Summary
Repair of tissue damaged during inflammatory processes is key to return of local homeostasis and restoration of epithelial integrity. Here we describe CD161+ regulatory T cells (Treg cells) as a distinct, highly suppressive, population of Tregs mediating wound-healing. These Tregs were enriched in intestinal lamina propria, particularly in Crohn’s disease. CD161+ Tregs had an all-trans retinoic acid (ATRA)-regulated gene signature and CD161 expression on Tregs was induced by ATRA, which directly regulated the CD161 gene. CD161 was co-stimulatory and ligation with the T cell receptor induced cytokines that accelerated wound-healing of intestinal epithelial cells. We identified a transcription factor network, including BACH2, RORγt, FOSL2, AP-1 and RUNX1, controlling expression of the wound-healing program and found that a CD161+ Treg signature in Crohn’s disease mucosa associated with reduced inflammation. These findings identify CD161+ Tregs as a population involved in controlling the balance between inflammation and epithelial barrier healing in the gut.
Introduction
Regulatory T cells (Treg cells) are a non-redundant, suppressive, subset of CD4+ helper T (TH) cells critical for preventing autoimmunity and ideal for cell-based immunotherapy of autoimmunity and prevention of transplant rejection1. Treg cells express the master transcription factor FOXP3, the IL-2 receptor component CD25, the inhibitory co-receptor CTLA42 and depend on the transcription factor BACH23. Treg cells are thymically (tTreg cells) and peripherally (pTreg cells) derived and can also be induced in vitro (iTreg cells). There are no universally accepted ways to differentiate these populations although expression of Helios, Neuropilin and methylation status of the Treg-specific demethylation region (TSDR) have been proposed4–8.
Conventional T cells (Tconv) express their own master transcription factors; these include T-BET+ TH1 cells, GATA3+ TH2 cells and RORγt+ TH17 cells, respectively9. These transcription factors were considered to antagonize Treg development: in mice, induction of high T-bet expression in Treg cells within inflamed bowel drives Tregs into a pro-inflammatory phenotype reminiscent of Th1 cells10. This view has been challenged by specific deletions of these factors specifically within Foxp3+ cells of mice11–14. For example, T-bet expression within Foxp3+ Treg cells is required for trafficking to and suppression of Th1-mediated inflammation13, and Gata3 is required for full Treg function in the gut14. These findings support a ‘compartmentalized’ view of Treg cells, suggesting multiple sub-populations defined by expression of transcription factors associated with Tconv lineages and by specialized functions. Indeed, the transcription factor circuitry of Treg cells is complex, with significant interplay between Foxp3 and other lineage-associated transcription factors15.
In humans, heterogeneous populations of Treg cells have been reported, although typically defined by surface markers (e.g. CD39, HLA-DR and CD45RA1) rather than transcription factors. Whether these sub-populations have the ability to suppress specific parts of the human immune system has yet to be fully elucidated. Conventional methods to delineate Treg subsets are limited by numbers of markers that can be concurrently used and by biased approaches to data analysis (gating of Treg subsets)16. This has led to conflicting results, with memory Treg cells reported as both non-suppressive17 and highly suppressive18. By contrast, unbiased multi-dimensional analysis can delineate the most suppressive Treg sub-populations, identify new ones and exclude those less likely to be regulatory16.
Inflammatory bowel disease (IBD) represents a complex collection of disorders where aberrant mucosal immune system activation, epithelial barrier dysfunction and microbial dysbiosis contribute to chronic inflammation and unregulated local TH1 and TH17 responses19. Bowel mucosa is a key site for pTreg cell induction from naive CD4+ precursors via instruction from environmental factors, e.g. transforming growth factor (TGF)-β, IL-2 and all-trans retinoic acid (ATRA)20. Treg cells mediate dominant tolerance in gut mucosa, preventing or ameliorating murine colitis on adoptive transfer21. Conversely, FOXP3 mutations or disruption of other key Treg cell molecules (e.g. CTLA-4, IL-10R, TGF-β) cause enteropathy in humans and mice2, demonstrating their key role in preventing gut inflammation. Lamina propria Treg cells increase in number in IBD, but it is unclear why they do not control local inflammation and what function(s) they perform in these diseases22. Treg cells can express RORγt together with IL-17A and, in humans, these factors are restricted to a Treg cell subset expressing CD16123. CD161 is a C-type lectin-like receptor expressed on human NK cells24 and various T lymphocyte subsets25. CD161+ Tconv cells are memory cells acting as TH17 precursors26. The CD161 cognate ligand is lectin-like transcript 1 (LLT1)27. Single nucleotide polymorphisms associate with IBD in genome-wide association studies28, suggesting that the CD161-LLT1 interaction is physiologically important.
Here, we delineate the biological repertoire of CD161+ Treg cells, their role in the immune system and their mechanisms of action. Our data show that CD161+ Treg cells are a highly suppressive, distinct subset of induced Treg cells that accelerate wound healing of colorectal epithelium via production of soluble factors in a BACH2-dependent manner.
Results
CD161-expressing Treg cells are a discrete population with a distinct TCRVβ repertoire
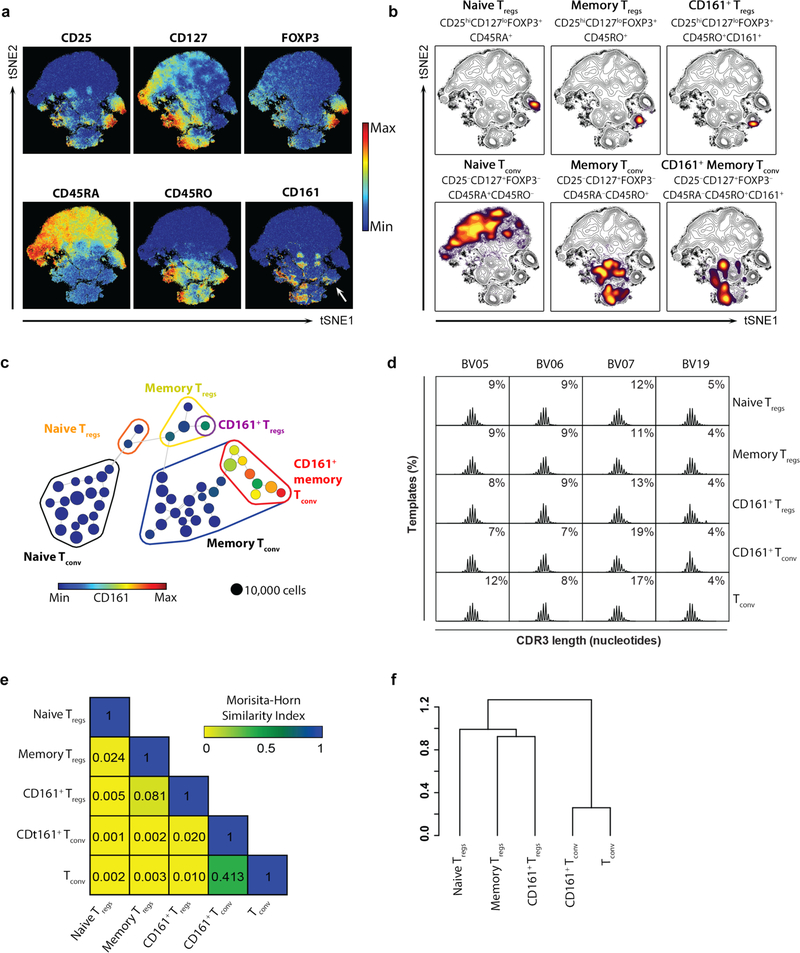
We used an unbiased multi-dimensional analysis pipeline via cytometry by time-of-flight (CyTOF) to identify and study biologically important human Treg sub-populations. Visualized stochastic neighbor embedding (viSNE) was used to create a map of CD4+ T cells from blood and arrange cells along t-distributed stochastic neighbor embedding (t-SNE) axes based on per-cell phenotypic similarity16,29 (Fig. 1a–b). Treg cells, identified by high expression of CD25 and FOXP3 and low expression of CD127, clustered together and could be resolved into naive and memory populations (Fig. 1a–b). Likewise, Tconv, identifiable by low CD25 and FOXP3 and high CD127 expression, clustered together and distinctly from Treg cells (Fig. 1a–b). In this unsupervised analysis, the C-type lectin CD161 was expressed by a sub-population of memory Treg cells (CD4+CD25+CD127loFOXP3+CD45RA–CD45RO+) and a group of memory Tconv (broadly CD4+CD25–CD127+FOXP3–CD45RAloCD45RO+) (Fig. 1a–b). To identify distinct T cell clusters, we performed a spanning-tree progression analysis of density-normalized events (SPADE)30 based on t-SNE values. Differential expression of markers within each identified SPADE node was used to further cluster T cells. This analysis resolved CD4+ T cells into 50 sub-populations, grouped into four main populations: naive Treg cells, memory Treg cells, naive Tconv and memory Tconv, characterized by different expression profiles (Fig. 1c and Supplementary Fig. 1a). CD161-expressing cells represented a distinct sub-population of memory Treg cells and several sub-populations of memory Tconv, which were all distinct from Treg cells (Fig. 1c). To further distinguish Treg cells from Tconv, we performed similar analysis of transcriptomes by single-cell RNA-seq using CD4+CD25+ cells as input (Supplementary Figs. 1b–e). This pipeline resolved cells into 9 clusters, of which two were Treg cells (clusters 0 and 3), five were Tconv (clusters 1, 2, 4, 5 and 6) and two were probably Treg cells although too small in number to sub-categorize (clusters 7 and 8) (Supplementary Figs. 1b–e). The two Treg clusters (clusters 0 and 3) were clearly separate from the Tconv clusters and one of them (cluster 3) expressed KLRB1, the gene encoding CD161 (Supplementary Figs. 1b–e). Consistent with CyTOF, expression of KLRB1 in CD161+ Tregs was lower than the CD161+ Tconv sub-population. A heatmap of Treg markers (IL2RA, IL7R, FOXP3), KLRB1 and naive or memory markers (CD62L and CCR7) confirmed clustering of the CD161+ Treg cells (cluster 3) independently from Tconv cells (Supplementary Fig. 1e) and was similar to heatmaps constructed from protein expression by CyTOF (Supplementary Fig. 1f). A similar unsupervised pipeline applied to CD4+ T cells flow-stained with just 5 markers, CD4, CD25, CD127, CD45RA and CD161 produced similar viSNE and SPADE plots to CyTOF (Supplementary Fig. 1g–h) and could be used to flow-sort peripheral blood Treg sub-populations for further analysis (Supplementary Figs. 1i–j). Henceforth, these Treg cells are referred to as ‘CD161+ Treg cells’ (CD4+CD25hiCD127loCD45RA–CD161+; denoted in purple), ‘naive Treg cells’ (CD4+CD25hiCD127loCD45RA+CD161–; denoted in orange) and CD161– memory Treg cells, abbreviated to ‘memory Treg cells’ (CD4+CD25hiCD127loCD45RA–CD161–; denoted in yellow).
Figure 1. CD161+ Treg cells are a discrete population of memory Treg cells.
(a) viSNE plots of CD4+ T cells clustered using surface and intracellular markers. Shown are heatmaps for expression of indicated markers. White arrow highlights expression of CD161 within Treg cells; (b) overlaid contour plots of T cell subsets, colored by density, to highlight sub-populations of Treg cells and Tconv; (c) 2D minimum spanning tree showing population nodes of CD4+ T cells. Node size represents cell number and color CD161 median intensity. Grouped together are naive (circled in orange), memory (circled in yellow) and CD161+ (circled in purple) Treg cells, as well as populations of naive (circled in black), memory (circled in black) and CD161+ (circled in red) Tconv; (a-c) show representative data from n=3 experiments; (d) representative spectratype histograms from n=3 experiments showing percentage of unique CDR3 sequences (templates) versus CDR3 length for the three highest (BV05, BV06 and BV07) and one of the lowest (BV19) TCRBV families contributing to the overall TCR repertoire in the indicated populations; (e) average Morisita-Horn Similarity Index of total TCRVB repertoire between the T cell populations (cumulative data from n=3 experiments); and (f) dendrogram showing linkage distance based on the Morisita-Horn Similarity Index.
T cell CD161 expression is associated with a restricted range of T cell receptors (TCRs): Vα7.2 in mucosal-associated invariant T (MAIT) cells25 and invariant Vα24-Jα18 in iNKT cells31. Neither TCR was significantly enriched in CD161+ Treg cells (Supplementary Fig. 1k). To determine clonality of CD161+ Treg cells and their relationship to other Treg cells and Tconv, we TCRVB sequenced the three Treg cells populations, as well as Tconv (defined as CD4+CD25–CD127+) and CD161+ Tconv (defined as CD4+CD25–CD127+CD45RO+CD161+) as controls. Among the TCRBV families, the three highest contributors to the total TCR repertoire (BV05, BV06 and BV07) and one of the lower contributors (BV19) were chosen to represent variability of the cellular repertoire by spectratyping. CD161+ Treg cells were polyclonal, had normally distributed CDR3 length in the different TCRBV families (Fig. 1d) and similar contribution to the overall TCR repertoire compared with other T cell subsets (Supplementary Fig. 1l). We asked whether the TCR repertoire of CD161+ Treg cells overlapped with either the Treg or Tconv populations and found limited clonality shared between the different subsets; in particular, CD161+ Treg cells shared only 4.5% and 4.8% of TCR sequences with Memory Treg cells and CD161+ Tconv cells, respectively (Supplementary Fig. 1m). We calculated the Morisita-Horn similarity index to measure TCR composition overlap between the different T cell populations32 - this index ranges between 0 (minimal similarity) and 1 (maximal similarity). Limited TCR repertoire overlap existed between any of the populations other than between CD161+ Tconv and Tconv (a value of 0.413) (Fig. 1e). Hierarchical clustering, based on the Morisita-Horn similarity index, indicated two subdivisions, separating the two Tconv populations from all three Treg populations; moreover, CD161+ Treg cells clustered with memory Treg cells within the regulatory T cell main branch (Fig. 1f). These data indicate that CD161+ Treg cells have distinct TCR repertoires from other Treg cells and do not represent a clonal expansion from a Tconv population.
CD161+ Treg cells share features with both CD161+ Tconv and classical Treg cells
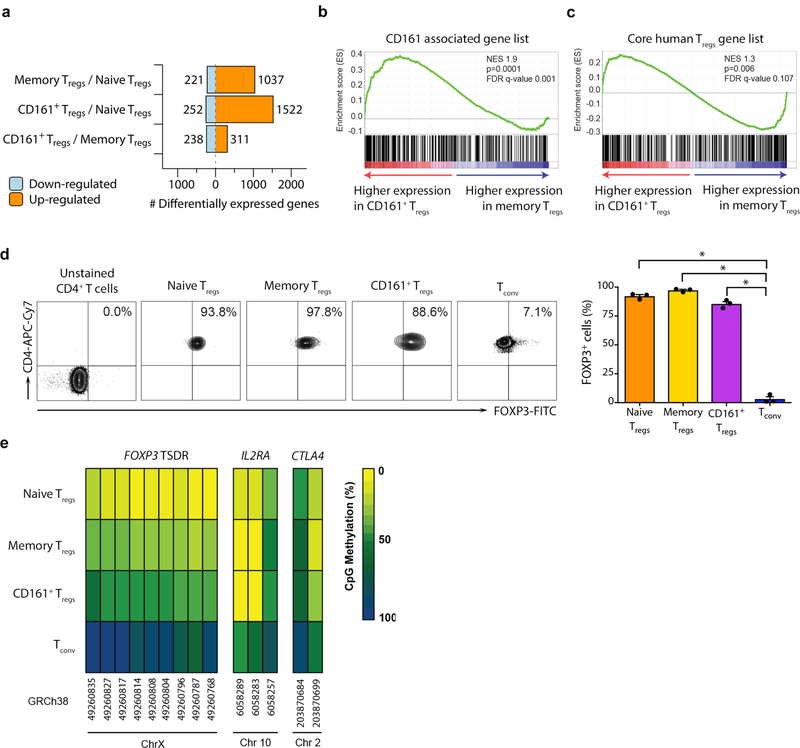
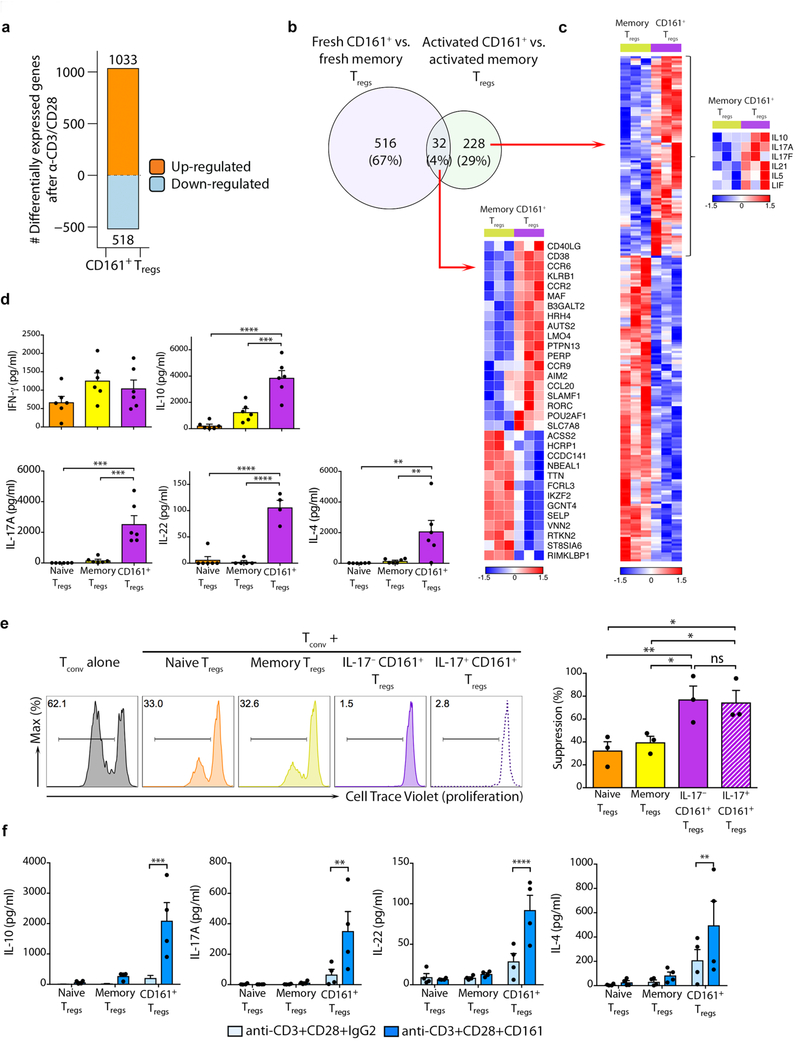
To identify similarities and differences between the 3 sub-populations, we compared their transcriptomes using microarray and Gene Set Enrichment Analysis (GSEA) (Supplementary Fig. 2a–d). Despite CD161+ Treg cells being a sub-population of memory Treg cells, there were 549 genes differentially expressed when comparing CD161+ to memory Treg cells (Fig. 2a, Supplementary Fig. 2b–d and Supplementary Table 1). CD161+ Treg cells were enriched in genes expressed by other CD161+ cells25 and also expressed the core transcriptional profile of Treg cells33 (Fig. 2b–c and Supplementary Fig. 2e–f). This data suggested that, in addition to core Treg genes they express a range of other transcripts related to CD161 induction or signaling.
Figure 2. CD161+ Treg cells have classical features of bona fide Treg cells.
(a) number of differentially expressed genes between the sub-populations of freshly isolated Treg cells; (b-c) Gene Set Enrichment Analysis (GSEA) plots for genes associated with other CD161+ cells (b) and core human Treg signature genes (c) comparing freshly isolated memory to CD161+ Treg cells (n=3 per group); NES = normalized enrichment score; empirical p- and multiple test adjusted q-values from GSEA are shown; (d) FOXP3 expression by sub-populations of Treg cells and Tconv, showing representative flow cytometry plots (left) and cumulative data (mean + sem; right); (e) mean percentage CpG methylation of conserved CpGs (with chromosomal coordinates) at the FOXP3 TSDR, IL2RA and CTLA4 loci of naive, memory, CD161+ Treg cells and Tconv of 3 male donors. n=3 independent experiments in a-e; *p<0.0001 by one-way ANOVA.
All three Treg populations had similar FOXP3 protein expression (Fig. 2d). We measured the methylation status of the FOXP3 TSDR and two other key Treg-associated genes, IL2RA and CTLA4, in the three Treg populations and, for comparison, Tconv (Supplementary Fig. 2g). Methylation at IL2RA and CTLA4 loci were similar between the three Treg populations but distinct from the more highly methylated Tconv (Fig. 2e and Supplementary Fig. 2h). At the FOXP3 TSDR we noted CD161+ Treg cells to have methylation intermediate (55%) between naive (25%) or memory (28%) Treg cells and Tconv (88%) (Fig. 2e and Supplementary Fig. 2h), a figure similar to that seen in studies that compared Helios+ Treg cells with Helios– Treg cells in humans34. In summary, CD161+ Treg cells express the Treg master transcription factor FOXP3, the same surface markers as Treg cells and the Treg gene transcription profile. Their methylation pattern at key gene loci are similar to other Treg cells, albeit with intermediate TSDR methylation.
ATRA directly regulates CD161 expression
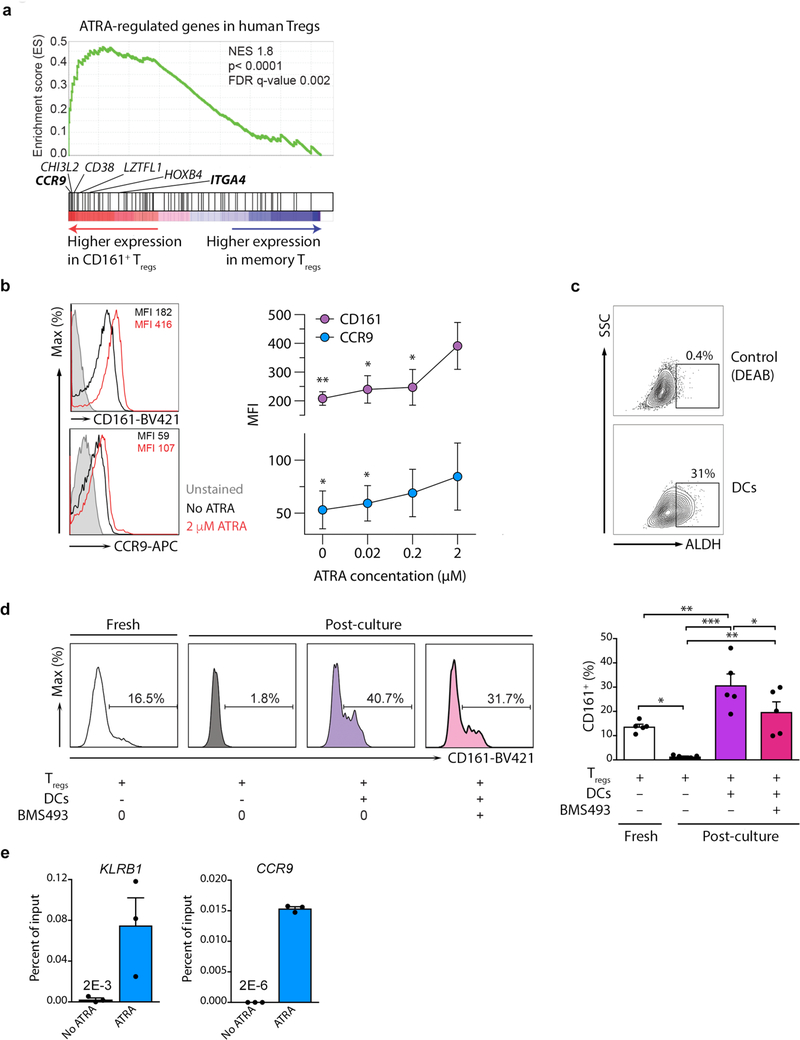
Long-term in vitro culture of Treg cells with ATRA supports persistence of cells expressing CD16135. CD161+ Treg cells were enriched in ATRA-regulated genes compared to memory Treg cells (Fig. 3a) by GSEA, including two gut homing markers, CCR9 and ITGA4, which are classical ATRA-regulated genes (Fig. 3a and Supplementary Fig. 3a); CCR9 was confirmed at the protein level by flow cytometry (Supplementary Fig. 3b). Activation of freshly-isolated Treg cells with ATRA induced expression of both CCR9 and CD161 (Fig. 3b). Since Treg cells interact with dendritic cells (DCs) in the gut, we determined if mature DCs generate ATRA via a functional assay for the key enzyme aldehyde dehydrogenase (ALDH). Lipopolysaccharride (LPS)-matured DCs expressed substantial ALDH enzyme (Fig. 3c) together with lectin like transcript 1 (LLT1), the natural ligand for CD161 (data not shown)27. In the absence of DCs, Treg cells lost CD161 expression; whereas co-culture of freshly-isolated Treg cells with DCs induced CD161. Addition of BMS493, a pan-retinoic acid receptor (RAR) inverse agonist blocked CD161 expression in a dose-dependent manner (Fig. 3d and Supplementary Fig. 3c). To see whether ATRA directly regulates CD161, we scanned the KLRB1, which encodes CD161, and CCR9 gene loci (as control) for retinoic acid receptor alpha (RARA) DNA binding motifs (Supplementary Fig. 3d). One potential RARA site in KLRB1 and multiple sites in CCR9 were identified (Supplementary Fig. 3e). By RARA ChIP-qPCR we found that ATRA enhanced RARA binding at both loci in T cells, evident as significant increment in percentage of input for both KLRB1 and CCR9 target sequences after culture with ATRA, compared to untreated cells (Fig. 3e). Collectively, these data indicate that ATRA can induce CD161 expression on Treg cells.
Figure 3. CD161 expression is regulated by retinoic acid.
(a) GSEA for ATRA-regulated genes in human Treg cells comparing freshly isolated memory to CD161+ Treg cells. Classic ATRA-regulated genes within the leading edge of core enriched genes (see Supplementary Fig. 3a) are annotated. In bold are gut homing receptors; n=3 per group; NES = normalized enrichment score; empirical p- and multiple-test adjusted q-values from GSEA are shown; (b) expression of CD161 and CCR9 on Treg cells cultured with and without ATRA for 2 days; shown are representative flow cytometry plots (left) and cumulative data from n=3 experiments (right); p-values indicate comparisons with 2μM ATRA; (c) representative assay of ALDH activity in DCs from n=3 independent experiments; (d) representative flow cytometry plots (left) and cumulative data (right) from n=5 independent experiments showing CD161 expression on Treg cells before (fresh) and after 5 days either with medium alone or co-culture with DCs in the presence or absence of the pan-RAR inverse agonist, BMS493; (e) RARA ChIP-qPCR for binding sites in KLRB1 and CCR9, showing percentage of input; shown are representative examples from n=2 independent experiments. Bar charts show mean + sem throughout; *p<0.05, **p<0.01, ***p<0.001 by one-way ANOVA.
CD161+ Treg cells are a highly suppressive Treg population
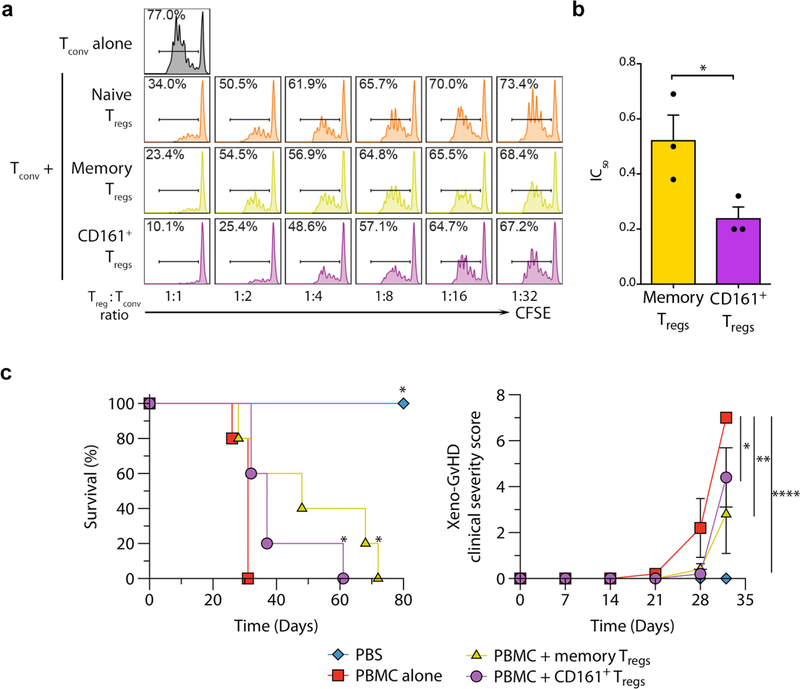
We next tested the ability of CD161+ Treg cells to suppress Tconv proliferation ex vivo by measuring the ratio of Treg cells to Tconv required to suppress proliferation by 50% (IC50) for each population36 (Fig. 4a–b and Supplementary Fig. 4a–b). CD161+ Treg cells had a lower mean IC50 compared to memory Treg cells, whereas naive Treg cells did not reach 50% suppression ex vivo (and therefore IC50 could not be calculated) (Fig. 4b and Supplementary Fig. 4b). CD161+ Treg cells remained stably regulatory even after in vitro proliferation for two weeks (Supplementary Fig. 4c). Separation of Treg cells from target cells by a transwell abrogated the majority of the suppressive function of CD161+ Treg cells (Supplementary Fig. 4d), suggesting contact-dependent effects and consistent with other Treg populations37. The presence of anti-CD161, anti-PD-L1, anti-TGFβRII or anti-IL-10R antibodies caused no significant impairment of CD161+ Treg suppressive function, indicating that suppressive function is likely not due to a single factor (Supplementary Fig. 4e). Furthermore, suppression assays under TH1 and TH17 skewing conditions did not impair regulatory function in CD161+ Treg cells (Supplementary Fig. 2f). CD161 is found on NK cells24 and cytotoxicity is a suppressive mechanism of some Treg cells38. However, we found neither perforin nor granzyme (A or B) in any of the Treg populations (Supplementary Fig. 4g) and no evidence of Tconv cytolysis after co-culture with CD161+ Treg cells (data not shown).
Figure 4. CD161+ Treg cells are regulatory both in vitro and in vivo.
(a-b) In vitro Treg suppression assay showing representative CFSE dilution histograms of Tconv co-cultured with and without Treg cells (a) and cumulative IC50 of memory and CD161+ Treg cells from n=3 experiments (b). Note that only one out of the 3 naive Treg donors tested reached 50% suppression, therefore the mean IC50 was not calculated for the naive Treg population. Bars show mean + sem. (c) Xeno-graft versus host disease with and without 2:1 PBMC : Treg cell injection, showing survival plots (left panel; * p<0.05 compared to PBMC alone) and clinical severity (right panel). Shown is one experiment from two independent experiments carried out with n=5 mice in each group. Error bars indicate mean + sem; *p<0.05, **p<0.01, ***p<0.0001 by t-test (b), long-rank (Mantel-Cox) test (c, left) and two-way ANOVA (c, right).
The mouse CD161 ortholog is not expressed on T cells (Supplementary Table 2). To confirm suppressive ability of CD161+ Treg cells in vivo, we used a humanized mouse model of severe xeno-graft versus host disease (GvHD) in NOD-SCID-Il2rg−/− (NSG) mice by injecting CD25-depleted human peripheral blood mononuclear cells (PBMC) with or without in vitro-expanded memory or CD161+ Treg cells. Mice receiving either memory or CD161+ Treg cells were protected from xeno-GvHD, surviving significantly longer than mice injected with PBMC alone and had significant reduction in clinical disease scores (Fig. 4c). Thus, CD161+ Treg cells had comparable suppressive activity to traditional Treg cells in vivo. These data indicate that CD161+ Treg cells are highly regulatory both in vitro and in vivo.
CD161 ligation stimulates cytokine production in CD161+ Treg cells
We next investigated the specific effects of TCR triggering by examining transcriptomes of Treg cells stimulated with anti-CD3+CD28 for 4 hours. Approximately 1500 transcripts showed changes from baseline in CD161+ Treg cells (Fig. 5a and Supplementary Fig. 5a–b). Although a core set of transcripts, including KLRB1, RORC and CCR9 differed between CD161+ and memory Treg cells both before and after these cells were activated, there was little overlap (Fig. 5b–c). Of note, a number of cytokine genes, including IL10, IL17A, IL17F and IL21 (Fig. 5c), were enriched in CD161+ Treg cells (Supplementary Fig. 5b). We confirmed preferential accumulation of IL-10, IL-17A, IL-22 and IL-4, but not IFN-γ, in three-day supernatants of αCD3+CD28-activated CD161+ Treg cells (Fig. 5d).
Figure 5. CD161 ligation is co-stimulatory and induces cytokine production from CD161+ Treg cells.
(a) differentially expressed genes following 4h stimulation of CD161+ Treg cells with anti-CD3/CD28; (b) venn diagram of transcriptional differences between freshly isolated and in vitro anti-CD3/CD28-activated CD161+ and memory Treg cells; (c) heatmaps of differentially expressed genes common to fresh and activated cells (lower panel) and specific to the activated condition (right panel), with cytokine genes highlighted in inset. Data in a-c are from 3 independent experiments; (d) concentration (pg/ml) of stated cytokines in supernatants of naive, memory and CD161+ Treg cells after 3 days of polyclonal activation with anti-CD3+CD28 (cumulative data from n=6 experiments); (e) suppression assay showing cell trace violet dilution in proliferating Tconv cultured alone or in co-culture with stated populations of Treg cells (Treg cell: Tconv ratio of 1:2). Shown are representative plots (left) and cumulative data from n=3 independent experiments (right); (f) concentrations of stated cytokines in supernatants of FACS-sorted CD161+ Treg cells stimulated for 3 days with either anti-CD3+CD28+IgG2 or anti-CD3+CD28+CD161-coated magnetic beads (cumulative data from n=4 experiments). Bar charts shown mean + sem throughout. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 by one-way (d-e) and two-way ANOVA (f).
CD161 in Tconv is a marker of TH17 cells. Furthermore, CD161+ Treg cells expressed RORC, the key transcription factor of TH17 cells, along with IL-17 itself. We next determined whether IL-17 production was compatible with suppressive function in CD161+ Treg cells, by isolating IL-17+CD161+ and IL-17–CD161+ Treg cells using an IL-17 capture assay (Supplementary Fig. 5c–d) and testing their respective suppressive functions. IL-17+CD161+ Treg cells remained highly suppressive despite producing IL-17 (Fig. 5e).
CD161 does not have classical immunoreceptor tyrosine-based activation or immunoreceptor tyrosine-based inhibitory domains but its ligation can have both activating and inhibitory functions, depending on cell type25,39. To assess the function of CD161 in Treg cells, we stimulated cells with anti-CD3+anti-CD28 magnetic beads additionally coated with anti-CD161 or an IgG2 isotype. After three days of culture CD161 cross-linking significantly enhanced cytokine production from CD161+ Treg cells (Fig. 5f), indicating that CD161 acts as a co-stimulatory molecule in CD161+ Treg cells. Thus, CD161+ Treg cells are a cytokine producing population of Treg cell, CD161 co-ligation is co-stimulatory to this process and IL-17 production is compatible with suppressive function.
Genome-wide chromatin landscapes define regulatory circuitry in CD161+ Treg cells
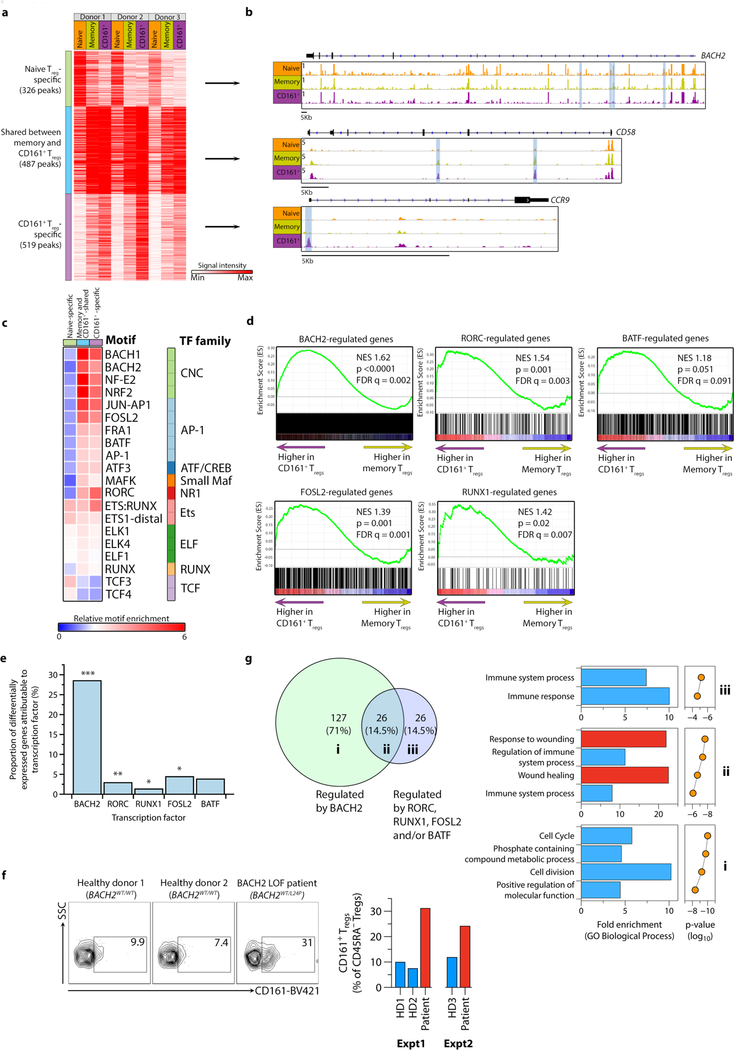
We next examined global chromatin landscapes of naive, memory and CD161+ Treg cells directly ex vivo using high-throughput sequencing ATAC-seq, hypothesizing that these dictate differences in biological function. Open chromatin regions (OCRs) surrounding the signature Treg-associated genes, notably FOXP3, CTLA4 and IL2RA were not significantly different between the three populations (Supplementary Fig. 6a). We focused on ~1300 OCRs that have consistent patterns among all three biological replicates but differ among the three Treg populations. These regions were in three clusters: those specific to naive Treg cells (25% of total), specific to CD161+ Treg cells (39% of the total) and those shared between CD161+ and memory Treg cells (37% of the total) (Fig. 6a–b and Supplementary Table 3). Examples of genes with OCRs specific to CD161+ Treg cells included cytokines, such as IL17A, transcription factors, such as PRDM1 and MAF, and chemokine receptors, including CCR9 (Supplementary Table 3). The genomic distributions of OCRs were similar in the three clusters, with the majority within intronic and intergenic regions (Supplementary Fig. 6b).
Figure 6. Genome-wide chromatin landscapes define the regulatory circuitry of CD161+ Treg cells.
(a-b) heatmap showing signal intensity of each ATAC peak and clustering of peaks into three groups (a) with representative examples of open chromatin regions (OCRs) from the three clusters (b) - highlighted in blue are OCRs corresponding to ATAC peaks in the heatmap; (c) transcription factor (TF) footprints enriched relative to background in each cluster of ATAC peaks, with corresponding TF family indicated; (d) GSEA plots for BACH2-, RORγt-, BATF-, FOSL2- and RUNX1-regulated genes, comparing memory to CD161+ Treg cells; n=3 per group; NES = normalized enrichment score; empirical p- and multiple-test adjusted q-values from GSEA are shown; (e) percentage of differentially expressed genes (DEG) between CD161+ and memory Treg cells that can be explained by each TF; (f) CD161 expression on CD4+CD25hiCD127loCD45RA– Treg cells of healthy age and sec-matched donors (BACH2WT/WT) and a patient with BACH2 haploinsufficiency (BACH2WT/L24P); shown are representative flow cytometry plots (left) and data from two independent experiments (right); (g) Venn diagram showing shared and unique DEGs regulated by TFs (see also Supplementary Fig. 6e) (left) and corresponding function for those DEGs (right). Data for Fig. 6a-e and g are from n=3 independent experiments. *p<0.05, **p<0.01, ***p<0.001 by Fisher exact test.
To determine potential transcription factors targeting distinct Treg regulomes, we searched for enrichment of known motifs above background within OCR clusters using HOMER (Fig. 6c and Supplementary Fig. 6c). CD161+-specific OCRs were enriched for motifs of RORγt, RUNX, AP-1 family (e.g. BATF, FOSL2), and Cap’n’collar (CNC) family members that include BACH2 (Fig. 6c and Supplementary Fig. 6c). We performed GSEA for genes regulated by BACH2, RORγt, BATF, FOSL2 and RUNX1. CD161+ Treg cells were enriched for BACH2, RORγt, FOSL2 and RUNX1 regulated genes, compared to memory Treg cells (Fig. 6d). Approximately 40% of the 549 genes differentially expressed between CD161+ and memory Treg cells could be attributed to regulation by Bach2, Runx1, Rorc, Fosl2 and Batf in mice using knockout models3,40,41, with BACH2 responsible for the majority of the transcriptional differences (Fig. 6e). Expression of BACH2 itself was significantly reduced and RORC significantly elevated in freshly isolated CD161+ Treg cells compared to other Treg populations (Supplementary Fig. 6d), suggesting that altered expression of these transcription factors could explain some of the transcriptional differences seen in their targets. CD4+ T cells from a patient heterozygous for a BACH2L24P mutation rendering her BACH2 haploinsufficient42 showed over-representation of the CD161+ Treg subset relative to controls (Fig. 6f), confirming that low BACH2 expression is important for development and/or persistence of these cells. An integrated network was constructed based on these effects to illustrate the transcriptional circuitry (Supplementary Fig. 6e). Genes controlled solely by BACH2 regulated cell division, whereas genes co-regulated by BACH2 and the other transcription factors in this model were especially involved in wound healing (Fig. 6f). These data point to distinct regulomes in CD161+ Treg cells imparting novel functions including wound-healing.
CD161+ Treg cells are enriched in IBD, enhance wound healing and associate with reduced inflammation
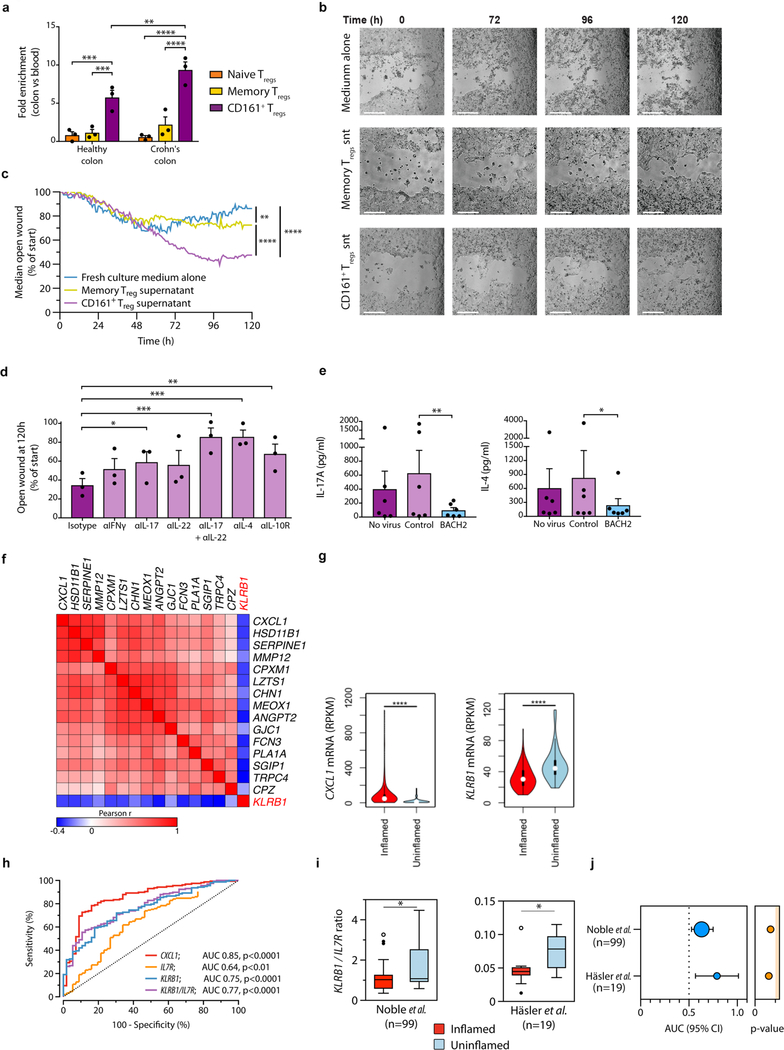
We confirmed by GSEA that activated CD161+ Treg cells were enriched for wound healing genes (e.g. PDGFA and CFS2), including soluble mediators (e.g. IL17A and IL22), compared to other Treg populations (Supplementary Fig. 7a–b). Since CD161+ Treg cells expressed CCR9 and ITGA4 (Fig. 3a and Supplementary Fig. 3a–b), we determined if they were enriched in the bowel during inflammation by comparing Treg populations in matched peripheral blood to colonic biopsies of healthy individuals and those with Crohn’s disease (CD). There was mild enrichment of naive and memory Treg cells in colonic mucosa compared to blood. By contrast, there was a significant enrichment of CD161+ Treg cells in healthy colons, which was even more pronounced in patients with CD (Fig. 7a). LLT1, the CD161 ligand, is expressed in inflamed areas27, which we speculated would drive cytokine expression in CD161+ Treg cells. We therefore tested the effect of CD161+ Treg supernatants on wound healing using a human epithelial colorectal adenocarcinoma cell line (Caco-2 cells) to approximate bowel epithelium. Supernatants from activated CD161+ Treg cells increased and accelerated closure of the wound by almost two-fold compared to supernatant from memory Treg cells or medium alone (Fig. 7b–c, Supplementary Fig. 7c and Supplementary Movies 1–3). Neutralization of IL-17 alone, IL17 together with IL-22, IL-4 or IL-10 impeded the wound healing capacity of supernatants of activated CD161+ Treg cells, implicating these cytokines as mediators of wound healing (Fig. 7d). Over-expression of BACH2 by lentiviral delivery in CD161+ Treg cells significantly inhibited production of IL-17 and IL-4, with a similar but not significant trend in IL-22 and IL-10 (Fig. 7e and Supplementary Fig. 7d), supporting the notion that the wound healing program of CD161+ Treg cells is dependent on reduced expression of the repressive BACH2 transcription factor in these cells.
Figure 7. CD161+ Treg cells accelerate wound healing and are associated with lower inflammation in IBD.
(a) enrichment of Treg cells sub-populations in colons relative to blood of healthy subjects and patients with Crohn’s disease (CD) (n=3 paired samples per group); (b-c) wound healing assay showing growth of Caco-2 cells cultured with medium alone or medium supplemented with culture supernatants (snt) of activated memory or CD161+ Treg cells; representative images captured over time (0, 72, 96 and 120h; white bars=0.5mm) (b) and percentage of open wound over time from n=3 experiments (c); (d) Open wound fraction at culture end for Caco-2 cells in the presence of culture supernatants from activated CD161+ Treg cells with blocking antibodies to stated cytokines or isotype control; cumulative data from n=3 experiments. (e) Concentration (pg/ml) of stated cytokines in supernatants of CD161+ Treg cells transduced, or not, with lentivirus encoding BACH2 or control virus (cumulative data from n=6 experiments); (f) correlation matrix of indicated transcripts in bowel specimens of active CD (n=171); (g) violin plots showing distribution of CXCL1 (left) and KLRB1 (right) expression in inflamed (n=157) and uninflamed CD or non-IBD tissue biopsies (n=56); (h) receiver operating characteristic (ROC) curves showing performance of CXCL1, IL7R, KLRB1 and KLRB1/IL7R ratio to discriminate inflamed (n=157) versus uninflamed CD or non-IBD tissue (n=56). Area under the ROC curve analyses (AUC) and their p-values indicated. (i-j) KLRB1/IL7R ratio in transcriptomes of inflamed and uninflamed sites of CD (i) and AUC + 95% CI of KLRB1/IL7R ROC curves to distinguish the two (j) from two additional, independent, datasets. Numbers of biologically independent samples (n) in i is indicated. In j, areas of circles indicate sample size. Source data for f-h: GSE57945; i-j: GSE20881 and Häsler et al., 2016. Bars show mean + sem throughout; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 by one-way (d-e) and two-way ANOVA (a and c), t-test (g and i) and AUC analysis (h and j).
The suppressive function of CD161+ Treg cells and their ability to accelerate wound healing suggested that they might be beneficial in IBD. To explore this possibility, we examined RNA-seq from a large dataset of patients with CD (GSE57945) for expression of KLRB1, which encodes CD161. KLRB1 correlated negatively with 15 genes previously shown to be upregulated in inflamed, compared to uninflamed, CD mucosa43 (Fig. 7f). Of these, CXCL1, a clinical biomarker of CD, was significantly more highly expressed in inflamed versus uninflamed CD mucosa, while KLRB1 expression was significantly higher in uninflamed CD mucosa (Fig. 7g). KLRB1 is expressed on both Treg cells, which are IL-7Rlo, and TH17 cells, that are IL-7Rhi. We used GSE57945 as a training dataset to test the performance of KLRB1, IL7R and KLRB1/IL7R ratio to distinguish inflamed from uninflamed CD. The receiver operating characteristic (ROC) curve for KLRB1/IL7R was highly significant (AUC=0.77; p-value<0.0001) and performed well in comparison with CXCL1, slightly better than KLRB1 and substantially better than IL7R alone (Fig. 7h) when predicting inflamed versus uninflamed mucosa. We used two additional transcriptome datasets from CD mucosa as validation datasets. In all cases the KLRB1/IL7R predictor was higher in uninflamed CD mucosa (Fig. 7i) and distinguished inflamed from uninflamed mucosa (AUC range 0.63 to 0.79; Fig. 7j), suggesting that tissue infiltration with CD161+ Treg cells is associated with lower inflammation in CD. These data suggest that CD161+ Treg cells are enriched in colonic mucosa, particularly in IBD, where they suppress inflammation and produce soluble factors that accelerate epithelial barrier healing, having a beneficial effect on outcomes.
Discussion
Understanding biologically important and clinically relevant Treg populations is key to elucidating disease mechanisms and tailoring immunotherapy. We delineated CD161+ Treg cells in humans, showing them to be a distinct, highly suppressive, bona fide retinoic-acid dependent sub-population enriched in lamina propria and associating with regions that have lower inflammation in CD. We show that they can enhance wound healing through soluble mediators and that transcriptional control involves a transcription factor network in which BACH2, RORγt, RUNX1, FOSL2 and BATF, all play a role. We show phenotypic distinction of CD161+ Tregs from Tconv, including a unique TCR repertoire that is different from Tconv and TH17 cells. Thus, it is highly unlikely that they are contaminating Tconv. We conclude that they are a distinct sub-population of Treg cells with stable regulatory function.
CD161+ Treg cells are either induced to develop or specialize in the periphery, consistent with absence of CD161+ Treg cells in cord blood23 and thymus (our unpublished observations) and relatively small circulating numbers. Murine RORγt+ Treg cells can either develop in the colon from naive CD4+ T cells by local microbiota to suppress intestinal inflammation, although these cells do not produce IL-1744, or from thymic Treg émigrés following immunization45. We identified that human CD161+ Treg cells have a retinoic acid-regulated gene signature and that retinoic acid directly regulates KLRB1, the gene encoding CD161. Since bowel is rich in retinoic acid, an area for Treg generation and since CD161+ Treg cells expressed CCR9, a gut-homing chemokine receptor, it was unsurprising that the lamina propria was enriched in CD161+ Treg cells, particularly in IBD. This is consistent with inflamed bowel as an enrichment site for FOXP3+IL-17+ T cells46. Thus, CD161+ Treg cells appear to be either induced or migrate to lamina propria.
Some open chromatin regions (OCRs) were broadly accessible in all three Treg populations we studied, including Treg lineage-associated loci, e.g. FOXP3, CTLA4 and IL2RA, consistent with similar protein expression in all the subsets. The regulomes of CD161+ Treg cells differed from memory Treg cells by only ~500 OCRs, congruous with relatively small transcriptional differences between them. The transcription factor circuitry explaining these differences was dominated by BACH2, which was itself expressed at lower level in CD161+ versus memory Treg cells. The importance of low BACH2 expression for development and/or persistence of these Treg cells was demonstrated by excess of CD161+ Tregs in a patient with genetic BACH2 haploinsufficiency. As BACH2 restricts T cell effector programs3, it is unsurprising that de-repression by BACH2 imparts functional properties to Treg cells resembling Tconv cells, notably cytokine production, without loss of suppressive phenotype. This is supported by significantly diminished cytokine production with BACH2 over-expression in these Tregs. Notably, CCR9 is a classic BACH2-repressed gene3 and one of the most differentially expressed genes in CD161+ Treg cells.
The CD161 ligand, LLT1, is expressed on activated DCs and in actively inflamed areas27, hence cross-linking of CD161 is likely in inflamed areas of bowel. Indeed, single nucleotide polymorphisms in LLT1 associate with IBD28. CD161 cross-linking synergized with anti-CD3+CD28-mediated signals to induce cytokine secretion in CD161+ Tregs, in agreement with CD161 stimulating proliferation and cytokine secretion on other T lymphocytes25 but contrasting with the inhibitory effect of CD161 ligation on NK cells39. This suggests that signals transduced by CD161 has cell-type and possibly context-dependent effects.
Although the association between CD161 and TH17 programs in Tconv is known, it was unexpected that transcriptomes of activated CD161+ Treg cells encode complex cytokine cocktails. Since IL-17 can be pro-inflammatory and is associated with autoimmunity, it was possible that IL-17 expression could have denoted a pro-inflammatory function. Indeed, in mice RORγt+ Treg cells can promote autoimmunity and cancer47,48. Our data, however, indicate that actively IL-17-producing Treg cells remain highly suppressive and we found no evidence that CD161+ Treg cells are a transitional or unstable population, consistent with mouse models where adoptive transfer of IL-17+Foxp3+RORγt+ T cells supports stable suppressive effects on gut inflammation49. The cytokine profile of CD161+ Treg cells could denote additional functions, in addition to cellular suppression, such as a potential role in wound healing50. In fact, several cytokines produced by CD161+ Treg cells, such as IL-17 and IL-22, have established wound healing roles, especially in the gut. IL-17 can promote epithelial repair and protect from excessive inflammation in colitis models51. Indeed, IL-17 blockade exacerbates human and murine colitis52. IL-22 also induces intestinal epithelial regeneration and protects from GvHD53, as well as dampening inflammation in models of IBD54. In conclusion, these data support the notion that enhancing CD161+ Treg cells at the site of inflammation, for example by cell therapy, might prove beneficial in IBD.
Online methods
T cell separation, sorting, and flow cytometry
Human PBMCs (peripheral blood mononuclear cells) and T cells subsets were purified from either anonymized leukodepletion cones (Blood Transfusion Service, NHS Blood and Transplantation, Tooting, London, UK) or fresh blood of healthy volunteers. Human studies were conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of Guy’s Hospital (reference 09/H0707/86). Informed consent was obtained from all healthy donors. Treg cells were isolated by initially enriching with CD4+ T Cell Isolation Kit II (Miltenyi Biotec), cells were subsequently stained with mouse anti human CD4 (OKT4), CD127 (eBioRDR5), CD45RA (HI100, all from eBiosciences) and CD25 in PE (both 2A3 and M-A251) and CD161 (DX12, all from BD Biosciences); PE-labelled cells were then captured using anti-PE MicroBeads (Miltenyi Biotec) to enrich for Treg cells, pre-sorting. Cells were FACS sorted for surface markers to separate naive (CD4+CD25hiCD127loCD45RA+CD161−), memory (CD4+CD25hiCD127loCD45RA−CD161−), and CD161+ (CD4+CD25hiCD127loCD45RA−CD161+) Treg cells. Sorting of naive Tconv (CD4+CD25−CD45RA+CD161−), total Tconv (CD4+CD25−) and TH17 (CD4+CD25−CD45RA−CD161+) cells was performed using the same panel. Intracellular staining for FOXP3 (PCH101, eBioscience) was carried out using the Foxp3 / Transcription Factor Staining Buffer Set Kit (eBioscience) according to manufacturer’s instructions. For intracellular staining of cytokines and transcription factors cells were activated for 4 hours with PMA (50 ng/mL) and ionomycin (1 mM, both from Sigma) with the addition of Brefeldin A (3 μg/mL; eBioscience) before staining. Additional staining of Treg cells were further stained for invariant chains TCRVα24-Jα18 (6B11) and TCR Vα7.2 (3C10, both from Biolegend), perforin (dG9, eBioscience), granzyme A (GB11, Biolegend) and granzyme B (CB9, Biolegend) using appropriate flurochrome-conjugated antibodies. Flow cytometry data were acquired on an LSR Fortessa (BD) and subsequenrly analyzed using FlowJo version 10.1 (TreeStar Inc.).
Mass Cytometry (CyTOF)
2×106 CD4+CD127lo cells were isolated using RosetteSep™ Human CD4+CD127low T Cell Enrichment Cocktail (STEMCELL Technologies) before extracellular and intracellular staining with metal conjugated antibodies. For full list of both antibody panels refer to Supplementary Table 4. CyTOF-2 mass cytometer (Fluidigm) was used for data acquisition and beads (Ce140) were used for normalization 29. 320,000 cells were proportionally sampled from all individuals to perform automated clustering. Data were initially processed and analysed using Cytobank 29; CD4 sample “cleanup” was performed by gating on intact (191Ir+ DNA stain), no beads (Ce140−), live (103Rh−), no B cells CD19−CD20− (Nd142), no neutrophils CD15− CD123− (Eu151), CD34− (Er166), CD45+(Y89), CD3+ (Sm154) and CD4+ (Nd145) T cells. Mass-cytometry complex data was analysed using viSNE, in combination with SPADE, to identify distinct subpopulations 30,55 using the following parameters: CCR6 (141Pr), CD45RA (143Nd), CCR4 (149Sm), CD161 (150Nd), CD103 (152Sm), CD62L (153Eu) Helios (156Gd), CCR7 (159Tb), Tbet (160Gd), CD95 (161Dy), CXCR3 (163Dy), CD45RO (164Dy), GATA3 (167Er), CCR9 (168Er), CD25 (169Tm), Foxp3 (171Yb), CXCR4 (173Yb), HLA-DR (174Yb), CD127 (176Yb). viSNE and SPADE plots were generated using Cytobank Inc. (CA, USA). When indicated in the figure legend, figures were overlaid for demonstration purpose.
T cell culture
Unless indicated otherwise, cells were cultured in X-Vivo 15 with Gentamycin and PR (Lonza) supplemented with Penicillin-Streptomycin Glutamine (PSG, Thermo Fisher) and 5% human AB serum (Biosera), henceforth shortened to X-Vivo 5% HS; cells were then seeded at 106cell/ml.
For the evaluation of the effect of ATRA on CD161 expression, 250,000 total Treg cells were isolated and stimulated for 48h with 1:1 ratio of αCD3/CD28 Dynabeads in X-Vivo 5% HS supplemented with 100IU/ml of IL-2 (Proleukin, Novartis), in the presence of 0, 0.02, 0.2 or 2μM ATRA (Sigma-Aldrich); cells were then stained for CD4, CD161 and CCR9 and the effects of ATRA on expression of CD161 (stained with anrtibody clone DX12 from BD) and CCR9 (stained with antibody clone L053E8 from Biolegend) was evaluated by flow cytometry.
For Treg cell expansion to facilitate in vivo models sorted memory and CD161+ Treg cells were cultured in X-Vivo 5% HS supplemented with 1,000 IU/ml IL-2 and stimulated with anti-CD3+CD28 Dynabeads at a 1:1 ratio for 2–3 rounds of expansion of 10–14 days each. Beads were removed by magnetic adherence following each round of stimulation and fresh anti-CD3+CD28 Dynabeads (1:1 ratio) added. After the last round of expansion, beads were removed and the cells were rested for 2 days before injection. Cell viability was close to 100% prior to each in vivo experiment.
To evaluate the effects of CD161 crosslinking on cytokine production, CD161+ Treg cells were cultured for 3 days in X-Vivo 5% HS, supplemented with 100 IU/ml IL-2 and stimulated 1:1 with microbeads from the T cell activation/expansion kit (Miltenyi Biotec) loaded with anti-CD3 and anti-CD28 following manufacturer’s instructions with the addition of anti-CD161 (191B8, MiItenyi) or IgG2a (eBM2a eBioscience).
Treg cell suppression assay
Cryopreserved Tconv cells (CD4+CD25−) were used as target cells throughout. These were isolated by negative selection of CD4+ T cells followed by positive selection of CD25+ T cells using miniMACS CD4+CD25+ T Regulatory Cell Isolation Kit (Miltenyi Biotec, UK) according to manufacturers’ instructions. Tconv cells were obtained from the CD25− negative fraction. Cryopreserved Tconv were thawed and labelled with either 2.5μM Carboxyfluorescein diacetate, sucinnimydil ester (CFSE) (Molecular Probes, USA) or 1μM CellTrace Violet (CTV) (Molecular Probes, USA) according to manufacturers’ instructions. Tconv cell viability was routinely greater than 95% prior to suppression assay. Suppression assays were conducted in X-Vivo 5% HS and U-bottom 96-well plates incubated at 37°C, 5% CO2 for 5 days, at constant Tconv cell number (105 cells) and Treg cells :Tconv ratio of 1:1 or 1:2, as indicated. Where indicated, Treg cell numbers were titrated to result in a Treg cells : Tconv ratio of 1:1 to 1:32 ratio. Cells were stimulated with αCD3/CD28 Dynabeads (bead:cell ratio of 1:40) and CFSE/CTV dilution was assessed by flow cytometry on day 5. Percentage of suppression and IC50 were calculated as previously described36. Where indicated, Treg cell suppression was also evaluated in the presence of either 10μg/ml anti-CD161 (BD Pharmingen), 10μg/ml anti-PDL-1 (eBioscience), 10μg/ml anti-IL-10R (Sigma), 1μg/ml αTGFBRII (R&D) or in the presence of TH1 (40ng/mL IL-12 (Biolegend) and 5μg/mL anti-IL-4 (R&D)) or TH17 (40ng/mL IL-1β (R&D) and IL-6, 10ng/mL TGF-β1 (both from Biolegend), 50ng/mL IL-23 and 5μg/mL anti-IL-4 and anti-IFNg, all from R&D) skewing conditions.
For transwell suppression assay the HTS Transwell-96 Permeable Support with a 0.4μm Pore Polycarbonate Membrane system (Corning) was used. Suppression was tested at a 1:1 Treg cells : Tconv ratio and Tconv were seeded in the lower compartment of the transwell plate, while Treg cells were seeded on to the upper compartment. Cells in both the upper and lower chambers were stimulated with anti-CD3+CD28 Dynabeads (bead:cell ratio of 1:40). As reference of standard suppression, Treg cells and Tconv were also co-seeded in the lower compartment.
Gene expression analysis
200,000 naive, memory and CD161+ Treg cells were either sorted from fresh blood directly into Trizol LS (Ambion) for baseline genetic profile or cells were polyclonally activated post-sorting for 4h with Dynabeads Human T-Activator CD3/CD28 (ratio 1:1) and then lyzed in Trizol LS. RNA was isolated using RNeasy mini kit (QIAGEN) and Ovation PicoSL WTA System V2 (NuGEN Technologies) was used for reverse transcription and cDNA amplification steps. Fragmentation and labelling was performed using the Encore Biotin Module (NuGEN Technologies); all kits were employed following manufacturer’s instructions. Samples were run on GeneChip® Human Gene 1.0 ST Array (Affymetrix Ltd). Data analysis was performed using Partek Genomic SuiteTM (Partek Incorporated). Thresholds for significance were set at 1.5-fold difference at p<0.05 for freshly isolated cells and 2-fold difference at p<0.05 for in vitro activated cells.
Gene set enrichment analysis (GSEA) was performed using GSEA version 2.2.2 56. Datasets used for GSEA are shown in Supplementary table 1. Core human Treg cell genes were sourced from 33; genes associated with CD161+ cells were obtained from25. The ATRA-regulated gene list was generated by microarray from human Treg cells treated, or not, with ATRA (Scottà et al. unpublished data). Genes regulated by RORC, BATF, and FOSL2 were curated from comparison of wild type and single gene knock-out Th17 cells from 41(GSE40918), RUNX1 from wild type and Cbfbeta knockout Treg cells40 (GPL1261) and BACH2 from wild type and Bach2 knockout Treg cells3 (GSE45975), all at a significance threshold of 1.5-fold change and p<0.05. Mouse gene symbols were converted to human homologs using the BioMart data-mining tool in ensemble: http://www.ensembl.org/biomart/martview. General wound healing associated genes were curated from published gene lists 57–59, the “resolve wound healing and fibrosis-related genes” dataset (http://www.resolve-whfg.appspot.com/list/Human/) and “wound healing RT2 profiler PCR array” (Qiagen). Wound healing associated soluble mediators were defined as the online dataset for “cytokines in wound healing (R&D Systems)” supplemented with cytokines and other soluble mediators from the general wound healing associated genes list.
Single cell RNA-sequencing
The single cell sequencing experiment was performed using the 10X Genomics’ Chromium Single Cell 3’ gene expression V2 kit following the manufacturer’s instruction. CD4+CD25+ T cells were freshly isolated from peripheral blood of 3 healthy donors. Cells were captured for each sample and libraries were sequenced on the Illumina Hiseq 3000 instrument. Raw reads from the 3 samples were combined and processed using 10x Genomics Cell Ranger v2.1 60. The result was summarized into an expression matrix with the unique molecular identifier (UMI) count for every cell and every gene. Genes expressed in < 3 genes and cells with < 200 genes detected or > 5% of the total UMI count in mitochondrial genes were removed, resulting a final matrix comprising 2,636 cells and 15,357 genes. Dropouts were imputed using DrImpute 61. We analyzed data using the R package Seurat 62 with default parameters if not specified. Top 15 principal components were used to compute the tSNE plot. Resolution was set to 0.4 for clustering.
Methylation analysis
For CpG methylation analysis 250,000 cells from each desired population were FACS sorted. CpG methylation analysis was determined by pyrosequencing of bisulphite-modified genomic DNA. Methylation analysis was conducted by EpigenDx, as previously described by 63. CpG methylation of FOXP3 TSDR (ADS783FS2), IL2RA (ADS4564FS) and CTLA4 (ADS3074FS2) loci shown in Supplementary Fig. 2g were evaluated.
TCRBV sequencing
For TCRBV sequencing 250,000 cells from each population were FACS sorted. Amplification and sequencing of TCRB CDR3 was performed by Adaptive Biotechnologies using the immunoSEQ Platform 64,65. Analysis was performed using ImmunoSEQ analyser for spectratyping analysis and clone sharing among samples; to assess the overlap in TCR composition between populations, the Morisita–Horn similarity index 32 was calculated using R-Studio. Only productive (in frame and without included STOP codon) amino acidic (VJ) sequences were analysed; the number of both common and specific sequences for each combination of populations was used to calculate the percentage of unique and shared sequences within the different populations.
DCs generation, MLR and CD161 induction
From PBMCs, CD14+ cells were isolated with CD14 microbeads (Miltenyi Biotec) and DCs generated by culturing for 5 days in X-Vivo 5% HS supplemented with 50ng/ml GM-CSF (Peprotech) and 800U/ml IL-4 (R&D). Maturation of the DCs was achieved by culturing for further 48h with 50ng/ml GM-CSF, 800U/ml IL-4, 10ng/ml of IL-1β, IL-6 (eBioscience), TNF-α (Biolegend) and 1μg/ml of PGE2 (BioVision) and LPS (Sigma-Aldrich). To assess the capacity of the DCs to produce ATRA the ALDEFLUOR kit (STEMCELL Technologies) was used to stain the cells for ALDH. Mixed lymphocyte reaction (MLR) was performed by co-culturing DCs with total Treg cells for 5 days in the presence or absence of 1μM pan RAR inverse agonist (BMS493, Tocris Bioscience).
RARA ChIP-qPCR
JASPAR 66 was used to scan the sequences of both KLRB1 and CCR9 gene loci (± 5kb from gene body) for predicted binding sites of ATRA. 15×106 cells were cultured for 4h in X-Vivo, not supplemented with human serum, in the presence or absence of 2μM of ATRA (Sigma-Aldrich). RARA ChIP-qPCR was then performed. Briefly, cells were fixed with 16% Formaldehyde (ThermoFisher) and harvested in PBS containing protease inhibitors Aprotinin (MP biochemicals), Leupeptin (Bachem), both at 1μg/mL and 1mM Phenylmethylsulfonyl fluoride (PMSF, Sigma). Frozen cell pellets were lyzed in SDS lysis buffer containing protease inhibitors and DNA sheared into lengths of 0.2–1Kbp using a Branson sonicator (11% power amplitude). Samples were resuspended in 2ml of ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100 (Sigma), 1.2mM EDTA, 16.7mM Tris HCl pH8.1, 167mM NaCl in dH2O) containing protease inhibitors; 100μl of samples were taken as input DNA and stored for downstream qPCR. To reduce non-specific binding, samples were pre-cleared by incubation with salmon sperm DNA/protein A agarose-50% slurry (EMD Millipore). Chromatin immunoprecipitations (ChIPs) were carried out using 10μl/IP of RARA polyclonal antibody (Diagenode C15310155) and salmon sperm DNA/protein A agarose- 50% slurry. Slurry was then serially washed with Low salt wash buffer (0.1% SDS, 1% Triton X-100, 2mM EDTA, 20mM Tris HCl pH 8.1, 150mM NaCl in dH2O) followed by High salt wash buffer (0.1% SDS, 1% Triton X-100, 2mM EDTA, 20mM Tris HCl pH 8.1, 500mM NaCl in dH2O), LiCl wash buffer (0.25M LiCl (Sigma), 1% NP40 (ThermoFisher), 1% deoxycholate (ThermoFisher), 1mM EDTA, 10mM Tris HCl pH 8.1 in dH2O) and finally twice with TE buffer pH 8 before elution form the beads with elution buffer (1% SDS, 0.1M NaHCO3). Cross-linking was reversed with 5M NaCl/ml and heating to 65°C for 4 hours; dissociated transcription factors and antibodies were digested by adding 0.5M EDTA, 1M Tris HCl (pH 6.5) and 20mg/ml Proteinase K (ThermoFisher) followed by overnight incubation at 55°C. DNA was precipitated with isopropanol and re-dissolved in dH2O before purification with MinElute PCR Purification Kit (QIAGEN). Primers for KLRB1 (forward: GTCCCCACCCACATACACTT; reverse: AGAACAAATGAGCCTCCCAGA) and CCR9 (forward: AGTTTCCCCTTATCCCAGCA; reverse: CAGCTACCCGATAACAACACG) were designed and used for quantification by qPCR. Percentage of input, normalising the signal obtained from the ChIP against the input sample, was calculated.
Cytokine secretion measurement
To evaluate the cytokine production of the different populations of Treg cells after activation, 106 naive, memory and CD161+ Treg cells were FACS sorted and activated for 3 days with 1:1 ratio of αCD3αCD28 Dynabeads and 100U/ml of IL-2. Cytokine protein levels were measured in the supernatant by either TH1/TH2/TH17 Cytometric Bead Array (CBA, BD) or LEGENDplex Human T helper Cytokine panel (Biolegend). To evaluate the effect of CD161 crosslinking on cytokine production 100,000 cells were stimulated for 3 days with ratio 1:1 microbeads coated with either αCD3αCD28IgG2 or αCD3αCD28αCD161 (T Cell Activation/Expansion Kit, Miltenyi Biotec, as previously described 25) and protein levels measured in the supernatant by LEGENDplex Human T helper Cytokine panel (Biolegend).
IL-17 capture assay
106 CD161+ Treg cells were FACS sorted and then stimulated with PMA and Ionomycin for 5h. IL-17 producing CD161+ Treg cells were then labelled using the IL-17 Secretion Assay Kit (Miltenyi Biotec) according to manufacturer’s instructions and then FACS sorted into IL-17 producing and non-producing cells. Cells were rested overnight and a standard suppression assay, including naive and memory Treg cells as controls, performed. Simultaneous staining of surface and intracellular IL-17 was carried out on 106 CD4+ T cells by incubating cells for an extra 2 hours in PMA/Io with supplementation of BD GolgiPlug, before washing and staining with surface IL-17 detection antibody (IL-17 Secretion Assay Kit, Miltenyi Biotec). Following extracellular staining and fixation/permeabilization, intracellular staining was then performed with a second αIL-17 antibody conjugated with a different fluorochrome. To validate the specificity of the double staining, staining with the antibody for intracellular IL-17 was also performed on cells without fixation/permeabilization step, but after surface staining with IL-17 Detection antibody; lack of double positive cells in the control sample demonstrated that the staining for intracellular IL- 17 was specific.
In vitro wound healing assay
The human intestinal cell line Caco-2 (clone HTB-37) was obtained from ECACC (European Collection of Cell Cultures, Health Protection Agency Culture Collection, Salisbury, UK) and grown in DMEM supplemented with 10% FBS (both from GIBCO) and 1% MEM Non-essential Amino Acid (NEAA, Sigma-Aldrich). Cells were routinely tested for contamination by mycoplasma. 250,000 Caco-2 cells were seeded into CytoSelect 24-Well Wound Healing Assay plates (Cell Biolabs) and grown until confluence for 7 days. Sorted memory and CD161+ Treg cells were stimulated for 3 days with 1:1 ratio of αCD3αCD28 Dynabeads and 100U/ml of IL-2 in complete RPMI (GIBCO) with 10% v/v HS and supernatant collected. Supernatant from the cells was then diluted 1:4 with fresh DMEM 10% FBS 1% NEAA and added to the Caco-2 cells, after insert removal. As negative control RPMI 10% HS diluted with 1:4 with DMEM 10% FBS 1% NEAA was used. Time-lapse image capture was recorded using Biostation CT (Nikon) over a period of 120h. Percentage of open wound was calculated using the “wound-healing” tool on NIS Elements Advanced Research Microscope Imaging Software (Nikon). For evaluation of the effect of cytokine blocking on wound healing either IgG1a (5μg/ml) together with IgG2b Isotype (5μg/ml), αIL-17 (5μg/ml), αIL-22 (5μg/ml), αIL-4 (5μg/ml), αIL-10R (10μg/ml), αIFNγ (5μg/ml), αIL-17 (5μg/ml) together with αIL-22 (5μg/ml) or αIL-10R (10μg/ml) together with αIL-4 (5μg/ml; all antibodies from R&D systems) were added to the wound healing assay; image capture was recorded at start and at 120h post culture.
Patient biopsies
Six consecutive patients who attended the endoscopy department at King’s College Hospital (London, UK) for a colonoscopy required in the context of their routine clinical care were consented for the study (REC 15/LO/1998). Three patients were recruited with an established diagnosis of Crohn’s disease (CD), based on conventional clinical criteria, and three without any history of chronic inflammatory bowel disease (IBD), but with gastrointestinal symptoms or anemia that required investigation; these latter patients were then considered as “healthy individuals” with no diagnosis of CD. Peripheral blood was collected in EDTA tubes on the day of attendance to endoscopy and prior to colonoscopy. During colonoscopy 2mm biopsies were collected from inflamed sites in patients with CD or randomly for those without a diagnosis of IBD. Endoscopic findings of inflammation and/ or normal macroscopic appearances were confirmed by histopathology.
PBMC were isolated following standard isolation protocol and colonic Lamina Propria Mononuclear cells (cLPMC) were isolated after digestion of the colonic tissue as previously described 67. Both PBMC and cLPMC were then stained for CD45, CD3, CD8, CD4, CD25, CD127, CD45RA and CD161. Total CD4+ T cells were initially defined as CD45+CD3+CD8-CD4+ and Treg cells subpopulations were then selected following standard gating strategy, as previously described. Treg cell populations were then compared in matched peripheral blood to colonic biopsy samples of healthy individuals and those with IBD.
Severe xeno-graft-versus-host-disease (GvHD) model
NOD/scid/IL-2Rγ−/− (NOD.cg-PrkdcscidIl2rgtm1Wjl/SzJ) mice (NSG; The Jackson Laboratory) were used between 8–10 weeks of age. Animals were bred and maintained in the Biological Services Unit of King’s College London. All mice were kept under specific-pathogen-free conditions, and procedures were conducted in accordance with institutional guidelines (PPL70/7302) and the Home Office Animals Scientific Procedures Act (1986). Sorted Memory and CD161+ Treg cells were cultured in X-Vivo 15 with Gentamycin and PR (Lonza) supplemented with 5% human AB serum (HS, Biosera), 1,000 IU/ml of IL-2 (Proleukin, Novartis) and stimulated with Dynabeads Human T-Activator CD3/CD28 (GIBCO) at a 1:1 ratio for 2–3 rounds of expansion (10–14 days each). After the last round of expansion, beads were removed and the cells were rested for 2 days before injection into the mice. Human PBMCs (10×106) depleted of CD25+ cells were injected intravenously to induce xeno-GvHD with or without in vitro –expanded Memory or CD161+ Treg cells (5×106) at a 2:1 PBMC : Treg cells ratio. Mice injected with PBS alone were used as negative control. Mice were monitored for symptoms of xeno-GvHD over time and experiment was carried on until either a clinical score greater than 6 or a weight loss greater than 15% from the initial weight was reached. Clinical GvHD score was calculated by applying a modified scoring system (adapted from 68: individual mice received a score of 0 to 2 for each criteria.
Grade 0 - <5% weight loss, normal posture, activity, fur texture and skin integrity;
Grade 1 – weight loss >5% to <10%, hunched posture only at rest, mild to moderately decreased activity, mild to moderate fur ruffling, scaling of paws/tail;
Grade 2 - >10% weight loss, severe hunching of posture impairing movement, severe ruffled fur/poor grooming, obvious areas of denuded skin.
Lentiviral preparation
Lentivirus was prepared by transfecting HEK293T cells (ATCC CRL-11268) and expanded in in DMEM 10% FCS and 1% PSG; cells were routinely tested for contamination by mycoplasma. 106 HEK293T cells were plated in DMEM 10% FCS and 1% PSG and transfected with 15μg of psPAX2 (Addgene), 5 μg of pMD2.G (Addgene) and 20μg of either Control or Bach2-expressing pLVX-EF1α-IRES-ZzGreen plasmids (Clonetech). The transfection mix was then added to an equal volume of CaCl2 0.5M. The resulting solution was added dropwise to an equal volume of 2x HEPES buffered saline (HBS; Fluka, Sigma-Aldrich Company Ltd., Dorset, England) to precipitate the DNA. After an incubation of 30 minutes at room temperature, the prepared solution was added evenly, drop wise, to the cells and the plate was incubated at 37°C overnight. Two collections of supernatant containing viral particles were performed at day 3 and 4 post transfection and concentration was performed by addition of PEG-it Virus concentration solution (SBI, System Bioscience, Mountain View, U.S.A) following manufacturer’s instructions. After allowing 48 hours for virus precipitation, the solution containing the viral particles was spun at 1500g for 30mins at 4°C to obtain a viral pellet that was resuspended in 300μl of cold medium and stored at −80°C until used.
CD161+ Treg cells transduction
Sorted CD161+ Treg cells were stimulated for 3 days with anti-CD3+CD28 dynabeads (1:1 cell to bead ratio) in RPMI 10% HS supplemented with 100U/ml of IL-2. Cells were then counted, washed and resuspended at 106/ml. 100,000 cells were seeded in a 96 well plate and left overnight at 37oC; the following day 1X Transdux and Max Enhancer (TransDux Max kit, SBI) were added to each well in the presence or absence of 10μl of Control or Bach2 lentivirus. 3 days post addition of the virus anti-CD3+CD28 dynabeads were magnetically removed, cells fed with fresh medium and left at 37oC for a further 3 days. At the end of the transduction protocol, cells were counted, washed and resuspended at 106/ml and stimulated for another 3 days with anti-CD3+CD28 dynabeads (1:1 cell to bead ratio) in RPMI 10% HS supplemented with 100U/ml of IL-2. Supernatant was then collected and cytokine production was measured.
CD161+ Treg cells transfection
Sorted CD161+ Treg cells were stimulated for 3 days with anti-CD3+CD28 dynabeads (1:1 cell to bead ratio) in RPMI 10% HS supplemented with 100U/ml of IL-2. Cells were then counted, washed and resuspended at 2×106/ml. 100,000 cells were then seeded in a 96 well plate and transfected with either All star Negative Control siRNA or HS_RORC_7 Flexitube siRNA (both from Qiagen) following manufacturer’s instruction; untransfected cells were used as baseline control. Cells were then cultured for 3 days at 37oC; supernatant was then collected and cytokine production measured.
ATAC-Seq
ATAC-seq was performed according to published protocol 69 with minor modification as described in 70. Paired-end libraries (50 cycles) were prepared according to ATAC-seq protocol (see above) with three biological replicates (i.e. cells from 3 different individuals) for each library. The sequencing was performed using Illumina 2000. To obtain the open chromatin regions, reads were aligned to hg19 using Bowtie v2.2.9 71 with parameters [--maxins 175 --no-discordant --no-mixed]. Properly paired and uniquely mapped alignments were extracted. The open chromatin regions were identified using Homer findPeaks tool 72 with parameters [-region -size 500 -minDist 50 -tbp 0]. All the open chromatin peaks were merged using bedtools 73 to obtain a set of all potential peaks. For each replicate, the differential set of open chromatin regions for each pair of samples (i.e. memory vs naive, memory vs CD161, naive v CD161) were extracted using homer getDifferentialPeaks with parameter –F 2 and the set of all potential peaks. For each pair of samples, the common peaks among three replicates were extracted using bedtools multiinter. All the common differential open chromatin regions were merged using bedtools and displayed using seqMINER 74 with three clusters. To identify motifs that are enriched in each cluster, we used homer findMotifsGenome on all known motifs with parameter [-size given].
Data analysis and statistical tools
Statistical analysis was carried out using GraphPad Prism 7 (GraphPad software Inc., USA). All measures of variance were expressed as mean ± standard error of the mean (SEM) and for data comparison t-test, one- or two-way RM ANOVA were used as indicated. All statistical tests were two-sided and data were considered statistically significant with p<0.05, p<0.01, p<0.001 or p<0.0001 and represented in figures as indicated. Hierarchical clustering of SPADE data was carried out using cytoClustR, available at https://github.com/kordastilab/cytoClustR. ROC curves were constructed in GraphPad Prism v7 (GraphPad software Inc) using the dataset from Haberman et al., 2014 (GSE57945) as a training set, taking inflamed versus uninflamed Crohn’s disease and healthy donor tissues as defined by the investigators. Test datasets were sourced from Noble et al., 2010 (GSE20881) and Häsler et al., 2016 75, classifying samples as either normal versus any inflammation (Noble et al., 2010) or non-inflamed versus inflamed as defined by the investigators (Häsler et al., 2016). The data generated for this study have been deposited at the Gene Expression Omnibus (GEO) under accession code GSE119375.
Supplementary Material
Supplementary Movies 1–3. Wound healing assay. Movies show growth of Caco-2 cells cultured with medium alone (Supplementary Movie 1) or medium supplemented with culture supernatants (snt) of activated memory (Supplementary Movie 2) or CD161+ Treg cells (Supplementary Movie 3). Time-lapse images were recorded from 0–120h. Shown are representative movies from n=3 independent experiments.
Supplementary Table 1. List of differentially expressed genes (DEGs). Listed are DEGs in comparisons of freshly isolated Treg cells to each other (tab 1) and transcriptional changes in each population when comparing CD3/CD28-activated cells to freshly isolated cells (tab 2).
Supplementary Table 2. Expression of killer cell lectin like receptors (Klr) in wild-type mouse Treg cells. Shown are RPKM values (n=6) for Klr gene transcripts, Foxp3 and Actin. Data from Feng et al., 2014, GSE58905.
Supplementary Table 3. Open chromatin regions (OCRs) from ATAC-seq. Listed are genes with OCRs conforming to cluster 1 (naive Treg cells-specific), cluster 2 (shared between CD161+ and memory Treg cells) and cluster 3 (CD161+ Treg cells-specific).
Supplementary Table 4. List of conjugated antibodies used for mass cytometry.
Supplementary Table 5. Gene lists used for gene set enrichment analyses.
Acknowledgements:
The authors wish to thank patients that contributed samples towards this study. This work was supported by the Wellcome Trust (grant 097261/Z/11/Z to B. Afzali and WT101159 to N. Powell), the Crohn’s & Colitis Foundation of America (grant CCFA #3765 – CCFA genetics initiative to A. Laurence), British Heart Foundation (grant RG/13/12/30395 to G. Lombardi), institutional startup fund from Purdue university and National Heart, Lung, and Blood Institute (grant 5K22HL125593–02 to M. Kazemian). Research was also supported by the National Institute for Health Research (NIHR) Biomedical Research Centre at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. This research was supported [in part] by the Intramural Research Programs of the National Institute of Arthritis and Musculoskeletal and Skin Diseases, the National Institute of Diabetes and Digestive and Kidney Diseases and the National Heart, Lung and Blood Institute of the National Institutes of Health. We thank John O’Shea (National Institutes of Health) for his support and for providing access to ATAC-seq, the National Heart, Lung and Blood Institute DNA Sequencing and Genomics Core for performing single cell sequencing experiment and acknowledge the assistance of Matt Arno (Genomics centre, King’s College London) with gene expression microarray studies as well as Suanne Heck and Richard Ellis (Biomedical research centre flow core facility, King’s College London) for CyTOF data acquisition. In addition, the authors thank Ewy Mathé (Ohio State University) for critically reading the manuscript.
Footnotes
Competing interests:
The authors have no competing interests to declare.
References
- 1.Edozie FC et al. Regulatory T-cell therapy in the induction of transplant tolerance: the issue of subpopulations. Transplantation 98, 370–379 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Sakaguchi S, Wing K, Onishi Y, Prieto-Martin P & Yamaguchi T Regulatory T cells: how do they suppress immune responses? Int Immunol 21, 1105–1111 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Roychoudhuri R et al. BACH2 represses effector programs to stabilize T(reg)-mediated immune homeostasis. Nature 498, 506–510 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yadav M et al. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J Exp Med 209, 1713–22– S1–19 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thornton AM et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol 184, 3433–3441 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Floess S et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol 5, e38 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohkura N et al. T Cell Receptor Stimulation-Induced Epigenetic Changes and Foxp3 Expression Are Independent and Complementary Events Required for Treg Cell Development. Immunity 37, 785–799 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Polansky JK et al. DNA methylation controls Foxp3 gene expression. Eur J Immunol 38, 1654–1663 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Shih H-Y et al. Transcriptional and epigenetic networks of helper T and innate lymphoid cells. Immunol Rev 261, 23–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oldenhove G et al. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity 31, 772–786 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaudhry A et al. CD4+ Regulatory T Cells Control TH17 Responses in a Stat3-Dependent Manner. Science 326, 986–991 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng Y et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control TH2 responses. Nature 458, 351–356 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koch MA et al. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol 10, 595–602 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wohlfert EA et al. GATA3 controls Foxp3⁺ regulatory T cell fate during inflammation in mice. J Clin Invest 121, 4503–4515 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu F, Sharma S, Edwards J, Feigenbaum L & Zhu J Dynamic expression of transcription factors T-bet and GATA-3 by regulatory T cells maintains immunotolerance. Nat Immunol 16, 197–206 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kordasti S et al. Deep phenotyping of Tregs identifies an immune signature for idiopathic aplastic anemia and predicts response to treatment. 128, 1193–1205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyara M et al. Functional delineation and differentiation dynamics of human CD4+ T cells expressing the FoxP3 transcription factor. Immunity 30, 899–911 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Booth NJ et al. Different proliferative potential and migratory characteristics of human CD4+ regulatory T cells that express either CD45RA or CD45RO. J Immunol 184, 4317–4326 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Maloy KJ & Powrie F Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature 474, 298–306 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Povoleri GAM et al. Thymic versus induced regulatory T cells - who regulates the regulators? Front. Immunol 4, 169 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maloy KJ et al. CD4+CD25+ T(R) cells suppress innate immune pathology through cytokine-dependent mechanisms. J Exp Med 197, 111–119 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maul J et al. Peripheral and intestinal regulatory CD4+ CD25(high) T cells in inflammatory bowel disease. Gastroenterology 128, 1868–1878 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Afzali B et al. CD161 expression characterizes a subpopulation of human regulatory T cells that produces IL-17 in a STAT3-dependent manner. Eur J Immunol 43, 2043–2054 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanier LL, Chang C & Phillips JH Human NKR-P1A. A disulfide-linked homodimer of the C-type lectin superfamily expressed by a subset of NK and T lymphocytes. J Immunol 153, 2417–2428 (1994). [PubMed] [Google Scholar]
- 25.Fergusson JR et al. CD161 Defines a Transcriptional and Functional Phenotype across Distinct Human T Cell Lineages. Cell Rep 9, 1075–1088 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cosmi L et al. Human interleukin 17-producing cells originate from a CD161+CD4+ T cell precursor. J Exp Med 205, 1903–1916 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Germain C et al. Induction of lectin-like transcript 1 (LLT1) protein cell surface expression by pathogens and interferon-γ contributes to modulate immune responses. J Biol Chem 286, 37964–37975 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolfkamp SCS et al. Single nucleotide polymorphisms in C-type lectin genes, clustered in the IBD2 and IBD6 susceptibility loci, may play a role in the pathogenesis of inflammatory bowel diseases. Eur J Gastroenterol Hepatol 24, 965–970 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Diggins KE, Ferrell PB & Irish JM Methods for discovery and characterization of cell subsets in high dimensional mass cytometry data. Methods 82, 55–63 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu P et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol 29, 886–891 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas SY et al. CD1d-restricted NKT cells express a chemokine receptor profile indicative of Th1-type inflammatory homing cells. J Immunol 171, 2571–2580 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Venturi V et al. Method for assessing the similarity between subsets of the T cell receptor repertoire. J Immunol Methods 329, 67–80 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Ferraro A et al. Interindividual variation in human T regulatory cells. Proc Natl Acad Sci 111, E1111–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim YC et al. Oligodeoxynucleotides stabilize Helios-expressing Foxp3+ human T regulatory cells during in vitro expansion. Blood 119, 2810–2818 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scotta C et al. Differential effects of rapamycin and retinoic acid on expansion, stability and suppressive qualities of human CD4(+)CD25(+)FOXP3(+) T regulatory cell subpopulations. Haematologica 98, 1291–1299 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Afzali B et al. Comparison of regulatory T cells in hemodialysis patients and healthy controls: implications for cell therapy in transplantation. Clin J Am Soc Nephrol 8, 1396–1405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thornton AM & Shevach EM CD4+CD25+ immunoregulatory T cells suppress polyclonal T cell activation in vitro by inhibiting interleukin 2 production. J Exp Med 188, 287–296 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao X et al. Granzyme B and Perforin Are Important for Regulatory T Cell-Mediated Suppression of Tumor Clearance. Immunity 27, 635–646 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Rosen DB et al. Functional consequences of interactions between human NKR-P1A and its ligand LLT1 expressed on activated dendritic cells and B cells. J Immunol 180, 6508–6517 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kitoh A et al. Indispensable Role of the Runx1-Cbfβ Transcription Complex for In Vivo-Suppressive Function of FoxP3+ Regulatory T Cells. Immunity 31, 609–620 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Ciofani M et al. A validated regulatory network for Th17 cell specification. Cell 151, 289–303 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Afzali B et al. BACH2 immunodeficiency illustrates an association between super-enhancers and haploinsufficiency. Nat Immunol 18, 813–823 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hong SN et al. RNA-seq Reveals Transcriptomic Differences in Inflamed and Noninflamed Intestinal Mucosa of Crohn’s Disease Patients Compared with Normal Mucosa of Healthy Controls. Inflamm. Bowel Dis 23, 1098–1108 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Sefik E et al. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORγ⁺ regulatory T cells. Science 349, 993–997 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim B-S et al. Generation of RORγt+ Antigen-Specific T Regulatory 17 Cells from Foxp3+ Precursors in Autoimmunity. Cell Rep 21, 195–207 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hovhannisyan Z, Treatman J, Littman DR & Mayer L Characterization of interleukin-17-producing regulatory T cells in inflamed intestinal mucosa from patients with inflammatory bowel diseases. Gastroenterology 140, 957–965 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blatner NR et al. Expression of RORγt marks a pathogenic regulatory T cell subset in human colon cancer. Sci Trans Med 4, 164ra159–164ra159 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Komatsu N et al. Pathogenic conversion of Foxp3+ T cells into TH17 cells in autoimmune arthritis. Nat Med 20, 62–68 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Yang B-H et al. Foxp3(+) T cells expressing RORγt represent a stable regulatory T-cell effector lineage with enhanced suppressive capacity during intestinal inflammation. Mucosal Immunology 9, 444–457 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Nosbaum A et al. Regulatory T Cells Facilitate Cutaneous Wound Healing. J Immunol 196, 2010–2014 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Connor W et al. A protective function for interleukin 17A in T cell-mediated intestinal inflammation. Nat Immunol 10, 603–609 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hueber W et al. Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn’s disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut 61, 1693–1700 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lindemans CA et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature 528, 560–564 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zenewicz LA et al. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity 29, 947–957 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Experimental procedures references
- 55.Amir E-AD et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol 31, 545–552 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barrientos S, Stojadinovic O, Golinko MS, Brem H & Tomic-Canic M Growth factors and cytokines in wound healing. Wound Repair Regen 16, 585–601 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Deonarine K et al. Gene expression profiling of cutaneous wound healing. J Transl Med 5, 11 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peake MA et al. Identification of a transcriptional signature for the wound healing continuum. Wound Repair Regen 22, 399–405 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng GXY et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gong W, Kwak I-Y, Pota P, Koyano-Nakagawa N & Garry DJ DrImpute: imputing dropout events in single cell RNA sequencing data. BMC Bioinformatics 19, 220 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng Y et al. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature 463, 808–812 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robins HS et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. 114, 4099–4107 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carlson CS et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun 4, 2680 (2013). [DOI] [PubMed] [Google Scholar]
- 66.Mathelier A et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 44, D110–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rovedatti L et al. Differential regulation of interleukin 17 and interferon gamma production in inflammatory bowel disease. Gut 58, 1629–1636 (2009). [DOI] [PubMed] [Google Scholar]
- 68.Cooke KR et al. An experimental model of idiopathic pneumonia syndrome after bone marrow transplantation: I. The roles of minor H antigens and endotoxin. 88, 3230–3239 (1996). [PubMed] [Google Scholar]
- 69.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shih H-Y et al. Developmental Acquisition of Regulomes Underlies Innate Lymphoid Cell Functionality. Cell 165, 1120–1133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ye T et al. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res 39, e35–e35 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Häsler R et al. Uncoupling of mucosal gene regulation, mRNA splicing and adherent microbiota signatures in inflammatory bowel disease. Gut (2016). doi: 10.1136/gutjnl-2016-311651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movies 1–3. Wound healing assay. Movies show growth of Caco-2 cells cultured with medium alone (Supplementary Movie 1) or medium supplemented with culture supernatants (snt) of activated memory (Supplementary Movie 2) or CD161+ Treg cells (Supplementary Movie 3). Time-lapse images were recorded from 0–120h. Shown are representative movies from n=3 independent experiments.
Supplementary Table 1. List of differentially expressed genes (DEGs). Listed are DEGs in comparisons of freshly isolated Treg cells to each other (tab 1) and transcriptional changes in each population when comparing CD3/CD28-activated cells to freshly isolated cells (tab 2).
Supplementary Table 2. Expression of killer cell lectin like receptors (Klr) in wild-type mouse Treg cells. Shown are RPKM values (n=6) for Klr gene transcripts, Foxp3 and Actin. Data from Feng et al., 2014, GSE58905.
Supplementary Table 3. Open chromatin regions (OCRs) from ATAC-seq. Listed are genes with OCRs conforming to cluster 1 (naive Treg cells-specific), cluster 2 (shared between CD161+ and memory Treg cells) and cluster 3 (CD161+ Treg cells-specific).
Supplementary Table 4. List of conjugated antibodies used for mass cytometry.
Supplementary Table 5. Gene lists used for gene set enrichment analyses.