Abstract
The Anopheles gambiae sensu lato species complex consists of a number of cryptic species with different habitats and behaviours. These morphologically indistinct species are identified by chromosome banding. Several molecular diagnostic techniques for distinguishing between An. coluzzii and An. gambiae are still under improvement. Although, the current SINE method for identification between An. coluzzii and An. gambiae works reliably, this study describes a refinement of the SINE method to increase sensitivity for identification of An. coluzzii, An. gambiae and An. arabiensis based on amplicon dissociation curve characteristics. Field-collected samples, laboratory-reared colonies and crossed specimens of the two species were used for the design of the protocol. An. gambiae, An. coluzzii, and hybrids of the two species were sampled from Ghana and An. arabiensis from Kenya. Samples were first characterised using conventional SINE PCR method, and further assayed using SYBR green, an intercalating fluorescent dye. The three species and hybrids were clearly differentiated using the melting temperature of the dissociation curves, with derivative peaks at 72°C for An. arabiensis, 75°C for An. gambiae and 86°C for An. coluzzii. The hybrids (An. gambiae / An. coluzzii) showed both peaks. This work is the first to describe a SYBR green real time PCR method for the characterization of An. arabiensis, An. gambiae and An. coluzzii and was purposely designed for basic melt-curve analysis (rather than high-resolution melt-curve) to allow it to be used on a wide range of real-time PCR machines.
Introduction
The Anopheles gambiae sensu lato (An. gambiae s.l.) complex comprises at least seven mosquito species originally defined by polytene chromosome analysis [1, 2]. All the An. gambiae s.l. species are currently identified by PCR-diagnostic assays based on specific DNA nucleotide differences in the intergenic spacer (IGS) of the ribosomal DNA (rDNA) [3–6]. Detailed analyses of the IGS region of rDNA further revealed nucleotide substitutions that differentiated between the two forms within the Anopheles gambiae s.s previously designated as S and M molecular forms [7], and were recently named An. gambiae and An. coluzzii [8]. These two species can be identified by PCR and gel electrophoresis showing the presence or absence of a diagnostic Short Interspersed Nuclear Element (SINE) on the X-chromosome. [9].
Hybrid forms of An. gambiae and An. coluzzii have been identified [10, 11]. Several countries have reported high proportion of hybrid population within wild adult mosquito collections. In Gambia, An. gambiae / An. coluzzii hybrids were identified from a number of sites at frequencies as high as 16.7% [12] while in Guinea-Bissau, over 20% of the individuals assayed were hybrids and later, more than 40% were observed in the same country and in Senegal [13–15]. However, correct hybrid species identification [16] and additional data on hybrid species distribution and potential survival rates of wild populations is still needed. It has been described that the first progeny (F1) of hybrids were fully fertile [17], and recent laboratory-based crossing experiments have shown that the hybrids of An. gambiae / An. coluzzii can be maintained over several generations [18] instigating the need to clearly differentiate hybrid An. gambiae / An. coluzzii within wild population of An. gambiae s.l.
Anopheles gambiae and An. coluzzii are characterised by a high degree of gene flow restriction, low level of genetic differentiation, and a largely overlapping geographical and temporal distribution [19]. Furthermore, the two species, together with An. arabiensis, can live in sympatry but show different living characteristics such as insecticide resistance profile with varying resistance allele distributions [20]. Therefore, the correct identification of An. arabiensis, An. gambiae and An. coluzzii mosquitoes constitutes an integral part of malaria vector control programmes, and insecticide resistance management.
A commonly used method for differential identification of An. arabiensis, An. gambiae and An. coluzzii involves a combination of protocols established by Scott et al. and Fanello et al.[4, 11, 21]. These methods are based on PCR-Restriction Fragment Length Polymorphism (PCR-RFLP) and make use of the presence of nucleotide substitutions within the 28S coding region, and part of the IGS region of rDNA [11]. More recently a Short Interspersed Nuclear Element (SINE) insertion (S200 X6.1) on the X chromosome of An. gambiae has been found to be fixed in all An. coluzzii and absent in An. gambiae and An. arabiensis [10]. Additionally, a 26 bp deletion in the same region defines An. arabiensis, allowing for the development of a novel PCR diagnostic assay that differentiates the three species [10]. Briefly, primers that flank the S200 X6.1 insertion were designed to amplify the genomic DNA isolated from Anopheles gambiae s.s. specimen. The PCR products are run on an agarose gel, and individuals (An. coluzzii) with the S200 X6.1 insertion show a single band at 479 bp while individuals (An. gambiae) with no S200 X6.1 insertion give a 249 bp band, and An. arabiensis 223 bp as a result of the deletion [10]. Either method for the identification of An. arabiensis, An. gambiae and An. coluzzii requires post-PCR analysis by gel electrophoresis, which is less sensitive and laborious than melt-curve analysis.
SYBR green DNA-based Real-Time PCR systems provide a good alternative to fluorescent probe-based Real-Time PCR techniques and are based on ability of SYBR green to produce a 100-fold increase in fluorescence when bound to double-stranded DNA. Even though SYBR green binds non-specifically to nucleic acids, the fluorescent signal produced when in complex with DNA is directly proportional to the length and amount of DNA copies synthesized during the reaction, making this technique very sensitive [22] and very precise when diagnostic primer sets are used.
The aim of this study is to demonstrate a time-efficient, highly sensitive and specific SYBR green-based real-time PCR diagnostic assay that differentiates between An. arabiensis, An. coluzzii and An. gambiae.
Materials and methods
Mosquito samples and DNA extraction
Anopheles gambiae mosquito samples were obtained from Vestergaard-NMIMR Vector Labs (VNVL). These consisted of more than 200 samples selected from the standard susceptible Kisumu strain originally from Kenya, and An. gambiae Tiassalé, a resistant strain from the village of Tiassalé in Côte d’Ivoire maintained in the VNVL insectary since 2010. Hundred and ninety An. coluzzii were collected from Okyereko, a rice irrigation field in the Central region of Ghana [23–25]. Hybrid An. gambiae / An. coluzzii (75 samples) were obtained from laboratory crossing of either An. gambiae Kisumu and An. coluzzii or An. gambiae Tiassalé and An. coluzzii. Fifty An. arabiensis samples were obtained from field collection in Kenya. The study was initiated at Noguchi Memorial Institute for Medical Research (NMIMR) in Ghana and the primer design, optimization, validation and high-throughput species identification were performed at Liverpool School of Tropical Medicine (LSTM), UK.
Whole mosquito DNA extraction was performed using a simplified version of the protocol designed by Collins and colleagues [26]. A single mosquito was homogenized in a 1.5 ml Eppendorf tube containing 200 μl of CTAB buffer and incubated at 65°C in a water bath for 5 minutes. Two hundred microliters (200 μl) of chloroform were added to the homogenate, mixed by inversion and centrifuged for 5 minutes at 12000 rpm 25°C. The supernatant was pipetted into new 1.5 ml Eppendorf tubes. 200 μl of isopropyl alcohol was added, mixed by inversion and then centrifuged at 12000 rpm for 15 minutes. The supernatant was then discarded gently and the DNA pellet was thereafter purified with 70% ethanol, dried overnight, and reconstituted in 20 μl of DNAse free water.
Mosquito species identification
Anopheles arabiensis and An. gambiae s.s. species were first determined using established protocols [4, 11]. Conventional PCR assays were performed with a 1 in40 dilution of the reconstituted DNA solution obtained from a single mosquito. PCR products were run on 2% agarose gels, stained with ethidium bromide and then visualized using UV Trans-illuminator (BioDoc-It Imaging System, Upland, USA).
SINE PCR was performed using primers designed by Santolamazza et al. [10] for the identification of An. arabiensis, An. coluzzii and An. gambiae using the primers; F6.1a (TCGCCTTAGACCTTGCGTTA) and R6.1b (CGCTTCAAGAATTCGAGATAC). Total PCR reaction volume of 25 μl containing 4 μl of 1 in 40 dilution of genomic DNA (used as template), 1 μl each of 10 μM of both forward and reverse primers, 6.5 μl of nuclease-free water and 12.5 μl of GoTaq master mix (Promega, Madison, WI, USA). Reaction conditions used were 94°C for 10 minutes; followed by 35 cycles of 94°C for 30 seconds, 59°C for 30 seconds72°C for 1 minute; and a final extension at 72°C for 10 minutes. PCR products were run on 2% agarose gels, stained with ethidium bromide and thereafter visualized using UV Trans-illuminator.
SYBR green-based Real-Time PCR for species identification
This designed melt curve method to distinguish between An. arabiensis, An. gambiae and An. coluzzii uses the diagnostic Short Interspersed Nuclear Element (SINE200). The original S200 X6.1 primers designed by [10] were unsuitable for melt curve analysis, firstly because they produce overlapping melt-peaks for An. coluzzii and An. gambiae /An. coluzzii hybrids and secondly they give indistinct melt peaks for An. gambiae and An. arabiensis. New primers were designed based on amplicon length and G/C content to produce distinct melt-curves for the three sibling species and the hybrids. A universal forward primer (SINE200Fa 5’-ATTGCTACCACCAAAATACATGAAA-3’) matching all three species is combined with An. gambiae / An. coluzzii specific reverse (SINE200Rd 5’-GGGGGGGGGAATAATAAGGAACTGCATTTAAT-3’) and for An. arabiensis reverse (SINE200Re 5’- GGATGTCTAATAGTCTCAATAGATG -3’).
SINE200Rd, the reverse primer for An. gambiae and An. coluzzii has a G8 stretch (GGGGGGGG) added to the 5’ end of the primer (as underlined) to increase the amplicon melting temperature (Tm), preventing the melt profiles of An. gambiae and An. arabiensis to overlap. SINE200Rd incorporates the SINE200 transposon in An. coluzzii into the amplicon and gives a distinct high Tm. The specificity of the An. arabiensis primer SINE200Re is based on only two mismatches with An. gambiae. Both mismatches are conveniently on the 3' end of the primer though a sufficiently high annealing temperature effectively prevents amplification of An. gambiae. The amplicon sizes are 60 bp, 103 bp and 333 bp for An. arabiensis, An. gambiae and An. coluzzii respectively.
A total of sixty (60) samples including 10 An. arabiensis, 20 of An. gambiae, 14 An. coluzzii, and 15 hybrids randomly selected among the individuals characterized by SINE PCR, and one artificial hybrid (pooled DNA of An. gambiae-An. coluzzii) were analysed using the designed melt-curve protocol.
A total reaction volume of 10 μl mixture was prepared by combining 3.75 μl of Nuclease-free PCR grade water, 5 μl of Luna Universal qPCR master mix (New England Biolabs, Ipswich, Ma, USA), 0.25 μl of each (10 μM) SINE200Fa, SINE200Rd and SINE200Re primers, and 0.5 μl mosquito DNA template (Table 1).
Table 1. SYBR green master mix.
| 1X (μl) | |
|---|---|
| Nuclease free water | 3.75 |
| Luna Universal qPCR Master Mix | 5 |
| SINE200Fa (10 μM) | 0.25 |
| SINE200Rd (10 μM) | 0.25 |
| SINE200Re (10 μM) | 0.25 |
| Genomic DNA template | 0.5 |
| Total | 10 |
The samples were run on AriaMx Real-time PCR system (Agilent, Santa Clara, Ca, USA) using the quantitative PCR DNA binding dye including standard melt curve program and 520 nm wavelength filter (FAM). Annealing temperature (Ta) and cycle number were optimised to eliminate non-target background melt-peaks. The optimized cycling conditions involved a denaturation of 95°C for 60 seconds followed by 33 cycles of 95°C for 15 seconds, 60°C for 20 seconds, and 72°C for 10 seconds with a final dissociation step of 95°C for 60 seconds, 55°C for 30 seconds and a melt ramp up to 95°C with 0.5°C increments.
The robustness of the assay was tested using SYBR green-based alternative reagents and an alternative real-time PCR machine. Eight samples of each group (An. arabiensis, An. gambiae, An. coluzzii, and hybrid) were repeated with Luna Universal qPCR Master Mix and with Brilliant III Ultra-Fast SYBR Green Low ROX QPCR Master Mix (Agilent) on a Stratagene Mx3005P real-time PCR machine (Agilent). Thermal cycling conditions and PCR mix were identical to those used for AriaMX. Furthermore, 88 samples from Sudan and 87 from Tanzania were run in both machines to check for consistency and degree of shift between machines.
The species identification was automated using a “nested if” statement in Microsoft Excel which assigns the melt peak temperatures to pre-defined temperature windows corresponding with the different species (S1 Text).
The peak temperature criteria are: >85°C = AC = An. coluzzii, between 85°C and 74°C = AG = An. gambiae, <74°C = AA = An. arabiensis, both >85°C and between 85°C and 74°C = HY = An. gambiae /An. coluzzii hybrid. This automation prevents the need for manually scoring species as is the case for gel-based species ID, thereby reducing time and scoring errors. A large number of samples (1075 mosquitoes) collected in western Kenya between 2011 and 2015 whose species ID was previously assigned using the 28S IGS gel-based method [11] were re-examined using the melt-curve technique. A subset of 24 individuals identified as An. gambiae with the 28S method and An. arabiensis according to the melt-curves plus all 10 An. arabiensis re-scored as An. gambiae were repeated with both the gel-based SINE200 method [10] and the 28S method [11]
The SINE200 region was later investigated in-silico to predict the performance of the assay for mosquitoes collected throughout Sub-Saharan Africa. For An. gambiae and An. coluzzii, the primer binding sites were examined in the phase 1 data of the Anopheles gambiae 1000 genome project (https://www.malariagen.net/apps/ag1000g/), covering West and East African populations. To predict whether the method can also be used for West-African An. arabiensis, sequence runs for 9 An. arabiensis individuals from Cameroon, and 10 from Burkina Faso were examined (S1 Table). Raw sequence reads were mapped against the SINE200 region of the AaraD1 assembly using Geneious 10 (Biomatters Ltd.) and examined for polymorphisms.
Results
Species identification
SINE PCR
The sixty samples selected, including 10 An. arabiensis, 20 An. gambiae, 14 An. coluzzii and 15 hybrids of both species and one pooled artificial hybrid were identified using SINE PCR. Gel electrophoresis analyses of SINE PCR products showed distinct diagnostic bands corresponding to each species. Anopheles arabiensis, An. coluzzii and An. gambiae showed specific band sizes of 315 bp, 479 bp and 249 bp respectively, while the An. gambiae/An. coluzzii hybrids showed both amplicon sizes (Fig 1).
Fig 1. 2.0% agarose gel for SINE PCR showing An. arabiensis, An. gambiae and An. coluzzii.
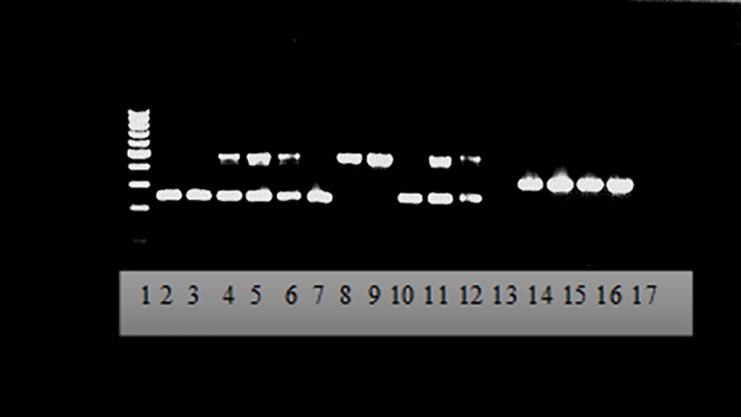
Lane 1: molecular weight ladder (100 bp); Lane 2; 3 & 10: An. gambiae, Lane 4–6 & 11–12: hybrids An. coluzzii / An. gambiae; Lane 8 & 9: An. coluzzii, Line 14–17: An. arabiensis. DNA fragments of 315 bp for An. arabiensis, 249 bp (An. gambiae) and 479 bp (An. coluzzii). Hybrid showed both band sizes at 249 bp and 479 bp.
SYBR green real-time PCR
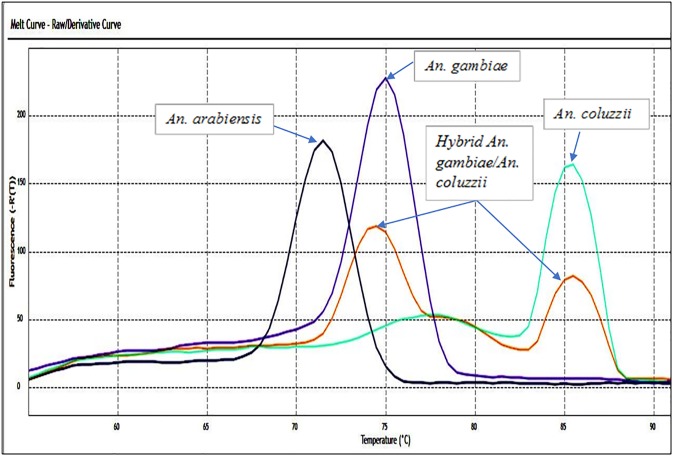
The identification of all the species and hybrid was based on specific melting temperatures (Tm) from the dissociation curves (Fig 2). Anopheles arabiensis showed a single peak at an average temperature of 72°C, An. gambiae at 75°C; whilst An. coluzzii peaks at 86°C (Fig 2). The hybrid showed the expected 86°C melting peak for An. coluzzii and shifted slightly for An. gambiae (74°C) instead of the expected 75°C. The pooled An. gambiae—An. coluzzii DNA produced a melt-curve identical to hybrids. Results were consistent between the two Real-time PCR machines and between alternative SYBR green qPCR master mixes, except for a slight shift (±1°C) in species-specific melt peaks between the machines. This “machine effect” demonstrates the importance of using positive controls for the three species to calibrate the species-specific peak temperature intervals when using different real-time machines (S1–S3 Figs).
Fig 2. Dissociation curves of all species (An. gambiae, An. coluzzii, hybrid An. gambiae / An. coluzzii and An. arabiensis) using SYBR green high throughput methods.
Re-evaluation of 1075 mosquitoes from western Kenya previously screened with 28S IGS revealed an inconsistency rate of 36% between the gel-based method and dissociation curve method. Out of 354 mosquitoes initially scored as An. arabiensis, 10 were identified as An. gambiae using the melt-curve technique, and 373 out of 721 assigned An. gambiae produced an An. arabiensis-specific melt peak. To investigate whether the inconsistently was caused by wrongly scored 28S IGS gels or by unreliable melt curves, a subset of inconsistent individuals was re-examined using both SINE200 and the 28S IGS gel methods allowing the gels to run sufficiently long to reliably distinguish the diagnostic bands (described in the Method section). The re-examined gel-based scores were now fully consistent with the melt-curve species identifications. Only long gel electrophoresis times showed distinct differences in band sizes between An. gambiae and An. arabiensis for both methods (S4 Fig).
In-silico validation of the assay showed two SNPs coinciding with SINE200Fa primer binding site and one in the SINE200Rd binding site based on the Anopheles gambiae 1000 genome data. The allele frequencies of these SNPs are 0.07%, 0.07%, and 0.20% and are therefore expected to have a negligible effect on assay performance. The primer sites as well as the sequence between them (i.e. the whole amplicon region) was completely conserved and identical to the AaraD1 assembly in the West-African An. arabiensis sequence runs. The in-silico validation therefore shows that the assay is expected to work across a wide range of populations and for all three species.
Discussion
Rapid and reliable identification of species and sub-species of malaria vector populations is an important part of malaria vector control programmes. PCR-RFLP and SINE PCR methods have been developed for this purpose and have both shown to successfully identify An. arabiensis, An. gambiae and An. coluzzii out of the complex of eight species [4, 10, 11]. However, both methods involve at least two PCR steps with gel staining that require precision and time to identify the species. In addition, inconsistent identification of An. gambiae, An. coluzzii and their hybrids has been reported by either PCR using form-specific primers or PCR-RFLP genotyping carried out in different laboratories [16]. Mis-identification of An. gambiae vs An. arabiensis based on gel-bands is even more likely since both the SINE200 and the 28S IGS techniques produce similar-sized amplicon sizes (223 vs 249 bp and 315 bp vs 390 bp respectively) which only separate when gels are run sufficiently long (the An. gambiae /An. coluzzii distinctive HhaI digest [11] is usually not performed in Eastern Africa). The 36% error rate in western Kenyan samples clearly demonstrates the risk of mis-interpreting bands on gels, and consequently the value of the melt-curve approach described here. From Scott et al. 1993 to date, SINE PCR has shown greater reliability among all the protocols allowing the differentiation of An. gambiae and An. coluzzii. Moreover, each species identification protocol allows partial identification of the whole An. gambiae complex (Table 2). In addition to the benefits outweighed given the fastest and more reliable results, the current SYBR green melt-curve technique allows the full identification of the three main malaria vectors, An. gambiae, An. coluzzii and An. arabiensis species simultaneously.
Table 2. Summary of the different protocols for the identification of An. gambiae s.l. complex.
| METHODS | STEPS | DIAGNOSIS | Identification Methods | REFERENCE | |
|---|---|---|---|---|---|
| STEP 1 | STEP 2 | ||||
| Sequencing ITS2 region | ITS2 (Internal transcribed spacers) & COI (cytochrome oxidase sub unit 1) | An. gambiae s.s. An. quadriannulatus, An. arabiensis & An. funestus s.s | DNA sequence | Lobo et.al 2015 | |
| MULTIPLEX RT-PCR | Taqman SNP genotyping + LNA(locked nucleic acid) | An. gambiae s.s., An. melas, An. merus, An. quadriannulatus & An. arabiensis | Fluorescent / Melting temperature | Bass et al, 2008. | |
| SINE PCR* | Conventional PCR | An. gambiae, An. coluzzii & An. arabiensis | Gel electrophoresis | Santolamazza et.al 2008 | |
| Taqman RT-PCR | RT-PCR 28S IGS Taqman | Taqman SNP genotyping using Walker et al, 2007 | An. gambiae s.s., An. melas, An. merus, An. quadriannulatus & An. arabiensis | Fluorescent | Bass et al, 2007 |
| RT-PCR | Taqman SNP genotyping | An. gambiae s.s, An. arabiensis An. quadriannulatus as a false positive for An. gambiae | Fluorescent/Melting temperature | Walker et.al 2007 | |
| Multiplex PCR* | Conventional PCR | An. quadriannulatus, An. melas/An. merus, An. gambiae, An. arabiensis & An. coluzzii | Gel electrophoresis | Wilkins et al, 2006 | |
| Multiplex PCR | Conventional PCR | An. quadriannulatus | Gel electrophoresis | Fettene et.al 2003 | |
| Normal PCR | Conventional PCR | An. quadriannulatus B | Gel electrophoresis | Fettene et al, 2002 | |
| PCR-RFLP* | Conventional PCR | RFLP | An. gambiae, An. coluzzii, An. melas, An. merus, An. quadriannulatus & An. arabiensis | Gel electrophoresis | Fanello et al, 2002 |
| Normal PCR* | Conventional PCR | An. gambiae, An. coluzzii, hybrid & Bamako form | Gel electrophoresis | Favia et al, 2001 | |
| PCR-RFLP* | Conventional PCR | RFLP; 28S IGS using Hha1 or Tru91 enzyme | An. gambiae, An. coluzzii & Bamako form | Gel electrophoresis | Favia et al, 1997 |
| Normal PCR | Conventional PCR | An. bwambae | Gel electrophoresis | Townson et al 1994 | |
| Normal PCR | Conventional PCR | An. gambiae s.s, An. melas, An. merus, An. arabiensis & An. quadriannulatus | Gel electrophoresis | Scott et.al 1993 | |
*Represents the protocols enabling the identification of An. gambiae and An. coluzzii
The aim of this study was to demonstrate a SYBR green, rapid, high throughput assay capable of identifying An. arabiensis, An. gambiae and An. coluzzii with high specificity and precision. The high throughput SYBR green rapid assay described here shows great specificity. Though the existing protocols have proven to be useful tools for species identification, this new high throughput assay does not require post PCR analyses such as restriction enzyme digestion and gel electrophoresis associated with PCR-RFLP and SINE PCR [10, 11].
Hence, this new assay showed the first use of SYBR green designed set of primer sequences to distinguish An. arabiensis, An. gambiae and An. coluzzii by Real-Time melt-curve analysis. The newly developed method together with Taqman qPCR methods [27] will allow complete characterization of mosquito specimen into species using Real-Time PCR. Compared to the existing method, this assay is faster, and the closed-tube reaction (without gel electrophoresis) reduces the risk of post-PCR contamination. The assay designed for basic melt-curve analysis (rather than high-resolution melt-curve) allows the use on a wide range of real-time PCR machines.
Conclusion
The SYBR green real time PCR techniques showed an additional option for the characterization of both sibling species and An. arabiensis while all the other real time PCR protocol of species differentiation could not allow the characterization of the sub-species such as An. gambiae and An. coluzzii. The assay designed in this study is a new tool to help researchers and particularly malaria vector control entities to identify clearly the subspecies within the An. gambiae s.l. complex in the context where each of the species has unique behaviour and impact on malaria.
Supporting information
(TIF)
(TIF)
A: Dissociation curves using Brilliant III Ultra-Fast SYBR Green Low ROX QPCR Master Mix. B: Dissociation curves using Luna Universal qPCR Master Mix “Any difference was observed using the same machine and different SYBR green master mix”.
(TIF)
“Samples with inconsistent species ID scores between the 28S gel analysis and the melt-curve technique were re-examined using both the SINE method and the 28S methods. A subset of 24 out of 373 potentially misidentified An. arabiensis, and all 10 potentially misidentified An. gambiae were included. The gel pictures presented here show 5 An. gambiae (Line 1–5) and 16 An. arabiensis (Line 6–21) according to the melt-curves (but scored different previously). Pictures were taken at two intervals; 33 minutes and 80 minutes for the 28S PCRs and 40 and 120 minutes for the SINE200 PCRs (electrophoresis at 120 volts). Size differences are very similar for An. gambiae and An. arabiensis at the shorter run times, making it difficult to reliably assign species ID. The longer gel runs confirm the melt-curve species ID”.
(TIF)
(DOCX)
(XLSX)
Acknowledgments
We would like to acknowledge the contribution of Godwin Amlalo, Dorothy Obuobi and Rebecca Pwalia of Noguchi Memorial Institute for Medical Research (NMIMR) for their support on part of the laboratory works undertaken in Ghana. We thank David Weetman for his valuable comments on an early version of the manuscript.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The work performed in Liverpool was supported by the National Institute of Allergy and Infectious Diseases funds (NIAID. R01-AI116811) and the Medical Research Council UK (MR/P02520X/1). Vestergaard SA supported the work in Ghana in the form of salaries for authors (JC, LKN, DO, AD) and provided laboratory spaces, equipment and reagents, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Coluzzi M, Sabatini A, Petrarca V, Di Deco MA. Chromosomal differentiation and adaptation to human environments in the Anopheles gambiae complex. Trans R Soc Trop Med Hyg. 1979;73(5):483–97. [DOI] [PubMed] [Google Scholar]
- 2.Coluzzi M, Sabatini A, della Torre A, Di Deco MA, Petrarca V. A polytene chromosome analysis of the Anopheles gambiae species complex. Science. 2002;298(5597):1415–8. 10.1126/science.1077769 [DOI] [PubMed] [Google Scholar]
- 3.Townson H, Onapa AW. Identification by rDNA-PCR of Anopheles bwambae, a geothermal spring species of the An. gambiae complex. Insect molecular biology. 1994;3(4):279–82. [DOI] [PubMed] [Google Scholar]
- 4.Scott JA, Brogdon WG, Collins FH. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993;49(4):520–9. [DOI] [PubMed] [Google Scholar]
- 5.Fettene M, Koekemoer LL, Hunt RH, Coetzee M. PCR assay for identification of Anopheles quadriannulatus species B from Ethiopia and other sibling species of the Anopheles gambiae complex. Medical and veterinary entomology. 2002;16(2):214–7. [DOI] [PubMed] [Google Scholar]
- 6.Wilkins EE, Howell PI, Benedict MQ. IMP PCR primers detect single nucleotide polymorphisms for Anopheles gambiae species identification, Mopti and Savanna rDNA types, and resistance to dieldrin in Anopheles arabiensis. Malaria journal. 2006;5:125 10.1186/1475-2875-5-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.della Torre A, Fanello C, Akogbeto M, Dossou-yovo J, Favia G, Petrarca V, et al. Molecular evidence of incipient speciation within Anopheles gambiae s.s. in West Africa. Insect Mol Biol. 2001;10(1):9–18. [DOI] [PubMed] [Google Scholar]
- 8.Coetzee M, Hunt RH, Wilkerson R, Della Torre A, Coulibaly MB, Besansky NJ. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa. 2013;3619:246–74. [PubMed] [Google Scholar]
- 9.Coetzee M. Distribution of the African malaria vectors of the Anopheles gambiae complex. Am J Trop Med Hyg. 2004;70(2):103–4. [PubMed] [Google Scholar]
- 10.Santolamazza F, Mancini E, Simard F, Qi Y, Tu Z, della Torre A. Insertion polymorphisms of SINE200 retrotransposons within speciation islands of Anopheles gambiae molecular forms. Malaria journal. 2008;7:163 10.1186/1475-2875-7-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fanello C, Santolamazza F, della Torre A. Simultaneous identification of species and molecular forms of the Anopheles gambiae complex by PCR-RFLP. Medical and veterinary entomology. 2002;16(4):461–4. [DOI] [PubMed] [Google Scholar]
- 12.Oliveira E, Salgueiro P, Palsson K, Vicente JL, Arez AP, Jaenson TG, et al. High levels of hybridization between molecular forms of Anopheles gambiae from Guinea Bissau. J Med Entomol. 2008;45(6):1057–63. [DOI] [PubMed] [Google Scholar]
- 13.Lanzaro GCL, Y. Speciation in Anopheles gambiae. The Distribution of Genetic Polymorphism and Patterns of Reproductive Isolation among Natural Populations. Infectious Diseases,. 2013; 10.5772/56232. [DOI]
- 14.Marsden CD, Lee Y, Nieman CC, Sanford MR, Dinis J, Martins C, et al. Asymmetric introgression between the M and S forms of the malaria vector, Anopheles gambiae, maintains divergence despite extensive hybridization. Molecular ecology. 2011;20(23):4983–94. 10.1111/j.1365-294X.2011.05339.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nwakanma DC, Neafsey DE, Jawara M, Adiamoh M, Lund E, Rodrigues A, et al. Breakdown in the process of incipient speciation in Anopheles gambiae. Genetics. 2013;193(4):1221–31. 10.1534/genetics.112.148718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santolamazza F, Caputo B, Calzetta M, Vicente JL, Mancini E, Petrarca V, et al. Comparative analyses reveal discrepancies among results of commonly used methods for Anopheles gambiae molecular form identification. Malaria journal. 2011;10:215 10.1186/1475-2875-10-215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diabate A, Dabire R.K., Millogo N. and Lehmann T. Evaluating the effects of postmating isolation between molecular forms of Anopheles gambiae (Diptera: Culicidae). J Med Entomol. 2007;44:60–4. [DOI] [PubMed] [Google Scholar]
- 18.Chabi J. Studies on hybridization, gene flow and resistance status of M and S molecular forms of Anopheles gambiae s.s. MPhil thesis. 2016; http://hdl.handle.net/123456789/8367(17-Mar-2016).
- 19.della Torre A, Tu Z, Petrarca V. On the distribution and genetic differentiation of Anopheles gambiae s.s. molecular forms. Insect Biochem Mol Biol. 2005;35(7):755–69. 10.1016/j.ibmb.2005.02.006 [DOI] [PubMed] [Google Scholar]
- 20.Awolola TS, Oyewole IO, Amajoh CN, Idowu ET, Ajayi MB, Oduola A, et al. Distribution of the molecular forms of Anopheles gambiae and pyrethroid knock down resistance gene in Nigeria. Acta tropica. 2005;95(3):204–9. 10.1016/j.actatropica.2005.06.002 [DOI] [PubMed] [Google Scholar]
- 21.Favia G, della Torre A, Bagayoko M, Lanfrancotti A, Sagnon N, Toure YT, et al. Molecular identification of sympatric chromosomal forms of Anopheles gambiae and further evidence of their reproductive isolation. Insect Mol Biol. 1997;6(4):377–83. [DOI] [PubMed] [Google Scholar]
- 22.Ho P, Ng M, Chu J. Establishment of one-step SYBR green-based real time-PCR assay for rapid detection and quantification of chikungunya virus infection. Virology Journal. 2010;7(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okoye PN, Wilson MD, Boakye DA, Brown CA. Impact of the okyereko irrigation project in Ghana on the risk of human malaria infection by anopheles species (diptera: Culicidae). African Entomology. 2005;13(2):249–53. [Google Scholar]
- 24.Charlwood JD, Tomas EV, Egyir-Yawson A, Kampango AA, Pitts RJ. Feeding frequency and survival of Anopheles gambiae in a rice-growing area in Ghana. Med Vet Entomol. 2012;26(3):263–70. 10.1111/j.1365-2915.2011.00987.x [DOI] [PubMed] [Google Scholar]
- 25.Chabi J, Baidoo PK, Datsomor AK, Okyere D, Ablorde A, Iddrisu A, et al. Insecticide susceptibility of natural populations of Anopheles coluzzii and Anopheles gambiae (sensu stricto) from Okyereko irrigation site, Ghana, West Africa. Parasites & vectors. 2016;9(1):182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collins FH, Mendez MA, Rasmussen MO, Mehaffey PC, Besansky NJ, Finnerty V. A Ribosomal RNA Gene Probe Differentiates Member Species of the Anopheles gambiae Complex. Am J Trop Med Hyg. 1987;37(1):37–41. [DOI] [PubMed] [Google Scholar]
- 27.Bass C, Williamson MS, Field LM. Development of a multiplex real-time PCR assay for identification of members of the Anopheles gambiae species complex. Acta Trop. 2008;107(1):50–3. 10.1016/j.actatropica.2008.04.009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
A: Dissociation curves using Brilliant III Ultra-Fast SYBR Green Low ROX QPCR Master Mix. B: Dissociation curves using Luna Universal qPCR Master Mix “Any difference was observed using the same machine and different SYBR green master mix”.
(TIF)
“Samples with inconsistent species ID scores between the 28S gel analysis and the melt-curve technique were re-examined using both the SINE method and the 28S methods. A subset of 24 out of 373 potentially misidentified An. arabiensis, and all 10 potentially misidentified An. gambiae were included. The gel pictures presented here show 5 An. gambiae (Line 1–5) and 16 An. arabiensis (Line 6–21) according to the melt-curves (but scored different previously). Pictures were taken at two intervals; 33 minutes and 80 minutes for the 28S PCRs and 40 and 120 minutes for the SINE200 PCRs (electrophoresis at 120 volts). Size differences are very similar for An. gambiae and An. arabiensis at the shorter run times, making it difficult to reliably assign species ID. The longer gel runs confirm the melt-curve species ID”.
(TIF)
(DOCX)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.