10, 306–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB & Bartel DP, 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell, 120, 15–20. [DOI] [PubMed] [Google Scholar]
- Lin CC, Liu LZ, Addison JB, Wonderlin WF, Ivanov AV & Ruppert JM, 2011. A KLF4-miRNA-206 autoregulatory feedback loop can promote or inhibit protein translation depending upon cell context. Mol Cell Biol, 31, 2513–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CC, Sharma SB, Farrugia MK, Mclaughlin SL, Ice RJ, Loskutov YV, Pugacheva EN, Brundage KM, Chen D & Ruppert JM, 2015. Kruppel-like factor 4 signals through microRNA-206 to promote tumor initiation and cell survival. Oncogenesis, 4, e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofgren DJ, Walley TL, Peters PM & Weis ML, 2003. MDI Exposure for Spray-On Truck Bed Lining. Appl Occup Environ Hyg, 18, 772–9. [DOI] [PubMed] [Google Scholar]
- Long CM, Lukomska E, Marshall NB, Nayak A & Anderson SE, 2016. Potential Inhibitory Influence of miRNA 210 on Regulatory T Cells during Epicutaneous Chemical Sensitization. Genes (Basel), 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TP, Lee CY, Tsai MH, Chiu YC, Hsiao CK, Lai LC & Chuang EY, 2012. miRSystem: an integrated system for characterizing enriched functions and pathways of microRNA targets. PLoS One, 7, e42390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle JR, Yario TA & Steitz JA, 2007. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5’ UTR as in the 3’ UTR. Proc Natl Acad Sci U S A, 104, 9667–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macian F, 2005. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol, 5, 472–84. [DOI] [PubMed] [Google Scholar]
- Macian F, Garcia-Rodriguez C & Rao A, 2000. Gene expression elicited by NFAT in the presence or absence of cooperative recruitment of Fos and Jun. EMBO J, 19, 4783–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macian F, Lopez-Rodriguez C & Rao A, 2001. Partners in transcription: NFAT and AP-1. Oncogene, 20, 2476–89. [DOI] [PubMed] [Google Scholar]
- Malo JL, Ghezzo H, L’archeveque J, Lagier F, Perrin B & Cartier A, 1991. Is the clinical history a satisfactory means of diagnosing occupational asthma? Am Rev Respir Dis, 143, 528–32. [DOI] [PubMed] [Google Scholar]
- Maragkakis M, Alexiou P, Papadopoulos GL, Reczko M, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, Simossis VA, Sethupathy P, Vergoulis T, Koziris N, Sellis T, Tsanakas P & Hatzigeorgiou AG, 2009a. Accurate microRNA target prediction correlates with protein repression levels. BMC Bioinformatics, 10, 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, Vergoulis T, Koziris N, Sellis T, Tsanakas P & Hatzigeorgiou AG, 2009b. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res, 37, W273–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, De Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, Kanapin A, Lewis S, Mahajan S, May B, Schmidt E, Vastrik I, Wu G, Birney E, Stein L & D’eustachio P, 2009. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res, 37, D619–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JT & Olson EN, 2012. MicroRNAs in stress signaling and human disease. Cell, 148, 1172–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milger K, Gotschke J, Krause L, Nathan P, Alessandrini F, Tufman A, Fischer R, Bartel S, Theis FJ, Behr J, Dehmel S, Mueller NS, Kneidinger N & Krauss-Etschmann S, 2017. Identification of a plasma miRNA biomarker signature for allergic asthma: A translational approach. Allergy. [DOI] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B & Rigoutsos I, 2006. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell, 126, 1203–17. [DOI] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB & Tewari M, 2008. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A, 105, 10513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli S, Solymar DC & Rao A, 2004. Role of NFAT proteins in IL13 gene transcription in mast cells. J Biol Chem, 279, 36210–8. [DOI] [PubMed] [Google Scholar]
- Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS & Sung JJ, 2009. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut, 58, 1375–81. [DOI] [PubMed] [Google Scholar]
- Niosh, 1994a. Letter from NIOSH to Distinctive Designs International Inc. with a study report. Cincinnati, OH, HETA 91–0386-2427, May. [Google Scholar]
- Niosh, 1994b. Letter from NIOSH to Jim Walter Resources, Inc. with a study report. Cincinnati, OH, Report No. HETA 94–0027, May 24. [Google Scholar]
- Niosh, 2004. A Summary of Health Hazard Evaluations: Issues Related to Occupational Exposure to Isocyanates, 1989 to 2002., DHHS (NIOSH) Publication No. 2004–116.
- Nishimura D, 2001. BioCarta. Biotech Softw Int Rep, 2, 117–20. [Google Scholar]
- Orom UA, Nielsen FC & Lund AH, 2008. MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell, 30, 460–71. [DOI] [PubMed] [Google Scholar]
- Ott MG, Jolly AT, Burkert AL & Brown WE, 2007. Issues in diisocyanate antibody testing. Crit Rev Toxicol, 37, 567–85. [DOI] [PubMed] [Google Scholar]
- Panganiban RP, Wang Y, Howrylak J, Chinchilli VM, Craig TJ, August A & Ishmael FT, 2016. Circulating microRNAs as biomarkers in patients with allergic rhinitis and asthma. J Allergy Clin Immunol, 137, 1423–32. [DOI] [PubMed] [Google Scholar]
- Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P & Hatzigeorgiou AG, 2009. The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res, 37, D155–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaia G, Gallelli L, Vatrella A, Grembiale RD, Maselli R, De Sarro GB & Marsico SA, 2002. Potential role of potassium channel openers in the treatment of asthma and chronic obstructive pulmonary disease. Life Sci, 70, 977–90. [DOI] [PubMed] [Google Scholar]
- Peng SL, Gerth AJ, Ranger AM & Glimcher LH, 2001. NFATc1 and NFATc2 together control both T and B cell activation and differentiation. Immunity, 14, 13–20. [DOI] [PubMed] [Google Scholar]
- Petsonk EL, Wang ML, Lewis DM, Siegel PD & Husberg BJ, 2000. Asthma-like symptoms in wood product plant workers exposed to methylene diphenyl diisocyanate. Chest, 118, 1183–93. [DOI] [PubMed] [Google Scholar]
- Place RF, Li LC, Pookot D, Noonan EJ & Dahiya R, 2008. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A, 105, 1608–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollaris L, Devos F, De Vooght V, Seys S, Nemery B, Hoet PH & Vanoirbeek JA, 2016. Toluene diisocyanate and methylene diphenyl diisocyanate: asthmatic response and cross-reactivity in a mouse model. Arch Toxicol, 90, 1709–17. [DOI] [PubMed] [Google Scholar]
- Rattray NJ, Botham PA, Hext PM, Woodcock DR, Fielding I, Dearman RJ & Kimber I, 1994. Induction of respiratory hypersensitivity to diphenylmethane-4,4’-diisocyanate (MDI) in guinea pigs. Influence of route of exposure. Toxicology, 88, 15–30. [DOI] [PubMed] [Google Scholar]
- Redlich CA & Karol MH, 2002. Diisocyanate asthma: clinical aspects and immunopathogenesis. Int Immunopharmacol, 2, 213–24. [DOI] [PubMed] [Google Scholar]
- Rengarajan J, Mowen KA, Mcbride KD, Smith ED, Singh H & Glimcher LH, 2002a. Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression. J Exp Med, 195, 1003–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengarajan J, Tang B & Glimcher LH, 2002b. NFATc2 and NFATc3 regulate T(H)2 differentiation and modulate TCR-responsiveness of naive T(H)cells. Nat Immunol, 3, 48–54. [DOI] [PubMed] [Google Scholar]
- Sastre J, Sastre B, Fernandez-Nieto M, Perez-Camo I, Sanchez JJ & Del Pozo V, 2010. Serum ferritin and transferrin levels are not serologic markers of toluene diisocyanate-induced occupational asthma. J Allergy Clin Immunol, 125, 762–4. [DOI] [PubMed] [Google Scholar]
- Schutze D, Sepai O, Lewalter J, Miksche L, Henschler D & Sabbioni G, 1995. Biomonitoring of workers exposed to 4,4’-methylenedianiline or 4,4’-methylenediphenyl diisocyanate. Carcinogenesis, 16, 573–82. [DOI] [PubMed] [Google Scholar]
- Seibold MA, Wang B, Eng C, Kumar G, Beckman KB, Sen S, Choudhry S, Meade K, Lenoir M, Watson HG, Thyne S, Williams LK, Kumar R, Weiss KB, Grammer LC, Avila PC, Schleimer RP, Burchard EG & Brenner R, 2008. An african-specific functional polymorphism in KCNMB1 shows sex-specific association with asthma severity. Hum Mol Genet, 17, 2681–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selgrade M, Boykin EH, Haykal-Coates N, Woolhiser MR, Wiescinski C, Andrews DL, Farraj AK, Doerfler DL & Gavett SH, 2006. Inconsistencies between cytokine profiles, antibody responses, and respiratory hyperresponsiveness following dermal exposure to isocyanates. Toxicol Sci, 94, 108–17. [DOI] [PubMed] [Google Scholar]
- Sharma SB, Lin CC, Farrugia MK, Mclaughlin SL, Ellis EJ, Brundage KM, Salkeni MA & Ruppert JM, 2014. MicroRNAs 206 and 21 cooperate to promote RAS-extracellular signal-regulated kinase signaling by suppressing the translation of RASA1 and SPRED1. Mol Cell Biol, 34, 4143–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarping G & Dalene M, 1995. Determination of 4,4’-methylenediphenyldianiline (MDA) and identification of isomers in technical-grade MDA in hydrolysed plasma and urine from workers exposed to methylene diphenyldiisocyanate by gas chromatography-mass spectrometry. J Chromatogr B Biomed Appl, 663, 209–16. [DOI] [PubMed] [Google Scholar]
- Skarping G, Dalene M & Littorin M, 1995. 4,4’-Methylenedianiline in hydrolysed serum and urine from a worker exposed to thermal degradation products of methylene diphenyl diisocyanate elastomers. Int Arch Occup Environ Health, 67, 73–7. [DOI] [PubMed] [Google Scholar]
- Srinivasan S, Selvan ST, Archunan G, Gulyas B & Padmanabhan P, 2013. MicroRNAs -the next generation therapeutic targets in human diseases. Theranostics, 3, 930–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Oikawa K, Takanashi M, Kudo M, Ohyashiki J, Ohyashiki K & Kuroda M, 2009. Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PLoS One, 4, e5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee RD, Cullinan P, Welch J, Burge PS & Newman-Taylor AJ, 1998. Specific IgE to isocyanates: a useful diagnostic role in occupational asthma. J Allergy Clin Immunol, 101, 709–15. [DOI] [PubMed] [Google Scholar]
- Tijsen AJ, Pinto YM & Creemers EE, 2012. Circulating microRNAs as diagnostic biomarkers for cardiovascular diseases. Am J Physiol Heart Circ Physiol, 303, H1085–95. [DOI] [PubMed] [Google Scholar]
- Truesdell SS, Mortensen RD, Seo M, Schroeder JC, Lee JH, Letonqueze O & Vasudevan S, 2012. MicroRNA-mediated mRNA translation activation in quiescent cells and oocytes involves recruitment of a nuclear microRNP. Sci Rep, 2, 842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang JS, Ebert MS & Van Oudenaarden A, 2010. Genome-wide dissection of microRNA functions and cotargeting networks using gene set signatures. Mol Cell, 38, 140–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchinovich A, Weiz L, Langheinz A & Burwinkel B, 2011. Characterization of extracellular circulating microRNA. Nucleic Acids Res, 39, 7223–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ & Lotvall JO, 2007. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol, 9, 654–9. [DOI] [PubMed] [Google Scholar]
- Van Rietschoten JG, Smits HH, Van De Wetering D, Westland R, Verweij CL, Den Hartog MT & Wierenga EA, 2001. Silencer activity of NFATc2 in the interleukin-12 receptor beta 2 proximal promoter in human T helper cells. J Biol Chem, 276, 34509–16. [DOI] [PubMed] [Google Scholar]
- Vanoirbeek JA, Tarkowski M, Ceuppens JL, Verbeken EK, Nemery B & Hoet PH, 2004. Respiratory response to toluene diisocyanate depends on prior frequency and concentration of dermal sensitization in mice. Toxicol Sci, 80, 310–21. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y & Steitz JA, 2007. Switching from repression to activation: microRNAs can up-regulate translation. Science, 318, 1931–4. [DOI] [PubMed] [Google Scholar]
- Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD & Remaley AT, 2011. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol, 13, 423–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chen J, Chang P, Leblanc A, Li D, Abbruzzesse JL, Frazier ML, Killary AM & Sen S, 2009. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila), 2, 807–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ML & Petsonk EL, 2004. Symptom onset in the first 2 years of employment at a wood products plant using diisocyanates: some observations relevant to occupational medical screening. Am J Ind Med, 46, 226–33. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yang L, Li P, Huang H, Liu T, He H, Lin Z, Jiang Y, Ren N, Wu B, Kamp DW, Tan J & Liu G, 2015. Circulating microRNA Signatures Associated with Childhood Asthma. Clin Lab, 61, 467–74. [DOI] [PubMed] [Google Scholar]
- Wass U & Belin L, 1989. Immunologic specificity of isocyanate-induced IgE antibodies in serum from 10 sensitized workers. J Allergy Clin Immunol, 83, 126–35. [DOI] [PubMed] [Google Scholar]
- Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ & Wang K, 2010. The microRNA spectrum in 12 body fluids. Clin Chem, 56, 1733–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winslow MM, Gallo EM, Neilson JR & Crabtree GR, 2006. The calcineurin phosphatase complex modulates immunogenic B cell responses. Immunity, 24, 141–52. [DOI] [PubMed] [Google Scholar]
- Wisnewski AV, Xu L, Robinson E, Liu J, Redlich CA & Herrick CA, 2011. Immune sensitization to methylene diphenyl diisocyanate (MDI) resulting from skin exposure: albumin as a carrier protein connecting skin exposure to subsequent respiratory responses. J Occup Med Toxicol, 6, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woellner RC, Hall S, Greaves I & Schoenwetter WF, 1997. Epidemic of asthma in a wood products plant using methylene diphenyl diisocyanate. Am J Ind Med, 31, 56–63. [DOI] [PubMed] [Google Scholar]
- Xiao F, Zuo Z, Cai G, Kang S, Gao X & Li T, 2009. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res, 37, D105–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zhao J, Evan G, Xiao C, Cheng Y & Xiao J, 2012. Circulating microRNAs: novel biomarkers for cardiovascular diseases. J Mol Med (Berl), 90, 865–75. [DOI] [PubMed] [Google Scholar]
- Xu P, Zhao Y, Liu M, Wang Y, Wang H, Li YX, Zhu X, Yao Y, Wang H, Qiao J, Ji L & Wang YL, 2014. Variations of microRNAs in human placentas and plasma from preeclamptic pregnancy. Hypertension, 63, 1276–84. [DOI] [PubMed] [Google Scholar]
- Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, Mayr A, Weger S, Oberhollenzer F, Bonora E, Shah A, Willeit J & Mayr M, 2010. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res, 107, 810–7. [DOI] [PubMed] [Google Scholar]
- Zanoni I, Ostuni R, Capuano G, Collini M, Caccia M, Ronchi AE, Rocchetti M, Mingozzi F, Foti M, Chirico G, Costa B, Zaza A, Ricciardi-Castagnoli P & Granucci F, 2009. CD14 regulates the dendritic cell life cycle after LPS exposure through NFAT activation. Nature, 460, 264–8. [DOI] [PubMed] [Google Scholar]
- Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, Hristov M, Koppel T, Jahantigh MN, Lutgens E, Wang S, Olson EN, Schober A & Weber C, 2009. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal, 2, ra81. [DOI] [PubMed] [Google Scholar]
- Zhang DH, Yang L, Cohn L, Parkyn L, Homer R, Ray P & Ray A, 1999. Inhibition of allergic inflammation in a murine model of asthma by expression of a dominant-negative mutant of GATA-3. Immunity, 11, 473–82. [DOI] [PubMed] [Google Scholar]
- Zhou H & Rigoutsos I, 2014. MiR-103a-3p targets the 5’ UTR of GPRC5A in pancreatic cells. RNA, 20, 1431–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Qin W, Atasoy U & Sauter ER, 2009. Circulating microRNAs in breast cancer and healthy subjects. BMC Res Notes, 2, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.