Abstract
In budding yeast, a collar of septin filaments at the neck between a mother cell and its bud marks the incipient site for cell division and serves as a scaffold that recruits proteins required for proper spatial and temporal execution of cytokinesis. A set of interacting proteins that localize at or near the bud neck, including Aim44/Gps1, Nba1 and Nis1, also has been implicated in preventing Cdc42‐dependent bud site re‐establishment at the division site. We found that, at their endogenous level, Aim44 and Nis1 robustly localize sequentially at the septin collar. Strikingly, however, when overproduced, both proteins shift their subcellular distribution predominantly to the nucleus. Aim44 localizes with the inner nuclear envelope, as well as at the plasma membrane, whereas Nis1 accumulates within the nucleus, indicating that these proteins normally undergo nucleocytoplasmic shuttling. Of the 14 yeast karyopherins, Kap123/Yrb4 is the primary importin for Aim44, whereas several importins mediate Nis1 nuclear entry. Conversely, Kap124/Xpo1/Crm1 is the primary exportin for Nis1, whereas both Xpo1 and Cse1/Kap109 likely contribute to Aim44 nuclear export. Even when endogenously expressed, Nis1 accumulates in the nucleus when Nba1 is absent. When either Aim44 or Nis1 are overexpressed, Nba1 is displaced from the bud neck, further consistent with the mutual interactions of these proteins. Collectively, our results indicate that a previously unappreciated level at which localization of septin‐associated proteins is controlled is via regulation of their nucleocytoplasmic shuttling, which places constraints on their availability for complex formation with other partners at the bud neck.
Keywords: cell cycle, karyopherin, mutants, scaffold, septin collar, septin filaments
1. INTRODUCTION
Successful completion of biological processes relies on accurate spatio‐temporal targeting of proteins and enzymes to specific subcellular sites within the cell. In budding yeast (Saccharomyces cerevisiae), a superstructure (“collar”) composed of filaments containing five members of the septin family of GTP‐binding proteins is assembled at the neck between a mother cell and its bud. Four of these septin subunits (Cdc3, Cdc10, Cdc11 and Cdc12) are encoded by genes originally identified in the now‐classical screen for cell division cycle (cdc) mutants conducted by Hartwell and colleagues (Hartwell, 1971; Hartwell, Culotti, Pringle, & Reid, 1974). The fifth septin subunit (Shs1) expressed in proliferating cells was identified via other means (Carroll, Altman, Schieltz, Yates, & Kellogg, 1998; Mino et al., 1998); it appears to be a recently evolved paralog of Cdc11 (Finnigan, Takagi, Cho, & Thorner, 2015; Garcia et al., 2011; Iwase, Luo, Bi, & Toh‐e, 2007). The septin‐based structures in mitotically dividing cells are assembled from two types of rod‐like, apolar hetero‐octamers that differ only in their terminal subunit: Cdc11‐Cdc12‐Cdc3‐Cdc10‐Cdc10‐Cdc3‐Cdc12‐Cdc11; and, Shs1‐Cdc12‐Cdc3‐Cdc10‐Cdc10‐Cdc3‐Cdc12‐Shs1 (Bertin et al., 2008; Finnigan, Takagi, et al., 2015; McMurray et al., 2011). In vitro, at physiological salt concentrations, Cdc11‐capped rods are able to polymerize end‐to‐end, and also associate laterally in register, thereby forming long filament pairs, especially on lipid layers containing phosphatidylinositol‐4,5‐bisphosphate that mimic the inner leaflet of the plasma membrane (PM) (Bertin et al., 2008; Bertin et al., 2010; Booth, Vane, Dovala, & Thorner, 2015; Bridges et al., 2014). By contrast, under the same conditions, Shs1‐capped rods alone cannot polymerize into filaments (Booth et al., 2015), but instead form more disorganized structures, which take the form of arcs, rings, spirals, bird's nest‐like assemblages, and gauze‐ or mesh‐like arrangements (Garcia et al., 2011). However, there is evidence that Cdc11‐capped rods and Shs1‐capped rods can form heterotypic junctions (Booth et al., 2015; Finnigan, Takagi, et al., 2015) and the relative proportions of these two types of rods influences the higher‐order assembly state of septin superstructures both in vivo and in vitro (Garcia et al., 2011; Khan, Newby, & Gladfelter, 2018). Indeed, complex lattice‐like filaments at the bud neck of dividing cells have been visualized at the ultrastructural level in spheroplasts “unroofed” by rapid freeze‐fracture and rotary coating with tantalum/tungsten (Rodal, Kozubowski, Goode, Drubin, & Hartwig, 2005) or platinum (Ong, Wloka, Okada, Svitkina, & Bi, 2014) and then examined by electron microscopy (EM), or in intact cells interrogated by cryo‐EM tomography (Bertin et al., 2012; Bertin & Nogales, 2016).
The septin collar plays an essential role in cell division in S. cerevisiae, for several reasons. First, because of their lipid‐binding properties, assembly of septin filaments at the cell cortex is thought to deform the PM (Bridges, Jentzsch, Oakes, Occhipinti, & Gladfelter, 2016; Cannon, Woods, & Gladfelter, 2017). Second, the septin filaments serve as a scaffold to recruit many other proteins required for the execution of cytokinesis, including assembly of the actomyosin contractile ring (Finnigan, Booth, Duvalyan, Liao, & Thorner, 2015; Marquardt, Chen, & Bi, 2019; Meitinger & Palani, 2016). Third, the collar also appears to be a physical barrier that restricts the diffusion or partitioning of various cellular constituents and organelles (Caudron & Barral, 2009; McMurray et al., 2011), as well as serving as a corral to constrain enzymes and other factors so that they act locally, such as the secretory vesicles and chitin synthases that are necessary for new membrane insertion and cell wall deposition at the division site (Dobbelaere & Barral, 2004; McMurray & Thorner, 2009). Fourth, among the proteins recruited to the bud neck via their physical association with the septins are regulatory proteins that ensure that the functions of the septin collar are properly coupled to the other events required for passage through the cell division cycle (Finnigan et al., 2016; Finnigan, Duvalyan, Liao, Sargsyan, & Thorner, 2016; Perez, Finnigan, Roelants, & Thorner, 2016; Sakchaisri et al., 2004; Shulewitz, Inouye, & Thorner, 1999).
Once a cell has committed to the cell cycle and initiated bud emergence, licensing of the next round of budding must be delayed until the completion of cell division. Bud site selection and bud emergence are dependent, in large part, on the local generation and action of the activated (GTP‐bound) state of the small GTPase Cdc42 (Chiou, Balasubramanian, & Lew, 2017). Therefore, in addition to promoting processes required for cell division, the septin collar must somehow prevent Cdc42 function within its immediate vicinity. Establishment of this exclusion zone at the division site that blocks subsequent polarization within that site had been attributed to localization and action at the collar of Rga1, a Cdc42‐specific GTPase‐activating protein (GAP) (Miller, Lo, Lee, Kang, & Park, 2017; Tong et al., 2007). However, more recent studies have indicated that an equally, if not more, important mechanism for imposing this zone of exclusion is mediated by a complex of collar‐associated proteins that act to squelch locally the activity of Cdc24, the Cdc42‐specific guanine nucleotide exchange factor (Meitinger et al., 2014; Meitinger & Pereira, 2017). This complex reportedly contains Aim44/Gps1 (758 residues), Nba1 (501 residues), Nap1 (417 residues) and Nis1 (407 residues).
Aim44 is thought to act as an organizing platform (Meitinger et al., 2013; Meitinger et al., 2014) because it is able to bind directly to all the other components (Nba1, Nap1 and Nis1), to Cdc42 itself, as well as to another small GTPase Rho1 involved in bud morphogenesis (Jonasson et al., 2016; Pruyne, Legesse‐Miller, Gao, Dong, & Bretscher, 2004). The available evidence indicates that collectively, the proteins in this complex sustain Rho1‐dependent polarization and inhibit premature Cdc42 activation at the site of cytokinesis.
Importantly, Aim44 is required for robust targeting to the bud neck of Nba1 and Nis1 (but not Nap1), a finding consistent with the observation that Nis1 does not interact intimately with any of the five septin subunits in the collar, as assessed using an in vivo tripartite split‐GFP assay (Finnigan, Duvalyan, et al., 2016). It is currently not known whether Aim44, Nba1 or Nap1 make direct physical contact with any septin in the collar. In this regard, however, it has been reported that physical association of Aim44 with the septin collar‐bound protein kinase Gin4 is required for efficient retention of Aim44 at the bud neck (Meitinger & Pereira, 2017).
Aim44 also has been implicated in somehow regulating phosphorylation of the bud neck‐localized, SH3‐ and F‐BAR domain‐containing protein Hof1 by the protein kinases Cdc5 and Dbf2 (Meitinger et al., 2011; Meitinger, S, Hub, & Pereira, 2013), modifications that reportedly enhance the efficiency of actomyosin ring contraction (Wolken, McInnes, & Pon, 2014), although phosphorylation does not influence interaction of Hof1 with its partner SH3 domain‐containing protein Cyk3 (Wang, Nishihama, Onishi, & Pringle, 2018). What seems better established now is that the Aim44‐dependent complex is able to prevent recruitment of Cdc24, thereby suppressing generation of active GTP‐bound Cdc42 in or near the existing septin collar (Meitinger et al., 2014). The blockade of Cdc24 appears to be mediated by Aim44‐bound Nba1. Available data (Meitinger et al., 2014) indicate that Nba1 binds directly to a complex of Cdc24 bound to another SH3 domain‐containing protein (Bem1) and thereby blocks binding of the Cdc24‐Bem1 complex to the small GTPase Rsr1/Bud1, a polarity landmark recruited to the previous budding site (Park, Bi, Pringle, & Herskowitz, 1997). On the other hand, a recent study indicates that Nba1 associates with the tandem LIM domains in Rga1, thus localizing this Cdc42 GAP and thereby squelching localized Cdc42 action for that reason (Miller et al., 2017). Nba1 (and Nis1) are retained at the previous sites of cytokinesis (Meitinger et al., 2014).
Clearly, further investigation of the localization, interrelationships, and dynamics of the Aim44‐Nba1‐Nap1‐Nis1 complex should help to refine our understanding of the mechanisms by which the septin collar serves to organize and integrate signals emanating from the bud neck. In this study, we used fluorescently‐tagged versions of these proteins to monitor their subcellular distribution, and both loss‐of‐function mutations and overexpression as a means to interrogate and glean new information about each of the components of the Aim44‐Nba1‐Nap1‐Nis1 complex and their interdependencies.
2. RESULTS
2.1. Aim44, Nba1, Nap1 and Nis1 display distinct localization patterns at the bud neck
To investigate the interplay among Aim44, Nis1, Nba1 and Nap1 and their relationship to the septin collar, we first examined asynchronous cultures of strains that each expressed from its native promoter at its normal chromosomal locus a derivative of the protein of interest to which the fluorophore mNeon Green (mNG) (Shaner et al., 2013) had been fused in‐frame to the C‐terminal end and also co‐expressed from its endogenous locus the septin Cdc10 tagged in the same way with the fluorophore mCherry (mCh) (Shaner et al., 2003). By examining cells at two different stages of the cell cycle (budded cells with an intact septin collar and budded cells in which the collar had split into two separate rings), we found that, although each of these proteins could be readily detected at the bud neck, the precise localization pattern for each protein was distinct (Table 1), suggesting that the Aim44‐Nba1‐Nap1‐Nis1 ensemble is not a stable complex in the typical sense.
Table 1.
Fraction of cells exhibiting bud neck localization for Aim44, Nba1, Nis1 and Nap1
| Asynchronous culture | Synchronized culture | ||||
|---|---|---|---|---|---|
| Cells with distinct septin collar | Cells with clearly split septin rings | Hydroxyurea arrest | Nocodazole arrest | cdc15ts arrest | |
| Aim44‐mNG | 81.7% (219)a | 81.5% (81) | 83.8% (198) | 83.9% (112) | 96.8% (95) |
| Nba1‐mNG | 29.5% (166) | 100% (105) | 24.2% (293) | 49.6% (242) | 97.9% (97) |
| Nis1‐mNG | 25.1% (187) | 100% (84) | 3.2% (188) | 21.7% (115) | 97.1% (105) |
| Nap1‐mNG | 23.6% (161) | 10.5% (38) | 81.0% (295) | 17.1% (275) | 90.5% (95) |
Number in parentheses represents the total number of cells scored in multiple fields in three independent experiments for each condition.
Aim44‐mNG localized predominantly at the bud neck in the majority of the cells with either an intact or split septin collar and co‐localized with it (Figure 1a, left column; Table 1), consistent with previous reports (Meitinger, Richter, et al., 2013; Wolken et al., 2014). In marked contrast, Nap1‐mNG displayed a diffuse cytoplasmic pattern and was only found prominently at the bud neck in ≤25% of the cells, where it too appeared to co‐localize with the septin collar (Figure 1a, right column; Table 1). Both Nis1‐mNG and Nba1‐mNG exhibited a third type of pattern. Each protein localized at the bud neck in about 25–30% of the cells with an intact septin collar, but both proteins were located at the bud neck in every cell in which the collar had split into two separate rings (Figure 1a, middle columns; Table 1). Even though the number of cells with an intact collar that displayed Nba1 or Nis1 at the bud neck was similar (Table 1), the intensity of the signal was always much weaker for Nis1 than Nba1, whereas for the cells with a split septin collar, the signal intensity was similar for Nis1 and Nba1 (Figure 1a). In addition, both proteins also accumulated at points on the mother cell cortex nearby the present division site, which likely correspond to the “cytokinetic remnants” or bud scars from previous divisions, as observed before (Meitinger et al., 2014). Together, these data are at least consistent with a sequential order of arrival of these proteins at the bud neck: Aim44 arrives at the septin collar first, followed by Nba1, and then Nis1. The role of Nap1 is more obscure, but our data suggest that it may arrive after Aim44, and only transiently. Furthermore, the fact that we found Nba1 and Nis1 at all split septin collars, some of which lack Aim44 (Table 1), suggests that Aim44 may be required for the initial recruitment of these proteins, but not for their retention at the bud neck.
Figure 1.
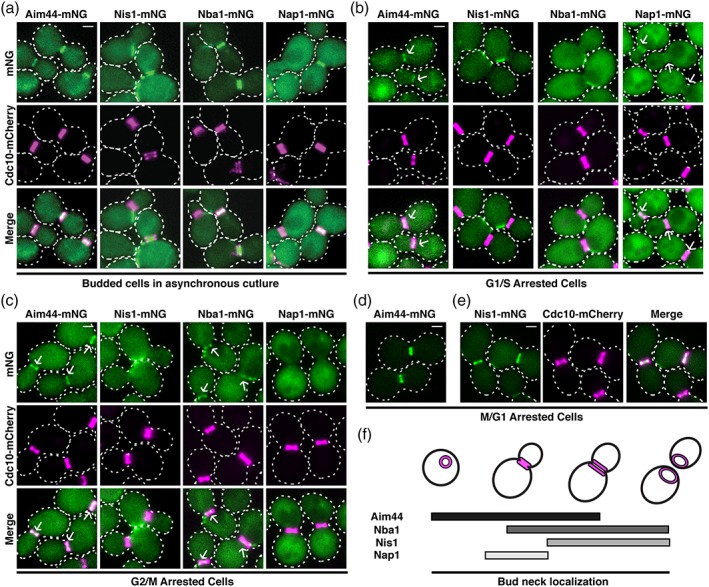
Effect of cell cycle stage on bud neck localization of endogenously‐expressed Aim44, Nap1, Nba1 and Nis1. (a) Cells expressing Cdc10‐mCh and Aim44 (APY185), Nis1 (APY104), Nba1 (APY252), or Nap1 (APY251) fusions to the N‐terminus of the fluorescent protein mNeonGreen (mNG) were grown to mid‐exponential phase and examined under the fluorescent microscope. (b) the same cells as in (a) were grown to early mid‐exponential phase, treated with 200 mM hydroxyurea for 2 h to arrest cells at the G1/S transition, and imaged. Arrows point to bud neck localization of the corresponding mNG‐tagged protein. (c) the same cells as in (a) were grown to early mid‐exponential phase, treated with 15 μg/mL nocodazole for 2 h to arrest cells at the G2/M transition, and imaged. Arrows point to bud neck localization of the corresponding mNG‐ tagged protein. (d) Aim44‐mNG was expressed from its native locus in a strain harboring the temperature‐sensitive cdc15‐2 allele (APY204). The cells were grown at 25° to early mid‐exponential phase, then shifted to 37° and cultured an additional 3 h to arrest cells at the M/G1 transition. (e) Nis1‐mNG and Cdc10‐mCh were expressed from their native loci in a strain harboring the temperature‐sensitive cdc15‐2 allele (APY112). Cells were grown as in (d) to induce cell cycle arrest. (f) Schematic diagram depicting the kinetic pattern of cell cycle‐dependent localization of Aim44, Nap1, Nba1 and Nis1 at the bud neck. The septins are depicted as magenta structures in the cells. Bars represent residency time at the bud neck of the indicated protein. Scale bars = 1 μm
2.2. Aim44, Nba1, Nap1 and Nis1 localize at the bud neck at different cell cycle stages
As a further means to assess the temporal and spatial relationships among this set of proteins, we examined the localization pattern for each protein in cells that had been arrested at three sequential stages in cell cycle progression by treatment with appropriate inhibitors or by shift of a cdc15 mutant to the restrictive temperature. In cells where completion of S phase was blocked by treatment with hydroxyurea, both Aim44 and Nap1 were associated predominantly with the septin collar at the bud neck, whereas Nba1 bud neck localization was only partial and Nis1 was, instead, restricted almost exclusively to the neighboring region of the cell cortex (Figure 1b; Table 1). In cells where completion of chromosome segregation was blocked by treatment with nocodazole, Aim44 was still robustly localized at the septin collar, whereas the number of cells with Nap1 at this location was markedly reduced (Figure 1c; Table 1). At this same stage, about half of the cells displayed Nba1 at the septin collar and, similarly, the number of cells with Nis1 at the bud neck increased sevenfold (Figure 1c; Table 1). Finally, in cells blocked in late anaphase by shift of the cdc15ts mutant to restrictive temperature (i.e., at a stage just prior to septin collar splitting), and just like Aim44 (Figure 1d; Table 1), Nis1 (Figure 1e; Table 1), as well as Nba1 and Nap1 (Table 1), are all associated with the septin collar in virtually all cells (Table 1). Thus, analysis of synchronized cells confirmed that Aim44 arrives first at the septin collar and remains there, followed by Nba1 after the completion of S phase, and then Nis1 is the last to arrive, just prior to ejection of Nap1, as summarized schematically in Figure 1f.
This order of function is further consistent with our observation that no Nis1‐mNG can be detected at the bud neck in aim44Δ, nap1Δ or nba1Δ cells (data not shown), indicating that the prior sequential deposition of Aim44, Nap1 and Nba1 is necessary for recruitment of Nis1. In fact, as presented later, in cells lacking Nba1, Nis1 accumulates in the nucleus (see Figure 5c). Moreover, in further agreement with our findings and conclusions, others have reported that Aim44 localizes to the septin collar in the absence of any of the other proteins (Nap1, Nba1 or Nis1), whereas Nba1 fails to localize at the bud neck in aim44Δ cells, but does localize to the bud neck in nap1Δ or nis1Δ cells (Meitinger et al., 2014). Only Nap1 displays its transient bud neck localization in the absence of the other components, revealing that its cell cycle‐specific recruitment to the bud neck, although necessary for Nis1 deposition there, occurs independently of the three other proteins (Meitinger et al., 2014).
Figure 5.
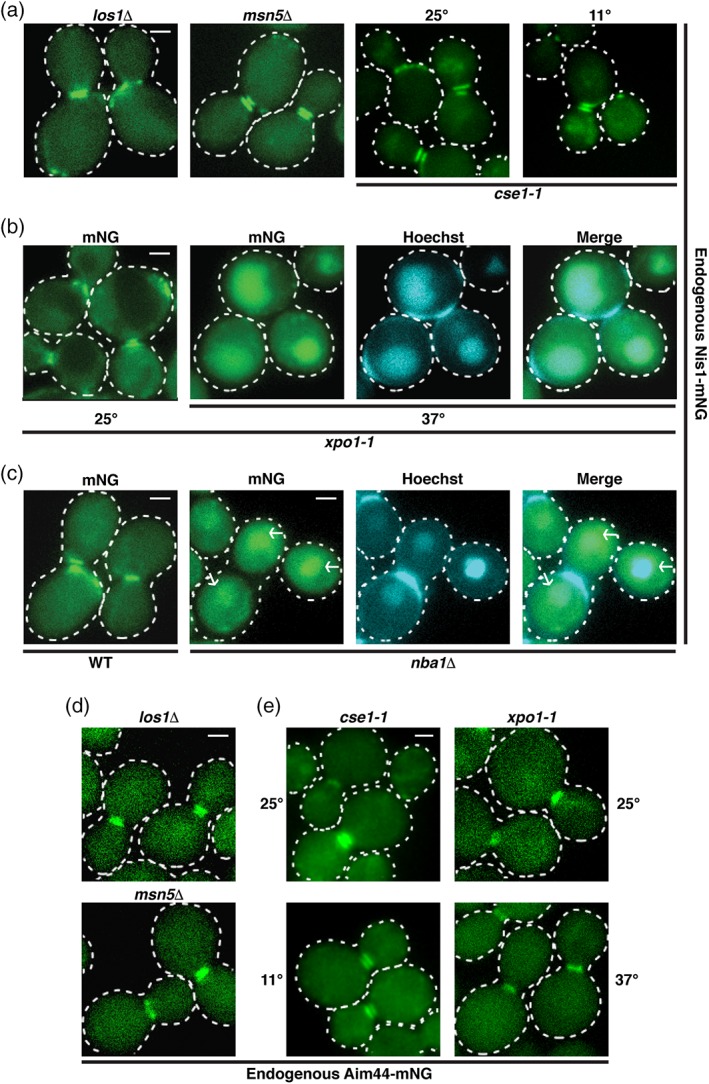
Bud neck localization of Nis1 requires its Xpo1‐dependent nuclear export and tethering to Nba1. (a) Nis1‐mNG was expressed from its native, chromosomal locus in the exportin knockout strains los1Δ (APY203) and msn5Δ (APY202; left panels), and in a strain harboring a temperature‐sensitive mutation of the exportin Cse1 (APY200; cse1‐1, right panels). The los1Δ and msn5Δ strains were grown at 30° to mid‐exponential phase before imaging. The cse1‐1 strain was grown at 25° to mid‐exponential phase and then either kept at 25° or shifted to 11° for 5 h before imaging. (b) Nis1‐mNG was expressed from its native, chromosomal locus in a strain harboring a temperature sensitive mutation of the exportin Xpo1 (xpo1‐1, APY201). The cells were grown at 25° to mid‐exponential phase and then either kept at 25° or shifted to 37° for 3 h before imaging. Cells grown at 37° were stained with Hoechst 33528 to visualize nuclei. (c) Localization of Nis1‐mNG expressed from its native, chromosomal locus was assayed in WT (left panel, APY80) or nba1Δ (right panels, APY191) genetic backgrounds. The nba1Δ cells were stained with Hoechst 33528 to visualize nuclei. Arrows point to nuclear accumulation of Nis1‐mNG. (d) Aim44‐mNG was expressed from its native, chromosomal locus in the exportin knockout strains los1Δ (APY243) and msn5Δ (APY242). Cells were grown to mid‐exponential phase and imaged. (e) Aim44‐mNG was expressed from its native, chromosomal locus in a strain harboring a temperature‐sensitive mutation in either the exportin Cse1 (cse1‐1, APY240) or the exportin Xpo1 (xpo1‐1, APY241). The cells were cultured as in panels (a) and (b), respectively. Scale bars = 1 μm
2.3. Aim44 and Nis1 localize to the nucleus when overexpressed
During our analysis of the subcellular distribution of these proteins expressed at their endogenous level, we noted that for Aim44‐mNG, Nap1‐mNG and Nis1‐mNG, but never for Nba1‐mNG, we could observe what appeared to be nuclear localization in a small fraction (≤5%) of the population (Supporting Information Figure S1A). This behavior is frequently indicative of proteins that undergo nucleocytoplasmic shuttling, but where the rate of export greatly exceeds the rate of import (Strahl, Hama, DeWald, & Thorner, 2005). Indeed, consistent with our observations, a prior study had reported that in addition to its localization at the bud neck, GFP‐Nis1 was readily detectable in the nucleus (Iwase & Toh‐e, 2001). Moreover, in a fragment‐based dissection of the Aim44 sequence, it was found that GFP‐Aim44(309–422) accumulated exclusively in the nucleus (Meitinger & Pereira, 2017). Furthermore, prior work had demonstrated that Nap1 undergoes nucleocytoplasmic shuttling because one of the roles attributed to this protein is as a chaperone for histones H2A and H2B (Miyaji‐Yamaguchi et al., 2003; Mosammaparast, Ewart, & Pemberton, 2002).
As one approach to verifying that these proteins indeed undergo nucleocytoplasmic shuttling, we reasoned that more robust nuclear accumulation would be observed when these proteins are overexpressed because, under that condition, we might be able to overwhelm their nuclear export machinery. Hence, we constructed a set of CEN plasmids in which the expression of Aim44‐mNG, Nba1‐mNG, Nap1‐mNG and Nis1‐mNG is each driven from the galactose‐inducible GAL1/10 promoter. The proteins of interest were overexpressed in a strain producing Cdc10‐mCh at its endogenous level to mark the septin collar and to permit monitoring of any possible effects on its structure. Strikingly, we found that overexpressed Aim44‐mNG localized robustly with the nuclear envelope and, albeit much more faintly, with the inner surface of the PM (most clearly in the bud) (Figure 2a, leftmost column), whereas overexpressed Nis1‐mNG localized prominently within the nucleus (Figure 2a, second column from the left). To confirm that the compartment occupied by overexpressed Aim44‐mNG and Nis1‐mNG is indeed the nucleus, these same proteins were overexpressed in a strain co‐expressing a fluorescent histone H2B marker, Htb2‐mCh. As expected, overexpressed Aim44‐mNG localized as a ring around the Htb2‐mCh signal, and overexpressed Nis1‐mNG signal localized congruent with Htb2‐mCh within the nucleus (Figure 2b). In marked contrast to Aim44 and Nis1, overexpressed Nba1‐mNG was localized diffusely throughout the cytoplasm (Figure 2a, second column from the right), but excluded from both the nucleus and the vacuole, whereas overexpressed Nap1‐mNG was also localized diffusely throughout the cytoplasm, but excluded only from the vacuole (Figure 2a, rightmost column).
Figure 2.
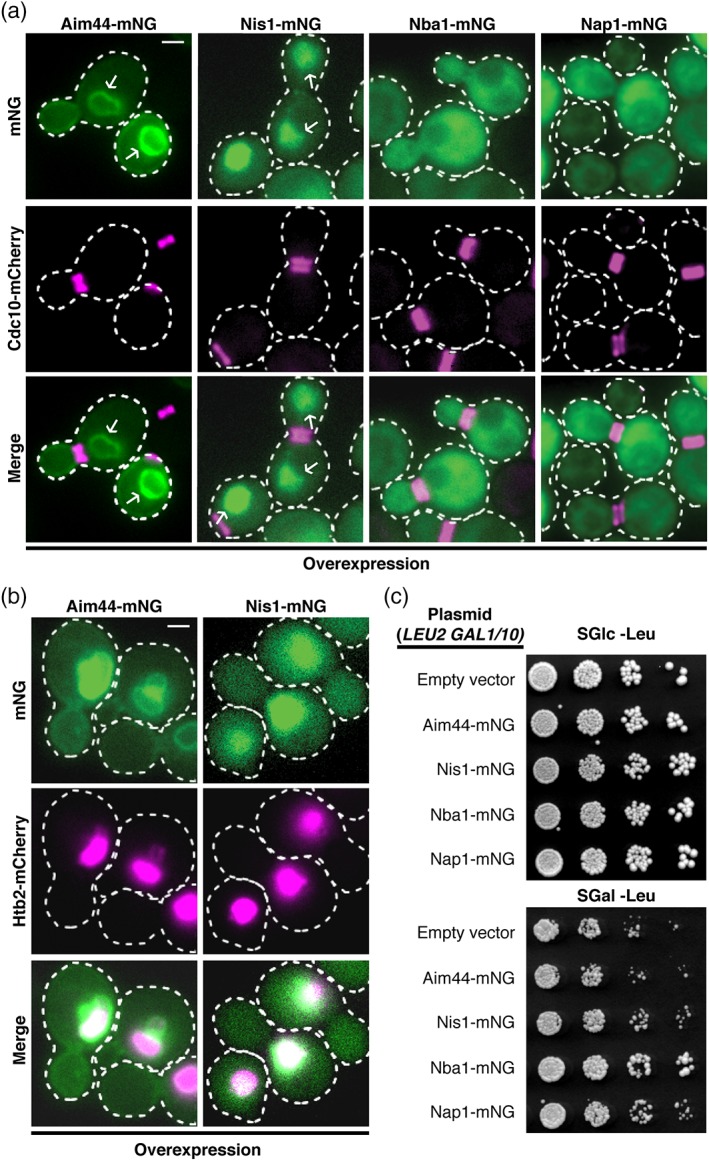
Aim44 and Nis1 accumulate in the nucleus when over‐expressed. (a) a strain expressing Cdc10‐mCh (GFY42) was transformed with a CEN plasmid expressing Aim44‐mNG (pAP101), Nis1‐mNG (pAP85), Nba1‐mNG (pAP103), or Nap1‐mNG (pAP102) under control of the GAL1/10 promoter. Cells were grown in a medium containing galactose for 3 h prior to imaging to induce expression of the mNG fusion proteins. Arrows point to sub‐cellular accumulation of corresponding mNG‐tagged proteins. (b) a strain expressing the Htb2‐mCh histone marker (JTY4202) was transformed with a CEN plasmid expressing Aim44‐mNG or Nis1‐mNG under control of the GAL1/10 promoter. Cells were grown as in (a) to induce expression of the corresponding mNG fusion protein. (c) the same cells as in (a) were grown to saturation and spotted as fivefold serial dilutions onto selective media containing either glucose or galactose. Scale bars = 1 μm
Although endogenous Aim44‐mNG localizes mainly at the septin collar (Figure 1), in agreement with prior studies (Meitinger, Richter, et al., 2013; Wolken et al., 2014), our observation that overexpressed Aim44‐mNG associated with the nuclear envelope and to a readily detectable extent with the PM, especially within the bud (Figure 2a,b), is consistent with the finding that another segment of Aim44 that contains an amphipathic helix, GFP‐Aim44(293–317), localized to the PM and represents a region that, in full‐length Aim44, is necessary for its localization to the septin collar (Meitinger & Pereira, 2017). Similarly, it has been demonstrated for the septin collar‐bound checkpoint kinase Hsl1 that the coordinate action of a septin‐binding element (residues 611–950) and a C‐terminal phosphatidylserine‐binding element (KA1 domain; residues 1,379–1,518) are necessary for optimal localization of this enzyme at the bud neck (Finnigan, Duvalyan, et al., 2016; Finnigan, Sterling, et al., 2016). These two cases illustrate what seems to be a emerging principle that, for efficient bud neck localization, certain proteins require both binding to the septins and recognition elements for PM association. In the case of overexpressed Aim44‐mNG, we presume that the excess amount of protein saturates the available binding sites at the septin collar, allowing the majority of the Aim44‐mNG to accumulate on the inner nuclear envelope and at the PM—sites for which it must clearly have affinity. In addition, the bias in the distribution of the PM‐associated pool of overexpressed Aim44‐mNG toward the bud is noteworthy because it resembles the localization of Rho1 (Yamochi et al., 1994), with which Aim44 reportedly interacts and regulates (Meitinger, Richter, et al., 2013). Thus, depending on its level of expression and the presence of factors that could influence its sequestration at different locations (and perhaps also on its state of post‐translational modification), Aim44 may be poised to respond to cues that direct it to different compartments.
2.4. Aim44, Nba1, Nap1 or Nis1 overexpression does not perturb growth or septin structure
Perhaps suprisingly, given their purported roles in regulating both Rho1 and Cdc42 function, overexpression of neither Aim44‐mNG, Nba1‐mNG, Nap1‐mNG nor Nis1‐mNG caused any discernable deleterious effect on cell morphology or the state of assembly of septins at the bud neck (Figure 2a), or on cell growth (Figure 2c). Although the majority of overexpressed Nap1‐mNG, Nba1‐mNG and Nis1‐mNG were distributed elsewhere, each of these proteins could be observed at the bud neck in a small fraction (% indicated) of the budded cells: Nap1, 1%; Nba1, 2%; and Nis1 9% (Figure S1B). Although overexpressed Aim44‐mNG was found at the nuclear envelope (and PM) in all cells examined, about 50% of the budded cells also contained Aim44 at the bud neck (Figure S1B). Together, these results indicate that the elevated levels of Aim44‐mNG, Nba1‐mNG and Nis1‐mNG do not preclude their recruitment to the bud neck, but reveal that the capacity of septin collar to associate with these components is limited. Normally, that is, when expressed at its endogenous level, Nap1‐mNG exhibited bud neck location in only a low percentage of the cells (Figure 1a; Table 1), apparently due to its only transient association with the other members of this network of proteins (Figure 1f). Therefore, we presume that when overexpressed, Nap1‐mNG association with the septin collar is likely prone to being obscured and hence the very small fraction of cells displaying Nap1‐mNG at the bud neck might be an underestimate (Figure 2a).
2.5. Nuclear localization of Aim44 and Nis1 is independent of Nap1, Nba1 and each other
We next asked whether the striking nuclear localization of overexpressed Aim44‐mNG or of overexpressed Nis1‐mNG was dependent on any of the other components. It seemed reasonable to postulate that such might be the case for three primary reasons: (a) Aim44 is required for bud neck localization of Nis1 and Nba1, and Nap1 and Nba1 are necessary for bud neck localization of Nis1 (Meitinger et al., 2014); (b) Aim44 directly interacts with Nap1, Nba1 and Nis1, as judged by co‐immunoprecitation and pull‐down assays (Meitinger et al., 2014) and, similarly, Nis1 has been found in complexes with Nap1 and Nba1, as judged by two‐hybrid interactions (Ito et al., 2001; Uetz et al., 2000; Yu et al., 2008), co‐immunoprecipitation (Iwase & Toh‐e, 2001), and affinity capture‐mass spectrometry (Calvert et al., 2008; Gavin et al., 2006; Krogan et al., 2006; Veisu, 2011); and, (c) Nap1 binds to histones H2A and H2B and promotes their nuclear import by presenting their nuclear localization sequence (NLS) to the importin Kap114 (Miyaji‐Yamaguchi et al., 2003; Mosammaparast, Ewart, & Pemberton, 2002).
Hence, to test whether nuclear localization of Aim44‐mNG required any of the other components, it was overexpressed in otherwise isogenic nap1Δ, nba1Δ or nis1Δ strains (Figure 3a) and, likewise, Nis1‐mNG was overexpressed in aim44Δ, nap1Δ, or nba1Δ strains (Figure 3b). Despite their reported physical associations, the nuclear localization of both Aim44‐mNG (Figure 3a,c) and Nis1‐mNG (Figure 3b,d) was unaffected in the absence of the other three interacting proteins. Thus, trafficking of Aim44 and Nis1 to the nucleus does not require any of the other members of these normally bud neck‐localized complexes. Interestingly, however, in cells lacking either Nap1 or Nis1, association of Aim44‐mNG with the PM was abrogated, even though its nuclear envelope association was not (Figure 3a), suggesting that binding of these proteins to Aim44 is needed to expose its PM‐targeting element (neither Nap1 nor Nba1 itself shows any propensity to associate with the PM in the absence of Aim44 [Meitinger et al., 2014]). Alternatively, phosphorylation of Aim44 by the bud neck‐localized protein kinase Gin4 might be involved in unmasking its PM‐binding element, given that both Nap1 and Nis1 interact with Gin4, that Nap1 reportedly activates Gin4 in a cell‐cycle dependent manner, and that efficient bud neck localization of Aim44 requires Gin4 catalytic activity (Altman & Kellogg, 1997; Calvert et al., 2008; Iwase & Toh‐e, 2001; Meitinger & Pereira, 2017). On the other hand, Nap1‐ and/or Nis1‐dependent Gin4 activity could promote Aim44 recruitment to the PM by affecting the local lipid composition because Gin4 controls (via its regulation of the flippase‐activating protein kinases Fpk1 and Fpk2) the PM‐localized aminoglycerophospholipid flippases Dnf1 and Dnf2 (Roelants et al., 2015).
Figure 3.
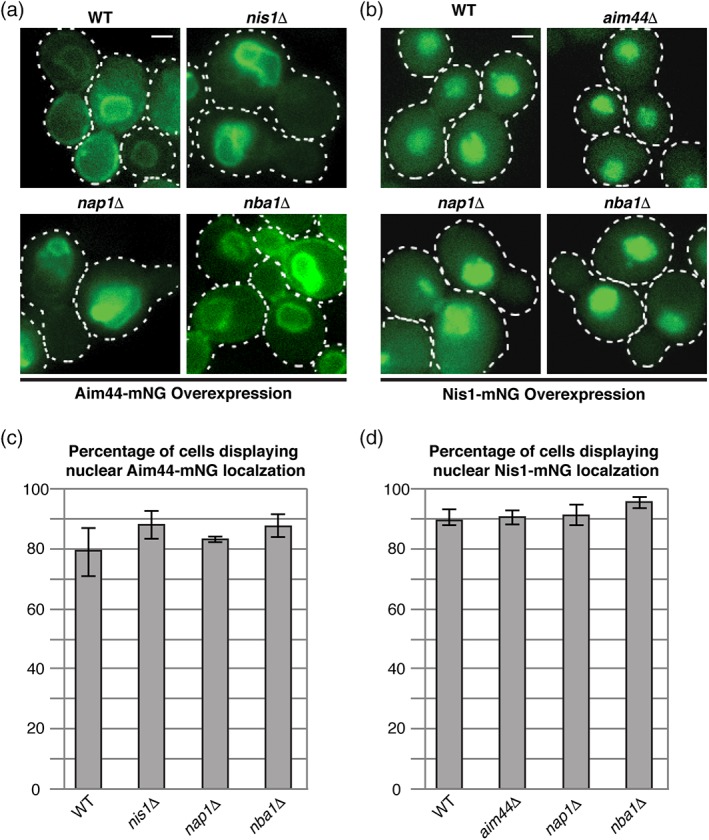
Nuclear accumulation of over‐expressed Nis1 and Aim44 occurs independently of each other and does not require either Nap1 or Nba1. (a) a CEN plasmid expressing Aim44‐mNG under control of the GAL1/10 promoter was transformed into BY4742 (WT) and otherwise isogenic nis1Δ, nap1Δ or nba1Δ strains. To induce expression of Aim44‐mNG, cells were grown in a selective medium containing galactose for 3 h prior to imaging. (b) a CEN plasmid driving expression of Nis1‐mNG under control of the GAL1/10 promoter was transformed into BY4742 (WT) and otherwise isogenic aim44Δ, nap1Δ or nba1Δ strains. Strains were grown as in (a) to induce expression of Nis1‐mNG. (c and d) cells from (a) and (B), respectively, were scored for nuclear localization of Aim44‐mNG (C) or Nis1‐mNG (D). The percentage of the population displaying fluorescent nuclear signal is plotted is the average of three independent experiments. Error bars represent the standard error of the mean. Scale bars = 1 μm
2.6. Distinct karyopherins mediate nuclear import of Aim44 and Nis1
Given our finding that nuclear localization of overexpressed Aim44‐mNG and overexpressed Nis1‐mNG did not require any of the other known interacting components, we next examined whether their nuclear entry required normal karyopherin‐dependent import or occurred via some novel mechanism. To this end, we overexpressed either Aim44‐mNG or Nis1‐mNG in deletion strains lacking each of nine different non‐essential importins: Kap104; Kap108/Sxm1; Kap111/Mtr10; Kap114; Kap119/Nmd5; Kap120/Lph2; Kap121/Pse1; Kap122/Pdr6; or, Kap123/Yrb4 (Soniat & Chook, 2015; Ström & Weis, 2001). To assess the role of the essential import factors, importin‐α (Kap60/Srp1) (Loeb et al., 1995) and importin‐β (Kap95/Rsl1) (Iovine & Wente, 1997), we used strains carrying temperature‐sensitive alleles (srp1‐3ts and kap95ts , respectively) and shifted the cultures to the non‐permissive temperature.
We found, first, that the absence of Kap104, Kap108/Sxm1, Kap114, Kap120/Lph2 or Kap122/Pdr6, or the loss of function of Kap60/importin‐α or Kap95/importin‐β, did not detectably reduce the nuclear entry of either overexpressed Aim44‐mNG or overexpressed Nis1‐mNG (Supporting Information Figure S2A, B and C). In contrast, for overexpressed Aim44‐mNG, absence of Kap111/Mtr10, Kap119/Nmd5 and Kap121/Pse1 all reduced the number of cells displaying a prominent nuclear signal (and, to some degree, the intensity of that signal), possibly due merely to indirect effects on the ability of the cells to maintain the Aim44‐mNG‐expressing CEN plasmid (Figure 4a). Most strikingly, in cells lacking Kap123/Yrb4, nuclear localization of Aim44‐mNG was almost completely abrogated (Figure 4a,c), even though AIm44‐mNG was expressed even more robustly in kap123Δ cells than in the wild‐type control or the other karyopherin mutants (Supporting Information Figure S3). We conclude that Kap123/Yrb4 (1,113 residues) is the primary importin for Aim44. In this regard, of the other karyopherins, Kap121/Pse1 (1,089 residues) is the most closely related to Kap123/Yrb4 (21% identity and 43% similarity over 457 residues); and, in those pse1Δ cells that clearly expressed Aim44‐mNG, we noted a distinct elevation of the cytosolic fluorescence (Figure 4a), suggesting that Kap121/Pse1 may make a minor contribution to Aim44 nuclear entry. Although overexpressed Aim44‐mNG was excluded from the nucleus in cells lacking Kap123/Yrb4, it remained diffuse throughout the cytoplasm and detectably decorated the PM (Figure 4a), as observed in wild‐type cells and the other karyopherin mutants.
Figure 4.

Assessment of the role of various karyopherins on nuclear import of over‐expressed Aim44 and Nis1. (a) a CEN plasmid expressing Aim44‐mNG under control of the GAL1/10 promoter was transformed into WT cells and otherwise isogenic importin knockout strains pse1Δ, mtr10Δ, nmd5Δ or kap123Δ. Cells were grown in a medium containing galactose to induce expression of Aim44‐mNG for 3 h prior to imaging. (b) a CEN plasmid expressing Nis1‐mNG under control of the GAL1/10 promoter was transformed into WT cells and otherwise isogenic importin knockout strains pse1Δ, mtr10Δ, nmd5Δ or kap123Δ. Cells were grown as in (a) to induce expression of Nis1‐mNG. (c) Quantitation of the experiment depicted in panel (a). Gray bars, percentage of cells displaying Aim44‐mNG nuclear localization for each strain; white bars, ratio of nuclear‐to‐cytoplasmic pixel intensity for those cells that displayed a fluorescent signal at the nucleus. Values represent the average from three independent experiments, in each of which ≥50 cells were examined; error bars represent standard error of the mean. Double asterisks (**), p < .05; triple asterisks (***), p < .001, as determined by a two‐tailed Student's t‐test. (d) Quantitation of the experiment depicted in panel (b), performed as described in panel (c). scale bars = 1 μm
For overexpressed Nis1‐mNG, the picture was less clear; absence of no single importin eliminated its nuclear entry (Supporting Information Figure S2A, B and C and Figure 4b). The greatest reduction in the nuclear intensity with a concomitant elevation in cytosolic intensity was observed in cells lacking Kap111/Mtr10 (Figure 4b); however, a similar pattern was also seen in cells lacking Kap121/Pse1 and, perhaps, Kap123/Yrb4, suggesting that multiple karyopherins are capable of mediating nuclear import of Nis1. Consistent with this viewpoint, Nis1(K179A R181A R183A)‐mNG, in which three residues were mutated in the sole region in Nis1 that resembles classical and bipartite NLS motifs (172‐RREDRQAKVRSRFRSKK‐188) (Lange et al., 2007; Soniat & Chook, 2015), exhibited no diminution in its nuclear accumulation (Supporting Information Figure S2D). To ascertain whether, in addition to Mtr10/Kap111, either Pse1/Kap121 or Yrb4/Kap123 contributes to the nuclear import of overexpressed Nis1, we attempted to construct (via genetic crosses of the corresponding haploids and tetrad dissection), double mutants in which the mtr10Δ/kap111Δ allele was combined with either pse1Δ/kap121Δ or yrb4Δ/kap123Δ mutations, and as a control with a nmd5Δ/kap119Δ because loss of Nmd5/Kap119 alone did not seem to affect nuclear entry of overexpressed Nis1‐mNG (Figure 4b). Unfortunately, the pse1Δ/kap121Δ mutation causes cells to grow extremely slowly and tetrad analysis indicated it was sufficient, by itself, to confer a tiny microcolony phenotype and, therefore, we could not isolate any viable mtr10Δ/kap111Δ pse1Δ/kap121Δ double mutants (or, for the same reason, any viable pse1Δ/kap121Δ nmd5Δ/kap119Δ double mutants; Supporting Information Figure S4A and B). Similarly, tetrad analysis indicated that nmd5Δ/kap119Δ yrb4Δ/kap123Δ double mutants are inviable because every such predicted spore in tetratype asci were unable to grow (Supporting Information Figure S4C). Nonetheless, by the same strategy, we were able to obtain both mtr10Δ/kap111Δ yrb4Δ/kap123Δ and mtr10Δ/kap111Δ nmd5Δ/kap119Δ double mutants. We observed, however, no enhancement in the defect in nuclear accumulation in either of these double mutants compared to that observed in mtr10Δ/kap111Δ cells alone (Supporting Information Figure S5). We conclude that although Kap111/Mtr10 mediates nuclear entry of Nis1, it is possible that Pse1/Kap121 may also contribute. In this regard, histones set a precedent for proteins whose nuclear import is mediated by multiple importins. In yeast, both Pse1/Kap121 and Yrb4/ Kap123 are responsible for nuclear entry of histones H3 and H4 (Keck & Pemberton, 2013); and, in humans, seven different importins are able to associate with H3 and H4, albeit with differential affinities (Soniat, Cağatay, & Chook, 2016). Given the well‐documented role for Nap1 in promoting Kap114‐dependent import of histones H2A and H2B (Mosammaparast et al., 2001; Mosammaparast, Guo, Shabanowitz, Hunt, & Pemberton, 2002), it seemed reasonable that the function of Nap1 in assembly of the Aim44‐Nba1‐Nis1 complex might be to help shuttle Aim44 and Nis1 into the nucleus. However, in nap1Δ cells, nuclear accumulation of overexpressed Aim44‐mNG and overexpressed Nis1‐mNG was unaffected (Figure 3a,b).
2.7. Nuclear export of Nis1 requires exportin Xpo1 and its bud neck retention requires sequestration by association with Aim44‐bound Nba1
We reasoned that if Aim44 and Nis1 normally undergo nucleocytoplasmic shuttling, then each should accumulate in the nucleus, even when expressed at its endogenous level, in cells in which the karyopherin responsible for its nuclear export is absent. To test this hypothesis, we expressed either Aim44‐mNG or Nis1‐mNG endogenously in deletion strains lacking each of the two non‐essential exportins: Kap142/Msn5 and Los1 (Ström & Weis, 2001). To assess the role of the two essential export factors, Kap124/Xpo1 (Stade, Ford, Guthrie, & Weis, 1997) and Kap109/Cse1 (Xiao, McGrew, Schroeder, & Fitzgerald‐Hayes, 1993), we used strains carrying a heat‐sensitive allele (xpo1‐1ts) and a cold‐sensitive allele (cse1‐1cs ), respectively, and shifted the cultures to the non‐permissive temperature.
For Nis1‐mNG, we found that unmistakable nuclear accumulation occurred only in the xpo1‐1ts mutant cells and only at the restrictive temperature (Figure 5a,b). Thus, Nis1 displays all of the expected hallmarks of a protein that undergoes nucleocytoplasmic shuttling and its nuclear export is mediated by Kap124/Xpo1. Remarkably, we also found that in cells lacking Nba1, Nis1‐mNG expressed at its endogenous level also accumulated prominently in the nucleus (Figure 5c). This observation indicates that once exported to the cytosol, for Nis1 to be retained at the bud neck, it must be captured at that location by binding to the Nba1 that is tethered there via its association with Aim44, in agreement with our analysis of the spatial and temporal relationships among these proteins (Figure 1f).
For Aim44‐mNG, we observed what appeared to be a hint of nuclear accumulation in cse1‐1cs mutant cells (Figure 5d,e). To ascertain whether multiple exportins contribute to the nuclear exit of Aim44‐mNG, we constructed, first, by genetic crosses and tetrad dissection, cse1cs los1Δ and cse1cs msn5Δ double mutants; however, contrary our initial observations, we did not find any detectable nuclear accumulation of endogenously expressed Aim44‐mNG when the cse1cs allele was combined with either los1Δ or msn5Δ (Supporting Information Figure S6A). In contrast, in both xpo1ts los1Δ and xpo1ts msn5Δ double mutants, there was some very modest, but detectable, nuclear accumulation of endogenously expressed Aim44‐mNG (Supporting Information Figure S6B), whereas this was not observed for a los1Δ msn5Δ double mutant (Supporting Information Figure S6C). Thus, Xpo1 seems to make some contribution to the nuclear export of Aim44‐mNG. For a cse1cs xpo1ts double mutant, there was no condition under which the function of each of these mutant exportins could be simultaneously blocked. Hence, we were unable to test whether both Xpo1 and Cse1 are able to mediate efficient Aim44 exit from the nucleus.
2.8. Effects of Aim44 or Nis1 overexpression on localization of Nba1, Nis1 and Nap1
As described above, overexpression of Aim44‐mNG and Nis1‐mNG led to a drastic change in their subcellular localization (Figure 2). If the association between Aim44 or Nis1 with each other or the remaining members (Nap1 or Nba1) of the ensemble is of sufficient affinity and stability, then it might be expected that such an interacting protein would also show concomitant re‐localization with Aim44 or Nis1. To explore this possibility, cells expressing endogenously either Nap1‐mCh, Nba1‐mCh or Nis1‐mCh were constructed, and these displayed (Figure 6a) the same pattern as observed for the mNG‐tagged derivatives (Figure 1) (although background fluorescence was also emitted by the vacuole which contains intrinsic compounds that absorb and emit fluorescence under the regime used to visualize mCh). In these experiments, localization at the bud neck of budded cells (rather than co‐localization with septins per se) was scored because no fluorescently tagged septin was expressed in the cells.
Figure 6.
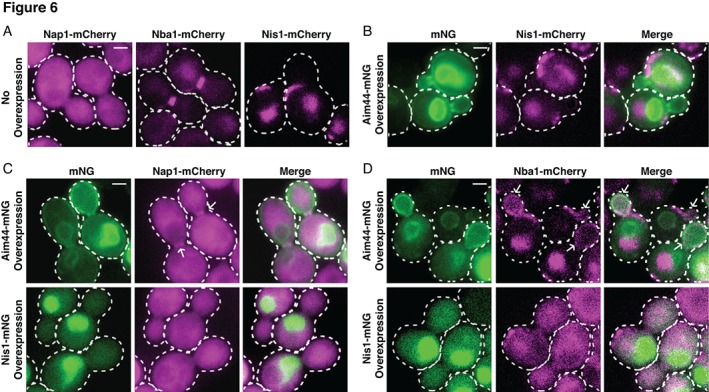
Over‐expression of either Aim44 or Nis1 displaces Nba1 from the bud‐neck. (a) Strains expressing Nap1‐mCh (APY211), Nba1‐mCh (APY212) or Nis1‐mCh (APY208) from their endogenous, chromosomal loci were transformed with empty vector (pRS315) and grown in a selective medium containing galactose for 3 h prior to imaging. (b) the Nis1‐mCh strain from (a) was transformed with a CEN plasmid expressing Aim44‐mNG under control of the GAL1/10 promoter and grown as in (a) to induce expression of Aim44‐mNG. (c) The Nap1‐mCh strain from (a) was transformed with a CEN plasmid expressing Aim44‐mNG or Nis1‐mNG under control of the GAL1/10 promoter and grown as in (a) to induce expression of the corresponding mNG fusion protein. Arrows point to bud neck localization of Nap1‐mCh. (d) The Nba1‐mCh strain from (a) was transformed with a CEN plasmid expressing Aim44‐mNG or Nis1‐mNG under control of the GAL1/10 promoter and grown as in (a) to induce expression of the corresponding mNG fusion protein. Arrows point to Nba1‐mCh localization at the plasma membrane of the bud. The cytosolic patches of fluorescence in the Nba1‐mCh and Nis1‐mCh images are the result of vacuolar background. Scale bars = 1 μm. Correction added on 29 April 2019 after first online publication: the image in Panel D was revised. No changes were made to the caption
In cells marked with Nba1‐mCh and overexpressing Aim44‐mNG, there was a significant loss of Nba1‐mCh from the bud neck (Figure 6d; Table 2); and, more strikingly, a substantial fraction of the Nba1‐mCh was recruited to the PM‐associated fraction of Aim44‐mNG, most notably in the bud, whereas no detectable Nba1‐mCh was associated with the majority of the Aim44‐mNG localized to the nucleus (Figure 6d). This finding suggests that Nba1 binds directly to Aim44, but that only the cytosolic pool of Aim44 is competent to bind Nba1. Aim44‐mNG overexpression also displaced Nis1‐mCh from the bud neck (Figure 6b; Table 2); but, unlike Nba1‐mCh, no Nis1‐mCh was recruited to the PM. Thus, the PM‐bound Aim44‐Nba1 complexes are not competent to bind Nis1. This observation further suggests that the Aim44‐ and Nba1‐dependent recruitment of Nis1 to the split septin rings late in the cell cycle must require some other factor present only at the bud neck and only at that stage in the cell division cycle. We suspect, but have not yet explored experimentally, that this additional factor may be the requirement for modification of the septins by SUMOylation because Cdc3, Cdc11 and Shs1 become SUMOylated at the onset of cytokinesis (Johnson & Blobel, 1999) and Nis1 is a protein that both sequence analysis (Hannich et al., 2005; Veisu, 2011) and direct binding experiments (Uzunova et al., 2007; Veisu, 2011) indicate contains at least one SUMO interaction motif. In cells marked with Nap1‐mCh and overexpressing Aim44‐mNG, in virtually all the cells that exhibited detectable Aim44‐mNG at the bud neck Nap1‐mCh was also present (Figure 6c; Table 2), whereas no detectable Nap1‐mCh was associated with either the fraction of Aim44‐mNG associated with the PM or the bulk of the Aim44‐mNG localized to the nucleus. Thus, the increase in Nap1 at the bud neck upon Aim44 overexpression appears to be indirect, that is, requires some additional factor present only at the septin collar.
Table 2.
Effect of over‐expression on fraction of cells exhibiting bud neck localization for Nba1, Nis1 and Nap1
| Aim44‐mNG | Nis1‐mNG | |||
|---|---|---|---|---|
| Overexpressed | Overexpressed | |||
| No | Yes | No | Yes | |
| Aim44‐mCh | ND | ND | ND | ND |
| Nba1‐mCh | 45.8% (120)a | 20% (110) | 45.8% (120) | 14.1% (92) |
| Nis1‐mCh | 33.3% (33) | 11.8% (59) | ND | ND |
| Nap1‐mCh | 2.4% (83) | 67.5% (117) | 2.4% (83) | 3.6% (83) |
Number in parentheses represents the total number of cells scored in multiple fields in three independent experiments for each condition.
Despite reports that Nis1 and Nap1 are able to physically associate, as judged by the two‐hybrid method (Ito et al., 2001; Uetz et al., 2000; Yu et al., 2008), by co‐immunoprecipitation (Iwase & Toh‐e, 2001), and by co‐purification (Calvert et al., 2008; Gavin et al., 2006; Krogan et al., 2006; Veisu, 2011), we found, first, that no detectable Nap1‐mCh co‐localized with overexpressed Nis1‐mNG, which was located almost exclusively in the nucleus (Figure 6c). Also, unlike Aim44‐mNG overexpression, overexpression of Nis1‐mNG did not affect the amount of Nap1‐mCh at the bud neck (Table 2). In marked contrast, in cells marked with Nba1‐mCh and overexpressing Nis1‐mNG, there was a substantial loss of Nba1‐mCh from the bud neck (Table 2), and a marked re‐localization of Nba1‐mCh to the cytosol, concomitant with a noticeable increase in the amount of Nis1‐mNG in the cytosol (Figure 6d). These findings suggest that Nis1 and Nba1 can physically interact directly; thus, their co‐localization to the split septin rings observed in normal cells may not be due solely to their independent tethering to Aim44. Why the nuclear pool of Nis1‐mNG appears to be incompetent to recruit Nba1‐mCh is unclear; perhaps the Nis1‐mNG that remains cytosolic does so simply because the binding of cytosolic Nba1‐mCh masks the NLS(s) in Nis1‐mNG.
3. DISCUSSION
The studies described here have shed considerable new light on the spatial and temporal relationships between Aim44, Nap1, Nba1 and Nis1, a set of proteins thought to be involved in maintaining Rho1‐dependent cell polarity and impeding ectopic Cdc42‐dependent bud formation after initial bud emergence each cell cycle (Meitinger et al., 2014). Most strikingly, our study revealed that two components of this system normally undergo nucleocytoplasmic shuttling. However, what regulates the nuclear trafficking of Aim44 and Nis1 and what the mechanistic and physiological roles of this sub‐cellular compartmentation are remain to be elucidated. Both Aim44 and Nis1 are phosphoproteins (Albuquerque et al., 2008; Holt et al., 2009; Swaney et al., 2013). Hence, their subcellular distribution might be under the control of bud neck‐associated or other protein kinases (Perez et al., 2016). In this regard, transit through the nucleus might be necessary for dephosphorylation of such regulatory sites, given that at least three of the S. cerevisiae phosphoprotein phosphatase 2C isoforms (Ptc1, Ptc2 and Ptc3) localize exclusively or predominantly to the nucleus (Ariño, Casamayor, & González, 2011), as does a significant fraction of both phosphoprotein phosphatase 1 (Glc7) (Gilbert & Guthrie, 2004) and the Cdc55‐bound isoform of phosphoprotein phosphatase 2A (Rossio et al., 2013).
Our data reinforce and extend the conclusion that Aim44 is a scaffold protein that early in the cell cycle, initiates and is required, directly or indirectly, for the eventual recruitment of the other three proteins to the septin collar, whereas Nis1 is the last component to join the ensemble just at the onset of cytokinesis. Thus, one intriguing possibility is that control of the nuclear sequestration and release of Aim44 and Nis1 are checkpoint‐like events that help fine‐tune properly organized and appropriately timed progression through the cell division cycle. Dynamic shuttling of Aim44 and Nis1 from the nucleus to the cytosol and then to the septin collar at the bud neck would be an effective way to communicate between different subcellular sites information about the status of transit through various cell cycle stages (such as, completion of DNA replication, assembly of bud neck structures necessary for cytokinesis, and readiness of the spindle to execute chromosome segregation).
There is precedent for roles of other septin‐associated proteins in such coordination, for example, in ensuring that licensing of cell division only occurs after successful nuclear division and proper septin collar assembly (for review, see [Perez et al., 2016]). A similar function for nucleocytoplasmic shuttling in controlling how the septins get SUMOylated only during anaphase and then abruptly deSUMOylated at cytokinesis has been described (Makhnevych, Ptak, Lusk, Aitchison, & Wozniak, 2007). During interphase, the major SUMO ligase Siz1 is imported into the nucleus in a Kap95‐dependent manner, but then gains access to its septin substrates in M phase because Siz1 is then ejected from the nucleus by the exportin Kap142/Msn5, presumably in response to modifications installed at that stage in the cell cycle by the cyclin B‐bound form of Cdk1 (Clb‐bound Cdc28) (Kõivomägi et al., 2011). Likewise, the SUMO isopeptidase Ulp1 is imported into the nucleus in a Kap121‐ and Kap95‐Kap60‐dependent manner and sequestered at the nuclear pore complex (NPC), but then abruptly released from the NPC at the end of mitosis (Makhnevych et al., 2007), presumably in response to changes in modification state imposed either by one or more of the protein kinases (Cdc15, Dbf2, Dbf20) or by the phosphatase (Cdc14) of the mitotic exit network (MEN) (Baro, Queralt, & Monje‐Casas, 2017), and thus Ulp1 is now free to interact with its substrates, the SUMOylated septins at the bud neck.
In this same regard, we discovered that exit of Nis1 from the nucleus is mediated by the exportin Kap124/Xpo1. Likewise, it has been demonstrated by others that Nap1 is also exported from the nucleus in an Xpo1‐dependent manner (Mosammaparast, Ewart, & Pemberton, 2002). Thus, Xpo1 function is presumably key in providing a sufficient cytoplasmic pool of either Nap1 or Nis1 (or both) to permit efficient assembly of the Aim44‐Nap1‐Nba1‐Nis1 complex at the bud neck. Given that binding of any given cargo molecule to Xpo1 will exclude the binding of another cargo molecule, another possibility is that the level of Nap1 regulates the rate of Nis1 export, or vice‐versa. Xpo1‐mediated export of Nap1 has a role in promoting mitotic progression (Altman & Kellogg, 1997; Kellogg & Murray, 1995; Miyaji‐Yamaguchi et al., 2003). However, in cells overexpressing Nis1‐mNG, the absence of Nap1 did not result in any detectable increase in the amount of Nis1‐mNG in the cytosol (Figure 3b). Hence, such a competition model remains purely speculative.
As described in this study, only in a small fraction (≤5%) of a population of cells in balanced growth did we detect endogenously expressed Nis1‐mNG in the nucleus. Thus, when Nis1 is expressed at its normal level, its rate of Xpo1‐mediated nuclear export exceeds its rate of nuclear entry. Therefore, one mechanism to exclude localization of Nis1 at the bud neck would be to reverse the balance of these rates. Notably, when endogenously expressed Nis1‐mNG was confined to the nucleus in xpo1‐1ts cells shifted to the restrictive temperature, we also did not observe any retention of Nis1‐mNG either at the bud neck or at bud‐scars (Figure 5b). This result is consistent with what is currently known about the cell cycle dynamics of Nis1 localization at the cell cortex—Nba1 and then Nis1 are first targeted to the bud neck by Aim44, as we have documented here (Figure 1), and then, on completion of cytokinesis and septum formation, both are recruited to the newly formed bud‐scar via interaction with Rax1 and Rax2 (Meitinger et al., 2014). Thus, when confined to the nucleus, no Nis1 is available to be deposited at either location.
Unexpectedly, but revealing, we found that the amount of endogenously expressed Nis1‐mNG located in the nucleus markedly increased in cells lacking Nba1 (Figure 5c). Thus, efficient Xpo1‐mediated export of Nis1 alone is not sufficient for its deposition at the bud neck, but Nis1 must also be retained there by its tethering to Nba1, which is itself anchored at the bud neck via its binding to Aim44. Indeed, in the nba1Δ cells, we also noted that the cytosolic pool of Nis1‐mNG was highly diffuse, consistent with the lack of a factor critical for localization of Nis1 at the bud neck, in agreement with prior results (Meitinger et al., 2014).
In conclusion, taken together, our data demonstrate that the nuclear‐to‐cytoplasmic ratio of Aim44 and Nis1, and perhaps Nap1, are critical for controlling the availability of these proteins for their subsequent localization to the bud neck. Only Nba1 appears not to undergo nucleocytoplasmic shuttling. Further mapping of the interaction surfaces that mediate the associations among these proteins and determination of which of these components makes direct physical interactions with the septins will be fruitful questions to address to further understand the assembly, dynamics and function of this ensemble of proteins.
4. MATERIALS AND METHODS
4.1. Yeast strains and plasmid construction
All yeast strains used in this study are listed in Table 3. Standard PCR‐based techniques were used to generate all yeast strains (Janke et al., 2004). We have documented previously that Cdc10‐mCh is fully functional, as judged by complementation of the inviability of cdc10Δ cells (Finnigan, Booth, et al., 2015; Finnigan, Takagi, et al., 2015). In contrast, aim44Δ, nba1Δ, nap1Δ and nis1Δ mutants are all viable; however, they do have other phenotypes. In this regard, cells expressing Aim44‐mNG do not exhibit the abnormal septa found in aim44Δ mutant cells. Likewise, cells expressing Nap1‐mNG do not exhibit the abnormal bud morphology found in nap1Δ mutant cells. Similarly, cells expressing Nba1‐mNG or Nis1‐mNG do not exhibit the abnormal vacuole morphology displayed by nba1Δ and nis1Δ cells. Furthermore, as judged by immunoblot analysis (data not shown), the mNG‐tagged versions of these proteins are stably expressed as intact fusions. Hence, by these criteria, the mNG‐tagged versions of all four proteins appear to be biologically functional.
Table 3.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| BY4741 | MAT a leu2Δ ura3Δ met15Δ his3Δ | Brachmann et al., 1998 |
| BY4742 | MATα leu2Δ ura3Δ lys2Δ his3Δ | Brachmann et al., 1998 |
| APY55 | BY4741; nis1Δ::Kan R | This study |
| APY80 | BY4741; nis1Δ::NIS1::mNG::ADH1(t)::Hyg R | This study |
| APY104 | BY4741; cdc10Δ::CDC10::mCherry::ADH1(t)::SpHIS5 nis1Δ::NIS1::mNG::ADH1(t)::Hyg R |
This study |
| APY112 | BY4741; cdc10Δ::CDC10::mCherry::ADH1(t)::SpHIS5 nis1Δ::NIS1::mNG::ADH1(t)::Hyg R cdc15‐2 |
This study |
| APY185 | BY4741; cdc10Δ::CDC10::mCherry::ADH1(t)::SpHIS5 aim44Δ::AIM44::mNG::ADH1(t)::Hyg R |
This study |
| APY191 | BY4742; nis1Δ::NIS1::mNG::ADH1(t)::Hyg
R
nba1Δ |
This study |
| APY200 | KWY125; nis1Δ::NIS1::mNG::ADH1(t)::Hyg
R
cse1‐1 |
This study |
| APY201 | KWY121; nis1Δ::NIS1::mNG::ADH1(t)::Hyg R | This study |
| APY202 | BY4742; nis1Δ::NIS1::mNG::ADH1(t)::Hyg
R
msn5Δ |
This study |
| APY203 | BY4742; nis1Δ::NIS1::mNG::ADH1(t)::Hyg
R
los1Δ |
This study |
| APY205 | BY4741; aim44Δ::AIM44::mNG::ADH1(t)::Hyg
R
cdc15‐2 |
This study |
| APY208 | BY4741; nis1Δ::NIS1::mCherry::ADH1(t)::SpHIS5 | This study |
| APY211 | BY4741; nap1Δ::NAP1::mCherry::ADH1(t)::SpHIS5 | This study |
| APY212 | BY4741; nba1Δ::NBA1::mCherry::ADH1(t)::SpHIS5 | This study |
| APY240 | KWY125; aim44Δ::AIM44::mNG::ADH1(t)::Hyg R cse1‐1 | This study |
| APY241 | KWY121; aim44Δ::AIM44::mNG::ADH1(t)::Hyg R | This study |
| APY242 | BY4742; aim44Δ::AIM44::mNG::ADH1(t)::Hyg
R
msn5Δ |
This study |
| APY243 | BY4742; aim44Δ::AIM44::mNG::ADH1(t)::Hyg
R
los1Δ |
This study |
| APY251 | BY4741; cdc10Δ::CDC10::mCherry::ADH1(t)::SpHIS5 nap1Δ::NAP1::mNG::ADH1(t)::Hyg R |
This study |
| APY252 | BY4741; cdc10Δ::CDC10::mCherry::ADH1(t)::SpHIS5 nba1Δ::NBA1::mNG::ADH1(t)::Hyg R |
This study |
| GFY42 | BY4741; cdc10Δ::CDC10::mCherry::ADH1(t)::SpHIS5 | Finnigan, Takagi, Cho, & Thorner, 2015 |
| KWY121 |
MATα ade2‐1 LYS2 xpo1Δ::LEU2 [pKW457/pRS313‐xpo1‐1ts] |
Stade et al., 1997 |
| KWY125 | MAT a ade2‐101 his3‐11,15 trp1‐Δ901 ura3‐52 cse1‐1 | Xiao et al., 1993 |
| JTY4902 | BY4742; htb2Δ::HTB2::mCHERRY::URA3 | Jake ma, this laboratory |
| JTY3405 | MAT a his3‐200 leu2‐3,112 lys2‐801 trp1‐1 ura3‐52 kap104Δ::URA3 | Aitchison, Blobel, & Rout, 1996 |
| JTY3488 | BY4741; mtr10Δ::Kan R | Strahl et al., 2005 |
| APY276 | BY4742; nmd5Δ::HIS3 | This study |
| JTY3110 | BY4741; kap120Δ::Kan R | Brachmann et al., 1998 |
| JTY3486 | BY4741; pse1Δ::Kan R | Strahl et al., 2005 |
| APY275 | BY4741; kap123Δ::HIS3 | This study |
| JTY2805 | MAT a ade2‐1 LYS2 srp1‐31 ts | Loeb et al., 1995 |
| JTY3387 | MAT a ade2‐1 can1‐100 his3‐11,15 leu2‐3,112 trp1‐1 ura3‐1 kap95Δ::HIS3 [pSW509/pRS315‐kap95(L63A) ts] | Iovine & Wente, 1997 |
|
S. cerevisiae
Deletion collection |
BY4742; aim44Δ::Kan R | Brachmann et al., 1998 |
|
S. cerevisiae
Deletion collection |
BY4742; nap1Δ::Kan R | Brachmann et al., 1998 |
|
S. cerevisiae
Deletion collection |
BY4742; nbaΔ::Kan R | Brachmann et al., 1998 |
|
S. cerevisiae
Deletion collection |
BY4742; sxm1Δ::Kan R | Brachmann et al., 1998 |
|
S. cerevisiae
Deletion collection |
BY4742; kap114Δ::Kan R | Brachmann et al., 1998 |
|
S. cerevisiae
Deletion collection |
BY4742; kap122Δ::Kan R | Brachmann et al., 1998 |
| APY277 | BY4741; mtr10Δ::Kan R kap123Δ::HIS3 | This study |
| APY278 | BY4741; mtr10Δ::Kan R nmd5Δ::HIS3 | This study |
| APY279 | KWY125; aim44Δ::AIM44::mNG::ADH1(t)::HygR cse1‐1 los1Δ::Kan R | This study |
| APY280 | KWY125; aim44Δ::AIM44::mNG::ADH1(t)::HygR cse1‐1 msn5Δ::Kan R | This study |
| APY281 | KWY121; aim44Δ::AIM44::mNG::ADH1(t)::HygR xpo1‐1 los1Δ::Kan R | This study |
| APY282 | KWY121; aim44Δ::AIM44::mNG::ADH1(t)::HygR xpo1‐1 msn5Δ::Kan R | This study |
| APY283 | BY4741; aim44Δ::AIM44::mNG::ADH1(t)::HygR los1Δ::SpHIS5 msn5Δ::Kan R | This study |
| APY285 | BY4741; cdc10Δ::CDC10::mCherry::ADH1(t)::SpHIS5 nap1Δ::NAP1::mNG::ADH1(t)::HygR cdc15‐2 | This study |
| APY286 | BY4741; cdc10Δ::CDC10::mCherry::ADH1(t)::SpHIS5 nba1Δ::NBA1::mNG::ADH1(t)::HygR cdc15‐2 | This study |
4.2. Culture conditions
Yeast were grown in a rich medium (YPD; 1% yeast extract, 2% peptone, 2% dextrose/glucose) or in a synthetic minimal medium (S) (Sherman, Fink, & Hicks, 1986) containing the appropriate amino acids and either 2% glucose, 2% galactose, or a mixture of 2% raffinose and 0.2% sucrose. For ade2 strains, excess adenine sulfate was added to the medium at a final concentration of 40 μg/mL (YPAD; YPD + adenine). All strains were cultured at 30 °C unless stated otherwise. For overexpression of fluorescently tagged proteins from the GAL promoter on CEN plasmids (Table 4), the cells were grown overnight to saturation in appropriately supplemented S medium containing raffinose and sucrose, back‐diluted 1:20 into S medium containing galactose, and cultured for the time indicated in the figure legends. To perform the growth assay, cultures were grown to saturation overnight in S medium containing raffinose and sucrose, then adjusted to A 600 nm ~1 in water, and fivefold serial dilutions of the cell suspension were spotted onto agar plates containing S‐Leu medium with either 2% glucose or galactose. The plates were imaged after incubation at 30 °C for 3 days. For the xpo1‐1ts and cdc15‐2ts strains, cultures were grown at 25 °C in YPAD medium to A 600 nm ~0.5 at 25 °C, then split into two equal portions, and cultured for an additional 3 h at either 25 °C or 37 °C before imaging. For the cse1‐1cs strain, cultures were grown at 25 °C in YPAD medium to A 600 nm ~0.5 at 25 °C, then split into two equal portions, and cultured for an additional 5 h at either 25 °C or 11 °C before imaging.
Table 4.
Plasmids used in this study
| Plasmid | Description | Reference |
|---|---|---|
| pAP85 | pRS315; prGAL1/10::NIS1‐mNeonGreen::ADH1(t) | This study |
| pAP101 | pRS315; prGAL1/10::AIM44‐mNeonGreen::ADH1(t)::SpHIS5 | This study |
| pAP102 | pRS315; prGAL1/10::NAP1‐mNeonGreen::ADH1(t) SpHIS5 | This study |
| pAP103 | pRS315; prGAL1/10::NBA1‐mNeonGreen::ADH1(t) SpHIS5 | This study |
| pAP109 | pRS315; prGAL1/10::nis1(K179A R181A R183A)‐mNeonGreen::ADH1(t) | This study |
4.3. Fluorescent microscopy and image analysis
For all imaging experiments, cultures were grown at 30 °C to mid‐exponential phase (A600 nm ~0.5–0.7), unless otherwise indicated. Samples of the cultures were diluted into phosphate‐buffered saline (PBS), spotted onto 1.5% agarose pads, and imaged using an upright epifluorescence microscope (Olympus, model BH‐2) equipped with a 100X oil immersion objective (Olympus) and a SOLA light source (Lumencore). Images were captured using a charge‐coupled device camera (CoolSNAP MYO, Photometrics) using Micro‐Manager software (Edelstein et al., 2014). Within any given experiment, each image was collected at the same exposure time and adjusted identically using ImageJ (National Institutes of Health) and processed using Photoshop (Adobe). Cell outlines were drawn on the basis of either an overexposed fluorescence image or a corresponding brightfield image. The cell images shown are representative of the patterns reproducibly observed in multiple experiments (n ≥ 3). To quantify the percentage of cells displaying fluorescent localization to the nucleus, cells containing fluorescent signal were scored for the visual accumulation of signal at the nucleus or nuclear envelope. For each strain, 50–150 cells were counted in three independent experiments. To quantify the ratio of nuclear to cytoplasmic fluorescent signal, only cells that displayed fluorescent nuclear localization were selected for analysis. Using ImageJ, the images were background subtracted using a rolling ball radius of 50 pixels. Next, the box tool was used to measure the mean pixel intensity in the nucleus or at the nuclear envelope. This number was divided by the mean cytoplasmic intensity in the same cell and measured using the same box size. An average of these values was taken for 50–100 cells of each genotype.
4.4. Identification of the nucleus
To visualize the nucleus, cells expressing histone H2B tagged with mCherry (Htb2‐mCh) and expressing another protein of interest tagged with mNeon Green were examined. Alternatively, samples of cells (1 mL) that were cultured in S medium to mid‐exponential phase were stained by addition of Hoechst 33258 (Invitrogen) at a final concentration of 1 μg/mL, followed by incubation at room temperature in the dark for 5 min. The resulting stained cells were washed once with growth medium anf then imaged.
4.4.1. Preparation of cell extracts and immunoblotting
Samples (2 mL) of exponentially‐growing cultures (A 600 nm = 0.7) of cells overexpressing from a GAL promoter on a CEN plasmid either Aim44‐mNG or Nis1‐mNG were harvested by brief centrifugation (1 min) in a microfuge at 6,000 rpm, immediately frozen in liquid N2, and then ruptured by resuspension in 150 μL of 1.85 M NaOH, 7.4% β‐mercaptoethanol. The resulting whole‐cell lysate was then mixed with 150 μL of 50% trichloroacetic acid (TCA), incubated on ice for 10 min, and the resulting precipitate collected by centrifugation (2 min) in a microfuge at maximum rpm. The resulting pellet was washed twice with acetone to remove excess TCA, and the protein solubilized by resuspension in 80 μL of 0.1 M Tris‐5% SDS, followed by 20 μL of 5X concentrated SDS‐PAGE sample buffer. After boiling for 5 min, aliquots (20 μL) of each sample were resolved by SDS‐PAGE in 8% gels, transferred electrophoretically to a nitrocellulose membrane, and the membranes then incubated in Odyssey™ blocking buffer (Li‐Cor Biosciences, Lincoln, NE) containing a 1:1,000 dilution of mouse monoclonal anti‐mNeonGreen antibody 32F6 (ChromoTek, Inc., Hauppauge, NY) and, as a loading control, a 1:20,000 dilution of rabbit polyclonal anti‐Pgk1 antibodies (Baum et al., 1978, 1:20,000). The membranes were then washed in Odyssey™ blocking buffer containing 0.1 Tween‐20, incubated with appropriate infrared fluorophore‐labeled secondary antibodies (CF770 dye‐conjugated goat anti‐mouse IgG and CF680‐dye‐conjugated goat anti‐rabbit IgG; Biotium, Inc., Fremont, CA) and the resulting immune complexes visualized using an Odyssey CLx™ infrared imager (Li‐Cor).
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
Supporting information
Fig. S1 Endogenously‐expressed Aim44‐mNG, Nap1‐mNG and Nis1‐mNG (but not Nba1‐ mNG) can be detected in the nucleus in a small fraction of the cells, whereas overexpression does not prevent localization of the same proteins to the bud neck. (A) Strains expressing Aim44‐mNG, Nap1‐mNG, Nis1‐mNG or Nba1‐mNG from their native promoters were grown to mid‐exponential phase and examined by fluorescence microscopy. A representative image is shown of a cell (≤5% of the population) that exhibited nuclear localization (arrows) of the indicated protein. (B) Cells bearing a CEN plasmid from which expression of Aim44‐mNG, Nap1‐mNG, Nis1‐mNG or Nba1‐mNG was driven from the GAL1/10 promoter by growth in medium containing 2% galactose for 3 h were examined by fluorescence microscopy. Representative cells displaying bud neck localization (arrows) are shown. Scale bars = 1 μm.
Figure S2. Assessment of the role of various karyopherins on nuclear import of overexpressed Aim44 and Nis1 and of a candidate NLS in Nis1. (A) Aim44‐mNG (top panels) or Nis1‐mNG (bottom panels) were overexpressed in the indicated importin knock‐out strains. Cells were grown as in Figure S1B and then examined. (B) Aim44‐mNG (top) or Nis1‐mNG (bottom) were over‐expressed in a strain bearing a temperature‐sensitive allele of Importin‐α /Kap60 (srp1‐31) by growing the cultures at 25°C to mid‐exponential phase in S medium containing 2% raffinose and 0.2% sucrose, then splitting the cultures into two equal portions, followed by incubation for 1 hr at either 25° or 37°C, as indicated, whereupon galactose was added (2% final concentration) and incubation continued for an additional 3 h before the cells were imaged. (C) As in (B), except that Nis1‐mNG overexpression was carried out a strain harboring a temperature‐sensitive allele of Importin‐β/Kap95 (kap95ts). (D) Nis1(K179A R181A R183A)‐mNG was over‐expressed, as in (A), in BY4741 (Table 4). Scale bars = 1 μm
Figure S3. Level of Nis1‐mNG and Aim44‐mNG expression in importin mutants. (A) Aim44‐ mNG or (B) Nis1‐mNG was overexpressed as in Figure S1B in the indicated importin deletion strains. Cells were harvested, lysed, and proteins precipitated with trichloroacetic acid. Precipitated proteins were resolved via SDS‐PAGE, transferred to a nitrocellulose membrane, and detected by immunoblotting with an anti‐mNeonGreen antibody or, as a control for equivalent loading, anti‐Pgk1 antibodies. MW, molecular weight markers.
Figure S4. Attempted construction of importin double mutants. Appropriate haploid strains containing single importin deletion mutations were mated, yielding the indicated heterozygous diploids, which were subjected to sporulation and the resulting tetrads dissected. (A) Regardless of the presence or absence of the mtr10Δ/kap111Δ mutation, any spore containing the pse1Δ/kap121Δ mutation produced only a barely visible colony (arrows), indicative of exceedingly poor growth. (B) Regardless of the presence or absence of the nmd5Δ/kap119Δ mutation, any spore containing the pse1Δ/kap121Δ mutation produced only a barely visible colony (arrows), indicative of exceedingly poor growth. (C) The expected tetratype asci were obtained, but the predicted kap123Δ/yrb4Δ nmd5Δ/kap119Δ double mutant spore was always inviable.
Figure S5. Neither Kap123 nor Nmd5 contribute to nuclear entry of overexpressed Nis1‐ mNG. (A) Nis1‐mNG was overexpressed as in Figure S1B in the indicated deletion strains. Scale bar = 1 μm. (B) Quantitation of the experiment depicted in panel (A). The percentage of cells displaying Nis1‐mNG localization for each strain (gray bars) plotted is the average of three independent experiments. For cells that display fluorescent signal at the nucleus, the mean ratio of nuclear to cytoplasmic pixel intensity is plotted (white bars; n ≥ 50 cells). Error bars represent standard error of the mean.
Figure S6. Suggestive evidence that Xpo1 contributes to nuclear export of Aim44‐mNG. All cultures were grown to mid‐exponential phase before the following imaging experiments were initiated. (A) Aim44‐mNG was expressed from its endogenous, chromosomal locus in strains harboring the cse1‐1cs allele in combination with deletions of either LOS1 or MSN5. Cells were grown at 25°, then half of the culture was shifted to 11°. Cells were cultured for an additional 5 h before imaging. (B) Aim44‐mNG was expressed from its endogenous, chromosomal locus in strains harboring the xpo1‐1ts allele in combination with deletions in either LOS1 or MSN5. Cells were grown at 25°, then half of the culture was shifted to 37° for 3 h. (C) Aim44‐mNG was expressed from its endogenous, chromosomal locus in a los1Δ msn5Δ strain. Scale bars = 1 μm
ACKNOWLEDGEMENTS
This work was supported by a Ford Foundation Postdoctoral Fellowship (to A.M.P.) and by NIH R01 Research Grants GM101314 and GM21841 (to J.T.). We thank Vikram Panse (Univ. of Zürich, Switzerland) for helpful discussion and the communication of unpublished results about Nis1 obtained by his former ETH Ph.D. student, Dr. Maria Veisu, Sarah M. Sterling (Harvard Med. Sch.) for thoughtful suggestions, Gregory C. Finnigan (Kansas State Univ.) for providing plasmids, William Y. Shin (this laboratory) for assistance in confirming the genotype of karyopherin knock‐out strains, and Gislene Pereira (Centre for Organismal Studies, Univ. of Heidelberg, Germany) for providing Aim44/Gps1‐related research material.
Perez AM, Thorner J. Septin‐associated proteins Aim44 and Nis1 traffic between the bud neck and the nucleus in the yeast Saccharomyces cerevisiae . Cytoskeleton. 2019;76:15–32. 10.1002/cm.21500
Funding information Ford Foundation, Grant/Award Numbers: Postdoctoral Fellowship to Dr. Adam M. Perez; National Institute of General Medical Sciences, Grant/Award Numbers: GM101314 and GM21841 to Jeremy Thorner.
References
- Aitchison, J. D. , Blobel, G. , & Rout, M. P. (1996). Kap104p: A karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science, 274, 624–627. [DOI] [PubMed] [Google Scholar]
- Albuquerque, C. P. , Smolka, M. B. , Payne, S. H. , Bafna, V. , Eng, J. , & Zhou, H. (2008). A multidimensional chromatography technology for in‐depth phosphoproteome analysis. Molecular & Cellular Proteomics, 7, 1389–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman, R. , & Kellogg, D. (1997). Control of mitotic events by Nap1 and the Gin4 kinase. The Journal of Cell Biology, 138, 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariño, J. , Casamayor, A. , & González, A. (2011). Type 2C protein phosphatases in fungi. Eukaryotic Cell, 10, 21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baro, B. , Queralt, E. , & Monje‐Casas, F. (2017). Regulation of mitotic exit in Saccharomyces cerevisiae . Methods in Molecular Biology, 1505, 3–17. [DOI] [PubMed] [Google Scholar]
- Baum, P., Thorner, J., & Honig, L. (1978). Identification of tubulin from the yeast Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States, 75, 8274–8279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin, A. , McMurray, M. A. , Grob, P. , Park, S. S. , Garcia, G. , Patanwala, I. , … Nogales, E. (2008). Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly. Proceedings of the National Academy of Sciences of the United States of America, 105, 8274–8279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin, A. , McMurray, M. A. , Pierson, J. , Thai, L. , McDonald, K. L. , Zehr, E. A. , … Nogales, E. (2012). Three‐dimensional ultrastructure of the septin filament network in Saccharomyces cerevisiae . Molecular Biology of the Cell, 23, 423–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin, A. , McMurray, M. A. , Thai, L. , Garcia, G. , Votin, V. , Grob, P. , … Nogales, E. (2010). Phosphatidylinositol‐4,5‐bisphosphate promotes budding yeast septin filament assembly and organization. Journal of Molecular Biology, 404, 711–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin, A. , & Nogales, E. (2016). Characterization of septin ultrastructure in budding yeast using electron tomography. Methods in Molecular Biology, 1369, 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth, E. A. , Vane, E. W. , Dovala, D. , & Thorner, J. (2015). A Förster resonance energy transfer (FRET)‐based system provides insight into the ordered assembly of yeast septin hetero‐octamers. The Journal of Biological Chemistry, 290, 28388–28401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann, C. B. , Davies, A. , Cost, G. J. , Caputo, E. , Li, J. , et al. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR‐mediated gene disruption and other applications. Yeast, 14, 115–132. [DOI] [PubMed] [Google Scholar]
- Bridges, A. A. , Jentzsch, M. S. , Oakes, P. W. , Occhipinti, P. , & Gladfelter, A. S. (2016). Micron‐scale plasma membrane curvature is recognized by the septin cytoskeleton. The Journal of Cell Biology, 213, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges, A. A. , Zhang, H. , Mehta, S. B. , Occhipinti, P. , Tani, T. , & Gladfelter, A. S. (2014). Septin assemblies form by diffusion‐driven annealing on membranes. Proceedings of the National Academy of Sciences of the United States of America, 111, 2146–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert, M. E. , Keck, K. M. , Ptak, C. , Shabanowitz, J. , Hunt, D. F. , et al. (2008). Phosphorylation by casein kinase 2 regulates Nap1 localization and function. Molecular and Cellular Biology, 28, 1313–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon, K. S. , Woods, B. L. , & Gladfelter, A. S. (2017). The unsolved problem of how cells sense micron‐scale curvature. Trends in Biochemical Sciences, 42, 961–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, C. W. , Altman, R. , Schieltz, D. , Yates, J. R. , & Kellogg, D. (1998). The septins are required for the mitosis‐specific activation of the Gin4 kinase. The Journal of Cell Biology, 143, 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudron, F. , & Barral, Y. (2009). Septins and the lateral compartmentalization of eukaryotic membranes. Developmental Cell, 16, 493–506. [DOI] [PubMed] [Google Scholar]
- Chiou, J. G. , Balasubramanian, M. K. , & Lew, D. J. (2017). Cell polarity in yeast. Annual Review of Cell and Developmental Biology, 33, 77–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere, J. , & Barral, Y. (2004). Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science, 305, 393–396. [DOI] [PubMed] [Google Scholar]
- Edelstein, A. D. , Tsuchida, M. A. , Amodaj, N. , Pinkard, H. , Vale, R. D. , & Stuurman, N. (2014). Advanced methods of microscope control using μManager software. Journal of Biological Methods, 1, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnigan, G. C. , Booth, E. A. , Duvalyan, A. , Liao, E. N. , & Thorner, J. (2015). The carboxy‐terminal tails of septins Cdc11 and Shs1 recruit myosin‐II binding factor Bni5 to the bud neck in Saccharomyces cerevisiae . Genetics, 200, 843–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnigan, G. C. , Duvalyan, A. , Liao, E. N. , Sargsyan, A. , & Thorner, J. (2016). Detection of protein‐protein interactions at the septin collar in Saccharomyces cerevisiae using a tripartite split‐GFP system. Molecular Biology of the Cell, 27, 2708–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnigan, G. C. , Sterling, S. M. , Duvalyan, A. , Liao, E. N. , Sargsyan, A. , Garcia, G., III , … Thorner, J. (2016). Coordinate action of distinct sequence elements localizes checkpoint kinase Hsl1 to the septin collar at the bud neck in Saccharomyces cerevisiae . Molecular Biology of the Cell, 27, 2213–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnigan, G. C. , Takagi, J. , Cho, C. , & Thorner, J. (2015). Comprehensive genetic analysis of paralogous terminal septin subunits Shs1 and Cdc11 in Saccharomyces cerevisiae . Genetics, 200, 821–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, G. , Bertin, A. , Li, Z. , Song, Y. , McMurray, M. A. , Thorner, J. , & Nogales, E. (2011). Subunit‐dependent modulation of septin assembly: budding yeast septin Shs1 promotes ring and gauze formation. The Journal of Cell Biology, 195, 993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin, A. C. , Aloy, P. , Grandi, P. , Krause, R. , Boesche, M. , Marzioch, M. , … Superti‐Furga, G. (2006). Proteome survey reveals modularity of the yeast cell machinery. Nature, 440, 631–636. [DOI] [PubMed] [Google Scholar]
- Gilbert, W. , & Guthrie, C. (2004). The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Molecular Cell, 13, 201–212. [DOI] [PubMed] [Google Scholar]
- Hannich, J. T. , Lewis, A. , Kroetz, M. B. , Li, S. J. , Heide, H. , Emili, A. , & Hochstrasser, M. (2005). Defining the SUMO‐modified proteome by multiple approaches in Saccharomyces cerevisiae . The Journal of Biological Chemistry, 280, 4102–4110. [DOI] [PubMed] [Google Scholar]
- Hartwell, L. H. (1971). Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Experimental Cell Research, 69, 265–276. [DOI] [PubMed] [Google Scholar]
- Hartwell, L. H. , Culotti, J. , Pringle, J. R. , & Reid, B. J. (1974). Genetic control of the cell division cycle in yeast. Science, 183, 46–51. [DOI] [PubMed] [Google Scholar]
- Holt, L. J. , Tuch, B. B. , Villen, J. , Johnson, A. D. , Gygi, S. P. , & Morgan, D. O. (2009). Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science, 325, 1682–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine, M. K. , & Wente, S. R. (1997). A nuclear export signal in Kap95p is required for both recycling the import factor and interaction with the nucleoporin GLFG repeat regions of Nup116p and Nup100p. The Journal of Cell Biology, 137, 797–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T. , Chiba, T. , Ozawa, R. , Yoshida, M. , Hattori, M. , & Sakaki, Y. (2001). A comprehensive two‐hybrid analysis to explore the yeast protein interactome. Proceedings of the National Academy of Sciences of the United States of America, 98, 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase, M. , Luo, J. , Bi, E. , & Toh‐e, A. (2007). Shs1 plays separable roles in septin organization and cytokinesis in Saccharomyces cerevisiae . Genetics, 177, 215–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase, M. , & Toh‐e, A. (2001). Nis1 encoded by YNL078W: a new neck protein of Saccharomyces cerevisiae . Genes & Genetic Systems, 76, 335–343. [DOI] [PubMed] [Google Scholar]
- Janke, C. , Magiera, M. M. , Rathfelder, N. , Taxis, C. , Reber, S. , Maekawa, H. , … Knop, M. (2004). A versatile toolbox for PCR‐based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast, 21, 947–962. [DOI] [PubMed] [Google Scholar]
- Johnson, E. S. , & Blobel, G. (1999). Cell cycle‐regulated attachment of the ubiquitin‐related protein SUMO to the yeast septins. The Journal of Cell Biology, 147, 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonasson, E. M. , Rossio, V. , Hatakeyama, R. , Abe, A. , Ohya, Y. , & Yoshida, S. (2016). Zds1/Zds2‐PP2ACdc55 complex specifies signaling output from Rho1 GTPase. The Journal of Cell Biology, 212, 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck, K. M. , & Pemberton, L. F. (2013). Histone chaperones link histone nuclear import and chromatin assembly. Biochimica et Biophysica Acta, 1819, 277–289. [PubMed] [Google Scholar]
- Kellogg, D. R. , & Murray, A. W. (1995). NAP1 acts with Clb1 to perform mitotic functions and to suppress polar bud growth in budding yeast. The Journal of Cell Biology, 130, 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, A. , Newby, J. , & Gladfelter, A. S. (2018). Control of septin filament flexibility and bundling by subunit composition and nucleotide interactions. Molecular Biology of the Cell, 29, 702–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kõivomägi, M. , Valk, E. , Venta, R. , Iofik, A. , Lepiku, M. , Morgan, D. O. , & Loog, M. (2011). Dynamics of Cdk1 substrate specificity during the cell cycle. Molecular Cell, 42, 610–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N. J. , Cagney, G. , Yu, H. , Zhong, G. , Guo, X. , Ignatchenko, A. , … Greenblatt, J. F. (2006). Global landscape of protein complexes in the yeast Saccharomyces cerevisiae . Nature, 440, 637–643. [DOI] [PubMed] [Google Scholar]
- Lange, A. , Mills, R. E. , Lange, C. J. , Stewart, M. , Devine, S. E. , & Corbett, A. H. (2007). Classical nuclear localization signals: definition, function and interaction with importin alpha. The Journal of Biological Chemistry, 282, 5101–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb, J. D. , Schlenstedt, G. , Pellman, D. , Kornitzer, D. , Silver, P. A. , & Fink, G. R. (1995). The yeast nuclear import receptor is required for mitosis. Proceedings of the National Academy of Sciences of the United States of America, 92, 7647–7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhnevych, T. , Ptak, C. , Lusk, C. P. , Aitchison, J. D. , & Wozniak, R. W. (2007). The role of karyopherins in the regulated sumoylation of septins. The Journal of Cell Biology, 177, 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt J, Chen X, Bi E (2019) Architecture, remodeling, and functions of the septin cytoskeleton. Cytoskeleton (Hoboken) 76, 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray, M. A. , Bertin, A. , Garcia, G. , Lam, L. , Nogales, E. , & Thorner, J. (2011). Septin filament formation is essential in budding yeast. Developmental Cell, 20, 540–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray, M. A. , & Thorner, J. (2009). Septins: molecular partitioning and the generation of cellular asymmetry. Cell Division, 4, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger, F. , Boehm, M. E. , Hofmann, A. , Hub, B. , Zentgraf, H. , Lehmann, W. D. , & Pereira, G. (2011). Phosphorylation‐dependent regulation of the F‐BAR protein Hof1 during cytokinesis. Genes & Development, 25, 875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger, F. , Khmelinskii, A. , Morlot, S. , Kurtulmus, B. , Palani, S. , Andres‐Pons, A. , … Pereira, G. (2014). A memory system of negative polarity cues prevents replicative aging. Cell, 159, 1056–1069. [DOI] [PubMed] [Google Scholar]
- Meitinger, F. , & Palani, S. (2016). Actomyosin ring driven cytokinesis in budding yeast. Seminars in Cell & Developmental Biology, 53, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger, F. , & Pereira, G. (2017). The septin‐associated kinase Gin4 recruits Gps1 to the site of cell division. Molecular Biology of the Cell, 28, 883–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger, F. , Richter, H. , Heisel, S. , Hub, B. , Seufert, W. , & Pereira, G. (2013). A safeguard mechanism regulates Rho GTPases to coordinate cytokinesis with the establishment of cell polarity. PLoS Biology, 11, e1001495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger, F. , S, P. , Hub, B. , & Pereira, G. G. (2013). Dual function of the NDR‐kinase Dbf2 in the regulation of the F‐BAR protein Hof1 during cytokinesis. Moll. Biol. Cell, 24, 1290–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, K. E. , Lo, W. C. , Lee, M. E. , Kang, P. J. , & Park, H. O. (2017). Fine‐tuning the orientation of the polarity axis by Rga1, a Cdc42 GTPase‐activating protein. Molecular Biology of the Cell, 28, 3773–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mino, A. , Tanaka, K. , Kamei, T. , Umikawa, M. , Fujiwara, T. , & Takai, Y. (1998). Shs1p: a novel member of septin that interacts with Spa2p, involved in polarized growth in Saccharomyces cerevisiae . Biochemical and Biophysical Research Communications, 251, 732–736. [DOI] [PubMed] [Google Scholar]
- Miyaji‐Yamaguchi, M. , Kato, K. , Nakano, R. , Akashi, T. , Kikuchi, A. , & Nagata, K. (2003). Involvement of nucleocytoplasmic shuttling of yeast Nap1 in mitotic progression. Molecular and Cellular Biology, 23, 6672–6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast, N. , Ewart, C. S. , & Pemberton, L. F. (2002). A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B. The EMBO Journal, 21, 6527–6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast, N. , Guo, Y. , Shabanowitz, J. , Hunt, D. F. , & Pemberton, L. F. (2002). Pathways mediating the nuclear import of histones H3 and H4 in yeast. The Journal of Biological Chemistry, 277, 862–868. [DOI] [PubMed] [Google Scholar]
- Mosammaparast, N. , Jackson, K. R. , Guo, Y. , Brame, C. J. , Shabanowitz, J. , Hunt, D. F. , & Pemberton, L. F. (2001). Nuclear import of histone H2A and H2B is mediated by a network of karyopherins. The Journal of Cell Biology, 153, 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong, K. , Wloka, C. , Okada, S. , Svitkina, T. , & Bi, E. (2014). Architecture and dynamic remodelling of the septin cytoskeleton during the cell cycle. Nature Communications, 5, 5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, H. O. , Bi, E. , Pringle, J. R. , & Herskowitz, I. (1997). Two active states of the Ras‐related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proceedings of the National Academy of Sciences of the United States of America, 94, 4463–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, A. M. , Finnigan, G. C. , Roelants, F. M. , & Thorner, J. (2016). Septin‐associated protein kinases in the yeast. Saccharomyces cerevisiae. Frontiers in Cell and Development Biology, 4, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne, D. , Legesse‐Miller, A. , Gao, L. , Dong, Y. , & Bretscher, A. (2004). Mechanisms of polarized growth and organelle segregation in yeast. Annual Review of Cell and Developmental Biology, 20, 559–591. [DOI] [PubMed] [Google Scholar]
- Rodal, A. A. , Kozubowski, L. , Goode, B. L. , Drubin, D. G. , & Hartwig, J. H. (2005). Actin and septin ultrastructures at the budding yeast cell cortex. Molecular Biology of the Cell, 16, 372–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants, F. M. , Su, B. M. , von Wulffen, J. , Ramachandran, S. , Sartorel, E. , Trott, A. E. , & Thorner, J. (2015). Protein kinase Gin4 negatively regulates flippase function and controls plasma membrane asymmetry. The Journal of Cell Biology, 208, 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossio, V. , Michimoto, T. , Sasaki, T. , Ohbayashi, I. , Kikuchi, Y. , & Yoshida, S. (2013). Nuclear PP2A‐Cdc55 prevents APC‐Cdc20 activation during the spindle assembly checkpoint. Journal of Cell Science, 126, 4396–4405. [DOI] [PubMed] [Google Scholar]
- Sakchaisri, K. , Asano, S. , Yu, L. R. , Shulewitz, M. J. , Park, C. J. , Park, J. E. , … Lee, K. S. (2004). Coupling morphogenesis to mitotic entry. Proceedings of the National Academy of Sciences of the United States of America, 101, 4124–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner, N. C. , Campbell, R. E. , Steinbach, P. A. , Giepmans, B. N. , Palmer, A. E. , & Tsien, R. Y. (2003). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature Biotechnology, 22, 1567–1572. [DOI] [PubMed] [Google Scholar]
- Shaner, N. C. , Lambert, G. G. , Chammas, A. , Ni, Y. , Cranfill, P. J. , Baird, M. A. , … Wang, J. (2013). A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum . Nature Methods, 10, 407–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F. , Fink, G. R. , & Hicks, J. B. (1986). Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Shulewitz, M. J. , Inouye, C. J. , & Thorner, J. (1999). Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae . Molecular and Cellular Biology, 19, 7123–7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soniat, M. , Cağatay, T. , & Chook, Y. M. (2016). Recognition elements in the histone H3 and H4 tails for seven different importins. The Journal of Biological Chemistry, 291, 21171–21183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soniat, M. , & Chook, Y. M. (2015). Nuclear localization signals for four distinct karyopherin‐β nuclear import systems. The Biochemical Journal, 468, 353–362. [DOI] [PubMed] [Google Scholar]
- Stade, K. , Ford, C. S. , Guthrie, C. , & Weis, K. (1997). Exportin 1 (Crm1p) is an essential nuclear export factor. Cell, 90, 1041–1050. [DOI] [PubMed] [Google Scholar]
- Strahl, T. , Hama, H. , DeWald, D. B. , & Thorner, J. (2005). Yeast phosphatidylinositol 4‐kinase, Pik1, has essential roles at the Golgi and in the nucleus. The Journal of Cell Biology, 171, 967–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ström, A. C. , & Weis, K. (2001). Importin‐beta‐like nuclear transport receptors. Genome Biology, 2, REVIEWS3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney, D. L. , Beltrao, P. , Starita, L. , Guo, A. , Rush, J. , Fields, S. , … Villén, J. (2013). Global analysis of phosphorylation and ubiquitylation cross‐talk in protein degradation. Nature Methods, 10, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, Z. , Gao, X. D. , Howell, A. S. , Bose, I. , Lew, D. J. , & Bi, E. (2007). Adjacent positioning of cellular structures enabled by a Cdc42 GTPase‐activating protein‐mediated zone of inhibition. The Journal of Cell Biology, 179, 1375–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz, P. , Giot, L. , Cagney, G. , Mansfield, T. A. , Judson, R. S. , Knight, J. R. , … Rothberg, J. M. (2000). A comprehensive analysis of protein‐protein interactions in Saccharomyces cerevisiae . Nature, 403, 623–627. [DOI] [PubMed] [Google Scholar]
- Uzunova, K. , Göttsche, K. , Miteva, M. , Weisshaar, S. R. , Glanemann, C. , et al. (2007). Ubiquitin‐dependent proteolytic control of SUMO conjugates. The Journal of Biological Chemistry, 282, 34167–34175. [DOI] [PubMed] [Google Scholar]
- Veisu, M. , 2011. Identification and characterization of SUMO interaction motif (SIM)‐containing proteins from Saccharomyces cerevisiae (Ph.D. Thesis, Institute of Biochemistry). Dept. of Biology, Swiss Federal Institute of Technology, Zürich, Switzerland, pp. 109.
- Wang, M. , Nishihama, R. , Onishi, M. , & Pringle, J. R. (2018). Role of the Hof1‐Cyk3 interaction in cleavage‐furrow ingression and primary‐septum formation during yeast cytokinesis. Molecular Biology of the Cell, 29, 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolken, D. M. , McInnes, J. , & Pon, L. A. (2014). Aim44p regulates phosphorylation of Hof1p to promote contractile ring closure during cytokinesis in budding yeast. Molecular Biology of the Cell, 25, 753–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Z. , McGrew, J. T. , Schroeder, A. J. , & Fitzgerald‐Hayes, M. (1993). CSE1 and CSE2, two new genes required for accurate mitotic chromosome segregation in Saccharomyces cerevisiae . Molecular and Cellular Biology, 13, 4691–4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamochi, W. , Tanaka, K. , Nonaka, H. , Maeda, A. , Musha, T. , & Takai, Y. (1994). Growth site localization of Rho1 small GTP‐binding protein and its involvement in bud formation in Saccharomyces cerevisiae . The Journal of Cell Biology, 125, 1077–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H. , Braun, P. , Yildirim, M. A. , Lemmens, I. , Venkatesan, K. , Sahalie, J. , … Vidal, M. (2008). High‐quality binary protein interaction map of the yeast interactome network. Science, 322, 104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Endogenously‐expressed Aim44‐mNG, Nap1‐mNG and Nis1‐mNG (but not Nba1‐ mNG) can be detected in the nucleus in a small fraction of the cells, whereas overexpression does not prevent localization of the same proteins to the bud neck. (A) Strains expressing Aim44‐mNG, Nap1‐mNG, Nis1‐mNG or Nba1‐mNG from their native promoters were grown to mid‐exponential phase and examined by fluorescence microscopy. A representative image is shown of a cell (≤5% of the population) that exhibited nuclear localization (arrows) of the indicated protein. (B) Cells bearing a CEN plasmid from which expression of Aim44‐mNG, Nap1‐mNG, Nis1‐mNG or Nba1‐mNG was driven from the GAL1/10 promoter by growth in medium containing 2% galactose for 3 h were examined by fluorescence microscopy. Representative cells displaying bud neck localization (arrows) are shown. Scale bars = 1 μm.
Figure S2. Assessment of the role of various karyopherins on nuclear import of overexpressed Aim44 and Nis1 and of a candidate NLS in Nis1. (A) Aim44‐mNG (top panels) or Nis1‐mNG (bottom panels) were overexpressed in the indicated importin knock‐out strains. Cells were grown as in Figure S1B and then examined. (B) Aim44‐mNG (top) or Nis1‐mNG (bottom) were over‐expressed in a strain bearing a temperature‐sensitive allele of Importin‐α /Kap60 (srp1‐31) by growing the cultures at 25°C to mid‐exponential phase in S medium containing 2% raffinose and 0.2% sucrose, then splitting the cultures into two equal portions, followed by incubation for 1 hr at either 25° or 37°C, as indicated, whereupon galactose was added (2% final concentration) and incubation continued for an additional 3 h before the cells were imaged. (C) As in (B), except that Nis1‐mNG overexpression was carried out a strain harboring a temperature‐sensitive allele of Importin‐β/Kap95 (kap95ts). (D) Nis1(K179A R181A R183A)‐mNG was over‐expressed, as in (A), in BY4741 (Table 4). Scale bars = 1 μm
Figure S3. Level of Nis1‐mNG and Aim44‐mNG expression in importin mutants. (A) Aim44‐ mNG or (B) Nis1‐mNG was overexpressed as in Figure S1B in the indicated importin deletion strains. Cells were harvested, lysed, and proteins precipitated with trichloroacetic acid. Precipitated proteins were resolved via SDS‐PAGE, transferred to a nitrocellulose membrane, and detected by immunoblotting with an anti‐mNeonGreen antibody or, as a control for equivalent loading, anti‐Pgk1 antibodies. MW, molecular weight markers.
Figure S4. Attempted construction of importin double mutants. Appropriate haploid strains containing single importin deletion mutations were mated, yielding the indicated heterozygous diploids, which were subjected to sporulation and the resulting tetrads dissected. (A) Regardless of the presence or absence of the mtr10Δ/kap111Δ mutation, any spore containing the pse1Δ/kap121Δ mutation produced only a barely visible colony (arrows), indicative of exceedingly poor growth. (B) Regardless of the presence or absence of the nmd5Δ/kap119Δ mutation, any spore containing the pse1Δ/kap121Δ mutation produced only a barely visible colony (arrows), indicative of exceedingly poor growth. (C) The expected tetratype asci were obtained, but the predicted kap123Δ/yrb4Δ nmd5Δ/kap119Δ double mutant spore was always inviable.
Figure S5. Neither Kap123 nor Nmd5 contribute to nuclear entry of overexpressed Nis1‐ mNG. (A) Nis1‐mNG was overexpressed as in Figure S1B in the indicated deletion strains. Scale bar = 1 μm. (B) Quantitation of the experiment depicted in panel (A). The percentage of cells displaying Nis1‐mNG localization for each strain (gray bars) plotted is the average of three independent experiments. For cells that display fluorescent signal at the nucleus, the mean ratio of nuclear to cytoplasmic pixel intensity is plotted (white bars; n ≥ 50 cells). Error bars represent standard error of the mean.
Figure S6. Suggestive evidence that Xpo1 contributes to nuclear export of Aim44‐mNG. All cultures were grown to mid‐exponential phase before the following imaging experiments were initiated. (A) Aim44‐mNG was expressed from its endogenous, chromosomal locus in strains harboring the cse1‐1cs allele in combination with deletions of either LOS1 or MSN5. Cells were grown at 25°, then half of the culture was shifted to 11°. Cells were cultured for an additional 5 h before imaging. (B) Aim44‐mNG was expressed from its endogenous, chromosomal locus in strains harboring the xpo1‐1ts allele in combination with deletions in either LOS1 or MSN5. Cells were grown at 25°, then half of the culture was shifted to 37° for 3 h. (C) Aim44‐mNG was expressed from its endogenous, chromosomal locus in a los1Δ msn5Δ strain. Scale bars = 1 μm


