Abstract
Two-photon excitation microscopy (TPEM) has revolutionized our understanding of adaptive immunity. However, TPEM usually requires animal models and is not amenable to the study of human disease. Recognition of antigen by T cells requires cell contact and is associated with changes in T cell shape. We postulated that by capturing these features in fixed-tissue samples, we could quantify in situ adaptive immunity. Therefore, we used a deep convolutional neural network to identify fundamental distance and cell shape features associated with cognate help (Cell Distance Mapping, CDM). In mice, CDM was comparable to TPEM for discriminating cognate from non-cognate T cell:dendritic cell (DC) interactions. In human lupus nephritis, CDM confirmed that myeloid DCs present antigen to CD4+ T cells and identified plasmacytoid DCs as an important antigen-presenting cell. These data reveal a new approach to study human in situ adaptive immunity broadly applicable to autoimmunity, infection, and cancer.
Introduction
Adaptive immunity depends upon both antigen-restricted cell-cell interactions and environmental niches, which enable and coordinate cellular communication. In mice, two-photon excitation microscopy (TPEM) has revolutionized our understanding of immune cell architectures and their contribution to normal immunity. By visualizing cells and structures in live hosts, TPEM provides both a quantitative and dynamic picture of immune processes1, 2, 3, 4, 5.
While the gold standard for understanding the organization of immunity, TPEM has several limitations. Cells must be fluorescently labeled to be visualized6, 7 and, therefore, manipulated systems must be used8. Only small volumes of tissue can be assessed and this must be done over sufficient time to capture cellular dynamics. These restraints limit the number of measurements that can be practically obtained using TPEM. Furthermore, only tissue that can be exposed in live mice is generally amenable to TPEM. While TPEM has a maximal effective depth of 1.6 mm9, most applications are limited to less than 500 μm. Therefore, immune processes occurring within the interior of some organs cannot be visualized. Finally, with few exceptions10, 11, TPEM cannot be used to directly study human disease.
Great strides have been made in multiparameter imaging of fixed-human tissue such that 36 or more markers can be assayed simultaneously12, 13, 14, 15. With these and other approaches16, 17 one can identify infiltrating cell subsets and describe their relative regional behaviors. Such studies have revealed that the cellular constituency of inflammation is very complex16,18 and the organization of immune cells can be both characteristic of disease states13 and define prognosis14. However, it is difficult to know why different cell populations appear together. Cells such as T cells and antigen-presenting cells (APCs), can engage in cognate interactions that drive local adaptive immunity and inflammation19,20. Alternatively, cells can just be bystanders of inflammation with different populations coalescing because they are responding to similar environmental cues such as chemokines21. There are limited tools to discriminate between these states in human tissue.
Previously, we demonstrated that quantitative analysis of human frozen tissue samples, imaged by multicolor confocal microscopy, could be used to characterize interactions between T follicular helper (TFH) cell populations and B cells19. In these investigations, we observed that when TFH cells formed cognate interactions with B cells, their nuclei became tightly apposed. These data indicate that distances between nuclear borders can discriminate between cognate interactions and when T and B cells are merely in close proximity. Therefore, by mapping relative distances between T and B cells in tissue (CDM), we could identify functional relationships.
However, the fixed filters and algorithms used in CDM to segment signals within tissue were insufficient for defining positions of larger complex objects such as stains associated with DCs. Furthermore, CDM did not accurately capture object shape. We postulated that this might be important, as T cells adopt different shapes when scanning for antigen and after recognizing peptides in the context of MHC22, 23, 24, 25, 26, 27, 28. In the latter case, T cells flatten against the APC to form a stable synapse. In contrast, T cells scanning for antigen or those engaged in brief antigen-specific interactions (kinapses), do not undergo the same changes in T cell shape and polarity29. We hypothesized that using computational tools that accurately captured T cell shape features and DC boundaries, we could identify stable synapses and, thus, discriminate between cognate and non-cognate T cell:APC interactions in human tissue.
Therefore, we implemented a deep convolutional neural network (DCNN) that precisely measured both cell position and shape. The DCNN output was then analyzed with a tuned neural network (TNN) to identify the combination of distance and cell shape features that best discriminated between different T cell populations relative to DCs. The use of a TNN allowed us to restrict our analysis to fundamental morphological features of T cell:DC interactions conserved in mice and humans22, 23, 24, 25, 26, 27, 28. We refer to this analysis pipeline as CDM version 3 (CDM3). Herein, we demonstrate that in both mice and humans, CDM3 can discriminate between in situ cognate and non-cognate T cell:DC interactions30.
Results
Identifying cognate T cell:dendritic cell interactions using TPEM
To develop better computational tools to study immunity in fixed tissue, we first used an established murine model of T cell and dendritic cell interactions30. Briefly, three cell populations were prepared (Fig. 1a): CD11c+ DCs from CD45.1+ mice stimulated in vitro with lipopolysaccharide (LPS), loaded with pigeon cytochrome C peptide (PCC, 10 μM), and labeled with the fluorescent dye CMF2HC (blue); polyclonal CD45.1+ CD4+ T cells (wild-type) labeled with CMFDA (green); TCR-transgenic 5CC7 CD45.2+ CD4+ T cells labeled with CMTPX (red). Previous studies have identified a peptide recognition rate of 0.1–0.3% for wild-type cells in this model system31,32. Cells were then transferred into wild-type recipient mice and, after 12 h, popliteal lymph nodes (LNs) were imaged by TPEM. These same LNs were then frozen, sectioned, stained for cell nuclei with TO-PRO-3, and imaged by confocal microscopy.
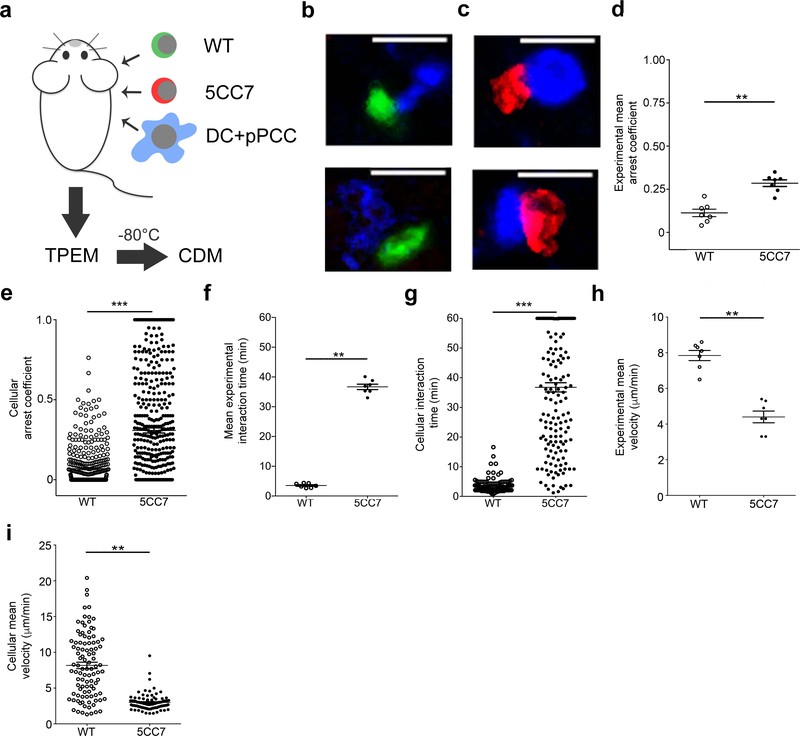
Figure 1. TPEM to measure antigen-specific CD4+ T cells interacting with dendritic cells.
(a) Indicated T cells (wild-type (WT) or 5CC7) and antigen-pulsed DCs were transferred into B10.A2 CD45.2– mice and, after 12 h, popliteal lymph nodes were first imaged by TPEM, frozen, and then imaged by confocal microscopy for CDM3. (b) Confocal microscopy examples of WT cells (green) interacting with DCs (blue). (c) Examples of 5CC7 cells (red) interacting with DCs. (d,e) Arrest coefficient plotted either as mean per mouse (n = 7) (d) or for individual cells, all experiments (e). (f,g) Interaction time plotted per mouse (f) or per cell, all experiments (g). (h,i) Cellular velocity plotted either per mouse (h) or per cell, all experiments (i). **P < 0.005, ***P < 0.0005, 2-sided Mann-Whitney U test. All center values denote the mean and error bars denote standard error of the mean. Scale bars, 10 μm. n = 3 independent experiments for (b,c) and n = 2 independent experiments for (d-i).
TPEM revealed clear differences in the behavior of 5CC7 antigen-specific and wild-type T cells relative to antigen-pulsed DCs (Supplementary Movie 1). Many of the PCC-specific 5CC7 T cells engaged in prolonged interactions with DCs, while wild-type T cells were more motile and only interacted briefly. Furthermore, wild-type T cells were spherical when interacting with DCs (Supplementary Movie 1 and Fig. 1b), while 5CC7 T cells tended to flatten against DCs (Fig. 1c).
Quantitative analysis of TPEM data (Supplementary Table 1) revealed that measures of cellular motion discriminated between wild-type and 5CC7 T cells relative to DCs. Plotting the mean T cell arrest coefficient per mouse (Fig. 1d) revealed that the motility of 5CC7 cells was less than that of wild-type cells. However, when plotting values per cell across mice there was substantial overlap between the two populations (Fig. 1e). Mean T cell interaction time provided better separation with larger relative differences between wild-type and 5CC7 T cells on a per mouse basis and less overlap when individual cells were plotted (Fig. 1f,g). The cellular mean velocity of T cells, plotted both per mouse and per cell, provided intermediate separation between groups with moderate overlap between individual cell values (Fig. 1h,i). In contrast, in response to very low doses of antigen (DCs pulsed with 0.01 μM PCC) there was relatively little difference between wild-type and 5CC7 cells by most TPEM measures (Supplementary Fig. 1). These data indicate that TPEM measures can accurately discriminate between wild-type and antigen-specific T cell populations based on their behavior relative to antigen-pulsed DCs.
CDM3 captures cell shape
We then used a custom three-dimensional deep convolutional neural network (DCNN) to analyze confocal images of LN frozen sections, extracting cell type, position, and shape. The DCNN was implemented in Tensorflow (https://www.tensorflow.org), with 10 convolutional layers, three maximum pooling layers, and 701,000 trainable variables (Fig. 2a and Supplementary Fig. 2). Rather than down-sampling the feature maps (FM) at each maximum pooling layer, we increased the sparsity of subsequent convolutional and maximum pooling layers. This allowed us to capture features at different scales yet retain information critical for accurate segmentation. Training was conducted on patches of pixels (184×184×5) with a model field of view (FOV) of 85×85×5, where FOV refers the size of the region around a single pixel that goes into predicting the cell type of that pixel. Each pixel was assigned the cell type with the maximum predicted probability from the DCNN output. This produced solid nuclear and dendritic cell segmentations on which shape-based object analysis was performed.
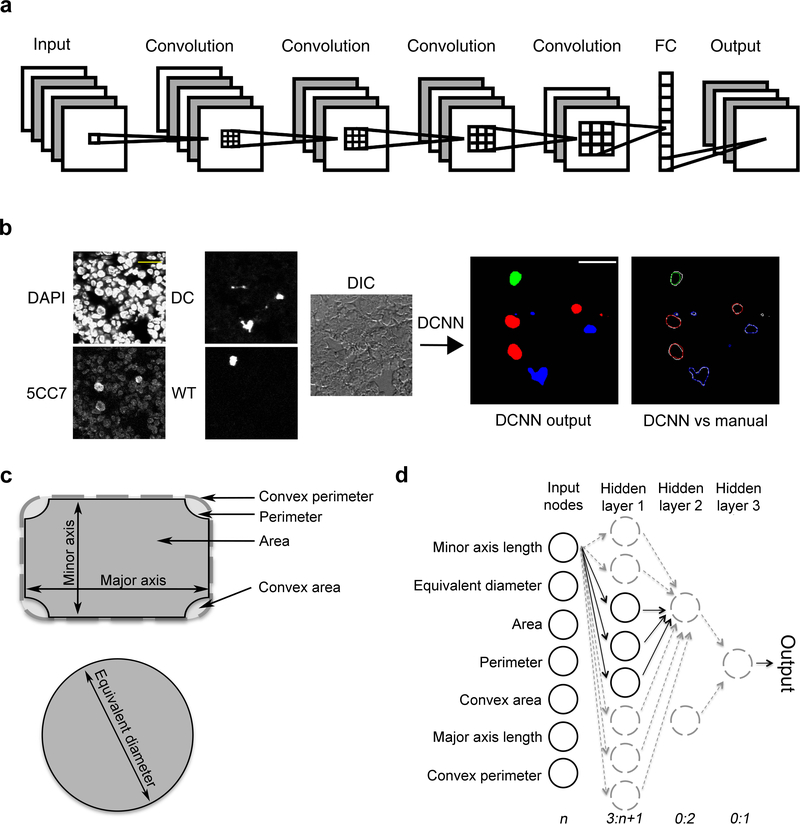
Figure 2. Development of CDM3.
(a) Abbreviated schematic of DCNN (also see Supplementary Figure 1) illustrating input of five layers (three fluorescent channels, TO-PRO-3 nuclei, and differential interference contrast [DIC]), four subsequent levels of convolution, and 4 classifier outputs (fluorescent signatures for WT T cell nuclei, 5CC7 T cell nuclei, DCs, and background). Across each convolution level, sparsity is increased, resulting in progressively larger kernel sizes (increasingly larger 3×3×3 boxes with spacing between elements). The ROI size stays constant throughout the DCNN. In the fully connected (FC) layer, all features (600) are integrated at each pixel, which are then ascribed classifier probabilities in the softmax layer (not shown). Each pixel is assigned to the cell type with maximum predicted probability to produce solid objects (segmentations). (b) Example of DCNN input and output. Five input channels are processed through the DCNN with an output of solid objects (middle right) for 5CC7 T cells (red), WT T cells (green), and DCs (blue). Far right, DCNN segmentations are shown in colored lines, and the corresponding manual segmentations shown in white lines. (c) Illustration of independent cell shape parameters. Convex measurements can be thought of as applying a theoretical rubber band around an object (dashed gray line) and are important in identifying local concave shape changes. The equivalent diameter represents the diameter of a circle, possessing an identical area as a non-circular object. (d) Outline of the neural network models used to analyze lymphocyte distance and cell shape data. After segregation by minimum distance, the indicated seven measures of cell shape were scaled, normalized, and used as input into three neural network models (simple, tuned, and linear output) in R statistical software, as described in Methods. The arrows and circles indicate data flow from a single input node throughout the network (applied weights omitted). The maximum potential number of hidden layers and nodes, used for active tuning, is indicated by dashed gray circles, with data flows in between steps denoted by dashed gray lines. Dark circles and lines denote obtained optimal parameters used at the completion of network tuning. n denotes the number of input nodes. Scale bars, 10 μm.
To train the DCNN, we manually segmented confocal images using ICY Bio Image Analysis software and ImageJ. All segmentations were independently validated by a blinded observer. For the murine experiment above, the total data set of 295 randomly collected high-powered fields (HPFs), corresponding to regions of interest (ROIs) containing all three cell types, was segmented for wild-type T cells, 5CC7 T cells, DCs, and corresponding cell nuclei (Supplementary Table 1). Training batches consisted of 4 image patches drawn randomly from the entire dataset, each belonging to four different classes (5CC7 T cells, wild-type T cells, DCs, and background). The DCNN was trained for 200,000 iterations at which point cross-entropy error was stable and small (Supplementary Figure 2b).
Confocal images segmented by the DCNN were compared for agreement against the manually segmented images using 5-fold DCNN model-based cross validation. The DCNN output as compared to manual segmentation of input images revealed excellent agreement between the two analysis methods (Fig. 2b). To assess the sensitivity and specificity of localization and segmentation, a segmented cell was considered detected if the intersection over union (IOU) of the manually segmented cell with the automatically segmented cell was greater than or equal to 0.5. Overall, across all cells, the DCNN had an average sensitivity of 88%, specificity of 92%, and an IOU of 0.85. (Supplementary Tables 1 and 2).
From the DCNN output, we extracted relative distances between T cell populations and DCs, as well as features of T cell shape. For the latter, we used seven independent measures of two-dimensional shape that include major and minor axis lengths, convex and regular perimeters, convex and regular areas, and equivalent diameter33 (Fig. 2c). These data were imported into R statistical software34 and analyzed to determine which combination of variables and variable weights best discriminated between the 5CC7 and wild-type T cell populations relative to DCs. Our approach included the use of the following algorithms: logistic regression, support vector machines (SVM), and neural networks. For the latter, three different neural network models (simple, tuned, and linear output) were generated. The performance of each algorithm was assessed as measured by parameters of classification accuracy, error, and receiver operator characteristic (ROC) curve performance for correctly predicting cell type (Supplementary Table 3a). An actively tuned NN (TNN) (Fig. 2d) consistently displayed the best performance among neural network models (Supplementary Table 3b) at the expense of increased number of steps and computation time (Supplementary Fig. 3a-c). Therefore, we used a DCNN followed by a TNN in the CDM3 pipeline.
Comparison of CDM3 to TPEM in mice
Random forest analysis revealed that minimum distance to a DC provided the best discrimination between 5CC7 and wild-type T cells (Fig. 3a). Simply plotting relative distance to closest DC provided excellent discrimination between wild-type and 5CC7 T cells with 5CC7 T cells being, on average, much closer to the nearest DC (Fig. 3b). We next plotted the true-positive rate (sensitivity) versus the false-positive rate (1-specificity) in a receiver-operator curve (ROC) to determine how distance performed as a test to discriminate between wild-type and 5CC7 T cells (Fig. 3c). This analysis revealed that cellular distance provides good discrimination with an area under the curve (AUC) of 0.70 (95% CI: 0.62–0.74, P < 5 × 10−5).
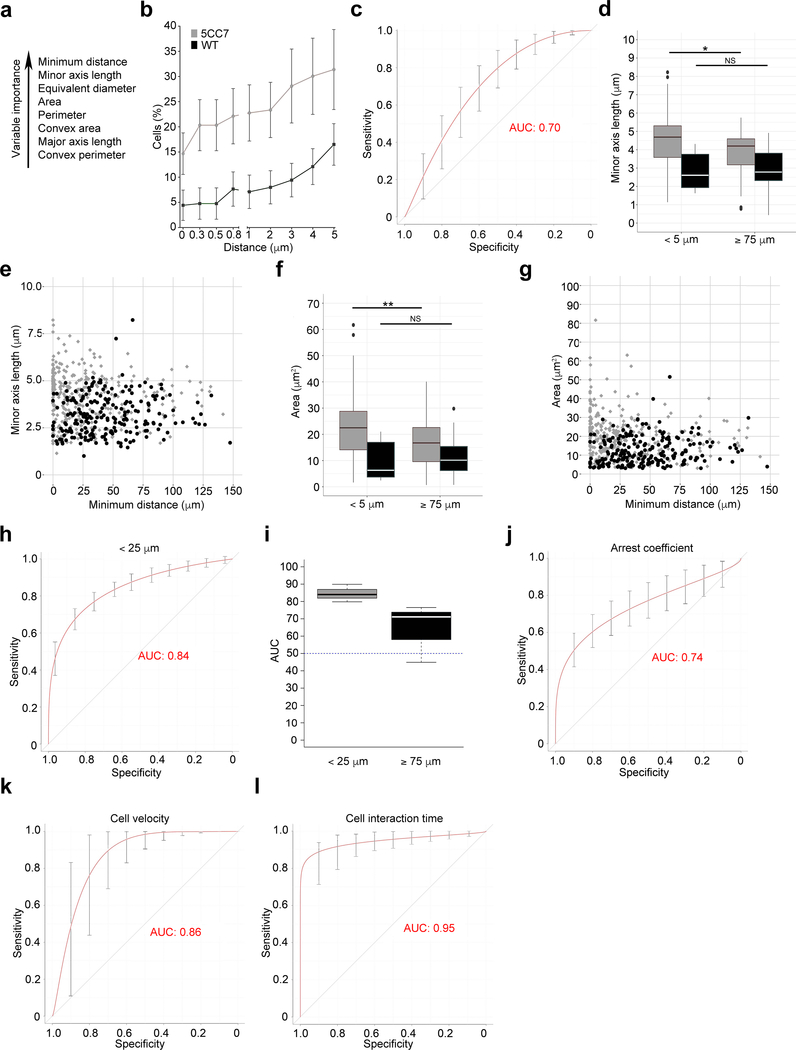
Figure 3. The sensitivity and specificity of TPEM and CDM3 are comparable.
(a) Hierarchy of contribution of distance and T cell shape parameters to accuracy, as determined by random forest analysis (see Methods). (b) Cumulative frequency of either 5CC7 (grey) or WT (black) T cells as a function of distance from antigen-pulsed DCs (P < 0.005). (c) Plot of sensitivity and specificity of distance alone in discriminating between 5CC7 and WT T cells (AUC 95% CI: 0.62–0.74, P < 5 × 10−5). (d) Comparison of minor axis length for 5CC7 (grey) or WT (black) T cells at indicated distances. (e) Minor axis length for each cell plotted as a function of distance from DCs. (f,g) 5CC7 (grey) and WT (black) T cell cross-sectional area as an average at indicated distances (f) or for each cell as a function of distance (g). (h) Plot of sensitivity and specificity of CDM3 for discriminating between 5CC7 and WT T cells (AUC 95% CI: 0.80–0.90, P < 5 × 10−5). (i) Comparison of AUC and 95% confidence interval performance shown in (h) with values derived from analysis of cells at distances ≥ 75 μm. (j,k) Plot of sensitivity and specificity of TPEM measures for discriminating between 5CC7 and WT T cells including cellular arrest coefficient (j, AUC 95% CI: 0.72–0.82 P < 5 × 10−5), cellular velocity (k, AUC 95% CI: 0.78–0.90, P < 5 × 10−5) and cellular interaction time (l, AUC 95% CI: 0.94–0.97, P < 5 × 10−5). * P < 0.05, ** P < 0.005, two-sided Mann-Whitney U test. Center values denote the mean (b,d,f,h,i) and error bars denote standard deviation (b), standard error of the mean (i), or cross-validation error (c,h,j-l). Center lines in box plots (d,f) denote median value with upper and lower hinges denoting first (Q1) and third (Q3) quartile values, vertical bars corresponding to values 1.5x of the inter-quartile range (IQR) for Q1 and Q3, respectively, and dots representing outlier values, not included in the above. Diagonal lines in (c,h,j-l) denote AUC of 0.5, which represents a random probability ( P = 0.5). n = 2 independent experiments for all panels.
Among the T cell shape variables, minor-axis length was the most promising for discriminating wild-type from 5CC7 T cells. Comparison of T cell minor axis length at distances of less than 5 versus 75 μm or greater from DCs, revealed that 5CC7 T cells had a longer minor axis compared to wild-type T cells at close distances (Fig. 3d). Plotting individual cell minor axes as a function of distance (Fig. 3e and Supplementary Fig. 4a) clearly revealed a subpopulation of 5CC7 cells displaying high values, especially when contacting a DC (0 μm). This difference was diminished by 25 μm and largely lost at distances greater than 50 μm. These changes are consistent with the 5CC7 cells flattening against the antigen-pulsed DCs resulting in an increase in minor cell axis for some cells in the two-dimensional plane of the confocal micrograph. In contrast, the minor axis of wild-type T cells in some cells decreased as a function of distance from DCs. This latter trend is consistent with wild-type T cells becoming more spherical upon contacting DCs.
Plotting T cell cross-sectional area as a function of distance revealed similar results (Fig. 3f,g and Supplementary Fig. 4b) with a subset of 5CC7 T cells having relatively large cross-sectional areas when very close to or abutting DCs. Wild-type T cell area decreased at close DC distances. In contrast, 5CC7 and wild-type T cells have similar shape characteristics at distances of 75 μm or greater from DCs. These data suggest that there were no substantial intrinsic differences between the T cell populations in shape or size. Rather, antigen-specific 5CC7 and wild-type cells diverge in their shape properties in proximity to antigen-pulsed DCs. These data suggest that CDM3 can capture changes in T cell shape that occur upon recognition of antigen presented by DCs.
We next determined how well CDM3 could discriminate between the 5CC7 and wild-type T cell populations by simultaneously incorporating both distance relationships and T cell shape. We focused our analysis to T cell populations within 25 μm of DCs, as this was the distance at which we observed substantial differences in T cell shape. Furthermore, we examined close distances because we were interested in discriminating between T cells that recognize antigen from those that are scanning peptide-MHC class II complexes, looking for antigen. The full CDM3 output, which integrates distance and T cell shape variables, provided an AUC of 0.84 (95% CI 0.80 – 0.90, P < 5 × 10−5) and was substantially better than distance alone for all measurements (Fig. 3h). In contrast, at distances of greater than 75 μm, the two T cell populations were indistinguishable (Fig. 3i). Within 25 μm, the use of the minimum distance variable by itself could also discriminate between 5CC7 and wild-type cells (data not shown). However, differences between populations were less robust (AUC=0.65, 95% CI: 0.59–0.72, P = 0.008). These data indicate that CDM3, by combining measurements of both cell distance and T cell shape, provides excellent discrimination between T cell populations that are scanning for antigen versus those that have recognized antigen.
We next examined how well measures obtained by TPEM discriminated between 5CC7 and wild-type cells interacting with antigen-pulsed DCs. Therefore, we took the TPEM outputs described above (Fig. 1 and Supplementary Table 1) and subjected them to the same statistical modeling by plotting their true-positive rate versus false-positive rate. The arrest coefficient provided good discrimination with an AUC of 0.74 (95% CI: 0.72–0.82 P < 5 × 10−5; Fig. 3j). Cell velocity was more robust with an AUC of 0.86 (95% CI: 0.78–0.90, P < 5 × 10−5, Fig. 3k). Only cell interaction time, with an AUC of 0.95 (95% CI: 0.94–0.97, P < 5 × 10−5), substantially outperformed CDM3 (Fig. 3l). These data indicate that CDM3 performs as well as many TPEM measures in identifying antigen-specific T cell interactions with DCs.
T cell nuclei as a surrogate for cell shape
We next sought to apply CDM3 to the analysis of multichannel confocal images of human tissue. However, immunofluorescence with antibodies to surface markers is often inadequate for identifying the exact boundaries of lymphocytes in dense infiltrates. In our original studies using CDM, nuclear stains were necessary to define lymphocyte position19. As the nucleus constitutes the majority of a lymphocyte’s volume35, we postulated that nuclear shape would approximate cell shape.
Therefore, 5CC7 and wild-type T cell nuclei were segmented and analyzed using CDM3. Evaluation of representative images of cell trackers compared to TO-PRO-3 nuclear staining revealed extensive overlap between the two staining signatures (Fig. 4a). Analysis of individual shape parameters revealed similar relationships between 5CC7 and wild-type T cell nuclear shape and distance to closest DCs as observed for their cell tracker counterparts (Fig. 4b-d, Supplementary Fig. 4d,e and Supplementary Table 1). That is, in a subset of 5CC7 cell nuclei, shape parameters increased close to DCs while in wild-type nuclei they did not. Interestingly, a small population of 5CC7 nuclei displayed a decrease in two-dimensional T cell shape parameters close to DCs. This distance-dependent increase in nuclear shape variability is consistent with 5CC7 cells becoming more irregular (less spherical) upon contacting DCs. This is expected when a cell (sphere) flattens against a DC (surface).
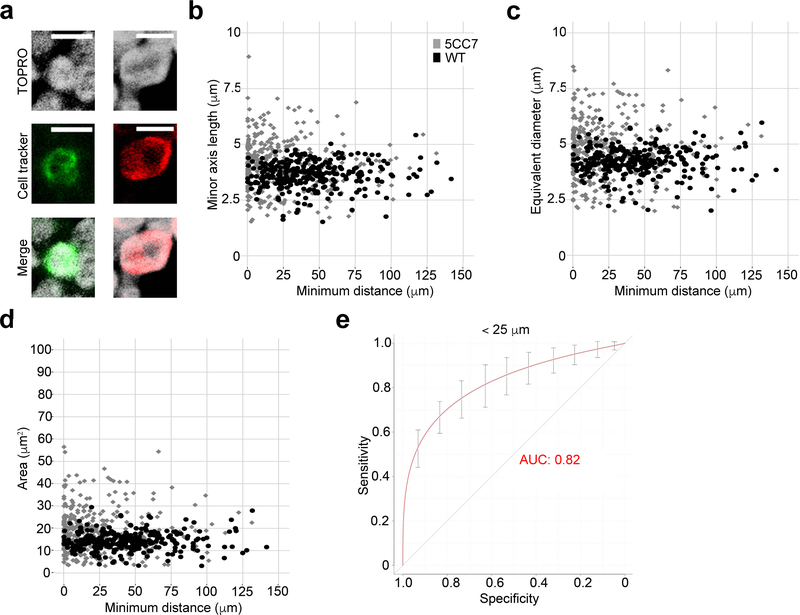
Figure 4. Segmentation and shape of T cell nuclei.
(a) Representative images of fluorescent cell tracker-labeled WT (left) and 5CC7 (right) T cells stained with TO-PRO-3 iodide (TOPRO). (b-d) Plots of 5CC7 (grey) and WT (black) T cell nuclear minor axis length (b), equivalent diameter (c), and nuclear area (d) as functions of minimum distance from antigen-pulsed DCs. (e) Curves denoting sensitivity and specificity of CDM3 for discriminating between 5CC7 versus WT cell nuclei at distances < 25 μm (AUC 95% CI: 0.77–0.91, P < 0.005). Scale bar: 5 μm. Diagonal lines in (e) denote AUC of 0.5, which represents a random probability (P = 0.5). n = 3 independent experiments for (a) and n = 2 independent experiments for (b-e).
Application of the composite distance and T cell nuclear shape scores revealed similar discrimination between 5CC7 and wild-type T cell interactions with DCs as that observed for whole cells (Fig. 4e, AUC=0.82, 95% CI: 0.77–0.91, P < 0.005). Similarly, at distances greater than 25 μm, the two T cell nuclei populations were indistinguishable (data not shown, AUC=0.52, 95% CI: 0.45–0.72). These data indicate that nuclear shape alone can be used to approximate T cell shape for the purpose of discriminating cognate from non-cognate T cell–DC interactions.
Finally, to validate our mouse experiments, we independently repeated the adoptive cell transfer experiment in mice and obtained 233 additional ROIsHere, DC and T cell nuclei were subjected to the same segmentation and analysis using CDM3 as described for the original experiment. These data revealed similar discrimination as before between 5CC7 and wild-type T cell nuclei with an AUC of 0.82 (95% CI: 0.72 to 0.87, P < 5 × 10−5) at distances less than 25 μm from DCs. Likewise, for distances greater than 25 μm, the two T cell nuclei populations were similar (AUC=0.54, 95% CI: 0.45–0.58).
Application of CDM3 to human lupus nephritis
We next sought to understand the relationships between DCs and T cells in human tissue. Specifically, we examined the relative abilities of mDCs and pDCs to present antigen to T cells in lupus nephritis tubulointerstitial inflammation (TII). Conventionally, mDCs are considered a professional APC36, 37, while the function of pDCs is thought to be the secretion of interferon-α and other cytokines38. However, some subpopulations of pDCs can present antigen to CD4+ T cells39, 40.
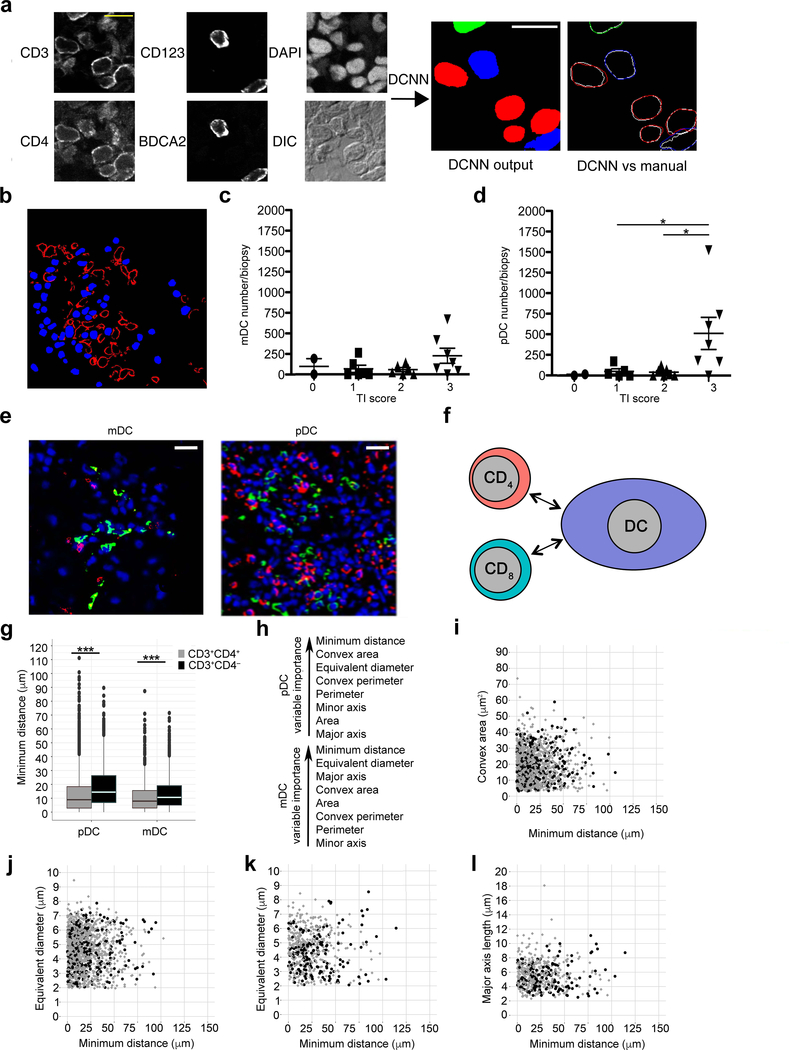
Single longitudinal sections were captured by either tiling HPFs across the entire renal biopsy (n = 10 biopsies), or by capturing those with at least one mDC or pDC (n = 12 biopsies). A total of 243 ROIs, from a data set of 687 ROIs, were manually segmented and used for training the DCNN, as described above (Supplementary Table 4). An example of raw input images, the output from the DCNN, and a comparison of cell segmentation using DCNN versus manual approaches is depicted (Fig. 5a). Overall, there was an excellent agreement between the cross-validated DCNN and manual segmentation with a sensitivity of 0.72, specificity of 0.86, and an IOU of 0.70 for all cell types (Supplementary Table 5). The lowest sensitivity was observed for CD3+CD4– T cells, which coincided with the most technically difficult staining, as compared to the other cell types. Another example of the DCNN output for pDCs and CD4+ T cell nuclei (Fig. 5b) illustrates the DC outlines and T cell nuclear edges used to calculate minimum distances. Cells that could be categorically assigned with 90% or greater confidence were used for the subsequent analysis.
Figure 5. Identification of pDCs as an antigen-presenting cell in lupus nephritis.
(a) Example of DCNN input and output. Utilizing a similar framework as described for the murine model, six input channels were processed through the DCNN with an output of solid objects (middle) for CD3+CD4+ T cells (red), CD3+CD4– T cells (green), and plasmacytoid DCs (blue). Right, DCNN segmentations are shown in colored lines and the corresponding manual segmentations shown in white lines. Scale bar = 10 μm. (b) Example of processed image showing outlines of pDCs (red outlines), CD4+, and CD4– T cell nuclei (blue). (c,d) Frequency of mDCs (c) and pDCs (d) per biopsy by TII grade (0 = none, 1 = mild, 2 = moderate, 3 = severe41). (e) Representative images of mDCs and pDC (green) with CD4+ T cells (red) in lupus TII. Nuclei are blue. Scale bar = 40 μm. (f), Schematic of CD3+CD4+ and CD3+CD4– T cells relative to DCs. (g) Minimum distances between indicated DC populations and CD3+CD4+ (grey) and CD3+CD4– (black) T cells. (h) Hierarchy of contribution of distance and T cell shape parameters to accuracy based on random forest analysis of pDC and mDC data sets. (i,j) Plot of convex area (i) and equivalent diameter (j) per T cell [CD3+CD4+ (grey) and CD3+CD4– (black)] as a function of distance from pDCs (P < 0.0005 for both groups at all distances). Random 10% of total values plotted for visualization. (k,l) Equivalent diameter (k, P < 0.05) and major axis (l, P < 0.005) per T cell as a function of distance from mDCs. Random 10% of total values plotted for visualization. * P < 0.05, *** P < 0.0005, 2-sided Mann-Whitney U test; Benjamini-Hochberg correction with FDR of 5% was used for (i-l). Center values denote the mean and error bars denote standard standard error of the mean (c,d). Center lines in box plot (g) denote median value with upper and lower hinges denoting first (Q1) and third (Q3) quartile values, vertical bars corresponding to values 1.5x of the inter-quartile range (IQR) for Q1 and Q3, respectively, and dots representing outlier values, not included in the above. n = 2 independent experiments for all panels.
We then compared the DCNN segmentation output to the output obtained using the original CDM19. With detections computed at an IOU of 0.50, the sensitivity and specificity for CD3+CD4+ nuclei was only 0.07 and 0.17, respectively. The sensitivity and specificity for CD3+CD4– nuclei was 0.29 and 0.05, respectively. The segmentation of DCs by CDM was so poor a sensitivity and specificity could not be calculated. Therefore, the original CDM, which used fixed segmentation filters, was poor at capturing nuclear and cell shape.
Twenty five lupus nephritis biopsies, scored for degree of TII (integer scale: 0–341), were stained with antibodies specific for CD11c, BDCA1, BDCA2, CD123, CD3, CD4, and DAPI. In individual longitudinal biopsy sections, the numbers of mDCs (CD11c+BDCA1+) and pDCs (CD123+BDCA2+) were determined using CDM3. Myeloid DCs were present in all degrees of TII with no statistically significant differences among the groups. In contrast, the majority of pDCs occurred in severe TII (P < 0.05)(Fig. 5c,d, Supplementary Table 6). CD4+ T cells appeared more frequent around pDCs than mDCs; CD4– T cells exhibited similar behavior, albeit they were less frequent (Fig. 5e and Supplementary Figure 5). However, the number of CD4+ and CD4– T cells per either DC population was similar when considering whole biopsies or HPFs (Supplementary Table 5). These data indicate that pDCs are more common than mDCs in severe TII and are associated with a proportional increase in local T cell infiltrates.
Both pDCs and mDCs could just be co-segregated with T cells in areas of active inflammation or they could be contributing to inflammation by locally presenting antigen to CD4+ T cells. To discriminate between these two possibilities, we analyzed the distance and shape characteristics of CD3+CD4+ T cell nuclei relative to each DC population in biopsies with severe TII (n = 8). Local CD3+CD4– T cells provide a non-MHC class II restricted bystander control population (Fig. 5f). Analysis of relative distances revealed that CD3+CD4+ T cells were, on average, closer to both pDCs and mDCs than CD3+CD4− T cells (Fig. 5g and Supplementary Fig. 6). Analysis of T cell nuclear shape revealed differences between CD3+CD4+ and CD3+CD4– cells relative to each DC population, with different measures of T cell shape making differential contributions to accuracy relative to either pDCs or mDCs (Fig. 5h). Examination of T cells relative to pDCs for both convex area and equivalent diameter revealed that some of those CD3+CD4+ cells close to pDCs tended to be larger on cross-section (Fig. 5i,j and Supplementary Fig. 4f,g). Similar relationships were observed for T cell equivalent diameter and major axis relative to mDCs (Fig. 5k,l and Supplementary Fig. 4h,i). In contrast, some CD3+CD4– cells tended to get smaller in cross-section as a function of distance from either DC population. These same trends were observed for 5CC7 and wild-type murine T cells relative to antigen pulsed DCs. However, in contrast to the murine experiment, CD3+CD4– cells were larger than CD3+CD4+ cells when not in proximity to a DC (Supplementary Fig. 4f–i). These data demonstrate that by examining changes in T cell shape as a function of distance, one can compare T cell populations that are intrinsically different in size.
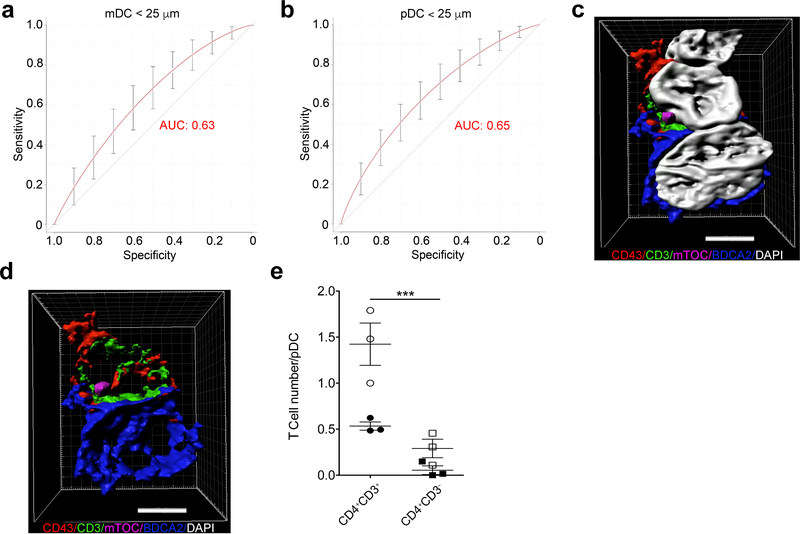
Plotting the composite CDM3 output of distance and T cell shape features revealed clear discrimination between CD3+CD4+ and CD3+CD4– T cells within 25 μm for both mDCs (Fig. 6a, AUC=0.63, 95% CI: 0.55–0.71, P < 0.01) and pDCs (Fig. 6b, AUC=0.65, 95% CI: 0.61 – 0.69, P < 0.0005). Use of minimum distance alone yielded inferior results in cell type discrimination (data not shown; AUC=0.63, 95% CI: 0.50–0.69, P = 0.05, and AUC=0.57, CI: 0.52–0.61, P = 0.03 for CD3+CD4+ and CD3+CD4– T cells within 25 μm of mDCs and pDCs, respectively). These data are consistent with distance and T cell shape relationships observed in murine cells and suggest that both mDCs and pDCs present antigen to CD4+ T cells. However, as there are more pDCs than mDCs in severe TII, pDCs appear to make a larger contribution to in situ CD4+ T cell activation.
Figure 6. Confirmation of pDCs as an antigen-presenting cell in lupus nephritis.
(a,b) Sensitivity and specificity of CDM3 for discriminating between CD3+CD4+ and CD3+CD4– cells for mDC (a, 95% CI: 0.55–0.71, P < 0.005) and pDC (b, 95% CI: 0.61–0.69, P < 0.0005) datasets at a minimum distance cutoff of < 25 μm. (c,d) Three-dimensional surface reconstructions from lupus TII, utilizing Imaris software. T cell (top) abutting a pDC is shown with (c) (left) and without (d) (right) nuclei, as stained by DAPI. Immunofluorescent antibody staining as indicated. Scale bar: 4 μm. (e) Analysis of mTOC localization in 3 highly inflamed (score: 3) lupus nephritis biopsies. z-stack acquisition was performed with examination of pDC:CD3+CD4+ and pDC:CD3+CD4– T cell pairs to determine mTOC localization. White circles denote all counted T cells, while black circles signify the number of T cells with mTOCs polarized towards the abutting DC, as indicated. *** P < 0.0005, 2-sided Mann-Whitney U test. Diagonal lines in (a,b) denote AUC of 0.5, which represents a random probability (P = 0.5). Error bars denote cross-validation error (a,b) or standard error of the mean (e). n = 2 independent experiments for all panels.
In peripheral blood, a subpopulation of pDCs expressing the surface markers AXL and SIGLEC6 can function as APCs40. However, the lupus intrarenal pDCs are AXL–SIGLEC6– (data not shown). Therefore, to confirm that intrarenal pDCs were an important APC in vivo, lupus TII biopsies were stained with antibodies specific for CD3, CD4, CD43, BDCA2, and tubulin to visualize the microtubule organizing center (mTOC), as well as DAPI. We then performed three-dimensional confocal imaging on representative T cell:pDC conjugates. As seen, there is polarization of the TCR:CD3 complex towards the interface with the pDC (Fig. 6c,d). Likewise, the T cell mTOC is oriented towards the interface with the pDC. In contrast, CD43 is accumulated at the distal pole complex - all consistent with a canonical, mature T cell:APC synapse42, 43.
To quantify mTOC polarization, lupus renal biopsies with pDCs were stained with antibodies specific for CD3, CD4, BDCA2, tubulin, and DAPI then subjected to confocal microscopy to obtain z-stack images as described in Methods. We scored the number of CD3+CD4+ and CD3+CD4– T cells abutting pDCs (186 pDCs across three biopsies), and how many of these T cells had their mTOCs polarized towards the interface with pDCs. CD3+CD4+ were almost 6-times more likely to be abutting pDCs (a two to three-fold enrichment compared to total T cell numbers (Fig. 6e, Supplementary Table 6). Of these, 40% had a mTOC polarized towards pDCs while only 10% of abutting CD3+CD4– cells did. Therefore, there was an overall 24-fold difference in conjugate rate between CD4+ and CD4– T cells (P < 0.0005). These data confirm that CDM3 accurately detects T cell:pDC conjugates.
Discussion
The study of immune cell dynamics in tissue is challenging and presents several trade-offs. Techniques such as TPEM provide direct visualization of precisely labeled and characterized cell populations and quantifies their interactions with other cells over time. However, TPEM can only be used on some tissues at limited organ depths. It is also difficult to apply directly to the study of human disease10, 11. In this work, we demonstrate that by utilizing multi-channel confocal microscopy and a novel analytic pipeline that we term CDM3, we approach the performance measures of TPEM in discriminating stable cognate from non-cognate T cell–DC interactions in mice. These data indicate that a quantitative analysis of many static two-dimensional images can approximate much of the information obtained from time-lapse 3D videos of the same phenomenon. Additionally, as CDM3 is performed on single plane images of fixed tissue, we could use CDM3 to study human disease and identify important in situ APCs.
Beyond applicability to the study of both human disease and animal models, not readily accessible to TPEM, CDM3 offers several additional advantages. It provides higher throughput, which enables larger sample sizes and robust statistical confidence. CDM3 does not require experimental manipulations to label cells and, therefore, can be performed on native systems, minimizing experimental artifact. Furthermore, it can be performed on any tissue and at any depth. CDM3 cannot assess the kinetics of cellular interactions and, hence, cannot be used to address some questions. Regardless, for many experimental applications in animal models, CDM3 might be a preferred approach.
However, the major advantage of CDM3 is that it can be performed on single sections from frozen tissue. Therefore, it is ideally suited to human studies and can be applied to biopsies that are routinely obtained as part of clinical care. In lupus nephritis, we were able to identify putative in situ cognate T cell–DC interactions and, furthermore, assign relative importance of different DC populations in presenting antigen to CD4+ T cells. Overall sensitivities and specificities in human disease were less than those observed in transgenic mice. This could reflect inherent differences in the imaging approaches used in mice and humans. More likely, the lower signal observed in human disease reflects underlying heterogeneity in the cells and antigens driving in situ adaptive immunity. However, by capturing more events than is practical with TPEM, we overcame this heterogeneity to make statistically robust conclusions.
The sequential use of a DCNN followed by a TNN in CDM3 is critical for application to human disease. In mice, we can dictate the antigen specificity of T cell populations and, therefore, use a DCNN to best discriminate between antigen-specific and non-antigen- specific interactions with DCs. We have an ideal training set. In humans, we cannot control T cell antigen specificity. However, by using mice to learn how to extract fundamental features of how T cells interact with DCs, that are conserved across mice and humans22, 23, 24, 25, 26, 27, 28, we can apply the CDM3 pipeline to human samples and identify stable cognate interactions. Thus, our sequential pipeline helps overcome major limitations of machine learning including transparency, in-depth understanding of how machine-learning algorithms function, and what newly created predictors or intermediate variables represent. These ambiguities often make it difficult to relate machine learning outputs to meaningful biological variables or behavior44,45.
In CDM3, we designed and trained a highly customized DCNN from scratch because our application was so different than mainstream applications designed for photographic images. Particularly, we were interested in making predictions on multichannel images where there was significant bleed-through between stain channels. We also wanted to classify nuclei based on surface stains that were not spatially co-localized between image channels. To do this we used 3D convolutional kernels to relate information across the channel dimension. This not only allowed us to relate non-spatially co-localized information between image channels, but it also allowed the DCNN to use features from one image channel to mitigate noise or ambiguity in another.
Additional customization of our DCNN included selecting an appropriate number of neural network parameters for the CNN such that it was complex enough to learn features in our labeled image data set, but not so complex that it overfit and simply memorized the training set. Training images were also carefully selected to be representative of the whole dataset and representative of variations in image quality. The classes of training examples used were balanced to ensure equal performance in classifying the different cell types. The scale of the dilated convolutions used in the layers of the neural network were selected such that they acted on spatial scales where features relevant to classifying the image data were most likely to exist. Data were normalized using the standard score (z-score) applied separately to each channel of each ROI. These design decisions were made so that learned features from the training data were robust and transferrable to other experimental data.
Our studies were limited to single plane confocal images and simple pair-wise cell:cell interactions. However, CDM3 is adaptable to three-dimensional images that would provide more definitive measures of in situ APC function. Furthermore, the general approach illustrated by CDM3 is applicable to the study of complex cellular networks containing three or more cell types. A quantitative understanding of the cellular architectures of in situ adaptive immunity and inflammation in human disease will provide new insights into the pathogenic mechanisms of autoimmunity and features of immunity effective against cancer.
Methods
Mice
B10.A CD45.2− and B10.A CD45.2+ 5CC7 TCR-transgenic Rag2−/− mice were obtained from Taconic Laboratories through a special NIAID contract. All animal experiments were conducted under a protocol approved by the NIAID Animal Care and Use Committee (LSB-1E) and the University of Chicago Animal Resource Center. All animal experiments were conducted in compliance with all relevant ethical regulations.
Adoptive transfer
Dendritic cells were purified from mouse spleens using anti-CD11c beads (Miltenyi). Purified dendritic cells were activated in vitro with LPS (1 μg/ml) and pulsed with pigeon cytochrome C peptide (Bachem, sequence corresponding to amino acids 88–104) at high (10 μM) or low (0.01 μM) concentration for 4 h at 37 °C. Activated DCs were labeled with Cell Tracker Blue (CMF2HC, Invitrogen), then injected (1 × 106 per recipient) into the right rear footpad of recipient mice. Polyclonal and 5CC7 TCR-transgenic T cells were isolated from the lymph nodes of B10.A CD45.2– wild-type and B10.A CD45.2+ 5CC7 TCR-transgenic Rag2–/– mice, respectively, and purified using a CD4+ T cell isolation kit (Miltenyi). Polyclonal T cells were then labeled with Cell Tracker Green (CMFDA, Invitrogen) and 5CC7 T cells with Cell Tracker Red (CMTPX, Invitrogen). 2 × 106 of each T cell population were then co-injected IV into recipient mice 18 h post transfer of DCs. 12 h post T cell transfer, mice were subjected to two-photon emission microscopy studies as previously described46. Isoflurane was used to anesthetize mice prior to exposure of popliteal LN (Baxter; 2.5% for induction, ∼1%–1.5% for maintenance, vaporized in an 80:20 mixture of O2 and air), and subsequent TPEM was performed as described46. Briefly, imaging was conducted on a Bio-Rad/Zeiss Radiance 2100MP, configured with a Nikon 600FN upright microscope equipped with a 203 water immersion lens (NA 0.95, Olympus) and LaserSharp acquisition control software. Anesthetized mice were maintained in environmental chambers warmed by heated air with the surgically exposed LN kept at 36–37 °C with warmed PBS. Upon completion, mice were euthanized and draining popliteal lymph nodes were isolated, cured overnight in 30% sucrose, and frozen at −80 °C. The tissue was subsequently sectioned at 5 μm thickness and prepared for confocal microscopy.
Imaging of mouse tissue
Mouse tissue sections were prepared and stained with TO-PRO-3 Iodide (Invitrogen) to visualize nuclei and avoid interference with the fluorescence spectrum of transferred cell trackers. Single-fluorochrome controls were utilized to ensure no cross-bleeding was present in between fluorescent channels. Images were acquired at 12-bit depth, 1024×1024 pixel size, at 400× and 630× magnifications utilizing either the SP5 Tandem Scanner Spectral 2-photon confocal microscope or the SP8 3D 3-color STED laser scanning confocal microscope with time gating (Leica). Each region of interest (ROI) was 144.74 μm/1024 pixels wide, corresponding to an average absolute resolution size of 0.28 μm, based on Nyquist sampling. Regions of interest, containing all three transferred cell populations, were selected for acquisition. Raw images were stored in manufacturer-specified .lif format. Lif files were converted to multi-channel .tif images and used as input for DCNN analysis.
Renal biopsy staining
This study used a total of 25 renal biopsies from de-identified patients, obtained from the University of Chicago Human Tissue Resource Center (HTRC), Department of Pathology. The tissue was fresh-frozen in OCT Tissue Plus (Thermo-Fischer) and stored at −80 °C. The study protocol was approved by the University of Chicago Institutional Review Board (IRB#15–0727) and did not require informed consent as no patient data were used. All human experiments were conducted in compliance with all relevant ethical regulations. Confirmation of the diagnosis of lupus nephritis as well as grading of the severity of tubulointerstitial inflammation was performed by a blinded reading nephropathologist (A.C.) as previously described47. In addition, de-identified tonsil samples were utilized from the University of Chicago Pathology Core Facility for antibody testing and validation. Two distinct antibody panels were utilized to stain 3–4 μm thick tissue sections; for pDC analysis – CD3 (clone SP7, Abcam or clone CD3–12, AbD Serotec), CD4 (clone YNB46.1.8, Abcam), BDCA2 (clone AC144, Miltenyi), and CD123 (clone 6H6, eBioscience); mDC analysis – CD3, CD4, BDCA1 (clone L161, Beckman Coulter), and CD11c (clone EP1347Y, Abcam). DAPI (Hoechst 33342, Invitrogen) was used with the above to visualize tissue nuclei. Images were acquired using a SP5 or SP8 confocal microscope as described above. In addition to selecting individual ROIs for analysis, selected biopsies underwent tiling, wherein the entire available tissue was imaged and a composite stitched image obtained based on default manufacturer settings (SP8). All images were stored in .lif format. Lif files were converted to multi-channel .tif images and used as input for DCNN analysis.
mTOC localization quantification
Three 3 μm-thick severely inflamed lupus nephritis biopsies (TII grade 3) were stained with antibodies to CD3, CD4, BDCA2, and DAPI as per the above protocol. Anti-tubulin (clone YL1/2, Abcam) was added to visualize the mTOC. Automated z-stack protocol images were obtained from regions containing pDCs using the Leica SP8 laser scanning confocal microscope. The images were processed in Fiji using the 3D viewer plug-in (https://imagej.net/3D_Viewer) to obtain a maximal projection in 2D. Manual counting of mTOCs was then performed in a blinded fashion by a single observer (J.A.). All CD3+CD4– and CD3+CD4+ T cells abutting a pDC were quantified along with those, featuring an mTOC at the junction between the T cell and APC.
DCNN
Manual segmentation of confocal images was conducted using ICY Bio Image Analysis software, version 1.8.6.0 (http://icy.bioimageanalysis.org/) and Fiji/ImageJ, version 2.0.0-rc-43/1.50i (http://imagej.nih.gov/ij/). Image channels were rigidly registered using the ‘multimodel’ configuration of the imregister function in Matlab48. All segmentations were independently validated by a blinded observer (V.M.L.). The total data set for the murine analysis consisted of 295 ROIs, which were segmented for wild-type T cells, 5CC7 T cells, transferred DCs, and corresponding cell nuclei. The standard score, z = (x – μ)/σ, of the ROIs was taken for every individual channel within every ROI independently. All training and inference was performed on z-score transformed ROIs.
The DCNN was trained with a batch size of four 184×184×6 image patches distributed across 4 Tesla GPUs with 12GB memory per card, system memory of 128 GB with 2 Intel E5–2680v4 CPUs @ 2.4 GHz on the University of Chicago Research Computing Center GPU2 nodes on Midway249. Image patches were sampled from the entire labeled dataset. Each of the four patches for a training iteration represented one of the four classes to be segmented. Class membership of a patch was determined by the class of the center pixel. Where this 184×184 patch around pixels extended beyond the ROI border, mirror padding was used. Sampling was implemented in Tensorflow (https://www.tensorflow.org)50 using a list of all pixel locations by class, stored in system memory. The image, label, and data for each ROI was stored in binary TFrecord format in 32-bit float format and accessed by Tensorflow queue runners for active data augmentation (rotation and mirroring) of training examples onto a queue. Error was computed for all classes within the patch.
Softmax binomial cross-entropy was used to compute neural network error over an output image patch of 100×100 pixels, reduced input size of 184×184. This dense output was important because it reduces training redundancy and increases training stability. All convolutional and pooling layers were allowed to shrink the input by filter overlap at each layer in x and y, known as valid padding. Convolutional layers were padded in the channel dimension to keep channel dimensions the same. All weights were initialized with Xavier initialization51 and all biases were initialized at zero. Gradients were averaged across GPUs for each variable at each iteration. Stochastic gradient descent was used for optimization with learning rate of 0.001 with no decay of the learning rate. The DCNN was trained for a total of 200,000 iterations.
Inference was performed on 1024×1024 ROIs with boundary mirroring to fit the field of view. The final segmentation result is taken by assigning each pixel the label of the class with the max predicted probability from the softmax output. Segmentation performance was assessed using 5-fold cross validation, in which a full DCNN model is trained on 4/5ths folds of the data set and tested on the 5th fold, for all 5 folds. We assessed the sensitivity and specificity of our cell detection and computed the mean intersection over union (IOU) - defined as: IOU(A, B) = |A∩B|/|A∪B|, for each class of detected cells. All object analysis in this work was conducted using scikit-image52. For the purposes of computing sensitivity and specificity, a cell was considered to be detected if the IOU with the truth, based on manual segmentation, was ≥ 0.5.
Following segmentation by the DCNN, minimum and mean minimum distances in between cells or nuclei of interest, the convex and regular areas, circularity and eccentricity, convex and regular perimeters, equivalent diameter, major and minor axis lengths, aspect ratio, pixel size, solidity, perimeter/circularity ratio, and probability of belonging to the designated class for each object were computed for detected CD3+CD4+ and CD3+CD4– T cells. In addition, for every T cell the minimum distance to the nearest DC was computed in two ways: the first was simply the minimum Euclidean distance found between all the pixels in the T cell and the nearest DC pixel; an additional mean minimum distance was computed by averaging the distances from all pixels in the T cell to the nearest dendrite.
Data Analysis and Algorithm Comparison
Original CDM analysis based on minimum distance48 and preliminary TPEM data was performed with GraphPad Prism 5.0a software for Mac (GraphPad Software). All subsequent dataset analysis was performed using R statistical software (version 3.4.1, The R Foundation for Statistical Computing) and RStudio (version 1.0.153, RStudio), running on MacOS 10.13.1 High Sierra, powered by a 3.5 GHz 6-core Intel Xeon E5 CPU with 32 GB of RAM. Mouse adoptive transfer and lupus nephritis data were subjected to multivariate logistic regression, support vector machine (SVM), random forest, and neural network analyses. The following R packages were used for modeling: randomforest (ver 4.6–14), e1071 (1.7–0), xgboost (0.71.2), rpart (4–1.13) and rpart.plot (3.0.4), neuralnet (1.33), glmnet (2.0–16). The caret package was utilized when multiple instances or tunes were required to be generated and compared. Each instance of SVM, neural network, and random forest model generation was preceded by explicitly invoking a specific kernel seed to allow for result reproducibility. Modeling was performed on log-transformed and normalized input data with a binary outcome variable, representing classification (either wild-type or 5CC7, or CD3+CD4+ or CD3+CD4– cells). To control for incomplete or incorrect segmentation, all objects with areas < 3 and ≥ 100 square pixels were removed. Test and train datasets were defined as random 1/3:2/3 selections of input data. All n-fold cross-validations were performed with an n = 5. SVM analysis was subjected to linear, radial, and sigmoid kernels when comparing among models. Random forest analysis was used to define the relative importance of predictors as follows: a default of 500 trees were generated for each datapoint using the randomforest package and the optimal Cp parameter was selected based on minimum square error (MSE) optimization. The resultant RF plots were visualized and the relative importance of each split versus mean decrease in accuracy was recorded. The features across each experiment were compared to determine predictor hierarchy. Results of logistic regression were cross-referenced to ensure data consistency and agreement. ROCR and pROC packages were used to generate AUC curves and confidence interval on all included ROC plot figures with the following parameters: 10,000 bootstrap replicates, stratification, curve smoothing, and a confidence interval alpha of 0.90, corresponding to a type I error of 0.05. P values for comparison of neural network algorithm output with the null hypothesis (AUC = 0.5) were obtained with the verification R package using the roc.area function, and are separate from the indicated confidence intervals. Mann-Whitney U-test (unpaired Wilcoxson rank sum test) was utilized whenever group comparisons were performed. A Benjamini-Hochberg correction was applied for large datasets exceeding 5,000 observations with an FDR set to 5%, which was applicable for human lupus nephritis analysis.
TNN generation
After segregating the data based on minimum distance cutoffs, neural network models were generated using the following independent predictors: convex and regular areas, convex and regular perimeters, equivalent diameter, and major and minor axis lengths. A total of 3 different models of neural networks were generated and their performance compared: a single-layer model with (n+1) nodes in the hidden layer, where n is number of predictors; an actively tuned model, with 3:(n−1) nodes in the first hidden layer with the addition of 0:2 additional hidden layers consisting of a maximum of 2 and 1 additional hidden nodes, respectively; and a linear output neural network model with the specification of no hidden layers, constant weights, and linear output. In order to avoid result skewing due to unbalanced numbers of cell types in relation to distance, each instance of neural network analysis was performed by taking the minimum number of cells (wild-type and 5CC7 cells for mouse, and CD3+CD4+ and CD3+CD4– cells for lupus nephritis), and randomly sampling an equal number of cells from the second population. Every network analysis performed was specified with a threshold of 0.1, stepmax of 1 × 108, default (logistic) activation function, cross entropy error differentiable function, and otherwise default package parameters for learning rate, starting weights, and number of repetitions. When possible, the performance of each algorithm was compared based on the parameters of total error, classification accuracy, and receiver operator curve performance of correctly predicting cell type.
Data and materials availability
The data that support the findings of this study are available from the corresponding authors upon request.
Supplementary Material
Acknowledgements
The research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases (NIH) under Award Number U19 AI082724 (M.R.C.) and the National Institute of Arthritis and Musculoskeletal and Skin Diseases under Award Number K08 AR068421 (V.M.L.). Funding was also provided by R01 AR055646 (M.R.C.), T32 EB002103 (A.S.), and U01 CA195564 (M.L.G.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Computational support was provided by the SIRAF cluster with special thanks to Chun-Wai Chan, M.Sc (Cancer Center Support Grant P30 CA014599). GPU computation was supported by the University of Chicago Research Computing Center. Computational support on the Beagle supercomputer was provided by the NIH through resources provided by the Computation Institute and the Biological Sciences Division of the University of Chicago and Argonne National Laboratory, under grant 1S10OD018495-01, with thanks to Lorenzo Pesce and Joe Urbanski. All imaging was performed at the University of Chicago Integrated Light Microscopy Facility, with thanks to Vytas Bindokas, and Christine Labno. We thank Rebecca Abraham for careful reading of this manuscript.
Footnotes
Competing Interests:
R.N.G. is a full-time employee of the National Institutes of Health. There are no other applicable disclosures for the authors.
Accession Codes
Not applicable. No data with mandated deposition were utilized in this work.
References
- 1.Miller MJ, Wei SH, Parker I & Cahalan MD Two-photon imaging of lymphocyte motility and antigen response in intact lymph node. Science 296, 1869–1873 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Miller MJ, Safrina O, Parker I & Cahalan MD Imaging the single cell dynamics of CD4+ T cell activation by dendritic cells in lymph nodes. J Exp Med 200, 847–856 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mempel TR, Henrickson SE & Von Andrian UH T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature 427, 154–159 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Stoll S, J. D, T.M. B & Germain RN Dynamic imaging of T cell-dendritic cell interactions in lymph nodes. Science 296, 1873–1876 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Germain RN, Robey EA & Cahalan MD A decade of imaging cellular motility and interaction dynamics in the immune system. Science 336, 1676–1681 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masedunskas A et al. Intravital microscopy: a practical guide on imaging intracellular structures in live animals. Bioarchitecture 2, 143–157 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Secklehner J, Lo Celso C & Carlin LM Intravital microscopy in historic and contemporary immunology. Immunol Cell Biol 95, 506–513 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.You S et al. Intravital imaging by simultaneous label-free autofluorescence-multiharmonic microscopy. Nat Commun 9, 2125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobat D, Horton NG & Xu C In vivo two-photon microscopy to 1.6-mm depth in mouse cortex. Journal of biomedical optics 16, 106014 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Yew E, Rowlands C & So PT Application of multiphoton microscopy in dermatological dtudies: A mini-review. Journal of innovative optical health sciences 7, 1330010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher DT et al. Intraoperative intravital microscopy permits the study of human tumour vessels. Nat Commun 7, 10684 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerner MY, Kastenmuller W, Ifrim I, Kabat J & Germain RN Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity 37, 364–376 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goltsev Y et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174, 968–981.e915 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keren L et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell 174, 1373–1387.e1319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giesen C et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods 11, 417–422 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Azizi E et al. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 174, 1293–1308.e1236 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arazi A et al. The immune cell landscape in kidneys of lupus nephritis patients. bioRxiv 10.1101/363051 (2018). [DOI]
- 18.Chevrier S et al. An Immune Atlas of Clear Cell Renal Cell Carcinoma. Cell 169, 736–749.e718 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liarski V et al. Quantitative cell distance mapping in human nephritis reveals organization of in situ adaptive immune responses. Sci Trans Med 6(230):230ra46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q et al. CD8+ effector T cell migration to pancreatic islet grafts is dependent on cognate antigen presentation by donor graft cells. J Immunol 197, 1471–1476 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simoni Y et al. Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 557, 575–579 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Martin-Cofreces NB, Baixauli F & Sanchez-Madrid F Immune synapse: conductor of orchestrated organelle movement. Trends Cell Biol 24, 61–72 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dustin ML & Groves JT Receptor signaling clusters in the immune synapse. Annu Rev Biophys 41, 543–556 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lesserre R & Alcover A Microtubule dynamics and signal transduction at the immunological synapse: new partners and new connections. EMBO J 31 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monks CR, Freiberg BA, Kupfer H, Sciaky N & Kupfer A Three-dimensional segregation of supramolecular activation clusters in T cells. Nature 395, 82–86 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Dustin ML et al. A novel adpator protein orchestrates receptor patterning and cytoskeletal polarity in T-cell contacts. Cell 94, 667–677 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Tourret M et al. T Cell polarity at the immunological synapse Is required for CD154-dependent IL-12 secretion by dendritic cells. J Immunol 185, 6809–6818 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Blanchard N et al. Strong and durable TCR clustering at the T/dendritic cell immune synapse is not required for NFAT activation and IFN-γ production in human CD4 T Cells. J Immunol 173, 3062–3072 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Dustin ML The cellular context of T cell signaling. Immunity 30, 482–492 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Panhuys N, Klauschen F & Germain RN T-cell-receptor-dependent signal intensity dominantly controls CD4(+) T cell polarization In Vivo. Immunity 41, 63–74 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malherbe L, Mark L, Fazilleau N, McHeyzer-Williams L & McHeyzer-Williams MG Vaccine adjuvants alter TCR-based selection thresholds. Immunity 28, 698–709 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baumbartner CK, Ferrante A, Nagaoka M, Gorski J & Malherbe LP Peptide-MHC class II complex stability governs CD4 T cell clonal selection. J Immunol 184, 573–581 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olson E Particle shape factors and their use in image analysis-Part 1: Theory. J GXP Compl 15, 85–90 (2011). [Google Scholar]
- 34.Team, R.C. R: A language and environment for statistical computing. R version 3.4.1 (2017–06-30) -- “Single Candle” ed. Vienna, Austria; 2017. [Google Scholar]
- 35.Polliack A et al. Identification of human B and T lymphocytes by scanning electron microscopy. J Exp Med 138, 607–624 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sallusto F & Lanzavecchia A Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J Exp Med 179, 1109–1118 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guermonprez P, Valladeau J, Zitvogel L, Thery C & Amigorena S Antigen presentation and T cell stimulation by dendritic cells. Annu Rev Immunol 20, 621–667 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Swiecki M & Colonna M The multifaceted biology of plasmacytoid dendritic cells. Nat Rev Immunol 15, 471–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villadangos JA & Young L Antigen-presentation properties of plasmacytoid dendritic cells. Immunity 29, 352–361 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Villani AC et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, 273–284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang A et al. In situ B cell-mediated immune responses and tubulointerstitial inflammation in human lupus nephritis. J Immunol 186, 1849–1860 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delon J, Kaibuchi K & Germain RN Exclusion of CD43 from the immunological synapse is mediated by phosphorylation-regulated relocation of the cytoskeletal adaptor moesin. Immunity 15, 691–701 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Allenspach EJ et al. ERM-dependent movement of CD43 defines a novel protein complex distal to the immunological synapse. Immunity 15, 739–750 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Hutton L & Larrie V Using statistics to assess the performance of neural network classifiers. Johns Hopkins APL technical digest 13, 291–299 (1992). [Google Scholar]
- 45.Razi M & Athappilly K A comparative predictive analysis of neural networks (NNs), nonlinear regression and classification and regression tree (CART) models. Expert systems with applications 29, 65–74 (2005). [Google Scholar]
References for Methods
- 46.van Panhuys N, Klauschen F & Germain RN T-cell-receptor-dependent signal intensity dominantly controls CD4(+) T cell polarization In Vivo. Immunity 41, 63–74 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang A et al. In situ B cell-mediated immune responses and tubulointerstitial inflammation in human lupus nephritis. J Immunol 186, 1849–1860 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MATLAB and Signal Processing Toolbox. The MathWorks Inc. [Google Scholar]
- 49.Midway2: University of Chicago Research Computing Center Cluster. [cited] Available from: https://rcc.uchicago.edu/support-and-services/midway2
- 50.Abadi M et al. TensorFlow: Large-scale machine learning on heterogeneous systems. 2015. [Google Scholar]
- 51.Glorot YB a.X. Understanding the difficulty of training deep feedforward neural networks. 13th International Conference on Artificial Intelligence and Statistics Sardinia, Italy; 2010. [Google Scholar]
- 52.van der Walt S et al. scikit-image: Image processing in Python. PeerJ 2, e453 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.