Abstract
As immune checkpoint inhibitors are rapidly developing into the standard of care for patients with advanced melanoma, the value of diagnostic metrics to predict response to immunotherapy is steadily increasing. Next-generation sequencing based parameters include tumor mutation burden (TMB) as well as genomic amplification of PD-L1/PD-L2/JAK2 at 9p24.1. At present, there are limited studies documenting response to checkpoint blockade in 9p24.1 amplified solid tumors. Herein, we have compared a cutaneous melanoma with a mucosal melanoma, both with 9p24.1 amplifications and durable response to immunotherapy. While the cutaneous melanoma had a high TMB, the mucosal melanoma had a lower TMB compared to the mean TMB for all melanomas within the institutional clinical sequencing cohort. In summary, PD-L1/PD-L2/JAK2 amplification was associated with durable response to therapy for both cases and this genomic signature is a potential valuable metric in predicting response to immunotherapy.
Keywords: Melanoma, JAK2, PD-L1, PD-L2, 9p24.1 Amplification, Pembrolizumab, Ipilimumab, Nivolumab, Immunotherapy
1. Introduction
FDA approved agents for the treatment of patients with advanced melanoma include BRAF/MEK-targeted therapeutics (BRAF: vemurafenib, dabrafenib; MEK: trametinib and cobimetinib) as well as immune checkpoint inhibitors (1, 2). Ligands such as programmed death ligand-1 (PD-L1, also known as CD274/ PDCD1LG1) and programmed death ligand-2 (PD-L2, also known as CD273/ PDCD1LG2) expressed on the tumor cell surface bind to specific immune cell receptors, such as programmed death 1 (PD-1), to evade the host immune response (2). Immunotherapeutic agents disrupt this interaction to restore the host immune response and currently approved agents include ipilimumab (anti- cytotoxic T-lymphocyte associated protein 4, CTLA4), nivolumab and pembrolizumab (anti-PD-1) and response rates as high as 55% have been reported with anti-CTLA4/PD-1 therapy (2, 3).
The vast majority of melanomas, including mucosal melanomas, elicit an adaptive immune response where host T-cells upregulate PD-1 expression in response to an increased load of tumor neoantigens, while cytokines such as interferon-gamma released by these activated T-cells promote the expression of PDL-1 on tumor cells, allowing them to escape the host immune response (2).
Biomarkers used to predict response to immunotherapy include metrics to assess the immune micro environment (CD8+ cell density), tumor neoantigen load (tumor mutational burden), as well as the expression of PD-L1 itself (2, 4). Constitutive PD-L1/PD-L2 expression, often secondary to genomic amplification at the 9p24.1 locus, is a rare event seen in <1% of melanomas (5, 6). The presence of JAK2 at the 9p24.1 locus is significant as prior studies have shown that JAK2 signaling can further upregulate PD-L1 expression (7). Hematolymphoid neoplasias with PD-L1/PD-L2/JAK2 co-amplifications such as classical Hodgkin lymphoma, primary central nervous system lymphoma and primary testicular lymphoma have shown good response to nivolumab, however, studies in solid tumors with a similar genomic signature are lacking (8–10).
A prior report documented an exceptional response to anti-PD1 therapy in a patient with a metastatic basal cell carcinoma with a 9p24.1 amplification but this patient also had a high tumor mutation burden (TMB) and therefore exhibited multiple genomic signatures that would predict response to immunotherapy (11). A more recent study involving various 9p24.1 amplified solid tumors with a low to intermediate TMB (based on sequencing of 1.2Mb of the genome; low TMB defined as ≤5 mutations/megabase, intermediate TMB defined as 6–19 mutations/megabase, high TMB defined as ≥20 mutations/megabase) documented objective responses to the administration of immune checkpoint blockade agents for 6 of 9 (66.7%) cases, emphasizing the importance of this genomic signature (6).
Herein, we report two patients with metastatic melanoma that had a 9p24.1 amplification that showed a durable response to immunotherapy.
2. Materials and Methods
2.1. Methods
Immunohistochemistry for PD-L1 (Cell Signaling Technology, Danvers, MA, clone E1L3N, 1:400 dilution) was performed on representative whole-tissue sections.
The Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) assay was used for molecular profiling and details of this assay have been previously reported (12). In brief, the MSK-IMPACT assay involves hybridization capture-based library preparation, followed by deep sequencing of up to 6,614 protein-coding exons of 468 genes in the current panel. Approximately 1.5 megabases of the human genome is targeted by this assay, with homogenous distribution of single nucleotide polymorphism tiling probes across the genome which allows for accurate genome-wide copy number assessment. This assay is currently FDA authorized as a class II in vitro diagnostic test.
Tumor mutation burden (TMB) is defined as the total number of nonsynonymous mutations that occur as a fraction of the total genomic target region (mutations per megabase, mt/MB) identified using MSK-IMPACT. In this report, the metastatic cutaneous melanoma involving the liver was sequenced using a 468 gene panel and the mucosal melanomas were sequenced using a prior version of the assay with a 410 gene panel.
2.2. Case Report: Case 1
An 88 year-old male with a remote history of prostatic adenocarcinoma and cutaneous melanoma presented with widely metastatic disease involving the liver, lung and bone. Biopsy of the 2.5cm hepatic lesion revealed metastatic cutaneous melanoma (Figure 1A, B). Immunohistochemistry for PD-L1 revealed strong, diffuse and membranous expression of PD-L1 in most neoplastic cells (Figure 1A, inset). Molecular profiling by MSK-IMPACT revealed a high TMB of 52.7 mt/MB (average TMB for melanomas in the MSK-IMPACT clinical sequencing cohort: 7.9 mt/MB), an abundance of G>A and C>T transitions, consistent with UV exposure, and no clear driver alteration (Table 1). A greater than threefold amplification at the 9p24.1 genomic locus including JAK2/PD-L1/PD-L2 was identified (Table 1, Figure 2A) and immunotherapy with an anti-PD1 agent (pembrolizumab) was initiated. The patient was also treated with radiation therapy to the sacrum. At last follow up (day 313), the patient had stable disease, including radiologic resolution of the hepatic mass (Figure 1C, 2B).
Figure 1: Histopathology & Imaging.
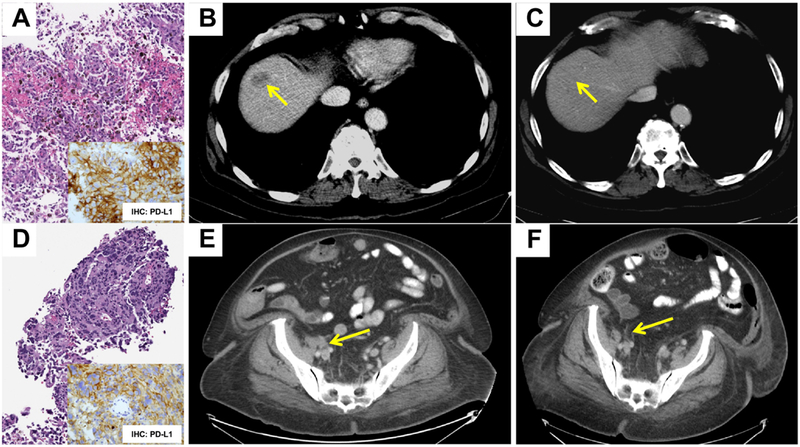
Representative histology of a liver biopsy showing metastatic cutaneous melanoma involving the liver (A, Case 1) and CT imaging showing response to therapy with Pembrolizumab (B: 2.5cm × 1.6cm on Day 1; C: 0.7cm × 0.6cm on Day 313). Representative immunohistochemistry for PD-L1 is depicted as well (A, inset). Histology of an inguinal mass biopsy showing involvement by metastatic mucosal melanoma (D, Case 2) and corresponding immunohistochemistry for PD-L1 (D, inset). CT imaging showing resolution of the lesion following therapy with Pembrolizumab (E: 2.4cm × 1.9cm, Day 256; F: no appreciable adenopathy, Day 1050) is depicted. Arrows demonstrate the lesion of interest.
Table 1.
Next-Generation Sequencing by MSK-IMPACT
| Case1. Metastatic Cutaneous Melanoma (to Liver) |
Case2. Mucosal (Urethral) Melanoma |
|
|---|---|---|
|
Tumor Purity (* Facets) & Coverage |
36% Purity; 810X Coverage |
36% Purity; 661X Coverage |
| TMB (mt/MB) | 52.7mt/MB (468 gene panel)† | 3.9mt/MB (410 gene panel)† |
| Mutations Include |
TERT (g.1295228C>T) & TP63 (p.E609K) |
TP53 (p.R175L & p.R333fs) & ATRX (p.C1122fs) |
| Copy Number Gain | 3.3 FC at 9p24.1 (JAK2/PD-L1/PD-L2) |
3.6 FC at 9p24.1 & 2.1 FC at 4q12 (PDGFRA/KIT) |
| Copy Number Loss | NA | NA |
| Structural Variants | Not Detected | Not Detected |
| UV Signature | Present | Absent |
MSK-IMPACT: Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT); TMB: Tumor Mutation Burden; mt/MB=mutations/Megabase; FC: Fold Change; NA: Not Applicable.
FACETS: Algorithm involving allele-specific copy number analysis from next-generation sequencing data, Nucleic Acids Res. 2016 Sep 19;44(16):e131.
Average TMB for Melanoma in MSKCC-IMPACT cohort: 7.9mt/MB.
Figure 2: Timeline and Copy Number Assessment in Melanomas with 9p24.1 Amplification.
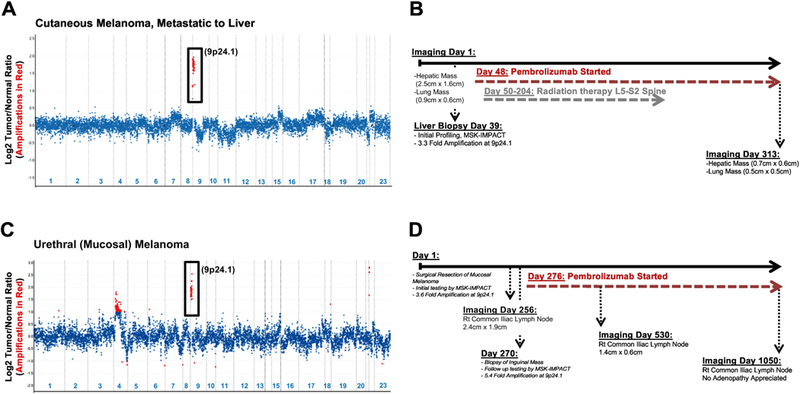
Copy number plots have been shown for both patients (A: Case 1, cutaneous melanoma metastatic to liver; C: Case 2, urethral melanoma). Relative (Log2) tumor/normal ratios (y-axis) and corresponding chromosomes (x-axis) are displayed, with each blue dot representing an individual probe region. Amplified regions are shown in red. Corresponding timelines detailing key dates pertaining to imaging, molecular testing and immunotherapy has been shown for the cutaneous metastatic melanoma involving the liver (B, Case 1) and the mucosal melanoma (D, Case 2).
2.3. Case Report: Case 2
A 73 year-old female presented with a urethral mass which was biopsied and revealed a mucosal melanoma. MSK-IMPACT testing revealed a low TMB of 3.9 mt/MB (Table 1), KIT/PDGFRA amplification at 4q12 likely serving as a driver alteration, and 3.6-fold amplification at 9p24.1 (Table 1, Figure 2C). On follow up, the patient developed a metastasis to the right common iliac lymph node (Figure 1D, 1E) and MSK-IMPACT testing revealed a similar genotype, TMB and copy number alterations, including a 5.4 fold amplification at 9p24.1. This 9p24.1 amplification was concordant with immunohistochemistry for PD-L1, which revealed strong, diffuse and membranous expression of PD-L1 in most neoplastic cells (Figure 1D, inset). This patient was treated with pembrolizumab as well, starting on day 276 and showed radiologic improvement by day 530 and resolution of the abdominopelvic lymphadenopathy at last follow-up (day 1050, Figure 1F, 2D).
3.0. Discussion
Immunotherapeutic options are steadily being incorporated into the standard of care for patients with advanced melanoma. The expression of ligands such as PD-L1, PD-L2 and CD80 (also known as B7.1/ CD28LG) on the tumor cell surface allow them to bind receptors on T-cells within the tumor microenvironment, such as PD-1 and CTLA4, to downregulate the host immune response (2, 13). FDA approved immune checkpoint inhibitors such as ipilimumab (anti-CTLA4), pembrolizumab and nivolumab (humanized and fully human anti-PD-1 monoclonal antibodies, respectively) counteract this immune evasion and have shown promising responses (2, 13). Response rates as high as 50% and durable response to therapy in many cases, with concurrent or sequential therapy with ipilimumab and nivolumab, in one study, highlight the success of these agents (2, 13).
The primary biomarker used to predict response to immunotherapeutic agents is PD-L1 protein expression assessed by immunohistochemistry, while others include CD8 positive T-cell density and specific transcriptional signatures (2, 13). Recently TMB, a surrogate metric for assessing tumor immunogenicity by determining the load of neo-epitopes, has emerged as a promising biomarker for predicting response to immunotherapy (4, 14). While cutaneous melanomas often have a high TMB secondary to UV exposure, characterized by an abundance of G>A and C>T transitions; non-cutaneous melanomas, such as mucosal melanomas, lack a similar mutational signature and tend to exhibit a lower TMB. A recent study of 9p24.1 amplifications, as determined by next-generation sequencing based methodology, in over 100,000 tumors showed that the majority of these tumors in fact have a low (defined as ≤5 mutations/Mb) to intermediate TMB (defined as 6–19 mutations/Mb) (6).
Comprehensive molecular profiling of tumors using panels such as MSK-IMPACT therefore provide metrics such as TMB and copy number status of genes such as PD-L1/PD-L2/JAK2 at 9p24.1 to help guide immunotherapeutic strategies (11). While the majority of melanomas, including mucosal melanomas, exhibit an induced pattern of PD-L1 expression secondary to signaling events within the tumor microenvironment, a small percentage of cases exhibit constitutive expression secondary to 9p24.1 amplifications (2, 5, 6). Although the literature is limited in regards to treatment outcomes for solid tumors with 9p24.1 amplifications, a few case series and reports suggest that such a signature is likely to predict favorable response to immunotherapy (6, 11).
Herein, we compared a case of metastatic cutaneous melanoma with a case of a metastatic mucosal melanoma. Both cases showed 9p24.1 amplifications and exhibited a durable response to immunotherapy. A high TMB, as defined by ≥20 mutations/Mb by Goodman et al, was associated with favorable response to immunotherapy for the metastatic cutaneous melanoma (4, 6). However, in the case of the mucosal melanoma it was lower than the mean TMB for all melanomas in the MSKCC clinical sequencing cohort. In contrast, PD-L1/PD-L2/JAK2 amplification events were correlated with durable response to therapy for both cases and suggest that such genomic signatures should be carefully documented and correlated with response to checkpoint blockade administration to guide future immunotherapeutic decision making.
Highlights.
PD-L1/PD-L2/JAK2 amplification can be identified using next generation sequencing
Durable response to immunotherapy is shown in 2 cases of metastatic melanoma
PD-L1/PD-L2/JAK2 amplification: a potential biomarker of response to immunotherapy
Acknowledgments
Sources of Support: This research was funded in part through the NIH/NCI Cancer Center Support Grant P30 CA008748 and supported by the Marie-Josée and Henry R. Kravis Center for Molecular Oncology.
Footnotes
Disclosures: The authors of this article have no relevant financial relationships with commercial interests to disclose.
References
- 1.Coit DG, Thompson JA, Algazi A, Andtbacka R, Bichakjian CK, Carson WE 3rd, Daniels GA, DiMaio D, Fields RC, Fleming MD, Gastman B, Gonzalez R, Guild V, Johnson D, Joseph RW, Lange JR, Martini MC, Materin MA, Olszanski AJ, Ott P, Gupta AP, Ross MI, Salama AK, Skitzki J, Swetter SM, Tanabe KK, Torres-Roca JF, Trisal V, Urist MM, McMillian N, Engh A. NCCN Guidelines Insights: Melanoma, Version 3.2016. J Natl Compr Canc Netw 2016; 14, 945–958. [DOI] [PubMed] [Google Scholar]
- 2.Taube JM, Galon J, Sholl LM, Rodig SJ, Cottrell TR, Giraldo NA, Baras AS, Patel SS, Anders RA, Rimm DL, Cimino-Mathews A. Implications of the tumor immune microenvironment for staging and therapeutics. Mod Pathol 2018; 31, 214–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M, Rutkowski P, Ferrucci PF, Hill A, Wagstaff J, Carlino MS, Haanen JB, Maio M, Marquez-Rodas I, McArthur GA, Ascierto PA, Long GV, Callahan MK, Postow MA, Grossmann K, Sznol M, Dreno B, Bastholt L, Yang A, Rollin LM, Horak C, Hodi FS, Wolchok JD. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med 2015; 373, 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, Stephens PJ, Daniels GA, Kurzrock R. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol Cancer Ther 2017; 16, 2598–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaunitz GJ, Cottrell TR, Lilo M, Muthappan V, Esandrio J, Berry S, Xu H, Ogurtsova A, Anders RA, Fischer AH, Kraft S, Gerstenblith MR, Thompson CL, Honda K, Cuda JD, Eberhart CG, Handa JT, Lipson EJ, Taube JM. Melanoma subtypes demonstrate distinct PD-L1 expression profiles. Lab Invest 2017; 97, 1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodman AM, Piccioni D, Kato S, Boichard A, Wang HY, Frampton G, Lippman SM, Connelly C, Fabrizio D, Miller V, Sicklick JK, Kurzrock R. Prevalence of PDL1 Amplification and Preliminary Response to Immune Checkpoint Blockade in Solid Tumors. JAMA Oncol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Green MR, Monti S, Rodig SJ, Juszczynski P, Currie T, O’Donnell E, Chapuy B, Takeyama K, Neuberg D, Golub TR, Kutok JL, Shipp MA. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood 2010; 116, 3268–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, Schuster SJ, Millenson MM, Cattry D, Freeman GJ, Rodig SJ, Chapuy B, Ligon AH, Zhu L, Grosso JF, Kim SY, Timmerman JM, Shipp MA, Armand P. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med 2015; 372, 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nayak L, Iwamoto FM, LaCasce A, Mukundan S, Roemer MGM, Chapuy B, Armand P, Rodig SJ, Shipp MA. PD-1 blockade with nivolumab in relapsed/refractory primary central nervous system and testicular lymphoma. Blood 2017; 129, 3071–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Younes A, Santoro A, Shipp M, Zinzani PL, Timmerman JM, Ansell S, Armand P, Fanale M, Ratanatharathorn V, Kuruvilla J, Cohen JB, Collins G, Savage KJ, Trneny M, Kato K, Farsaci B, Parker SM, Rodig S, Roemer MG, Ligon AH, Engert A. Nivolumab for classical Hodgkin’s lymphoma after failure of both autologous stem-cell transplantation and brentuximab vedotin: a multicentre, multicohort, singlearm phase 2 trial. Lancet Oncol 2016; 17, 1283–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikeda S, Goodman AM, Cohen PR, Jensen TJ, Ellison CK, Frampton G, Miller V, Patel SP, Kurzrock R. Metastatic basal cell carcinoma with amplification of PD-L1: exceptional response to anti-PD1 therapy. NPJ Genom Med 2016; 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM, Hellmann MD, Barron DA, Schram AM, Hameed M, Dogan S, Ross DS, Hechtman JF, DeLair DF, Yao J, Mandelker DL, Cheng DT, Chandramohan R, Mohanty AS, Ptashkin RN, Jayakumaran G, Prasad M, Syed MH, Rema AB, Liu ZY, Nafa K, Borsu L, Sadowska J, Casanova J, Bacares R, Kiecka IJ, Razumova A, Son JB, Stewart L, Baldi T, Mullaney KA, Al-Ahmadie H, Vakiani E, Abeshouse AA, Penson AV, Jonsson P, Camacho N, Chang MT, Won HH, Gross BE, Kundra R, Heins ZJ, Chen HW, Phillips S, Zhang H, Wang J, Ochoa A, Wills J, Eubank M, Thomas SB, Gardos SM, Reales DN, Galle J, Durany R, Cambria R, Abida W, Cercek A, Feldman DR, Gounder MM, Hakimi AA, Harding JJ, Iyer G, Janjigian YY, Jordan EJ, Kelly CM, Lowery MA, Morris LGT, Omuro AM, Raj N, Razavi P, Shoushtari AN, Shukla N, Soumerai TE, Varghese AM, Yaeger R, Coleman J, Bochner B, Riely GJ, Saltz LB, Scher HI, Sabbatini PJ, Robson ME, Klimstra DS, Taylor BS, Baselga J, Schultz N, Hyman DM, Arcila ME, Solit DB, Ladanyi M, Berger MF. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017; 23, 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Achkar T, Ali SM, Welsh A, Dhir R, Gollin SM, Parikh RA. A prolonged response to platinum-based therapy in a patient with metastatic urothelial carcinoma harboring a single rearranged and truncated NF2 gene. Genes Chromosomes Cancer 2018. [DOI] [PubMed] [Google Scholar]
- 14.Hellmann MD, Callahan MK, Awad MM, Calvo E, Ascierto PA, Atmaca A, Rizvi NA, Hirsch FR, Selvaggi G, Szustakowski JD, Sasson A, Golhar R, Vitazka P, Chang H, Geese WJ, Antonia SJ. Tumor Mutational Burden and Efficacy of Nivolumab Monotherapy and in Combination with Ipilimumab in Small-Cell Lung Cancer. Cancer Cell 2018; 33, 853–861e854. [DOI] [PMC free article] [PubMed] [Google Scholar]


