Abstract
When expressed in tobacco cells, the catalytic subunit of the dimeric ribosome inactivating protein, ricin, is first inserted into the endoplasmic reticulum (ER) and then degraded in a manner that can be partially inhibited by the proteasome inhibitor clasto-lactacystin β-lactone. Consistent with the implication of cytosolic proteasomes, degradation of ricin A chain is brefeldin A-insensitive and the polypeptides that accumulate in the presence of the proteasome inhibitor are not processed in a vacuole-specific fashion. Rather, these stabilized polypeptides are in part deglycosylated by a peptide:N-glycanase-like activity. Taken together, these results indicate that ricin A chain, albeit a structurally native protein, can behave as a substrate for ER to cytosol export, deglycosylation in the cytosol, and proteasomal degradation. Furthermore, retrotranslocation of this protein is not tightly coupled to proteasomal activity. These data are consistent with the hypothesis that ricin A chain can exploit the ER-associated protein degradation pathway to reach the cytosol. Although well characterized in mammalian and yeast cells, the operation of a similar pathway to the cytosol of plant cells has not previously been demonstrated.
Ricin is a dimeric cytotoxin produced and stored in the developing endosperm tissue of Ricinus communis seeds. The mature toxin is composed of an enzymatic ribosome inactivating A chain (RTA) disulfide-bonded to a galactose-binding B chain (RTB). Each subunit is N-glycosylated, giving apparent molecular masses for RTA and RTB of 32 kDa and 34 kDa, respectively. During seed biogenesis, ricin is synthesized from a catalytically inactive precursor, preproricin. From N to C terminus, this polypeptide comprises a 35-aminoacyl residue presequence of which the first 26 residues comprise a signal peptide (1), the mature RTA, a 12-residue linker containing a vacuolar targeting signal (2), and RTB. Nascent preproricin is targeted to the endoplasmic reticulum (ER) and then core-glycosylated and disulfide-bonded before being transported by means of the Golgi to storage vacuoles of the endosperm cells (3–5). There, the N-terminal and internal propeptides are proteolytically removed (5–9), leaving active, mature holotoxin inert within a compartment from which it cannot escape to damage sensitive Ricinus ribosomes.
We previously have demonstrated that these biosynthetic events can be reproduced in tobacco protoplasts (10). In this heterologous system the proricin precursor was quantitatively targeted to vacuoles, as determined by the appearance of processed RTA and RTB with time (10). However, during this earlier investigation, we observed that the RTA precursor (comprising the 35-residue presequence in front of mature RTA), when synthesized without the intervening linker or RTB, did not travel along the secretory pathway but disappeared in a time-dependent, brefeldin A (BFA)-insensitive fashion. We also showed that RTA disappearance was tightly coupled to a reduction in protein synthesis, suggesting that RTA might be able to access the cytosol at some stage during its degradation (10), possibly by exploiting an ER quality-control mechanism that normally deals with aberrant proteins.
Quality control of newly synthesized proteins in the ER ensures that only correctly folded and assembled proteins exit this compartment for transport through the secretory pathway (11). Those proteins that are not allowed to proceed along the secretory pathway are degraded in a nonlysosomal process, aspects of which have been elucidated in recent years. For aberrant glycoproteins, one ER quality-control mechanism operating in mammalian cells is the calnexin-calreticulin cycle, which can be interrupted by mannose trimming events that somehow target the protein for destruction (11–13). Other chaperones (e.g., BiP) can be involved in retaining malfolded or orphan polypeptides (14, 15), but eventually, if not correctly folded or assembled, these, too, are degraded. This degradation process is commonly referred to as ER-associated protein degradation (ERAD). The current model proposes that ERAD substrates are recognized by chaperones in the ER, retrotranslocated via Sec61 translocons, and degraded by proteasomes in the cytosol (for recent reviews see refs. 16 and 17). In most cases, degradation is preceded by ubiquitination and, for glycoproteins, deglycosylation by cytosolic peptide:N-glycanase (PNGase). The driving force for retrotranslocation may be substrate-dependent because in different cases chaperones (18), ubiquitination (19, 20), and the proteasome itself (14) have been implicated.
In plants, there is evidence for a quality-control system within the secretory pathway (21), although little is known about the way defective proteins are actually removed. Although proteasomal degradation is the normal route of disposal for cytosolic proteins (22, 23), it is as yet unknown whether plants use proteasomes to degrade malfolded proteins detected and translocated from the ER. Orphan subunits in the secretory system can be targeted to plant vacuoles (24, 25), and it is evident that many plant cells are well equipped to dispose of unwanted proteins within this compartment that is rich in hydrolytic enzymes. Indeed, the plant ER quality-control system may well work in concert with vacuolar delivery because of this potent alternative site for protein degradation. Vacuolar degradation of misfolded proteins has also been shown in yeast where it appears to depend on the vacuolar sorting receptor Vps10p (26). However, no homologs of Vps10p have yet been found in plants.
Using the RTA precursor (mature RTA with a 26-residue signal peptide and nine-residue propeptide) as a model protein, we have investigated in some detail the previously reported disappearance of newly synthesized RTA in tobacco leaf cells. In particular, we have focused on the cellular site of this degradation. We demonstrate that there is no detectable mislocalization of RTA to the cytosol during its biosynthesis. Rather, a significant fraction of the newly synthesized protein becomes posttranslationally retrotranslocated to the cytosol from the ER lumen and is degraded at this site and not within vacuoles. Furthermore, we show that RTA can be deglycosylated in the cytosol before its degradation. These data demonstrate the existence of an operational pathway from the ER lumen to the cytosol in plant cells and support the hypothesis that free ricin A chain can behave as an ERAD substrate.
Materials and Methods
Materials.
Preproricin, RTA, and phaseolin DNA constructs in the expression vector pDHA have been described (10). For constitutive expression in transgenic tobacco, pDHA-containing RTAE177D (27) was inserted into the HindIII site of the binary vector pGA470 (28), which was then used to transform Agrobacterium tumefaciens EHA105 (29) by electroporation. 35S Pro-Mix, immobilized pH gradient (IPG) strips, and IPG buffer 3–10 NL were purchased from Amersham Pharmacia. clasto-Lactacystin β-lactone (β-lactone, 20 mM stock in DMSO) and MG132 (100 mM stock in DMSO) were obtained from Calbiochem. Flavobacterium meningosepticum PNGase F was from New England Biolabs. BFA (2 mg/ml stock in ethanol), tunicamycin (TM, 5 mg/ml stock in 10 mM NaOH), and 1-deoxymannojirimycin (DMM, 200 mM stock in water) were from Sigma.
Transient Transformation of Leaf Protoplasts and Production of Transgenic Tobacco Plants.
Protoplasts were prepared from axenic leaves (4–7 cm long) of N. tabacum cv. Petit Havana SR1. Protoplasts were subjected to polyethylene glycol-mediated transfection as described (30). The agrobacterium containing RTAE177D was used to produce transgenic plants as described (21).
Pulse–Chase Analysis.
Cells were radiolabeled with Pro-Mix and chase was performed exactly as described (10). In some experiments, before radioactive labeling, protoplasts were incubated for 1 h at 25°C in K3 medium supplemented with either 10 μg/ml BFA, 25 μg/ml TM, or 5 mM DMM. β-Lactone and MG132 were added, at the indicated final concentration, at the beginning of the labeling period. At the desired time points, 3 vol of W5 medium (10) were added and protoplasts were pelleted by centrifugation at 60 g for 5 min. Cells were frozen in liquid nitrogen and stored at −80°C.
Preparation of Protein Extracts and Immunoprecipitation.
Frozen samples were homogenized by adding 2 vol of protoplast homogenization buffer supplemented with Complete protease inhibitor mixture (Roche) (21). Homogenates were used for immunoprecipitation with rabbit anti-RTA, anti-BiP (21), or anti-phaseolin antisera. Immunoselected polypeptides were analyzed by 15% SDS/PAGE. Gels were treated with DMSO-PPO (31) and radioactive polypeptides were revealed by fluorography.
Cell Fractionation.
Protoplast pellets (from 500,000 cells) obtained at the desired time points during pulse–chase were resuspended in 400 μl sucrose buffer [100 mM Tris⋅HCl, pH 7.6/10 mM KCl/1 mM EDTA/12% (wt/wt) sucrose] and homogenized by repeated passage through a 23-gauge syringe needle. Intact cells and debris were removed by centrifugation for 5 min at 500 g. An aliquot of the supernatants was saved and directly used for immunoprecipitations. The rest was loaded on top of a 17% (wt/wt) sucrose pad and centrifuged at 100,000 g for 30 min at 4°C. Pellets (microsomes) and supernatants (soluble proteins) were diluted in protoplast homogenization buffer and immunoprecipitated as described above.
PNGase F Treatment.
Protein A Sepharose beads carrying the immunoprecipitated protein were washed twice in water and the proteins were denatured by adding 1/10 vol of 5% SDS and 10% β-mercaptoethanol, and boiling for 5 min. Supernatants were split into two aliquots, one of which was treated at 37°C for 60 min with 1,000 units of PNGase F (or water) in 50 mM sodium phosphate buffer (pH 7.5), 1% Nonidet P-40 in a total volume of 20 μl. The second aliquot was mock-treated.
Two-Dimensional Electrophoresis.
Samples (15 μl) from PNGase treatments (or mock-treated controls) were solubilized for 1 h at room temperature in a final volume of 100 μl in 9 M urea, 4% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, 65 mM DTT, 0.7% IPG buffer 3–10 NL. In the first dimension, samples were applied by gel rehydration, and proteins were separated by isoelectric focusing (IEF) (20,000 V-h) in the nonlinear pH range of 3 to 10 by using 7-cm strips and the IPGphor IEF System (Amersham Pharmacia). IPG strips were equilibrated before the second dimension by incubating for 15 min at room temperature in 6 M urea, 30% glycerol, 2% SDS, 50 mM Tris⋅Cl (pH 6.8), 2% DTT, followed by a 5-min incubation in the same buffer but with 2.5% iodoacetamide in place of DTT. Strips were analyzed for the second dimension by 15% SDS/PAGE, and radioactive polypeptides were revealed by autoradiography.
Results and Discussion
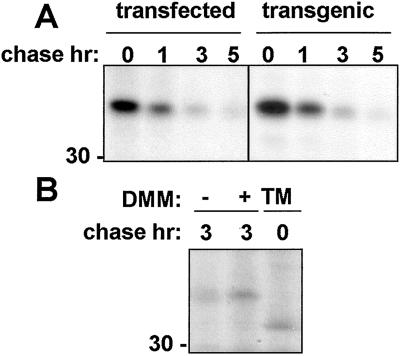
We have previously shown that the RTA precursor, when transiently expressed in tobacco protoplasts, is degraded intracellularly (10). Fig. 1 shows that the time course of degradation of ER-segregated RTA in plant cells after a 1-h pulse with radiolabeled amino acids is similar in both transfected protoplasts and protoplasts derived from leaves of stably transformed plants expressing a RTA transgene (Fig. 1A). This transgene encodes a well-characterized catalytically defective, but structurally native, variant known as RTAE177D (27). Thus, data from either transfected or transgenic sources can be regarded as comparable. It is noticeable from Fig. 1A that the RTA band acquires a smeared appearance during the chase (also evident in Fig. 2B, lanes 2, 4, 6, and 8) apparently caused by the conversion of a fraction of the molecules to faster migrating forms, whereas Fig. 1B reveals that all of the detectable RTA synthesized became N-glycosylated. Here, in cells treated with tunicamycin, all detectable RTA had a faster gel mobility consistent with its naked polypeptide molecular mass of ≈31 kDa (10, 32). That the smearing of the RTA band during the chase is caused by mannose trimming events (33) is indicated in Fig. 1B where, in the presence of the mannosidase inhibitor DMM, RTA with the mobility of fully core-glycosylated protein is detected.
Figure 1.
Ricin A chain is degraded in transfected and transgenic tobacco protoplasts. (A) Tobacco mesophyll protoplasts transfected with the construct encoding RTA or from transgenic plants stably expressing RTAE177D were subjected to pulse labeling and chased for the indicated periods of time. RTA was immunoprecipitated and polypeptides were visualized by SDS/PAGE and fluorography. (B) Protoplasts transiently expressing RTA were pulse-labeled and chased for the indicated periods, in the presence or in the absence of DMM or presence of TM. RTA was immunoprecipitated and polypeptides were visualized by SDS/PAGE and fluorography. The number at the left indicates molecular mass in kDa.
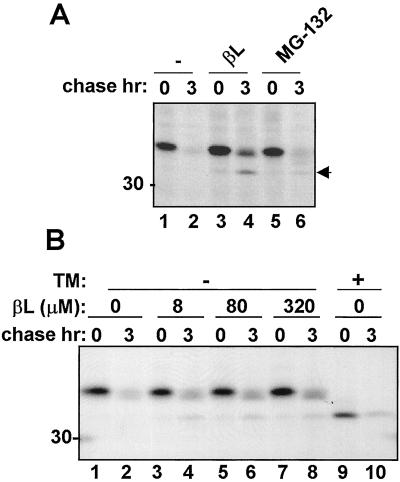
Figure 2.
The proteasomal inhibitor β-lactone retards the degradation of RTA and leads to the accumulation of a degradation intermediate. (A) Protoplasts transiently expressing RTA were subjected to pulse labeling (1 h) in the presence of 80 μM β-lactone (βL), or 1 mM MG132, and then chased for the indicated periods. (B) Protoplasts transiently expressing RTA were subjected to pulse labeling in the presence of the indicated concentrations of β-lactone, either in the presence (+) or absence (−) of TM, and chased for the indicated periods. The arrow at the right indicates the RTA degradation intermediate. The number at the left indicates molecular mass in kDa.
Because RTA is not secreted into the incubation medium (not shown; ref. 10), degradation must occur within the cell. In plant cells, proteolysis is one function of lytic vacuoles. Despite this fact, one appealing possibility is that RTA within the ER is able to masquerade as a substrate for ERAD. Exploitation of such a physiological process in mammalian cells is a hallmark of several protein toxins. By subverting this pathway, endocytosed toxins are able to cross the membrane barrier that separates them from their targets in the cytosol (34). Although the destiny of more typical ERAD substrates in animal cells is proteasomal degradation, a lethal dose of toxin somehow escapes this fate and instead refolds, possibly on ribosomes in the case of RTA (35), and goes on to kill the cell. However, such a pathway to the cytosol has not been previously shown to exist in plant cells. Indeed, in the absence of a complete itinerary of the proteolytic systems in plant cells, there remained a strong possibility that RTA in this system was being degraded within the early secretory pathway itself or within vacuolar compartments.
One of the most direct ways to biochemically detect whether the disappearance of an ER protein is by means of the ERAD pathway is to block the activity of downstream cytosolic proteasomes by using inhibitors (36) to thereby stabilize or at least retard degradation of the ERAD substrate. The specific proteasomal inhibitor, β-lactone, (37) and the nonspecific proteasomal inhibitor, MG132 (36), have been previously used to affect proteasomal degradation of normally short-lived cytosolic proteins in plant cells (23, 38, 39). Fig. 2A shows the effects of treating tobacco protoplasts with the two inhibitors. It is clear that proteasomal inhibitors were able to partially stabilize RTA, permitting visualization of a faster migrating form (Fig. 2A, arrowhead). This effect was particularly evident with the specific inhibitor, β-lactone. Fig. 2B shows that in tobacco protoplasts 8 μM β-lactone was sufficient to stabilize a proportion of the radiolabeled RTA. Furthermore, Fig. 2B clearly shows that the faster moving species seen when β-lactone is present has the same mobility as nonglycosylated RTA made in the presence of tunicamycin (compare lane 8 with lane 9). Treatment with proteasomal inhibitor for 1 or 2 h before the radioactive pulse did not increase the amount of stabilized RTA (data not shown). It therefore appeared that blocking cytosolic proteasomes protected a fraction of RTA from disappearance, including an immunoprecipitable species migrating with the mobility of nonglycosylated RTA. Because this species of RTA is particularly evident when proteasomes are inhibited, we shall refer to it as a degradation intermediate.
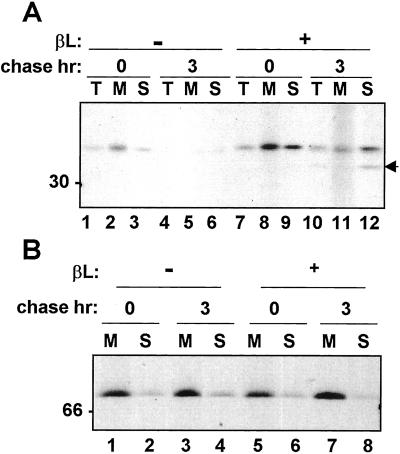
Because proteasomes are found in the cytosol and yet all of the RTA we normally see is clearly N-glycosylated, it was important to confirm the location of the toxin and its degradation intermediate upon proteasome inhibition. Cell fractionation revealed that a proportion of glycosylated RTA but strikingly all of the putative deglycosylated RTA degradation intermediate were in the soluble fraction (Fig. 3A). The ER resident chaperone BiP was predominantly detected in the microsomal pellets (Fig. 3B), indicating microsomal integrity. On closer examination it can be seen that glycosylated RTA present in the microsomes during the chase when proteasomes are inhibited had a more diffuse appearance typical of the mannose-trimmed species seen for example in Fig. 1B (DMM). By contrast, glycosylated RTA in the soluble fraction was resolved as a much sharper band (Fig. 3, compare lanes 11 and 12). This finding suggests the existence of two populations of RTA distinguishable by a differential trimming of mannose residues (Fig. 1B) and that we are not observing leakage of RTA from microsomes.
Figure 3.
The RTA degradation intermediate is not found within the ER. Protoplasts transiently expressing RTA were subjected to pulse labeling either in the presence (+) or absence (−) of β-lactone (βL) and chased for the indicated periods. Protoplasts were fractionated as described in Materials and Methods. Total cell homogenates (T), microsome pellets (M), and supernatants (S) were immunoprecipitated with (A) anti-RTA antiserum or (B) anti-BiP antiserum. The arrow at the right indicates the RTA degradation intermediate, and the numbers at the left indicate molecular mass in kDa.
Because the soluble fraction could represent cytosol or vacuoles, which break during homogenization (21), we carried out a series of controls that are shown in Fig. 4. First, we examined whether the accumulation of the RTA degradation intermediate could be blocked by BFA, a drug that inhibits Golgi-mediated trafficking to the vacuole (40) (Fig. 4A). To monitor the efficacy of BFA treatment, we also followed the transport of the bean storage protein phaseolin, whose arrival in the vacuoles is followed by proteolytic processing and generation of a series of fragments in the 22- to 28-kDa range (21). Whereas BFA alone did not block the degradation of RTA during the chase (Fig. 4A, compare lanes 7 and 8, and ref. 10), it did block the generation of phaseolin vacuolar processing products (Fig. 4A, lane 16). β-Lactone itself did not compromise vacuolar processing because cells treated with the inhibitor were still able to accumulate vacuolar fragments of phaseolin (Fig. 4A, lane 12). However, in β-lactone-treated protoplasts, BFA efficiently inhibited phaseolin fragmentation (Fig. 4A, compare lanes 14 and 12) but did not block the accumulation of the RTA degradation intermediate (Fig. 4A, compare lane 4 with lane 6).
Figure 4.
RTA is not degraded within the vacuole. (A) Protoplast obtained from plants expressing RTAE177D and phaseolin T343F (21) were mixed at a 7:3 ratio, pulse-labeled in the presence of the indicated inhibitors (βL: β-lactone; BFA), and chased for the indicated periods of time. Aliquots of the protoplast homogenates were immunoprecipitated with anti-RTA or anti-phaseolin antiserum, as indicated. The arrow at the left indicates the RTA degradation intermediate. The numbers at the left indicate molecular mass in kDa. Open arrowhead to the right indicates intact phaseolin. Bar indicates phaseolin fragmentation products. (B) Transiently transfected protoplasts expressing RTA or preproricin (ppricin) were subjected to pulse labeling in the presence (+) or absence (−) of β-lactone (βL), in the presence (+) or absence (−) of DMM, and chased for 5 h. Polypeptides immunoselected with anti-RTA antiserum were incubated with PNGase (+) or buffer (−) as a control. A and B refer to glycosylated and deglycosylated RTA and RTB, respectively. The arrow at the left indicates the RTA degradation intermediate. Note the size difference by comparing lanes 2 and 6 with lane 8. In lane 8, deglycosylated RTB comigrates with RTA.
Although these data are consistent with nonvacuolar events being responsible for the proteolysis of RTA, it is known that some membrane proteins can be routed to vacuoles in a nonconventional pathway that bypasses the Golgi and is therefore BFA-insensitive (40). To examine this possibility we took advantage of the fact that transport of RTA to the vacuole as part of proricin is accompanied by a processing event that leads to a clear decrease in molecular weight. This decrease is consistent with removal of the 9-aminoacyl residue N-terminal propeptide that remains after signal peptide removal is cleaved in the ER (1, 2, 10). We therefore compared the molecular sizes of free RTA targeted to the ER via the full 35-residue presequence, with that of RTA generated in vacuoles from the proricin precursor (Fig. 4B). To eliminate any distortion caused by the presence of sugars, the glycans on the polypeptides were removed after cell lysis. This was achieved by using commercial PNGase F on immunoprecipitated samples prepared from cells treated in the presence or absence of DMM, as indicated. DMM blocks mannose trimming and has the effect of precluding the known addition of fucose to the RTA glycan within the Golgi (41), which in turn would render the protein resistant to PNGase. In fact, in the absence of DMM treatment, whereas B chain could be efficiently deglycosylated, proricin-derived A chain was resistant to PNGase treatment (Fig. 4B, compare lanes 3 and 4). In contrast, proricin-derived A chain synthesized in DMM-treated protoplasts was sensitive to PNGase treatment and was converted to a deglycosylated form that comigrated with deglycosylated B chain (Fig. 4B, compare lanes 7 and 8). These polypeptides provide a size marker for bona fide deglycosylated vacuolar RTA.
In protoplasts expressing just RTA, the degradation intermediate accumulating in the presence of β-lactone had a slower mobility than deglycosylated vacuolar RTA (Fig. 4B, compare lanes 1 and 5 with lane 8), strongly suggesting that the two proteins are located in different compartments. Treatment of RTA immunoprecipitates with PNGase (regardless of DMM treatment) did not convert the degradation intermediate to a faster migrating species, confirming that these polypeptides do indeed lack N-linked glycan chains (Fig. 4B, compare lane 1 with lane 2 and lane 5 with lane 6). The fact that the PNGase-treated RTA band comigrated with the natural degradation intermediate indicates a lack of vacuole-specific processing. These data are consistent with the presence of the 9-aa N-terminal propeptide when RTA was expressed in the ER as a free polypeptide, but the absence of it when RTA was generated from the natural precursor that is processed only within vacuoles (5, 10). In addition, Golgi-specific modifications that render A chain resistant to PNGase treatment (fucosylation) did not occur on glycosylated RTA accumulated in the presence of β-lactone, further supporting the conclusion that when expressed as an individual subunit, RTA is not transported through the Golgi complex before degradation.
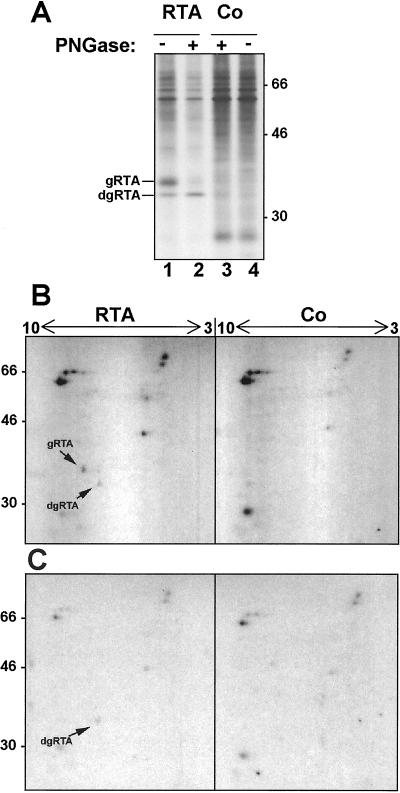
Altogether, the data presented so far indicate that RTA stabilized by β-lactone accumulates in the cytosol. In addition, the visualization of a putative deglycosylated RTA in a soluble fraction when cells are treated with proteasomal inhibitors is reminiscent of the accumulation of deglycosylated ERAD substrates in the cytosol of mammalian and yeast cells under similar conditions (42). Cytosolic PNGases have been shown responsible for this processing (43–46). To assess whether the faster migrating RTA seen in plant cells on inhibition of proteasomes truly represented a deglycosylated species, rather than a nonglycosylated but signal peptide-cleaved RTA that has been prematurely aborted from the import translocons (47), we analyzed the isoelectric point of RTA and its degradation intermediate. When a cytosolic glycanase removes a N-linked glycan, it concomitantly converts the glycan-bearing Asn to an Asp residue, thus rendering the protein more acidic. To compare the isoelectric points of the two RTA forms accumulated in the presence of β-lactone, protoplasts expressing RTA and control protoplasts were radiolabeled for 1 h in the presence of the protease inhibitor and chased for 3 h. Polypeptides selected with anti-RTA antibodies were analyzed by two-dimensional electrophoresis, before and after treatment with PNGase. Fig. 5A shows aliquots of the prepared samples run on SDS/PAGE only, confirming that the PNGase treatment successfully converted glycosylated RTA to a deglycosylated form. It has been reported that RTA is extremely difficult to resolve by IEF (48), and indeed we found that recovery of RTA on standard two-dimensional analysis was very poor. Still, Fig. 5B shows that the two species of RTA seen at 3-h chase in the presence of β-lactone have different isoelectric points, with the faster migrating species being more acidic, and that upon treatment with PNGase, only the faster migrating, more acidic species is recovered. This finding is compatible with the degradation intermediate representing a deglycosylated form of RTA.
Figure 5.
The RTA degradation intermediate is deglycosylated by PNGase. (A) Protoplasts transiently expressing RTA or mock-transfected protoplasts (Co) were labeled in the presence of 80 μM β-lactone (βL) and chased for 3 h. RTA was immunoprecipitated and immunoprecipitates were treated (+) or not treated (−) with PNGase. Samples were analyzed by SDS/PAGE. (B) Aliquots of the same samples shown in A were analyzed by two-dimensional electrophoresis. (C) As in B but samples were treated with PNGase before two-dimensional electrophoresis. Numbers at the left indicate molecular mass in kDa. Numbers at the top indicate the pH range of the first-dimension IEF. gRTA is glycosylated RTA, dgRTA is deglycosylated RTA.
We have previously shown that, when expressed in tobacco protoplasts, RTA is toxic and can readily assemble with its companion B chain, indicating that the protein is structurally mature (10). The data presented here suggest that RTA expressed alone, despite being a native protein, is exploiting the plant ERAD pathway to reach the cytosol where a proportion of it is subjected to deglycosylation and proteasomal degradation in the cytosol. This finding provides evidence for the existence of a functional retrotranslocation pathway in plant cells.
Although RTA is a structurally native protein, there is evidence that in the absence, or upon reduction of its partner B chain, it is able to retrotranslocate from the ER of Saccharomyces cerevisiae (49) and mammalian cells (50). It is plausible that reduced RTA has inherent properties that allow it to hijack the ERAD machinery in a way that is currently unclear. Here we provide compelling evidence that in vivo, RTA acquires the expected glycan within the ER lumen and subsequently loses this upon emergence into the cytosol, a modification that can be visualized when proteasomal degradation is blocked. Although a complete analysis of the mechanism and driving force for toxin retrotranslocation is beyond the scope of this study, our observations suggest that the process is not tightly coupled with proteasomal activity in contrast to what has been observed for some ERAD substrates (14, 51). For a number of mammalian glycoprotein ERAD substrates, mannose trimming in the ER lumen condemns them to the retrotranslocation apparatus (12, 13) in a process that can be blocked by the inhibition of mannosidases (15, 52, 53). By contrast for RTA, it appears that the nontrimmed version can be degraded (Fig. 1B) and indeed, this version is found in the cytosol fraction (Fig. 4). This puzzling observation may reflect differences in the way the ER quality control system perceives this particular protein as compared with other, more structurally abnormal substrates, or it may reflect peculiarities of the plant cell ER.
One striking feature of the behavior of RTA in plant cells is the magnitude of its degradation. It is conceivable that conventional proteasomal inhibitors characterized for use in other systems may not be completely effective in plant cells. Alternatively, other proteolytic pathways may be operating that will require further analysis. Whatever the dominant degradation system, the present study has demonstrated that at least a proportion of RTA that had at one time been segregated in the ER lumen was subsequently detected in the cytosol. This was determined by a combination of cell fractionation and IEF analyses. The presence of a deglycosylated RTA species is suggestive of an operational ERAD pathway in plants, the mechanistic details of which now require rigorous investigation.
Acknowledgments
We thank Massimo Alessio and Emanuela Pedrazzini for advice on IEF. This work was supported by United Kingdom Biotechnology and Biological Sciences Research Council Grant C08612 (to L.M.R. and J.M.L.) and Consiglio Nazionale delle Ricerche Target Project “Biotechnology” (to A.C.).
Abbreviations
- RTA
ribosome inactivating A chain
- RTB
galactose-binding B chain
- ER
endoplasmic reticulum
- BFA
brefeldin A
- ERAD
ER-associated protein degradation
- IPG
immobilized pH gradient
- TM
tunicamycin
- DMM
1-deoxymannojirimycin
- PNGase
peptide:N-glycanase
- β-lactone
clasto-lactacystin β-lactone
- IEF
isoelectric focusing
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Ferrini J B, Martin M, Taupiac M P, Beaumelle B. Eur J Biochem. 1995;233:772–777. doi: 10.1111/j.1432-1033.1995.772_3.x. [DOI] [PubMed] [Google Scholar]
- 2.Frigerio L, Jolliffe N A, Di Cola A, Hernández Felipe D, Paris N, Neuhaus J-M, Lord J M, Ceriotti A, Roberts L M. Plant Physiol. 2001;126:167–165. doi: 10.1104/pp.126.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butterworth A G, Lord J M. Eur J Biochem. 1983;137:57–65. doi: 10.1111/j.1432-1033.1983.tb07795.x. [DOI] [PubMed] [Google Scholar]
- 4.Lord J M. Eur J Biochem. 1985;146:403–409. doi: 10.1111/j.1432-1033.1985.tb08666.x. [DOI] [PubMed] [Google Scholar]
- 5.Lord J M. Eur J Biochem. 1985;146:411–416. doi: 10.1111/j.1432-1033.1985.tb08667.x. [DOI] [PubMed] [Google Scholar]
- 6.Harley S M, Lord J M. Plant Sci. 1985;41:111–116. [Google Scholar]
- 7.Hara-Nishimura I, Inoue K, Nishimura M. FEBS Lett. 1991;294:89–93. doi: 10.1016/0014-5793(91)81349-d. [DOI] [PubMed] [Google Scholar]
- 8.Hara-Nishimura I, Shimada T, Hiraiwa N, Nishimura M. J Plant Physiol. 1995;145:632–640. [Google Scholar]
- 9.Yamada K, Shimada T, Kondo M, Nishimura M, Hara-Nishimura I. J Biol Chem. 1999;274:2563–2570. doi: 10.1074/jbc.274.4.2563. [DOI] [PubMed] [Google Scholar]
- 10.Frigerio L, Vitale A, Lord J M, Ceriotti A, Roberts L M. J Biol Chem. 1998;273:14194–14199. doi: 10.1074/jbc.273.23.14194. [DOI] [PubMed] [Google Scholar]
- 11.Ellgaard L, Molinari M, Helenius A. Science. 1999;286:1882–1888. doi: 10.1126/science.286.5446.1882. [DOI] [PubMed] [Google Scholar]
- 12.Frigerio L, Lord J M. Curr Biol. 2000;10:R674–R677. doi: 10.1016/s0960-9822(00)00680-1. [DOI] [PubMed] [Google Scholar]
- 13.Helenius A, Aebi M. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 14.Chillarón J, Haas I G. Mol Biol Cell. 2000;11:217–226. doi: 10.1091/mbc.11.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Virgilio M, Kitzmüller C, Schwaiger E, Klein M, Kreibrich G, Ivessa N E. Mol Biol Cell. 1999;10:4059–4073. doi: 10.1091/mbc.10.12.4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lord J M, Davey J, Frigerio L, Roberts L M. Semin Cell Dev Biol. 2000;11:159–164. doi: 10.1006/scdb.2000.0160. [DOI] [PubMed] [Google Scholar]
- 17.Brodsky J L, McCracken A A. Cell Dev Biol. 1999;10:507–513. doi: 10.1006/scdb.1999.0321. [DOI] [PubMed] [Google Scholar]
- 18.Plemper R K, Bohmler S, Bordallo J, Sommer T, Wolf D H. Nature (London) 1997;388:891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- 19.Biederer T, Volkwein C, Sommer T. Science. 1997;278:1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- 20.de Virgilio M, Weninger H, Ivessa N E. J Biol Chem. 1998;273:9734–9743. doi: 10.1074/jbc.273.16.9734. [DOI] [PubMed] [Google Scholar]
- 21.Pedrazzini E, Giovinazzo G, Bielli A, de Virgilio M, Frigerio L, Pesca M, Faoro F, Bollini R, Ceriotti A, Vitale A. Plant Cell. 1997;9:1869–1880. doi: 10.1105/tpc.9.10.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vierstra R D. Plant Mol Biol. 1996;32:275–302. doi: 10.1007/BF00039386. [DOI] [PubMed] [Google Scholar]
- 23.Genschik P, Criqui M C, Parmentier Y, Derevier A, Fleck J. Plant Cell. 1998;10:2063–2076. doi: 10.1105/tpc.10.12.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coleman C, Herman E M, Takasaki K, Larkins B A. Plant Cell. 1996;8:2335–2345. doi: 10.1105/tpc.8.12.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bagga S, Adams H P, Rodriguez F D, Kemp J D, Sengupta-Gopalan C. Plant Cell. 1997;9:1683–1696. doi: 10.1105/tpc.9.9.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong E, Davidson A R, Kaiser C A. J Cell Biol. 1996;135:623–633. doi: 10.1083/jcb.135.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaddock J A, Roberts L M. Protein Eng. 1993;6:425–431. doi: 10.1093/protein/6.4.425. [DOI] [PubMed] [Google Scholar]
- 28.An G, Watson B D, Stachel S, Gordon M P, Nester E W. EMBO J. 1985;4:277–284. doi: 10.1002/j.1460-2075.1985.tb03626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hood E E, Helmer G L, Fraley R T, Chilton M D. J Bacteriol. 1986;168:1291–1301. doi: 10.1128/jb.168.3.1291-1301.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedrazzini E, Giovinazzo G, Bollini R, Ceriotti A, Vitale A. Plant J. 1994;5:103–110. [Google Scholar]
- 31.Bonner W M, Laskey R A. Eur J Biochem. 1974;46:83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- 32.Lamb F I, Roberts L M, Lord J M. Eur J Biochem. 1985;148:265–270. doi: 10.1111/j.1432-1033.1985.tb08834.x. [DOI] [PubMed] [Google Scholar]
- 33.Lerouge P, Cabanes-Macheteau M, Rayon C, Fitchette-Lainé A C, Gomord V, Faye L. Plant Mol Biol. 1998;38:31–48. [PubMed] [Google Scholar]
- 34.Lord J M, Roberts L M. J Cell Biol. 1998;140:733–736. doi: 10.1083/jcb.140.4.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Argent R H, Parrott A M, Day P J, Roberts L M, Stockley P G, Lord J M, Radford S E. J Biol Chem. 2000;275:9263–9269. doi: 10.1074/jbc.275.13.9263. [DOI] [PubMed] [Google Scholar]
- 36.Lee D H, Goldberg A L. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 37.Fenteany G, Standaert R F, Lane W S, Choi S, Corey E J, Schreiber S L. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 38.Potushak T, Stary S, Schlögerhofer P, Becker F, Nejinskaia V, Bachmair A. Proc Natl Acad Sci USA. 1998;95:7904–7908. doi: 10.1073/pnas.95.14.7904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Osterlund M T, Hardtke C S, Wei N, Deng X W. Nature (London) 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- 40.Gomez L, Chrispeels M J. Plant Cell. 1993;5:1113–1124. doi: 10.1105/tpc.5.9.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lord J, Harley S. FEBS Lett. 1985;189:72–76. [Google Scholar]
- 42.Wiertz E J H J, Tortorella D, Bogyo M, Yu J, Mothes W, Jones T R, Rapoport T A, Ploegh H L. Nature (London) 1996;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- 43.Cresswell P, Hughes E A. Curr Biol. 1997;7:R552–R555. doi: 10.1016/s0960-9822(06)00279-x. [DOI] [PubMed] [Google Scholar]
- 44.Kopito R R. Cell. 1997;88:427–430. doi: 10.1016/s0092-8674(00)81881-4. [DOI] [PubMed] [Google Scholar]
- 45.Ploegh H L. Immunol Today. 1997;18:269–271. doi: 10.1016/s0167-5699(97)80021-7. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki T, Park H, Kitajima K, Lennarz W J. J Biol Chem. 1998;273:21526–21530. doi: 10.1074/jbc.273.34.21526. [DOI] [PubMed] [Google Scholar]
- 47.Theuer C P, Buchner J, FitzGerald D, Pastan I. Proc Natl Acad Sci USA. 1993;90:7774–7778. doi: 10.1073/pnas.90.16.7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hegde R, Podder S K. Eur J Biochem. 1998;254:596–601. doi: 10.1046/j.1432-1327.1998.2540596.x. [DOI] [PubMed] [Google Scholar]
- 49.Simpson J C, Roberts L M, Romisch K, Davey J, Wolf D H, Lord J M. FEBS Lett. 1999;459:80–84. doi: 10.1016/s0014-5793(99)01222-3. [DOI] [PubMed] [Google Scholar]
- 50.Rapak A, Falnes P O, Olsnes S. Proc Natl Acad Sci USA. 1997;94:3783–3788. doi: 10.1073/pnas.94.8.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kamhi-Nesher S, Shenkman M, Tolchinsky S, Vigodman Fromm S, Ehrlich R, Lederkremerm G Z. Mol Biol Cell. 2001;10:1711–1723. doi: 10.1091/mbc.12.6.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang M, Omura S, Bonifacino J S, Weissman A M. J Exp Med. 1998;187:835–846. doi: 10.1084/jem.187.6.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson C M, Farmery M, Bulleid N J. J Biol Chem. 2000;275:21224–21232. doi: 10.1074/jbc.M000567200. [DOI] [PubMed] [Google Scholar]