Abstract
Alzheimer’s disease (AD) is characterized by the accumulation of neurotoxic amyloid-β (Aβ) peptides consisting of 39-43 amino acids, proteolytically derived fragments of the amyloid-β protein precursor (AβPP), and the accumulation of the hyperphosphorylated microtubule-associated protein tau. Inhibiting Aβ production may reduce neurodegeneration and cognitive dysfunction associated with AD. We have previously used an AβPP-firefly luciferase enzyme complementation assay to conduct a high throughput screen of a compound library for inhibitors of AβPP dimerization, and identified a compound that reduces Aβ levels. In the present study, we have identified an analog, compound Y10, which also reduced Aβ. Initial kinase profiling assays identified the receptor tyrosine kinase cKit as a putative Y10 target. To elucidate the precise mechanism involved, AβPP phosphorylation was examined by IP-western blotting. We found that Y10 inhibits cKit phosphorylation and increases AβPP phosphorylation mainly on tyrosine residue Y743, according to AβPP751 numbering. A known cKit inhibitor and siRNA specific to cKit were also found to increase AβPP phosphorylation and lower Aβ levels. We also investigated a cKit downstream signaling molecule, the Shp2 phosphatase, and found that known Shp2 inhibitors and siRNA specific to Shp2 also increase AβPP phosphorylation, suggesting that the cKit signaling pathway is also involved in AβPP phosphorylation and Aβ production. We further found that inhibitors of both cKit and Shp2 enhance AβPP surface localization. Thus, regulation of AβPP phosphorylation by small molecules should be considered as a novel therapeutic intervention for AD.
Keywords: High throughput screening, kinase, neurodegeneration, phosphatase, phosphorylation
INTRODUCTION
Amyloid-β protein precursor (AβPP) is a singlepass transmembrane glycoprotein that is ubiquitously expressed in many cell types, including neurons. Amyloidogenic processing of AβPP by β- and γ-secretases leads to the production of neurotoxic amyloid-β (Aβ) peptides that can oligomerize and aggregate into amyloid plaques, a characteristic hallmark of AD brains [1].
We have previously reported the results of a high throughput screen (HTS) performed using a firefly luciferase (Fluc) fragment complementation assay in a stable cell line that expresses luciferase-tagged AβPP fragments. The HTS campaign resulted in the identification of a compound, “Compound Y”, which significantly inhibited AβPP dimerization and reduced the production of both Aβ40 and Aβ42 [2].
Following the procurement and testing of additional Compound Y analogs, we have identified compound Y10, which also shows inhibition in the firefly luciferase fragment complementation assay and results in a modest reduction of Aβ. Compounds Y and Y10 have in common a pyrazolopyrimidine motif found in known kinase inhibitors. Accordingly, we profiled Y10 against a small panel of kinases targeted by this chemotype, as well as kinases known to be implicated in neurodegenerative diseases such as AD. Of this panel, one kinase inhibited by Y10 to a significant degree was the receptor tyrosine kinase cKit. Despite the putative kinase inhibitory activity of compound Y10, we subsequently found that Y10 increases the level of AβPP phosphorylation, suggesting that the phosphorylation state of AβPP may be regulated by cKit-mediated modulation of a downstream signaling molecule.
A number of reports support the role of the phosphorylation of the AβPP cytosolic domain on AβPP processing and function. Phosphorylated AβPP has been shown to accumulate in large vesicular structures in hippocampal lysates from AD patients [3]. It has also been demonstrated that the short cytoplasmic domain of AβPP contains a canonical endocytic signal motif for membrane-associated receptors (Y682ENPTY687 (AβPP695 numbering)) and that mutations of Y682, N684, or P685 inhibit the internalization of AβPP and decrease the generation of secreted sAβPPβ and Aβ [3, 4]. Phosphorylation of the AβPP intracellular domain on Thr668, Tyr653, Tyr682, and Tyr687 has been shown to play a determining role in the fate of AβPP processing and the production of Aβ [5-7].
Herein, we wished to decipher the mechanism of action of the Y compounds in reducing Aβ. We report the involvement of cKit and its downstream phosphatase Shp2 in the dephosphorylation of AβPP tyrosine 743 (AβPP751 numbering). Moreover, we demonstrate that the inhibition of this pathway leads to an increase in AβPP phosphorylation and a decrease in Aβ levels. Thus, our findings suggest a new regulatory mechanism of Aβ production through the modulation of AβPP phosphorylation. Small molecules that affect the cKit/Shp2 pathway may be useful for novel therapeutics aimed at reducing Aβ levels in AD.
MATERIALS AND METHODS
Reagents
Compounds Y, Y1, Y6, Y10, and Y11 were purchased from Key Organics, Ltd. PHPS1 was purchased from Santa Cruz Biotechnology, PKC412 from Sigma-Aldrich, NSC87877 from EMD Millipore, and masitinib and tandutinib from Selleck Chemicals.
Enzyme-Linked ImmunoSorbent Assay (ELISA) Aβ40 and Aβ42
Aβ40 and Aβ42 were measured as described [2] using HEK293 cells stably transfected with wtAPP751, a kind gift from Dr. D. Selkoe. ELISAs were carried out using the human Aβ40 and Aβ42 ELISA kits (Invitrogen) in accordance with manufacturer’s protocol with samples diluted 1: 2 in diluent buffer.
Crystal violet staining
Kinase profiling
Protein kinase assays were conducted using the KinaseProfiler™ service of Eurofins Pharma Discovery Services UK Limited [10]. The kinase of interest was incubated with the test compound in assay buffer containing substrate, 10 mM magnesium acetate and [γ-33P-ATP]. The reaction was initiated by the addition of the Mg/ATP mix. After incubation at room temperature, the reaction was stopped by the addition of a 3% phosphoric acid solution. An aliquot of the reaction was then spotted onto a filtermat and washed in phosphoric acid followed by a rinse in methanol prior to drying and scintillation counting. Results were expressed in relation to controls containing DMSO only in place of test compound. The ATP concentration in each assay was within 15 μM of the determined apparent Km for ATP.
Mutagenesis and plasmids construction
Human cKit in pCMV-XL5 plasmid was obtained from Origene (Rockville, MD). To construct a V5-tag into the C-terminus of cKit cDNA, a XhoI site was introduced into the cKit-pCMV-XL5 vector by replacing the stop codon using QuickChange Site-Directed mutagenesis kit (Stratagene, La Jolla, CA). The V5-tag was ligated into cKit-pCMV-XL5 digested with XhoI/SacII with the following primers:
cKit XhoI sense: 5’- TGTGCACGACGATGTCTCGAGAGAATCAGTGTTTGGG-3’
cKit XhoI anti-sense: 5’- CCCAAACACTGATTCTCTCGAGACATCGTCGTGCACA-3’
cKit-V5-tag linker sense:5’-TCGAGA-GGCAAACCCATACCAAATCCACTGCTGGGACTGGACTCAACCCGTACCGGTCATCATCACCATCACCATTGACCGC-3’ and cKit-V5-tag linker anti-sense: 5’- GGTCAATGGTGATGGTGATGATGACCGGTACGGGTTGAGTCCAGTCCCAGCAGTGGATTTGGTATGGGTTTGCCTC-3’
The mutagenesis of cKit GNNK- isoform to delete the GNNK tetra-amino acid residues in cKit-V5 plasmid was done using QuickChange Site-Directed mutagenesis kit (Stratagene, La Jolla, CA) using the following primers:
GNNK- sense: 5’-ACTTTGCATTTAAAGAGCAAATCCATCCCCACA-3’
GNNK- anti-sense: 5’-TGTGGGGATGGATTTGCTCTTTAAATGCAAAGT-3’
The mutagenesis of AβPP tyrosine mutants were performed using the following primers:
Y709G sense: 5’-CTGAAGAAGAAACAGGGCACATCCATTCATCATG-3’
Y709G anti-sense: 5’-CATGATGAATGGATGTGCCCTGTTTCTTCTTCAG-3’
Y738G sense: 5’-ATGCAGCAGAACGGCGGCGAAAATCCAACCTAC-3’
Y738G anti-sense: 5’-GTAGGTTGGATTTTCGCCGCCGTTCTGCTGCAT-3’
Y743G sense: 5’-TACGAAAATCCAACCGGCAAGTTCTTTGAGCAG-3’
Y743G anti-sense: 5’-CTGCTCAAAGAACTTGCCGGTTGGATTTTCGTA-3’
Cell culture, transfections, and protein sample collection
Details of cell culture, transfections and protein sample collection were described previously [11]. Mouse primary neuronal cultures were described previously [12].
SDS-PAGE and western blotting
SDS-PAGE and western blotting were described previously [11]. Primary antibodies for western blots were: mouse monoclonal 6E10 (Covance, 1: 1000) against amino acids 1–17 of Aβ that also recognize AβPP; total ERK and p-ERK from phosphoERK pathway kit (1: 1000, Cell Signaling, Danvers, MA) and were used according to the manufacturer’s protocol; mouse anti-V5 antibody (1 : 5,000, Invitrogen, NY, NY), a monoclonal antibody against cKit, mAb AB81 (1 : 1000, Cell signaling #3308), a polyclonal antibody against Shp2 (1 : 1000, Cell Signaling #3752), a rabbit monoclonal antibody against CD71 (1 : 1000, Cell signaling #13113), and anti phosphotyrosine (1 : 2000, Cell Signaling, #9416). Secondary antibody used for western blots was 1 : 5000 peroxidase labeled goat anti-mouse or anti-rabbit IgG (H+L) (KPL). Protein expression in western blots was assessed and normalized by densitometry using ImageJ.
cKit signaling assay
HEK293 cells were transfected with either V5-tagged cKit GNNK+ or GNNK− plasmids. Forty-eight hours after transfection, cells were incubated in serum-free medium for 2 h and then either 100 or 200 ng/mL of stem cell factor (PeproTech, Rocky Hill, NJ), the activator for cKit, was added to the wells. The cells were incubated for 15 min at 37°C and were then immediately washed in PBS and lysed in Lysis buffer (1% Triton X-100, 0.01M Tris-HCl, 0.01 M EDTA, 0.05 M NaCl, 0.05 M NaF, pH 7.2) containing protease and phosphatase inhibitors (Roche, Mannheim GE). After lysis, samples were prepared for SDS-PAGE as described previously [11].
c-Kit and AβPP phosphorylation
Protein levels in extracts from cell lysates were measured using the BCA reagent. Equal amounts of protein from each sample were used for immunoprecipitation using V5-beads or anti-AβPP mAb 6E10, 22C11 (Sigma, St Louis) or mAb 4.1 (a gift from W. Van Nostrand) followed by immunoprecipitation with protein G agarose and western blotting with phosphotyrosine antibody (Cell Signaling, #9416).
cKit and Shp2 knockdown with siRNA
For cKit and Shp2 siRNA knockdown experiments, either scrambled negative control siRNA, cKit or Shp2 specific siRNA (Origene) were transfected in Opti-MEM with TransIT-X2 (Mirus) according to the manufacturer’s protocol overnight. Cells were washed once with medium, medium replaced, and cells incubated overnight at 37°C. Forty-eight hours after transfection, cells were incubated in serum-free medium for 2 h and then washed in PBS and lysed in Lysis buffer. The lysates were used for western blotting to detect cKit, Shp2 and AβPP.
BACE inhibition assay
BACE inhibition assay was performed using BACE1 (β-secretase) FRET assay kit (Thermo Fisher #P2985) according to manufacturer’s protocol. A substrate analog peptide was used as control BACE inhibitor (Calbiochem, #171601).
AβPP Cell-surface biotinylation assay
HEK293 cells stably expressing AβPP were seeded in 6-well plates and grown to 90% confluency. The cells were treated with DMSO, Y10 or PHPS1 at 1 μM for 2 h and washed twice with PBS and incubated with a 1 mM solution of cell impermeable NHS-SS-Biotin (Pierce, Rockford IL) in PBS for 30 min on ice. The cells were then washed twice with ice-cold PBS, lysed in lysis buffer and the cell lysates were collected. Ten μL of lysate were saved to serve as AβPP input control, while the rest of the lysates were incubated at 4°C with 20 μL neutravidin beads (Thermo Scientific, Rockford IL) overnight. After three washes with Lysis buffer, bound protein was eluted from the beads by incubating in 2× Laemmli sample buffer and separated on 10% acrylamide SDS-PAGE.
Data analysis
Quantitative data are expressed as the means ± SD. Statistical comparisons between experimental groups were made using the two-tailed, unpaired Student’s t-test. Multi-group data were analyzed using one-way ANOVA followed by Tukey post hoc test to detect significant differences among treatments. Statistical analyses were performed with Excel and GraphPad Prism 6.0 (Graphpad Software Inc.). Probability values of p < 0.05 were considered significant.
RESULTS
Effects of Y and Y analog on Aβ reduction and AβPP dimerization inhibition
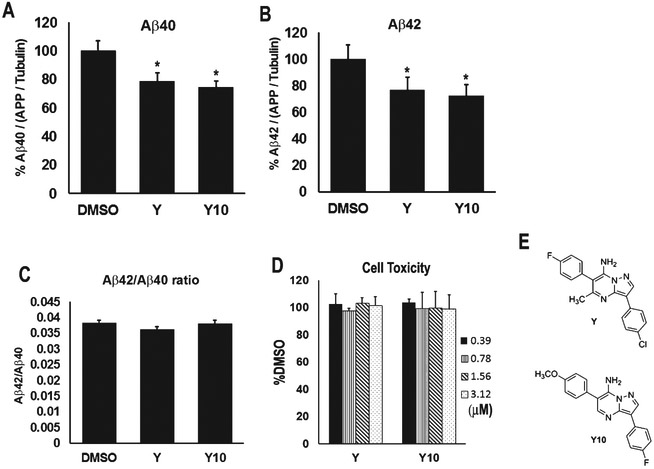
We have previously reported that Compound Y reduces production of both Aβ40 and Aβ42 [2]. We next tested a set of close structural analogs and found that a Y analog, Y10, also reduces Aβ40 and Aβ42 production (Fig. 1). Y10 was found to reduce Aβ40 and Aβ42 by 25.1% at 1 μM concentration, slightly better than Y (21.2% reduction) (Fig. 1A, B), with the Aβ42/Aβ40 ratio remaining unchanged (Fig. 1C). The compounds showed no cell toxicity at the concentrations tested (Fig. 1D). Initial assays indicated that Y10 inhibited AβPP dimerization in a dose-dependent manner. However, consistent with data reported for this compound in PubChem [13] we determined that Y10 inhibits firefly luciferase at low micromolar concentrations, and thus may interfere with our AβPP-Fluc enzyme complementation assay. Given this uncertainty, this study describes our efforts to identify other potential mechanisms by which the Y compounds may reduce Aβ production.
Fig. 1.
Effects of Y and its analog, Y10, on Aβ40 and Aβ42 production. The pyrazolopyrimidines compounds, Y and Y10, (F) were assayed at 1 μM for 18 h for their effects on Aβ40 level (A), Aβ42 level (B), and Aβ42/Aβ40 ratio (C), as measured by ELISA in HEK293 cells stably overexpressing AβPP751. Results were normalized to AβPP/tubulin ratio. Results are mean ± standard error, n=4-11. Asterisks (*) indicate statistical significance compared to DMSO of p<0.05 by t-test. D) Cell viability assay by crystal violet staining. The concentrations of Y and Y10 used were indicated. E) Structures of Y and Y10.
The Y series of compounds have in common a pyrazolopyrimidine heterocyclic motif that is found in many known kinase inhibitors [14-19]. We thus hypothesized that the mechanism of action of the Y series may involve inhibition of a critical kinase (or kinases) affecting AβPP processing. In order to identify possible target kinase(s), Y10 was profiled for inhibitory activity against a small panel of 69 kinases at a concentration of 1 μM. This panel was comprised of kinases reported to be inhibited by pyrazolopyrimidines in the patent and journal literature, as well as kinases known to be implicated in AD, including those known to phosphorylate AβPP [17-19]. In this particular panel, the sole kinase that was reproducibly and significantly inhibited was the receptor tyrosine kinase (RTK) cKit (68% inhibition, Table 1).
Table 1.
Kinase inhibition profiling of Y10 at 1 μM concentration.
| Kinase | Percent activity @ 1 μM Y-10 |
|---|---|
| Abl(h) | 83 |
| Abl(T315I)(h) | 96 |
| ASK1(h) | 107 |
| Aurora-A(h) | 91 |
| Aurora-B(h) | 148 |
| Aurora-C(h) | 96 |
| CaMKK2(h) | 102 |
| CDK1/cyclinB(h) | 98 |
| CDK2/cyclinA(h) | 107 |
| CDK2/cyclinE(h) | 101 |
| CDK3/cyclinE(h) | 100 |
| CDK5/p25(h) | 88 |
| CDK5/p35(h) | 108 |
| CDK6/cyclinD3(h) | 117 |
| CDK7/cyclinH/MAT1(h) | 96 |
| CDK9/cyclin T1(h) | 103 |
| CHK1(h) | 112 |
| CHK2(h) | 98 |
| CK1(y) | 97 |
| CK2(h) | 101 |
| cKit(h) | 32 |
| CSK(h) | 104 |
| EGFR(h) | 111 |
| EphB4(h) | 90 |
| FGFR1(h) | 87 |
| Flt1(h) | 129 |
| Fms(h) | 90 |
| Fyn(h) | 80 |
| GSK3α(h) | 93 |
| GSK3β(h) | 102 |
| IRE1(h) | 97 |
| JNK3(h) | 101 |
| KDR(h) | 85 |
| Lck(h) | 49 |
| LRRK2(h) | 110 |
| MAPK1(h) | 103 |
| MAPK2(h) | 100 |
| MAPKAP-K2(h) | 100 |
| MKK4(m) | 109 |
| mTOR(h) | 88 |
| PDGFRα(h) | 75 |
| PDGFRβ(h) | 87 |
| PDK1(h) | 109 |
| Pim-1(h) | 106 |
| Pim-2(h) | 107 |
| Pim-3(h) | 98 |
| PKA(h) | 111 |
| PKBα(h) | 98 |
| PKBβ(h) | 94 |
| PKCα(h) | 95 |
| PKC-II(h) | 107 |
| PKCγ (h) | 89 |
| PKCδ(h) | 102 |
| PKCϵ(h) | 107 |
| PKCη(h) | 101 |
| PKCι(h) | 102 |
| PKCμ(h) | 100 |
| PKCθ(h) | 104 |
| PKCζ(h) | 115 |
| Ret(h) | 118 |
| ROCK-II(h) | 102 |
| Ros(h) | 93 |
| Rsk2(h) | 96 |
| Src(1-530)(h) | 85 |
| SRPK2(h) | 105 |
| Tie2(h) | 97 |
| TrkB(h) | 107 |
| PI3 Kinase (p110b/p85a)(h) | 100 |
| PI3 Kinase (p110a/p65a)(h) | 98 |
Effects of Y10 and cKit on AβPP phosphorylation
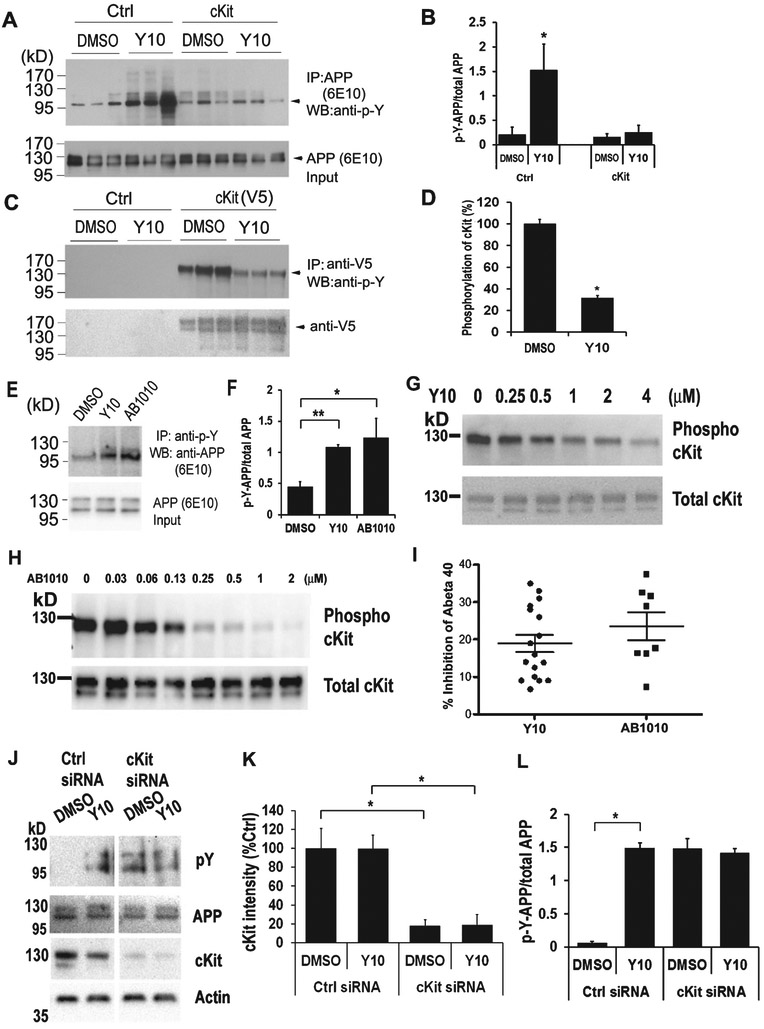
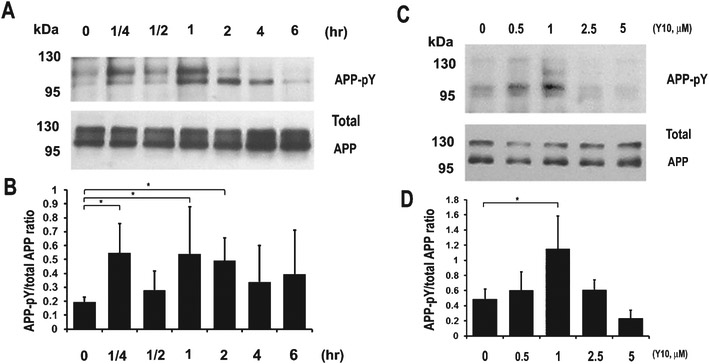
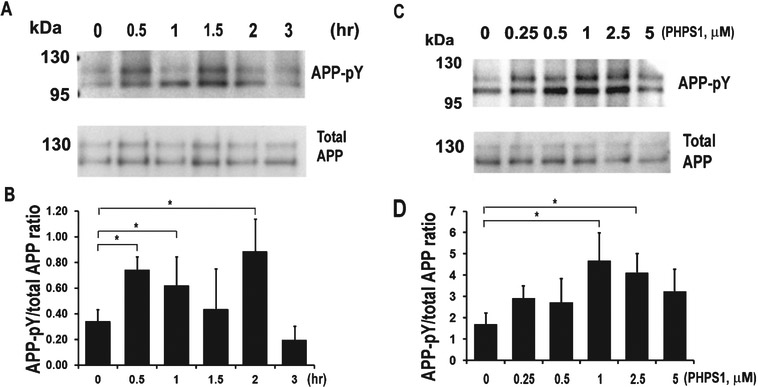
Having validated the kinase inhibitory activity of Y10, we next examined its effects on AβPP phosphorylation. Interestingly, we found that Y10 enhanced AβPP tyrosine phosphorylation, and also inhibited cKit autophosphorylation (Fig. 2A-D). Furthermore, overexpression of cKit reversed the effects of Y10 on AβPP phosphorylation (Fig. 2A); another known cKit inhibitor of a completely different structural class (AB1010, “masitinib”) also enhanced AβPP phosphorylation (Fig. 2E, F). In our gel system AβPP typically appears as two major bands at approximately 110 kD and slightly above 130 kD (Fig. 2A, E, J). Major phosphorylated AβPP bands were found around 110 kD and slightly below 110 kD (Fig. 2A, E, J); protein phosphorylation changes AβPP’s running position in SDS-PAGE. Our observation of the apparent molecular weight of AβPP phosphorylated on tyrosine residues is consistent with AβPP phosphorylation patterns at T668 [20]. Follow up experiments revealed that Y10 and AB1010 inhibit cKit tyrosine autophosphorylation in a dose dependent manner (Fig. 2G, H), with an IC50 of 0.47 μM and maximal inhibition of 72.6% (Y10), and IC50 of 0.15 μM and a 98.2% maximal inhibition (AB1010). AB1010 was found to reduce Aβ40 by 22% (N=8) at 1 μM concentration, slightly better than Y10 (19% reduction, N=17) (Fig. 2I). Specific knockdown of cKit with siRNA enhanced AβPP phosphorylation regardless of Y10 treatment (Fig. 2J-L). We also examined the kinetic and dose-dependent effects of Y10 on AβPP phosphorylation. The results showed that AβPP phosphorylation peaks approximately 1 h after Y10 treatment at the Y10 optimal concentration of 1 μM (Fig. 3).
Fig. 2.
Y10 enhances AβPP phosphorylation via cKit. A) IP-WB analysis of the effects of Y10 at 1 μM on AβPP phosphorylation of empty vector or cKit-transfected AβPP stable cells. B) Statistical analysis of (A). Results normalized to AβPP input. C) IP-WB analysis of the effects of Y10 on inhibition of cKit phosphorylation. D) Statistical analysis of (C). Results normalized to cKit input. E) In addition to Y10, a specific inhibitor of cKit, AB1010, also enhances AβPP phosphorylation on Tyr residue. F) Statistical analysis of (E). Results normalized to AβPP input. p-Y, phosphorylated Tyr. G, H) IP-WB analysis of the dose-dependent effects of Y10 and AB1010 on AβPP phosphorylation of cKit-transfected HEK cells stably expressing AβPP. I) Y10 and AB1010 were assayed at 1 μM for 18 h for their effects on Aβ40 level, as measured by ELISA in HEK293 cells stably overexpressing AβPP751. N=17 for Y10 and N=8 for AB1010. Standard error indicated. J) IP-WB analysis of the effects of Y10 at 1 μM on AβPP phosphorylation of control siRNA or cKit siRNA transfected HEK293 cells stably expressing AβPP. K) Statistical analysis of J for cKit intensity. L) Statistical analysis of G for AβPP phosphorylation. Results are mean ± standard deviation, n=3. Asterisks (*) indicate statistical significance of p<0.05 by t-test.
Fig. 3.
Kinetics and dose-response experiments for Y10-induced AβPP phosphorylation in HEK cells stably expressing AβPP751. A) Phosphorylation kinetics. IP-WB analysis of the effects of Y10 at 1 μM on AβPP phosphorylation at various time points. Results are from an average of 3-5 experiments. B) Dose-response experiments. IP-WB analysis of the effects of Y10 on AβPP phosphorylation at various concentrations at 2 h time point. Results are mean ± standard deviation, n=3. Asterisks (*) indicate statistical significance of p<0.05 by one-way ANOVA followed by Tukey post hoc test.
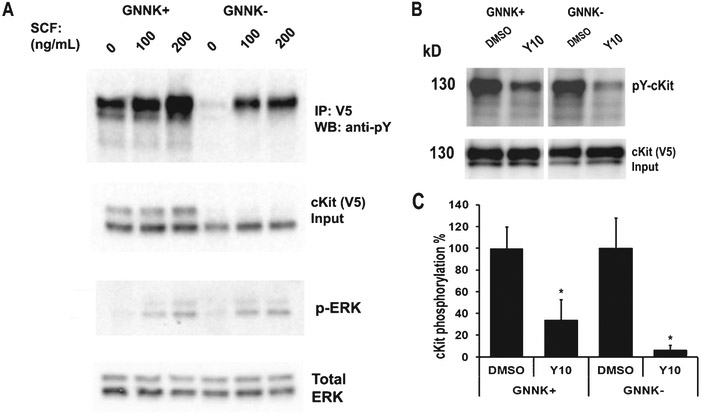
cKit is expressed as two different functional isoforms due to alternative splicing [21]. The two isoforms are characterized by the presence or absence of a tetrapeptide sequence (GNNK) in the extracellular juxtamembrane region. The GNNK− isoform is the predominant form found in most tissues and mediates stronger Src activation, leading to stronger recruitment of downstream signaling molecules [22]. However, the GNNK+ isoform, also responsible for critical, albeit weaker, signaling responses is expressed in elevated quantities in the brain [23]. We have shown that overexpression of the cKit GNNK+ isoform reversed the effects of Y10 on AβPP phosphorylation and cKit autophosphorylation (Fig. 2A). We then evaluated the effects of Y10 on both cKit isoforms. Upon stimulation with the cKit ligand stem cell factor (SCF), both cKit isoforms were phosphorylated with similar signaling effects on ERK phosphorylation in HEK cells (Fig. 4A). cKit has two major bands likely due to glycosylation and phosphorylation (Fig. 4A, B). In some cases, the GNNK− isoform showed a higher molecular weight band compared to GNNK+ (Fig. 4A). This could be due to the degree of post-translational modifications occurring in the two different isoforms. Y10 inhibited the autophosphorylation of both isoforms of cKit with a 70-95% reduction compared to the DMSO control (Fig. 4B). SCF, by activating cKit activity, should reduce AβPP phosphorylation. The addition of SCF slightly, but not significantly, reduced AβPP phosphorylation (data not shown), since the basal levels of AβPP phosphorylation are low.
Fig. 4.
Y10 inhibits autophosphorylation of both cKit isoforms. A) cKit signaling assay in HEK293 cells. cKit GNNK+ and GNNK− isoforms were transfected into HEK293 cells and 48 h post-transfection cells were treated with SCF as indicated for 15 min, and cell lysates were collected for WB analysis with the antibodies indicated. B) IP-WB analysis of the effects of Y10 at 1 μM on cKit phosphorylation of GNNK+ or GNNK− isoforms-transfected HEK293 cells. Cells were treated with Y10 for 90 min and then 200 ng/mL of SCF for 15 min, and cell lysates were collected for IP-WB analysis. C) Statistical analysis of (B). Results normalized to cKit input. p-Y, phosphorylated Tyr. Results are mean ± standard deviation, n=3. Asterisks (*) indicate statistical significance of p<0.05 by t-test.
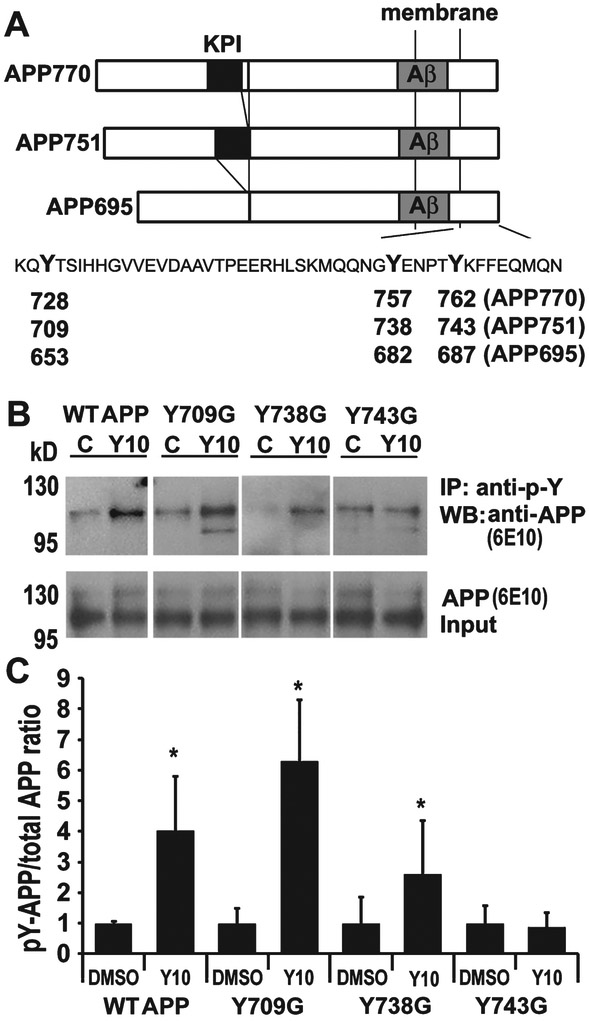
Tyrosine residue responsible for the effect of Y10 on AβPP phosphorylation
There are eight potential phosphorylation sites within the AβPP cytoplasmic domain, and seven of these potential sites have been shown to be phosphorylated in AD brains, Y653, S655, T668, S675, Y682, T686, and Y687 (AβPP695 isoform numbering) as shown in Figure 5A [3]. The phosphorylation state of AβPP, specifically at the cytoplasmic YENPTY motif, is important for the sorting of AβPP and Aβ production. We have demonstrated that Y10 enhances AβPP phosphorylation on a Tyr residue (Fig. 2 and 3). There are three Tyr residues in the cytoplasmic domain of AβPP: Y653, Y682, and Y687 according to AβPP695 numbering (Y709, Y738, and Y743 using AβPP751 numbering). To identify the Tyr residue(s) affected by Y10 treatment, we transfected HEK293 cells with three AβPP mutants (Y709G, Y738G, and Y743G) [24, 25] and determined the effects of Y10 treatment on the phosphorylation levels of each mutant. As depicted in Figure 5, mutation of solely the third Tyr residue (Y743) to Gly, resulted in a loss of Y10 effects on AβPP phosphorylation.
Fig. 5.
Y10 enhances phosphorylation at AβPP Y743. A) Schematic representation of the three isoforms of AβPP and the tyrosine residues in the cytosolic C-terminus. Kunitz protease inhibitor (KPI), transmembrane and Aβ domains are indicated. The C-terminal sequence is shown with the three tyrosine (Y) residues in bold. The numbering corresponding to the three isoforms of AβPP is indicated. B) IP-WB analysis of the effects of Y10 at 1 μM on AβPP phosphorylation of WT AβPP751 or AβPP751 mutants (Y709G, Y738G, and Y743G) on each tyrosine residue in the C-terminal domain on transfected HEK293 cells. C) Statistical analysis of (A). Results normalized to AβPP input. p-Y, phosphorylated Tyr; C, control DMSO treated; Y10, Y10 treated. Results are mean ± standard deviation, n=3. Asterisks (*) indicate statistical significance of p<0.05 by t-test.
Effects of Shp2 inhibitors on AβPP phosphorylation
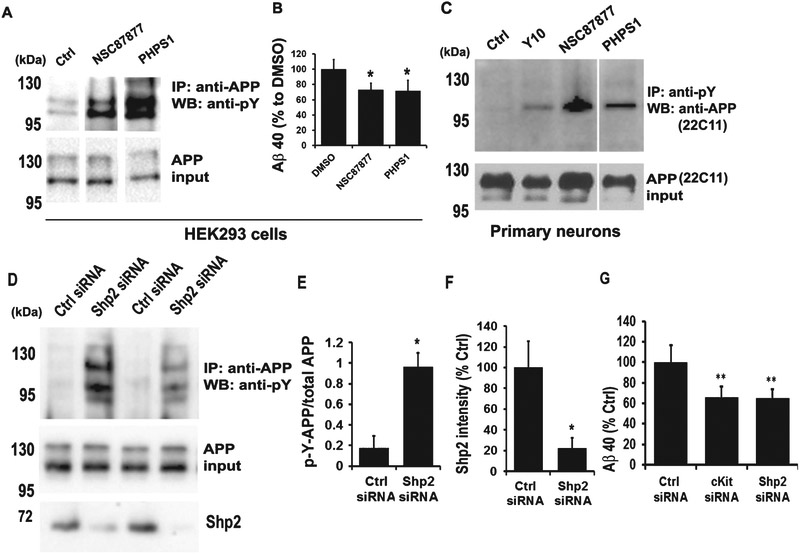
After identifying cKit as a relevant target for Y10, we searched for known cKit interactors that could also work downstream cKit and upstream of AβPP phosphorylation. Among cKit-interacting proteins are the SH protein tyrosine phosphatase-2 (Shp2; gene known as PTPN11), and the scaffolding protein Gab2, both of which bind to cKit via growth factor receptor binding protein-2 (Grb2) and SRC homology adaptor protein (Shc). Interestingly, the scaffolding protein Gab2, has been reported in a GWAS study to be associated with AD and involved in Aβ reduction [26]. In order to probe whether Shp2 is a potential modulator of AβPP phosphorylation, we examined the kinetic and dosedependent effects of the Shp2 inhibitor PHPS1 [27] on AβPP phosphorylation. PHPS1 increased AβPP phosphorylation, peaking between 1.5-2 h with an optimal concentration range of 1-2.5 μM (Fig. 6). We observed a biphasic behavior of AβPP phosphorylation with two significant peaks at 15 min and 2 h time points (Fig. 6A, B). A similar phenomenon was also observed in the phosphorylation kinetics of GSK2β [9] and ERK [28]. An additional known Shp2 inhibitor, NSC87877 [29], also increased AβPP phosphorylation and reduced Aβ production in HEK293 cells (Fig. 7A, B). Inhibition of cKit by Y10 and of Shp2 by either NSC87877 or PHPS1 resulted in increased AβPP phosphorylation in primary mouse hippocampal neurons (Fig. 7C). However, the increase in AβPP phosphorylation by Y10 in primary neurons is not as strong in comparison to the effects of the Shp2 inhibitors (Fig. 7C). The possible explanations include: 1) lower cKit expression in primary cells, and 2) Shp2 inhibition may directly affect AβPP phosphorylation, whereas cKit is a cell surface receptor, and there are many signaling molecules downstream that may affect AβPP phosphorylation. Furthermore, as a more direct assessment of the involvement of Shp2 in the phosphorylation state of AβPP, we performed Shp2 knockdown experiments. We found that a siRNA specific to Shp2 reduces Shp2 protein expression by 80% (Fig. 7D, F) and, concomitantly, AβPP phosphorylation was significantly increased (Fig. 7D, E). Finally, siRNA specific to cKit and Shp2 significantly reduced Aβ production in HEK293 cells (Fig. 7G). Taken together, these results strongly suggest that the phosphatase Shp2 is involved in AβPP dephosphorylation.
Fig. 6.
Kinetics and dose-response experiments for Shp2 inhibitor PHPS1-induced AβPP phosphorylation in HEK cells stably expressing AβPP751. A) Phosphorylation kinetics. IP-WB analysis of the effects of PHPS1 at 1 μM on AβPP phosphorylation at various time points. B) Dose-response experiments. IP-WB analysis of the effects of PHPS1 on AβPP phosphorylation at various concentrations at 2 h time point. All results are mean ± standard deviation, n=3. Asterisks (*) indicate statistical significance of p<0.05 by one-way ANOVA followed by Tukey post hoc test.
Fig. 7.
Shp2 inhibitors enhance AβPP phosphorylation and reduce Aβ production. A) IP-WB analysis of the effects of two Shp2 inhibitors at 1 μM for 2 h on AβPP phosphorylation in AβPP stable cells. B) Two Shp-2 inhibitors as indicated at 1 μM for 18 h were tested for effects on Aβ40 level as measured by ELISA. Results are normalized to total AβPP. Results are mean ± standard deviation, n=3. Asterisks (*) indicate statistical significance of p<0.05 by t-test. C) Y10 and Shp2 inhibitors at 1 μM for 2 h also increase AβPP phosphorylation in primary mouse hippocampal neurons. D) Shp2 siRNA knockdown enhances AβPP phosphorylation. IP-WB analysis of the effect of control (Ctrl) or Shp2 siRNA on AβPP phosphorylation. E) Statistical analysis of AβPP phosphorylation in (D). Results normalized to AβPP input. p-Y, phosphotyrosine. F) Statistical analysis of Shp2 protein intensity in (D). All results are mean ± standard deviation, n=3. Asterisks (*) indicate statistical significance of p<0.05 by t-test. G) The effect of control (Ctrl), cKit siRNA, or Shp2 siRNA on Aβ40 level as measured by ELISA. Media collected 24-48 h post-transfection. Results are normalized to total AβPP. Results are mean ± standard deviation, n=3. Asterisks (**) indicate statistical significance of p<0.05 by t-test.
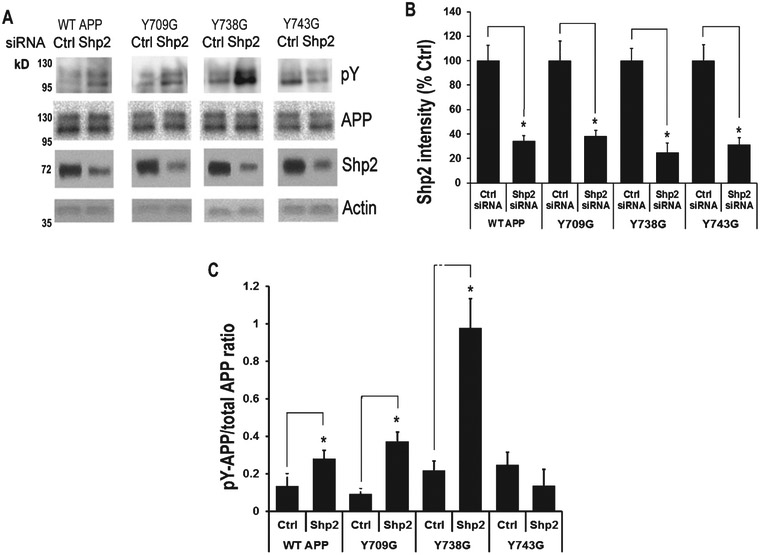
To investigate which tyrosine residue in AβPP is affected by Shp2 knockdown, we once again tested HEK293 cells transfected with three AβPP mutants (Y709G, Y738G, and Y743G) for changes in the effects of Shp2 knockdown by siRNA on AβPP phosphorylation. Similar to what we observed with Y10 treatment, the data showed that when the third tyrosine residue (Y743) was mutated to glycine, it resulted in a loss of Shp2 effects on AβPP phosphorylation (Fig. 8).
Fig. 8.
Shp2 siRNA knockdown enhances AβPP phosphorylation on Y743. A) IP-WB analysis of the effects of Shp2 siRNA on AβPP phosphorylation of WT AβPP751 or AβPP751 mutants (Y709G, Y738G, and Y743G) on each tyrosine residue in the C-terminal domain of transfected HEK293 cells. B, C) Statistical analysis of (A) showing Shp2 knockdown (B) and increased AβPP phosphorylation (C). Results normalized to Actin for Shp2 and to AβPP input for AβPP phosphorylation. p-Y, phosphotyrosine. Results are mean ± standard deviation, n=3. Asterisks (*) indicate statistical significance of p<0.05 by t-test.
We showed previously that compound Y lowered sAβPPβ levels, suggesting that it may affect the β-secretase cleavage of AβPP. We therefore examined whether the compounds could inhibit β-secretase activity. We found that neither Y nor Y10 inhibited β-secretase activity (Fig. 9A).
Fig. 9.
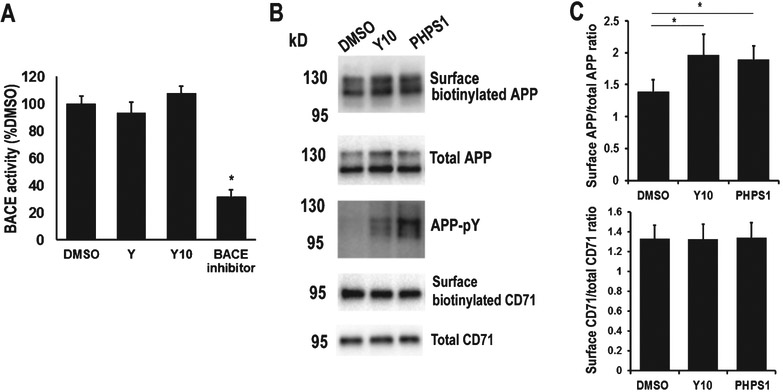
cKit and Shp2 inhibitors have no effect on BACE activity but enhance AβPP surface localization. A) BACE activity assay of cKit and Shp2 inhibitors. A BACE substrate analog peptide (0.1 μM) was used as inhibition control. B) The HEK293 cells stably expressing AβPP751 were treated with 1 μM of DMSO control, Y10 or PHPS1 for 2 h, and surface biotinylation assay was performed to analyze AβPP and a control membrane protein CD71 (transferrin receptor) surface localization. Biotinylated proteins were pulled-down by Neutravidin beads and detected by WB with anti-AβPP antibody (6E10), and anti-CD71 antibody. C) Statistical analysis of AβPP surface localization in (B). Results normalized to AβPP and CD71 inputs. p-Y, phosphotyrosine. Results are mean ± standard deviation, n=3. Asterisks (*) indicate statistical significance of p<0.05 by t-test.
AβPP surface localization and processing is regulated by cytoplasmic phosphorylation [30]. Given our findings that Y10, the cKit inhibitor, and Shp2 inhibitors both enhance AβPP phosphorylation, we next examined whether these inhibitors affect AβPP surface localization. We found that both Y10 and PHPS1, a Shp2 inhibitor, enhanced AβPP surface localization but not a control membrane protein CD71 (transferrin receptor) (Fig. 9B, C). These results suggest that the effects of Y10 and PHPS1 are specific to AβPP. Even though we detected a significant increase in AβPP surface localization, the enhancement was not dramatic, and it may only account partially for the effects of Y10 and PHPS1 on Aβ reduction.
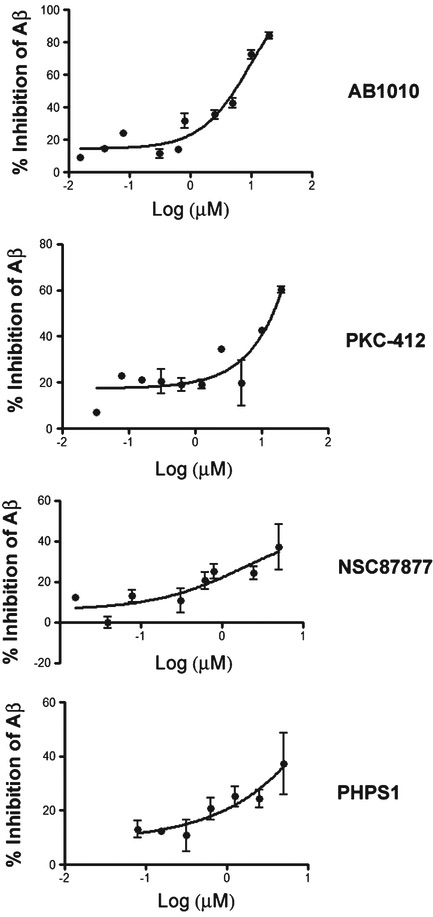
We next performed a 10-point dose response assessment of cKit inhibitors (Y, Y10, AB1010, PKC412, and Tandutinib) and Shp2 inhibitors (NSC87877 and PHPS1) to examine effects on Aβ40 reduction. While two cKit inhibitors (PKC412 and AB1010) did exhibit modest dose dependent Aβ reduction, there was significant cytotoxicity observed at the concentrations necessary to lower Aβ (Fig. 10). These results are not surprising, as PKC412 is a nonspecific tyrosine kinase inhibitor and c-Kit itself is involved in multiple critical cellular pathways. Y, Y10 and Tandutinib failed to show a dose dependent reduction in Aβ40. The inhibition patterns of the two Shp2 inhibitors (NSC87877 and PHPS1) are similar to each other with an optimal inhibition concentration of 5 μM (Fig. 10).
Fig. 10.
Dose response curve of cKit and Shp2 inhibitors on Aβ40 production. Concentration response curve of cKit inhibitors (PKC412, and AB1010) and Shp2 inhibitors (NSC87877 and PHPS1) on Aβ40 production as measured by ELISA in HEK293 cells stably overexpressing AβPP751. N=4 for each dose. Results were normalized to values obtained from cell viability assay by crystal violet staining. AβPP protein level is found to be linearly correlated to crystal violet staining (not shown). Results were normalized to % DMSO control.
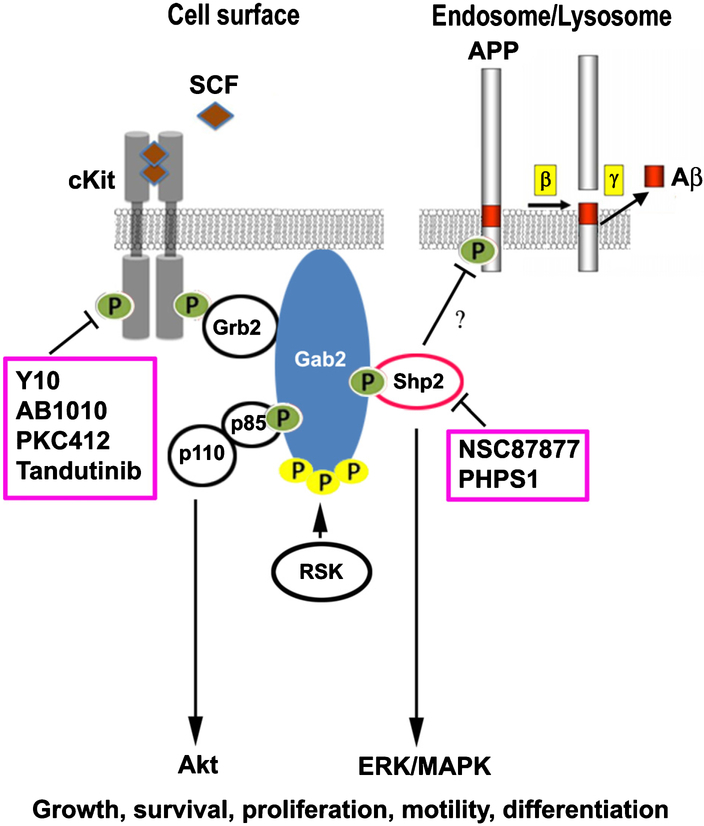
A proposed model for cKit and Shp2-mediated AβPP phosphorylation and Aβ reduction is shown in Figure 11. cKit plays important roles in the cell growth, survival, proliferation, and differentiation. When cKit binds its ligand, stem cell factor (SCF), cKit is autophosphorylated, and signaling commences via the adaptor Grb2 and scaffold protein Gab2 that recruits Shp2, p85 (the regulatory subunit of PI3-kinase), and other signaling molecules that activate Akt, and ERK/MAPK signaling pathways. RSK (p90 ribosomal S6 kinase) phosphorylates Gab2 and regulates its recruitment of signaling molecules. The basal level activity of cKit may have occurred from SCF stimulation found in the fetal calf serum. The basal activity would be required to recruit adapter proteins such as Grb2 and Gab2, which in turn would recruit active Shp2 to dephosphorylate AβPP and other target proteins to a basal level. cKit inhibitors, including Y, Y10, AB1010 and PKC412 [31] all were found to reduce Aβ. The Shp2 inhibitors PHPS1 and NSC87877 also reduced Aβ production (Figs. 7B and 10). We hypothesize that cKit-mediated recruitment of Shp2 is crucial for AβPP dephosphorylation. In summary, cKit and Shp2 inhibitors both increase AβPP phosphorylation in HEK293 cells leading to a reduction in Aβ production.
Fig. 11.
Current model for potential interactions among AβPP, cKit, Shp2, and Gab2. Gab2 is a scaffold protein found by GWAS to be associated with AD. Inhibition of cKit and Shp2 increases AβPP phosphorylation on the Y743 residue resulting in a reduction in Aβ production. Kinase and phosphatase inhibitors that increased AβPP phosphorylation and reduced Aβ formation are in pink boxes. Yellow P, pSer; Green P, pTyr; SCF, stem cell factor.
DISCUSSION
Protein phosphorylation is a highly dynamic process. In the cell, protein kinases, phosphatases, and their respective substrates must be precisely coordinated in cellular space and time in order to achieve signaling specificity in response to a given environmental stimulus. In terms of AβPP phosphorylation, there is very little basal phosphorylation at steady-state level (Fig. 2A, B, E, and F, DMSO controls). Upon drug treatment, AβPP is phosphorylated and quickly dephosphorylated back to a basal level (Figs. 3 and 6).
In this study, we found that the Y-series of compounds, originally discovered in a screen for AβPP dimerization modulators and shown to modestly lower Aβ levels, also increase levels of AβPP phosphorylation. We determined that a single tyrosine residue is responsible for Y10-mediated AβPP phosphorylation enhancement (Y743). We also identified the tyrosine receptor kinase cKit as a relevant Y10 target. Given the known promiscuity among kinase inhibitors, further profiling will be required to assess for the presence of other relevant kinases or targets for the chemotype.
The receptor tyrosine kinase cKit is involved in several types of cancer, such as gastrointestinal tumors, systemic mastocytosis, and subsets of acute myeloid leukemia and melanoma [32]. Signaling through cKit is crucial for normal hematopoiesis, pigmentation, fertility and gut movement (reviewed in [33]). cKit involvement in some aspects of the nervous system is of greater relevance to our study. Earlier evidence implicated cKit signaling in neurogenesis through regulation of neurite outgrowth in dorsal root ganglia neurons [34] and crest-derived melanocyte precursors [35]. Later studies implicated the cKit pathway in the migration of neural stem/progenitor cell to sites of brain injury, when SCF, a cKit ligand, was applied exogenously [36]. cKit protein expression was demonstrated in the dendrites of pyramidal cells in the stratum radiatum area of the hippocampus [37], raising the possibility that cKit downstream signaling is involved in synaptic transmission in the hippocampus [38]. Indeed, impaired spatial learning and memory was demonstrated in homozygous cKit mutant rats with very low cKit kinase activity due to a deletion in the tyrosine kinase domain [38].
Our results suggest that dephosphorylation of AβPP Y743 is regulated by either cKit-mediated deactivation of a second, unknown kinase or, conversely, cKit-mediated activation of a downstream phosphatase. One phosphatase directly linked to cKit signaling is the tyrosine phosphatase Shp2. We have demonstrated that known Shp2 inhibitors also increase AβPP phosphorylation and lower Aβ, further supporting our hypothesis that cKit is an upstream modulator of AβPP phosphorylation. In addition, siRNA specific to Shp2 increased AβPP phosphorylation, suggesting that Shp2 is involved in AβPP dephosphorylation (Fig. 7D-F). The involvement of protein phosphatases in AD has been recognized (reviewed in [39]). Protein kinases that phosphorylate tau and AβPP and affect the AD pathology present a viable drug target, and the role of protein phosphatases that reverse the effect of the kinases should be considered as equally influential (reviewed in [7]). Protein phosphatases are distinguished based on their potential to dephosphorylate serine, threonine or tyrosine residues. Protein phosphatase 2A (PP2A) is a threonine phosphatase that is linked to AD pathology through its involvement in tau dephosphorylation [40, 41], and dephosphorylation of AβPP on Thr668, leading to decreased levels of Aβ [42].
In this report, we provide the first link between protein tyrosine phosphatase (PTP) Shp2 inhibition and AβPP processing. Recent studies have suggested a role for other PTPs in AD. For example, striatal-enriched tyrosine phosphatase (STEP) has been shown to promote the effect of Aβ on glutamate receptor endocytosis [43], while increased levels of STEP have been detected in the frontal cortex of AD patients [44]. The levels of another PTP, PTP1B, have been shown to be increased in the mouse model of AD (AβPP(SWE/PSEN1A246E)) and are associated with glucose intolerance [45]. This increasing evidence supports a crucial role of protein phosphorylation in the normal and abnormal processing of AβPP and Aβ production. Hence, further investigation of the role of various phosphatases in the regulation of these processes and the development of their potential inhibitors is warranted.
There are three potential tyrosine phosphorylation sites within the AβPP cytoplasmic domain, and all three have been shown to be phosphorylated in AD brains [3]. Phosphorylation of Y682 and T668 (AβPP695 isoform numbering, part of the YENPTY sequence) increases Aβ production [3], while phosphorylation of Y687 decreases it. These differences in phosphorylation outcomes are likely a result of altered cell surface localization [30]. In this study, we demonstrate that the Y10 treatment and down-regulation of Shp2 expression by siRNA, which lead to an increase in AβPP phosphorylation, is specific to Y743 (equivalent to Y687) (Figs. 5 and 8). We have further found that Aβ production is reduced with siRNA to cKit and Shp2 treatment (Fig. 7G), which is consistent with studies published by other researchers who reported a decrease in Aβ, when Y687 in AβPP is phosphorylated [30]. AβPP phosphorylation regulates AβPP endocytosis, AβPP processing, and Aβ production [3, 30, 46] and can facilitate the interaction with adaptor proteins such as Shc, NUMB, DAB, X11, and JIP [7]. Phosphorylation of AβPP at Y687 affects processing and subcellular distribution. An AβPP mutant mimicking constitutive dephosphorylation of Y687 (Y687F) was shown to have a faster turnover rate when compared to both WT AβPP and the mutant mimicking constitutive phosphorylation (Y687E) [30]. In addition, the Y687E mutant was not incorporated into visible vesicular structures, resulting in a dramatic decrease in Aβ production [30]. In contrast, the Y687F mutant was endocytosed similarly to WT AβPP, but was relatively favored for β-secretase cleavage [30]. Another study using mutagenesis found that Y687 in AβPP was the only Tyr residue that contributed to AβPP binding to AP-4 complex involved in transport of AβPP from the trans-Golgi network (TGN) to endosomes [47]. Disruption of the AβPP-AP-4 interaction decreases AβPP localization to endosomes and affects Aβ production [47]. In the present study, we found that inhibitors of cKit phosphorylation and Shp2 activity both resulted in enhancement of AβPP surface localization (Fig. 9B, C). However, the increase in AβPP surface localization may account only partially for the reduction in Aβ production. The detailed mechanisms leading to the reduction in Aβ and the potential adaptor proteins known to bind to the intracellular C-terminus of AβPP and reported to be involved in AβPP processing and Aβ production would require further investigation. Whether AβPP dimerization leads to reduction or enhancement of Aβ production is still controversial [48]. We previous hypothesized that inhibition of AβPP dimerization results in Aβ reduction, which led us to the discovery of the Y compound [2]. AβPP may dimerize via the adapter proteins that bind to phosphorylated AβPP in the C-terminal domain. The particular phosphorylation site may control the different subcellular localization for AβPP processing, leading to either an increase or decrease in Aβ production.
In this study, we identified a novel regulatory mechanism of Aβ production through the modulation of AβPP phosphorylation at Y687. The consequence of phosphorylation at Y687 is opposite of what was observed for AβPP phosphorylation at Y682 and T668. Increasing phosphorylation of Y687 by either inhibition of cKit or Shp2 resulted in Aβ reduction, while increasing phosphorylation at Y682 and T668 resulted in a higher level of Aβ production [3]. The Shp2 inhibitor, NSC87877, that reduced Aβ in our study, was also tested in vivo and demonstrated Aβ reduction [49]. However, NSC87877 is not a specific Shp2 inhibitor, as it also inhibits DUSP26, and Shp1 which is involved in AβPP axonal transport and inflammatory response by macrophages [49, 50]. Searching for a specific and potent Shp2 inhibitor is required for further investigating its role in vivo in an AD mouse model.
We did not find a correlation in relative activities between cKit phosphorylation inhibitors and Aβ production inhibitors, although we clearly demonstrated that cKit phosphorylation was inhibited by Y10 and AB1010 (Figs. 2 and 4) and knockdown of cKit and Shp2 reduced Aβ production (Fig. 7G). Several possibilities may explain the lack of correlation between cKit inhibition and Aβ reduction: 1) multiple steps are involved from cKit inhibition to kinase/phosphatase signaling and AβPP phosphorylation, trafficking and processing making it somewhat difficult to envision a linear relationship with so many steps and limiting factors involved including other endogenous kinases and phosphatases we cannot control; 2) AβPP interacting proteins/binding partners and trafficking molecules do not change although cKit is inhibited; 3) there may be off-target effects of the inhibitors that also contribute to AβPP trafficking/processing; and finally 4) the variation in Aβ reduction (noise) is large even with the gamma-secretase inhibitor DAPT (data not shown). Because of the reasons mentioned above, it is unlikely that one can use cKit inhibition to screen for better analogs that reduce Aβ production. Nevertheless, it may be beneficial to test existing cKit inhibitors such as AB1010 and Shp2 inhibitors such as NSC87877, PHPS1 for Aβ-lowering in vivo in AβPP mouse models.
Small molecule kinase inhibitors have been successfully used in treatment of gastrointestinal stromal tumors [32]. Masitinib (AB1010) is currently in Phase III clinical trials for the treatment of many cancers, and it is also in Phase II/III clinical trials for the treatment of multiple sclerosis, rheumatoid arthritis, and AD [51]. In the case of AD, the authors hypothesize that masitinib’s targeted inhibitory action on anti-inflammation of mast cells may reduce the symptoms of AD. Likewise, there has been an interest in developing Shp2 inhibitors for cancer treatment, as studies implicate Shp2 as a key factor in breast cancer progression [52, 53]. In this study, we identified cKit and Shp2 as novel targets in a pathway that involves AβPP phosphorylation and Aβ production. Overall, this study presents a new direction toward the characterization of the precise roles of cKit and Shp2 in Aβ reduction, and the determination of their utility as potential therapeutic targets for AD.
ACKNOWLEDGEMENTS
This work was supported by grants from the Alzheimer’s Association, the Cure Alzheimer’s Fund and the Boston University Alzheimer’s Disease Center. Work at the BU-CMD is supported by R24-GM111625.
Footnotes
Authors’ disclosures available online (https://www.j-alz.com/manuscript-disclosures/18-0923r2).
REFERENCES
- [1].Selkoe DJ, Hardy J (2016) The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med 8, 595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].So PP, Zeldich E, Seyb KI, Huang MM, Concannon JB, King GD, Chen CD, Cuny GD, Glicksman MA, Abraham CR (2012) Lowering of amyloid beta peptide production with a small molecule inhibitor of amyloid-βeta precursor protein dimerization. Am J Neurodegener Dis 1, 75–87. [PMC free article] [PubMed] [Google Scholar]
- [3].Lee MS, Kao SC, Lemere CA, Xia W, Tseng HC, Zhou Y, Neve R, Ahlijanian MK, Tsai LH (2003) APP processing is regulated by cytoplasmic phosphorylation. J Cell Biol 163, 83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Steinhilb ML, Turner RS, Gaut JR (2002) ELISA analysis of beta-secretase cleavage of the Swedish amyloid precursor protein in the secretory and endocytic pathways. J Neurochem 80, 1019–1028. [DOI] [PubMed] [Google Scholar]
- [5].Takahashi K, Niidome T, Akaike A, Kihara T, Sugimoto H (2008) Phosphorylation of amyloid precursor protein (APP) at Tyr687 regulates APP processing by alpha- and gamma-secretase. Biochem Biophys Res Commun 377, 544–549. [DOI] [PubMed] [Google Scholar]
- [6].Matrone C, Luvisetto S, La Rosa LR, Tamayev R, Pignataro A, Canu N, Yang L, Barbagallo AP, Biundo F, Lombino F, Zheng H, Ammassari-Teule M, D’Adamio L (2012) Tyr682 in the Abeta-precursor protein intracellular domain regulates synaptic connectivity, cholinergic function, and cognitive performance. Aging Cell 11, 1084–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tamayev R, Zhou D, D’Adamio L (2009) The interactome of the amyloid beta precursor protein family members is shaped by phosphorylation of their intracellular domains. Mol Neurodegene 4, 28-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chen CD, Sloane JA, Li H, Aytan N, Giannaris EL, Zeldich E, Hinman JD, Dedeoglu A, Rosene DL, Bansal R, Luebke JI, Kuro-o M, Abraham CR (2013) The antiaging protein Klotho enhances oligodendrocyte maturation and myelination of the CNS. J Neurosci 33, 1927–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chen CD, Sloane JA, Li H, Aytan N, Giannaris EL, Zeldich E, Hinman JD, Dedeoglu A, Rosene DL, Bansal R, Luebke JI, Kuro-o M, Abraham CR (2013) The antiaging protein Klotho enhances oligodendrocyte maturation and myelination of the CNS. J. Neurosci 33, 1927–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Davies SP, Reddy H, Caivano M, Cohen P (2000) Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J 351, 95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chen CD, Podvin S, Gillespie E, Leeman SE, Abraham CR (2007) Insulin stimulates the cleavage and release of the extracellular domain of Klotho by ADAM10 and ADAM17. Proc Natl Acad Sci USA 104, 19796–19801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zeldich E, Chen CD, Colvin TA, Bove-Fenderson EA, Liang J, Tucker Zhou TB, Harris DA, Abraham CR (2014) The neuroprotective effect of Klotho is mediated via regulation of members of the redox system. J Biol Chem 289, 24700–24715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Thorne N, Inglese J, Auld DS (2010) Illuminating insights into firefly luciferase and other bioluminescent reporters used in chemical biology. Chem Biol 17, 646–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Frey RR, Curtin ML, Albert DH, Glaser KB, Pease LJ, Soni NB, Bouska JJ, Reuter D, Stewart KD, Marcotte P, Bukofzer G, Li J, Davidsen SK, Michaelides MR (2008) 7-Aminopyrazolo[1,5-a]pyrimidines as potent multitargeted receptor tyrosine kinase inhibitors. J Med Chem 51, 3777–3787. [DOI] [PubMed] [Google Scholar]
- [15].Montano R, Chung I, Garner KM, Parry D, Eastman A (2012) Preclinical development of the novel Chk1 inhibitor SCH900776 in combination with DNA-damaging agents and antimetabolites. Mol Cancer Ther 11, 427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dwyer MP, Paruch K, Labroli M, Alvarez C, Keertikar KM, Poker C, Rossman R, Fischmann TO, Duca JS, Madison V, Parry D, Davis N, Seghezzi W, Wiswell D, Guzi TJ (2011) Discovery of pyrazolo[1,5-a]pyrimidine-based CHK1 inhibitors: a template-based approach-part 1. Bioorg Med Chem Lett 21, 467–470. [DOI] [PubMed] [Google Scholar]
- [17].Yang LL, Li GB, Yan HX, Sun QZ, Ma S, Ji P, Wang ZR, Feng S, Zou J, Yang SY (2012) Discovery of N6-phenyl-1H-pyrazolo[3,4-d]pyrimidine-3,6-diamine derivatives as novel CK1 inhibitors using common-feature pharmacophore model based virtual screening and hit-to-lead optimization. Eur J Med Chem 56, 30–38. [DOI] [PubMed] [Google Scholar]
- [18].Dowling JE, Chuaqui C, Pontz TW, Lyne PD, Larsen NA, Block MH, Chen H, Su N, Wu A, Russell D, Pollard H, Lee JW, Peng B, Thakur K, Ye Q, Zhang T, Brassil P, Racicot V, Bao L, Denz CR, Cooke E (2012) Potent and selective inhibitors of CK2 kinase identified through structure-guided hybridization. ACS Med Chem Lett 3, 278–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gommermann N, Buehlmayer P, von Matt A, Breitenstein W, Masuya K, Pirard B, Furet P, Cowan-Jacob SW, Weckbecker G (2010) New pyrazolo 1,5a pyrimidines as orally active inhibitors of Lck. Bioorg Med Chem Lett 20, 3628–3631. [DOI] [PubMed] [Google Scholar]
- [20].Bukhari H, Kolbe K, Leonhardt G, Loosse C, Schroder E, Knauer S, Marcus K, Muller T (2016) Membrane tethering of APP c-terminal fragments is a prerequisite for T668 phosphorylation preventing nuclear sphere generation. Cell Signal 28, 1725–1734. [DOI] [PubMed] [Google Scholar]
- [21].Reith AD, Ellis C, Lyman SD, Anderson DM, Williams DE, Bernstein A, Pawson T (1991) Signal transduction by normal isoforms and W mutant variants of the Kit receptor tyrosine kinase. EMBO J 10, 2451–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sun J, Pedersen M, Ronnstrand L (2008) Gab2 is involved in differential phosphoinositide 3-kinase signaling by two splice forms of c-Kit. J Biol Chem 283, 27444–27451. [DOI] [PubMed] [Google Scholar]
- [23].Piao XH, Curtis JE, Minkin S, Minden MD, Bernstein A (1994) Expression of the Kit and KitA receptor isoforms in human acute myelogenous leukemia. Blood 83, 476–481. [PubMed] [Google Scholar]
- [24].Hicks DA, Makova NZ, Gough M, Parkin ET, Nalivaeva NN, Turner AJ (2013) The amyloid precursor protein represses expression of acetylcholinesterase in neuronal cell lines. J Biol Chem 288, 26039–26051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gough M, Blanthorn-Hazell S, Delury C, Parkin E (2014) The E1 copper binding domain of full-length amyloid precursor protein mitigates copper-induced growth inhibition in brain metastatic prostate cancer DU145 cells. Biochem Biophys Res Commun 453, 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zou F, Belbin O, Carrasquillo MM, Culley OJ, Hunter TA, Ma L, Bisceglio GD, Allen M, Dickson DW, Graff-Radford NR, Petersen RC, Genetic and Environmental Risk for Alzheimer’s disease (GERAD1) Consortium, Morgan K, Younkin SG (2013). Linking protective GAB2 variants, increased cortical GAB2 expression and decreased Alzheimer’s disease pathology. PLoS One 8, e64802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hellmuth K, Grosskopf S, Lum CT, Wurtele M, Roder N, von Kries JP, Rosario M, Rademann J, Birchmeier W (2008) Specific inhibitors of the protein tyrosine phosphatase Shp2 identified by high-throughput docking. Proc NatlAcadSci USA 105, 7275–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zeldich E, Koren R, Nemcovsky C, Weinreb M (2007) Enamel matrix derivative stimulates human gingival fibroblast proliferation via ERK. J Dent Res 86, 41–46. [DOI] [PubMed] [Google Scholar]
- [29].Cooke M, Orlando U, Maloberti P, Podesta EJ, Cornejo Maciel F (2011) Tyrosine phosphatase SHP2 regulates the expression of acyl-CoA synthetase ACSL4. J Lipid Res 52, 1936–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Rebelo S, Vieira SI, Esselmann H, Wiltfang J, da Cruz e Silva EF, da Cruz e Silva OA (2007) Tyrosine 687 phosphorylated Alzheimer’s amyloid precursor protein is retained intracellularly and exhibits a decreased turnover rate. Neu- rodegener Dis 4, 78–87. [DOI] [PubMed] [Google Scholar]
- [31].Fabbro D, Ruetz S, Bodis S, Pruschy M, Csermak K, Man A, Campochiaro P, Wood J, O’Reilly T, Meyer T (2000) PKC412-a protein kinase inhibitor with a broad therapeutic potential. Anticancer Drug Des 15, 17–28. [PubMed] [Google Scholar]
- [32].Ashman LK, Griffith R (2013) Therapeutic targeting of c-KIT in cancer. Expert Opin Investig Drugs 22, 103–115. [DOI] [PubMed] [Google Scholar]
- [33].Lennartsson J, Ronnstrand L (2012) Stem cell factor receptor/c-Kit: from basic science to clinical implications. Physiol Rev 92, 1619–1649. [DOI] [PubMed] [Google Scholar]
- [34].Langtimm-Sedlak CJ, Schroeder B, Saskowski JL, Carnahan JF, Sieber-Blum M (1996) Multiple actions of stem cell factor in neural crest cell differentiation in vitro. Dev Biol 174, 345–359. [DOI] [PubMed] [Google Scholar]
- [35].Hirata T, Morii E, Morimoto M, Kasugai T, Tsujimura T, Hirota S, Kanakura Y, Nomura S, Kitamura Y (1993) Stem cell factor induces outgrowth of c-kit-positive neurites and supports the survival of c-kit-positive neurons in dorsal root ganglia of mouse embryos. Development 119, 49–56. [DOI] [PubMed] [Google Scholar]
- [36].Sun L, Lee J, Fine HA (2004) Neuronally expressed stem cell factor induces neural stem cell migration to areas of brain injury. J Clin Invest 113, 1364–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhang SC, Fedoroff S (1997) Cellular localization of stem cell factor and c-kit receptor in the mouse nervous system. J Neurosci Res 47, 1–15. [PubMed] [Google Scholar]
- [38].Katafuchi T, Li AJ, Hirota S, Kitamura Y, Hori T (2000) Impairment of spatial learning and hippocampal synaptic potentiation in c-kit mutant rats. Learn Mem 7, 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Braithwaite SP, Stock JB, Lombroso PJ, Nairn AC (2012) Protein phosphatases and Alzheimer’s disease. Prog Mol Biol Transl Sci 106, 343–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Liu F, Grundke-Iqbal I, Iqbal K, Gong CX (2005) Contributions of protein phosphatases PP1, PP2A, PP2B and PP5 to the regulation of tau phosphorylation. Eur J Neurosci 22, 1942–1950. [DOI] [PubMed] [Google Scholar]
- [41].Wang JZ, Grundke-Iqbal I, Iqbal K (2007) Kinases and phosphatases and tau sites involved in Alzheimer neurofibrillary degeneration. Eur J Neurosci 25, 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sontag E, Nunbhakdi-Craig V, Sontag JM, Diaz-Arrastia R, Ogris E, Dayal S, Lentz SR, Arning E, Bottiglieri T (2007) Protein phosphatase 2A methyltransferase links homocysteine metabolism with tau and amyloid precursor protein regulation. J Neurosci 27, 2751–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhang Y, Kurup P, Xu J, Carty N, Fernandez SM, Nygaard HB, Pittenger C, Greengard P, Strittmatter SM, Nairn AC, Lombroso PJ (2010) Genetic reduction of striatal-enriched tyrosine phosphatase (STEP) reverses cognitive and cellular deficits in an Alzheimer’s disease mouse model. Proc Natl Acad Sci USA 107, 19014–19019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kurup P, Zhang Y, Xu J, Venkitaramani DV, Haroutunian V, Greengard P, Nairn AC, Lombroso PJ (2010) Abetamediated NMDA receptor endocytosis in Alzheimer’s disease involves ubiquitination of the tyrosine phosphatase STEP61. J Neurosci 30, 5948–5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Mody N, Agouni A, McIlroy GD, Platt B, Delibegovic M (2011) Susceptibility to diet-induced obesity and glucose intolerance in the APP (SWE)/PSEN1 (A246E) mouse model of Alzheimer’s disease is associated with increased brain levels of protein tyrosine phosphatase 1B (PTP1B) and retinol-binding protein 4 (RBP4), and basal phosphorylation of S6 ribosomal protein. Diabetologia 54, 2143–2151. [DOI] [PubMed] [Google Scholar]
- [46].Suzuki T, Nakaya T (2008) Regulation of amyloid beta-protein precursor by phosphorylation and protein interactions. J Biol Chem 283, 29633–29637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Burgos PV, Mardones GA, Rojas AL, daSilva LL, Prabhu Y, Hurley JH, Bonifacino JS (2010) Sorting of the Alzheimer’s disease amyloid precursor protein mediated by the AP-4 complex. Dev Cell 18, 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Eggert S, Midthune B, Cottrell B, Koo EH (2009) Induced dimerization of the amyloid precursor protein leads to decreased amyloid-βeta protein production. J Biol Chem 284, 28943–28952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Jung S, Nah J, Han J, Choi SG, Kim H, Park J, Pyo HK, Jung YK (2016) Dual-specificity phosphatase 26 (DUSP26) stimulates Abeta42 generation by promoting amyloid precursor protein axonal transport during hypoxia. J Neurochem 137, 770–781. [DOI] [PubMed] [Google Scholar]
- [50].Capiralla H, Vingtdeux V, Venkatesh J, Dreses-Werringloer U, Zhao H, Davies P, Marambaud P (2012) Identification of potent small-molecule inhibitors of STAT3 with anti-inflammatory properties in RAW 264.7 macrophages. FEBS J 279, 3791–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Folch J, Petrov D, Ettcheto M, Pedros I, Abad S, Beas- Zarate C, Lazarowski A, Marin M, Olloquequi J, Auladell C, Camins A (2015) Masitinib for the treatment of mild to moderate Alzheimer’s disease. Expert Rev Neurother 15, 587–596. [DOI] [PubMed] [Google Scholar]
- [52].Butterworth S, Overduin M, Barr AJ (2014) Targeting protein tyrosine phosphatase SHP2 for therapeutic intervention. Future Med Chem 6, 1423–1437. [DOI] [PubMed] [Google Scholar]
- [53].Aceto N, Sausgruber N, Brinkhaus H, Gaidatzis D, Martiny-Baron G, Mazzarol G, Confalonieri S, Quarto M, Hu G, Balwierz PJ, Pachkov M, Elledge SJ, van Nimwegen E, Stadler MB, Bentires-Alj M (2012) Tyrosine phosphatase SHP2 promotes breast cancer progression and maintains tumor-initiating cells via activation of key transcription factors and a positive feedback signaling loop. Nat Med 18, 529–537. [DOI] [PubMed] [Google Scholar]