Abstract
Neonatal molecular biomarkers of neurobehavioral responses (measures of brain-behavior relationships), when combined with neurobehavioral performance measures, could lead to better predictions of long-term developmental outcomes. To this end, we examined whether variability in buccal cell DNA methylation (DNAm) associated with neurobehavioral profiles in a cohort of infants born less than 30 weeks postmenstrual age (PMA) and participating in the Neonatal Neurobehavior and Outcomes in Very Preterm Infants (NOVI) Study (N = 536). We tested whether epigenetic age, age acceleration, or DNAm levels at individual loci differed between infants based on their NICU Network Neurobehavioral Scale (NNNS) profile classifications. We adjusted for recruitment site, infant sex, PMA, and tissue heterogeneity. Infants with an optimally well-regulated NNNS profile had older epigenetic age compared to other NOVI infants (β1 = 0.201, p-value = 0.026), but no significant difference in age acceleration. In contrast, infants with an atypical NNNS profile had differential methylation at 29 CpG sites (FDR < 10%). Some of the genes annotated to these CpGs included PLA2G4E, TRIM9, GRIK3, and MACROD2, which have previously been associated with neurological structure and function, or with neurobehavioral disorders. These findings contribute to the existing evidence that neonatal epigenetic variations may be informative for infant neurobehavior.
Subject terms: Neurodevelopmental disorders, DNA methylation
Introduction
Preterm birth is a significant global public health problem. In the United States one in eight children are born less than 37 weeks of gestation1. Survival of infants born very preterm, prior to 30 weeks postmenstrual age (PMA), has improved due to technological and medical advancements2,3. These youngest infants are more likely to suffer from chronic illnesses, potentially devastating brain injuries, and adverse neuromotor, cognitive, and behavioral outcomes that persist through adulthood4–15. These consequences of premature birth often require extensive healthcare, educational and psychosocial community resources, in addition to increased burden on the families and caregivers of these children, emotionally and financially.
In addition to immaturity, premature infants vary widely in the health complications they experience. As such, assessments prior to discharge from the neonatal intensive care unit (NICU) are needed to identify the earliest risks for adverse neurodevelopmental outcomes, and to maximize the potential benefits of interventions aimed at ameliorating long term deficits. There is growing evidence that neonatal neurobehavior (the relationships between the nervous system and behavior), as measured by the NICU Network Neurobehavioral Scale (NNNS)16, predicts developmental deficits in infants born preterm and others at risk, beyond what can be predicted based on the assessment of medical risk factors throughout the newborn’s hospital stay17–20. Latent profiles of NNNS summary scores have been used to classify infants into groups with similar responses across the overall NNNS assessment. These neurobehavioral profiles in infants have been associated with prenatal exposures (prenatal drugs21,22 and perfluorooctanoic acid23), birth outcomes (gestational age and birth weight21), and with behavioral and cognitive outcomes in childhood21,22,24. Poorer performance on the NNNS has also been shown to be predictive of non-optimal developmental outcomes through early childhood21. Beyond the neurobehavioral and medical assessments, molecular biomarkers may provide insights into how the environment and experiences of the preterm newborn are internalized and may hold additional value as predictive tools useful in risk stratification.
Epigenetics refers to mitotically and meiotically heritable changes in gene expression potential that are not explained by changes in DNA sequence. The most thoroughly studied epigenetic mechanism is DNA methylation (DNAm), particularly in the context of cytosine-phosphate-guanine (CpG) motifs and islands. These methylation marks can be inherited across cell divisions, established in-utero and/or affected by the environment throughout life, thus representing a truly integrated measure of exposure and disease susceptibility. In preterm infants, variability in DNAm of candidate genes have been related to medical complications such as sepsis25, pain related stress26,27, medical and neurobehavioral risk28,29, and as a potential moderator of NICU environment stress on serotonergic tone and temperament30. We have also used an epigenome-wide scan of DNAm in the placenta to demonstrate relationships between methylation of the FHIT and ANDKR11 genes, which had been previously linked to neurodevelopmental and behavioral outcomes, and performance on the NNNS attention scale in a cohort of term newborns31. Additionally, DNAm can be used to estimate epigenetic age, which is thought to be a marker of underlying biological aging32. In children and adolescents, epigenetic age acceleration has been associated with physical development33, pubertal development, internalization and thought problems34, and increased cortisol production34,35. These studies have begun to elucidate the potential role for epigenetic aging in developmental processes but has not been thoroughly studied in relation to neonatal neurobehavioral responses.
Incorporating molecular biomarkers, such as DNAm and epigenetic age, with performance measures may improve long-term predictions of health outcomes in preterm infants. Before this can be done, it is important to demonstrate whether variability in DNAm and epigenetic age, measured from accessible tissues, are associated with behavioral measures in newborns. In this study, we hypothesized that infants with the most atypical or optimal neurobehavioral profiles, measured via the NNNS, will exhibit unique patterns of DNAm. In a U.S. multisite cohort of infants born less than 30 weeks PMA, we profiled genome-wide DNAm from buccal swab samples using the Illumina MethylationEPIC array platform. We estimated epigenetic age and tested for differences in epigenetic age and in DNAm among infants with neurobehavioral profiles, ranging from most optimal to atypical, as determined via NNNS latent profile classification.
Methods
Study Population
The Neonatal Neurobehavior and Outcomes in Very Preterm Infants (NOVI) Study was conducted at 9 university-affiliated NICUs in Providence, RI, Grand Rapids, MI, Kansas City, MO, Honolulu, HI, Winston-Salem, NC, and Torrance and Long Beach CA from April 2014 through June 2016. These NICUs were also Vermont Oxford Network (VON) participants. All participating mothers provided written informed consent. Enrollment and consent procedures for this study were approved by the institutional review boards of Women and Infants Hospital, Spectrum Health, Children’s Mercy Office of Research Integrity, Wake Forest University Health Sciences, John F. Wolf, MD Human Subjects Committee at Los Angeles BioMed, Emory University and Western Institutional Review Board (WIRB); all methods employed in the study were performed in accordance with the relevant guidelines and regulations. Eligibility was determined based on the following inclusion criteria: (1) birth at <30 weeks’ gestation; (2) parental ability to read and speak English, Spanish, Japanese, or Chinese; and (3) residence within 3 hours of the NICU and follow-up clinic. Infants were excluded if their medical record indicated presence of a major congenital anomaly, including central nervous system, cardiovascular, gastrointestinal, genitourinary, chromosomal, and nonspecific anomalies36. Parents of eligible infants were invited to participate in the study at 31–32 weeks PMA, or when survival to discharge was determined to be likely by the attending neonatologist. Demographic variables, including infant gender, race, ethnicity, maternal education and partner status were collected from the maternal interview. Socioeconomic status (SES) was assessed using the Hollingshead Index, with Hollingshead level V indicating low SES37. Neonatal medical variables including birthweight, gestational age, length of NICU stay, weight at discharge, and gestational age at discharge were abstracted from medical records. Overall, 709 infants were enrolled, 679 from whom complete neurobehavioral assessment data were obtained, and buccal cells were collected on 624 of these infants for epigenomic screening.
NICU Network Neurobehavioral Scale (NNNS)
Neonatal neurobehavior was assessed using the NNNS. The NNNS is a 20–30 minute standardized procedure that includes measures of active and passive tone, primitive reflexes, items that reflect physical maturity, social and behavioral functioning including visual and auditory tracking, cuddling and soothability, and a checklist of stress signs organized by organ system38. The NNNS was administered during the week of NICU discharge (+/− 3 days) by site examiners who were trained and certified by a central NOVI NNNS trainer. The exam was conducted 45 minutes prior to a scheduled feeding or routine care in order to maximize alertness and avoid disrupting NICU routines that facilitate sleep patterns. Individual items were converted to 12 summary scores: attention, handling, self-regulation, arousal, excitability, lethargy, hypertonicity, hypotonicity, non-optimal reflexes, asymmetric reflexes, quality of movement and stress abstinence. A higher summary score does not necessarily reflect better performance, but instead more of the construct. Summary scores were converted to NNNS profiles, which are mutually exclusive, discrete categories representing the infant’s pattern of performance across the summary scores, and which have been shown to be related to future behavioral and cognitive performance21,22,24.
DNA Methylation (DNAm) Analysis
Genomic DNA was extracted from buccal swab samples, collected near term-equivalent age, using the Isohelix Buccal Swab system (Boca Scientific), quantified using the Quibit Fluorometer (Thermo Fisher, Waltham, MA, USA) and aliquoted into a standardized concentration for subsequent analyses. DNA samples were plated randomly across 96-well plates and provided to the Emory University Integrated Genomics Core for bisulfite modification using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA), and subsequent assessment of genome-wide DNAm using the Illumina MethylationEPIC Beadarray (Illumina, San Diego, CA) following standardized methods based on the manufacturer’s protocol. Samples with more than 5% of probes yielding detection p-values > 1.0E-5 (74 samples), with mismatch between reported and predicted sex (7 samples), or incomplete covariate data (7 samples) were excluded. Additionally, probes with median detection p-values < 0.05 were excluded. Array data were normalized via functional normalization, then standardized across Type-I and Type-II probe designs with beta-mixture quantile normalization39. Probes that measured methylation on the X and Y chromosomes, probes that had single nucleotide polymorphisms (SNP) within the binding region, that could cross-hybridize to other regions of the genome40, or probes that had low variability (range of beta-values < 0.05)41 were excluded. After exclusions, 690,781 probes were available from 536 samples for this study. We used gaphunter to flag probes that had outliers or distributional issues that may be related to genetic effects on DNAm measurement42. These data are accessible through NCBI Gene Expression Omnibus (GEO) via accession series GSE128821.
Estimate of Epigenetic Age
We estimated epigenetic age using the online (https://horvath.genetics.ucla.edu/html/dnamage/) [Accessed 01/15/2019] epigenetic clock calculator43. This method utilizes DNAm levels at previously identified CpGs that are predictive of chronological age and has been shown to be highly accurate across a wide range of different cell and tissue types32. This clock also calculates two measures of age acceleration: the difference between epigenetic and chronological age, and the residuals when epigenetic age is regressed on chronological age in a linear model. We investigated the age acceleration residuals and epigenetic age in this study.
Estimates of Tissue Heterogeneity
DNAm differs between cell-types, and cellular heterogeneity presents a likely source of confounding in epigenome-wide association studies of mixed cell samples44. Thus, we estimated the proportions of epithelial, fibroblast, and immune cells (including B-cells, natural killers, CD4+ T-cells, CD8+ T-cells, monocytes, neutrophils, and eosinophils) in our cheek swab samples using reference methylomes45. For 95% of our samples, epithelial cells made up 95.7% of the cells (Supplemental Fig. 1), while immune cells made up the majority of the remaining cell types. Due to very strong inverse correlations between immune cell proportions and epithelial cell proportions (Supplemental Fig. 2), we adjusted for cellular heterogeneity by including the proportions of epithelial cells as covariates in the statistical models.
Statistical Analyses
Latent profile analysis (LPA, Mplus version 8.1) was used to group infants into mutually exclusive categories using 12 NNNS summary scores based on previous work21. Membership in categorical latent profiles that represent heterogeneous subgroups was inferred from the 12 NNNS variables. LPA models with different numbers of profiles were fitted. We identified the model containing the optimal number of profiles using criteria outlined by Nylund et al.46. Determination of the best model fit was assessed via Bayesian information criteria (BIC) adjusted for sample size, whereby the smallest BIC value indicates the best fit as well as minimization of cross classification probabilities, the bootstrapped likelihood ratio test, and the number of cases in each profile. As the number of profiles increased from 2 to 6, the sample-size adjusted BIC values decreased, suggesting improvement in the goodness of fit (Supplemental Table 1). Infants in Profile 6 had a pattern of responses (lowest scores for attention, self-regulation and quality of movement, average scores for lethargy, hypotonicity, nonoptimal reflexes and asymmetric reflexes, combined with the highest scores for arousal, excitability, hypertonicity and the most stress abstinence) that was consistent with a latent profile identified by Liu et al. (2010) that had the most extreme negative scores and was most predictive of nonoptimal developmental outcomes in childhood21. Whereas infants in Profile 1 exhibited the most optimal responses, and provided the greatest contrast to Profile 6, thus we focused on these two profiles for the EWAS and epigenetic age analyses.
Statistical analyses of epigenomic data were performed in R version 3.5. We tested for differences in epigenetic age and age acceleration between the atypical NNNS profile (Profile 6) and optimal NNNS profile (Profile 1) versus those in the other NNNS profiles using robust linear models via the MASS package. Standard errors and p-values for robust regressions were estimated using White’s sandwich estimator to protect against potential heteroscedasticity. Epigenetic age and age acceleration were included as continuous dependent variables, the NNNS profiles were included as a three-level factor for the independent variable, while adjusting for sex, recruitment site, and cellular heterogeneity. The epigenome-wide association study was performed with robust linear regressions for each CpG site that passed QC, regressing methylation beta-values (dependent variable) on the NNNS profiles (independent variable), while adjusting for sex, PMA, and proportions of epithelial cells and fibroblasts. QQ-plots and Manhattan plots were produced using the qqman package. To account for multiple testing, we implemented a false discovery rate (FDR) and considered those associations that were within a 10% FDR to be statistically significant. We report all results from models that yielded suggestive associations (p-value < 0.0001) in the Supplemental Materials.
We performed sensitivity analyses to examine the impacts of other potential confounders on the associations between NNNS profiles and DNAm. For these analyses, we included additional variables in the linear models, and compared the parameter estimates before and after these additional adjustments. We assessed the confounding effects of sample plate as a batch variable (7-level factor), maternal socioeconomic status (2-level factor), maternal educational attainment (2-level factor), proportions of fibroblasts and immune cells (B-cells, natural killers, CD4+ T-cells, CD8+ T-cells, monocytes, neutrophils, and eosinophils), race (white, black, Asian, Hawaiian/Pacific Islander, and other), and birth weight (grams).
To gain insights into the biological functions of the NNNS-associated CpG sites, we performed over-representation analyses with ConsensusPathDB (CPDB)47,48. We utilized CPDB to examine our gene lists for enrichment with neighborhood-based entity sets (NESTs) with a radius of one, pathway-based gene sets from KEGG, Biocarta, and Reactome with minimum overlap with our gene-set of 2, and gene-ontology (GO) terms. Over-representation results within a 10% FDR were determined to be statistically significant. For over-representation analyses, we utilized a gene-list containing the genes annotated to the top 250 CpG sites from the EWAS that were associated with the atypical NNNS profile. We also aimed to examine whether our NNNS-associated CpGs were within genes that have been linked with phenotypes related to neurodevelopment or neurodegeneration. Thus, we annotated the top 250 CpGs with traits that have been linked to genes via the NHGRI-EBI genome-wide association study catalog (GWAS catalog)49.
Results
Study Population and NNNS Profile Results
We identified six distinct NNNS profiles representing groups of infants with similar neurobehavioral responses (Fig. 1 and Table 1); two of these profiles stood out as particularly distinctive. Infants in Profile 1 had the most optimal performance, with the best attention and regulations scores, an average requirement for handling, typical motor tone and movement, and few signs of stress. Infants in Profile 6 showed atypical performance with poor attention, a substantial requirement for handling, poor regulation, exceptionally high arousal and excitability, hypertonia, poor quality of movement, and substantial signs of stress. Thus, Profile 1 represents an optimal profile characterized by generally positive, well-modulated neurobehavioral responses, while Profile 6 represents infants with atypical neurobehavioral responses. These findings are similar to profiles observed previously by others21 and in our own research29. To limit the number of tests being performed, the current study focused on the most optimal (Profile 1) and atypical (Profile 6) profiles, while using the combination of Profiles 2–5 as the referent category in downstream analyses. Average PMA at birth, PMA at buccal cell collection, and maternal age did not substantially differ between the different NNNS profile groupings (Table 2). On the other hand, we did find that a larger proportion of infants with atypical profiles had caregivers with lower socioeconomic status (SES) (16.7%) and lower educational attainment (22.2%), compared to those in the optimal group (4.8% and 6.5% respectively). We also observed significant differences in NNNS profile assignment by recruitment site, and thus recruitment site was controlled for in all downstream analyses.
Figure 1.
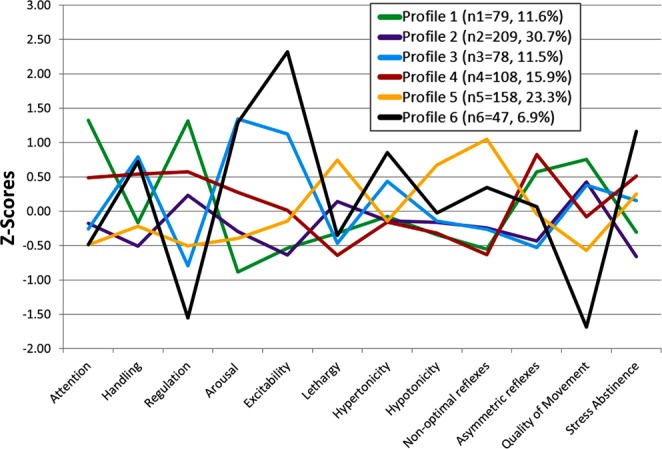
Patterns of NNNS z-scores across individual assessments for all six latent profiles among all NOVI infants that were assessed for the NNNS (N = 679); Profile 6 represents the atypical profile (black) and Profile 1 represents the optimal profile (green).
Table 1.
Means and standard errors of individual NNNS assessment scores across the NNNS profile groupings identified by LPA (N = 536).
| Assessment | Profiles | ANOVA p-val. | |||||
|---|---|---|---|---|---|---|---|
| 1 (n = 62) | 2 (n = 176) | 3 (n = 61) | 4 (n = 83) | 5 (n = 118) | 6 (n = 36) | ||
| Attention | 7.27 (1.14) | 5.04 (1.22) | 4.89 (0.90) | 6.04 (1.18) | 4.43 (1.36) | 4.36 (1.25) | <0.001 |
| Handling | 0.37 (0.25) | 0.29 (0.22) | 0.61 (0.25) | 0.55 (0.22) | 0.37 (0.26) | 0.62 (0.27) | <0.001 |
| Quality of Movement | 5.06 (0.55) | 4.89 (0.45) | 4.9 (0.59) | 4.58 (0.58) | 4.23 (0.49) | 3.4 (0.64) | <0.001 |
| Self Regulation | 6.69 (0.60) | 5.82 (0.45) | 5.04 (0.53) | 6.09 (0.58) | 5.21 (0.51) | 4.44 (0.72) | <0.001 |
| Non-Optimal Reflexes | 4.18 (1.61) | 4.83 (1.52) | 4.84 (1.67) | 4.1 (1.50) | 7.47 (1.70) | 6.22 (1.93) | <0.001 |
| Stress Abstinence | 0.11 (0.06) | 0.09 (0.06) | 0.14 (0.07) | 0.17 (0.06) | 0.16 (0.06) | 0.22 (0.08) | <0.001 |
| Arousal | 3.18 (0.48) | 3.55 (0.37) | 4.59 (0.51) | 3.91 (0.50) | 3.47 (0.50) | 4.61 (0.51) | <0.001 |
| Hypertonicity | 0.34 (0.75) | 0.27 (0.55) | 0.8 (1.15) | 0.25 (0.49) | 0.23 (0.53) | 0.97 (1.18) | <0.001 |
| Hypotonicity | 0.05 (0.22) | 0.15 (0.36) | 0.13 (0.39) | 0.06 (0.29) | 0.52 (0.72) | 0.25 (0.44) | <0.001 |
| Asymmetrical Reflexes | 1.69 (1.52) | 0.43 (0.73) | 0.25 (0.54) | 2.05 (1.51) | 0.94 (1.15) | 0.97 (1.44) | <0.001 |
| Excitability | 1.42 (1.15) | 1.16 (0.93) | 4.77 (1.27) | 2.52 (1.35) | 2.18 (1.13) | 7.22 (1.61) | <0.001 |
| Lethargy | 3.77 (1.58) | 4.8 (1.87) | 3.64 (1.53) | 3.17 (1.58) | 6.17 (2.15) | 3.97 (1.92) | <0.001 |
ANOVA: analysis of variance, LPA: latent profile analysis, NNNS: NICU network neurobehavioral scale.
Table 2.
Means ± standard deviations (continuous) or percentages and frequencies (categorical) of covariates by NNNS profile groupings.
| Sample Characteristics | Profiles 2,3,4,5 (N = 438) | Profile 1 (Optimal) (N = 62) | Profile 6 (Atypical) (N = 36) |
|---|---|---|---|
| Infant Gender: Male | 55.3% (242/438) | 56.5% (35/62) | 61.1% (22/36) |
| Recruitment Site: | |||
| WIH | 19.4% (85/438) | 3.2% (2/62) | 19.4% (7/36) |
| SHD | 22.6% (99/438) | 1.6% (1/62) | 0.0% (0/36) |
| KMC | 16.0% (70/438) | 17.7% (11/62) | 19.4% (7/36) |
| CMH* | 14.8% (65/438) | 8.1% (5/62) | 0.0% (0/36) |
| WFU* | 14.6% (64/438) | 67.7% (42/62) | 22.2% (8/36) |
| LAB* | 12.6% (55/438) | 1.6% (1/62) | 38.9% (14/36) |
| PMA at Buccal Collection (weeks) | 38.930 ± 3.150 | 39.415 ± 4.012 | 40.210 ± 3.462 |
| PMA at Birth (weeks) | 27.091 ± 1.881 | 26.756 ± 1.954 | 26.452 ± 2.154 |
| Birth Weight (grams) | 969.0 ± 281.4 | 903.8 ± 265.0 | 861.9 ± 291.4 |
| Maternal Age at childbirth (years) | 29.171 ± 6.297 | 29.095 ± 6.972 | 27.972 ± 6.396 |
| Maternal Smoking During Pregnancy | 16.3% (71/436) | 14.8% (9/61) | 8.3% (3/36) |
| Education: less than High School/GED | 14.6% (62/426) | 6.5% (4/62) | 22.2% (8/36) |
| Low SES | 8.2% (35/427) | 4.8% (3/62) | 16.7% (6/36) |
| Maternal Race: | |||
| White | 55.0% (236/429) | 52.5% (32/61) | 27.8% (10/36) |
| Black | 21.9% (94/429) | 29.5% (18/61) | 19.4% (7/36) |
| Asian | 7.0% (30/429) | 9.8% (6/61) | 11.1% (4/36) |
| Hawaiian or Pacific Islander | 6.8% (29/429) | 6.6% (4/61) | 13.9% (5/36) |
| Other | 9.3% (40/429) | 1.6% (1/61) | 27.8% (10/36) |
PMA: Postmenstrual Age, SES: socioeconomic status, GED: General Equivalency Diploma, NNNS: NICU network neurobehavioral scale, *enrolled from 2 university-affiliated NICUs.
Epigenetic Age and Age Acceleration Associations with NNNS Profiles
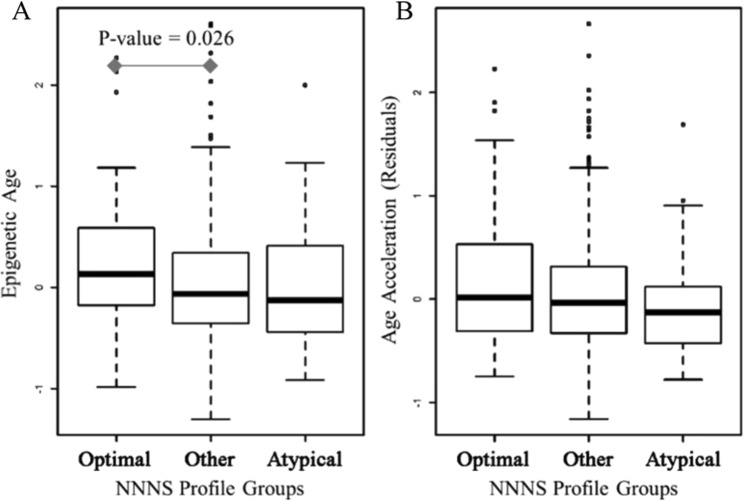
Epigenetic age negatively correlated with gestational age at birth (R2 = 0.06, p-value < 0.0001), but positively correlated with age since birth (R2 = 0.15, p-value < 0.0001) and PMA (R2 = 0.14, p-value < 0.0001) (Supplemental Fig. 3). In a linear model with epigenetic age regressed on both gestational age at birth and PMA at exam date, the relationship with PMA remained strong while the associations with gestational age was heavily attenuated. Thus, PMA appeared to be the appropriate age variable to include in models when testing for age acceleration. We examined differences in epigenetic age and age acceleration that associated with the optimal (n = 62) and atypical (n = 36) NNNS profiles, by comparing them to the rest of the NOVI infants (n = 438). We found that the infants in the optimal profile tended to have significantly older epigenetic age (β1 = 0.201, p-value = 0.026) whereas the atypical profile exhibited no difference in epigenetic age (β1 = −0.022, p-value = 0.84) when compared to the rest of the NOVI infants (Fig. 2A). However, age acceleration did not significantly differ when comparing the optimal or atypical profiles to the rest of the NOVI infants (Fig. 2B). These models were adjusted for sex, recruitment site, postmenstrual age, and estimated proportions of epithelial cells and fibroblast cells.
Figure 2.
Relationships between epigenetic age (A) and age acceleration (B) with NNNS profile groupings; the x-axis includes the optimal profile (Profile 1), the atypical profile (Profile 6), as well as other NOVI infants (Profiles 2–5), while the y-axis represents epigenetic age and age acceleration after adjusting for sex, recruitment site, and cell-mixture.
Epigenome-Wide Association Study of NNNS Profiles
We then performed an EWAS of these two NNNS profiles to examine whether underlying patterns of epigenetic regulation measured in buccal cells differed among infants within the NNNS profiles. We report all results for the EWAS of the optimal and atypical NNNS profiles that yielded associations with p-values < 0.0001 in the Supplemental Materials (Supplemental Tables 2, 3, respectively). Thirty CpGs were differentially methylated (at a 10% FDR) with either of the NNNS profile groupings (Table 3). The only CpG associated with the optimal NNNS profile was cg03046148. Infants with this profile had higher DNAm levels (β1 = 0.0145, p-value = 1.43E-07). On the other hand, we identified 29 epigenetic loci that were associated with the atypical NNNS profile after FDR-adjustment, which were located throughout the genome (Fig. 3). The most statistically significant relationship was observed at cg23172057 (β1 = 0.0299, p-value = 5.43E-08) which is within the body of the coagulation factor X (F10) gene. The magnitudes of effect among the FDR-significant hits tended to be small, with differential methylation ranging between 0.42% to 6.53% lower and between 0.31% to 10.61% higher methylation among the atypical NNNS group. The CpGs with the largest magnitudes of association among the FDR-significant hits were observed at cg14792155 (β1 = 0.1061, p-value = 4.04E-06) which is within the body of the phospholipase A2 group IVE (PLA2G4E) gene and at cg07850633 (β1 = −0.0653, p-value = 1.49E-06) which is within the body of the MACRO domain containing 2 (MACROD2) gene.
Table 3.
Epigenome-wide association study results for CpG sites that yielded associations within a 5% (**) or 10% (*) FDR for either the optimal or atypical NNNS profile groupings; beta coefficients (β1) represent the mean difference in methylation proportion when comparing the optimal or atypical NNNS profiles to the rest of the NOVI sample.
| CpG Annotations | Optimal NNNS Profile | Atypical NNNS Profile | ||||
|---|---|---|---|---|---|---|
| CpG | Location | Gene | β1 | P-value | β1 | P-value |
| cg25755851 | chr1:9335794 | — | −0.013 | 1.89E-01 | −0.032 | 2.53E-06* |
| cg22960767 | chr1:37422679 | GRIK3 (Body) | 0.022 | 1.39E-02 | 0.036 | 3.55E-06* |
| cg26057754 | chr1:183774231 | RGL1 (Body) | −0.001 | 7.00E-01 | −0.004 | 7.38E-07* |
| cg23264395 | chr1:207096239 | FCMR (TSS1500) | 0.001 | 8.50E-01 | 0.034 | 2.48E-06* |
| cg01479768 | chr2:731298 | — | −0.006 | 6.91E-01 | 0.045 | 2.20E-06* |
| cg08902894 | chr2:3142407 | — | 0.002 | 7.22E-01 | 0.029 | 1.10E-07* |
| cg05696361 | chr2:107108978 | — | 0.003 | 7.78E-01 | 0.048 | 2.79E-07* |
| cg17505883 | chr2:130552292 | — | 0.006 | 2.59E-01 | 0.021 | 1.94E-06* |
| cg03046148 | chr3:13695666 | LOC285375 (Body) | 0.015 | 1.43E-07* | 0.003 | 4.56E-01 |
| cg07193729 | chr4:176031198 | — | 0.006 | 5.03E-02 | 0.014 | 3.07E-07* |
| cg02236672 | chr5:132449216 | — | −0.001 | 9.31E-01 | 0.037 | 9.98E-07* |
| cg00210856 | chr5:158466226 | EBF1 (Body) | 0.001 | 9.23E-01 | 0.028 | 4.29E-07* |
| cg02057469 | chr7:95951623 | SLC25A13 (TSS200) | −0.001 | 1.19E-01 | −0.004 | 3.05E-06* |
| cg04524088 | chr7:127847835 | MIR129-1 (TSS200) | −0.016 | 1.68E-01 | 0.045 | 2.27E-06* |
| cg17287134 | chr7:154959606 | — | −0.008 | 4.52E-01 | 0.040 | 1.01E-06* |
| cg21672855 | chr8:135614777 | ZFAT (Body) | −0.002 | 1.73E-01 | 0.005 | 1.71E-06* |
| cg14632902 | chr9:139017648 | — | 0.012 | 1.91E-02 | 0.026 | 3.49E-06* |
| cg06846137 | chr10:131682939 | EBF3 (Body) | 0.004 | 4.20E-01 | 0.019 | 1.36E-06* |
| cg13716458 | chr11:28997975 | — | 0.006 | 6.38E-02 | 0.020 | 5.51E-07* |
| cg07895260 | chr12:55537168 | — | −0.001 | 9.37E-01 | 0.034 | 3.19E-06* |
| cg27361636 | chr12:120502417 | BICDL1 (Body) | −0.001 | 9.20E-01 | 0.044 | 2.93E-06* |
| cg23172057 | chr13:113800351 | F10 (Body) | 0.010 | 1.55E-01 | 0.030 | 5.43E-08** |
| cg11042421 | chr14:42881184 | — | 0.006 | 4.29E-02 | 0.015 | 2.71E-06* |
| cg14354244 | chr14:51446038 | TRIM9 (Body) | 0.006 | 5.04E-01 | 0.034 | 3.66E-06* |
| cg03444659 | chr14:94834215 | SERPINA2 (TSS1500) | −0.002 | 7.14E-01 | 0.015 | 1.43E-06* |
| cg14792155 | chr15:42289618 | PLA2G4E (Body) | −0.007 | 7.50E-01 | 0.106 | 4.04E-06* |
| cg02187389 | chr16:1247777 | CACNA1H (Body) | 0.000 | 9.97E-01 | 0.024 | 7.96E-07* |
| cg02234314 | chr19:55986224 | ZNF628 (TSS1500) | 0.003 | 7.85E-01 | 0.059 | 2.08E-06* |
| cg07850633 | chr20:15795880 | MACROD2 (Body) | 0.017 | 3.49E-01 | −0.065 | 1.49E-06* |
| cg09772858 | chr22:49549729 | — | 0.006 | 4.15E-01 | 0.030 | 2.52E-06* |
CpG = cytosine-phosphate-guanine methylation site, FDR = False Discover Rate, NNNS = NICU Network Neurobehavioral Scale; For CpGs with no annotated genes, we annotated this with the nearest genes within 2500 bp of the CpG.
Figure 3.
Manhattan plot of epigenetic loci associated with the atypical NNNS profile; the x-axis represents the genomic location of the individual probes and the y-axis represents the -log10(p-values) from related to the Atypical NNNS profile, adjusted for sex, recruitment site, postmenstrual age, and cell-mixture; gene annotations for the CpGs yielding associations within the 10% FDR threshold have been added to the plot.
Based on observed differences in SES, maternal educational attainment, and maternal smoking during pregnancy among those with optimal or atypical NNNS profiles, as well as hypothesized potential confounding effects of batch (sample plate), immune cell proportions, race, and birth weight, we performed a sensitivity analyses with additional adjustment for these variables in our linear models. Additional adjustments for these potential confounders did not alter the observed associations between DNAm and atypical NNNS profiles (Supplemental Fig. 4).
Functional and Phenotype Enrichment
We then used enrichment analyses to examine whether the genes annotated to NNNS-associated CpGs have a higher than expected proportion of genes that interact with each other, are involved in known biological pathways, or are linked to specific gene ontology terms. For this analysis, we utilized the top 250 CpGs that associated with the atypical NNNS profiles. We found that this gene-set was enriched for one neighborhood-based entity sets (NESTs) (FDR q-value = 0.031), centered on the CRIM1 gene which has physical interactions with four genes, two of which were also in our gene-set: ATXN7 and MEGF6. We also identified 54 pathway-based gene-sets (Supplemental Table 4) many of which may be relevant for neurodevelopment, including synaptic activity, neurotransmitters, and nerve growth factors (Table 4). Additionally, our gene-set was enriched for nine gene-ontology (G0) terms (Supplemental Table 5), including neuron projection (FDR q-value = 0.0371) and neuron part (FDR q-value = 0.0505). Multiple pathway and GO-term enrichments included GRIK3, TRIM9, and PLA2G4E, genes that were annotated to CpGs that yielded FDR-significant associations from our EWAS.
Table 4.
Pathways involved in neurodevelopment and/or neuronal activity that were significantly (FDR < 0.10) over-represented among the genes annotated to the top 250 CpGs that associated with the Atypical NNNS profile.
| Pathway ID | Pathway Description | Total Genes | NNNS-Associated Genes | P-val. | FDR Q-val. |
|---|---|---|---|---|---|
| path:hsa04724 | Glutamatergic synapse - Homo sapiens (human) | 114 | PLCB1; GRIK3; PRKCB; PLA2G4E; ADCY3 | 0.0020 | 0.026 |
| R-HSA-112314 | Neurotransmitter receptors and postsynaptic signal transmission | 152 | PLCB1; GRIK3; AP2A2; PRKCB; ADCY3 | 0.0069 | 0.040 |
| R-HSA-416993 | Trafficking of GluR2-containing AMPA receptors | 17 | PRKCB; AP2A2 | 0.0077 | 0.040 |
| R-HSA-112316 | Neuronal System | 367 | PLCB1; PRKCB; PTPRS; KCNK9; ADCY3; GRIK3; AP2A2; KCNA3 | 0.0082 | 0.041 |
| ngfpathway | nerve growth factor pathway (ngf) | 18 | PLCG1; NGFR | 0.0086 | 0.043 |
NNNS = NICU Network Neurobehavioral Scale; FDR = False Discover Rate.
Since only one of our CpGs yielded an association with the NNNS profiles at a stricter threshold of statistical significance (5% FDR), we focused on this CpG for follow-up analyses examining whether the atypical NNNS profile exhibited differential methylation with each of the NNNS profiles. We regressed cg23172057 DNAm levels on a six level factor variable (using the atypical profile as the referent) while adjusting for sex, site, PMA, proportions of epithelial cells, and proportions of fibroblasts. The atypical NNNS profile exhibited significantly higher DNAm (p-values < 0.05) than each of the NNNS profiles, with the most substantial differential DNAm when comparing to Profile 3 (4.0% lower; p-value = 1.242E-07) and Profile 5 (3.4% lower; p-value = 9.76E-08) (Supplemental Fig. 5).
CpG Annotation
Relatively few genes have been studied for their associations with neonatal neurobehavioral characteristics. However, it is plausible that the genes that are linked to cognition, neurobehavior, or neurodegeneration at other life stages may also be important in neurobehavioral function in very early life. We identified phenotypes or traits that have been associated with the genes annotated to the CpGs associated with NNNS profiles at a 10% FDR (Table 5). Importantly, Table 5 also presents 7 of the 11 genes that have been linked to neurobehavioral or neurodegenerative traits including autism, attention deficit hyperactivity disorder, cognitive impairment, depression, and psychosis.
Table 5.
Genes annotated to our atypical NNNS-associated CpGs that have been linked to traits from the GWASdb.
| Gene | N | Trait Types | Specific Cognitive, Neurobehavioral or Neurological Traits |
|---|---|---|---|
| F10 | 4 | Hematopoietic | — |
| EBF1 | 34 | Hematopoietic, Cardiovascular, Immune, Growth & Metabolism, Birth Outcomes, Neurobehavior, Cognition | Psychosocial stress measurement, cognitive impairment, cognitive decline measurement |
| RGL1 | 7 | Neurobehavior, Cardiovascular, Environmental Exposures, Hematopoietic, Liver | Attention deficit hyperactivity disorder, conduct disorder, schizophrenia, response to antipsychotic drug, response to antidepressant |
| CACNA1H | 1 | Hematopoietic | — |
| EBF3 | 3 | Cancer, Bone, Neurological, Sleep | Peripheral neuropathy |
| SERPINA2 | 6 | Growth, Respiratory, Cardiovascular, Hematopoietic | — |
| MACROD2 | 31 | Neurobehavior, Neurological, Liver, Growth & Metabolism, Hematopoietic, Bone, Birth Outcomes, Respiratory, Cardiovascular, Environmental Exposures, Bone, Immune | Autism, eating disorder, brain connectivity measurement, sporadic amyotrophic lateral sclerosis, prion disease, mood disorder |
| ZFAT | 12 | Growth & Metabolism, Cardiovascular, Cognitive, Environmental Exposures, Immune, Respiratory, Other | Self-reported educational attainment |
| GRIK3 | 6 | Neurobehavior, Immune, Respiratory, Environmental Exposures | Unipolar depression, neuroticism measurement, depressive symptom |
| TRIM9 | 5 | Growth & Metabolism, Neurobehavior, Immune, Cancer | Psychosis |
| PLA2G4E | 1 | Environmental Exposures | — |
N = number of traits linked to that gene.
Discussion
Our study focused on a comparison of neurobehavioral profiles with DNAm levels and epigenetic age. We used the NNNS summary scores to identify a group of infants with an optimal profile and a group with an atypical profile, which are similar to what has been observed previously by others and in our own research including preterm infants21,24,29. The infants in the atypical profile had the lowest scores for attention, self-regulation and quality of movement, average scores for lethargy, hypotonicity, nonoptimal reflexes and asymmetric reflexes, combined with the highest scores for arousal, excitability, hypertonicity and the most stress abstinence. This pattern of NNNS responses in our atypical profile was consistent with a latent profile identified by Liu et al. (2010) that was most predictive of non-optimal developmental outcomes in childhood21. Whereas the infants in our optimal profiles had the most positive responses across the NNNS and provided the greatest contrast to compare with the atypical profile. We found that very premature infants in the NOVI cohort with an optimal neurobehavioral profile had older epigenetic age than other very premature infants. We also found that age acceleration followed a stepwise trend in which infants with the optimal profile had the greatest age acceleration and infants with the atypical profile had the least age acceleration, though this finding was not statistically significant and may be driven by the differences in age at exam across the NNNS profile groups. Epigenetic age is an estimate of the state of underlying physiologic processes, as they relate to biological development and maintenance32, and has been studied in the context of health conditions that are linked to the aging process, including frailty50, physical capability51,52, cognitive fitness51, decreased cognitive function and neuropathologies in persons suffering from Alzheimer’s Disease53, and all-cause mortality54. In these studies, age acceleration is analogous to biological decline.
These relationships may differ in early life, however, when children are still undergoing substantial growth and development, and it is unclear whether accelerated aging would be expected to be associated with positive or negative developmental characteristics. In fact, if epigenetic aging captures, or is a surrogate for, the activity of developmental processes, epigenetic age acceleration throughout early development may be an indicator through which to track developmental “catch-up”. An epigenetic clock has also been developed to estimate epigenetic gestational age acceleration from cord blood DNAm55. Interestingly, gestational age acceleration has been associated with reduced infant respiratory morbidities56, which provides some evidence that older epigenetic age in infancy may correlate with positive developmental characteristics. In our analysis, we did not observe a significant relationships between age acceleration with the optimal or atypical NNNS profiles.
A handful of studies have examined the relationships between epigenetic age and development in children. For instance, epigenetic age positively correlates with measures of physical development such as fat mass, height, Tanner stages33, pubertal development, internalization and thought problems34, and increased cortisol34,35. One study observed interrelationships between age acceleration, cortisol production, and hippocampal volume, potentially linking hypothalamic-pituitary-adrenal (HPA) axis activity, neuroanatomy, and epigenetic aging35. This topic requires additional study and should ideally be investigated in a longitudinal manner, in which both epigenetic age and neurobehavioral assessments are tracked in parallel through early-life development.
Though the infants with the atypical NNNS profile did not have significantly different epigenetic age from the other NOVI infants, we did identify multiple differentially methylated CpGs throughout the genome that were associated with the group with atypical neurobehavioral responses. Our EWAS revealed 29 epigenetic loci that significantly associated (<10% FDR) with the atypical profile. The CpG with the smallest p-value was annotated to the (cg2317205) F10 gene and was the only CpG that yielded an association at a stricter threshold for statistical significance (<5% FDR). We also found that the average DNAm at cg23172057 was significantly higher among the atypical NNNS profile even when comparing to each of the 5 other NNNS profiles individually. Thus, this CpG holds promise as a potential marker of problematic neurobehavioral responses in preterm infants. However, the gene that this CpG is annotated to (F10) has not been associated with behavioral or cognitive function. Instead, F10 is involved in the blood coagulation cascade and primarily associated with hematologic disorders57. On the other hand, the other 28 CpGs only met a 10% FDR threshold of statistical significance, and likely do include some false positives, but a number of the genes annotated to these CpGs have been linked to cognition or educational attainment (EBF1 & ZFAT), and neurobehavioral or neurological disorders in GWAS studies (EBF1, RGL1, EBF3, MACROD2, GRIK3, and TRIM9). We also found that the top 250 CpGs from this analysis were enriched for genes within pathways involving neurotransmitters and synaptic activity, as well as GO-terms related to neuron projection and structure. Of particular note, three genes annotated to the FDR-significant CpGs were within these neuronal-associated GO-terms and pathways: phospholipase A2 group IVE (PLAG2G4E), glutamate ionotropic receptor kainate type subunit 3 (GRIK3), and tripartite motif containing 9 (TRIM9). The CpG site that yielded the largest magnitude of association among our statistically significant hits, cg14792155, was within the body of the PLA2G4E gene. This gene encodes for a calcium-dependent N-acyltransferase; experimental mouse models have implicated that it likely plays a critical role in endocannabinoid signaling in the nervous system58 and thus differential regulation of this gene has implications for neurodevelopment and neurodegenerative disorders59. In human observational studies, placental CpGs within PLA2G4E have been observed to be differentially methylated in association with extremely preterm births60 and genetic variants within the PLA2G4E gene have been implicated as a potential risk factor for neurodevelopmental problems such as panic disorder61. The protein encoded by GRIK3 is involved in presynaptic neurotransmission, and has been associated with developmental delay62, schizophrenia63, obsessive-compulsive disorder64, and depression65. The protein encoded by TRIM9 regulates axon guidance and neural outgrowth66,67, while deletion of TRIM9 has been associated with structural and functional abnormalities and impaired learning and memory in mice68. The CpG site that yielded the largest statistically significant inverse association, cg07850633, was within the body of the MACRO domain-containing protein 2 (MACROD2) gene; which has been associated with autism spectrum disorder (ASD)69, though other studies have yielded potentially contradictory evidence70, autism-like traits71and has been implicated in other neurological disorders72,73, and temporal lobe volume74. Variants in EBF3, which encode for the early B Cell Factor 3, may contribute to developmental delay and intellectual impairment75. Genetic variation in the Ral Guanine Nucleotide Dissociation Stimulator Like 1 (RGL1) gene, has been associated with attention76 and conduct problems among children with attention deficit hyperactivity disorder77. Additionally, genetic mutations within CACNA1H, which encodes for a subunit of a voltage gated calcium channel, lead to decreased calcium channel activity in neuronal cells, and have been linked to ASD78, to epilepsy79, and to amyotrophic lateral sclerosis80. Overall, these findings suggest that our NNNS-associated epigenetic variations occurred at numerous genomic regions with recognized roles in neurodevelopmental or neurodegenerative disorders. Interestingly, Sparrow et al. (2016) performed an EWAS of preterm birth using saliva samples to identify a number of genes that were differentially methylated in association with preterm birth that are also involved in neuronal function and/or neurobehavioral traits81. Their findings lead them to speculate that preterm-associated variations in DNAm may contribute to the neural and behavioral phenotypes that are linked to preterm birth. Thus, differential epigenetic regulation in babies that are born preterm may provide a link between preterm birth and poorer neurodevelopmental outcomes.
There were some limitations to this study. We used a false discovery rate of 10% to identify significantly differentially methylated CpG sites. Only one of our models yielded an FDR < 5%, and none of the models would survive Bonferroni adjustment. Thus, it is probable that some of identified epigenetic loci are false-positives. We encourage additional investigation of infant DNAm, epigenetic age, and neurobehavior to determine whether similar relationships can be observed in independent populations. There is also the possibility of residual confounding, though our findings were robust to adjustments for numerous potential confounders. We utilized buccal cells as a surrogate tissue to examine the relationships between neurobehavioral profiles and DNAm, as it is not possible to perform such examinations in the neuronal tissues. However, for studies of children, buccal cell collection leads to greater compliance82, and evidenced here in our 93% consent rate for parents who gave overall consent for NOVI. Recent evidence also suggests that buccal samples may be very appropriate for epigenetic analyses of neurodevelopmental outcomes, as they arise from the same germ cell layer as the brain and thus may share similar early epigenetic patterning and susceptibility83–85, and have demonstrated DNAm variability associated with later neurobehavioral outcomes.
We nevertheless remain cautious in the interpretation of these observations in terms of mechanism. These data were collected and analyzed cross-sectionally, so we cannot infer directionality of the observed relationships between NNNS profiles with DNAm or epigenetic age. It is notable, however, that the atypical profile observed by us and others in different populations21,24,29, has also been related to differential DNAm in other tissues29 and predicted developmental outcomes in childhood21. Thus, it is possible that the combination of epigenetic measures and NNNS profiles may lead to the early identification of which individual children are most at risk for adverse developmental outcome. Longitudinal studies of epigenomics and neurobehavioral outcomes are needed to establish whether epigenetic variations are detectable prior to the presentation of neurobehavioral impairments, and to examine whether and how these potential predictors vary throughout early life development.
Conclusions
We found that among very preterm infants (<30 weeks PMA), those with an optimal neurobehavioral profile had slightly older epigenetic age, while infants with a poorly regulated neurobehavioral profile had differentially methylated CpGs at multiple genes linked to neural structure, function, or different neurobehavioral or neurodegenerative conditions. These relationships were detected using buccal cell DNAm, building upon the existing evidence that buccal cells may be a suitable surrogate tissue for studying neurobehavioral conditions in human observational studies. One CpG within the F10 gene had the strongest association (<5% FDR) with the NNNS, while three other CpGs (<10% FDR) were within genes yielding multiple levels of evidence for plausible roles in neurobehavioral health, annotated to PLA2G4E, TRIM9, and GRIK3, all of which were among the significantly enriched GO-terms or neuronal pathways, and linked to neurobehavioral disorders. The combination of epigenomics and neurobehavior holds promise for a personalized medicine approach to the early detection of children most at risk for poor developmental outcome.
Supplementary information
Acknowledgements
We would like to thank the Emory & WIH lab teams, NOVI Study Coordinators and NNNS examiners that made this work possible. This work was supported by NIH Grants NICHD R01HD072267 (Lester and O’Shea), R01HD084515 and UH3OD023347 (Lester and Marsit).
Author Contributions
C.J.M., T.M.O. and B.M.L. initiated, acquired the funding for, and designed this investigation. K.H. and J.F.P. coordinated laboratory analyses. B.S.C., J.H., J.A.H., E.C.M., C.R.N., S.L.P., L.M.S., A.S. and S.A.D. coordinated data collection. T.M.E. and A.B. performed the statistical analyses. T.M.E., C.J.M., B.M.L., S.A.D. and L.M.D. drafted the manuscript. All authors contributed to interpretation of the results, revisions to the manuscript, and approval of the final version.
Data Availability
The microarray data generated and/or analyzed in the current study are available in the NCBI GEO [Accession series GSE128821]. R codes used for the analyses presented in the paper are available upon request to the corresponding author.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-42654-4.
References
- 1.Purisch SE, Gyamfi-Bannerman C. Epidemiology of preterm birth. Semin Perinatol. 2017;41:387–391. doi: 10.1053/j.semperi.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 2.Martin JA. Preterm births - United States, 2007. MMWR Surveill Summ. 2011;60(Suppl):78–79. [PubMed] [Google Scholar]
- 3.Institute of Medicine. Preterm Birth: Causes, Consequences and Prevention. (National Academic Press, 2006).
- 4.Hack M, et al. Poor Predictive Validity of the Bayley Scales of Infant Development for Cognitive Function of Extremely Low Birth Weight Children at School Age. Pediatrics. 2005;116:333–341. doi: 10.1542/peds.2005-0173. [DOI] [PubMed] [Google Scholar]
- 5.Aarnoudse-Moens CS, Weisglas-Kuperus N, van Goudoever JB, Oosterlaan J. Meta-analysis of neurobehavioral outcomes in very preterm and/or very low birth weight children. Pediatrics. 2009;124:717–728. doi: 10.1542/peds.2008-2816. [DOI] [PubMed] [Google Scholar]
- 6.Vohr BR, Wright LL, Poole WK, McDonald SA. Neurodevelopmental outcomes of extremely low birth weight infants <32 weeks’ gestation between 1993 and 1998. Pediatrics. 2005;116:635–643. doi: 10.1542/peds.2004-2247. [DOI] [PubMed] [Google Scholar]
- 7.Aylward G. Neurodevelopmental Outcomes of Infants Born Prematurely. Journal of Developmental and Behavioral Pediatrics. 2005;25:427–440. doi: 10.1097/01.DBP.0000452240.39511.d4. [DOI] [PubMed] [Google Scholar]
- 8.Grunau RE, Whitfield MF, Davis C. Pattern of learning disabilities in children with extremely low birth weight and broadly average intelligence. Arch Pediatr Adolesc Med. 2002;156:615–620. doi: 10.1001/archpedi.156.6.615. [DOI] [PubMed] [Google Scholar]
- 9.Hack M, et al. Behavioral outcomes and evidence of psychopathology among very low birth weight infants at age 20 years. Pediatrics. 2004;114:932–940. doi: 10.1542/peds.2003-1017-L. [DOI] [PubMed] [Google Scholar]
- 10.Hille ET, et al. Social lifestyle, risk-taking behavior, and psychopathology in young adults born very preterm or with a very low birthweight. J Pediatr. 2008;152:793–800. doi: 10.1016/j.jpeds.2007.11.041. [DOI] [PubMed] [Google Scholar]
- 11.Taylor HG, Klein N, Hack M. School-age consequences of birth weight less than 750 g: a review and update. Dev Neuropsychol. 2000;17:289–321. doi: 10.1207/S15326942DN1703_2. [DOI] [PubMed] [Google Scholar]
- 12.Hack M, et al. Behavioral outcomes of extremely low birth weight children at age 8 years. J Dev Behav Pediatr. 2009;30:122–130. doi: 10.1097/DBP.0b013e31819e6a16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephens BE, Vohr BR. Neurodevelopmental outcome of the premature infant. Pediatr Clin North Am. 2009;56:631–646. doi: 10.1016/j.pcl.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Allen MC. Neurodevelopmental outcomes of preterm infants. Curr Opin Neurol. 2008;21:123–128. doi: 10.1097/WCO.0b013e3282f88bb4. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt B, et al. Impact of bronchopulmonary dysplasia, brain injury, and severe retinopathy on the outcome of extremely low-birth-weight infants at 18 months: results from the trial of indomethacin prophylaxis in preterms. JAMA. 2003;289:1124–1129. doi: 10.1001/jama.289.9.1124. [DOI] [PubMed] [Google Scholar]
- 16.Lester, B. & Tronick, E. The Neonatal Intensive Care Unit Network Neurobehavioral Scale. Pediatrics113 (Suppl. 3 Pt. 2), 631–695, PubMed PMID: 14993524 (2004). [PubMed]
- 17.Pineda RG, et al. Patterns of altered neurobehavior in preterm infants within the neonatal intensive care unit. J Pediatr. 2013;162:470–476 e471. doi: 10.1016/j.jpeds.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coleman MB, et al. Neonatal neurobehavioral abnormalities and MRI brain injury in encephalopathic newborns treated with hypothermia. Early human development. 2013;89:733–737. doi: 10.1016/j.earlhumdev.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheong Jeanie Ling Yoong, Thompson Deanne Kim, Olsen Joy Elizabeth, Spittle Alicia Jane. Late preterm births: New insights from neonatal neuroimaging and neurobehaviour. Seminars in Fetal and Neonatal Medicine. 2019;24(1):60–65. doi: 10.1016/j.siny.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Brown Nisha C., Inder Terrie E., Bear Merilyn J., Hunt Rod W., Anderson Peter J., Doyle Lex W. Neurobehavior at Term and White and Gray Matter Abnormalities in Very Preterm Infants. The Journal of Pediatrics. 2009;155(1):32-38.e1. doi: 10.1016/j.jpeds.2009.01.038. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, et al. Neonatal neurobehavior predicts medical and behavioral outcome. Pediatrics. 2010;125:e90–98. doi: 10.1542/peds.2009-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lester BM, et al. Infant neurobehavioral dysregulation: behavior problems in children with prenatal substance exposure. Pediatrics. 2009;124:1355–1362. doi: 10.1542/peds.2008-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donauer S, et al. Prenatal exposure to polybrominated diphenyl ethers and polyfluoroalkyl chemicals and infant neurobehavior. J Pediatr. 2015;166:736–742. doi: 10.1016/j.jpeds.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sucharew H, Khoury JC, Xu Y, Succop P, Yolton K. NICU Network Neurobehavioral Scale profiles predict developmental outcomes in a low-risk sample. Paediatric and perinatal epidemiology. 2012;26:344–352. doi: 10.1111/j.1365-3016.2012.01288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tendl KA, et al. DNA methylation pattern of CALCA in preterm neonates with bacterial sepsis as a putative epigenetic biomarker. Epigenetics. 2013;8:1261–1267. doi: 10.4161/epi.26645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chau CM, et al. Neonatal pain and COMT Val158Met genotype in relation to serotonin transporter (SLC6A4) promoter methylation in very preterm children at school age. Frontiers in behavioral neuroscience. 2014;8:409. doi: 10.3389/fnbeh.2014.00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Provenzi L, et al. Pain-related stress during the Neonatal Intensive Care Unit stay and SLC6A4 methylation in very preterm infants. Frontiers in behavioral neuroscience. 2015;9:99. doi: 10.3389/fnbeh.2015.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lester BM, Marsit CJ. Epigenetic mechanisms in the placenta related to infant neurodevelopment. Epigenomics. 2018;10:321–333. doi: 10.2217/epi-2016-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lester BM, et al. Neurobehavior related to epigenetic differences in preterm infants. Epigenomics. 2015;7:1123–1136. doi: 10.2217/epi.15.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montirosso R, et al. Serotonin Transporter Gene (SLC6A4) Methylation Associates With Neonatal Intensive Care Unit Stay and 3-Month-Old Temperament in Preterm Infants. Child development. 2016;87:38–48. doi: 10.1111/cdev.12492. [DOI] [PubMed] [Google Scholar]
- 31.Paquette AG, et al. Regions of variable DNA methylation in human placenta associated with newborn neurobehavior. Epigenetics. 2016;11:603–613. doi: 10.1080/15592294.2016.1195534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nature reviews. Genetics. 2018;19:371–384. doi: 10.1038/s41576-018-0004-3. [DOI] [PubMed] [Google Scholar]
- 33.Simpkin AJ, et al. The epigenetic clock and physical development during childhood and adolescence: longitudinal analysis from a UK birth cohort. International journal of epidemiology. 2017;46:549–558. doi: 10.1093/ije/dyw307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suarez A, et al. The epigenetic clock and pubertal, neuroendocrine, psychiatric, and cognitive outcomes in adolescents. Clinical epigenetics. 2018;10:96. doi: 10.1186/s13148-018-0528-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis EG, et al. Accelerated DNA methylation age in adolescent girls: associations with elevated diurnal cortisol and reduced hippocampal volume. Translational psychiatry. 2017;7:e1223. doi: 10.1038/tp.2017.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walden RV, et al. Major congenital anomalies place extremely low birth weight infants at higher risk for poor growth and developmental outcomes. Pediatrics. 2007;120:e1512–1519. doi: 10.1542/peds.2007-0354. [DOI] [PubMed] [Google Scholar]
- 37.Hollinshead, A. B. Four factor index of social status., Unpublished manuscript, Yale University, New Haven, CT (1975).
- 38.Lester BM, Tronick EZ, Brazelton TB. The Neonatal Intensive Care Unit Network Neurobehavioral Scale procedures. Pediatrics. 2004;113:641–667. [PubMed] [Google Scholar]
- 39.Teschendorff AE, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics (Oxford, England) 2013;29:189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pidsley R, et al. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome biology. 2016;17:208. doi: 10.1186/s13059-016-1066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Logue MW, et al. The correlation of methylation levels measured using Illumina 450 K and EPIC BeadChips in blood samples. Epigenomics. 2017;9:1363–1371. doi: 10.2217/epi-2017-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andrews SV, Ladd-Acosta C, Feinberg AP, Hansen KD, Fallin MD. “Gap hunting” to characterize clustered probe signals in Illumina methylation array data. Epigenetics & chromatin. 2016;9:56. doi: 10.1186/s13072-016-0107-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horvath S. DNA methylation age of human tissues and cell types. Genome biology. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome biology. 2014;15:R31. doi: 10.1186/gb-2014-15-2-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng SC, et al. A novel cell-type deconvolution algorithm reveals substantial contamination by immune cells in saliva, buccal and cervix. Epigenomics. 2018;10:925–940. doi: 10.2217/epi-2018-0037. [DOI] [PubMed] [Google Scholar]
- 46.Nylund K, Asparouhov T, Muthén B. Deciding on the Number of Classes in Latent Class Analysis and Growth Mixture Modeling: A Monte Carlo Simulation Study. Structural Equation Modeling: A Multidisciplinary Journal. 2007;14:535–569. doi: 10.1080/10705510701575396. [DOI] [Google Scholar]
- 47.Kamburov A, et al. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Research. 2011;39:D712–D717. doi: 10.1093/nar/gkq1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kamburov A, Wierling C, Lehrach H, Herwig R. ConsensusPathDB—a database for integrating human functional interaction networks. Nucleic Acids Research. 2009;37:D623–D628. doi: 10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MacArthur J, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Research. 2017;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Breitling LP, et al. Frailty is associated with the epigenetic clock but not with telomere length in a German cohort. Clinical epigenetics. 2016;8:21. doi: 10.1186/s13148-016-0186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marioni RE, et al. The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. International journal of epidemiology. 2015;44:1388–1396. doi: 10.1093/ije/dyu277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simpkin AJ, et al. Are objective measures of physical capability related to accelerated epigenetic age? Findings from a British birth cohort. BMJ open. 2017;7:e016708. doi: 10.1136/bmjopen-2017-016708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levine ME, Lu AT, Bennett DA, Horvath S. Epigenetic age of the pre-frontal cortex is associated with neuritic plaques, amyloid load, and Alzheimer’s disease related cognitive functioning. Aging. 2015;7:1198–1211. doi: 10.18632/aging.100864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marioni RE, et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome biology. 2015;16:25. doi: 10.1186/s13059-015-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Knight AK, et al. An epigenetic clock for gestational age at birth based on blood methylation data. Genome biology. 2016;17:206. doi: 10.1186/s13059-016-1068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Knight AK, et al. Relationship between Epigenetic Maturity and Respiratory Morbidity in Preterm Infants. J Pediatr. 2018;198:168–173 e162. doi: 10.1016/j.jpeds.2018.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Menegatti M, Peyvandi F. Factor X deficiency. Semin Thromb Hemost. 2009;35:407–415. doi: 10.1055/s-0029-1225763. [DOI] [PubMed] [Google Scholar]
- 58.Ogura Y, Parsons WH, Kamat SS, Cravatt BF. A calcium-dependent acyltransferase that produces N-acyl phosphatidylethanolamines. Nature chemical biology. 2016;12:669–671. doi: 10.1038/nchembio.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Viveros MP, et al. The endocannabinoid system in critical neurodevelopmental periods: sex differences and neuropsychiatric implications. Journal of psychopharmacology (Oxford, England) 2012;26:164–176. doi: 10.1177/0269881111408956. [DOI] [PubMed] [Google Scholar]
- 60.Tilley SK, et al. Placental CpG methylation of infants born extremely preterm predicts cognitive impairment later in life. PloS one. 2018;13:e0193271. doi: 10.1371/journal.pone.0193271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morimoto Y, et al. Whole-exome sequencing and gene-based rare variant association tests suggest that PLA2G4E might be a risk gene for panic disorder. Translational psychiatry. 2018;8:41. doi: 10.1038/s41398-017-0088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takenouchi T, et al. 1p34.3 deletion involving GRIK3: Further clinical implication of GRIK family glutamate receptors in the pathogenesis of developmental delay. American journal of medical genetics. Part A. 2014;164a:456–460. doi: 10.1002/ajmg.a.36240. [DOI] [PubMed] [Google Scholar]
- 63.Dai D, et al. Meta-analyses of 10 polymorphisms associated with the risk of schizophrenia. Biomedical reports. 2014;2:729–736. doi: 10.3892/br.2014.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Delorme R, et al. Frequency and transmission of glutamate receptors GRIK2 and GRIK3 polymorphisms in patients with obsessive compulsive disorder. Neuroreport. 2004;15:699–702. doi: 10.1097/00001756-200403220-00025. [DOI] [PubMed] [Google Scholar]
- 65.Schiffer HH, Heinemann SF. Association of the human kainate receptor GluR7 gene (GRIK3) with recurrent major depressive disorder. American journal of medical genetics. Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2007;144b:20–26. doi: 10.1002/ajmg.b.30374. [DOI] [PubMed] [Google Scholar]
- 66.Menon S, et al. The E3 Ubiquitin Ligase TRIM9 Is a Filopodia Off Switch Required for Netrin-Dependent Axon Guidance. Developmental cell. 2015;35:698–712. doi: 10.1016/j.devcel.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Plooster M, et al. TRIM9-dependent ubiquitination of DCC constrains kinase signaling, exocytosis, and axon branching. Molecular biology of the cell. 2017;28:2374–2385. doi: 10.1091/mbc.E16-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Winkle CC, et al. Trim9 Deletion Alters the Morphogenesis of Developing and Adult-Born Hippocampal Neurons and Impairs Spatial Learning and Memory. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2016;36:4940–4958. doi: 10.1523/jneurosci.3876-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anney R, et al. A genome-wide scan for common alleles affecting risk for autism. Human molecular genetics. 2010;19:4072–4082. doi: 10.1093/hmg/ddq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Curran S, et al. No association between a common single nucleotide polymorphism, rs4141463, in the MACROD2 gene and autism spectrum disorder. American journal of medical genetics. Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2011;156b:633–639. doi: 10.1002/ajmg.b.31201. [DOI] [PubMed] [Google Scholar]
- 71.Jones RM, et al. MACROD2 gene associated with autistic-like traits in a general population sample. Psychiatric genetics. 2014;24:241–248. doi: 10.1097/ypg.0000000000000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lionel AC, et al. Rare copy number variation discovery and cross-disorder comparisons identify risk genes for ADHD. Science translational medicine. 2011;3:95ra75. doi: 10.1126/scitranslmed.3002464. [DOI] [PubMed] [Google Scholar]
- 73.Xu B, et al. Elucidating the genetic architecture of familial schizophrenia using rare copy number variant and linkage scans. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16746–16751. doi: 10.1073/pnas.0908584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kohannim O, et al. Discovery and Replication of Gene Influences on Brain Structure Using LASSO Regression. Frontiers in neuroscience. 2012;6:115. doi: 10.3389/fnins.2012.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Harms FL, et al. Mutations in EBF3 Disturb Transcriptional Profiles and Cause Intellectual Disability, Ataxia, and Facial Dysmorphism. American journal of human genetics. 2017;100:117–127. doi: 10.1016/j.ajhg.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alemany S, et al. A Genome-Wide Association Study of Attention Function in a Population-Based Sample of Children. PloS one. 2016;11:e0163048. doi: 10.1371/journal.pone.0163048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Anney RJ, et al. Conduct disorder and ADHD: evaluation of conduct problems as a categorical and quantitative trait in the international multicentre ADHD genetics study. American journal of medical genetics. Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2008;147b:1369–1378. doi: 10.1002/ajmg.b.30871. [DOI] [PubMed] [Google Scholar]
- 78.Splawski I, et al. CACNA1H mutations in autism spectrum disorders. J Biol Chem. 2006;281:22085–22091. doi: 10.1074/jbc.M603316200. [DOI] [PubMed] [Google Scholar]
- 79.Eckle VS, et al. Mechanisms by which a CACNA1H mutation in epilepsy patients increases seizure susceptibility. J Physiol. 2014;592:795–809. doi: 10.1113/jphysiol.2013.264176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rzhepetskyy Y, Lazniewska J, Blesneac I, Pamphlett R, Weiss N. CACNA1H missense mutations associated with amyotrophic lateral sclerosis alter Cav3.2 T-type calcium channel activity and reticular thalamic neuron firing. Channels (Austin) 2016;10:466–477. doi: 10.1080/19336950.2016.1204497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sparrow S, et al. Epigenomic profiling of preterm infants reveals DNA methylation differences at sites associated with neural function. Translational psychiatry. 2016;6:e716. doi: 10.1038/tp.2015.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dlugos DJ, Scattergood TM, Ferraro TN, Berrettinni WH, Buono RJ. Recruitment rates and fear of phlebotomy in pediatric patients in a genetic study of epilepsy. Epilepsy & behavior: E&B. 2005;6:444–446. doi: 10.1016/j.yebeh.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 83.Berko ER, et al. Mosaic epigenetic dysregulation of ectodermal cells in autism spectrum disorder. PLoS Genet. 2014;10:e1004402. doi: 10.1371/journal.pgen.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lin X, et al. Choice of surrogate tissue influences neonatal EWAS findings. BMC medicine. 2017;15:211. doi: 10.1186/s12916-017-0970-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lowe R, et al. Buccals are likely to be a more informative surrogate tissue than blood for epigenome-wide association studies. Epigenetics. 2013;8:445–454. doi: 10.4161/epi.24362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray data generated and/or analyzed in the current study are available in the NCBI GEO [Accession series GSE128821]. R codes used for the analyses presented in the paper are available upon request to the corresponding author.